Abstract
The global rapid emergence of azithromycin/ceftriaxone resistant Neisseria gonorrhoeae threatens current recommend azithromycin/ceftriaxone dual therapy for gonorrhea to ensure effective treatment. Here, we identified the first two N. gonorrhoeae isolates with decreased ceftriaxone susceptibility in Thailand. Among 134 N. gonorrhoeae isolates collected from Thai Red Cross Anonymous Clinic, Bangkok, two isolates (NG-083 and NG-091) from urethral swab in male heterosexual patients had reduced susceptibility to ceftriaxone (MICs of 0.125 mg/L). Both were multidrug resistant and strong biofilm producers with ceftriaxone tolerance (MBEC > 128 mg/L). NG-083 and NG-091 remained susceptible to azithromycin (MIC of 1 mg/L and 0.5 mg/L, respectively). Reduced susceptibility to ceftriaxone was associated with alterations in PBP2, PBP1, PorB, MtrR, and mtrR promoter region. NG-083 belonged to sequence type (ST) 7235 and NG-091 has new allele number of tbpB with new ST. Molecular docking revealed ceftriaxone weakly occupied the active site of mosaic XXXIV penicillin-binding protein 2 variant in both isolates. Molecular epidemiology results revealed that both isolates display similarities with isolates from UK, USA, and The Netherlands. These first two genetically related gonococcal isolates with decreased ceftriaxone susceptibility heralds the threat of treatment failure in Thailand, and importance of careful surveillance.
Subject terms: Microbiology, Molecular biology, Molecular medicine, Infectious diseases, Urogenital diseases, Infection
Introduction
Neisseria gonorrhoeae is the second most common bacterial sexually transmitted infection and results in substantial morbidity1. The World Health Organization (WHO) estimates that in 2016, 86.9 million gonococcal cases occurred among adolescents and adults worldwide2. Most causes are asymptomatic and predominantly in male and female under the age of 251,3. Complication of untreated gonorrhoea can be leads to major morbidities such as pelvic inflammatory disease, ectopic pregnancies, and infertility in women1. It also increases the risk of human immunodeficiency virus (HIV) transmission and acquisition among man who having sex with man and heterosexual individuals1,3. The youngrace or ethnicity, urban residence, multiple, sequential, or concurrent sexual relationships, inconsistent use of condoms, and frequent use of alcohol, illicit substances4 and antibiotic resistance are related to increased transmission of gonorrhoea infections3.
In N. gonorrhoeae, resistance to penicillin, tetracycline, and fluoroquinolones has successively developed, limiting the therapeutic use of these antibiotics1,5,6. Currently, the injectable third generation cephalosporin (ceftriaxone) in combination with a macrolide (azithromycin) are recommended by the World Health Organization (WHO) as the antibiotics of choice to treat most gonococcal infections7 while the Centre for Disease Control and Prevention (CDC) recommend the injectable ceftriaxone2. However, increasing reports of N. gonorrhoeae infections with azithromycin resistance, and ceftriaxone reduced susceptibility or resistantance, threatens this regimen1,8–10. N. gonorrhoeae strains with resistance or reduced susceptibility to ceftriaxone have now been reported in several other countries1,11–19. In N. gonorrhoeae, mutations within the penicillin-binding protein either PBP1 or PBP2 lead to elevated minimum inhibitory concentrations (MICs) via decreased affinity to β-lactams1,14,20–22. An alternative mechanism of β-lactam resistance involves overexpression of the MtrCDE efflux pump or reduced outer membrane permeability by mutations of the mtrR and por genes1,21,22. Mosaic-like structures in the penA gene which encodes PBP2 are known to be associated with high-level resistance to penicillin and reduced susceptibility to ceftriaxone, cefipime and other cephems1,11,14–16. Additionally, PBP2 the amino acid substitutions G542S, P551S, and P551L in nonmosaic penA alleles have been reported in isolates with reduced susceptibility to extended spectrum cephalosporins1,11,14–16.
No confirmed reports of gonorrhoea resistance or reduced susceptibility to ceftriaxone have been documented in Thailand to date. In current gonococcal treatment in Thailand, azithromycin is added to ceftriaxone in dual therapy as a method of limiting the selection of ceftriaxone resistant mutants. Thailand is a major risk area for gonorrhea because it is a key destination for the sex tourism industry, and where antibiotic resistant gonorrhea can appear and spread easily and quickly across the region. Also in Thailand, asymptomatic gonorrhea prevalence was found to be higher among HIV-infected individuals who are less likely to have the infection diagnosed and treated effectively23,24. This may compromise early diagnosis and treatment of multi drug resistance N. gonorrhoeae infection.
In 2016, the Antimicrobial Resistance and Stewardship Research Unit, at the Faculty of Medicine, Chulalongkorn University in collaboration with the Thai Red Cross AIDS Research Centre began investigating a possible N. gonorrhoeae infection with resistance or reduce susceptibility to ceftriaxone in Thailand. Here, we first report two nearly identical isolates from heterosexual men with gonococcal infections, each sharing the similar mutation in the penA alleles, that demonstrate decreased susceptibility to ceftriaxone and remain susceptible to azithromycin.
Results
N. gonorrhoeae clinical isolates from urethral swabs in male patients had reduced susceptibility to ceftriaxone
Among the 134 N. gonorrhoeae clinical isolates collected during 2016–2019, two isolates (NG-083 and NG-091) which were isolated in 2017 from urethral swabs in male heterosexual patients at Thai Red Cross AIDS Research Centre, Anonymous Clinic, had reduced susceptibility to ceftriaxone (agar dilution MICs of 0.125 mg/L). Both isolates were resistant to penicillin G, tetracycline, ciprofloxacin, and gentamicin, while remaining susceptible to azithromycin (Table 1). The patients were a 30 and 32-year-old long term foreign resident men (South African and Australian) in Thailand, symptomatic with dysuria, or penile discharge at the time of specimen collection, and negative for HIV. Both were isolated cases and no relation to each other epidemiologically. One patient develops symptoms one week after unprotected oral sex with a female, while the second patients origin of infection was not disclosed. Both were treated for gonorrhea with Ceftriaxone 250 mg, and Azithromycin 250 mg as per current CDC guidelines5. Patients di not return to the clinical follow-up and therefore unable to be verified as having been cured or not.
Table 1.
