Abstract
Red palm weevil [Rhynchophorus ferrugineus (Olivier)], is native to South Asia and expanding its distribution range globally. Recent invasions of red palm weevil around the world, including Saudi Arabia, has become a global constraint for the production of palm species. Although, several control measures have been tested, none of them seemed successful against this invasive species. Therefore, we focused on silencing the reproduction control gene vitellogenin (Vg) based on RNA interference (RNAi) strategy for its possible application in the management of R. ferrugineus. The Vg is a major yolk protein precursor critical for oogenesis. To do this, fat body transcriptome of R. ferrugineus female adults was sequenced, which provided partial Vg gene transcript (FPKM 5731.60). A complete RfVg gene transcript of 5504 bp encoding 1787 amino acids was then sequenced using RCAE-PCR strategy and characterized. Phylogenetic analysis suggested that RfVg has closer ancestry to the coleopteran insects. The RfVg-based RNAi significantly suppressed the expressions of Vg gene. The 15, 20 and 25 days post-injection periods suppressed Vg expressions by 95, 96.6 and 99%, respectively. The suppressed Vg expressions resulted in the dramatic failure of Vg protein expression, which caused atrophied ovaries or no oogenesis and ultimately eggs were not hatched. These results suggest that knockdown of Vg gene involved in R. ferrugineus reproduction is a promising target for RNAi-based management of R. ferrugineus.
Subject terms: Biotechnology, Molecular biology, Plant sciences
Introduction
Date palm, [Phoenix dactylifera (Linnaeus, 1753)] is one of the oldest fruit trees in the Arabian Peninsula and major economic fruit crop in Saudi Arabia (SA). Date palm is cultivated on an estimated area of 156,848 hectares1 in SA with an annual production of 1,050,000 MT2. Unfortunately, palm trees are at high risk of red palm weevil [Rhynchophorus ferrugineus (Olivier) (Coleoptera: Dryophthoridae)] infestation, which is the most noxious pest of palm trees.
Rhynchophorus ferrugineus causes severe damages to palm trees while spending its entire life cycle inside the tree3. This species commonly attacks and feeds on young date palm trees, which are less than 20 years of age4. Heavy economic losses have been caused by R. ferrugineus infestation throughout the world during last few decades4. Females of R. ferrugineus can lay 270–396 eggs5. Generally, eggs are hatched within 3 to 5 days and emerging larvae start boring into the interior of the palms. The advanced damage to the palms results in the mortality of trees3. Egg production in R. ferrugineus depends on major yolk protein precursor -vitellogenin (Vg)- and its uptake by developing oocytes6. Although, several strategies, i.e., chemicals, entomopathogens, and pheromone traps7–10 have been used to control R. ferrugineus, all of these were unable to control the pest. The hidden nature of the pest is the possible reason of failed control by these management practices. Continuous and non-judicious application of synthetic chemicals against insect pests have posed adverse effects to human health, caused environmental pollution, and resulted in the evolution of resistant populations of several pests11,12. Nonetheless, excessive pesticides’ use in the field negatively affect the populations of predators and parasites of harmful insects11,13 through non-target effects. Consequently, technologies capable of suppressing insect pest populations in an environment-friendly manner, such as molecular approaches are direly needed for environment-friendly and sustainable management of insect pests. Although R. ferrugineus is a crucial pest of date palm, its reproduction mechanism at molecular level remains elusive. The Vg gene encodes major yolk protein precursor -vitellogenin (Vg)- and plays a critical role in effective reproduction of all oviparous organisms, including insects. In insects, the Vg is expressed in the female fat body cells6,14–16, which is then translated and Vg is secreted into the hemolymph and finally sequestered by oocytes through a particular receptor called Vg receptor (VgR) by endocytosis17–21. Once Vg enters the oocytes, it is stored in crystalline form as vitellin (Vn) and used as a major protein for developing embryo22–24. The Vg genes have been characterized at molecular and genetic levels from several insect species, including cockroaches14,15,25, giant water bug 26 and rice brown planthopper27. It is proposed that Vg could be an ideal target for future pest control strategies.
Since Vg gene plays a critical role in the reproduction and enhance population of insects, disruption of Vg gene expression is believed to hold a significant potential as an effective control measure for insects. Therefore, RNA interference (RNAi)-based species-specific insecticide could be convenient and effective. The RNAi is a versatile technique that can inhibit the expression of target messenger RNA (mRNA) and provides tremendous opportunity to investigate gene functions28. Principally, RNAi is an idiosyncratic gene silencing mechanism that employs double-stranded RNA (dsRNA) to degrade precise mRNAs29. This technique can be extensively used in crop protection; however, it is still limited to the laboratory30–39. The effectiveness of RNAi varies between species, selection of target genes, and the mode of dsRNA delivery40,41. The RNAi technology could efficiently silence the targeted genes in the form of feeding and transgenic plants37,42,43. Therefore, it is of vital importance for the management of pests resistant to pesticide or those having hidden nature such as R. ferrugineus.
The present study addressed several issues beginning with the characterization and temporal expression profile of the RfVg gene transcript and use of RfVg-based RNAi. Finally, RNAi application was investigated using quantitative real-time polymerase chain reaction (qRT-PCR), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and observation of oogenesis, ovarian development, fecundity, and eggs’ hatchability. Silencing of RfVg gene significantly reduced its expression, which lead to the failure of R. ferrugineus reproduction confirmed by oogenesis, ovarian development, fecundity, and egg hatchability. The RNAi results demonstrate that knockdown of RfVg gene has the potential to halt reproduction in R. ferrugineus, which warrants the development of novel control strategies against the pest.
Results
Full-length sequencing of RfVg and structural analysis
The complete RfVg gene transcript was of 5504 bp, which encoded 1787 residues mature protein representing all conserved structures typical of insects’ Vgs. In the Vg protein analysis, first twenty amino acid residues were predicted as signal peptide [analyzed with the SignalP program (www.cbs.dtu.dk/services/SignalP/)]. The deduced R. ferrugineus Vg protein contained five putative cleavage recognition sites, i.e., RRSR (361–364), RSRR (362–365), REGR (593–596), RLAR (666–669) and RPQR (1679–1682). In addition, all conserved motifs such as DGXR and GL/ICG, which are usually found at C-terminus region of Vgs also existed in the Vg of R. ferrugineus. The sequence of DGXR motif was DGKR (amino acids position 1,622–1,625), while of GL/ICG motif was GLCG (amino acids position 1,641–1,644). The RfVg contained 27 cysteine residues, of which seven were located at the C-terminus. As predicted by the NetNGlyc 1.0 program (www.cbs.dtu.dk/services/NetNGlyc/), RfVg contained 10 putative glycosylation sites (NXT/S). Moreover, RfVg contained 149 putative phosphorylation sites, including 90 serine (S), 26 threonine (T) and 33 tyrosine (Y). Moreover, three conserved domains were identified in the RfVg by using NCBI conserved domain database (CDD) search (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Among these conserved domains, one was Vg_N spanning from amino acids 21 to 735. The second domain was unknown function 1943 (DUF1943) spanning from amino acid residues 769 to 1059. The third was Von Willebrand factor domain (VWD) located at the C-terminus and spanned from amino acid residues 1467 to 1657 (Fig. S1). To elucidate the evolutionary relationship of RfVg, a neighbor joining phylogenetic tree was generated based on 99 insect and non-insect Vgs sequences present in the NCBI database (Fig. 1). The sequence of RfVg was grouped with other coleopteran Vgs as expected. The evolutionary relationship based on the current phylogenetic tree analysis showed that Vgs from insects are closer to the Vg of nematode and arachnids as compared to Vg of vertebrate and crustacean.
