Abstract
Three hundred and twenty isolates of Staphylococcus aureus were typed by DNA sequence analysis of the X region of the protein A gene (spa). spa typing was compared to both phenotypic and molecular techniques for the ability to differentiate and categorize S. aureus strains into groups that correlate with epidemiological information. Two previously characterized study populations were examined. A collection of 59 isolates (F. C. Tenover, R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hébert, B. Hill, R. Hollis, W. R. Jarvis, B. Kreiswirth, W. Eisner, J. Maslow, L. K. McDougal, J. M. Miller, M. Mulligan, and M. A. Pfaller, J. Clin. Microbiol. 32:407–415, 1994) from the Centers for Disease Control and Prevention (CDC) was used to test for the ability to discriminate outbreak from epidemiologically unrelated strains. A separate collection of 261 isolates form a multicenter study (R. B. Roberts, A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, and A. Tomasz, J. Infect. Dis. 178:164–171, 1998) of methicillin-resistant S. aureus in New York City (NYC) was used to compare the ability of spa typing to group strains along clonal lines to that of the combination of pulsed-field gel electrophoresis and Southern hybridization. In the 320 isolates studied, spa typing identified 24 distinct repeat types and 33 different strain types. spa typing distinguished 27 of 29 related strains and did not provide a unique fingerprint for 4 unrelated strains from the four outbreaks of the CDC collection. In the NYC collection, spa typing provided a clonal assignment for 185 of 195 strains within the five major groups previously described. spa sequencing appears to be a highly effective rapid typing tool for S. aureus that, despite some expense of specificity, has significant advantages in terms of speed, ease of use, ease of interpretation, and standardization among laboratories.
Staphylococcus aureus is the leading cause of nosocomial infection in the United States (6). In New York City (NYC) methicillin-resistant S. aureus (MRSA) accounts for approximately 29% of these infections and 50% of associated deaths (20). Increasingly, S. aureus typing has become an important tool in the study of strain origin, clonal relatedness, and the epidemiology of outbreaks. Typing also plays an important role in hospital investigations (1), as MRSA is now endemic or epidemic in many institutions (17). Although several different phenotypic and, more recently, molecular techniques are available for differentiating S. aureus, no method is clearly superior under all conditions. Currently, macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) is the standard at the Centers for Disease Control and Prevention (CDC) for S. aureus strain typing and has been used successfully to study strain dissemination, especially in the identification of nosocomial outbreaks (3, 18, 19, 28). However, while PFGE has excellent discriminatory power, it is labor-intensive and difficult to standardize among different laboratories (33). As with other gel-based typing systems, the interpretation of PFGE results is often subjective (27). These problems make the exchange of strain typing information difficult and complicate the creation of an S. aureus and MRSA typing database.
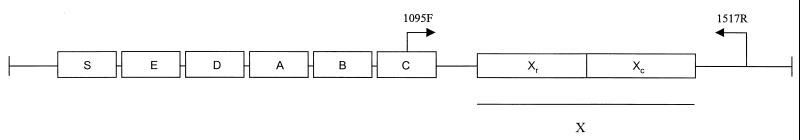
DNA sequencing is a powerful approach to strain typing with advantages in speed, unambiguous data interpretation, and simplicity of large-scale database creation. These advantages have been described by Maiden et al. (13), who developed the multilocus sequence typing (MLST) approach. This technique combines sequence information from several housekeeping genes to compare strains in a manner similar to multilocus enzyme electrophoresis (MLEE) (22). Recently, DNA sequencing of the polymorphic X, or short sequence repeat (SSR), region (32) of the protein A gene (spa) has been proposed as an alternative to current techniques for the typing of S. aureus (8). The polymorphic X region (Fig. 1) consists of a variable number of 24-bp repeats and is located immediately upstream of the region encoding the C-terminal cell wall attachment sequence (9, 21, 29). The diversity of the SSR region seems to arise from deletion and duplication of the repetitive units and also by point mutation (4). While the biological function is not known, the protein A domain encoded by the X region may serve to extend the N-terminal immunoglobulin G binding portion of the protein through the cell wall (34). The existence of well-conserved regions flanking the X region coding sequence in spa allows the use of primers for PCR amplification and direct sequence typing (Fig. 1). The sequencing of the spa SSR region combines many of the advantages of a sequencing-based system such as MLST but may be more rapid and convenient for outbreak investigation in the hospital setting since spa typing involves a single locus. Inasmuch as the protein A X region has a high degree of polymorphism, it may have a variation rate (or clock speed) that provides suitable discrimination for outbreak investigation.
FIG. 1.
Protein A gene map. Boxes indicate segments of the gene coding for the signal sequence (S), the immunoglobulin G-binding regions (A–D), a region homologous to A–D (E), and the COOH terminus (X), which includes the SSRs (Xr) and the cell wall attachment sequence (Xc). Primers are numbered from the 5′ end of the primer on the forward strand of S. aureus (GenBank accession no. J01786 [29]).
Despite the potential of spa typing, the clinical and epidemiological validity of protein A polymorphism analysis has not been clearly established (31, 32). The present study was undertaken to evaluate spa typing based on typeability, discriminatory power, reproducibility, ease of interpretation, and ease of use (14, 27). Fifty-nine isolates from the CDC were used to compare spa typing to a broad range of techniques in terms of their abilities to correctly group outbreak-related strains of S. aureus. In addition, we compared the utility of spa typing with that of two current molecular techniques for the identification of major MRSA clusters and specifically the clone I:A:A (type determined by restriction fragment length polymorphism (RFLP) typing with mecA and Tn554 combined with PFGE [mecA/Tn554/PFGE type]) in a larger study of MRSA in NYC hospitals (18). spa typing displayed a high degree of reproducibility and typeability and adequate resolving power and was simple to use and interpret.
MATERIALS AND METHODS
Bacterial strains.
The three hundred and twenty MRSA and methicillin-susceptible S. aureus isolates used in this study for the validation of spa typing were obtained from two previously characterized collections (26).
(i) CDC collection.
Fifty-nine strains were from a CDC collection previously analyzed by several typing techniques (24, 27, 30). Typing results and key features of these isolates are presented in Table 1 (adapted from Tenover et al. [27]). This collection included 29 isolates from four well-documented outbreaks (I–IV), 30 epidemiologically unlinked isolates, and one Staphylococcus intermedius isolate (Table 1). Isolates are divided into three sets, SA, SB, and SC (26).
TABLE 1.
