Abstract
Proteostasis is primarily a function of protein synthesis and degradation. Although the components and processes involved in intracellular proteostasis have been studied extensively, it is apparent that extracellular proteostasis is equitably crucial for the viability of organisms. The 26S proteasome, a unique ATP-dependent proteolytic complex in eukaryotic cells, contributes to the majority of intracellular proteolysis. Accumulating evidence suggests the presence of intact 20S proteasomes in the circulatory system (c-proteasomes), and similar to other plasma proteins, the levels of these c-proteasomes may vary, potentially reflecting specific pathophysiological conditions. Under normal conditions, the concentration of c-proteasomes has been reported to be in the range of ~0.2–2 μg/mL, which is ~2–4-fold lower than that of functional plasma proteins but markedly higher than that of signaling proteins. The characterization of c-proteasomes, such as their origin, structure, role, and clearance, has been delayed mainly due to technical limitations. In this review, we summarize the current perspectives pertaining to c-proteasomes, focusing on the methodology, including our experimental understanding. We believe that once the pathological relevance of c-proteasomes is revealed, these unique components may be utilized in the diagnosis and prognosis of diverse human diseases.
Subject terms: Diagnostic markers, Proteasome
Disease biomarkers: Circulating protein complexes
Protein complexes called proteasomes degrade other proteins inside cells as part of cellular maintenance, but accumulating evidence suggests that proteasomes also circulate in the blood at levels that may vary in association with several diseases and also with normal aging. Researchers do not fully understand the cellular origins, biological functions and control of these circulating ‘c-proteasomes’, partly due to technical difficulties in analyzing their presence and their activity in blood samples. Min Jae Lee and colleagues at Seoul National University in South Korea review current understanding of c-proteasomes and their relevance to diseases including various cancers of blood cells and solid tissues, autoimmune disorders, sepsis and mild cognitive impairment. Emerging evidence suggests that c-proteasome levels and activity could serve to diagnose diseases and perhaps also to monitor and predict disease progression.
Introduction: Proteasomes
Protein homeostasis, or proteostasis, is achieved both quantitatively (via translation and proteolysis) and qualitatively (by folding, cleavage, and posttranslational modification). In eukaryotic cells, the ubiquitin–proteasome system (UPS) is known to be involved in the degradation of the majority of regulatory proteins and the elimination of aberrant protein products, thereby critically contributing to the maintenance of the functional protein pool1. Polyubiquitylated UPS substrates are hydrolyzed by proteasomes in both the cytoplasm and nucleus into small peptide fragments. The 26S proteasome is an ~2.5 MDa holoenzyme comprising two distinct and dissociable components: the 20S catalytic particle and the 19S regulatory particle2. The cylindrical 20S proteasome is composed of four heteroheptameric rings (α7β7β7α7): the α1−7 outer ring forms a part of the substrate translocation channel, while the β1−7 inner ring retains the proteolytic active sites (caspase-like, trypsin-like, and chymotrypsin-like activities in the β1, β2, and β5 subunits, respectively). The 19S complex (with a molecular weight of ~930 kDa) recognizes, unfolds, and translocates the target substrates into the interior of the 20S proteasome (~730 kDa) and simultaneously deubiquitylates these substrates, thereby allowing ubiquitin recycling3. Marked advances in the structural and mechanical details of the 26S proteasome were recently made through cryo-electron microscopy4–6.
The archetype 20S proteasome was once considered a latent enzyme, partly because the gate in the substrate translocation channel was identified to be closed7,8. However, a growing number of endogenous substrates of the 20S proteasome with diverse physiological functions have been identified over the last decade, and now, this ubiquitin- and ATP-independent mode of proteasomal degradation is generally perceived as an alternative key regulator of the cellular proteome. Notably, the 20S proteasome degrades either fully or partially disordered proteins, including amyloid-β peptides9, tau10,11, α-synuclein12,13, and the cell-cycle-regulating proteins p21, p53, and p7314–16. In addition, oxidatively modified proteins, which largely affect the intracellular signaling pathway and cell viability when they accumulate, also form a class of 20S substrates17,18. This larger than anticipated physiological and pathological role of the 20S proteasome appears to be mediated by its conformational change upon engagement with substrates (from a resting form to a processing form)19. The Glickman group recently demonstrated that the binding of an unstructured model substrate was sufficient to induce conformational changes in α subunits and subsequently 20S gate opening, facilitating substrate hydrolysis without the involvement of the 19S proteasome or other activators20.
Several other chambered proteases with structural and mechanical similarities to the eukaryotic 20S proteasome have been identified in prokaryotes, which lack ubiquitin. For example, bacterial ClpP and HslV comprise 14 and 12 identical subunits, respectively, and sequester their active sites inside their gated chambers21. Direct binding of specific sequence motifs (in bacteria) or disordered regions (in eukaryotes) to chambered protease complexes may be evolutionarily analogous biochemical features in controlled proteolysis. Considering that more than 30% of the total proteins identified in eukaryotes are disordered-sregion-containing proteins22, the contribution of ubiquitin-independent 20S proteasome-mediated degradation in global proteostasis must be more prevalent than what is currently characterized. Fabre et al. used in vivo cross-linking prior to quantitative mass spectrometry to evaluate the steady-state ratio between the 20S and 26S proteasomes and found that 47–74% of these proteasomes exist in their free 20S form across a wide range of mammalian cell lines23,24. The ratio between the proteasome subtypes is expected to be dynamically switched due to changes in the cellular environment, allowing these proteasome subtypes to actively participate in cellular surveillance against pathological stress, such as reactive oxygen species25–27.