Patients summery and antimicrobial susceptibility profile against the N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone (n = 2), Thailand, 2016–2018.
| Patient Number |
Isolate Number |
Demographics | Sexual Preference | Healthcare Setting | Symptoms | Treatment | HIV Status | MIC (mg/L) | Biofilm Category | MBEC (mg/L) |
|---|---|---|---|---|---|---|---|---|---|---|
| 83 | NG-083 | 30-years-old South African Male | Heterosexual | Anonymous Clinic | Dysuria and Penile Discharge | Ceftriaxone 250 mg IM, + Azithromycin 250 mg orally twice daily × 14 d | Negative |
Penicillin G 2 Tetracycline 4 Ciprofloxacin 4 Azithromycin 1 Cefixime 0.125 Ceftriaxone 0.125 Gentamicin 16 Fosfomycin 16 Ertapenem 1 |
Strong |
Ceftriaxone 128 Azithromycin 256 |
| 91 | NG-091 | 32-years-old Australian Male | Heterosexual | Anonymous Clinic | Penile Discharge | Ceftriaxone 250 mg IM, + Azithromycin 250 mg orally twice daily × 14 d | Negative |
Penicillin G 2 Tetracycline 4 Ciprofloxacin 4 Azithromycin 0.5 Cefixime 0.125 Ceftriaxone 0.125 Gentamicin. 32 Fosfomycin 16 Ertapenem. 1 |
Strong |
Ceftriaxone 128 Azithromycin 256 |
HIV human immunodeficiency viruses, MIC minimum inhibitory concentration, IM intramuscular, MBEC minimal biofilm eradication concentrations were categorized as responsive reaching about 90% of the total non-viable cells within a given antibiotic concentration range.
N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone are associated with alterations in PBP2, PBP1, PorB, MtrR, and mtrR promoter region
The isolates NG-083 and NG-091 with reduced susceptibility to ceftriaxone harbor specific ceftriaxone resistance patterns (Table 2). NG-083 and NG-091 had an L421P substitution in PBP1, the mosaic PBP2 patterns XXXIV (Fig. 1) (Supplementary Figs. S2 and S3), an adenine deletion in the 13-bp inverted repeat sequence of the mtrR promoter region, an H105Y substitution in the MtrR repressor, and G120K and A121N substitutions in PorB porin. Mosaic XXXIV PBP2 variants contain up to 52 amino acid alterations compared with the wild-type PBP2 (Table 3). Figure 2 summarises the mosaic XXXIV PBP2 variant showing the location of all amino acid alterations drawn in Pymol using the crystal structure of a soluble form of N. gonorrhoeae wild-type PBP2 (extracted from rcsb.org/PDB: 3EQU).
Table 2.
Mutations in resistance determinants and NG-MAST sequence types of the N. gonorrhoeae clinical isolates with reduced susceptibility to ceftriaxone (n = 2), Thailand, 2016–2018.
| Patient number | Isolate number | Sexual Preference | Culture Site | Resistance Determinants | NG-MAST | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PBP2 | PBP1 | mtrR promoter | MtrR | PorB | por | tbpB | ST | ||||
| 83 | NG-083* | Male Heterosexual | Urethra | XXXIV | L421P | A del | H105Y | G120K, A121N | 908 | 1180 | ST7235 |
| 91 | NG-091* | Male Heterosexual | Urethra | XXXIV | L421P | A del | H105Y | G120K, A121N | 1914 | New allele | New ST |
MIC minimum inhibitory concentration, WT wild type, New new mosaic pattern, PBP2 penicillin binding protein 2, PBP1 penicillin binding protein 1, mtrR, mtrR promoter, MtrR MtrR repressor, PorB PorB porin, NG-MAST N. gonorrhoeae multi-antigen sequence typing, NG-STAR Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance ST, sequence type, A del adenine deletion.
*N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone.
Figure 1.
The amino acid sequence alignments of different PBP2 patterns of 9 N. gonorrhoeae isolates including nonmosaic PBP2 pattern II, VII, XIV, XIX, XVI, XVIII, XXXXIV, mosaic XXXIV, and new mosaic pattern (GC-013) were compared with the wild type PBP2 of N. gonorrhoeae strain LM306 (GenBank accession no. AAA25463).
Table 3.
The amino acid residues in wild type N. gonorrhoeae Penicillin- binding proteins (PBPs) and mutative in Penicillin- binding proteins (mPBPs) in N. gonorrhoeae clinical isolates NG-083 and NG-091 with reduced susceptibility to ceftriaxone (n = 2), Thailand, 2016–2018.
| Residue number | Original PDB:3EQU | Mutative PDB: 3EQU | Residue number | Original PDB:3EQU | Mutative PDB: 3EQU |
|---|---|---|---|---|---|
| 101 | ASP | GLU | 374 | AGR | MET |
| 160 | VLA | ALA | 376 | GLY | THR |
| 173 | ASN | SER | 377 | ALA | PRO |
| 201 | TYR | HIS | 378 | GLU | LYS |
| 202 | GLY | ALA | 386 | GLU | ASP |
| 203 | GLU | GLY | 389 | ILE | VAL |
| 204 | ASP | GLU | 406 | ASN | SER |
| 214 | GLN | GLU | 412 | ARG | GLN |
| 279 | ALA | VAL | 413 | PRO | LYS |
| 285 | ASP | GLU | 438 | ALA | VAL |
| 288 | ARG | LYS | 444 | VAL | GLU |
| 289 | ARG | GLN | 448 | LEU | VAL |
| 312 | ILE | MET | 458 | GLN | LYS |
| 316 | VAL | THR | 462 | ILE | VAL |
| 323 | ALA | SER | 463 | PHE | ILE |
| 326 | THR | VAL | 465 | GLU | ALA |
| 328 | LEU | ALA | 469 | ARG | LYS |
| 329 | ASN | THR | 470 | GLU | LYS |
| 330 | GLU | ASP | 473 | ASN | GLU |
| 331 | ARG | THR | 481 | PRO | ALA |
| 332 | LEU | PHE | 505 | PHE | LEU |
| 335 | GLN | LEU | 511 | ALA | VAL |
| 341 | PRO | SER | 513 | ASN | TRY |
| 342 | SER | ALA | 541 | HIS | ASN |
| 343 | PRO | THR | 545 | GLY | SER |
| 345 | ARG | GLN | |||
| 353 | SER | THR |
Figure 2.
The structure of mosaic XXXIV PBP2 variant of N. gonorrhoeae clinical isolate (a) showing the location of important mutations and the active site sequence motifs, enlarge of (b) C terminal domain (c) N terminal domain (d) Site of phosphorylation. Mutations were modeled by PyMOL [DeLano, 2002] (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.).
N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone belonged to different sequence types (STs)
Isolate NG-083 belonged to ST7235 (Table 2) and NG-091 had a new allele in tbpB and new ST according to the NG-MAST. However, both NG-083 and NG-091 have same ST-90 sequence types according to the NG-STAR (Table 4).
Table 4.
