Abstract
Climate change with altered pest-disease dynamics and rising abiotic stresses threatens resource-constrained agricultural production systems worldwide. Genomics-assisted breeding (GAB) approaches have greatly contributed to enhancing crop breeding efficiency and delivering better varieties. Fast-growing capacity and affordability of DNA sequencing has motivated large-scale germplasm sequencing projects, thus opening exciting avenues for mining haplotypes for breeding applications. This review article highlights ways to mine haplotypes and apply them for complex trait dissection and in GAB approaches including haplotype-GWAS, haplotype-based breeding, haplotype-assisted genomic selection. Improvement strategies that efficiently deploy superior haplotypes to hasten breeding progress will be key to safeguarding global food security.
Subject terms: Agricultural genetics, Plant breeding, Plant genetics
In this Review, Bhat et al. highlight ways to mine crop haplotypes and apply them for dissecting complex traits and genomics-assisted breeding (GAB) approaches. This Review presents new avenues to discover superior haplotypes and assemble them in targeted manner in crop breeding for faster delivery of high-yielding cultivars with better adaptation to future climates.
Introduction
Crop plants are subjected to a variety of biotic and abiotic stresses that impair normal crop growth and cause substantial losses in crop yields worldwide1,2. Amid these stresses, developing climate smart and nutritious crop varieties that remain vital to securing food security of the incessantly growing human population, presents a daunting challenge to the agricultural scientists worldwide. Although conventional breeding has made great success in the development of high-yielding crop varieties3, it is important to accelerate the pace of crop improvement programmes especially for the complex traits such as yield under stress conditions. In this regard, the genomics-assisted breeding (GAB) by implementing genomics tools in breeding was proposed by Varshney et al.4. This approach has delivered several high-yielding, stress-tolerant and better nutrition varieties5,6. For instance, the low-throughput sequence-based markers, such as simple sequence repeats (SSRs), were extensively used in the molecular breeding programmes; however, these marker systems have limitations such as low density across the genome, low coverage, expensiveness. Application of these second-generation DNA marker systems resulted in poor resolution of gene mapping and relatively low efficiency of plant selections and breeding7,8. Fortunately, recent advances in the next generation sequencing (NGS) and the genotyping platforms have considerably alleviated this bottleneck in crop breeding. These NGS-based platforms have provided remarkable marker-density and coverage at reduced cost9, and are now commercially available for both model and non-model crop species10,11. These high-throughput platforms make hundreds of millions of DNA polymorphisms accessible for use in genetic and genomics research12,13; and their application in crop breeding has considerably increased the gene mapping resolution and prediction accuracy in genomic selection (GS)14,15. Majority of the economically important crop traits, such as yield, quality and stress tolerance, are of complex quantitative nature, which are influenced by several small effect QTL/genes and manifest substantial genotype x environment (G x E) interactions16. Although efforts to understand the complex genetic makeup of these agriculturally relevant traits have been successful in the identification of major-effect genomic regions, conventional experimental populations faced the problem of limited genetic diversity, low resolution and limited recombination events17,18. Hence, the genome-wide association study (GWAS) has emerged as a powerful tool for dissecting complex quantitative traits in crop plants with enhanced resolution and allelic richness19,20. Furthermore, due to the availability of cost-effective and high-density genotyping platforms, it has been possible now to screen larger breeding populations for estimating and using the breeding value in crop improvement programmes by using GS, another breeding approach21.
In recent years, the NGS-based genotyping methods such as genotyping-by-sequencing, restriction site-associated DNA sequencing, whole-genome resequencing as well as fixed SNP arrays have greatly facilitated genotyping of large germplasm collections for GWAS and GS analyses8,22. However, the major limitations for the use of SNPs in these analyses include their biallelic nature, the presence of rare alleles, and abundant levels of linkage drag16,23. Therefore, the candidate genomic loci identified by GWAS often do not represent the causative locus; but correspond to the loci that are in linkage drag with a gene or a regulatory element, eventually affecting the trait of interest24,25. In this regard, an effective approach to overcome the limitations of SNPs and increase the resolution of candidate genomic regions is to consider haplotypes for genome-wide analyses26. Haplotype is a specific combination of jointly inherited nucleotides or DNA markers from polymorphic sites in the same chromosomal segment27,28.
In the present review, we discusses the potential and need of haplotypes in the crop breeding for the development of improved varieties. We have also compared the efficiency of haplotype- and individual SNP-based markers in the GWAS and GS analyses. Besides, the challenges associated with the use of haplotypes in crop breeding at the commercial level are also addressed. We conclude by highlighting the scope of haplotypes in the future crop breeding programs.
Crop improvement: conventional breeding to genomics-assisted breeding
Development of improved crop varieties for food, feed and industrial purposes can be accomplished mainly by plant breeding29. The science of plant breeding has evolved from conventional to present day GAB6,30. In the last century, tremendous efforts have been made by plant breeders across the globe to develop improved varieties in different crop species by using the conventional breeding approaches31–45. It is estimated that the undernourished proportion of the human population has been reduced from 40% in the 1960s to <11% now, which is principally attributable to the improved high-yielding and stress-tolerant crop varieties produced mainly through conventional breeding44. The conventional plant breeding for crop yield enhancement progressed consistently over time. The high-yielding varieties/hybrids were mostly responsible for this increase in both area and productivity, and the large-scale adoption of these varieties/hybrids provides strong evidence for contributions by plant breeding innovations over the last century.
In recent years, the plant breeding community has recognized the need of introducing genetic variability in breeding programs to enhance the genetic base of elite gene pool, enhancing precision and efficiency in selection and reducing the breeding cycle4,6,46. In this context, the GAB approach proposed by Varshney et al.4 outlined the use of genomics tools and technologies to identify markers, candidate genes associated with target traits and integration of genomics approaches in breeding. Several GAB approaches including marker-assisted backcrossing (MABC), marker-assisted selection (MAS), marker-assisted recurrent selection (MARS) and advanced backcross QTL (AB-QTL) were suggested for crop improvement. In recent years, GS approach has also been added to GAB portfolio6,21. For MAS, the first step is the identification of molecular markers that are strongly associated with genomic regions/quantitative trait loci (QTLs) regulating the traits of interest. Eventually, these QTLs, either individually or in multiple numbers, can be pyramided into elite breeding material through MABC. Some success stories of MABC include the introgression of a ‘QTL-hotspot’ into elite chickpea varieties for improved yield under drought conditions47,48, improving the yield and stress tolerance of mega rice variety IR64 (Developed by IRRI, IR 64 was released in Phillipines in 1987. The rice variety registered a widespread acceptance owing to its multiple beneficial traits including better cooking quality, earliness, disease resistance and high yield)49,50, transferring QTLs (qDTY2.2 and qDTY4.1) into IR64 for reproductive stage drought tolerance51,52, and the improvement of different yield and stress-related traits in several major crop species6,53–55. Despite the aforementioned utilities of MABC, it is efficient only for the major-effect QTLs, while most of the genetic variations for yield, quality and stress tolerance traits in crop plants are governed by a large number of minor QTLs. Alternatively, the frequency of many beneficial alleles can be increased in a given population through the MARS scheme. Unlike MABC, the MARS has been applied for improving a breeding population with respect to QTLs exerting smaller effects on the phenotype. MARS has been successful in improving drought tolerance in multiple crop species viz., maize, soybean, sunflower, wheat, sorghum, and rice56–60. To capture minor effect QTLs scattered throughout the genome, the plant breeding community has recently started to use GS approach. GS estimates the genetic worth of an individual based on the large set of marker information distributed across the whole genome, rather than a few markers as in the case of MAS21. In this approach, a prediction model based on the genotypic and phenotypic data of training population (TP) is developed and then genomic estimated breeding values (GEBVs) for the individuals of breeding population (BP) are computed from their genome-wide marker profiles61. The GEBVs allow one to predict individuals that will perform better and are suitable either as a parent for the next breeding cycle or can directly enter into the variety release pipeline21. Unlike MAS, GS does not necessarily require a prior knowledge of significant marker-trait associations62. However, inclusion of the significant set of markers, such as resulting from GWAS, into GS models has been found to improve prediction accuracies63. GS has started gaining profound interest in plant breeding, with the recent studies establishing its superiority over other selection methods64–70. With the availability of a range of cost-effective genotyping platforms and advances in the development of prediction models, GS is expected to be a routine breeding approach, like MABC/MAS in crop improvement programmes.