Circulating proteasomes
For decades, proteostasis has commonly been referred to as an intracellular protein quality control mechanism. Our insight into how extracellular proteostasis operates and functions remains unclear, although more than 10% of the protein-encoding gene products are secreted28 and the majority of proteopathies, a group of human diseases associated with toxic protein accumulation and aggregation, occur in extracellular fluids29. Since the discovery of circulating proteasomes (hereafter referred to as c-proteasomes) in human blood ~30 years ago30, the clinical relevance of these components has been examined in various diseases, including myeloid and lymphoid malignancies, solid tumors, autoimmune disorders, sepsis, mild cognitive impairment, and other clinical conditions31,32. However, many of these studies used limited cohort sizes without further validation with independent reproduction. Here, we summarize the findings of previous studies on c-proteasomes (principally in chronological order) and discuss some of the key technical aspects of c-proteasome research. The availability of related reviews precludes the discussion on the extracellular proteasomes found in alveolar, cerebrospinal, and epididymal fluids, as well as other extracellular spaces31,33.
c-Proteasomes in serum
The c-proteasome was first identified in the serum in 1993, not long after proteasomes were first purified from rabbit reticulocyte lysates30,34 (Table 1). Wada et al. used a customized enzyme-linked immunosorbent assay (ELISA) and self-raised monoclonal anti-α6 (then described as C2, the first proteasome subunit cloned in 198935) antibody to evaluate the concentration of c-proteasomes in the serum of healthy individuals (N = 20) and patients. They determined that the average concentration of these proteins in the serum was 359.6 ng/mL and that these values were significantly elevated in patients with various hematological malignancies and in patients with liver disease (N = 175). They observed the highest c-proteasome level increase in patients with adult T cell leukemia (13.0 μg/mL, N = 6). They separated the serum proteins using gel filtration and enriched an ~650 kDa multimeric protein complex30, which was later characterized as the potential 20S proteasome. The levels of c-proteasomes in patients with various liver diseases were also elevated (1.34 μg/mL in patients with hepatocellular carcinoma; N = 16), and these values were positively correlated with those of serum alanine aminotransferase30.
Table 1.
Level and activity of c-proteasomes in human serum.
| First author | Year | Disease | Disease in detail | N | Serum c-proteasome concentration (ng/mL) | Chymotrypsin-like activity | Reference |
|---|---|---|---|---|---|---|---|
| Wadaa | 1993 | Healthy | 20 | 359.6 | N/A | 30 | |
| Hematological malignancies | Acute leukemia | 12 | 2900.4 | ||||
| Chronic myelogenous leukemia | 7 | 1964.6 | |||||
| Myelodysplastic syndrome | 3 | 1366.7 | |||||
| Non-Hodgkin’s lymphoma | 16 | 866.3 | |||||
| Adult T cell leukemia | 6 | 12,955.0 | |||||
| Multiple myeloma | 12 | 577.8 | |||||
| Chronic lymphocytic leukemia | 2 | 546.5 | |||||
| Waldenstrom’s macroglobulinemia | 3 | 373.3 | |||||
| Liver diseases | Acute hepatitis | 4 | 20,589.0 | ||||
| Chronic hepatitis | 55 | 735,9 | |||||
| Liver cirrhosis | 23 | 608.0 | |||||
| Hepatocellular carcinoma | 16 | 1340 | |||||
| Fatty liver | 16 | 577.9 | |||||
| Egerera | 2002 | Healthy | 85 | 221.4 | N/A | 36 | |
| Systemic autoimmune diseases | Autoimmune myositis | 10 | 1598.4 | ||||
| Jo-1 syndrome | 6 | 693.0 | |||||
| Systemic lupus erythematosus | 76 | 681.3 | |||||
| Autoimmune hepatitis | 37 | 669.8 | |||||
| Primary Sjögren syndrome | 56 | 598.6 | |||||
| Antiphospholipid syndrome | 11 | 565.9 | |||||
| Rheumatoid arthritis | 66 | 531.6 | |||||
| Vasculitis | 21 | 522.2 | |||||
| Systemic scleroderma | 14 | 499.4 | |||||
| CREST syndrome | 7 | 353.8 | |||||
| Myasthenia gravis | 10 | 293.0 | |||||
| Jakobb | 2007 | Healthy | 50 | 224.1 | N/A | 37 | |
| Multiple myeloma (MM) | Monoclonal gammopathy of undetermined significance | 20 | 378.1 | ||||
| Total MM | 141 | 599.6 | |||||
| Smoldering MM | 40 | 314.7 | |||||
| Active MM | 101 | 744.3 | |||||
| Majetschakb | 2008 | Healthy | 22 | 445.5 | N/A | 38 | |
| Systemic autoimmune diseases | Connective tissue disease | 35 | 831 | ||||
| Systemic lupus erythematous | 56 | 889 | |||||
| de Martinob | 2012 | Healthy | 15 | 1520 | N/A | 39 | |
| CRCC | Clear cell renal cell carcinoma (CRCC) | 113 | 4660 | ||||
| Rotha | 2004 | Healthy | 15 | 2157 | N/A | 40 | |
| Sepsis | 15 | 33,551 | |||||
| Abdominal surgery | 15 | 4661 | |||||
| Trauma in the intensive care unit | 13 | 29,669 | |||||
| Kakurinab | 2017 | Healthy | 15 | N/A | 1150 U/ml | 41 | |
| HNSCC | Head and neck squamous cell carcinoma (HNSCC) | 48 | N/A | 1166–1500 U/ml | |||
aThe values are presented as the mean.
bThe values are presented as the median.