Properties of staphylococcal strains from CDC collectiona
| Strain | Outbreak | Ox | Phage type | Antibiogram type | Biotype | Plasmid type | Hind ribo | Cla ribo | IS type | RFLP type | PCR | PFGE type | FIGE type | Immuno | MLEE type | spa type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SC-20 | NO | S | NR | I | INTER | NP | D | e | NH | NH:NH:NH:NH | 0.0 | I | VII | K | F | NA |
| SA-4 | NO | S | 6/47/54/75 | B | A-2b | B | F | i | NH | NH:X:4:NH | 2.1 | E | IV | D | E | 49 |
| SA-12 | NO | R | 47/54/75/77/83A | G | A-3b | NP | B | b | C | I:A:1:NH | 9.0 | J | IC2 | A | A5 | 2 |
| SA-18 | NO | R | 47/54/75/77/83A | J | A-3b | I | B | b | C | I:A:1:NH | 9.0 | J | IC3 | A2 | A3 | 51 |
| SA-20 | NO | R | 47/54/75/77/83A | K | A-3b | J | B | b | C | I:A:1:NH | 9.0 | J | IC1 | A1 | A1 | 2 |
| SA-6 | NO | I | NR | C | A-3b | C | A | a | B | II:NH:1:a | 9.0 | C | III | E1 | A4 | 2 |
| SA-7 | NO | R | 53/+ | D | H-4 | D | B | Ci | NH | NH:NH:1:NH | 9.0 | B | V | C | A2 | 2 |
| SA-8 | NO | R | 54/75/77/81 | E | 1-2b | E | E | d | D | I:NH:6:NH | 7.0 | G | IIA | E1 | D1 | 17 |
| SA-11 | NO | R | NR | F | A-2b | E | G | d | G | II:NH:6:NH | 7.0 | F | IIB | E2 | D2 | 17 |
| SA-1 | NH1 | R | 54/77 | A1 | A-1b | A | A | a.1 | A | I:A:5:a | 9.0 | K.1 | IB | A1 | A1 | 2 |
| SA-9 | NH1 | R | 54/77 | A | A-1b | NP | A | a.1 | A | I:A:5:a | 9.0 | K.2 | JB | A1 | A1 | 2 |
| SA-3 | NH1 | R | 47/54/75/77 | A2 | A-3b | NP | A | a | C | I:A:1:NH | 9.0 | A | IA | A | A1 | 2 |
| SA-13 | NH1 | R | 54/77 | A3 | A-1b | G | A | a | A | I:A:1:a | 9.0 | A | IA | A3 | A2 | 2 |
| SA-14 | NH1 | S | 54/75/77 | H | B-1b | H | C | I | NH | NH:NH:1:NH | 9.0 | H | VI | E3 | C | 50 |
| SA-19 | NH1 | R | 54/77 | A4 | G-1b | A | A | a.1 | A | I:A:1:a | 9.0 | K.3 | IB | A1 | A1 | 57 |
| SA-17 | NH2 | R | 54/75/77 | A | C-3b | A | A | a | A | I:A:1:a | 9.0 | A | IA | A | A1 | 2 |
| SA-2 | NH2 | R | 75/77 | A | A-3b | A | A | a | A1 | I:A:1:b | 9.0 | A | IA | A | A1 | 2 |
| SA-15 | NH2 | R | 77 | A | A-3b | A | A | a | A1 | I:A:1:a | 9.0 | A | IA | A1 | A5 | 2 |
| SA-5 | NH2 | R | 77 | A | A-3b | A | A | a | A | I:A:1:a | 9.0 | A | IA | A | A1 | 2 |
| SA-10 | NH2 | R | 77 | A | A-3b | A | A | a | A | I:A:1:a | 9.0 | D | ID | A1 | B | 2 |
| SB-7 | NO | S | 6/47/54/75 | C | A-2b | D | C | i | NH | NH:X:4:NH | 2.1 | D | IIB3 | D′ | B3 | 49 |
| SB-3 | I | R | 75/+ | A | C-4 | C | A | a | E | I:A:1:a | 9.0 | A | IA | A6 | A1 | 2 |
| SB-5 | I | R | 75/+ | A | A-4 | C | A | a | E | I:A:1:a | 9.0 | A | IA | A6 | A1 | 2 |
| SB-10 | I | R | 75/+ | A | A-4 | C | A | a | E | I:A:1:a | 9.0 | A | IA | A6 | A1 | 2 |
| SB-12 | I | R | 75/+ | A | C-4 | A | A | a | E | I:A:1:a | 9.0 | A.1 | IA | A6 | A1 | 2 |
| SB-15 | I | R | 75/77/83A | A | C-4 | C | A | a | E | I:A:1:a | 9.0 | A | IA | A6 | A1 | 2 |
| SB-19 | I | R | 75/+ | A | A-4 | C | A | a | E | I:A:1:a | 9.0 | A | IA | A5 | A1 | 2 |
| SB-20 | I | R | 75/+ | A | A-4 | C | A | a | E | I:A:1:a | 9.0 | A | IA | A5 | A1 | 2 |
| SB-1 | NO | R | 75/55 | A | A-4 | A | A | a | E | I:Y:1:a | 9.0 | A.1 | IB1 | A5 | A1 | 2 |
| SB-16 | NO | R | 75/77/83A | A | A-4 | A | A | a | E | I:Y:1:a | 9.0 | A.1 | IB1 | A5 | A1 | 2 |
| SB-18 | NO | R | 75/+ | A | C-4 | J | A | a | E1 | I:A:1:a | 9.0 | A.1 | IA | A7 | A1 | 2 |
| SB-17 | NO | I | 96 | E | B-3b | I | F | i | NH | NH:NH:1:NH | 6.0 | E | IV | G | A2 | 54 |
| SB-14 | NO | R | 47/54/75/77/83A | A1 | A-3b | H | E | a | D | I:A:1:NH | 9.0 | A.2 | IB2 | A5 | A3 | 2 |
| SB-8 | NO | S | 95 | B1 | C-4 | E | D | d.1 | NH | NH:NH:7:NH | 2.0 | F | III | E5 | C | 42 |
| SB-2 | II | S | 3A/55 | B | B-1b | B | B | b | NH | NH:NH:7:NH | 6.0 | B | IIA | D1 | B1 | 52 |
| SB-4 | II | S | 3A/55 | B | D-1b | B | B | b | NH | NH:NH:7:NH | 6.0 | B | IIA | D1 | B1 | 52 |
| SB-6 | II | S | 3A/55 | B | B-1b | B | B | b | NH | NH:NH:7:NH | 6.0 | B | IIA | D1 | B1 | 52 |
| SB-11 | II | S | 3A/55 | B | B-3b | G | B1 | b | NH | NH:NH:7:NH | 14.0 | C | IIB2 | D2 | B1 | 21 |
| SB-9 | NO | S | 3A | D | D-3b | G | B | b | NH | NH:Z:7:NH | 6.0 | B | IIA | D1 | B1 | 21 |
| SB-13 | NO | S | 3A | B2 | D-3b | G | B | b | NH | NH:NH:7:NH | 6.0 | B.1 | IIB1 | E6 | B2 | 53 |
| SC-3 | NO | S | 6/47/54/75 | C | A-2b | C | A | i | NH | NH:NH:4:NH | 2.1 | C | III | D | B | 49 |
| SC-1 | III | R | 75 | A | A-1b | A | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-4 | III | S | 75 | A | A-1b | D | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-5 | III | R | NR | A1 | A-1b | D | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-9 | III | R | 75 | A | A-1b | D | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-11 | III | R | 75 | E | A-1b | NP | A | b | NH | I:A:4:NH | 10.0 | A | IB | F | A1 | 7 |
| SC-12 | III | R | 75 | A2 | A-1b | A | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-14 | III | R | 75 | A2 | B-2b | A | A | b | F | I:A:4:a | 10.0 | A | IA | F | A2 | 7 |