Figure 1.
Neighbor-joining phylogenetic tree of 99 insects and non-insect Vg protein sequences. The Clustal W program was used for distance analysis of amino acid sequences and the neighbor-joining tree was constructed by using MEGA 6 software program74. Scale 0.2 indicates distance (number of amino acid substitutions per site)45 Species belonging to different orders have been indicated with different colors.
Expression pattern and developmental traits of RfVg gene
The RT-PCR was conducted to determine the sex, tissue, and stage-specific expression and probe the temporal profiles of RfVg gene transcription. The RT-PCR studies were performed by amplifying RfVg-specific region by using gene-specific primers (RfVgRTF2, RfVgRTR2) (Table S1). The RfVg gene was exclusively expressed in the female fat body cells as demonstrated by a single band, whereas no expression was observed for other tissue (Fig. 2A). The expression of tubulin gene -an internal control- in all tissues confirms the quality of the cDNAs used in these studies. To analyze the temporal expression profile of RfVg gene, total RNAs were extracted from the fat body of adult R. ferrugineus females up to three weeks. The RT-PCR was conducted to determine the developmental profile of RfVg gene transcription. The expression of RfVg gene was detected from the first day of R. ferrugineus female adults, which was still present inside the cocoons but with light bands. The expression of RfVg gene and intensity of the bands gradually increased after the emergence of adult females from the cocoons. The expression level of RfVg gene remained almost the same from day 10 to 21(Fig. 2B).
Figure 2.
Expression pattern and temporal traits of RfVg gene transcription. (A) Expression pattern of RfVg and tubulin genes from different tissues of Rhynchophorus ferrugineus was analyzed by RT-PCR. Agarose gels (2%) were used to analyze the amplified PCR products. The M is a molecular marker (bp), while 640 bp and 196 bp on right side are amplified products of Vg and tubulin genes, respectively. (B) The RfVg and tubulin genes expression profile up to three weeks in the adult R. ferrugineus females analyzed by RT-PCR. The amplified bands were visualized under the UV light and photographed by using gel documentation BioDocAnalyze system (Biometra). d days.
Silencing of RfVg gene and qRT-PCR validation
For RNAi-mediated silencing of RfVg gene function, dsRNA targeting an inimitable region (position 3538–3938 bp) showing very low or no homology with other insect Vgs was injected (2-μg/weevil) dorsally in the second abdominal segment of the female pupae. The qRT-PCR was executed to validate the impact of Vg-based RNAi on Vg gene transcription. Tubulin was used as an endogenous gene to fulfill the requirements of 2−∆∆CT calculation method. The qRT-PCR revealed that the level of Vg gene expression was drastically declined in RfVg-dsRNA injected females as compared to nucleus free water (NFW)-injected and no injection (NI) groups (Fig. 3). In RNAi-treated females, the Vg gene expression was suppressed by 95.3% 15 days after injection, whereas 96.6 and 99.4% downregulation in RfVg gene expression was recorded on 20 and 25 days post-injection, respectively.
Figure 3.
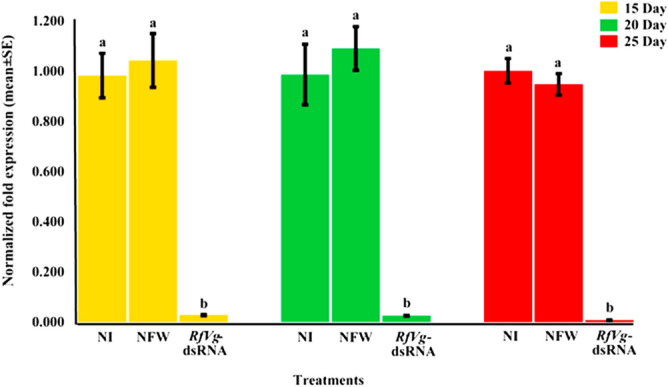
The RNAi-based silencing of RfVg gene results in significant down regulation of Vg gene. Expression levels of the RfVg gene were quantified by quantitative real time PCR. Normalized fold expression of the RfVg-dsRNA-injected group were compared with no injection (NI) and nucleus free water (NFW) groups on day 15, 20, and 25 of post-injection periods. Different letters above the bars (a,b) show significant differences among the groups at (α = 0.05). The expression levels of RfVg gene were significantly reduced in RfVg-dsRNA injected group in comparison to the NI injection and NFW injected groups. All biological groups contained three replicates. Each replicate has a single animal, while there were three technical replicates.
Validation of RfVg gene silencing through SDS-PAGE
A drastic reduction in Vg protein expression was observed in RfVg-dsRNA-treated females as compared to NFW and NI groups. The SDS-PAGE was executed to assess the impact of Vg-based RNAi on the expression pattern of Vg protein. The SDS-PAGE exhibited an obvious difference in expression of the Vg protein among RfVg-dsRNA, NFW, and NI groups (Fig. 4). Both identified Vg bands (~ 175 kDa and ~ 45 kDa) were observed in NI and NFW groups, while these bands were almost missing in RfVg-dsRNA-injected group.
Figure 4.

Validation of RNAi and expression analysis of RfVgs by SDS-PAGE. To clarify the RfVg protein expression, hemolymph samples (10 μl/lane) prepared from NI, NFW, and RfVg-dsRNA injected groups on 15, 20, and 25 days (d) post-injection periods were loaded on SDS-PAGE (8%) and compared. All biological groups contained three replicates; each replicate was an individual R. ferrugineus, while there were three technical replicates. The gel was stained with Coomassie blue and washed with destaining solution. M is the molecular marker (kDa) while, arrows on right side indicate the identified Vg polypeptides (175 and 45 kDa) in R. ferrugineus. A highly downregulated expression of Vg protein in RfVg-dsRNA injected group as compared to NI and NFW injected groups has been shown with the red circles. The protein bands were visualized under the white light and photographed by using gel documentation BioDocAnalyze system (Biometra). d days.
