Abstract
In Western countries, breast cancer tends to occur in older postmenopausal women. However, in Asian countries, the proportion of younger premenopausal breast cancer patients is increasing. Increasing evidence suggests that the gut microbiota plays a critical role in breast cancer. However, studies on the gut microbiota in the context of breast cancer have mainly focused on postmenopausal breast cancer. Little is known about the gut microbiota in the context of premenopausal breast cancer. This study aimed to comprehensively explore the gut microbial profiles, diagnostic value, and functional pathways in premenopausal breast cancer patients. Here, we analyzed 267 breast cancer patients with different menopausal statuses and age-matched female controls. The α-diversity was significantly reduced in premenopausal breast cancer patients, and the β-diversity differed significantly between breast cancer patients and controls. By performing multiple analyses and classification, 14 microbial markers were identified in the different menopausal statuses of breast cancer. Bacteroides fragilis was specifically found in young women of premenopausal statuses and Klebsiella pneumoniae in older women of postmenopausal statuses. In addition, menopausal-specific microbial markers could exhibit excellent discriminatory ability in distinguishing breast cancer patients from controls. Finally, the functional pathways differed between breast cancer patients and controls. Our findings provide the first evidence that the gut microbiota in premenopausal breast cancer patients differs from that in postmenopausal breast cancer patients and shed light on menopausal-specific microbial markers for diagnosis and investigation, ultimately providing a noninvasive approach for breast cancer detection and a novel strategy for preventing premenopausal breast cancer.
Subject terms: Bacterial genetics, Breast cancer
Breast cancer: gut microbes before and after menopause
Women developing breast cancer before the menopause have less diverse gut microbe populations than their healthy peers, and significantly different gut microbial profiles than postmenopausal breast cancer patients. There is increasing evidence of a link between gut microbes and breast cancer, but most studies have focused on women after the menopause. The incidence of breast cancer in premenopausal women is increasing, especially in Asia. Researchers in Taiwan led by Chih-Po Chiang at Kaohsiung Medical University Hospital compared gut microbial populations in 267 breast cancer patients at different menopausal stages to matched controls. The results suggest that gut microbe profiling might offer a novel and noninvasive method for diagnosing and monitoring breast cancer. It may also lead to new strategies for prevention and treatment, as evidence emerges of a link between gut microbes and cancer.
Introduction
Breast cancer is the most commonly diagnosed cancer worldwide, with an estimated 2.3 million incident cases (11.7%) reported in 2020. Breast cancer is also the fifth leading cause of cancer mortality worldwide and the leading cause of cancer-associated death in women1. Compared to the incidence of breast cancer in Western countries, the incidence of breast cancer in Asia is relatively low; however, the proportion of younger women diagnosed with breast cancer is increasing in Asian countries. The median onset age of breast cancer in Asia (40–50 years) is approximately 20 years younger than that in Western countries (60–70 years)2,3. In Taiwan, more than 30% of women with breast cancer are younger than 50 years of premenopausal breast cancer. This high proportion of premenopausal breast cancer in Taiwan differs from that in Western countries, where the proportion is usually less than 20%4,5.
Genetic background is one of the potential explanations of this premenopausal status of young breast cancer patients. Germline mutations in BRCA1 and BRCA2 are the dominant targets in this subtype of young breast cancer patients. Based on multiple gene analyses, TP53 and RAD50 are also involved in the pathogenesis of breast cancer in young patients6. However, BRCA and TP53 mutations are present in only 20% of young breast cancer patients, especially in those with the familial type. Westernization might play an important role in Asia of young breast cancer patients, including environmental (e.g., endocrine-disrupting chemicals, EDCs), dietary (e.g., high fat and alcohol intake), and reproductive factors (e.g., delayed childbearing)7,8. However, the role of the gut microbiota in premenopausal breast cancer has often been neglected.