| SC-15 | III | R | 75 | A | A-1b | D | B | 2b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-17 | III | R | 75 | A | A-1b | A | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-20 | III | R | 75 | A | A-1b | D | A | b | F | I:A:4:a | 10.0 | A | IA | F | A1 | 7 |
| SC-8 | NO | S | NR | B | B-3a | E | B1 | g | NH | NH:NH:1:NH | 2.0 | B.1 | II | E7 | A3 | 58 |
| SC-2 | IV | S | 52/52A/80/47/54/83A/84/95 | B | E-1b | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | C1 | 57 |
| SC-6 | IV | S | 95 | B | J-1b | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | C1 | 57 |
| SC-7 | IV | S | 95 | D | I-1a | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | C1 | 57 |
| SC-10 | IV | S | 52A/79/80/47/54/75/77/83A/95 | B | I-2a | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | C1 | 57 |
| SC-13 | IV | S | 95 | B1 | I-1b | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | C1 | 57 |
| SC-16 | IV | S | 95 | B1 | I-1b | B | A | g | NH | NH:NH:1:NH | 2.0 | B | II | H | D1 | 2 |
| SC-18 | IV | S | 95 | F | I-3b | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | C1 | 57 |
| SC-19 | IV | S | 95 | B1 | D-1a | B | B | g | NH | NH:NH:1:NH | 2.0 | B | II | E7 | D2 | 57 |
Adapted from reference 27; for further characterization see references 24 and 30. Ox, oxacillin susceptibility test results; plasmid, plasmid restriction profile; Hind ribo, ribotyping result with Hind; Cla ribo, ribotyping result with Cla; PCR, coagulase gene PCR typing result; immuno, immunoblot typing result; S, susceptible; R, resistant, I, intermediate; INTER, S. intermedius biotype; NO, not in an epidemiologically related cluster; NP, no plasmids; NH, no hybridization; NA, not applicable (protein A gene is not present in S. intermedius SA-16).
S. aureus ATCC 12600 (SA-4, SB-7, SC-3) was included as a control in all groups. Group SA contains nine isolates from two nursing homes (labeled NH1 and NH2) that did not have a clear epidemiological link but were characterized as belonging to a pseudo-outbreak (26). Seven additional isolates from seven states and S. intermedius isolate ATCC 49052 (SA-16) were included in the SA group and are labeled NO. Among the SA isolates, SA-1 and SA-9 are duplicates as are SA-2 and SA-15. Strains SA-12, SA-18, and SA-20 all have the same bacteriophage type but are epidemiologically unlinked.
SB contains eight unrelated isolates labeled NO and isolates obtained during two outbreaks, identified as I and II. Strains SB-3, -5, -10, -12, -15, -19, and -20 from outbreak I were obtained from the Iowa Veterans Affairs Medical Center. Outbreak II strains (SB-2, -4, -6, and -11) were isolated from a contaminated anesthetic.
SC contains isolates from outbreaks III and IV, strain SC-3 (ATCC 12600), and an unrelated control (SC-8) labeled NO. Outbreak III strains were from the Sepulveda Veterans Affairs Medical Center, Sepulveda, Calif. Outbreak IV strains were from an anesthetic contamination unrelated to outbreak II. Strains SC-17 and -20 were duplicates included as internal controls.
(ii) NYC collection.
Two hundred and sixty-one isolates were from a consecutive single-patient MRSA study and were collected over a 6-month period from 12 NYC hospitals (18). These isolates had been typed by our laboratory previously via Southern blot hybridization with the two gene probes mecA and Tn554 following ClaI digestion (18). Isolates were also analyzed by macrorestriction analysis using PFGE of SmaI-digested chromosomal DNA. Both RFLP and PFGE were performed as described previously (5, 12). Five major clones were identified and assigned the codes I:A:A, I:D:C, V:NH:E, IV:M:B and I:E:F (mecA/Tn554/PFGE type).
spa sequencing.
Amplification and sequencing of the SSR region of the spa gene were performed with chromosomal DNA purified from each isolate as a template (18); several rapid techniques proved sufficient (26). Primer sites for PCR amplification were designed according to published sequence data (29) (Fig. 1).
PCR amplification of the SSR region of the spa gene was accomplished by adding 1 μl of a 1:200 dilution of genomic DNA and 24 μl of water to 25 μl of PCR master mixture in 0.2-ml PCR tubes (Perkin-Elmer Applied Biosystems Division [PE-ABI]). Master mixture buffer contains 0.5 μM PA 1095F forward PCR primer, 0.5 μM PA 1517R reverse primer, 2.5 U of AmpliTaq Gold DNA polymerase, 2 mM MgCl2, 350 μM total dNTPs, and 25 mM KCl. A negative control (sterile deionized water) and a positive control (from our laboratory's S. aureus collection) were included. Tubes were capped and placed in a GeneAmp PCR System 9600 Thermocycler (PE-ABI). Thermal cycling parameters included an initial 10 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, and 45 s at 72°C; and a final extension at 72°C for 10 min. PCR products were purified and concentrated twofold prior to sequencing in distilled water with a Microcon-100 (Millipore) microconcentrator. Completed reaction mixtures were stored at −20°C.