Biological studies to assess the impact of RfVg gene silencing in knockdown phenotypes
The phenotypic reflection of Vg gene knockdown in adult females was evaluated by analyzing pre-oviposition period to assess a possible delay in egg laying, oviposition period, post-oviposition period, total number of eggs laid per female, eggs hatchability percentage, and female lifespan (Fig. 5A–F) in addition to ovarian development. The RfVg knockdown greatly influenced pre-oviposition period in dsRNA-treated females as compared to control groups. The data represented that pre-oviposition periods were significantly different and delayed in dsRNA-treated females (df = 2, F = 14.9, P ˂ 0.0001) in RfVg-dsRNA-treated females as compared to the females in NFW and NI groups (Fig. 5A). Likewise, a significant difference (df = 2, F = 6.08, P ˂ 0.007) was recorded in the post-oviposition period of RNAi-treated females and those in NFW and NI groups (Fig. 5C). A significant difference was also noted in total number of eggs laid per female (df = 2, F = 13.3, P ˂ 0.0001) where RfVg-dsRNA-injected group laid lesser number of eggs as compared to NFW and NI groups (Fig. 5D). Moreover, no eggs hatchability (or 0%) was observed in RfVg-dsRNA-injected group, whereas NFW and NI groups recorded 75 and 79% eggs hatchability, respectively (Fig. 5E). However, non-significant differences were recorded for oviposition period (df = 2, F = 1.2, P = 0.3) (Fig. 5B) and adult female lifespan (df = 2, F = 1.1, P = 0.3) (Fig. 5F).
Figure 5.
Effects of RfVg RNAi on reproductive traits of adult females of Rhynchophorus ferrugineus, pre-oviposition (A), oviposition (B), post-oviposition periods (C), mean number of eggs laid per female (D). Effects of RfVg-dsRNA injection on eggs hatchability % (E). Effects of RfVg-dsRNA injection on adult female life span (F). To test the statistical significance among different biological parameters, one-way analysis of variance was performed at (α = 0.05). Different letters above the bars show significant difference among groups.
The investigations on ovarian development in RfVg-dsRNA-injected females also revealed remarkable phenotypic repercussions. The ovaries were decreased with lesser yolk compared the ovaries from NFW and NI groups where oocytes were well-developed and ovaries were larger in size (Fig. 6A). Moreover, a drastic decrease in the egg size was observed in RNAi-treated females as compared to the control groups (Fig. 6B).
Figure 6.
Phenotypic effects of RfVg-based RNAi on ovarian/egg development of Rhynchophorus ferrugineus. (A) To observe the impact of RNAi on ovarian development, ovaries from three biological groups (RfVg-dsRNA, NFW, and NI) were dissected 20 days post injection period, observed under a stereomicroscope (DM 165 C, Leica, Wetzlar, Germany) and compared by using auto-montage software system (Syncroscopy, Cambridge, UK). (B) The eggs laid by NI, NFW, and RfVg-dsRNA-injected females were observed under the stereomicroscope (Leica MZ 125, Germany), and compared by using the auto-montage software (Helicon focus 6). Scale bars: 10 mm and 1.0 mm for ovaries and eggs, respectively.
The length (df = 2, F = 170.4, P ˂ 0.0001) and width (df = 2, F = 73.7, P ˂ 0.0001) of eggs significantly differed among treatment groups (Fig. 7). Overall, low egg production, delayed oviposition periods, smaller-sized eggs with no hatchability were recorded for RfVg-dsRNA-injected females suggesting that RNAi-targeting Vg gene has the ability to inhibit reproduction in R. ferrugineus.
Figure 7.

Effects of RfVg-based RNAi on eggs size of Rhynchophorus ferrugineus. To identify the impact of RfVg based RNAi on egg size, 15 eggs from each group (NI, NFW, and RfVg-dsRNA) were measured by using Dino-Lite Digital Microscope AM4815ZT (AnMo Electronic Corp, USA). To test the statistical significance among different biological parameters a one-way analysis of variance was performed at (α = 0.05). Different letters (a,b) above the bars show significant difference among the treatment groups.
Discussion
Red palm weevil [Rhynchophorus ferrugineus (Olivier)] has become a noxious pest of palm trees around the world. It has gained significant importance due to its global invasion and associated economic costs. Recent molecular studies have revealed that Vg structures and functions seemed conserved across diverse insect taxa, although some deviations exist in post-translational processing/number of cleavage sites, number of Vg genes, and in the hormonal system regulating these genes6,44,45. The Vgs proteins have been sequenced and characterized from several insect species representing different orders, including dictyoptera14,15,25,46, hemiptera27, lepidoptera16, hymenoptera47,48, diptera49, and coleoptera50–52 due to their prime importance in insect reproduction. However, there is no information available on molecular mechanism of R. ferrugineus reproduction. The present study, therefore, focused on molecular characterization, expression profiling, and silencing of the reproduction control gene Vg in R. ferrugineus.
The RfVg gene transcript comprised of 5,504 bp nucleotides that encoded deduced protein of 1,787 amino acids. The molecular weight of RfVg protein was 210 kDa, which is almost similar to other insects’ Vgs, including coleopterans50,51,53. Like other coleopterans, conserved domains were present in RfVg (Fig. S1). Conserved domains play critical role in organisms’ physiology54,55. The RfVg protein have 5 post-translational cleavage sites without polyserine clusters, which shows robust structural similarity with known Vgs of coleopteran50–52 and some other insects including cotton leafworm (Spodoptera litura)16, parasitoid wasp (Encarsia formosa)56, and fire ant (Solenopsis invicta)57. The precursor Vg in most of the insects are post-translationally modified and proteolytically cleaved at a consent cleavage site (RXXR) into subunits by dibasic endoproteases58. Moreover, presence of 149 phosphorylation sites (S, T and Y) in RfVg protein sequence (Fig. S1) indicated that RfVg is highly phosphorylated similar to several other insects50,51,59. In addition, the presence of GL/ICG motif and cysteine residues at C-terminus are essential for oligomerization60,61. Furthermore, this study first time reports the data, which support potential use of RfVg gene silencing as a tool for the management of R. ferrugineus. The gene silencing approach has been successfully demonstrated against several specific genes in targeted insects, including Vg using different delivery methods, i.e., injection, feeding, and drops30–39,62,63.