It was not until the early 2000s that studies on c-proteasomes were reported again. Thereafter, the Feist group determined that the average concentration of c-proteasomes in the serum was 221.4 ng/mL in healthy individuals (N = 85), and they found that c-proteasome concentrations were significantly elevated, in a range of 300–700 ng/mL, in patients with a variety of systemic autoimmune diseases (total N = 314)36. Notably, they also detected 20S proteasome subunits in serum samples using ion-exchange chromatography, ammonium sulfate precipitation, and subsequent sucrose-gradient ultracentrifugation36. The serum c-proteasome levels in patients with multiple myeloma were found to be significantly elevated (median 744.3 ng/mL, N = 101) compared with those in healthy controls (median 224.1 ng/mL, N = 50)37. After chemotherapy, a significant decrease in the levels of c-proteasomes was observed in patients with a positive response to chemotherapy but not in nonresponders37.
Only a few other studies reported serum levels of c-proteasomes (Table 1). Several studies used an in-house ELISA and commercial anti-α6 antibodies to demonstrate that c-proteasome levels were significantly higher in patients with autoimmune diseases, such as systemic lupus erythematous (median 889 ng/mL [N = 56] vs. 446 ng/mL in controls [N = 22])38 and clear cell renal cell carcinoma (median 4.66 μg/mL [N = 113] vs. 1.52 μg/mL [N = 15] in controls)39. A dramatic increase in serum c-proteasome levels was observed in patients with sepsis resulting from peritonitis and pneumonia (33.6 μg/mL [N = 15] vs. 2.16 μg/mL [N = 15] in healthy controls)40. In addition to the c-proteasome concentration, the activity of the c-proteasomes in the sera was also evaluated and reported. These evaluations revealed that the activity of c-proteasomes was largely correlated with the size and degree of the differentiation of tumors in patients with head and neck squamous cell carcinoma (N = 48)41.
c-Proteasomes in plasma
The majority of extracellular proteasome studies were performed using human plasma, usually using a small cohort (N ≤ 100). One of the earliest plasma c-proteasome studies included a total of 317 patients, with only 14–44 patients in each neoplastic disease group. The Bureau group performed an α6-targeting ELISA to evaluate the concentrations of c-proteasomes in the plasma of healthy individuals (2.36 μg/mL, N = 73) and demonstrated that these values were increased in patients with hemopoietic malignancies, including myeloproliferative disorder (4.10 μg/mL, N = 37) and myelodysplastic syndromes (2.92 μg/mL, N = 19). The c-proteasome levels were generally higher in plasma samples than in serum samples (Table 2). The changes in the plasma c-proteasome profile were correlated with the prognoses of some patients42,43. In a similar manner, the c-proteasome levels were measured in patients with metastatic malignant melanoma, and the highest c-proteasome levels were found to be correlated with the most advanced stages of melanoma (median 8.55 μg/mL in stage IV patients [N = 10] vs. 1.96 μg/mL in healthy individuals [N = 14])44. They also found that plasma c-proteasome levels were significantly higher in patients with hepatocellular carcinoma (3.74 μg/mL, N = 50) than in patients with cirrhosis without malignant transformation (1.81 μg/mL, N = 33) and controls (2.3 μg/mL, N = 40)45. In a follow-up study, the levels of c-proteasomes were significantly and positively correlated with the progression of melanoma, suggesting that these enzyme complexes could be used in the diagnosis of metastatic melanoma46.
Table 2.
Level and activity of c-proteasomes in human plasma.
| First author | Year | Disease | Disease in detail | N | Plasma proteasome concentration (ng/mL) | Chymotrypsin-like activity | Ref |
|---|---|---|---|---|---|---|---|
| Lavabre-Bertranda | 2001 | Healthy | 73 | 2356 | N/A | 42 | |
| Hemopoietic malignancies | Solid tumor | 20 | 7589 | ||||
| Myeloproliferative disorder | 37 | 4099 | |||||
| Myelodysplastic syndromes | 19 | 2922 | |||||
| Stoebnerb | 2005 | Healthy | 14 | 1957 | N/A | 44 | |
| Metastatic malignant melanoma | Stage I/II | 13 | 2515 | ||||
| Stage III | 6 | 3725 | |||||
| Stage IV | 10 | 8554 | |||||
| Severe psoriasis | 13 | 2981 | |||||
| Chronic idiopathic urticaria | 6 | 3190 | |||||
| Henryb | 2009 | Healthy | 40 | 2302 | N/A | 45 | |
| Liver cirrhosis | with hepatocellular carcinoma (HCC) | 50 | 3737 | ||||
| without HCC | 33 | 1808 | |||||
| Henryb | 2013 | Metastatic melanoma | Stage I/II | 53 | 184 | N/A | 46 |
| Stage III | 41 | 228 | |||||
| Stage IV | 27 | 499 | |||||
| Mab | 2008 | Healthy | 40 | N/A | 0.80 pmol/s/mL | 47 | |
| Chronic lymphocytic leukemia | 225 | N/A | 1.84 pmol/s/mL | ||||
| Mab | 2009 | Healthy | 97 | N/A | 0.8 pmol/s/mL | 48 | |
| Acute myeloid leukemia | 174 | N/A | 2.0 pmol/s/mL | ||||
| Myelodysplastic syndrome | 52 | N/A | 1.4 pmol/s/mL | ||||
| Majetschakb | 2010 | Healthy | 40 | 195 | N/A | 54 | |
| Burn | Day 0 | 50 | 673 | ||||
| Day 30 | 40 | 116.5 | |||||
| Heubnerb | 2011 | Healthy | 55 | 290 | N/A | 55 | |
| Epithelial ovarian cancer | Patient | 120 | 595 | ||||
| Patient, after therapy | 68 | 457.5 | |||||
| Hoffmannb | 2011 | Healthy | 50 | 305 | N/A | 56 | |
| Nonmetastasized breast cancer | 224 | 397.5 | |||||
| Fukasawab | 2015 | Healthy | 76 | 1,340 | N/A | 57 | |
| Hemodialysis patient | 76 | 1.381 | |||||
| Manasanchb | 2017 | Multiple myeloma | Patient | 45 | N/A | 0.83 pmol/s/ml | 58 |
| After carfilzomib treatment | N/A | 0.23 pmol/s/ml | |||||
| Oldziejb | 2014 | Healthy | 30 | 2010 | 1.02 U/mg | 59 | |
| Multiple myeloma | 64 | 4380 | 1.32 U/mg | ||||
aThe values are presented as the mean.
bThe values are presented as the median.