Features of haplotypes
Defining haplotypes: harnessing the wealth of whole-genome sequencing data
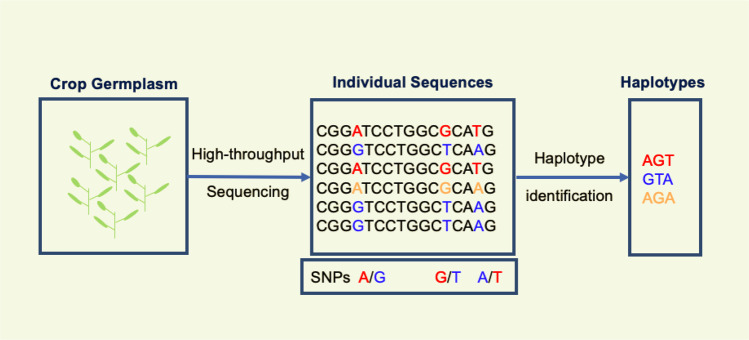
Haplotype is a combination of alleles for different polymorphisms (such as SNPs, insertions/deletions and other markers or variants) present on the same chromosome, which are inherited together with minimum chance of contemporary recombination71,72. Any individual has two haplotypes for a given stretch of chromosomal DNA; while at the population level, many haplotypes can be found for the same stretch73. In other words, a haplotype is defined as a set of nearby genomic structural variations, such as polymorphic SNPs, with a strong linkage disequilibrium (LD) between them74. As shown in Fig. 1, two or more polymorphic SNPs of the haploid sequences inherited together as a unit constitute a haplotype71. The haplotypes are defined/assigned in three principal ways: (a) by using the haplotype diversity in a given chromosomal segment, (b) by using the pairwise LD between the jointly inherited markers that show lack of evidence for historical recombination, it is measured by r2 (measure of LD)75,76 and (c) by grouping of SNPs through sliding-windows of fixed or variable length77. Evidence suggests that the LD-based approaches are more efficient for defining the haplotypes in the genomic/chromosome regions26,74. This is because (a) historical recombination identification is the direct focus in a particular population through the haplotype detection, (b) visualization of the LD coefficients is very easy, (c) for diploid data with unknown haplotype phase, it is applicable. The LD in the given population is determined by many factors such as mode of pollination, population size and structure, mutation rate, genetic drift, recombination frequency, and the type of selection on a given chromosomal fragment78.
Fig. 1. Formation and development of haplotypes from haploid sequences.
Resequencing of the crop germplasm is done to identify the polymorphic SNPs to be subsequently used in the development of haplotypes.
During the evolution of the important crop species such as rice, maize, wheat, sorghum, cassava and rapeseed, the selection of genes/alleles regulating desirable phenotype for the trait of interest is the major factor responsible for the formation of signatures of selection26. The signatures of selection (also known as conserved haplotype blocks and selective sweeps) possess multiple genes, which are regulated together by many regulatory genes. The correlation among different traits as reflected from the selection signatures is either due to the true linkage among the genes or resulting from the pleiotropic effect of the same genes34,79. Therefore, the crop breeders should preferably target these genomic regions to elucidate their effect on the traits of interest. Besides, the integration of genomics to identify the recombinants produced by crossing of contrasting parents will greatly assist in resolving the complexity of quantitative traits. This will enhance the efficiency to improve the specific traits in modern varieties for their better adaptation to extreme environments80.
Due to the availability of sequencing data from large number of individuals for a given crop species it has been easier to define the haplotype. By using the whole genome sequencing data, Bevan et al.81 defined the concept of the haplotype assembly. Together with the phenotyping data of germplasm/breeding lines, it is possible to assess and validate phenotypic effects of the ‘component’ haplotypes. Based on this premise, and by using large-scale whole-genome resequencing datasets in combination with haplo-pheno analysis, Abbai et al.82 identified useful haplotypes for future breeding in rice and Sinha et al.46 followed the similar approach in pigeonpea. High-density SNP data generated from multiple genotypes via NGS-based or array-based approaches have been used for the development of haplotypes in many plant species. These haplotypes have also been used for various applications in research and breeding in different crop species (see details in Tables 1, 2).
Table 1.
The use of haplotype markers in genome-wide association mapping (GWAS) analyses in different crop species.
| Crop Species | Trait | Population size | Haplotype markers | Haplotype-trait associations | PVE (%) | Reference |
|---|---|---|---|---|---|---|
| Soybean | 100-seed weight; plant height; seed yield | 169 | 941 | 87 | 9.14-15.83 | 134 |
| Soybean | Agronomic and yield-related traits | 296 | – | 10 | >10.0 | 153 |
| Wheat | Heading date; plant height; 1000-grain weight; grain number per spike; fruiting efficiency at harvest | 102 | 4516 | 97 | – | 121 |
| Wheat | Grain yield; days to heading; plant height | 6461 | 519 | 36 | 2.2-5.6 | 154 |
| Barley | Deoxynivalenol content in kernels; heading time; days to maturity; grain yield; plant height; specific weight; 1000-kernel weight | 277 | 14,400 | – | 2.0-14.0 | 135 |
| Barley | Yield and quality-related traits | 106 | 2770 | 23 | >10.0 | 131 |
| Rice | Grain shape | 372 | – | 30 | – | 155 |
| Rice | Agronomic traits | 414 | 15,275 | – | – | 109 |
| Maize | Agronomic and reproductive traits | 322 | 53,403 | 44 | 5.6-17.0 | 156 |
| Maize | Total plant height; ear height; ear height/plant height | 183 | 7,831 | 40 | 7.0-22.0 | 74 |
| Maize | Agronomic and reproductive traits | >1000 | 154,104 | – | >10.0% | 157 |
| Oat | Heading date | 4657 | 164741 | 184 | – | 158 |
| Rapeseed | Days to flowering; seed glucosinolate content | 950 | – | 15 | – | 152 |
Table 2.
The use of haplotype markers in genomic selection in different crop species.
| Crop Species | Trait | Training Population size | Haplotype markers | GS prediction accuracy | Reference |
|---|---|---|---|---|---|
| Bluegum | Traits related to wood quality and tree growth | 646 | ~3000 | 0.58 | 105 |
| Soybean | Plant height & grain yield per plant | 235 | 357 | >0.80 & >0.45 | 159 |
| Sorghum | Agronomic and yield-related traits | 207 | 1,974 | 0.57-0.73 | 160 |
| Wheat | Yield, test weight, and protein content | 383 | 1400 | >0.40 | 151 |
| Wheat | Grain yield and related traits | 4,302 | 1162 | 0.39-0.48 | 154 |
| Oat | Heading date | 635 | 13954 | 0.42-0.67 | 158 |
Third-generation sequencing: alleviating the bottlenecks in haplotype identification
The long-term goal of genetics is to elucidate the effect of DNA sequence variations on the plant traits, and how these variations have led to the evolution of different populations and species83,84. In genetics, linkage is a core concept on which molecular mapping of genetic determinants relies. For example, in the linkage or association mapping, the individual genetic markers/variants are used to determine their association(s) with the trait(s) of interest, instead of pinpointing the causal mutation3. The trait-associated DNA markers are then used as surrogates for the selection of the desirable phenotypes5. As we mentioned in the previous section, fast-tracking the process of targeted trait improvement will require a paradigm shift from individual SNP markers to haplotypes. The information on haplotypes regulating the important phenotypes is currently limited in the genetic studies85, which prevents the accurate determination of ancestry reconstruction, rearrangements of chromosomes, allele-specific expression, and detection of selective sweeps86,87.
However, the availability of the high-throughput sequencing platforms has made a tremendous impact on the identification of haplotypes and their application in the genetic studies. Although, the second-generation sequencing techniques produce short reads of 150 bp, these small reads normally do not possess more than a single variant88. Hence, the haplotypes are constructed indirectly from this data and this needs specific statistical inferences from population genotyping data, which in turn increases the time and cost for the haplotype construction88,89. In contrast, third-generation sequencing (TGS), such as the Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), produce long reads from which the haplotypes can be directly constructed88. In comparison to the second-generation sequencing methods, analysis of DNA molecules can be performed directly via long-read sequencing platforms90. However, the ‘phasing’ is used for some adjustment of the long-read sequencing data to increase the efficiency for haplotype identification. Construction of the haplotypes from the sequence data through haplotype estimation is known as phasing; which is very important to elucidate the sequence-specific variations such as the effect of methylation, specific expression of alleles and compound heterozygosity91. Fixing of higher error rate (~10%) in the long-read sequencing technologies compared to short-read sequencing methods (NGS methods) needs specific bioinformatics-mediated adjustments92. In this regard, many different phasing methods enabling haplotype construction/reconstruction from long-read sequencing data have been recently developed, such as reference-based phasing (molecular haplotyping, single-cell phasing, and polyploid phasing), de novo genome assembly (such as diploid and polyploid assembly) and strain-resolved metagenome assembly (de novo re-assembly, single nucleotide variant-based assembly, read and contig binning)72. Combination of these haplotype analysis methods with various computational tools such as WhatsHap, HapCut2, HapTree, WhatsHap- polyphase, Falcon phase, Hifiasm, SDip, POLYTE, DESMAN, MetaMaps, and ProxiMeta, has greatly enhanced the efficiency and precision in the identification of do novo and rare variants from the long-read sequencing data72. Therefore, integrating the various phasing and bioinformatics tools with the long-read sequencing technologies has allowed us to fully exploit the potential of these sequencing approaches in haplotype construction91. For example, Ammar et al.73 showed that MinION nanopore sequencer efficiently resolved the variants/haplotypes of HLA-A, HLA-B and CYP2D6 genes by producing the long reads without even using the statistical phasing. Similarly, Zhang et al.93 also demonstrated the higher accuracy of Nanopore sequencing in the identification of haplotypes across the genomes. Besides, recent advances in the PacBio’s HiFi technology have allowed to produce long reads in the range of 15-20 Kb, with an error rate comparable to the second-generation sequencing i.e., more than 99% accuracy was achieved94. These advancements have allowed reconstruction of the previously impossible near-complete human haplotypes that include microsatellites, repetitive elements, and other complex structural variations95. Moreover, Sun et al.96 used the PacBio HiFi reads (30x per haplotype) and hifiasm to produce the assembly of the autotetraploid genome of potato. This was the first study demonstrating the haplotype-resolved assembly of potato crop. Through single-cell genotyping and high-quality long-read sequencing of the tetraploid plants, the authors successfully reconstructed all four haplotypes showing considerably higher diversity among themselves. This haplotype diversity is significantly higher than the diversity commonly found within a given species. This evidenced that successful haplotype reconstruction in the polyploid species has a huge impact on breeding these crops in the future96. Recent research demonstrates the enormous potential of the TGS in resolving the accuracy issues in the haplotype identification, thereby increasing the scope of haplotypes for genetic studies in both animals and plants72. Hence, the TGS platforms offer promising alternative to obtain haplotype-related information from the genomes, and future affordability of these sequencing platforms will have a profound impact on plant research and breeding.