Mutations in resistance determinants and NG-STAR sequence types of N. gonorrhoeae clinical isolates with reduced susceptibility to ceftriaxone (n = 2), Thailand, 2016–2018.
| Patient number | Isolate number | Sexual preference | Culture site | Locus | Allele type | Amino acid alteration | NG-STAR Type |
|---|---|---|---|---|---|---|---|
| 83,91 | NG-083, NG-091 | Male Heterosexual | Urethra | penA | 34.001 | penA Type XXXIV Mosaic with N513Y | 90 |
| mtrR | 1 | -35A Del with H105Y | |||||
| porB | 11 | G120K, A121N | |||||
| ponA | 1 | L421P | |||||
| gyrA | 1 | S91F, D95G | |||||
| parC | 3 | S87R | |||||
| 23S rRNA | 100 | Wild Type |
MIC minimum inhibitory concentration, WT wild type, New new mosaic pattern, PBP2 penicillin binding protein 2, PBP1 penicillin binding protein 1, mtrR mtrR promoter, MtrR MtrR repressor, PorB PorB porin, NG-STAR Neisseria gonorrhoeae sequence typing for antimicrobial resistance ST, sequence type, A Del adenine deletion.
*N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone.
Ceftriaxone weakly occupies the active site of mosaic XXXIV penicillin-binding protein 2 (PBP2) variant in N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone
Ceftriaxone (binding energy: − 6.64 kcl/mol, Ki: 13.6 μM) has formed three hydrogen bonds with three different amino acids (Fig. 3). The NH of Thr500 has formed an H-bond with 1,2,4-triazine ring of ceftriaxone at a distance of 1.828 Å, within the β3–β4 loop of standard PBP2 binding site. The OH of Tyr350 has formed a hydrogen bond with NH group near the cephalosporin ring (O–HN) at distance of 2.219 Å. The C=O group of carboxylic acid has formed an acyl-enzyme complex by making an H-bond with OH of Tyr 544 with a distance of 2.019 Å and completely occupied the catalytic triad located at the N-terminal end of helix α11. In case of mosaic XXXIV PBP2 variant, the binding energy of ceftriaxone was found to be -5.92 kcl/mol with 45.63 μM as Ki value, which is far less than the standard PBP2. The ceftriaxone has formed two hydrogen bonds and one π–π interaction with the altered protein (Fig. 4). The OH phenol group of tyr350 has formed an h-bond with ceftriaxone (OH—- -O) at a distance of 1.894 Å. The C=O group near the cephalosporin ring has formed a H-bond with the NH group of Lys313 at a distance of 1.894 Å, which confirms the binding in the active site. The π-µ interaction was observed between 1,2,4-triazine ring of ceftriaxone and the phenolic ring of Tyr422 at a distance of 14.323 Å, which helps to understand the rotation of the molecule within the active site.
Figure 3.
Molecular docking of ceftriaxone in the active site of a) wild type PBP2 and b) mosaic XXXIV PBP2 variant. The standard macromolecule was extracted from rcsb.org/ (PDB: 3EQU) and the mutations were modeled by PyMOL [DeLano, 2002] (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.).
Figure 4.
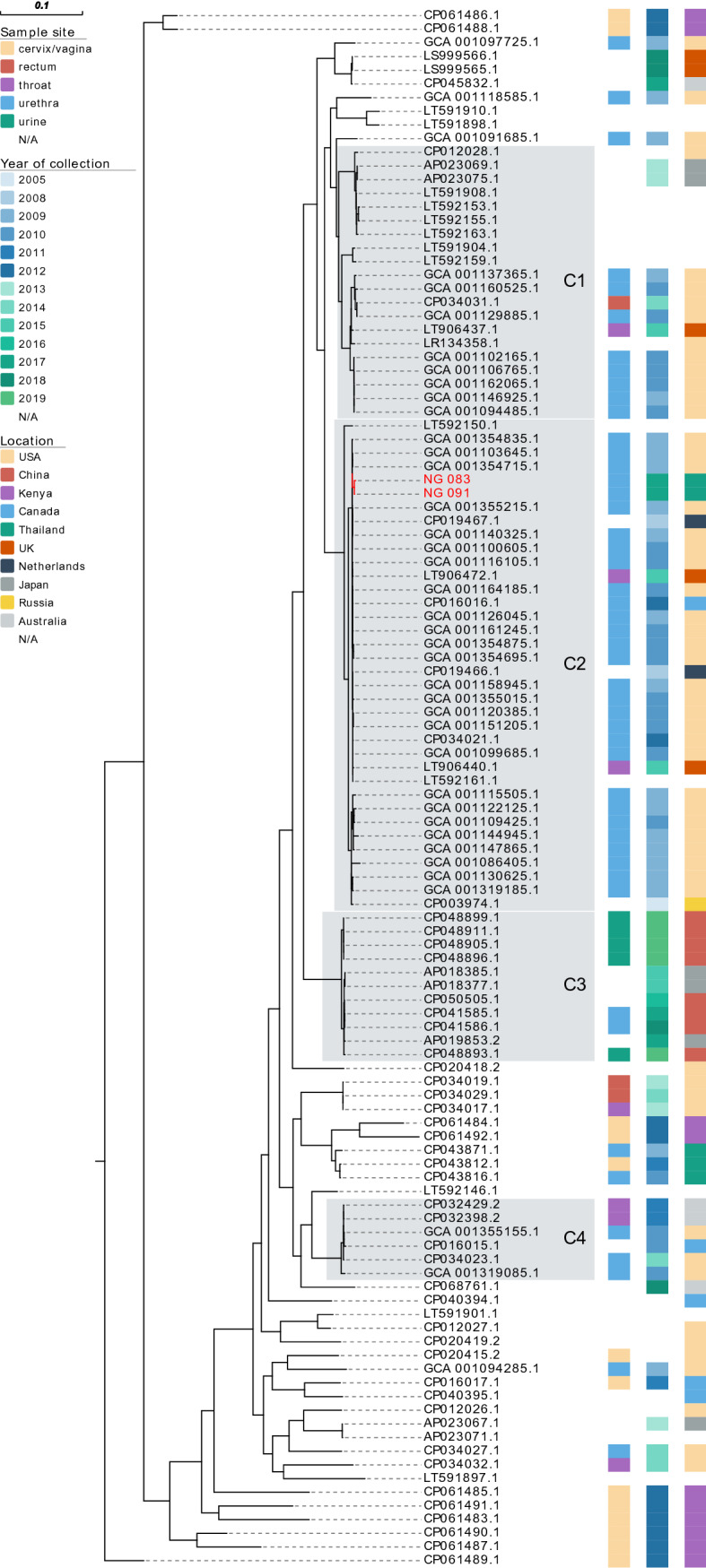
Phylogenetic analyses based on core genome from draft genomes of Neisseria gonorrhoeae NG-083 and NG-091 clinical isolated in Thailand and from varying N. gonorrhoeae WGS investigations conducted elsewhere (available on NCBI database) were determined for the number of single nucleotide polymorphisms (SNPs) by Core-Genome SNP Analysis. C1-C4 represent major clusters.