DNA cycle sequencing reaction mixtures had a total volume of 20 μl, and reactions were carried out with a GeneAmp PCR System 9600 with the following reaction conditions: 13 μl of AmpliTaq FS Dye Terminator ready reaction mixture and sequencing primer (PA 1095F and PA 1517R), 3 μl of purified PCR product, and 4 μl of deionized water. The cycle sequencing profile consisted of a 96°C denaturation step for 10 s followed by an annealing/extension step starting at 65°C and decreasing 1°C every six cycles until a touchdown temperature of 55°C was reached, for a total of 66 cycles. Dye terminator cycle sequencing reaction products were purified with a Sephadex column (Pharmacia) and a Silent Screen filter plate (Nalge Nunc). The column filtrate was evaporated in a vacuum centrifuge and resuspended in 2 μl of blue dextran-EDTA–deionized formamide (1:5). Sequences were determined by electrophoresis with the ABI PRISM 377 DNA sequencer. Consensus sequences were assembled from both orientations with PE-ABI software.
Identification and classification of spa types.
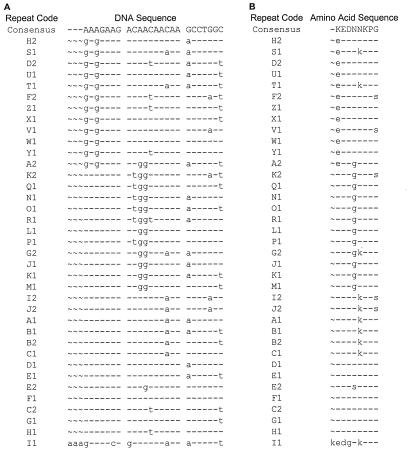
Since the major source of variation in the X region seems to be duplication or deletion of the repetitive units, strain lineages cannot be obtained by comparing the sequences with an algorithm based on sequence alignment. This precludes the use of a dendrogram to visually represent typing results because dendrograms rely on sequence alignments. Therefore, we attempted to establish strain relatedness by first identifying all possible variations of the repeat units and then assessing how these repeat units were organized in the X regions of the different isolates. The program FINDPATTERNS from the Genetics Computer Group (GCG) Wisconsin Package 9.1 was used to identify repeat units matching the ambiguous patterns AAAGAAGAXXXXAAXAAX {1,4} CCXXXX and GAGGAAGAXXXXAAXAAX {1,4} CCXXXX. This strategy allowed the identification of all SSRs contained in the spa regions analyzed from all isolates. A customized Perl script (NEWREPEATS; M. Bergman) was used to identify, from the output of FINDPATTERNS, unique repeat sequences that were assigned a type based on a random alphabetical code (Fig. 2A). Another Perl script (CLEANFP; M. Bergman) was used to assign, to each of the spa regions analyzed, a spa repeat code based on the order of SSR codes as defined by the output of NEWREPEATS. The spa repeat code, therefore, represents the structure of the SSR region of the S. aureus isolates studied (Tables 2 and 6). All unique spa repeat codes were assigned a random numerical code, or spa type, for identification.
FIG. 2.
DNA (A) and amino acid (B) sequences of individual spa repeats and their letter codes based on variation from a DNA consensus sequence. Repeats were obtained by a GCG FINDPATTERNS search with the ambiguous search patterns AAAGAAGAXXXXAAXAAX {1,4} CCXXXX and GAGGAAGAXXXXAAXAAX {1,4} CCXXXX. Identical residues are identified by dashes, and gaps are identified by tildes.
TABLE 2.
spa repeat patterns for CDC collectiona
| spa type-repeat code | No. of strains |
|---|---|
| 42-A2AKBEMBKB | 1 |
| 51-TJMBMDMGKK | 1 |
| 2-TJMBMDMGMK | 21 |
| 21-UJGBBGGJAGJ | 2 |
| 52-UJGBBGGJAGJ | 3 |
| 53-UJGBBIGGJ | 1 |
| 50-UJGFMGGM | 1 |
| 58-XKAKEMEKB | 1 |
| 57-XKBBMBKB | 7 |
| 7-YHGCMBQBLO | 8 |
| 49-YHGGFCBQBLO | 3 |
| 17-ZDMDMNKB | 2 |
| 54-ZFGDMGGM | 1 |
| Total | 52 |
Repeat codes are derived from the organization of individual repeats (random alphabetical code) seen in Fig. 2. There are 13 distinct repeat types for the CDC collection. To make the notation shorter, letter codes ending with 1 have been simplified by removing the number.
TABLE 6.
spa repeat patterns for NYC collection
| spa type-repeat codea | No. of minor-clone strains | Major clones
|
No. of strains not hybridizing with mecA gene | |
|---|---|---|---|---|
| No. of strains | Type | |||
| 15-A2AKEEMBMK | 4 | 1 | ||
| 60-TJMAMGMK (A) | 1 | 1 | I:A:A | |
| 29-TJMBMDMGGMK (A) | 2 | I:A:A | ||
| 23-TJMBMDMGK (A) | 1 | I:A:A | ||
| 2-TJMBMDMGMK (A) | 25 | 98 | I:A:A | 1 |
| 14-TJMBMDMGMKK (A) | 2 | |||
| 26-TJMBMGMK (A) | 3 | I:A:A | ||
| 25-TJMDMGMK (A) | 3 | |||
| 24-TJMEMDMGMK (A) | 1 | 2 | I:A:A | |
| 28-TKJMBMDMGMKK (A) | 1 | |||
| 38-TLMDMGMMK (A) | 1 | 1 | ||
| 27-TMDGMMK (A) | 1 | |||
| 21-UJGBBGGJABJ | 1 | 1 | ||
| 37-UKGJB | 1 | 1 | ||
| 18-WFKAOMQ (B) | 1 | 1 | ||
| 16-WGKAKAOMQQQ (B) | 11 | 10 | I:D:C | |
| 3-WGKAOMQ (B) | 3 | 26 | IV:M:B | |
| 19-XKAKAOMQ | 1 | |||
| 20-YHB2CMBQBLO (C) | 1 | V:NH:E | ||
| 4-YHFGFMBQBLO (C) | 1 | 14 | I:E:F | |
| 7-YHGCMBQBLO (C) | 3 | 38 | V:NH:E | |
| 1-YHGFMBQBLO (C) | 2 | |||
| 22-ZDKB | 1 | 1 | ||
| 17-ZDMDMGGM | 2 | |||
| Total | 65 | 196 | 7 | |
spa types and their distribution for major and minor clones of the NYC collection (n = 261). spa types are arbitrarily assigned for each unique sequence identified; there are 24 distinct types for the NYC collection. Repeat codes are derived from the organization of individual repeats (random alphabetical code) seen in Fig. 1. To make the notation shorter, letter codes have been simplified as noted for Table 2. Strains with similar repeat patterns are indicated by the same letter in parentheses (A, B, or C).