The gut microbiota is a collection of information on the bacteria in our gut, including the number, type, diversity, etc. The gastrointestinal tract contains >1014 microorganisms, which is ten times the number of our own cells, and accounts for 1–2 kg of body weight. The gut microbiota has been well recognized as a symbiotic organ that maintains the normal function of the gut and affects metabolic, protective, and structural functions9. Dysbiosis, a condition that reflects the imbalance of the gut microbiota, has been recognized to be closely associated with many human diseases, including colorectal cancer10, metabolic syndrome11, inflammatory bowel disease12, and brain nerve diseases13. The gut microbiota is also involved in cancer formation, and is termed the “oncobiome”, which induces the transformation of host cells and accounts for 20% of human malignancies14.
To date, the best-known mechanism by which the gut microbiota affects breast cancer is through the “estrobolome”, which refers to gut bacterial genes (gut microbiota) whose products can metabolize estrogens15,16. The metabolism of conjugated estrogens occurs in the liver, and the compounds are excreted in urine, bile, and feces; however, conjugated estrogens can be deconjugated by gut bacteria via β-glucuronidase activity, and estrogen can be reabsorbed into circulation. The accumulation of endogenous estrogens increases the risk of breast cancer, especially in postmenopausal women17. In addition to the estrobolome, bacterial metabolites are transferred to distant sites through circulation and influence the occurrence of breast cancer18. Thus, the gut microbiota secretes and synthesizes bioactive metabolites that affect breast cancer formation.
However, the concept of the estrobolome mainly exists in postmenopausal breast cancer, and bacterial metabolites were verified in a cell line model. In addition, several human studies have focused on the gender or menopausal status of healthy controls, but not of breast cancer patients. Moreover, only a few studies have assessed the gut microbiota in premenopausal breast cancer patients. Thus, in this study, we aimed to comprehensively explore the gut microbial profiles between premenopausal and postmenopausal breast cancer patients to elucidate the critical microbial markers, diagnostic value, and related functional pathways in premenopausal breast cancer patients, ultimately providing a noninvasive approach for breast cancer detection and a novel strategy for preventing premenopausal breast cancer in the future.
Materials and methods
Patient recruitment
From October 2018 to December 2020, 267 subjects were recruited from the Division of Breast Oncology and Surgery, Department of Surgery, Kaohsiung Medical University Chung-Ho Memorial Hospital. The subjects included 67 age-matched female controls (premenopausal, Pre-C = 50; postmenopausal, Post-C = 17) and 200 breast cancer patients (premenopausal, Pre-BC = 100; postmenopausal, Post-BC = 100). All patients with de novo breast cancer were diagnosed with stage I–II disease by pathological examination. Subjects were excluded if they were diagnosed with malignancies other than breast cancer or administered steroids, antibiotics, or probiotics within 4 weeks before the screening visit. Fecal samples were collected for DNA extraction before patients received any chemo/hormone/target therapy, radiation, or surgery. Furthermore, clinical characteristics such as age, body mass index, grade, stage, tumor size, and receptor status were recorded (Table 1). The study was approved by the Internal Review Board, and informed consent was obtained from all subjects [KMUHIRB-G(II)-20180018 and KMUHIRB-E(I)-20200285].
Table 1.
Clinical characteristics of subjects according to the menopausal status.