The Albitar group demonstrated an increase in plasma c-proteasome activity in patients with chronic lymphocytic leukemia (N = 225)47, acute myeloid leukemia (N = 174), and advanced-stage myelodysplastic syndrome (N = 52)48. All three proteolytic activities in patients were found to be elevated compared with those in healthy controls, and they showed significant correlations with prognosis, therapeutic response, and survival prediction. In both cases, the plasma samples (collected in EDTA tubes) were incubated with a 1% final concentration of sodium dodecyl sulfate (SDS) for 15 min to “activate” the plasma47,48. To clinically employ c-proteasome activity as a reliable biomarker, it seems to be important to simultaneously quantify the c-proteasome absolute (rather than relative) activity and concentration.
The hydrolysis of peptidyl fluorogenic substrates has been widely used to determine c-proteasome activity in human blood. These substrates include succinyl-Leu-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (suc-LLVY-AMC), ter-butyloxycarbonyl-Leu-Arg-Arg-AMC (Boc-LRR-AMC), and carbonylbenzyl-Leu-Leu-Glu-AMC (Z-LLE-AMC), which are specific for chymotrypsin-like β5, trypsin-like β2, and caspase-like β1 activities, respectively, in the 20S catalytic chamber49. The fluorescence intensities must be normalized to the values obtained in the presence of proteasome inhibitors as the identical principle applies to assess the purified proteasomes and whole-cell/tissue lysates. The hydrolysis of suc-LLVY-AMC, which was used as a primary substrate during the first proteasome assays in the late 1980s34, is largely regarded to represent the overall proteasome activity50. Using human plasma, we also observed that the changes in the activity of the three catalytic sites in c-proteasomes were highly correlated (Fig. 1a). In addition, suc-LLVY-AMC hydrolysis was not affected by a number of protease inhibitors; however, it was effectively abolished by reversible or irreversible proteasome inhibitors (Fig. 1b, c). In vitro reconstituted polyubiquitylated proteins, such as polyubiquitylated sic1 or DHFR51,52, which act as more physiologically relevant substrates for proteasomal degradation, have rarely been evaluated in studies related to c-proteasome activity.
Fig. 1. Activity of circulating proteasomes (c-proteasomes) in human plasma.
a Plasma samples were collected from four individuals (plasma A–D) in EDTA tubes, and their c-proteasome activity (in 20 μL of plasma) was evaluated by monitoring the hydrolysis of the fluorogenic reporter substrates (final concentration of 250 μM in a total of 100 μL reaction), such as suc-LLVY-AMC (for chymotrypsin-like activity), Boc-LRR-AMC (for trypsin-like activity), and Z-LLE-AMC (for caspase-like activity) in the presence or absence of the proteasome inhibitor MG132 (10 μM). These reactions were performed using assay buffer (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mg/mL BSA, 1 mM ATP, and 1 mM DTT). Sodium dodecyl sulfate (SDS) was not added to the reaction unless otherwise described. The graphs (left) represent the results obtained in three independent experiments, and the mean of the raw fluorescence values (right) at 60 min are plotted with their standard deviations (N = 3). b Human c-proteasome activity was analyzed using suc-LLVY-AMC as the substrate, along with a wide range of protease inhibitors, including aprotinin (trypsin inhibitor), pepstatin A (aspartyl protease inhibitor), and leupeptin (serine/cysteine protease inhibitor). c As in (b), but using different proteasome inhibitors (10 μM MG132, 2 μM bortezomib, 2 μM epoxomicin, or 100 nM carfilzomib). d The plasma samples were preincubated with SDS at the indicated final concentrations for 10 min before initiating the suc-LLVY-AMC hydrolysis reactions. Relative fluorescence values after 30-min reactions were normalized to those obtained in the presence of 10 μM MG132. The values represent the mean ± standard deviation (N = 3). e As in (a), except that assay buffer without ATP was used. No significant changes were observed.
A critical concern for the assessment of c-proteasome activity using human blood and fluorogenic substrates is the addition of varying concentrations of SDS (0.1–1.0% final concentration), which is intended for the “preactivation” of c-proteasomes. Although mildly elevated proteasome activity after SDS incubation was reported34,53, it has also been shown that the 20S component exhibits strong suc-LLVY-AMC hydrolysis activity even in the absence of SDS and that a high concentration of SDS may inadvertently inhibit c-proteasome activity (Fig. 1d). It is possible that certain concentrations of SDS may result in the dissolution of phospholipid membrane-like extracellular vesicles encapsulating c-proteasomes, leading to an increase in c-proteasome activity in these assays. Therefore, the application of SDS to mediate c-proteasome activation requires further validation. In addition, we found that the plasma samples prepared in EDTA tubes showed much higher c-proteasome activity than those prepared in heparin tubes or the serum samples that underwent a clotting process (data not shown). The exact mechanism underlying these differences remains unclear, but it seems to be critical to resolve this issue to establish a standardized protocol and improve experimental reproducibility.
c-Proteasome levels and activities were both found to be elevated in burn patients on the day of admission compared to those in healthy volunteers54. The highest median 20S concentration (673 ng/mL, N = 50) was reported on Day 0, but the levels gradually decreased within the first week following burn injury, eventually returning to baseline levels (195 ng/mL, N = 40) after 30 days. The 26S form of c-proteasomes was virtually undetectable in the ELISA-based assay54. A standard proteasome peptidase assay was performed using the plasma samples collected in EDTA or sodium citrate tubes: 100 μM suc-LLVY-AMC and 35 μL of plasma in 10 mM Tris-HCl (pH 7.5) buffer at 37 °C for 60 min, without SDS activation. Basal proteasome activity was determined upon the addition of an irreversible proteasome inhibitor, epoxomicin (7 μM), to the mixture. The relatively weak c-proteasome activity was detected in their samples: among the nine randomly selected samples, only three specimens exhibited detectable fluorescence signals corresponding to the reporter peptides54. Nevertheless, the findings of this study strongly supported the notion that c-proteasomes are primarily expressed in the 20S form and are enzymatically active even without further activation.