Haplotagging: A novel sequencing strategy for rapid discovery of haplotypes
Recently, a simple, rapid and promising technique for linked-read (LR) sequencing (called ‘haplotagging’) has emerged97,98. In this technique, molecular barcoding of long DNA molecules is carried out prior to sequencing, which in turn retains the long-range information by preserving the linked variants85. The shared barcode is then used to link the individual short reads for constructing the original haplotype98. However, currently the commercial utilization of haplotagging in the genetic studies is prevented by certain factors, which include the requirement of custom sequencing primers, and cost-ineffectiveness, and poor scalability of the current techniques98. Nevertheless, if managing these factors, especially the lower cost and more scalability, becomes possible in near future, the haplotagging will be greatly used in the genetic studies. For instance, it will enable the haplotyping of the larger plant and animal populations, and allow the sequencing and systematic discovery of haplotypes in tens of thousands of samples, that too in both model and non-model plant species. It has been documented that both standard Illumina sequencing and haplotagging maintain full compatibility, and there is no extra cost in the haplotagging98,99. The utility of haplotagging technique, for the identification of the haplotypes in the genome, has not yet been demonstrated in the plants, but recently, the haplotagging has been demonstrated in the two butterfly species85. For example, Meier et al.85 applied haplotagging approach to generate the haplotypes of megabase-size for the case of around six hundred butterflies’ individuals belonging to the two species viz., Heliconius erato and H. melpomene, and these two species were identified to form hybrid zones that are overlapping across an elevational gradient in Ecuador. Besides, Meier et al.85 also showed that haplotagging was able to detect the genetic loci regulating the distinct wing color patterns, namely, high- and low-land. In both the species the different haplotype alleles were detected at the same major loci; however, the chromosome rearrangements show no parallelism. To this end, this study demonstrated that technique of the “haplotagging” was successful to identify the distinct haplotype allele classes regulating the different phenotypes of the wing color patterns. Hence, these results suggested the enhanced power of the efficient haplotyping methods when combined with large-scale sequencing data from natural populations85.
The above findings suggest the potential role of haplotagging in the identification of haplotype alleles regulating different phenotypes for a particular trait of interest. Hence, the haplotagging technique might be a promising strategy to identify the superior haplotype alleles in the diverse plant populations/germplasm for their ultimate use in the breeding for the development of improved crop varieties. This technique will be crucial to harness the true potential of the haplotype-based breeding for crop improvement.
Haplotype vs. individual markers: Comparative efficiency for crop breeding
Variations in the complex phenotypes are associated with the presence of SNPs, insertion–deletions and copy number variations in certain genomic loci100–102. Currently, most of the plant breeders are using SNP markers to tag novel genetic variations underlying different phenotypes, and introgress these variations into the elite crop cultivars. However, the superiority of haplotype markers compared to individual SNP markers in addressing complex traits has been demonstrated through efficient gene identification and GS26. For example, the use of haplotypes has been reported to considerably increase the prediction accuracy of the low-heritable quantitative traits as compared to the individual SNP markers103–107. Besides, the use of haplotypes in gene mapping analyses has emerged as a more efficient approach for the identification of genomic loci and candidate genes regulating traits of interest72,108,109. The latest evidence suggests that the haplotype-based approach can improve not only the predictive abilities of GS models but also the precision with which genomic loci are detected in GWAS109–111.
The higher efficiency of the haplotypes over individual SNP is due to some important reasons. For example, SNPs tiled on arrays are usually chosen for their moderate to high minor allele frequency (MAF). Therefore, most of the SNPs in the commercial chips are expected to be the old mutations, given that all new mutations remain at a low frequency in the beginning and a large part of them may disappear before reaching considerable frequency112. Since the single-nucleotide-based genomic relationship matrix (GSNP) is based on SNPs with relatively high MAF, this may imply that GSNP traces old relationships from distant relatives and, therefore, may trace less accurately the changes due to recent selection as compared to the multi-locus haplotype-based relationship matrix, GHAP112. Meuwissen et al.112 suggested that building the relationship matrix using haplotypes instead of single SNPs may improve the accuracy of genomic predictions. Another potential limitation of GSNP is that the SNPs are biallelic and, therefore, their polymorphism information content (PIC) value is not high. This restricts the ability to effectively capture LD between SNPs and multi-allelic QTLs. On the other hand, haplotype blocks are generally “multi-allelic” and may therefore better capture LD with multi-allelic QTLs compared to individual SNPs112. It is also worth noting that longer haplotype blocks provide more information about possible recent mutations and close relationships than the shorter ones113,114. Furthermore, haplotype effects could also factor in local epistatic effects among QTLs located within the haplotype blocks113. In addition, GHAP can differentiate between identical by descent (IBD) and identical by state (IBS), while GSNP cannot. This is because long shared haplotype blocks are likely to come from common ancestors. Therefore, long haplotype blocks can better capture information on IBD regions than individual SNPs in GS experiments115.
Applications of haplotypes in genetic analysis and breeding
Gene mapping
Recent studies elucidate the great potential of GWAS for the genetic dissection of important traits in major crop species. Researchers have mostly used SNP markers for the GWAS analysis116, because of the ability of the NGS-based genotyping systems to provide genome-wide marker data in cost- and time-efficient manner11. As mentioned earlier, SNP markers are biallelic in nature having low informativeness and mutational rate117. Besides, the SNP arrays possess the inherent ascertainment biases, and thus in the GWAS analyses, the significant SNPs often do not represent the causal molecular variants5,8. It can be explained by the fact that rare alleles often determine the extreme phenotypes23. The existence of LD between true molecular variant and the non-causative markers causes stronger marker-trait linkage than that of causal variant itself25,118.
Several researchers advocate for using haplotypes for conducting GWAS (Fig. 2). Recent GWA studies based on empirical and simulation data have revealed higher mapping accuracy and power of haplotype blocks over individual SNPs for the detection of QTLs/genes76,119–122. A variety of reasons explain this superiority of haplotypes (Fig. 2). For example, Stephens et al.27 demonstrated that the multi-allelic nature of haplotype blocks makes them more informative compared to SNP markers (biallelic in nature). The authors reported higher abundance of haplotype variants than SNPs, indicating recombination and recurrent mutation events within and among the genes in the haplotype. Moreover, the haplotype-based analysis is expected to control false positives and reveal the complex mechanism of causal haplotypes in a better way as compared to individual SNPs. For example, the repulsion states between two causal QTLs located close to each other26. In particular, haplotype-based analysis can capture epistatic interactions between SNPs at a locus123,124, provide more information to estimate whether two alleles are IBD125, assess the biological role played by neighboring amino-acids on a protein structure123, reduce the number of tests and hence the type I error rate126, capture information from evolutionary history127, and can provide more power than single marker system to analyze an allelic series existing at a particular locus128–131. To this end, Hamblin and Jannink129 reported that as compared to individual-based SNP markers, the haplotype approach increased the allelic effect and phenotypic variation explained (PVE) by 34% and 50%, respectively. N’Diaye et al.120 observed that by combining multiple SNPs into haplotype blocks, the average PIC increased from 0.27 per SNP to 0.50 per haplotype in wheat. Over the last few years, haplotype-based GWAS analyses have identified important QTLs and candidate genes for various crop traits (Table 1). Greater power of haplotype-based mapping compared to SNP-based GWAS in the detection of genetic loci associated with the plant height and biomass was evident in maize119. It is interesting to note that in comparison to single SNP-based mapping the haplotype-based mapping detected fewer significant associations and candidate genes for drought tolerance in maize; however, with higher PVE values132. Recently, applications of haplotype-based GWAS for various traits including yield, quality and stress tolerance in different plant species such as Arabidopsis133, soybean134, wheat121, barley131,135, rice136 and maize137 have shown great promise for trait discovery and crop improvement.