N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone are closely related with urethra isolates from male patients from Las Vegas
A rooted-ML phylogenetic tree demonstrated four major cluster (C1–C4) (Fig. 4 and Supplementary Fig. S4), each of which contained at least six isolates, using 3% substitutions per site as a cluster genetic distance (8). Cluster C2, the largest cluster, involved 36 N. gonorrhoeae isolates from the USA, Netherlands, the UK, and Canada, and included two ceftriaxone-reduced susceptible isolates found in our study (NG-083 and NG-091) from two foreign male patients in Thailand in 2017. The isolates are closely related with two N. gonorrhoeae isolated from the urethra of male patients from Las Vegas in the USA in 2009 under BioProject accession no PRJEB2999, which differed by 2 core SNPs (accession no GCA_001103645.1 and GCA_001354715.1).
Ceftriaxone plus azithromycin combination display no synergistic but bactericidal activity against N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone
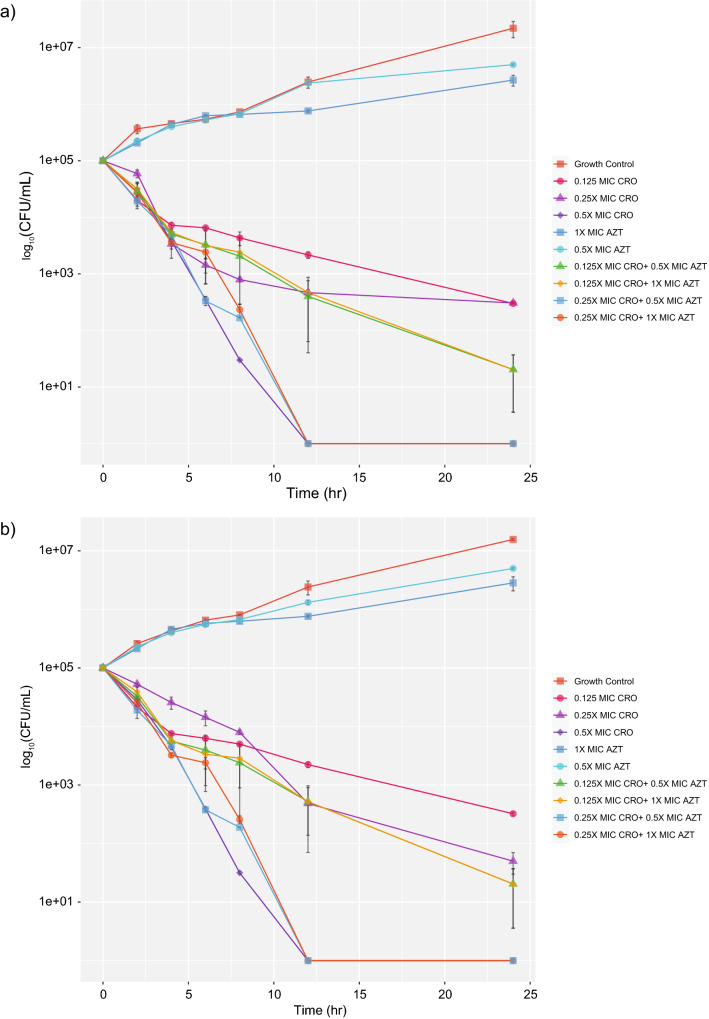
Synergistic and antagonistic effects were not found in all antibiotic combinations against both isolates (Table 5). Additive effects were found with ceftriaxone plus azithromycin, fosfomycin, or gentamicin. Indifferent was found in ceftriaxone plus ertapenem. At 0.5X MIC of ceftriaxone, bacterial growth was rapidly inhibited within 8 to 12 h in both isolates, but the concentration at 0.125X and 0.25X MIC was unable to inhibit bacterial growth within 24 h (Fig. 5). However, 0.125X MIC of ceftriaxone plus 0.5X or 1X MIC of azithromycin, and 0.25X MIC of ceftriaxone plus 0.5X or 1X MIC of azithromycin showed effective killing against NG-083 and NG-091.
Table 5.
Summary of antibiotic combinations against N. gonorrhoeae clinical isolates NG-083 and NG-091 with reduced susceptibility to ceftriaxone (n = 2), Thailand, 2016–2018.
| Isolate no | Resistance Determinants | NG-MAST | FICI index | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PBP2 pattern | PBP1 | mtrR promoter | MtrR | PorB porin | CRO:AZT | CRO:FOS | CRO:GEN | CRO:ETP | ||
| NG-083 | XXXIV | L421P | A del | H105Y | G120K, A121N | ST7235 | 0.75 (A) | 1.00 (A) | 1.00 (A) | 1.50 (I) |
| NG-091 | XXXIV | L421P | A del | H105Y | G120K, A121N | New ST | 0.74 (A) | 1.00 (A) | 1.00 (A) | 1.25 (I) |
AZT azithromycin, CRO ceftriaxone, ETP ertapenem, FOS fosfomycin, GEN Gentamicin, A additive, I indifferent, FICI Fractional Inhibitory Concentration Index.
Figure 5.
Time-kill curve of ceftriaxone plus azithromycin against N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone (a) NG-083 and (b) NG-091.
N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone produce strong biofilms with ceftriaxone tolerance
The isolates NG-083 biofilm had significantly more biomass than the NG-091 biofilm (P = 0.004) (Fig. 6). Both isolates were strong biofilm producers with an atypical ellipse-shaped structure or clumps, and well tolerated to a ceftriaxone concentration up to 128 mg/L (Fig. 7). The analysis showed that the live/dead bacterial ratio of NG-083 and NG-091 biofilms treated with ceftriaxone MIC (0.125 mg/L) concentration was not different in comparison with non-treated group (P > 0.05) (Fig. 7). Both isolates displayed minimal biofilm eradication concentration for ceftriaxone around 128 mg/L concentration (Table 1).
Figure 6.
Confocal imaging analysis (3D and cross sectional) of biofilm structure of N. gonorrhoeae isolates (a,b) NG-083 and (c,d) NG-091treated with MIC concentration (0.125 mg/L) of ceftriaxone.
Figure 7.
Effects of different concentration of ceftriaxone on N. gonorrhoeae isolates (n = 2). Isolate NG-083; (a) in vitro biomass (b) in vitro Live/Dead cell ratio (c) in vitro bio-volume inhibition and isolate NG-091; (d) in vitro biomass (e) in vitro bio-volume inhibition (f) in vitro Live/Dead cell ratio.