In a large-scale study, the Heubner group observed elevated plasma levels of c-proteasomes in patients with epithelial ovarian cancer (595 ng/mL, N = 120) and nonmetastatic breast cancer (397.5 ng/mL, N = 224) compared to the levels in healthy controls (290 ng/mL, N = 55)55,56. Fukasawa et al. thoroughly assessed plasma c-proteasome levels and various other clinical parameters in 76 patients undergoing hemodialysis and found a significant negative correlation between c-proteasome levels and abdominal muscle area57. In patients with multiple myeloma, treatment with a single dose of the proteasome inhibitor carfilzomib (20 mg/m2), which irreversibly targets the β5 (chymotrypsin-like) proteolytic site, led to drastically reduced chymotrypsin-like activity, but it did not affect caspase- or trypsin-like activities58. In a similar study conducted by Oldziej et al.59, both the concentration and the activity of c-proteasomes in plasma samples were shown to be significantly higher in patients with multiple myeloma (4.38 μg/mL and 1.32 U/mg [N = 64]) than in healthy controls (2.01 μg/mL and 1.02 U/mg [N = 30]). The Matuszczak group analyzed the plasma levels of c-proteasomes in pediatric patients with mild head injury60, acute appendectomy61, and moderate to major burns62. In general, the group found that c-proteasome activity and concentration increased initially following an acute onset and reduced gradually after treatment. Most recently, a significant correlation was reported between the c-proteasome levels and the decline in the lean tissue indices in hemodialysis patients over two years, although c-proteasome levels could not be used to predict patient survival in that particular time period63.
A series of biochemical purifications, including albumin removal, ammonium sulfate precipitation, anion exchange column fractionation, and affinity purification, followed by negative-staining electron microscopy, revealed that most c-proteasomes in the plasma exist in the 20S form64. ATP-independent proteolytic activity of c-proteasomes was observed in both mouse and human plasma, consistent with the general principle of the 20 S c-proteasomes established in previous studies (Fig. 1e). Considering the ATP-depleted environment of the blood and the role of ATP in the association between the 20S and 19S subunits, it is conceivable that c-proteasomes circulate in the free 20S form in human plasma. This possibility would suggest a protective function of 20S c-proteasomes in the clearance of potentially harmful misfolded proteins in the extracellular space, which is highly oxidizing (in comparison with the reducing nature of the intracellular compartment). We postulate that the c-proteasomes in the plasma may exhibit different enzymatic activities than those exhibited by intracellular proteasomes. For example, 20S c-proteasomes with limited proteolytic activity may catalyze the partial cleavage of disordered (or oxidized) plasma proteins, similar to the function of the serine proteases involved in hemostasis. Certain translation initiation factors, transcription factors, and heat shock proteins have already been described to be endoproteolytically processed by intracellular proteasomes65. Therefore, the physiological role of the c-proteasomes in extracellular proteostasis may be to assist in untangling the aggregation-prone proteins (rather than complete protein degradation) and in facilitating their cell-surface receptor-mediated lysosomal degradation.
Conclusions and clinical perspectives
In this review, we have summarized the current knowledge of c-proteasomes in the serum and plasma, focusing on methodological aspects. Many studies discussed herein reported a strong correlation between disease status and the level (or activity) of 20S c-proteasomes. However, c-proteasomes are still an orphan biological entity for which the origin, function, substrate, and regulatory mechanism have yet to be elucidated. Our understanding of the potential of c-proteasomes as diagnostic and prognostic factors at the cellular level is far from comprehensive. The delays in this field are primarily attributed to technical issues associated with blood chemistry and the lack of a standardized protocol. The implementation of a wide variety of laboratory procedures, from sample acquisition procedures to assay protocols, also makes it difficult to obtain consistency and concordance between the results of independent c-proteasome studies. Since researchers are reluctant to publish negative or nonsignificant findings, a possible explanation for the very small number of total c-proteasome studies (approximately one paper per year) might be attributed to the negative outcomes in the unreported studies.
Despite many unresolved issues around c-proteasomes, we believe that it is still possible to establish a validated biomarker based on c-proteasomes by improving both research and clinical practices. Biomarkers of Alzheimer’s disease may serve as a good example: only a few years ago, the development of these markers was regarded practically unachievable, but they are now cross-validated and almost ready for clinical implementation due to biological and technical advances66. Considering that cellular proteasome activity has been found to decrease with age67, it seems interesting to examine whether the changes in c-proteasome activity are correlated with the aging process. It seems possible that insufficient renal function, such as low urinary filtration, could lead to elevated levels of c-proteasomes. Notably, the assessment of the enzymatic activity of c-proteasomes is much less expensive (in the cent range) and faster (<30 min per assay) than the performance of antibody-based methodologies. At present, the activity of c-proteasomes can be considered to be positively correlated with their plasma concentration in humans32, and most FDA-approved blood tests are decades old. Therefore, slight advancements in c-proteasome biology and the technology for monitoring c-proteasome activity may fundamentally transform the procedural approaches and allow broader applications of c-proteasomes in routine clinical evaluations in the near future.