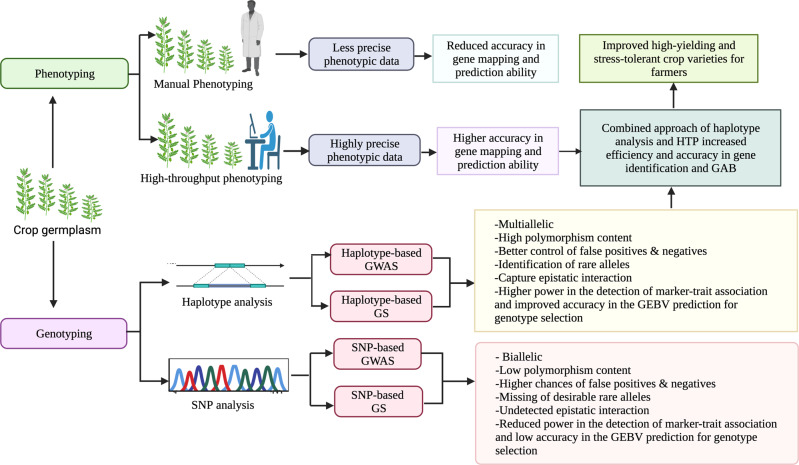
Fig. 2. Mining of SNPs and construction of haplotypes for detecting marker-trait associations (GWAS) and computing genomic estimated breeding values (GS).
This diagram describes the comparative potential of the Haplotype-Based GWAS/Haplotype-Based GS in relation to SNP-Based GWAS/SNP-Based GS for the development of improved crop cultivars via genomics-assisted breeding (GAB). It showed that Haplotype-Based GWAS/Haplotype-Based GS in combination with the high-throughput phenotyping (HTP) has great potential to enhance the precision and accuracy in the gene identification and GAB. The image was created using BioRender (https://biorender.com/).
However, the presence of non-informative SNPs in a given haplotype block (either small or long block) masks the effect of adjacent informative SNPs, which in turn leads to spurious associations, decreasing the effectiveness of the GWAS analysis138. Hence, the haplotype-based GWAS and GS analyses uses the approaches such as sliding windows of fixed/variable length, haplotypes diversity among samples, LD between adjacent SNPs, and SNP number within haplotype to construct the haplotype blocks139. All these approaches have one thing in common i.e., they all use the consecutive SNPs that possess high LD for the development of haplotypes. Therefore, under many circumstances, the haplotypes generated via these approaches’ have been observed to show no difference in the information provided by the haplotype and single SNP, because the SNPs in high LD provide redundant information140. To this end, recently a new haplotype-based GWAS approach called FH-GWAS has been introduced76. This approach uses a different method to generate haplotypes i.e., only those SNPs are combined into functional haplotypes that possess true contribution to the haplotype effects via additive and/or epistatic effects. Thus, FH-GWAS is able to overcome the constraints of combining redundant SNPs (in high LD) into haplotypes and avoids the highly time-consuming process of selecting optimal combinations of SNPs. It is therefore expected to be more powerful than SNP-based and other haplotype-based GWAS approaches.
FH-GWAS analysis: an efficient substitute for discovering superior haplotype alleles
Notwithstanding the superiority of GWAS based on haplotypes over SNPs, the use of haplotypes in the GWAS faces some challenges141. For instance, the contrasting effects of different haplotype allele classes will be diluted if the irrelevant markers are added to a possible causal genetic variant123. Theoretically, in the case of a haplotype with m SNPs, the total number of different haplotype alleles will be equal to 2 m. This will increase the degree of freedom (this holds good for the estimation of population structure but not for GWAS, especially in the estimation of means and variance if the haplotypes are identified only once or twice), and that in turn will diminish the power of association analysis131. However, the 2m formula for determining the number of haplotype alleles do not always work in practice because haplotype diversity is affected by a variety of factors including genetic structure and size of the population, mutation, recombination, marker ascertainment and demography142. For example, Scott et al.143 by analyzing a panel of 16 wheat genotypes, representing the founders of MAGIC population, established that by using the SNPs of the promoter and genic regions, at most of the genes no greater than three haplotypes are identified, and most of the genes were biallelic. Besides, the most critical factor affecting the haplotype-based GWAS analysis is the method(s) used for the construction of haplotypes, as discussed in the previous section. Only the consecutive SNPs in high LD are grouped into the haplotypes in all these methods. Sometimes the redundant information is provided by the SNPs that are in high LD, and as a result the use of these haplotypes does not provide more information than the individual SNPs140. This explains the contradictions reported in recent studies regarding the efficiency of haplotype- and SNP-based GWAS approaches76. As discussed above, the alternative approaches have been proposed for the identification of the haplotypes with non-consecutive SNPs that provide more information than the haplotypes with consecutive SNPs74,140,144. Also, high computational burden associated with these approaches, further limits their use in the association studies74.
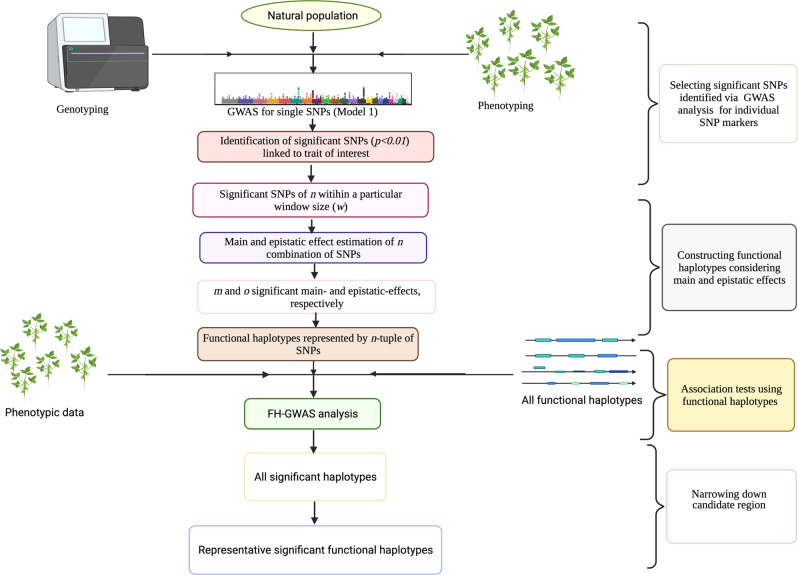
To alleviate the limitations of the haplotype-based GWAS, an alternative efficient approach based on functional haplotype-based-GWAS (FH-GWAS) has been introduced to identify the superior haplotype alleles for the trait of interest76 (Fig. 3). Given the significant role that the epistasis plays in the regulation of complex trait variations, FH-GWAS takes the associated epistatic effects of SNPs into consideration for trait discovery24,145,146. Hence, in FH-GWAS, the SNPs possessing mild threshold for the main effects are first selected, followed by the identification of consecutive and/or non-consecutive combinations of SNPs (having significant epistatic effects) in a chromosomal region of defined size (Fig. 3). This approach combines only those SNPs into a functional haplotype that really contribute to the haplotype effects via additive and/or epistatic effects, thus preventing the redundant SNPs (with high LD) from combining into a haplotype. Besides, it prevents the laborious and time-consuming search for the detection of the optimal combinations of SNPs. In this regard, FH-GWAS is more powerful and efficient compared to haplotype-based and SNP-based approaches.
Fig. 3. Functional haplotype-GWAS (FH-GWAS) analysis for identification of the superior haplotypes for traits of interest.
FH-GWAS approach first involves the individual SNP-marker based GWAS analysis (Model 1), that allows the identification of the candidate SNPs (SNP-trait association). This step is followed by the identification of the closely linked adjacent SNPs within a specific range in a chromosome region; and the SNPs within this specific region possessing additive and/or epistatic effects as well as have true contribution to the haplotype effects are combined into the functional haplotype. Lastly, the GWAS analysis was again performed by combining the functional haplotypes and phenotypic data, that ultimately leads to the identification of significant haplotypes associated with the trait of interest. The image was created using BioRender (https://biorender.com/).
FH-GWAS outperformed SNP-based approach in a simulation study unless the SNPs of the haplotypes possess low MAF and the LD of haplotype SNPs is high76. Analysis of flowering-time trait in a large population of Arabidopsis thaliana using FH-GWAS has revealed its great potential and efficiency in the association studies76. Importantly, FH-GWAS detected all the genomic/candidate regions that were also identified via the SNP-based and haplotype-based GWAS approaches; however, it was only the FH-GWAS that could find a novel genomic region for flowering time on chromosome 4 of A. thaliana76. In view of the evidences available from both simulation and empirical studies, FH-GWAS arguably holds a great promise for trait mapping in crop breeding. Further, this approach can be used for any crop species, particularly the homozygous ones, where sufficient coverage and suitable size of SNPs are available76. However, if the FH-GWAS is to be used for the improvement of multiple traits, the construction of functional haplotypes for each individual trait must be done separately, as the tests of main and epistatic effects of markers are trait-dependent. Although FH-GWAS can improve the efficiency of the gene-trait association studies, this approach is computationally demanding in comparison to the other haplotype-based approaches76.