Discussion
We identified a pair of gonococcal isolates with decreased susceptibility to ceftriaxone, ciprofloxacin, tetracycline, penicillin G, gentamicin, and ertapenem, while remaining susceptible to azithromycin. Both isolates appeared to be unrelated, with for example two different sequence types according to the NG-MAST but same sequence type according to Ng-STAR. The genome sequencing analysis revealed alterations in PBP2, PBP1, PorB, MtrR, and mtrR promoter region in both isolates. Molecular docking studies suggested ceftriaxone weakly occupies the active site of mosaic XXXIV PBP2 variant, which may explain the decreased ceftriaxone activity. Interestingly, both isolates with reduced susceptibility to ceftriaxone demonstrated strong biofilm production that was associated with ceftriaxone tolerance at concentrations higher than the MIC. Molecular epidemiology results revealed that both isolates display similarities with isolates from the UK, USA, and the Netherlands. The most effective combination was ceftriaxone plus azithromycin which showed bactericidal activity for both isolates.
This Thai isolates is concerning because two isolates from heterosexual male patients (NG-083 and NG-091) demonstrated decreased susceptibility to ceftriaxone with mosaic PBP2 pattern XXXIV, similar to isolates previously reported from USA25, Austria26, Japan27, France17, Spain28, and Canada29. Moreover, these two isolates are genetically closely related to two N. gonorrhoeae isolated from the urethra of male patients from Las Vegas in USA in 2009, which also exhibited reduced susceptibility to cefixime, with most of the isolates having mosaic penA allele XXXIV30. As given the 8 years between specimen collection of the Las Vegas isolates and two isolates in our study, it’s difficult to relate an epidemiological connection between these two and the Las Vegas isolates. However, Thailand is a popular getaway for sex tourism3, there is the possibility that these two isolates (NG-083 and NG-091) may have transmitted through travelers. However, because we did not have samples from other sexual partners of these two men to provide broader context, is difficult to established clear epidemiological relations.
To our knowledge, no isolates have previously been reported in Thailand with decreased ceftriaxone susceptibility. The data from the National Antimicrobial Resistance Surveillance Center, Thailand (NARST)31, and other studies from Thailand32 showed that 100% of N. gonorrhoeae isolates were susceptible to cefixime and ceftriaxone. A report from the United Kingdom documented a gonorrhea treatment failure with the isolate exhibiting a ceftriaxone MIC = 0.25 µg/mL and azithromycin MIC = 1 µg/mL12,33. In comparison, our isolates demonstrate similar susceptibility to azithromycin, while their ceftriaxone MICs were only a single dilution lower than the ceftriaxone MIC from the UK isolate, suggesting that if the strains identified in Thailand develop higher ceftriaxone MICs, they may be capable of causing treatment failure. It is uncertain whether current gonococcal treatment of ceftriaxone plus azithromycin may have contributed to the development of higher ceftriaxone MICs and widespread transmission of such strains34. In addition, the improper use or misuse of antibiotics, and over-the-counter drug usage in Thailand, may have contributed to the development of higher ceftriaxone MICs35. It is possible that the persistence of such isolates within biofilm, seen with infections involving higher MICs, reflected a slower killing of N. gonorrhoeae and may severely complicate gonorrhea treatment.
The threat of multidrug-resistant gonorrhea leading to treatment failure in Thailand with the current recommended dual therapy remains present. Our findings emphasized the value of the CDC recommendations for laboratories to maintain culture-based methods to detect antimicrobial-resistant N. gonorrhoeae, particularly for patients with possible treatment failure. In addition, we also emphasized the potential value of genomic monitoring to detect antimicrobial-resistant N. gonorrhoeae. It is important that clinicians be on high alert so that treatment failures can be identified and reported promptly to the local department of health. Rapid detection and effective treatment may prevent sequelae, allow partners to be identified and treated in a timely manner, and prevent or slow further transmission of resistant strains. Furthermore, given the study findings, we advised continued practice of dual therapy with ceftriaxone 250 mg plus azithromycin 1gm in Thailand. However, it is necessary to have new strategies such as resistance guided therapy for N. gonorrhoeae infections and contact tracing to identify the origin of the gonococcal infections as preventive measures. Finally, there are urgent need for development of new or combination therapies to tackle the multidrug resistant N. gonorrhoeae infections.
The strengths of this study are that all infections were diagnosed by culture, which has 100% specificity for gonorrhea.
Methods
N. gonorrhoeae clinical isolates
134 isolates collected from urethral swabs of patients with positive N. gonorrhoeae infections at Thai Red Cross Anonymous Clinic, Thai Red Cross AIDS Research Centre, and King Chulalongkorn Memorial Hospital, Bangkok, Thailand, from patients with gonococcal infections during 2016–2019. For culture preservation, all isolates were grown on TM medium (GC agar base supplemented with 1% haemoglobin, 1% IsoVitaleX, and vancomycin, colistin sulfate, and nystatin selective supplement (Oxoid, United Kingdom). All N. gonorrhoeae isolates were preserved at − 80 °C. N. gonorrhoeae ATCC 49226 reference strain was used for quality control of all phenotypic and molecular characterizations.
Phenotypic antimicrobial susceptibility testing
The isolates obtained from patients were analysed using both broth micro-dilution and agar plate dilution methods (Supplementary methods). MIC interpretive criteria were in accordance with the Clinical and Laboratory Standards Institute (CLSI)36or European Committee on Antimicrobial Susceptibility Testing (EUCAST)37 when available. In the present study in vitro decreased susceptibility to ceftriaxone was defined as having an MIC of > 0.064–0.125 mg/L.
Molecular epidemiological typing
The presence of carA and orf1 genes were detected by specific PCR primers (Supplementary methods Table S1) (BioDesign, Thailand) to confirm N. gonorrhoeae identification as described previously38,39.
Detection and characterization of ceftriaxone resistance mechanisms
The ceftriaxone resistance mechanisms in N. gonorrhoeae isolates were investigated by penA, mtrR, ponA, and porB genes using PCR and automated DNA sequencing (1st BASE Inc, Malaysia) (Supplementary methods Table S3). The PCR products of genes were purified using HiYieldTMGel/PCR fragments extraction kit as described by the manufacturer (RBC Bioscience, Taiwan). Sequencing was conducted using BigDye terminator cycling conditions.
Clonal studies of N. gonorrhoeae
The clonality of N. gonorrhoeae was initially determined by NG-MAST (http://www.ng-mast.net/) as previously described40. The internal fragments of 2 highly polymorphic antigen-encoding loci including outer membrane proteins (por) and transferrin binding protein unit B (tbpB)40 were amplified, purified (HiYieldTMGel/PCR) and sequenced (Supplementary methods Table S4) as previously described40. However, because NG-MAST no longer assigns new STs, the two N. gonorrhoeae with reduced susceptibility to ceftriaxone were further analyzed with NG-STAR. Sequence of 7 genes (penA, mtrR, porB, ponA, gyrA, parC and 23S rRNA) were extract from genome sequence of NG-083 and NG-091 isolates, then the sequence were analyzed using NG-STAR database (https://ngstar.canada.ca/alleles/query?lang=en). Preparation of sequencing reactions were performed by the 1st BASE Inc, Malaysia, as described above.