The origin of c-proteasomes is one of the major unresolved questions in c-proteasome research. While some studies support the notion that c-proteasomes are passively released from ruptured cells, others suggest that c-proteasomes are transported into the blood via an active secretory mechanism31,33 (references therein). It seems plausible that the 20S proteasome, which is ~10 nm long, is packaged into a membrane-bound organelle and released in a 30–100 nm extracellular vesicle such as exosomes (Fig. 2; route 2). In cultured T lymphocytes, significant numbers of proteasomes are reported to be secreted into the media via an exosomal pathway68,69. We hypothesize that the encapsulated 20S proteasomes originate from not only dissociation from the 19S complex but also de novo synthesis (and assembly) of 20S immunoproteasomes under stress conditions. Alternatively, 20S proteasomes may be shed into the extracellular spaces via microvesicles (Fig. 2; route 1), as the Vidal group recently identified70 or the autophagy–lysosome system may affect the quantity and quality of extracellular components through autophagosome-mediated unconventional secretion (Fig. 2; route 3) instead of the turnover of intracellular proteasomes71,72. Once this information is available, we may have a better biological framework to study the subunit composition, subtypes, and, most of all, the functional relevance of c-proteasomes. Because the concentration of proteasomes is >100-fold higher in the cytosol than in the extracellular milieu73, pharmacological modulations of their secretion system may result in dynamic alterations in the concentration of c-proteasomes in human blood, thereby affecting the pathophysiology of proteopathies characterized by the deposition of extracellular proteins.
Fig. 2. Schematic representation of proteasome secretory pathways.
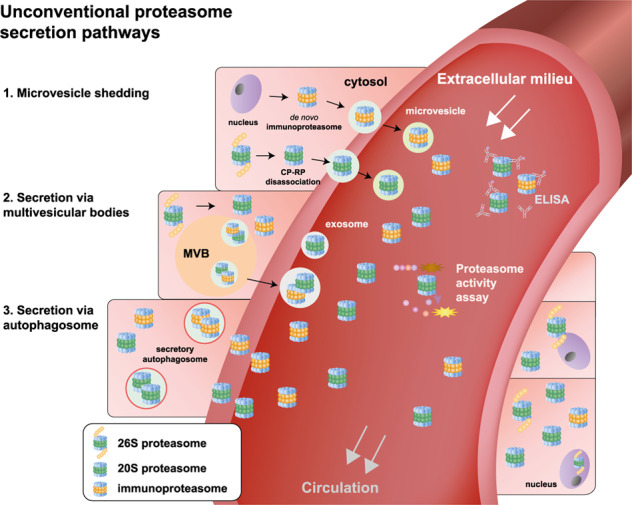
Unlike classic protein secretion involving ER-to-Golgi transport, cytosolic proteasomes are expected to be directed to the extracellular space through unconventional secretory pathways. Under stress or pathological conditions, the 26S proteasome dissociates into 20S and 19S particles, and simultaneously, de novo synthesis of the 20S immunoproteasome may be induced. The 20S forms of proteasomes can be released through microvesicle shedding (route 1). Alternatively, the 20S forms of proteasomes are first packaged into endocytic compartments, such as multivesicular bodies and autophagosomes (routes 2 and 3, respectively), which later fuse with the plasma membrane and are then released as exosomes. The level and activity of 20S c-proteasomes can be readily detected using ELISA and fluorogenic reporter peptides, respectively. The physiological and pathological significance of these pathways remains to be investigated.
Acknowledgements
W.H.C. and S.K. were supported by the BK21 FOUR program funded by the Ministry of Education and National Research Foundation (NRF) of Korea. This work was also supported by other grants from the NRF (2020R1A5A1019023 and 2021R1A2C2008023 to M.J.L.), the Korea Health Industry Development Institute and Korea Dementia Research Center (HU21C0071 to M.J.L.), the Korea Toray Science Foundation, and the Creative-Pioneering Researchers Program through Seoul National University.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Won Hoon Choi, Sumin Kim.
References
- 1.Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka K. The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck F, et al. Near-atomic resolution structural model of the yeast 26S proteasome. Proc. Natl Acad. Sci. USA. 2012;109:14870–14875. doi: 10.1073/pnas.1213333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 7.Choi WH, et al. Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nat. Commun. 2016;7:10963. doi: 10.1038/ncomms10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groll M, et al. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Yang J. Amyloid-beta peptide is a substrate of the human 20S proteasome. ACS Chem. Neurosci. 2010;1:655–660. doi: 10.1021/cn100067e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ukmar-Godec T, et al. Proteasomal degradation of the intrinsically disordered protein tau at single-residue resolution. Sci. Adv. 2020;6:eaba3916. doi: 10.1126/sciadv.aba3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David DC, et al. Proteasomal degradation of tau protein. J. Neurochem. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 12.Olshina MA, et al. Regulation of the 20S Proteasome by a Novel Family of Inhibitory Proteins. Antioxid. Redox Signal. 2020;32:636–655. doi: 10.1089/ars.2019.7816. [DOI] [PubMed] [Google Scholar]
- 13.Moscovitz O, et al. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015;6:6609. doi: 10.1038/ncomms7609. [DOI] [PubMed] [Google Scholar]
- 14.Li X, et al. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Touitou R, et al. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83:301–310. doi: 10.1016/S0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 18.Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 19.Sahu I, Glickman MH. Structural Insights into Substrate Recognition and Processing by the 20S Proteasome. Biomolecules. 2021;11:148. doi: 10.3390/biom11020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahu, I. et al. Signature activities of 20 S proteasome include degradation of the ubiquitin-tag with the protein under hypoxia. Preprint at https://www.biorxiv.org/content/10.1101/2019.12.20.883942v1 (2019).