Haplotype-based breeding (HBB)
The development of stress-tolerant crop varieties with improved yield potential is one of the major challenges for breeders, especially in the face of global climate change3,124. As discussed earlier, GS has emerged as an efficient approach for addressing complex polygenic traits, population improvement and developing improved varieties. The germplasm pool of the most crop species possesses complex genome structure; hence, the use of haplotypes in GS has been proposed as a powerful approach to improve the accuracy and efficiency in the prediction ability26. This is because the comprehensive haplotype maps allow the identification and utilization of genomic regions linked to a particular trait at higher accuracy in populations with pronounced LD structures4.
Implementation of haplotypes in crop improvement is accomplished through two approaches, viz., retrospective and prospective81. During the long-term selection process, the plant breeders have selected the favorable haplotypes that lead to desirable phenotype(s) for the trait(s) of interest. Hence, by using the genome resequencing approach to sequence an elite gene pool, these favorable haplotypes can be identified in the elite crop germplasm26. Furthermore, the molecular markers that define these favorable haplotypes can be developed and then all these haplotype-defining markers can be used to select the most desirable combination of haplotypes governing the specific phenotype. Besides, these haplotype-related markers can be used to separate favorable and unfavorable genetic variation by identifying lines with novel recombination in chromosomal blocks of interest. On the other hand, the haplotypes can also be used in the prospective manner, in which the large collection of ancestral and wild germplasm of particular crop species (not only the elite breeding pools) can be re-sequenced to identify haplotypes with a broader range of genetic variation81. In this approach, the genome-wide haplotypes are used to identify the novel haplotypes present in the wide range of natural germplasm. Hence, the main objective of this approach is to identify the new, desirable and superior haplotypes. In summary, based on information/utility of various haplotypes, it is possible for assembling desirable haplotype combinations to develop optimal parents in breeding programmes. Deployment of haplotypes in breeding as mentioned above has been referred as haplotype-based breeding (HBB)6,20.
Haplotype-assisted genomic selection
The prediction accuracies of GS models for yield and stress-related traits have outperformed the classical selection models, implying that GS is particularly suitable for the improvement of high-yielding and stress-tolerant crop cultivars3,147. For example, Zhang et al.148 demonstrated higher prediction accuracy of GS (0.75–0.87) as compared to MAS (0.62–0.75) for important agronomic traits in soybean. Similarly, GS was found superior to phenotypic selection for improving multiple agronomic traits related to yield and stress tolerance in different crop species147. Besides, GS can reduce the time required to complete a selection cycle in crop plants, which can lead to increased production of the commercially important crops7,149. Because of their high PIC value, fitting haplotypes with statistically significant associations to phenotypes as fixed effects in GS models could further improve prediction accuracies150,151. The haplotype-assisted GS depicts the complex relationships between genotypic information and phenotypes more accurately than individual SNPs. Hence, this approach could ultimately help further increasing selection gain per unit of time. The use of haplotypes may improve the accuracy of genomic prediction because haplotypes can better capture LD and genomic similarity in different lines and may capture local high-order allelic interactions109. Additionally, prediction accuracy could be improved by portraying population structure in the calibration set. A recent GS study that compared the prediction ability computed from haplotypes and SNPs in a set of 383 advanced lines and cultivars of wheat established the superiority of haplotype-based predictions over SNP-based predictions for all studied traits i.e., yield, test weight and protein content152. As compared to the individual SNPs, the combined use of haplotypes of 15 adjacent markers and training population optimization significantly improved the predictive ability for yield and protein content by 14.3% (four percentage points) and 16.8% (seven percentage points), respectively. Similar results were reported by other researchers in different crops such as maize151, Brassica napus152, and sorghum80. Recent examples on the use of haplotype markers for genomic selection/prediction analysis in different crop species are presented in Table 2. Taken together, these studies underscore better performance of haplotypes in comparison to individual markers in improving prediction accuracies of GS for complex traits. Hence, the use of haplotypes in GS will definitely increase the prediction ability and greatly assist in harnessing the true potential of GAB in crop improvement.
Conclusion
GAB approaches aim to accelerate the pace of genetic gain and contribute to the global food and nutrition security. Several GAB approaches such as MABC, MARS and more recently GS have been successfully utilized for developing superior varieties. However, in the context of large-scale genome resequencing projects of germplasm accessions and breeding lines, it is possible to define new haplotypes. The availability of long-read sequencing technologies is also accelerating the discovery of haplotypes that are helpful to improve genome assembly. From applications perspective, these haplotypes can be used for a variety of purposes. Instead of using SNPs, haplotype-based GWAS analysis identifies causal polymorphism in a precise manner. Similarly, evidence demonstrating higher genomic prediction efficiency, based on haplotypes as compared to SNPs, encourages researchers to increasingly embrace haplotypes-assisted genomic prediction in crop improvement programmes. Furthermore, advances in high-throughput phenotyping would enhance discovery and subsequent applications of superior haplotypes in crop breeding. We believe that haplotype-based research and their applications will be routine to develop improved cultivars for future food security.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
R.K.V. thanks Bill and Melinda Gates Foundation, USA (Grants ID# OPP1130244, OPP114827), Department of Cooperation and Farmers Welfare of the Ministry of Agriculture and Farmers Welfare, and JC Bose National Fellowship of Science & Engineering Research Board of Department of Science & Technology, Government of India for financial support. Authors are thankful to Rutwik Barmukh from ICRISAT for his help.
Author contributions
R.K.V. and S.A.G. conceptualized the idea and planned manuscript content. J.A.B. and D.Y. developed the manuscript draft. A.B. contributed special sections. A.B. and R.K.V. edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks Hon-Ming Lam and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Leena Tripathi and Caitlin Karniski.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Showkat Ahmad Ganie, Email: showkatmanzoorforever@gmail.com.
Rajeev K. Varshney, Email: rajeev.varshney@murdoch.edu.au
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02782-y.