Whole-genome sequencing and genome assembly
The genomes of two N. gonorrhoeae with reduced susceptibility to ceftriaxone were sequenced using Illumina sequencing platform (2 × 150 paired-end, Illumina, Inc., USA). The genomic DNA was extracted with the PureLink PCR Purification kit and quantified in a Qubit Fluorometer (Invitrogen), following the manufacturer's recommendations. Libraries were sequenced on a NovaSeq 6000. The low-quality and adapter bases were trimmed using Skewer v0.2.2 (https://github.com/relipmoc/skewer). The quality of pre- and postprocessed reads was assessed with the FastQC tool, v0.11.8 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The resulting high-quality reads were assembled de novo using Unicycler v0.4.8 (PMID: 28594827). The quality of genome sequences was checked using QUAST v5.0.2 (PMID: 23422339) and annotated by DFAST v1.2.6 (PMID: 29106469).
Phylogenetic analyses
The core genome from draft genomes of N. gonorrhoeae NG_83 and NG_91 clinical isolated in Thailand and from other N. gonorrhoeae WGS investigations conducted elsewhere (available on NCBI database) were determined for the number of single nucleotide polymorphisms (SNPs) by Core-Genome SNP Analysis. A reference was randomly selected among the genome sequences to generate a core genome alignment and a phylogenetic tree was constructed using a core SNP alignment. Draft genomes of N. gonorrhoeae were aligned following the detection and filtration of recombinant regions using Parsnp v1.241 and Gubbins v2.4.142. Maximum-likelihood (ML) trees were generated by RAxML v8.2.1243 using ASC_GTRGAMMA model of rate heterogeneity with the Lewis correction for ascertainment bias44,45. Best-scoring ML tree was visualized and annotated as a phylogenetic tree using FigTree v1.4.4 and Evolview v246–48 (Supplementary File GC_114RV2parsnp).
Sequence analysis
The nucleotide sequences and deduced amino acid sequences were analyzed with the online software available at the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/BLAST), and ExPASy (www.expasy.org). Multiple sequence alignment was analyzed by Multalin (http://multalin.toulouse.inra.fr/multalin). The nucleotide and deduced amino acid sequences identified in this study including penA, ponA, mtrR, and porB genes were compared with the corresponding sequences in the genome sequenced of N. gonorrhoeae reference strain FA1090 (GenBank accession number AE004969). The nucleotide sequences of 2 highly polymorphic antigen-encoding loci were analyzed and uploaded onto the NG-MAST database (http://www.ng-mast.net) to obtain the allele number and the sequence type (ST). Furthermore, two N. gonorrhoeae with reduced susceptibility to ceftriaxone were analyzed with NG-STAR (https://ngstar.canada.ca).
Molecular docking
Molecular docking studies were carried out to understand binding mode analysis and orientation of ceftriaxone in the active site of PBP2 and compared with mosaic XXXIV PBP2 variant. The standard macromolecule was extracted from rcsb.org/ (PDB: 3EQU) and the mutations were modeled by PyMOL [DeLano, 2002] (The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.) (Supplementary methods).
Antibiotic synergy analysis using microdilution checkerboard assay
The synergistic activities of antibiotics were screened against 2 strains of N. gonorrhoeae isolates with reduced susceptibility to ceftriaxone using checkerboard method (Supplementary methods).
Time kill assay
Two strains of N. gonorrhoeae with reduced susceptibility to ceftriaxone that showed the best synergistic activity by checkerboard method was confirmed using time-kill assay. All experiments were performed at least three times (Supplementary methods).
Biofilm formation, quantification and classification
Biofilm formation in a 96-well-microtitre-plate format was performed as described previously49–51 (Supplementary methods). Two methods52,53 were used to quantify and classify the biofilm by Crystal Violet staining and and confocal laser scanning microscopy, performed in triplicate, and repeated three times.
Minimal biofilm eradication concentrations (MBEC)
Minimal biofilm eradication concentrations (MBEC) were established as described previously49–51 by adding the serially diluted antibiotics to mature biofilms and incubating at 37 °C for 24 h and then staining with PrestoBlue (Thermo Fisher Scientific). All experiments were performed in triplicate and repeated three times.
Statistical analyses
Fisher's exact test (two-tailed) was used to verify the association between mutations in resistance determinants and penicillin resistance in N. gonorrhoeae isolates by using the R statistical package. Biofilm experiments were analyzed by confocal laser scanning microscope (biomass, Live/Dead ratio and biovolume inhibition). MATLAB-based tool PHLIP (without connected volume filtration) were used to calculate descriptive parameters of biofilms (including biovolume, substratum coverage, area-to-volume ratio, spatial spreading and 3D colocalization) from the integrated total of each individual slice of a thresholded z-stack as described previously49,50,54. The calculation of the different proportions of green (live bacteria) as well as red and yellow/colocalized (dead bacteria) biovolumes from the analyzed stacks were using the 'colocalization in 3D' value and the parameters 'red', 'green', and 'total biovolume' (in μm3) generated by the PHLIP software as described previously48,49,53. A biofilm was considered affected by an antibiotic within the given concentration range when there is a constant increase in the red + yellow (RY) biovolume fraction within the given antibiotic concentration range and this fraction is at least 80% of the total biovolume. The data were compared by either unpaired two-tailed Student’s t-test or unpaired two-tailed Mann–Whitney’s U test. Statistical significance was accepted at p < 0.05, p < 0.01, p < 0.001, and p < 0.0001.
Nucleotide sequence accession numbers
Genome assemblies of NG-083 and NG-091 isolates were submitted to the NCBI Database under BioSamples of SAMN20286673 and SAMN20286674. These two BioSamples were submitted under BioProject PRJNA747638 (https://www.ncbi.nlm.nih.gov/bioproject/747638), which the author sequence were the same with the author manuscript.
Ethics approval
The study protocol was approved (IRB No. 396/60, COA No. 715/2017) by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand, and was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments and comparable ethical standards.
Informed consent
For this retrospective study of anonymous clinical isolates, the requirement for informed consent from patients was waived by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No. 396/60, COA No. 715/2017).
Supplementary Information
Acknowledgements
We thank the staff of the bacteriology division, Department of Microbiology at King Chulalongkorn Memorial Hospital, and Thai Red Cross Anonymous Clinic, Thai Red Cross AIDS Research Centre, Bangkok, Thailand for providing the N. gonorrhoeae clinical isolates.