The results of RfVg-based RNAi revealed a drastic decrease in the expressions of Vg gene in RfVg-dsRNA group as compared to females in NFW and NI groups (Fig. 3). Almost 95.3% suppression was noted in the expressions of Vg mRNA for RfVg-dsRNA injected group 15 days after injection, whereas 96.6% and 99.4% suppression was recorded on 20 and 25 days after injection, respectively. These findings clearly demonstrate that expressions of RfVg gene in R. ferrugineus females was strongly affected by RfVg-dsRNA treatment. The drastic reduction in Vg mRNA level also confirm the sensitivity of R. ferrugineus to RNAi. Generally, literature indicates that coleopteran species are more sensitive to RNAi than other insect groups as shown in western corn rootworm (Diabrotica virgifera)37, red flour beetle (Tribolium castaneum)64, Colorado potato beetle (Leptinotarsa decemlineata)65, and cotton boll weevil (Anthonomus grandis)62. For example, Vg gene expressions in cotton boll weevil were reduced by 97% just within 72 h after injection62. The persistence of RfVg-dsRNA was examined on 15, 20, and 25 days after injection in the current study in addition to reproduction performance and a drastic reduction in RfVg expression (99%) was observed within 25 days after injection. Likewise, results of western corn rootworm neonates fed on V-ATPase-A gene based transgenic plants indicated the silencing of a particular gene, which lead to death of treated neonates37. Likewise, feeding of Actin and Copβ gene based dsRNA expressed bacteria to Colorado potato beetle reduced growth and caused mortality65. In contrast, RNAi efficiency is less supported in other insect groups, i.e., lepidopteran insect species such as light brown apple moth (Epiphyas postvittana) where third instar larvae were fed with carboxylesterase-based dsRNA through droplets and < 50% mRNA silencing was achieved66. The major factor between coleopteran and lepidoptera RNAi efficiency includes the uptake of dsRNA and its processing to siRNA. The cells and tissues in coleopteron insects uptake dsRNA quickly and process it into siRNA faster than lepidopteran species41. Similarly, RNAi indicated 50 and 40% efficiency against mid-gut protein tsetse EP and nitroporin-2 in Glossina morsitans morsitans and Rhodnius prolixus, respectively67,68.
Additionally, a striking reduction in the expressions of RfVg gene resulted in dramatic failure of Vg protein expression in the RfVg-dsRNA-injected group as compared to NFW and NI (Fig. 4). Silencing of Vg gene not only caused failure of Vg protein expression, but also affected the transport of other associated nutrients63,69.
Furthermore, present study also confirmed the consequences of RfVg-dsRNA on reproductive performance of R. ferrugineus females, where several parameters were observed, including pre-oviposition and oviposition periods, fecundity, egg hatchability, post-oviposition period, and female lifespan. No egg hatchability was recorded for RfVg-dsRNA-injected group, 75.2 and 78.6% egg hatchability was found in NFW-treated and NI groups, respectively (Fig. 5E). No egg hatchability was the result of significant reduction in expression level of RfVg gene, which caused inadequate production of vitellogenin protein to confirm normal egg size and hatchability. Moreover, this study revealed a significant difference in number of eggs laid per female among the treatment groups (Fig. 5D) in addition to no egg hatchability. Likewise, eggs’ viability was dramatically decreased (˂1%) in Vg dsRNA-injected females of cotton boll weevil and eggs were unable to hatch. However, injection of Vg-dsRNA had no effect on fecundity of this pest62. Moreover, in bedbug (Cimex lectularius) Vg-dsRNA injection radically suppressed egg production compared to control group and egg production was entirely ceased two weeks after Vg-based dsRNA injection63. Additionally, results of Vg dsRNA injection also decreased the eggs production in lubber grasshopper (Romalea microptera)69.
The pre-oviposition periods were delayed in dsRNA-treated females. The effect of Vg knockdown on pre-oviposition period has been reported earlier63; however, no effect of RNAi on pre-oviposition period of cotton boll weevil has also been reported62. Besides, present study revealed that ovaries were rigorously deformed with short and unorganized eggs and ovary size (Fig. 6A,B). The size of eggs was significantly decreased in RfVg-dsRNA-treated group as compared to NFW and NI groups (Fig. 7). Similarly, eggs development in cotton boll weevil was severely affected in Vg dsRNA-injected group62. Moreover, results of RNAi injection revealed shrunken ovaries in bedbug with no developed oocytes in Vg dsRNA-treated female as compared to control where ovaries were normal in size with mature oocytes63. Likewise, ovaries were rigorously deformed in rice moth having small ovarioles and disorganized egg sizes in Vg dsRNA-treated females relative to un-injected females31.
The present findings along with previous reports conclusively demonstrate a high potential of Vg-based RNAi technology for pest management. Silencing of RfVg gene provides evidence that RNAi technology could be a smart alternative to traditional management methods for coleopteran pests, particularly for R. ferrugineus. Undoubtedly, the success and effectiveness of RNAi varies with species, selection of target genes, and the mode of dsRNA delivery40,41,70. The present study has provided evidence that Vg gene is the best target for RNAi-based management of R. ferrugineus. Choosing a suitable tactic to deliver the dsRNA successfully after selection and identifying the target gene is a main challenge in RNAi-based plant protection method. Although microinjection is a suitable approach for functional genomic studies, this strategy is not appropriate to manage the pest in the field. However, numerous developments on dsRNA delivery made this technique more efficient in the field. For example, delivering dsRNA to insect pests through transgenic plant has been tried37. Moreover, successful feeding of insects pest via bacterially expressed dsRNA65 and application of dsRNA through nanoparticles71 have also been practiced. The silencing of RfVg gene by RNAi may have the potential to stop the reproduction of R. ferrugineus and RfVg could be an auspicious target candidate gene for developing an alternative pest management strategy for the pest at molecular level. Therefore, the future research should be focused on the delivery of RfVg-dsRNA for the management of R. ferrugineus in the field. This study supports the potential use of emerging RNAi technology for pest control and might provide an alternative to the conventional methods being used for the management of R. ferrugineus.
Materials and methods
Rearing of the red palm weevil
Red palm weevils were originally collected from infested date palm trees in Dirab, Kingdom of Saudi Arabia (24.4164°N, 46.5765°E). The adults were provided a piece of cotton saturated with 10% sugar solution72 and kept in plastic box (L: 17 cm; W: 11 cm; H: 7 cm). The laid eggs were collected with the help of forceps and shifted to wet filter papers placed in small plastic cup (d: 6 cm; h: 2.5 cm). The larvae were fed with artificial diet (250 g/5 larvae) for further development in the plastic box (L: 17 cm; W: 11 cm; H: 7 cm). Finally, the last instar larvae were shifted into a sugarcane set (10 cm) for pupation in plastic boxes (L: 17 cm; W: 11 cm; H: 7 cm). The R. ferrugineus culture was maintained in the growth chamber at 25 ± 1 °C, 70 ± 5% relative humidity72.