References
- 1.Bhat JA, et al. Role of silicon in mitigation of heavy metal stresses in cplants. Plants. 2019;8:71. doi: 10.3390/plants8030071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganie SA, Reddy ASN. Stress-induced changes in alternative splicing landscape in rice: Functional significance of splice isoforms in stress tolerance. Biology. 2021;10:309. doi: 10.3390/biology10040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat JA, et al. Harnessing high-throughput phenotyping and genotyping for enhanced drought tolerance in crop plants. J. Biotechnol. 2020;324:248–260. doi: 10.1016/j.jbiotec.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Varshney RK, Graner A, Sorrells ME. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005;10:621–630. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Bhat JA, et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016;7:221. doi: 10.3389/fgene.2016.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varshney RK, et al. Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 2021;26:631–649. doi: 10.1016/j.tplants.2021.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Varshney RK, Terauchi R, McCouch SR. Harvesting the promising fruits of genomics: Applying genome sequencing technologies to crop breeding. PLoS Biol. 2014;12:e1001883. doi: 10.1371/journal.pbio.1001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zargar SM, et al. Recent advances in molecular marker techniques: insight into QTL mapping, GWAS and genomic selection in plants. J. Crop Sci. Biotechnol. 2015;18:293–308. doi: 10.1007/s12892-015-0037-5. [DOI] [Google Scholar]
- 9.Przewieslik-Allen AM, et al. Developing a high-throughput SNP-based marker system to facilitate the introgression of traits from Aegilops species into bread wheat (Triticum aestivum) Front. Plant Sci. 2019;9:1993. doi: 10.3389/fpls.2018.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Ann. Rev. Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- 11.Rasheed A, et al. Crop breeding chips and genotyping platforms: progress, challenges, and perspectives. Mol. Plant. 2017;10:1047–1064. doi: 10.1016/j.molp.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Ganal MW, et al. Large SNP arrays for genotyping in crop plants. J. Biosci. 2012;37:821–828. doi: 10.1007/s12038-012-9225-3. [DOI] [PubMed] [Google Scholar]
- 13.Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.) Plant Sci. 2016;242:23–36. doi: 10.1016/j.plantsci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Yu Z, et al. Identification of QTN and candidate gene for seed-flooding tolerance in soybean [Glycine max (L.) Merr.] using genome-wide association study (GWAS) Genes. 2019;10:957. doi: 10.3390/genes10120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertsen CD, Hjortshøj RL, Janss LL. Genomic selection in cereal breeding. Agronomy. 2019;9:95. doi: 10.3390/agronomy9020095. [DOI] [Google Scholar]
- 16.Voss-Fels K, Snowdon RJ. Understanding and utilizing crop genome diversity via high‐resolution genotyping. Plant Biotechnol. J. 2016;14:1086–1094. doi: 10.1111/pbi.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collard BC, Jahufer MZZ, Brouwer JB, Pang ECK. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142:169–196. doi: 10.1007/s10681-005-1681-5. [DOI] [Google Scholar]
- 18.Brachi B, Morris GP, Borevitz JO. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 2011;12:1–8. doi: 10.1186/gb-2011-12-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. Plant Genome. 2008;1:5–20. doi: 10.3835/plantgenome2008.02.0089. [DOI] [Google Scholar]
- 20.Varshney RK, et al. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020;56:190–196. doi: 10.1016/j.pbi.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crossa J, et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trend. Plant Sci. 2017;22:961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Annicchiarico, P. et al. GBS‐based genomic selection for pea grain yield under severe terminal drought. Plant Genome10, 10.3835/plantgeonme2016.07.0072 (2017). [DOI] [PubMed]
- 23.Wray NR, et al. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 2013;14:507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 25.Korte A, Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Meth. 2013;9:1–9. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian L, et al. Exploring and harnessing haplotype diversity to improve yield stability in crops. Front. Plant Sci. 2017;8:1534. doi: 10.3389/fpls.2017.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, et al. Mitochondrial haplotypes may modulate the phenotypic manifestation of the deafness-associated 12S rRNA 1555A> G mutation. Mitochondrion. 2010;10:69–81. doi: 10.1016/j.mito.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganie SA, Wani SH, Henry R, Hensel G. Improving rice salt tolerance by Precision Breeding in a New era. Curr. Opin. Plant Biol. 2021;60:101996. doi: 10.1016/j.pbi.2020.101996. [DOI] [PubMed] [Google Scholar]
- 30.Ceballos H, Kawuki RS, Gracen VE, Yencho GC, Hershey CH. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: a case study for cassava. Theor. Appl. Genet. 2015;128:1647–1667. doi: 10.1007/s00122-015-2555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradshaw JE. Plant breeding: past, present and future. Euphytica. 2017;213:60. doi: 10.1007/s10681-016-1815-y. [DOI] [Google Scholar]
- 32.Lenaerts B, Collard BCY, Demont M. Review: improving global food security through accelerated plant breeding. Plant Sci. 2019;287:110207. doi: 10.1016/j.plantsci.2019.110207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banziger, M. & Diallo, A. O. Progress in developing drought and N stress tolerant maize cultivars for eastern and southern Africa in Integrated approaches to higher maize productivity in the new millennium. In Proc. 7th Eastern and Southern Africa Regional Maize Conference, CIMMYT/KARI, Nairobi, Kenya (eds Friesen, D. K. & Palmer, A. F. E.) 189–194 (CIMMYT (International Maize and Wheat Improvement Center) and KARI (Kenya Agricultural Research Institute, 2004).
- 34.Qian L, Qian W, Snowdon RJ. Haplotype hitchhiking promotes trait co-selection in Brassica napus. Plant Biotechnol. J. 2016;14:1578–1588. doi: 10.1111/pbi.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mühleisen J, Maurer HP, Stiewe G, Bury P, Reif JC. Hybrid breeding in barley. Crop Sci. 2013;53:819. doi: 10.2135/cropsci2012.07.0411. [DOI] [Google Scholar]
- 36.Dong H, Li W, Tang W, Zhang D. Development of hybrid Bt cotton in China—a successful integration of transgenic technology and conventional techniques. Curr. Sci. 2004;86:778–782. [Google Scholar]
- 37.Atlin GN, Cairns JE, Das B. Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Glob. Food Sec. 2017;12:31–37. doi: 10.1016/j.gfs.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labroo MR, Studer AJ, Rutkoski JE. Heterosis and hybrid crop breeding: a multidisciplinary review. Front. Genet. 2021;12:643761. doi: 10.3389/fgene.2021.643761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khush GS. Rice breeding: past, present and future. J. Genet. 1987;66:195–216. doi: 10.1007/BF02927713. [DOI] [Google Scholar]
- 40.Ashraf M. Inducing drought tolerance in plants: recent advances. Biotechnol. Adv. 2010;28:169–183. doi: 10.1016/j.biotechadv.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Glenn KC, et al. Bringing new plant varieties to market: Plant breeding and selection practices advance beneficial characteristics while minimizing unintended changes. Crop Sci. 2017;57:2906. doi: 10.2135/cropsci2017.03.0199. [DOI] [Google Scholar]
- 42.Bradshaw JE. Review and analysis of limitations in ways to improve conventional potato breeding. Potato Res. 2017;60:171–193. doi: 10.1007/s11540-017-9346-z. [DOI] [Google Scholar]
- 43.Saxena RK, et al. Genomics for greater efficiency in pigeonpea hybrid breeding. Front. Plant Sci. 2015;6:793. doi: 10.3389/fpls.2015.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qaim M. Role of new plant breeding technologies for food security and sustainable agricultural development. Appl. Econom. Pers. Policy. 2020;42:129–150. doi: 10.1002/aepp.13044. [DOI] [Google Scholar]
- 45.Evenson RE. Assessing the impact of the green revolution, 1960 to 2000. Science. 2003;300:758–762. doi: 10.1126/science.1078710. [DOI] [PubMed] [Google Scholar]
- 46.Sinha P, et al. Superior haplotypes for haplotype‐based breeding for drought tolerance in pigeonpea (Cajanus cajan L.) Plant Biotechnol. J. 2020;18:2482–2490. doi: 10.1111/pbi.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varshney, R. K. et al. Fast-track introgression of root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome6, 10.3835/plantgenome2013.07.0022 (2013).
- 48.Bharadwaj C, et al. Introgression of “QTL-hotspot” region enhances drought tolerance and grain yield in three elite chickpea cultivars. Plant Genome. 2021;14:e20076. doi: 10.1002/tpg2.20076. [DOI] [PubMed] [Google Scholar]
- 49.Henry A, Gowda VR, Torres RO, McNally KL, Serraj R. Variation in root system architecture and drought response in rice (Oryza sativa): phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crop Res. 2011;120:205–214. doi: 10.1016/j.fcr.2010.10.003. [DOI] [Google Scholar]
- 50.Kumar A, et al. Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J. Exp. Bot. 2014;65:6265–6278. doi: 10.1093/jxb/eru363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahmed HU, et al. Genetic, physiological, and gene expression analyses reveal that multiple QTL enhance yield of rice mega-variety IR64 under drought. PloS ONE. 2013;8:e62795. doi: 10.1371/journal.pone.0062795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry A, et al. Physiological mechanisms contributing to the QTL-combination effects on improved performance of IR64 rice NILs under drought. J. Exp. Bot. 2015;66:1787–1799. doi: 10.1093/jxb/eru506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasan MM, et al. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 2015;29:237–254. doi: 10.1080/13102818.2014.995920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cobb JN, Biswas PS, Platten JD. Back to the future: revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019;132:647–667. doi: 10.1007/s00122-018-3266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dormatey R, et al. Gene pyramiding for sustainable crop improvement against biotic and abiotic stresses. Agronomy. 2020;10:1255. doi: 10.3390/agronomy10091255. [DOI] [Google Scholar]
- 56.Gokidi Y, Bhanu AN, Singh MN. Marker assisted recurrent selection: an overview. Adv. Life Sci. 2016;5:6493–6499. [Google Scholar]
- 57.Khan A, Sovero V, Gemenet D. Genome-assisted breeding for drought resistance. Curr. Genomics. 2016;17:330–342. doi: 10.2174/1389202917999160211101417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali M, et al. Modeling and simulation of recurrent phenotypic and genomic selections in plant breeding under the presence of epistasis. Crop J. 2020;8:866–877. doi: 10.1016/j.cj.2020.04.002. [DOI] [Google Scholar]
- 59.Borrell AK, et al. Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Exp. Bot. 2014;65:6251–6263. doi: 10.1093/jxb/eru232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy NRR, Ragimasalawada M, Sabbavarapu MM, Nadoor S, Patil JV. Detection and validation of stay-green QTL in post-rainy sorghum involving widely adapted cultivar, M35-1 and a popular stay-green genotype B35. BMC Genomics. 2014;15:1–16. doi: 10.1186/1471-2164-15-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varshney RK, et al. Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor. Appl. Genet. 2019;132:797–816. doi: 10.1007/s00122-018-3252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, et al. Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front. Plant Sci. 2018;9:190. doi: 10.3389/fpls.2018.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beyene Y, et al. Genetic gains in grain yield through genomic selection in eight bi‐parental maize populations under drought stress. Crop Sci. 2015;55:154–163. doi: 10.2135/cropsci2014.07.0460. [DOI] [Google Scholar]