In this paper, isolates are considered members of the same clone based on identical spa sequences (spa types). The relatively large number of isolates (n = 261) from the NYC collection were also grouped based on similarity of the patterns of repeat sequences at both the DNA and amino acid levels (Table 6). This association is based on the assumption that accumulated point mutations are consistent between strains (4) and that, as such, similar repeat patterns may be an indication of genetic relatedness and a more recent common ancestor.
RESULTS
spa typing results.
The spa SSR regions of 320 isolates from two strain collections was sequenced. The analysis of the sequences resulted in the identification of 24 unique SSR types that were assigned letter codes (as suggested by Frenay et al. [8]). In the two collections analyzed in this study, repeat types were 24 bp long with the exception of one (repeat I1), which was 27 bp. The analysis of the SSR regions from additional S. aureus strains from our laboratory's collections (unpublished data) allowed the identification of 13 new unique SSRs. The combined list of 37 unique SSRs and their amino acid conversion are presented in Fig. 2A and B, respectively. Most nucleotide changes in the repeats result in synonymous mutations, indicating evolutionary pressure to conserve amino acid sequence. Therefore, positive Darwinian selection does not appear to be driving sequence diversity, as has been described for other highly polymorphic loci (25). This may account for the observation that, while variable enough to provide adequate strain discrimination, this region has the stability to group related strains for use as a typing tool (this study).
The organization of the repeats in the spa SSR region from each of the isolates was represented as a spa type repeat code that was used as a typing criterion. Thirty-three different spa (strain) types were defined. The strain types and their frequencies in both the CDC and NYC collections appear in Tables 2 and 5, respectively. In the 59-strain CDC collection there were 22 unique short repeats that defined 13 different strain types (Table 2). spa types 21 and 52 have the same repeat organization but have sequence alterations in the normally conserved adjacent region and are assigned a unique type. In the 260-isolate NYC collection, there were 20 repeat types and 24 different strain types (Table 6). From all strains studied, SSR regions composed of 4, 5, 7, 8, 9, 10, and 11 repeats were characterized by PCR products that ranged from 250 to 637 bp in length. All SSRs regardless of length were successfully sequenced and assembled. The length of the variable region was not an accurate indicator of strain type, as many had the same number of repeats (for example, 11 unique strain types had eight repeat elements). Type 2, the most common spa type, had 10 repeats and a 556-bp PCR product. Strain types 2, 7, 17, and 21 were present in both collections.
TABLE 5.
Major clones of NYC collection
| mecA/Tn554/ PFGE result | No. of isolates | No. of isolates spa classified together | spa typeb | % Agreementa |
|---|---|---|---|---|
| I/A/A | 107 | 98 | 2 | 92 |
| I/D/C | 10 | 10 | 16 | 100 |
| I/E/F | 14 | 14 | 4 | 100 |
| IV/M/B | 26 | 26 | 3 | 100 |
| V/NH/E | 39 | 38 | 7 | 92 |
| Total | 196 | 186 | 95 |
Agreement between major clonal groupings of the NYC collection as defined by spa typing and the combination of PFGE and Southern hybridization with the two gene probes mecA and Tn554 (n = 196).
For spa repeat codes and additional major clone spa types see Table 6.
Typeability and reproducibility.
All 320 isolates of S. aureus were typeable by spa sequencing. As indicated in Table 3, the 100% typeability of resistant and susceptible isolates of the 59-strain CDC collection by spa sequencing is an advantage over phage typing, plasmid typing, insertion sequence mapping, and most of the RFLP methods. The reproducibility of spa typing was tested by including duplicate isolates twice in set SA (SA-1 and SA-9 and SA-2 and SA-15) and in set SC (SC-17 and SC-20) of the CDC collection. In addition, strain ATCC 12600 was included in all three sets. There were no variations in the results for the duplicate isolates by spa sequencing, which is not the case for 5 of the 13 other methods including PFGE, (Table 1) (18).
TABLE 3.
spa typing of CDC collection: typed, subtyped, and nontypeable isolates by set and number of isolates correctly identified and misclassified by each method from four outbreaks (n = 29)a
| Method | No. of isolates in set
|
Total no. of types | No. classified correctly | No. misclassified | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SA
|
SB
|
SC
|
||||||||||
| Typed | Subtyped | Nontypeable | Typed | Subtyped | Nontypeable | Typed | Subtyped | Nontypeable | ||||
| Phage typing | 9 | 3 | 7 | 5 | 2 | 18 | 25 | 4 | ||||
| Antibiogram | 11 | 4 | 5 | 3 | 6 | 3 | 21 | 26 | 6 | |||
| Biotype | 6 | 8 | 4 | 8 | 6 | 11 | 23 | 17 | 2 | |||
| Plasmids | 9 | 4 | 10 | 5 | 1 | 20 | 23 | 0 | ||||
| HindIII ribotyping | 7 | 6 | 1 | 2 | 2 | 16 | 27 | 7 | ||||
| ClaI ribotyping | 6 | 1 | 5 | 1 | 3 | 9 | 29 | 7 | ||||
| IS typing | 5 | 1 | 4 | 2 | 1 | 9 | 1 | 11 | 9 | 16 | 3 | |
| RFLP typing | 10 | 1 | 7 | 4 | 17 | 28 | 3 | |||||
| Coagulase gene PCR | 3 | 1 | 4 | 1 | 2 | 1 | 7 | 28 | 8 | |||
| PFGE | 11 | 3 | 6 | 3 | 3 | 1 | 25 | 28 | 7 | |||
| FIGE | 11 | 3 | 6 | 5 | 4 | 25 | 27 | 3 | ||||
| Immunoblotting | 5 | 7 | 4 | 8 | 4 | 23 | 28 | 6 | ||||
| MLEE | 11 | 7 | 7 | 21 | 26 | 4 | ||||||
| Average | 8 | 6 | 4 | 18 | 25 | 5 | ||||||
| spa typing | 6 | 7 | 3 | 16 | 27 | 4 | ||||||
Adapted from reference 27.
The in vitro and in vivo stability of spa typing results was investigated previously (8). We independently confirmed in vitro stability by examining the effects of multiple passages on blood agar plates. Ten different strains were subcultured on a daily basis for 6 weeks. All 10 strains had unique spa types that ranged in size from 7 to 11 repeats with virtually all repeat groups (defined by organization of repeat sequence code) represented. No changes in either PFGE pattern or spa type were seen (data not shown). Previously, in vivo stability was assessed by using three isolates collected over a 5-year period in a chronic MRSA carrier cystic fibrosis patient. The spa type remained the same over this time period (8).
Discriminatory power with the CDC collection.