PCR amplification and sequencing to obtain full length RfVg and phylogenetic analysis
The partial sequence of RfVg gene transcript was obtained through the next-generation sequencing (NGS) of R. ferrugineus fat body tissues from Beijing Genomics Institute (BGI), China. The gene-specific primers (RfVgF1), which were designed based on the partial RfVg sequence and the adopter primer 1 (AP1) (Clontech) (Table S1) were used for 3ʹ RACE-PCR in order to get the full-length sequence of RfVg gene. The ds cDNA library was subjected to PCR by using the Gene Amp PCR system 9700 thermo cycler (Applied Biosystems, USA). The PCR conditions were; initial denaturation at 94 ºC for 1 min followed by 35 cycles of denaturation at 94 ºC for 30 s, annealing at 68 ºC for 3 min, and a final extension of 68 ºC for 5 min. The amplified PCR products were purified by using illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare Life Sciences, USA). The purified PCR products were sequenced from BGI, China. The obtained sequences were analyzed and checked for homology with other insects Vg sequences by using basic local alignment search tool (BLAST) of National Center for Biotechnology Information (NCBI). Finally, RfVg sequence was submitted to the NCBI GenBank database (accession number ALN38803) after confirmation. The RfVg sequence was aligned to other known insects Vgs sequences available in the NCBI database using the clustalW program73. Phylogenetic tree was constructed using neighbor-joining method on MEGA version 674.
Expression pattern and developmental traits of RfVg gene
To investigate the tissue, gender-specific expression, and developmental profile of RfVg gene transcription, total RNAs were extracted from the female fat body, ovary, mid-gut, muscle, male fat body, and female pupa. Total RNA was extracted from fat body of adult R. ferrugineus females up to three weeks (for one week when the female adults were inside the cocoons and two weeks after the eclosion) by using Tri-RNA reagent (Favorgen Biotech Corp, Taiwan) to analyze the developmental expression profile of RfVg gene.The RNA samples were treated with DNase I (Invitrogen, USA) to remove DNA contamination. A 2-μg of total RNA from each sample was reverse transcribed to cDNA using ReverTra Ace cDNA synthesis kit (Toyobo Co. Ltd, Japan). The reverse transcription polymerase chain reaction (RT-PCR) was performed by using gene-specific primers (RfVgRTF1, RfVgRTR1, RfVgRTF2, RfVgRTR2) and tubulin was used as internal control (TubulinRfer-F, TubulinRfer-R) (Table S1). The cDNA was subjected to RT-PCR by using Gene Amp PCR system 9700 thermo cycler (Applied Biosystem, USA). The following thermal programs were applied; 94 ºC for 1 min for denaturation followed by 30 cycles of 94 ºC for 30 s, 68 ºC for 30 s, and 72 ºC for 1 min. The PCR amplified products were run on 2% agarose gel, stained with ethidium bromide, and visually confirmed under ultra violet (UV) light by using gel documentation BioDocAnalyze system (Biometra, Germany).
RNAi-based silencing of RfVg gene and tissue collection for RNAi validation
A unique target region of 400 bp showing very low or no homology with other insect Vgs was selected from the C-terminus of the RfVg gene transcript. The RfVg-based dsRNA was synthesized using a MEGAscript® RNAi Kit (Life Technology, USA) according to the manufacturer’s protocol and used to knockdown the function of RfVg gene. The experiment consisted of three biological treatment groups, which were RfVg-dsRNA-injected, nuclease free water (NFW)-injected, and no injection (NI)31,39,76–78. A total 2-μg (10-μl) of RfVg-dsRNA was injected dorsally in the second abdominal segment of each 10–12 days-old female pupae by using 0.5 ml BD Micro-Finetm Plus syringe (Becton, Dick-inson Co, USA), whereas 10-μl of NFW was injected as control. The pupae used for injection were taken from the cocoons, which were placed again in the cocoon after injection for further development and kept in growth chamber at 25 ± 1 °C, 70 ± 5% relative humidity and photoperiod of 12:12 (L:D)5. When pupae converted to adults, they remained inside the cocoon for almost one week before eclosion. The female pupae were selected on the base of snout as R. ferrugineus female has smooth narrow long snout, while male has shorter and wider snout with some tuft hairs. Furthermore, females were also confirmed after adult eclosion. The newly emerged females (from cocoon) were shifted to a separate box having a piece of cotton saturated with 10% sugar solution. The females were harvested on three post-injection periods (i.e., 15, 20, and 25 days) for the collection of fat body (for cDNA synthesis to analyze Vg transcript levels), hemolymph (for Vg protein expression analysis) and ovaries. The RNAi experiments were validated using quantitative real time polymerase chain reaction (qRT-PCR), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and by observing phenotypic effects of RNAi on ovarian development. The cDNA and hemolymph samples were prepared for all biological groups. Total RNAs were extracted from the fat body by using Tri-RNA reagent (Favorgen Biotech Corp, Taiwan) to make cDNA. The RNA samples were treated with DNase I (Invitrogen, USA) to remove the DNA contamination. A 2-μg of total RNA from each sample was reverse transcribed to cDNA using ReverTra Ace cDNA synthesis kit (Toyobo Co. Ltd, Japan). Hemolymph was collected with the help of micropipette after the snout of weevil was amputated using a fine scissors and diluted to 1:50 with the sample buffer.
Validation of RfVg gene silencing through qRT-PCR
A qRT (quantitative real time)-PCR, analysis was performed by using the RfVg-gene-specific primers (RfVgRTF3 and RfVgRTR3) to corroborate the impact of RfVg-based RNAi on Vg gene expression (Table S1). Expression levels of the RfVg gene were normalized by quantifying the expression levels of tubulin (a housekeeping gene) using TubulinRfer-F and TubulinRfer-R primers (Table S1). The qRT-PCR trials were designed based on three biological groups, which were RfVg-dsRNA, NFW, and NI. All groups contained three replicates, each replicate had a single animal, while there were three technical replicates. The qRT-PCR was accomplished using CFX-96 Touch™ Real-Time PCR Detection System (BioRad, USA), while reactions (each contained a volume of 20-μl) were performed using SsoAdvanced™ Universal SYBR® Green Supermix (BioRad, USA). The following qRT-PCR conditions were applied for amplifying the cDNA; 95 ºC for 30 s, 40 cycles of 95 ºC for 15 s, 60ºC for 60 s, followed by melting curve analysis at 65–95 ºC with an increment of 0.5 ºC every 5 s. The 2−∆∆CT method was used to analyze the relative expression levels of RfVg gene by normalizing them to tubulin and control (NI) group.
Validation of RfVg gene silencing through SDS-PAGE
The Vg protein expression levels in dsRNA-injected females were analyzed through SDS-PAGE as reported previously15,27 to affirm the efficiency of RNAi. Protein analyses were conducted using samples of hemolymph and egg extracts (each 10 μl) run on 8% polyacrylamide gels. Three post-injection periods, i.e., 15, 20 and 25 days (after injection with dsRNA) with three biological groups (RfVg-dsRNA, NI, and NFW) were analyzed. All groups contained three replicates; each replicate was an individual of R. ferrugineus, while there were three technical replicates. The Vg protein expression levels in RfVg-dsRNA injected-weevils were compared with the NI and NFW groups. The gels were stained with Coomassie blue and washed with de-staining solution. The protein bands were visualized under the white light and photographed by using gel documentation BioDocAnalyze system (Biometra, Germany).