- 21.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 22.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 23.Fabre B, et al. Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics. Mol. Cell. Proteom. 2013;12:687–699. doi: 10.1074/mcp.M112.023317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabre B, et al. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014;13:3027–3037. doi: 10.1021/pr500193k. [DOI] [PubMed] [Google Scholar]
- 25.Taggart CC, et al. Cathepsin B, L, and S cleave and inactivate secretory leucoprotease inhibitor. J. Biol. Chem. 2001;276:33345–33352. doi: 10.1074/jbc.M103220200. [DOI] [PubMed] [Google Scholar]
- 26.Mayor T, Sharon M, Glickman MH. Tuning the proteasome to brighten the end of the journey. Am. J. Physiol. Cell Physiol. 2016;311:C793–C804. doi: 10.1152/ajpcell.00198.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baugh JM, Viktorova EG, Pilipenko EV. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt AR, Yerbury JJ, Poon S, Wilson MR. Therapeutic targets in extracellular protein deposition diseases. Curr. Med. Chem. 2009;16:2855–2866. doi: 10.2174/092986709788803187. [DOI] [PubMed] [Google Scholar]
- 30.Wada M, et al. Serum concentration and localization in tumor cells of proteasomes in patients with hematologic malignancy and their pathophysiologic significance. J. Lab Clin. Med. 1993;121:215–223. [PubMed] [Google Scholar]
- 31.Dwivedi V, Yaniv K, Sharon M. Beyond cells: the extracellular circulating 20S proteasomes. Biochim. Biophys. Acta Mol. Basis Dis. 2021;1867:166041. doi: 10.1016/j.bbadis.2020.166041. [DOI] [PubMed] [Google Scholar]
- 32.Yun Y, et al. Proteasome activity in the plasma as a novel biomarker in mild cognitive impairment with chronic tinnitus. J. Alzheimers Dis. 2020;78:195–205. doi: 10.3233/JAD-200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sixt SU, Beiderlinden M, Jennissen HP, Peters J. Extracellular proteasome in the human alveolar space: a new housekeeping enzyme? Am. J. Physiol. Lung Cell. Mol. Physiol. 2007;292:L1280–L1288. doi: 10.1152/ajplung.00140.2006. [DOI] [PubMed] [Google Scholar]
- 34.Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1987;262:8303–8313. doi: 10.1016/S0021-9258(18)47564-3. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara T, et al. Molecular cloning of cDNA for proteasomes (multicatalytic proteinase complexes) from rat liver: primary structure of the largest component (C2) Biochemistry. 1989;28:7332–7340. doi: 10.1021/bi00444a028. [DOI] [PubMed] [Google Scholar]
- 36.Egerer K, et al. Circulating proteasomes are markers of cell damage and immunologic activity in autoimmune diseases. J. Rheumatol. 2002;29:2045–2052. [PubMed] [Google Scholar]
- 37.Jakob C, et al. Circulating proteasome levels are an independent prognostic factor for survival in multiple myeloma. Blood. 2007;109:2100–2105. doi: 10.1182/blood-2006-04-016360. [DOI] [PubMed] [Google Scholar]
- 38.Majetschak M, et al. Circulating 20S proteasome levels in patients with mixed connective tissue disease and systemic lupus erythematosus. Clin. Vaccin. Immunol. 2008;15:1489–1493. doi: 10.1128/CVI.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Martino M, et al. Serum 20S proteasome is elevated in patients with renal cell carcinoma and associated with poor prognosis. Br. J. Cancer. 2012;106:904–908. doi: 10.1038/bjc.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth GA, et al. Heightened levels of circulating 20S proteasome in critically ill patients. Eur. J. Clin. Invest. 2005;35:399–403. doi: 10.1111/j.1365-2362.2005.01508.x. [DOI] [PubMed] [Google Scholar]
- 41.Kakurina GV, Cheremisina OV, Choinzonov EL, Kondakova IV. Circulating proteasomes in the pathogenesis of head and neck squamous cell carcinoma. Bull. Exp. Biol. Med. 2017;163:92–94. doi: 10.1007/s10517-017-3745-7. [DOI] [PubMed] [Google Scholar]
- 42.Lavabre-Bertrand T, et al. Plasma proteasome level is a potential marker in patients with solid tumors and hemopoietic malignancies. Cancer. 2001;92:2493–2500. doi: 10.1002/1097-0142(20011115)92:10<2493::AID-CNCR1599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Dutaud D, et al. Development and evaluation of a sandwich ELISA for quantification of the 20S proteasome in human plasma. J. Immunol. Methods. 2002;260:183–193. doi: 10.1016/S0022-1759(01)00555-5. [DOI] [PubMed] [Google Scholar]
- 44.Stoebner PE, et al. High plasma proteasome levels are detected in patients with metastatic malignant melanoma. Br. J. Dermatol. 2005;152:948–953. doi: 10.1111/j.1365-2133.2005.06487.x. [DOI] [PubMed] [Google Scholar]
- 45.Henry L, et al. Plasma proteasome level is a reliable early marker of malignant transformation of liver cirrhosis. Gut. 2009;58:833–838. doi: 10.1136/gut.2008.157016. [DOI] [PubMed] [Google Scholar]
- 46.Henry L, et al. Clinical use of p-proteasome in discriminating metastatic melanoma patients: comparative study with LDH, MIA and S100B protein. Int. J. Cancer. 2013;133:142–148. doi: 10.1002/ijc.27991. [DOI] [PubMed] [Google Scholar]
- 47.Ma W, et al. Enzymatic activity of circulating proteasomes correlates with clinical behavior in patients with chronic lymphocytic leukemia. Cancer. 2008;112:1306–1312. doi: 10.1002/cncr.23301. [DOI] [PubMed] [Google Scholar]
- 48.Ma W, et al. Proteasome enzymatic activities in plasma as risk stratification of patients with acute myeloid leukemia and advanced-stage myelodysplastic syndrome. Clin. Cancer Res. 2009;15:3820–3826. doi: 10.1158/1078-0432.CCR-08-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 50.Lee JH, Lee MJ. Isolation and characterization of RNA aptamers against a proteasome-associated deubiquitylating enzyme UCH37. Chembiochem. 2017;18:171–175. doi: 10.1002/cbic.201600515. [DOI] [PubMed] [Google Scholar]