Many of the pseudo-outbreak strains of the SA group had an identical spa type (type 2). It has been suggested by Smeltzer et al. (24) that this may represent the correct grouping of related strains (similarly, Smeltzer et al. found a unique type for pseudo-outbreak strain SA-14, in agreement with spa typing). Three strains, SA-12, SA-18, and SA-20, that were included in the SA group are known to be epidemiologically unrelated but have a common phage type. spa typing was more sensitive than phage typing and was able to classify SA-18 as different. Immunotyping, MLEE, field inversion gel electrophoresis (FIGE) and plasmid restriction analysis were able to distinguish all three. All other typing methods were unable to discern differences between these strains. This observation is in agreement with the previous results of Frenay et al. (8) that spa typing has sufficient stability to group strains in concordance with phage typing but with greater discrimination.
In the SB group, outbreak II strains SB-2, -4, and -6 had an identical type (type 52). This did not include outbreak II strain SB-11 (type 21), which was therefore misclassified. It has been noted that strains SB-2, -4, and -6 came from the same patient and that 8 of 13 other techniques distinguished SB-11 as different (24). All outbreak I strains were correctly grouped together as type 2, but four strains (SB-1, -14, -16, and -18) that were originally judged to be unrelated to this outbreak based on epidemiological information were also assigned this type. This result is in agreement with the typing of Smeltzer et al. (24), who have also shown that strains SB-1 and -16 may be related to each other and outbreak I strains. In the SC study group, without exception, strains from outbreak III were grouped together as type 7. Outbreak IV strains were grouped as type 57 with the exception of strain SC-16 (type 2), which was also incorrectly classified by immunotyping, HindIII ribotyping, and MLEE. In light of the epidemiological link between SC-16 and outbreak IV strains (25), this appears to be the only unambiguous example of a failure of spa typing to group strains appropriately.
A summary of the total number of types, subtypes, and nontypeable strains identified by each method and of the number of isolates correctly identified and misclassified by each typing system is given in Table 3. The information in this table is derived from the 29 strains that were epidemiologically linked to four different outbreaks (I–IV) from strain sets SB and SC from the CDC collection. SB and SC also contained isolates that were not outbreak related and that therefore were omitted from Table 3. Similarly, SA strains were not included since an outbreak was not clearly established for these isolates.
spa typing was able to group 27 of the 29 outbreak strains into four clusters corresponding to the outbreaks (outbreaks I–IV corresponded to spa types 2, 52, 7, and 57, respectively). Four additional strains from sets SB and SC that were unrelated, based on epidemiological information, to these outbreaks had the same spa type and are deemed misclassified. Therefore, the sensitivity and specificity of this method compare favorably with those of current techniques (Table 3) (average: 25 correct and 5 misclassified; PFGE: 28 correct and 7 misclassified).
Discriminatory power with the NYC collection.
Table 4 describes the ability of the three different molecular methods to differentiate the NYC isolates. spa typing identified 24 unique strain types. The combination of mecA and Tn554 hybridization patterns with PFGE subtype and spa type produced a total of 107 types for the 261 isolates. There were 39 mecA/Tn554/PFGE types, and there were 53 types when spa typing was added. Similarly, there were 21 mecA/Tn554 types and 47 mecA/Tn554/spa types. PFGE subtyping provides the greatest discrimination of the four methods, describing 80 subtypes. Typing with spa appears to provide discrimination similar to that of mecA/Tn554 polymorphism. However, spa typing was able to provide a clonal assessment for the isolates (Table 5) that was previously provided only with by mecA/Tn554/PFGE information (18). All five major clonal groups in the NYC collection were identified by spa typing with greater than 95% agreement for 196 isolates (Table 5; mecA/Tn554/PFGE) types I:A:A, I:D:C, I:E:F, IV:M:B, and V:NH:E correspond to spa types 2, 16, 4, 3, and 7, respectively). spa types for both major and minor clones (all other isolates) of the NYC collection are presented in Table 6. The minor clones represent 67 strains or 25% of the NYC study total (n = 261). The minor clones consist of isolates that did not display a prevalent clonal type (less than 10 isolates with one PFGE/hybridization pattern, including a few isolates that did not hybridize with the mecA probe) and that were categorized separately from the major clones in the NYC study (18). Interestingly, the 20 spa repeat patterns seen among the minor clones are the same as or similar to those of the five major clonal groups, indicating a relatedness which would otherwise have been overlooked by PFGE and hybridization alone.
TABLE 4.
Resolution of typing methods for NYC collectiona
| Typing method(s) | No. of different patterns |
|---|---|
| mecA/Tn554/PFGE subtype/spa | 107 |
| mecA/Tn554/PFGE subtype | 96 |
| mecA/Tn554/PFGE:spa | 53 |
| mecA/Tn554/PFGE | 39 |
| mecA/Tn554/spa | 47 |
| mecA/Tn554 | 21 |
| mecA/PFGE subtype | 84 |
| mecA/PFGE | 27 |
| mecA/spa | 34 |
| mecA | 8 |
| Tn554/PFGE subtype | 90 |
| Tn554/PFGE | 32 |
| Tn554/spa | 40 |
| Tn554 | 9 |
| PFGE subtype/spa | 92 |
| PFGE/spa | 32 |
| PFGE subtype | 78 |
| PFGE | 18 |
| spa typing | 24 |
Adapted from reference 18 with permission of the publisher.
In addition to clonal assessment, by analyzing the pattern of repeats, spa typing allowed the grouping of NYC strains that have similar repeat organizations and therefore may have a relatively recent common ancestor. This grouping (Table 6; types labeled A–C) is more subjective than the strict and simple criteria used for clonal assessment (based on identical spa SSR sequence as indicated in Materials and Methods). However, by doing so we observed three major repeat pattern groups for all but 10 NYC strains (additional sequence information from coagulase repeat regions confirms these groupings [unpublished data]. Of the six repeat types that do not fall into the three repeat similarity groups, four of them (types 21, 37, 15, and 22) are unique among strains that do not hybridize with the mecA gene, which confers methicillin resistance (MRSA phenotype). The apparent limited variation of repeat patterns (see A, B, and C groups in Table 6) for the MRSA strains may be a reflection of their clonal origin (2, 10, 11, 15).
Ease of use and interpretation.