- 65.Juliana P, et al. Retrospective quantitative genetic analysis and genomic prediction of global wheat yields. Front. Plant Sci. 2020;11:580136. doi: 10.3389/fpls.2020.580136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Y, et al. Genomic selection of agronomic traits in hybrid rice using an NCII population. Rice. 2018;11:32. doi: 10.1186/s12284-018-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui Z, et al. Assessment of the potential for genomic selection to improve husk traits in maize. G3. 2020;10:3741–3749. doi: 10.1534/g3.120.401600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stewart-Brown BB, Song Q, Vaughn JN, Li Z. Genomic selection for yield and seed composition traits within an applied soybean breeding program. G3. 2019;9:2253–2265. doi: 10.1534/g3.118.200917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roorkiwal M, et al. Genomic-enabled prediction models using multi-environment trials to estimate the effect of genotype × environment interaction on prediction accuracy in chickpea. Sci. Rep. 2018;8:11701. doi: 10.1038/s41598-018-30027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandey MK, et al. Genome-based trait prediction in multi-environment breeding trials in groundnut. Theor. Appl. Genet. 2020;133:3101–3117. doi: 10.1007/s00122-020-03658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stram, D. O. Multi-SNP haplotype analysis methods for association analysis. In Statistical Human Genetics. Methods Mol. Biol. (ed. Elston, R.) vol 1666, 485–504 (Humana Press, New York, NY, 2017). [DOI] [PubMed]
- 72.Garg S. Computational methods for chromosome-scale haplotype reconstruction. Genome Biol. 2021;22:1–24. doi: 10.1186/s13059-021-02328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ammar, R., Paton, T. A., Torti, D., Shlien, A. & Bader, G. D. Long read nanopore sequencing for detection of HLA and CYP2D6 variants and haplotypes. F1000Research4, 17 (2015). [DOI] [PMC free article] [PubMed]
- 74.Maldonado C, Mora F, Scapim CA, Coan M. Genome-wide haplotype-based association analysis of key traits of plant lodging and architecture of maize identifies major determinants for leaf angle: Hap LA4. PloS ONE. 2019;14:e0212925. doi: 10.1371/journal.pone.0212925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu F, Schmidt RH, Reif JC, Jiang Y. Selecting closely-linked SNPs based on local epistatic effects for haplotype construction improves power of association mapping. G3. 2019;9:4115–4126. doi: 10.1534/g3.119.400451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang BE, Amos CI, Lin DY. Detecting haplotype effects in genome wide association studies. Genet. Epidemiol. 2007;31:803–812. doi: 10.1002/gepi.20242. [DOI] [PubMed] [Google Scholar]
- 78.Gupta PK, Rustgi S, Kulwal PL. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol. Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- 79.Dixon LE, Pasquariello M, Boden SA. TEOSINTE BRANCHED1 regulates height and stem internode length in bread wheat. J. Exp. Bot. 2020;71:4742–4750. doi: 10.1093/jxb/eraa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jensen SM, Svensgaard J, Ritz C. Estimation of the harvest index and the relative water content–Two examples of composite variables in agronomy. Eur. J. Agron. 2020;112:125962. doi: 10.1016/j.eja.2019.125962. [DOI] [Google Scholar]
- 81.Bevan MW, et al. Genomic innovation for crop improvement. Nature. 2017;543:346–354. doi: 10.1038/nature22011. [DOI] [PubMed] [Google Scholar]
- 82.Abbai R, et al. Haplotype analysis of key genes governing grain yield and quality traits across 3K RG panel reveals scope for the development of tailor-made rice with enhanced genetic gains. Plant Biotechnol. J. 2019;17:1612–1622. doi: 10.1111/pbi.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalisz S, Kramer EM. Variation and constraint in plant evolution and development. Heredity. 2008;100:171–177. doi: 10.1038/sj.hdy.6800939. [DOI] [PubMed] [Google Scholar]
- 84.Sella G, Barton NH. Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu. Rev. Genomics Hum. Genet. 2019;20:461–493. doi: 10.1146/annurev-genom-083115-022316. [DOI] [PubMed] [Google Scholar]
- 85.Meier, J. I. et al. Haplotype tagging reveals parallel formation of hybrid races in two butterfly species. bioRxiv10.1073/pnas.2015005118 (2020). [DOI] [PMC free article] [PubMed]
- 86.Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. The importance of phase information for human genomics. Nat. Rev. Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garud NR, Rosenberg NA. Enhancing the mathematical properties of new haplotype homozygosity statistics for the detection of selective sweeps. Theor. Popul. Biol. 2015;102:94–101. doi: 10.1016/j.tpb.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maestri S, et al. A long-read sequencing approach for direct haplotype phasing in clinical settings. Int. J. Mol. Sci. 2020;21:9177. doi: 10.3390/ijms21239177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Delaneau O, et al. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-13225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amarasinghe SL, et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:1–16. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Al Bkhetan Z, Zobel J, Kowalczyk A, Verspoor K, Goudey B. 2019. Exploring effective approaches for haplotype block phasing. BMC Bioinform. 2019;20:1–14. doi: 10.1186/s12859-019-3095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laver TW, et al. Pitfalls of haplotype phasing from amplicon-based long-read sequencing. Sci. Rep. 2016;6:1–6. doi: 10.1038/srep21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang S, et al. Long-read sequencing and haplotype linkage analysis enabled preimplantation genetic testing for patients carrying pathogenic inversions. J. Med. Genet. 2019;56:741–749. doi: 10.1136/jmedgenet-2018-105976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. Direct determination of diploid genome sequences. Genome Res. 2017;27:757–767. doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wenger AM, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019;37:1155–1162. doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun, H. et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. bioRxiv10.1101/2021.05.15.444292 (2021). [DOI] [PMC free article] [PubMed]
- 97.Amini S, et al. Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet. 2014;46:1343–1349. doi: 10.1038/ng.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang O, et al. Efficient and unique cobarcoding of second-generation sequencing reads from long DNA molecules enabling cost-effective and accurate sequencing, haplotyping, and de novo assembly. Genome Res. 2019;29:798–808. doi: 10.1101/gr.245126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang F, et al. Haplotype phasing of whole human genomes using bead-based barcode partitioning in a single tube. Nat. Biotechnol. 2017;35:852–857. doi: 10.1038/nbt.3897. [DOI] [PubMed] [Google Scholar]
- 100.Ganie SA, Molla KA, Henry RJ, Bhat KV, Mondal TK. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019;132:851–870. doi: 10.1007/s00122-019-03301-8. [DOI] [PubMed] [Google Scholar]
- 101.Khanzada H, et al. Differentially evolved drought stress indices determine the genetic variation of Brassica napus at seedling traits by genome-wide association mapping. J. Adv. Res. 2020;24:447–461. doi: 10.1016/j.jare.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang X, et al. Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize. Plant Biotechnol. J. 2020;18:1271–1283. doi: 10.1111/pbi.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Calus MP, et al. Effects of the number of markers per haplotype and clustering of haplotypes on the accuracy of QTL mapping and prediction of genomic breeding values. Genet. Sel. Evol. 2009;41:1–10. doi: 10.1186/1297-9686-41-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cuyabano BC, Su G, Lund MS. Genomic prediction of genetic merit using LD-based haplotypes in the Nordic Holstein population. BMC Genomics. 2014;15:1–11. doi: 10.1186/1471-2164-15-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ballesta P, Maldonado C, Pérez-Rodríguez P, Mora F. SNP and haplotype-based genomic selection of quantitative traits in Eucalyptus globulus. Plants. 2019;8:331. doi: 10.3390/plants8090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Won S, et al. Genomic prediction accuracy using haplotypes defined by size and hierarchical clustering based on linkage disequilibrium. Front. Genet. 2020;11:134. doi: 10.3389/fgene.2020.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Matias FI, Galli G, Correia Granato IS, Fritsche-Neto R. Genomic prediction of autogamous and allogamous plants by SNPs and haplotypes. Crop Sci. 2017;57:2951–2958. doi: 10.2135/cropsci2017.01.0022. [DOI] [Google Scholar]
- 108.Zhang X, Zhang S, Zhao Q, Ming R, Tang H. Assembly of allele-aware, chromosomal-scale autopolyploid genomes based on Hi-C data. Nat. Plants. 2019;5:833–845. doi: 10.1038/s41477-019-0487-8. [DOI] [PubMed] [Google Scholar]
- 109.Hamazaki K, Iwata H. RAINBOW: Haplotype-based genome-wide association study using a novel SNP-set method. PLoS Comput. Biol. 2020;16:e1007663. doi: 10.1371/journal.pcbi.1007663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vinholes P, Rosado R, Roberts P, Borém A, Schuster I. Single nucleotide polymorphism‐based haplotypes associated with charcoal rot resistance in Brazilian soybean germplasm. Agron. J. 2019;111:182–192. doi: 10.2134/agronj2018.07.0429. [DOI] [Google Scholar]
- 111.Nyine M, et al. Association genetics of bunch weight and its component traits in East African highland banana (Musa spp. AAA group) Theor. Appl. Genet. 2019;132:3295–3308. doi: 10.1007/s00122-019-03425-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meuwissen TH, Odegard J, Andersen-Ranberg I, Grindflek E. On the distance of genetic relationships and the accuracy of genomic prediction in pig breeding. Genet. Sel. Evol. 2014;46:1–8. doi: 10.1186/1297-9686-46-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hickey JM, et al. Sequencing millions of animals for genomic selection 2.0. J. Anim. Breed. Genet. 2013;130:331–332. doi: 10.1111/jbg.12054. [DOI] [PubMed] [Google Scholar]
- 114.Sargolzaei M, Chesnais JP, Schenkel FS. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 2014;15:1–12. doi: 10.1186/1471-2164-15-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Broman KW, Weber JL. Long homozygous chromosomal segments in reference families from the centre d’Etude du polymorphisme humain. Am. J. Hum. Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Würschum T, Maurer HP, Dreyer F, Reif JC. Effect of inter-and intragenic epistasis on the heritability of oil content in rapeseed (Brassica napus L.) Theor. Appl. Genet. 2013;126:435–441. doi: 10.1007/s00122-012-1991-7. [DOI] [PubMed] [Google Scholar]
- 118.Platt A, Vilhjálmsson BJ, Nordborg M. Conditions under which genome-wide association studies will be positively misleading. Genetics. 2010;186:1045–1052. doi: 10.1534/genetics.110.121665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu X, et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.N’Diaye A, et al. Single marker and haplotype-based association analysis of semolina and pasta colour in elite durum wheat breeding lines using a high-density consensus map. PLoS ONE. 2017;12:e0170941. doi: 10.1371/journal.pone.0170941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Basile SML, et al. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019;19:1–16. doi: 10.1186/s12870-018-1600-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Srivastava RK, et al. Genome-wide association studies and genomic selection in Pearl Millet: Advances and prospects. Front. Genet. 2020;10:1389. doi: 10.3389/fgene.2019.01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Clark AG. The role of haplotypes in candidate gene studies. Genet. Epidemiol. 2004;27:321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- 124.Bardel C, Danjean V, Hugot JP, Darlu P, Génin E. On the use of haplotype phylogeny to detect disease susceptibility loci. BMC Genet. 2005;6:1–13. doi: 10.1186/1471-2156-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meuwissen THE, Goddard ME. Fine mapping of quantitative trait loci using linkage disequilibria with closely linked marker loci. Genetics. 2000;155:421–430. doi: 10.1093/genetics/155.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Templeton AR, Boerwinkle E, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and an analysis of alcohol dehydrogenase activity in Drosophila. Genetics. 1987;117:343–351. doi: 10.1093/genetics/117.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Akey J, Jin L, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Euro. J. Hum. Genet. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- 129.Hamblin, M. T. & Jannink, J. L. Factors affecting the power of haplotype markers in association studies. Plant Genome4, 10.3835/plantgenome2011.03.0008 (2011).