Biological studies to assess the impact of RfVg gene silencing in knockdown phenotypes
The phenotypic manifestation of RfVg gene silencing in RNAi-treated R. ferrugineus females was assessed based on the biological markers including pre-oviposition periods, oviposition periods, total number of eggs laid per female, eggs hatchability %, post-oviposition periods, and female life span in addition to the ovarian development. To inspect these biological traits, newly emerged adult females (dsRNA-injected) were paired with the normal males, transferred to a separate plastic box (1 kg) having a piece of cotton saturated with 10% sugar solution and kept in growth chamber at 25 ± 1 °C, 70 ± 5% relative humidity72. There were again three biological groups, i.e., RfVg dsRNA-injected (dsRNA), NFW-injected, and NI. All groups contained nine replicates and each replicate was an individual pair (normal male + dsRNA-treated female) of R. ferrugineus. All pairs were allowed to mate and lay eggs until females died. The oviposition periods were observed and the number of eggs and hatchability percentage were scored. Completely randomized design (CRD) was used in this experiment.
To investigate the impact of Vg-based RNAi on ovarian development, ovaries from all three groups (RfVg-dsRNA, NFW and NI) were isolated in the phosphate buffered saline (PBS) 20 days post-injection periods, viewed under stereomicroscope (DM 165 C, Leica, Wetzlar, Germany) and photographed using auto-montage software system (Syncroscopy, Cambridge, UK) to measure the ovarian development. Eggs from dsRNA-injected groups were compared with NFW and NI groups, observed under the stereomicroscope and photographed (Leica MZ 125, Helicon focus 6 software, Germany). Furthermore, 15 eggs from each group (NI, NFW, and RfVg-dsRNA injected groups) were measured using Dino-Lite Digital Microscope AM4815ZT (AnMo Electronic Corp, USA) to clarify the impact of RfVg based RNAi on egg size (length and width).
Statistical analysis
The qRT-PCR quantification results were analyzed following 2−∆∆CT method (Livak and Schmittgen, 2001). One-way analysis of variance was performed at (α = 0.05) by using SAS program ver. 9.2. To analyze the statistical significant differences among three experimental groups (RfVg-dsRNA, NI and NFW) for qRT-PCR data and biological studies75.
Ethics approval
The work is original. Moreover, no legal permission was required to conduct the experiments.
Supplementary Information
Acknowledgements
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia (Award Number 13-BIO 1407-02). The authors also thank the Research Support Service Unit at King Saud University for their technical support.
Author contributions
K.G.R., K.M., M.T. and A.S.A. participated in the planning, design and coordination of the study. K.M., M.H., and W.S.A. participated in the red palm weevil rearing, conducted Lab experiments. K.G.R., K.M., and M.H., collected and analyzed data, and participated in write up. A.S.A. and K.G.R. supervised the work. M.T. critically reviewed the manuscript and provided all technical guidance. All authors have read the final version of the manuscript carefully and approved it.
Data availability
All the data is present in the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-01159-9.
References
- 1.MOA. Central Department of Statistics and Information, Annual Statistics Book. Saudi Arabia. (2012).
- 2.FAO. FAO Statistical Year Book 2012. (2012).
- 3.Abraham VA, Al-Shuaibi M, Faleiro JR, Abozuhairah RA. An integrated approach for the management of red palm weevil Rhynchophorus ferrugineus Olivier—A key pest of date palm in the Middle East. Sultan Qaboos Univ. J. Sci. Res. Agric. Sci. 1998;3:77–83. [Google Scholar]
- 4.Faleiro JR. Insight into the management of red palm weevil Rhynchophorus ferrugineus Olivier: Based on experiences on coconut in India and date palm in Saudi Arabia. Fund. Agroalimed. 2006;06:35–57. [Google Scholar]
- 5.Wai YK, Bakar AA, Azmi WA. Fecundity, fertility and survival of red palm weevil (Rhynchophorus ferrugineus) larvae reared on sago palm. Sains Malays. 2015;44:1371–1375. doi: 10.17576/jsm-2015-4410-01. [DOI] [Google Scholar]
- 6.Tufail M, Takeda M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008;54:1447–1458. doi: 10.1016/j.jinsphys.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Abbas MST, Hanounik SB, Shahdad AS, Ai-Bagham SA. Aggregation pheromone traps, a major component of IPM strategy for the red palm weevil, Rhynchophorus ferrugineus in date palms (Coleoptera: Curculionidae) J. Pest. Sci. 2006;79:69–73. doi: 10.1007/s10340-005-0113-6. [DOI] [Google Scholar]
- 8.Aldawood, A. S., Alsagan, F., Altuwariqi, H., ALmuteri, A. & Rasool, G. K. Red palm weevil chemical treatments on date palms on date palms in Saudi Arabia: Results of extensive experiment. In AFPP: Colloque Mediterranean Sur Les Ravageurs Des Palmiers (2013).
- 9.Atwa AA, Hegazi EM. Comparative susceptibilities of different life stages of the red palm weevil (Coleoptera: Curculionidae) treated by entomopathogenic nematodes. J. Econ. Entomol. 2014;107:1339–1347. doi: 10.1603/EC13438. [DOI] [PubMed] [Google Scholar]
- 10.Abraham VA, Abdulla Koya KM, Kurian C. Evaluation of seven insecticides for control of red palm weevil Rhynchophorus ferrugineus FABR. J. Plant. Crops. 1975;3:71–72. [Google Scholar]
- 11.Wilson C, Tisdell C. Why farmers continue to use pesticides despite environmental, health and sustainability costs. Ecol. Econ. 2001;39:449–462. doi: 10.1016/S0921-8009(01)00238-5. [DOI] [Google Scholar]
- 12.Sharpe RM, Irvine DS. How strong is the evidence of a link between environmental chemicals and adverse effects on human reproductive health? BMJ Br. Med. J. 2004;328:447. doi: 10.1136/bmj.328.7437.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reissig WH, Weires RW, Forshey CG. Effects of gracillariid leafminers on apple tree growth and production. Environ. Entomol. 1982;11:958–963. doi: 10.1093/ee/11.4.958. [DOI] [Google Scholar]
- 14.Tufail M, Lee J, Hatakeyama M, Oishi K, Takeda M. Cloning of vitellogenin cDNA of the American cockroach, Periplaneta americana (Dictyoptera), and its structural and expression analyses. Arch. Insect Biochem. Physiol. 2000;45:37–46. doi: 10.1002/1520-6327(200009)45:1<37::AID-ARCH4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Tufail M, Takeda M. Vitellogenin of the cockroach, Leucophaea maderae: nucleotide sequence, structure and analysis of processing in the fat body and oocytes. Insect Biochem. Mol. Biol. 2002;32:1469–1476. doi: 10.1016/S0965-1748(02)00067-X. [DOI] [PubMed] [Google Scholar]
- 16.Shu Y, Zhou J, Tang W, Zhou Q, Zhang G. Molecular characterization and expression pattern of Spodoptera litura (Lepidoptera: Noctuidae) vitellogenin, and its response to lead stress. J. Insect Physiol. 2009;55:608–616. doi: 10.1016/j.jinsphys.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Snigirevskaya ES, Raikhel AS. Receptor-mediated endocytosis of yolk proteins in insect oocytes. Progress in vitellogenesis. Reprod. Biol. Invertebr. 2005;12:199–228. [Google Scholar]
- 18.Sappington TW, Raikhel AS. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem. Mol. Biol. 1998;28:277–300. doi: 10.1016/S0965-1748(97)00110-0. [DOI] [PubMed] [Google Scholar]
- 19.Tufail M, Takeda M. Molecular cloning, characterization and regulation of the cockroach vitellogenin receptor during oogenesis. Insect Mol. Biol. 2005;14:389–401. doi: 10.1111/j.1365-2583.2005.00570.x. [DOI] [PubMed] [Google Scholar]
- 20.Tufail M, Takeda M. Molecular cloning and developmental expression pattern of the vitellogenin receptor from the cockroach, Leucophaea maderae. Insect Biochem. Mol. Biol. 2007;37:235–245. doi: 10.1016/j.ibmb.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Tufail, M. & Takeda, M. Hemolymph Proteins and Functional Peptides: Recent Advances in Insects and Other Arthropods. Vol. 1 (Bentham Science Publishers, 2012).