The typing systems used to analyze the CDC collection have been previously assessed for ease of use and interpretation (27). We compared the ease of use and interpretation of spa typing with that of the molecular techniques of RFLP and PFGE by using the isolates from the NYC collection (18). PFGE demanded the use of special electrophoretic equipment and the preparation of DNA in agarose disks, which required delicate handling to provide the distinct banding patterns necessary for analysis. Similarly, a fair amount of expertise and time for analysis was required for the interpretation of the complex patterns produced by PFGE and the application of grouping criteria to relate them along clonal lines. The original analysis of the NYC strains was a collaborative effort at two institutions (Public Health Research Institute and Rockefeller University) (18). We found that it was necessary to standardize virtually all reaction conditions and apparatuses to permit a unified interpretation of results. Interpretation of PFGE also required the standardization of parameters for the computer software (Bioimage Whole Band Analyzer) used to analyze and compare banding patterns. The complex PFGE patterns, doublet bands, and partial digests also made the interpretation of results moderately subjective. RFLP typing with the gene probes mecA and Tn554 required separate electrophoretic equipment in addition to a transfer apparatus and labeling products for the probes. The patterns were more easily interpreted than PFGE patterns and were compared visually. The amount of time required to obtain, analyze, and compare typing information for the original 270 strains by RFLP and PFGE was approximately 1 year. spa typing required DNA preparation (possible by several rapid techniques) for PCR amplification and sequencing with a PE-ABD TC9600 sequencer. The identification of unique sequences required computer software from PE-ABD (sequence assembly) and GCG (analysis of repeats). The interpretation of repeats for assessing strain types is unambiguous because only isolates with the identical repeat region sequences are considered clonal, allowing rapid identification of related and unrelated strains. Sequencing and analysis of all strains in this study were accomplished in approximately 3 weeks. Cost analysis of spa sequencing was not performed and may vary depending on the apparatus and reagents chosen. However, in light of rapid improvements in automated sequencing technology, the cost (less than $10 per sample) will likely continue to decrease.
DISCUSSION
S. aureus is the most frequent cause of hospital-acquired infection in the United States (6). A number of different phenotypic and, recently, genotypic techniques are available to classify strains for epidemiological investigation in the detection and tracking of nosocomial outbreaks (27, 28). The goal of this study was to evaluate a new typing system for S. aureus based on the DNA sequencing of the variable region of protein A as an alternative to current techniques for use in research and clinical applications. Sequencing this variable region has been described as a typing tool by Frenay et al. (8) but has not been rigorously compared to current methods. As previously suggested, the availability of well-described collections such as those in this study can be used to establish the value of novel typing tools (30).
We found that spa typing compared favorably to other techniques and was able to identify and differentiate 27 of 29 epidemiologically related strains and misclassified only 4 unrelated strains in the four outbreaks of the CDC collection (27). Significantly, spa typing exhibited only one unambiguous discrepancy with epidemiological information. All strains included as internal controls were accurately identified. spa typing was able to distinguish the five major clones of a 261-strain NYC hospital MRSA collection originally described by a combination of PFGE and RFLP analysis (18). This included the correct clustering (14 of 14 with no misclassifications) of I/E/F isolates recently reported as the first outbreak of the “Iberian clone” in the United States (19). The observation that spa typing can group isolates in congruence with other methods in the two collections directly addresses concerns over the instability of this region for use in epidemiological studies (31).
For the 320 isolates in this study, there were 24 repeats, which were organized to describe a total of 33 different strain types. Unambiguous spa types were achieved for all isolates. The ability of spa typing to discriminate strains was similar to that of Southern blot hybridization with the gene probes mecA and Tn554 for the NYC study isolates. While spa sequencing does not have the resolving power of PFGE subtyping, it has several advantages in terms of speed, ease of use, ease of interpretation, and database creation. Significantly, spa typing also provides clonal groupings that RFLP and PFGE techniques cannot individually identify. This is accomplished without the use of subtypes, which are difficult to clearly define and which introduce a high degree of subjectivity that affects reproducibility among laboratories. The difficulties we encountered in coordinating PFGE typing of the NYC strains between two laboratories confirm the conclusions of a recent study of intercenter PFGE typing reproducibility, which stated that due to variability and bias, true standardization may never be achieved in this system (33). The cost of sequencing also compares favorably to that of techniques such as PFGE.
The main advantage of spa typing over current methods may be the unambiguity and portability of sequence data. This greatly simplifies the sharing of information between laboratories and facilitates the creation of a large-scale database for the study of global as well as local epidemiology (the electronic portability of sequence data allows rapid exchange of strain typing information without having to transfer bacterial strains). This is especially important in light of recent observations that MRSA outbreak strains from intercontinental sources have been documented within and among hospitals (19). Sequencing can facilitate the creation of an Internet Web site for the downloading of spa typing sequences, which could then be analyzed by software available at the site and added to a database. Such advantages have been ascribed to other systems, such as MLST (13), which was recently used to describe Streptococcus pneumoniae strains (7, 23).
The requirements for sequence typing are the ability to perform PCR and access to an automated sequencer, both of which are increasingly available to clinical laboratories and public health facilities worldwide. An additional advantage of spa typing is that adequate typing information is obtained from a single locus, as opposed to MLST, which requires the combination of allelic information from many genes (7) (seven loci for S. pneumoniae). This is because spa typing utilizes a single hypervariable SSR locus as opposed to the several housekeeping genes used in MLST and MLEE. Interpretation of the sequence information from spa sequencing does not require sophisticated algorithms and utilizes readily available sequence analysis software (GCG Wisconsin Package 9.1) that allows the description of strain types by a simple number code and alphabetical repeat designation. Thus, spa typing lends itself to use in a wide range of laboratories as well as the clinical environment.
While MLST provides information on strain lineage that is important for research, this may not be relevant from a clinical point of view, where the main goal is to rapidly identify if an outbreak is occurring. Even so, it is possible that groupings based on spa repeat sequence similarity could reflect chromosomal relationships (Table 6) and therefore allow the strain lineage to be inferred. To validate this hypothesis, we will assess whether spa repeat types (either alone or in combination with other alleles) can be accurately compared to the described S. aureus population genetic framework (unpublished data) characterized by MLEE (15, 16). A temporally and geographically diverse collection of worldwide MRSA isolates and a comparison group of MRSA and methicillin-susceptible S. aureus isolates that represent a wide breadth of genetically diverse S. aureus strains previously analyzed by MLEE will be sequenced. For S. aureus, identification of lineages may be simplified by the clonal nature of MRSA, which could limit the diversity of chromosomal backgrounds seen in clinical isolates (2, 10, 11, 15). In this way, guidelines to define an S. aureus strain type and assign a clonal grouping for an isolate with the use of the protein A repeat region alone may be established (as suggested by Maiden et al. (13), not all MLST genes may be necessary). Thus, in addition to its use for outbreak investigation, spa typing may prove useful as a practical method for describing a natural population of S. aureus organisms. This may aid in the identification of strains that have special virulence properties or drug resistance since in many bacteria these are believed to be nonrandomly distributed along clonal lines (16).