- 51.Lee JH, et al. Facilitated tau degradation by USP14 aptamers via enhanced proteasome activity. Sci. Rep. 2015;5:10757. doi: 10.1038/srep10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lam YA, Huang JW, Showole O. The synthesis and proteasomal degradation of a model substrate Ub5DHFR. Methods Enzymol. 2005;398:379–390. doi: 10.1016/S0076-6879(05)98031-2. [DOI] [PubMed] [Google Scholar]
- 53.Tylicka M, et al. Proteasome activity and C-reactive protein concentration in the course of inflammatory reaction in relation to the type of abdominal operation and the surgical technique used. Mediators Inflamm. 2018;2018:2469098. doi: 10.1155/2018/2469098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majetschak M, et al. Circulating proteasomes after burn injury. J. Burn Care Res. 2010;31:243–250. doi: 10.1097/BCR.0b013e3181d0f55d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heubner M, et al. The prognostic impact of circulating proteasome concentrations in patients with epithelial ovarian cancer. Gynecol. Oncol. 2011;120:233–238. doi: 10.1016/j.ygyno.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 56.Hoffmann O, et al. Circulating 20S proteasome in patients with non-metastasized breast cancer. Anticancer Res. 2011;31:2197–2201. [PubMed] [Google Scholar]
- 57.Fukasawa H, et al. Circulating 20S proteasome is independently associated with abdominal muscle mass in hemodialysis patients. PLoS ONE. 2015;10:e0121352. doi: 10.1371/journal.pone.0121352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manasanch EE, et al. Enzymatic activities of circulating plasma proteasomes in newly diagnosed multiple myeloma patients treated with carfilzomib, lenalidomide and dexamethasone. Leuk. Lymphoma. 2017;58:639–645. doi: 10.1080/10428194.2016.1214953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oldziej A, et al. Assessment of proteasome concentration and chymotrypsin-like activity in plasma of patients with newly diagnosed multiple myeloma. Leuk. Res. 2014;38:925–930. doi: 10.1016/j.leukres.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Tylicka M, Matuszczak E, Debek W, Hermanowicz A, Ostrowska H. Circulating proteasome activity following mild head injury in children. Childs Nerv. Syst. 2014;30:1191–1196. doi: 10.1007/s00381-014-2409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matuszczak E, et al. Concentration of proteasome in the blood plasma of children with acute appendicitis, before and after surgery, and its correlation with CRP. World J. Surg. 2018;42:2259–2264. doi: 10.1007/s00268-017-4407-7. [DOI] [PubMed] [Google Scholar]
- 62.Matuszczak E, et al. Immunoproteasome in the plasma of pediatric patients with moderate and major burns, and its correlation with proteasome and UCHL1 measured by SPR IMaging Biosensors. J. Burn Care Res. 2018;39:948–953. doi: 10.1093/jbcr/iry011. [DOI] [PubMed] [Google Scholar]
- 63.Aniort J, et al. Circulating 20S proteasome for assessing protein energy wasting syndrome in hemodialysis patients. PLoS ONE. 2020;15:e0236948. doi: 10.1371/journal.pone.0236948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zoeger A, Blau M, Egerer K, Feist E, Dahlmann B. Circulating proteasomes are functional and have a subtype pattern distinct from 20S proteasomes in major blood cells. Clin. Chem. 2006;52:2079–2086. doi: 10.1373/clinchem.2006.072496. [DOI] [PubMed] [Google Scholar]
- 65.Kumar Deshmukh F, Yaffe D, Olshina MA, Ben-Nissan G, Sharon M. The contribution of the 20S proteasome to proteostasis. Biomolecules. 2019;9:190. doi: 10.3390/biom9050190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zetterberg H, Blennow K. Moving fluid biomarkers for Alzheimer’s disease from research tools to routine clinical diagnostics. Mol. Neurodegener. 2021;16:10. doi: 10.1186/s13024-021-00430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saez I, Vilchez D. The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genomics. 2014;15:38–51. doi: 10.2174/138920291501140306113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bochmann I, et al. T lymphocytes export proteasomes by way of microparticles: a possible mechanism for generation of extracellular proteasomes. J. Cell. Mol. Med. 2014;18:59–68. doi: 10.1111/jcmm.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai RC, et al. Proteolytic potential of the MSC exosome proteome: implications for an exosome-mediated delivery of therapeutic proteasome. Int. J. Proteom. 2012;2012:971907. doi: 10.1155/2012/971907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bec N, et al. Proteasome 19S RP and translation preinitiation complexes are secreted within exosomes upon serum starvation. Traffic. 2019;20:516–536. doi: 10.1111/tra.12653. [DOI] [PubMed] [Google Scholar]
- 71.Choi WH, et al. Aggresomal sequestration and STUB1-mediated ubiquitylation during mammalian proteaphagy of inhibited proteasomes. Proc. Natl Acad. Sci. USA. 2020;117:19190–19200. doi: 10.1073/pnas.1920327117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JH, Park S, Kim E, Lee MJ. Negative-feedback coordination between proteasomal activity and autophagic flux. Autophagy. 2019;15:726–728. doi: 10.1080/15548627.2019.1569917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pack CG, et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]