- 22.Bell WJ. Continuous and rhythmic reproductive cycle observed in Periplaneta americana (L.) Biol. Bull. 1969;137:239–249. doi: 10.2307/1540096. [DOI] [Google Scholar]
- 23.Hagedorn H, Judson C. Purification and site of synthesis of Aedes aegypti yolk proteins. J. Exp. Zool. 1972;182:367–377. doi: 10.1002/jez.1401820308. [DOI] [PubMed] [Google Scholar]
- 24.Oie M, Takahashi S, IshazakiI H. Vitellogenin in the eggs of the cockroach, Blattella germanica: Purification and characterization. Dev. Growth Differ. 1975;17:237–246. doi: 10.1111/j.1440-169X.1975.00237.x. [DOI] [PubMed] [Google Scholar]
- 25.Tufail M, Hatakeyama M, Takeda M. Molecular evidence for two vitellogenin genes and processing of vitellogenins in the American cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 2001;48:72–80. doi: 10.1002/arch.1059. [DOI] [PubMed] [Google Scholar]
- 26.Nagaba Y, Tufail M, Inui H, Takeda M. Hormonal regulation and effects of four environmental pollutants on vitellogenin gene transcription in the giant water bFfigug, Lethocerus deyrollei (Hemiptera: Belostomatidae) J. Insect Conserv. 2011;15:421–431. doi: 10.1007/s10841-010-9315-1. [DOI] [Google Scholar]
- 27.Tufail M, et al. Molecular cloning, transcriptional regulation, and differential expression profiling of vitellogenin in two wing-morphs of the brown planthopper, Nilaparvata lugens Stål (Hemiptera: Delphacidae) Insect Mol. Biol. 2010;19:787–798. doi: 10.1111/j.1365-2583.2010.01035.x. [DOI] [PubMed] [Google Scholar]
- 28.Mansoor S, Amin I, Hussain M, Zafar Y, Briddon RW. Engineering novel traits in plants through RNA interference. Trends Plant Sci. 2006;11:559–565. doi: 10.1016/j.tplants.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Mello CC, Conte D. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 30.Price DR, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400. doi: 10.1016/j.tibtech.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Veerana M, Kubera A, Ngernsiri L. Analysis of the vitellogenin gene of rice moth, Corcyra cephalonica Stainton. Arch. Insect Biochem. Physiol. 2014;87:126–147. doi: 10.1002/arch.21185. [DOI] [PubMed] [Google Scholar]
- 32.Rützler M, Zwiebel L. Molecular biology of insect olfaction: Recent progress and conceptual models. J. Comp. Physiol. A. 2005;191:777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 33.Kola VSR, Renuka P, Madhav MS, Mangrauthia SK. Key enzymes and proteins of crop insects as candidate for RNAi based gene silencing. Front. Physiol. 2015;6:119. doi: 10.3389/fphys.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonio DSM, Guidugli-Lazzarini KR, Do Nascimento AM, Simões ZLP, Hartfelder K. RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften. 2008;95:953–961. doi: 10.1007/s00114-008-0413-9. [DOI] [PubMed] [Google Scholar]
- 35.Raza A, et al. RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): Potential technology for the control of whitefly. PLoS ONE. 2016;11:e0153883. doi: 10.1371/journal.pone.0153883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Guan R, Guo H, Miao X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015;38:2277–2285. doi: 10.1111/pce.12546. [DOI] [PubMed] [Google Scholar]
- 37.Baum JA, et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 38.Upadhyay SK, Singh H, Dixit S, Mendu V, Verma PC. Molecular characterization of vitellogenin and vitellogenin receptor of Bemisia tabaci. PLoS ONE. 2016;11:e0155306. doi: 10.1371/journal.pone.0155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soffan A, et al. Silencing the olfactory co-receptor RferOrco reduces the response to pheromones in the red palm weevil, Rhynchophorus ferrugineus. PLoS ONE. 2016;11:e0162203. doi: 10.1371/journal.pone.0162203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott JG, et al. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013;59:1212–1221. doi: 10.1016/j.jinsphys.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla JN, et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016;13:656–669. doi: 10.1080/15476286.2016.1191728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Y-B, et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Yang G, Wang-Pruski G, You M. Phyllotreta striolata (Coleoptera: Chrysomelidae): Arginine kinase cloning and RNAi-based pest control. Eur. J. Entomol. 2008;105:815. doi: 10.14411/eje.2008.108. [DOI] [Google Scholar]
- 44.Tufail M, Raikhel AS, Takeda M. Biosynthesis and processing of insect vitellogenins. Progress in vitellogenesis. Reprod. Biol. Invertebr. 2005;12:1–32. [Google Scholar]
- 45.Tufail M, Nagaba Y, Elgendy AM, Takeda M. Regulation of vitellogenin genes in insects. Entomol. Sci. 2014;17:269–282. doi: 10.1111/ens.12086. [DOI] [Google Scholar]
- 46.Tufail M, Bembenek J, Elgendy AM, Takeda M. Evidence for two vitellogenin-related genes in Leucophaea maderae: The protein primary structure and its processing. Arch. Insect Biochem. Physiol. 2007;66:190–203. doi: 10.1002/arch.20212. [DOI] [PubMed] [Google Scholar]
- 47.Guidugli KR, et al. Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 2005;579:4961–4965. doi: 10.1016/j.febslet.2005.07.085. [DOI] [PubMed] [Google Scholar]
- 48.Nelson CM, Ihle KE, Fondrk MK, Page RE, Jr, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5:e62. doi: 10.1371/journal.pbio.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J-S, Cho W-L, Raikhel AS. Analysis of mosquito vitellogenin cDNA: Similarity with vertebrate phosvitins and arthropod serum proteins. J. Mol. Biol. 1994;237:641–647. doi: 10.1006/jmbi.1994.1261. [DOI] [PubMed] [Google Scholar]
- 50.Trewitt PM, Heilmann LJ, Degrugillier S, Kumaran AK. The boll weevil vitellogenin gene: Nucleotide sequence, structure, and evolutionary relationship to nematode and vertebrate vitellogenin genes. J. Mol. Evol. 1992;34:478–492. doi: 10.1007/BF00160462. [DOI] [PubMed] [Google Scholar]