In summary, we have evaluated spa typing by comparing it to several currently utilized techniques for the ability to differentiate well-defined collections of S. aureus strains. Spa typing appears to have significant advantages over many existing techniques in terms of speed, ease of use, ease of interpretation, standardization, and data management and dissemination. As mentioned by Tenover et al. (27), no single typing method appears to be clearly superior in all cases. However, the current ability of spa typing to distinguish both molecularly and epidemiologically linked strains rapidly and easily makes it particularly well suited for the initial screening that may be used to identify and direct epidemiological studies.
ACKNOWLEDGMENTS
We thank Mark Bergman (PHRI) for computer programming, Harriet Marasco (PHRI) for help in preparing the manuscript, and Nicole Ellis (PE) for her technical support.
REFERENCES
- 1.Arbeit R D. Laboratory procedures for epidemiologic analysis. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone, Inc.; 1997. pp. 253–286. [Google Scholar]
- 2.Archer G L, Niemeyer D M, Thanassi J A, Pucci M J. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob Agents Chemother. 1994;38:447–454. doi: 10.1128/aac.38.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigido M D M, Barardi C R, Bonjardin C A, Santos C L, Junqueira M L, Brentani R R. Nucleotide sequence of a variant protein A of Staphylococcus aureus suggests molecular heterogeneity among strains. J Basic Microbiol. 1991;31:337–345. doi: 10.1002/jobm.3620310508. [DOI] [PubMed] [Google Scholar]
- 5.de Lencastre H, Couto I, Santos I, Melo-Cristino J, Torres-Pereira A, Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994;13:64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- 6.Emori T G, Gaynes R P. An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev. 1993;6:428–442. doi: 10.1128/cmr.6.4.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright M C, Spratt B G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 8.Frenay H M, Bunschoten A E, Schouls L M, van Leeuwen W J, Vandenbroucke-Grauls C M, Verhoef J, Mooi F R. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur J Clin Microbiol Infect Dis. 1996;15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- 9.Guss B, Uhlen M, Nilsson B, Lindberg M, Sjoquist J, Sjodahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 10.Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996;2:117–129. doi: 10.3412/jsb.52.417. [DOI] [PubMed] [Google Scholar]
- 11.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 12.Kreiswirth B N, Lutwick S M, Chapnick E K, Gradon J D, Lutwick L I, Sepkowitz D V, Eisner W, Levi M H. Tracing the spread of methicillin-resistant Staphylococcus aureus by Southern blot hybridization using gene-specific probes of mec and Tn554. Microb Drug Resist. 1995;1:307–313. doi: 10.1089/mdr.1995.1.307. [DOI] [PubMed] [Google Scholar]
- 13.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed-field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 15.Musser J M, Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musser J M, Selander R K. Genetic analysis of natural populations of Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 59–68. [Google Scholar]
- 17.Panlilio A L, Culver D H, Gaynes R P, Banerjee S, Henderson T S, Tolson J S, Martone W J. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975–1991. Infect Control Hosp Epidemiol. 1992;13:582–586. doi: 10.1086/646432. [DOI] [PubMed] [Google Scholar]
- 18.Roberts R B, de Lencastre A, Eisner W, Severina E P, Shopsin B, Kreiswirth B N, Tomasz A. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. MRSA Collaborative Study Group. J Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- 19.Roberts R B, Tennenberg A M, Eisner W, Hargrave J, Drusin L M, Yurt R, Kreiswirth B N. Outbreak in a New York City teaching hospital burn center caused by the Iberian epidemic clone of MRSA. Microb Drug Resist. 1998;4:175–183. doi: 10.1089/mdr.1998.4.175. [DOI] [PubMed] [Google Scholar]
- 20.Rubin R J, Harrington C A, Poon A, Dietrich K, Greene J A, Molduddin A. The economic impact of Staphylococcus aureus infection in New York City hospitals. Emerg Infect Dis. 1999;5:9–17. doi: 10.3201/eid0501.990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 22.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Z Y, Enright M C, Wilkinson P, Griffiths D, Spratt B G. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J Clin Microbiol. 1998;36:3514–3519. doi: 10.1128/jcm.36.12.3514-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smeltzer M S, Gillaspy A F, Pratt F L, Thames M D. Comparative evaluation of use of cna, fnbA, fnbB, and hlb for genomic fingerprinting in the epidemiological typing of Staphylococcus aureus. J Clin Microbiol. 1997;35:2444–2449. doi: 10.1128/jcm.35.10.2444-2449.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stockbauer K E, Grigsby D, Pan X, Fu Y X, Mejia L M, Cravioto A, Musser J M. Hypervariability generated by natural selection in an extracellular complement-inhibiting protein of serotype M1 strains of group A Streptococcus. Proc Natl Acad Sci USA. 1998;95:3128–3133. doi: 10.1073/pnas.95.6.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y W, Ellis N M, Hopkins M K, Smith D H, Dodge D E, Persing D H. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol. 1998;36:3674–3679. doi: 10.1128/jcm.36.12.3674-3679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hebert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhlen M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 30.van Belkum A, Kluytmans J, van Leeuwen W, Bax R, Quint W, Peters E, Fluit A, Vandenbroucke-Grauls C, van den Brule A, Koeleman H, Melchers W, Meis J, Elaichouni A, Vaneechoutte M, Moouens F, Maes N, Struelens M, Tenover F, Verbrugh H. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J Clin Microbiol. 1995;33:1537–1547. doi: 10.1128/jcm.33.6.1537-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Belkum A, Eriksen N R, Sijmons M, van Leeuwen W, VandenBergh M, Kluytmans J, Espersen F, Verbrugh H. Are variable repeats in the spa gene suitable targets for epidemiological studies of methicillin-resistant Staphylococcus aureus strains? Eur J Clin Microbiol Infect Dis. 1996;15:768–770. doi: 10.1007/BF01691972. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 32.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Belkum A, van Leeuwen W, Kaufmann M E, Cookson B, Forey F, Etienne J, Goering R, Tenover F, Steward C, O'Brien F, Grubb W, Tassios P, Legakis N, Morvan A, El Solh N, de Ryck R, Struelens M, Salmenlinna S, Vuopio-Varkila J, Kooistra M, Talens A, Witte W, Verbrugh H. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J Clin Microbiol. 1998;36:1653–1659. doi: 10.1128/jcm.36.6.1653-1659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Heijne G, Uhlen M. Homology to region X from staphylococcal protein A is not unique to cell surface proteins. J Theor Biol. 1987;127:373–376. doi: 10.1016/s0022-5193(87)80113-3. [DOI] [PubMed] [Google Scholar]