- 130.Morris RW, Kaplan NL. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet. Epidemiol. 2002;23:221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- 131.Gawenda I, Thorwarth P, Günther T, Ordon F, Schmid KJ. Genome‐wide association studies in elite varieties of German winter barley using single‐marker and haplotype‐based methods. Plant Breed. 2015;134:28–39. doi: 10.1111/pbr.12237. [DOI] [Google Scholar]
- 132.Yuan X, Biswas S. Bivariate logistic Bayesian LASSO for detecting rare haplotype association with two correlated phenotypes. Genet. Epidemiol. 2019;43:996–1017. doi: 10.1002/gepi.22258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu X, et al. Resequencing of cv CRI‐12 family reveals haplotype block inheritance and recombination of agronomically important genes in artificial selection. Plant Biotechnol. J. 2019;17:945–955. doi: 10.1111/pbi.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Contreras-Soto RI, et al. A genome-wide association study for agronomic traits in soybean using SNP markers and SNP-based haplotype analysis. PLoS ONE. 2017;12:e0171105. doi: 10.1371/journal.pone.0171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abed A, Belzile F. Comparing single-SNP, multi-SNP, and haplotype-based approaches in association studies for major traits in Barley. Plant Genome. 2019;12:190036. doi: 10.3835/plantgenome2019.05.0036. [DOI] [PubMed] [Google Scholar]
- 136.Wang X, et al. Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan Y, et al. Genome-wide association mapping and genomic prediction analyses reveal the genetic architecture of grain yield and flowering time under drought and heat stress conditions in maize. Front. Plant Sci. 2019;9:1919. doi: 10.3389/fpls.2018.01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mathias RA, et al. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet. 2006;7:1–11. doi: 10.1186/1471-2156-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Srivastava A, et al. Most frequent South Asian haplotypes of ACE2 share identity by descent with East Eurasian populations. PLoS One. 2020;15:e0238255. doi: 10.1371/journal.pone.0238255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laramie JM, Wilk JB, DeStefano AL, Myers RH. HaploBuild: an algorithm to construct non-contiguous associated haplotypes in family based genetic studies. Bioinformatics. 2007;23:2190–2192. doi: 10.1093/bioinformatics/btm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lorenz AJ, Hamblin MT, Jannink JL. Performance of single nucleotide polymorphisms versus haplotypes for genome-wide association analysis in barley. PLoS ONE. 2010;5:e14079. doi: 10.1371/journal.pone.0014079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Stumpf MP. Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur. J. Hum. Gene. 2004;12:469–477. doi: 10.1038/sj.ejhg.5201179. [DOI] [PubMed] [Google Scholar]
- 143.Scott MF, et al. Limited haplotype diversity underlies polygenic trait architecture across 70 years of wheat breeding. Genome Biol. 2021;22:1–30. doi: 10.1186/s13059-021-02354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Knüppel S, et al. Multi-locus stepwise regression: a haplotype-based algorithm for finding genetic associations applied to atopic dermatitis. BMC Med Genet. 2012;13:8. doi: 10.1186/1471-2350-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- 146.Massawe F, Mayes S, Cheng A. Crop diversity: an unexploited treasure trove for food security. Trend Plant Sci. 2016;21:365–368. doi: 10.1016/j.tplants.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 147.Matei G, et al. Genomic selection in soybean: accuracy and time gain in relation to phenotypic selection. Mol. Breed. 2018;38:1–13. doi: 10.1007/s11032-018-0872-4. [DOI] [Google Scholar]
- 148.Zhang J, Song Q, Cregan PB, Jiang GL. Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max) Theor. Appl. Genet. 2016;129:117–130. doi: 10.1007/s00122-015-2614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qin J, et al. Genome wide association study and genomic selection of amino acid concentrations in soybean seeds. Front. Plant Sci. 2019;10:1445. doi: 10.3389/fpls.2019.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jiang Y, Schmidt RH, Reif JC. Haplotype-based genome-wide prediction models exploit local epistatic interactions among markers. G3. 2018;8:1687–1699. doi: 10.1534/g3.117.300548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sallam AH, Conley E, Prakapenka D, Da Y, Anderson JA. Improving prediction accuracy using multi-allelic haplotype prediction and training population optimization in wheat. G3. 2020;10:2265–2273. doi: 10.1534/g3.120.401165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jan HU, et al. Genome-wide haplotype analysis improves trait predictions in Brassica napus hybrids. Plant Sci. 2019;283:157–164. doi: 10.1016/j.plantsci.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 153.Bruce RW, et al. Haplotype diversity underlying quantitative traits in Canadian soybean breeding germplasm. Theor. Appl. Genet. 2020;133:1967–1976. doi: 10.1007/s00122-020-03569-1. [DOI] [PubMed] [Google Scholar]
- 154.Sehgal D, et al. Haplotype-based, genome-wide association study reveals stable genomic regions for grain yield in CIMMYT spring bread wheat. Front. Genet. 2020;11:589490. doi: 10.3389/fgene.2020.589490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ogawa D, et al. Haplotype analysis from unmanned aerial vehicle imagery of rice MAGIC population for the trait dissection of biomass and plant architecture. J. Exp. Bot. 2021;72:2371–2382. doi: 10.1093/jxb/eraa605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Maldonado C, Mora F, Bertagna FAB, Kuki MC, Scapim CA. SNP- and haplotype-based GWAS of flowering-related traits in maize with network-assisted gene prioritization. Agronomy. 2019;9:725. doi: 10.3390/agronomy9110725. [DOI] [Google Scholar]
- 157.Mayer M, et al. Discovery of beneficial haplotypes for complex traits in maize landraces. Nat. Commun. 2020;11:4954. doi: 10.1038/s41467-020-18683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bekele WA, Wight CP, Chao S, Howarth CJ, Tinker NA. Haplotype-based genotyping-by-sequencing in oat genome research. Plant Biotechnol. J. 2018;16:1452–1463. doi: 10.1111/pbi.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ma Y, et al. Potential of marker selection to increase prediction accuracy of genomic selection in soybean (Glycine max L.) Mol. Breed. 2016;36:113. doi: 10.1007/s11032-016-0504-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jensen SE, et al. A sorghum practical haplotype graph facilitates genome-wide imputation and cost-effective genomic prediction. Plant Genome. 2020;13:e20009. doi: 10.1002/tpg2.20009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.