- 51.Warr E, et al. A tapeworm molecule manipulates vitellogenin expression in the beetle Tenebrio molitor. Insect Mol. Biol. 2006;15:497–505. doi: 10.1111/j.1365-2583.2006.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J-L, Tang B-Z, Hou Y-M, Xie Y-X. Molecular cloning and expression of the vitellogenin gene and its correlation with ovarian development in an invasive pest Octodonta nipae on two host plants. Bull. Entomol. Res. 2016;106:642–650. doi: 10.1017/S0007485316000353. [DOI] [PubMed] [Google Scholar]
- 53.Yokoyama MN, et al. Storage proteins, vitellogenin and vitellin of wild silkworms, Antheraea yamamai, Antheraea pernyi and their hybrids. Comp. Biochem. Physiol. Part B Comp. Biochem. 1993;106:163–172. doi: 10.1016/0305-0491(93)90023-X. [DOI] [Google Scholar]
- 54.Baker M. Invertebrate vitellogenin is homologous to human von Willebrand factor. Biochem. J. 1988;256:1059. doi: 10.1042/bj2561059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akasaka M, Harada Y, Sawada H. Vitellogenin C-terminal fragments participate in fertilization as egg-coat binding partners of sperm trypsin-like proteases in the ascidian Halocynthia roretzi. Biochem. Biophys. Res. Commun. 2010;392:479–484. doi: 10.1016/j.bbrc.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 56.Donnell DM. Vitellogenin of the parasitoid wasp, Encarsia formosa (Hymenoptera: Aphelinidae): Gene organization and differential use by members of the genus. Insect Biochem. Mol. Biol. 2004;34:951–961. doi: 10.1016/j.ibmb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Lewis DK, et al. Characterization of vitellogenin in the red imported fire ant, Solenopsis invicta (Hymenoptera: Apocrita: Formicidae) J. Insect Physiol. 2001;47:543–551. doi: 10.1016/S0022-1910(00)00155-4. [DOI] [PubMed] [Google Scholar]
- 58.Barr PJ. Mammalian subtilisins: The long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-M. [DOI] [PubMed] [Google Scholar]
- 59.Liu W, et al. Juvenile hormone facilitates the antagonism between adult reproduction and diapause through the methoprene-tolerant gene in the female Colaphellus bowringi. Insect Biochem. Mol. Biol. 2016;74:50–60. doi: 10.1016/j.ibmb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Mayadas TN, Wagner DD. Vicinal cysteines in the prosequence play a role in von Willebrand factor multimer assembly. Proc. Natl. Acad. Sci. 1992;89:3531–3535. doi: 10.1073/pnas.89.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mouchel N, Trichet V, Betz A, Le Pennec J-P, Wolff J. Characterization of vitellogenin from rainbow trout (Oncorhynchus mykiss) Gene. 1996;174:59–64. doi: 10.1016/0378-1119(96)00336-8. [DOI] [PubMed] [Google Scholar]
- 62.Coelho RR, et al. Vitellogenin knockdown strongly affects cotton boll weevil egg viability but not the number of eggs laid by females. Meta Gene. 2016;9:173–180. doi: 10.1016/j.mgene.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moriyama M, Hosokawa T, Tanahashi M, Nikoh N, Fukatsu T. Suppression of Bedbug’s reproduction by RNA interference of vitellogenin. PLoS ONE. 2016;11:e0153984. doi: 10.1371/journal.pone.0153984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009;39:824–832. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 65.Zhu F, Xu J, Palli R, Ferguson J, Palli SR. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011;67:175–182. doi: 10.1002/ps.2048. [DOI] [PubMed] [Google Scholar]
- 66.Turner C, et al. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol. 2006;15:383–391. doi: 10.1111/j.1365-2583.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- 67.Walshe D, Lehane S, Lehane M, Haines L. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 2009;18:11–19. doi: 10.1111/j.1365-2583.2008.00839.x. [DOI] [PubMed] [Google Scholar]
- 68.Araujo R, et al. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol. 2006;36:683–693. doi: 10.1016/j.ibmb.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tokar, D. R., Veleta, K. A., Canzano, J., Hahn, D. A., & Hatle, J. D.. Vitellogenin RNAi halts ovarian growth and diverts reproductive proteins and lipids in young grasshoppers.Am. Zool.54, 931–941 (2014). [DOI] [PMC free article] [PubMed]
- 70.Chu C-C, Sun W, Spencer JL, Pittendrigh BR, Seufferheld MJ. Differential effects of RNAi treatments on field populations of the western corn rootworm. Pestic. Biochem. Physiol. 2014;110:1–6. doi: 10.1016/j.pestbp.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Mysore K, Flannery E, Michel K, Severson DW, Zhu KY, Duman-Scheel M. Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J. vis. Exp. JoVE. 2015 doi: 10.3791/52523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaakeh, W. Longevity, fecundity, and fertility of the red palm weevil, Rynchophorus ferrugineus Olivier (Coleoptera: Curculionidae) on natural and artificial diets. Emirates J. Food Agric.17 (2005).
- 73.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetic analysis version 6.0. Mol. Biol. Evolut. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.SAS, I. SAS/STAT 9.2 User’s Guide. (SAS Institute, 2008).
- 76.Husain M, Rasool KG, Tufail M, Aldawood AS. Molecular characterization, expression pattern and RNAi-mediated silencing of vitellogenin receptor gene in almond moth, Cadra cautella. Insect Mol. Biol. 2020;29(4):417–430. doi: 10.1111/imb.12646. [DOI] [PubMed] [Google Scholar]
- 77.Husain M, Rasool KG, Tufail M, Alwaneen WS, Aldawood AS. RNAi-mediated silencing of vitellogenin gene curtails oogenesis in the almond moth Cadra cautella. PLoS ONE. 2021;16(2):e0245928. doi: 10.1371/journal.pone.0245928. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Ye C, An X, Jiang YD, Ding BY, Shang F, Christiaens O, Wang JJ. Induction of RNAi core machinery’s gene expression by exogenous dsRNA and the effects of pre-exposure to dsRNA on the gene silencing efficiency in the pea aphid (Acyrthosiphon pisum) Front. Physiol. 2019;9:1906. doi: 10.3389/fphys.2018.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data is present in the manuscript.