Author contributions
N.K.: investigation, data curation, formal analysis. D.L.W.: conception, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript. S.L.-in: data acquisition, curation, formal analysis, methodology, validation, critical review and editing of the manuscript. P.H.: formal analysis, supervision, methodology, validation, critical review and editing of the manuscript. C.H.: formal analysis, supervision, critical review and editing of the manuscript. V.N.B.: data curation, formal analysis, methodology, validation, critical review and editing of the manuscript. P.J.: Bioinformatics analysis, methodology, validation. T.W.: Bioinformatics analysis, methodology, validation. N.T.: clinical sample collection, supervision. S.J.K.: Supervision, and advisory. S.A.: formal analysis, supervision, methodology, validation, critical review and editing of the manuscript. P.P.: supervision, critical review and editing of the manuscript. A.M.S.S.: Biofilm experiments data curation, formal analysis. T.S.: data curation, formal analysis, methodology, validation. N.C.: confocal laser scanning microscopy data acquisition, curation, methodology, validation. M.A.: supervision, critical review and editing of the manuscript. P.G.H.: supervision, critical review and editing of the manuscript. T.C.: conception, funding acquisition, supervision, critical review and editing of the manuscript.
Funding
Naris Kueakulpattana was supported by a grant from the 90th Year Anniversary Ratchadapiseksompotch Endowment Fund from the Faculty of Medicine and Graduate School, Chulalongkorn University, Bangkok, Thailand (batch No. 40(3/2561)). Dhammika Leshan Wannigama was supported by Second Century Fund (C2F) Fellowship, Chulalongkorn University, Bangkok, Thailand, and the University of Western Australia (Overseas Research Experience Fellowship). The sponsor(s) had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Data availability
Data generated and analysed during this study are included in this published article and its Supplemental information file. Additional clinical details available upon reasonable request from corresponding author TC.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Naris Kueakulpattana, Dhammika Leshan Wannigama, Sirirat Luk-in, Parichart Hongsing, Cameron Hurst, Paul G. Higgins.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-00675-y.
References
- 1.Unemo M, et al. Gonorrhoea. Nat. Rev. Dis. Primers. 2019;5:79. doi: 10.1038/s41572-019-0128-6. [DOI] [PubMed] [Google Scholar]
- 2.Cyr SS, Barbee L, Workowski KA, Bachmann LH, Pham C, Schlanger K, Torrone E, Weinstock H, Kersh EN, Thorpe P. Update to CDC’s treatment guidelines for gonococcal infection, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1911–1916. doi: 10.15585/mmwr.mm6950a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkcaldy RD, Weston E, Segurado AC, Hughes G. Epidemiology of gonorrhoea: A global perspective. Sex. Health. 2019;16:401–411. doi: 10.1071/SH19061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer CB, et al. Sociodemographic markers and behavioral correlates of sexually transmitted infections in a nonclinical sample of adolescent and young adult women. J. Infect. Dis. 2006;194:307–315. doi: 10.1086/506328. [DOI] [PubMed] [Google Scholar]
- 5.St Cyr S, et al. Update to CDC's treatment guidelines for gonococcal infection, 2020. MMWR Morb. Mortal Wkly. Rep. 2020;69:1911–1916. doi: 10.15585/mmwr.mm6950a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehn BM. Updated recommendations for gonorrhea treatment. JAMA. 2021;325:523–523. doi: 10.1001/jama.2021.0241. [DOI] [PubMed] [Google Scholar]
- 7.WHO . WHO Guidelines for the Treatment of Neisseria gonorrhoeae. WHO; 2016. [PubMed] [Google Scholar]
- 8.Cristillo AD, et al. Antimicrobial resistance in Neisseria gonorrhoeae: Proceedings of the STAR sexually transmitted infection—clinical trial group programmatic meeting. Sex. Transm. Dis. 2019;46:e18–e25. doi: 10.1097/OLQ.0000000000000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong FYS, et al. Treatment efficacy for pharyngeal Neisseria gonorrhoeae: A systematic review and meta-analysis of randomized controlled trials. J. Antimicrob. Chemother. 2020;75:3109–3119. doi: 10.1093/jac/dkaa300. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y-P, et al. Susceptibility of Neisseria gonorrhoeae to azithromycin and ceftriaxone in China: A retrospective study of national surveillance data from 2013 to 2016. PLoS Med. 2018;15:e1002499. doi: 10.1371/journal.pmed.1002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aniskevich A, et al. Antimicrobial resistance in Neisseria gonorrhoeae isolates and gonorrhoea treatment in the Republic of Belarus, Eastern Europe, 2009–2019. BMC Infect. Dis. 2021;21:520. doi: 10.1186/s12879-021-06184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyre DW, et al. Gonorrhoea treatment failure caused by a Neisseria gonorrhoeae strain with combined ceftriaxone and high-level azithromycin resistance, England, February 2018. Eurosurveillance. 2018;23:1800323. doi: 10.2807/1560-7917.ES.2018.23.27.1800323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianecini R, Oviedo C, Stafforini G, Galarza P. Neisseria gonorrhoeae resistant to ceftriaxone and cefixime, Argentina. Emerg Infect. Dis. 2016;22:1139–1141. doi: 10.3201/eid2206.152091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz AR, et al. Cluster of Neisseria gonorrhoeae isolates with high-level azithromycin resistance and decreased ceftriaxone susceptibility, Hawaii, 2016. Clin. Infect. Dis. 2017;65:918–923. doi: 10.1093/cid/cix485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebvre B, et al. Ceftriaxone-resistant Neisseria gonorrhoeae, Canada, 2017. Emerg. Infect. Dis. J. 2018;24:381. doi: 10.3201/eid2402.171756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohnishi M, et al. Ceftriaxone-resistant Neisseria gonorrhoeae, Japan. Emerg Infect. Dis. 2011;. 17:148–149. doi: 10.3201/eid1701.100397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unemo M, et al. High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: Novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrob. Agents Chemother. 2012;56:1273–1280. doi: 10.1128/aac.05760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adamson PC, et al. Trends in antimicrobial resistance in Neisseria gonorrhoeae in Hanoi, Vietnam, 2017–2019. BMC Infect. Dis. 2020;20:809. doi: 10.1186/s12879-020-05532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, et al. High percentage of the ceftriaxone-resistant Neisseria gonorrhoeae FC428 clone among isolates from a single hospital in Hangzhou, China. J. Antimicrob. Chemother. 2021;76:936–939. doi: 10.1093/jac/dkaa526. [DOI] [PubMed] [Google Scholar]
- 20.Powell AJ, Tomberg J, Deacon AM, Nicholas RA, Davies C. Crystal structures of penicillin-binding protein 2 from penicillin-susceptible and -resistant strains of Neisseria gonorrhoeae reveal an unexpectedly subtle mechanism for antibiotic resistance*. J. Biol. Chem. 2009;284:1202–1212. doi: 10.1074/jbc.M805761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quillin SJ, Seifert HS. Neisseria gonorrhoeae host adaptation and pathogenesis. Nat. Rev. Microbiol. 2018;16:226–240. doi: 10.1038/nrmicro.2017.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa-Lourenço APRD, Barros Dos Santos KT, Moreira BM, Fracalanzza SEL, Bonelli RR. Antimicrobial resistance in Neisseria gonorrhoeae: History, molecular mechanisms and epidemiological aspects of an emerging global threat. Braz. J. Microbiol. 2017;48:617–628. doi: 10.1016/j.bjm.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asavapiriyanont S, et al. Sexually transmitted infections among HIV-infected women in Thailand. BMC Public Health. 2013;13:373–373. doi: 10.1186/1471-2458-13-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Budkaew J, Chumworathayi B, Pientong C, Ekalaksananan T. Prevalence and factors associated with gonorrhea infection with respect to anatomic distributions among men who have sex with men. PLoS ONE. 2019;14:e0211682. doi: 10.1371/journal.pone.0211682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandori M, et al. Mosaic penicillin-binding protein 2 in Neisseria gonorrhoeae isolates collected in 2008 in San Francisco, California. Antimicrob. Agents Chemother. 2009;53:4032–4034. doi: 10.1128/aac.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unemo M, Golparian D, Stary A, Eigentler A. First Neisseria gonorrhoeae strain with resistance to cefixime causing gonorrhoea treatment failure in Austria, 2011. Euro Surveill. 2011;16:19998. [PubMed] [Google Scholar]
- 27.Seike K, et al. Novel penA mutations identified in Neisseria gonorrhoeae with decreased susceptibility to ceftriaxone isolated between 2000 and 2014 in Japan. J. Antimicrob. Chemother. 2016;71:2466–2470. doi: 10.1093/jac/dkw161. [DOI] [PubMed] [Google Scholar]
- 28.Cámara J, et al. Molecular characterization of two high-level ceftriaxone-resistant Neisseria gonorrhoeae isolates detected in Catalonia, Spain. J. Antimicrob. Chemother. 2012;67:1858–1860. doi: 10.1093/jac/dks162. [DOI] [PubMed] [Google Scholar]
- 29.Allen Vanessa G, et al. Molecular analysis of antimicrobial resistance mechanisms in Neisseria gonorrhoeae isolates from Ontario, Canada. Antimicrob. Agents Chemother. 2011;55:703–712. doi: 10.1128/AAC.00788-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grad YH, et al. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: A retrospective observational study. Lancet Infect. Dis. 2014;14:220–226. doi: 10.1016/s1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thailand, N. A. R. S. C. I. Antibiotic susceptibility of Neisseria gonorrhoeae in 2012–2019. NARST, http://www.cdc.gov/std/gonorrhea/lab/ngon.htm (2019).
- 32.Tribuddharat C, et al. Gonococcal antimicrobial susceptibility and the prevalence of bla(TEM-1) and bla(TEM-135) genes in Neisseria gonorrhoeae isolates from Thailand. Jpn. J. Infect. Dis. 2017;70:213–215. doi: 10.7883/yoken.JJID.2016.209. [DOI] [PubMed] [Google Scholar]
- 33.Fifer H, et al. Failure of dual antimicrobial therapy in treatment of gonorrhea. N. Engl. J. Med. 2016;374:2504–2506. doi: 10.1056/NEJMc1512757. [DOI] [PubMed] [Google Scholar]
- 34.Kenyon C. Dual azithromycin/ceftriaxone therapy for gonorrhea in PrEP cohorts results in levels of macrolide consumption that exceed resistance thresholds by up to 7-fold. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab178. [DOI] [PubMed] [Google Scholar]
- 35.Chanvatik S, et al. Knowledge and use of antibiotics in Thailand: A 2017 national household survey. PLoS ONE. 2019;14:e0220990. doi: 10.1371/journal.pone.0220990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 31. Clinical and Laboratory Standards Institute; 2021. [Google Scholar]
- 37.EUCAST. Clinical breakpoints and dosing of antibiotics European Committee for Antimicrobial Susceptibility Testing v 11.0 (2021).
- 38.Mayta H, et al. Use of a reliable PCR assay for the detection of Neisseria gonorrhoeae in Peruvian patients. Clin. Microbiol. Infect. 2006;12:809–812. doi: 10.1111/j.1469-0691.2006.01452.x. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhry U, Saluja D. Detection of Neisseria gonorrhoeae by PCR using orf1 gene as target. Sex. Transm. Infect. 2002;78:72. doi: 10.1136/sti.78.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin IM, Ison CA, Aanensen DM, Fenton KA, Spratt BG. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J. Infect. Dis. 2004;189:1497–1505. doi: 10.1086/383047. [DOI] [PubMed] [Google Scholar]
- 41.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 45.Leaché AD, Banbury BL, Felsenstein J, de Oca AN-M, Stamatakis A. Short tree, long tree, right tree, wrong tree: New acquisition bias corrections for inferring SNP phylogenies. Syst. Biol. 2015;64:1032–1047. doi: 10.1093/sysbio/syv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Z, et al. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44:W236–W241. doi: 10.1093/nar/gkw370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaye M, Chibo D, Birch C. Phylogenetic investigation of transmission pathways of drug-resistant HIV-1 utilizing pol sequences derived from resistance genotyping. J. Acquir. Immune Defic. Syndr. 2008;49:9–16. doi: 10.1097/QAI.0b013e318180c8af. [DOI] [PubMed] [Google Scholar]
- 49.Wannigama DL, et al. A rapid and simple method for routine determination of antibiotic sensitivity to biofilm populations of Pseudomonas aeruginosa. Ann. Clin. Microbiol. Antimicrob. 2020;19:8. doi: 10.1186/s12941-020-00350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wannigama DL, et al. Simple fluorometric-based assay of antibiotic effectiveness for Acinetobacter baumannii biofilms. Sci. Rep. 2019;9:6300. doi: 10.1038/s41598-019-42353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wannigama DL, et al. A simple antibiotic susceptibility assay for Pseudomonas aeruginosa and Acinetobacter baumannii biofilm could lead to effective treatment selection for chronic lung infections. Chest. 2019;155:77A. doi: 10.1016/j.chest.2019.02.351. [DOI] [Google Scholar]
- 52.O'Toole GA. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stepanović S, et al. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 54.Müsken M, Di Fiore S, Römling U, Häussler S. A 96-well-plate–based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat. Protoc. 2010;5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated and analysed during this study are included in this published article and its Supplemental information file. Additional clinical details available upon reasonable request from corresponding author TC.