| Characteristic | Pre-C N = 50 | Pre-BC N = 100 | Post-C N = 17 | Post-BC N = 100 | p |
|---|---|---|---|---|---|
| Age (m ± sd) | 35.4 ± 6 | 41.5 ± 5.2 | 61.6 ± 8.9 | 60.08 ± 5.8 | <0.001 |
| BMI | 23.6 ± 4.4 | 24.3 ± 4.2 | 0.209 | ||
| Grade | 0.261 | ||||
| Grade 1 | – | 10 (10.0%) | – | 7 (7.0%) | |
| Grade 2 | – | 60 (60.0%) | – | 71 (71.0%) | |
| Grade 3 | – | 30 (30.0%) | – | 22 (22.0%) | |
| Stage | 0.657 | ||||
| Stage I | – | 63 (63.0%) | – | 66 (66.0%) | |
| Stage II | – | 37 (37.0%) | – | 34 (34.0%) | |
| Tumor size (mm) | – | 24.9 ± 15.4 | – | 21.3 ± 10.2 | 0.049 |
| ER | 0.182 | ||||
| – | – | 13 (13.0%) | – | 20 (20.0%) | |
| + | – | 87 (87.0%) | – | 80 (80.0%) | |
| PR | 0.008 | ||||
| – | – | 20 (20.0%) | – | 37 (37.0%) | |
| + | – | 80 (80.0%) | – | 63 (63.0%) | |
| HER2 | 0.102 | ||||
| – | – | 80 (80.0%) | – | 70 (70.0%) | |
| + | – | 20 (20.0%) | – | 30 (30.0%) | |
| Ki67 | 0.199 | ||||
| – | – | 39 (39.0%) | – | 48 (48.0%) | |
| + | – | 61 (61.0%) | – | 52 (52.0%) |
p value: Comparison between Pre-BC and Post-BC with t-test and Chi-squared test.
Fecal DNA Extraction and 16S sequencing
DNA was extracted according to the manufacturer’s instructions using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Valencia, CA, USA) to obtain an OD 260/280 ratio between 1.8 and 2.0 by NanoDrop 2000 spectrophotometer. The V3-V4 hypervariable region of 16S rDNA was amplified using the bacterial-specific forward (5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG 3′) and reverse (5′ GTCTCGTGGGCTCGGAGATG TGTATAAGAGACAGGACTACHVGGGTATCTAATCC 3′) primer sets. The PCR product was purified with AMPure XP magnetic beads (Beckman Coulter, Brea, IN, USA) and indexed adapters were added to the amplicons using the Nextera XT Index Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The amplified DNA sizing accuracy was verified using the 4200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA). After library construction, samples were mixed with MiSeq Reagent Kit v3 (600-cycle) and loaded onto a MiSeq cartridge. Then, a 2 × 300 bp paired-end sequencing run was performed using the MiSeq platform (Illumina, San Diego, CA, USA).
Taxonomic composition and diversity analysis
The raw paired-end reads were trimmed and passed through quality filters (quality trimming, discarding short read length, and removing chimeras) and were assigned to operational taxonomic units (OTUs) that shared ≥97% similarity with the Greengene 13.8 database. Raw paired-end reads were also analyzed using the BaseSpace Ribosomal Database Project (RDP) classifier. OTUs (relative abundance), α-diversity (Shannon entropy and Venn diagram), and β-diversity (PCoA-D_0.5 UniFrac) were determined using BaseSpace (Illumina, San Diego, CA, USA), CLC Genomics Workbench 21 with Microbial Genomics Module (Qiagen, Germantown, MD, USA) and GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA)19.
Linear discriminant analysis (LDA) effect size (LEfSe) algorithm
The LDA effect size (LEfSe) algorithm with α = 0.05 (Kruskal–Wallis and Wilcoxon tests) and effect size threshold of 2 on LDA was used to identify taxa of contrasting abundance between groups. The LEfSe identified taxa that (1) were of nonzero abundance in more than half of the samples in at least one comparison group, and (2) had an abundance difference > 0.0001 between the maximum and minimum group means were labeled on the LEfSe cladogram. Significant genera and species were further investigated for LDA scores and relative abundance by bar plots and box plots, respectively, if available.
Heatmap analysis
To visualize how bacterial abundance and samples were clustered, heatmaps were constructed using the R package ‘pheatmap’ at the genus and species levels, including genera with an average relative abundance > 0.5% and species > 0.05%. Taxa with “uncultured” or “unidentified” names were excluded. Hierarchical clustering in the heatmap was performed using rows and columns based on Euclidean distances with the complete linkage algorithm.
Correlation matrix analysis
Pairwise correlation analysis was performed among taxonomic groups at the genus level. Genera with an average relative abundance > 0.5% were included, and those with “uncultured” or “unidentified” taxa were excluded. The correlation was evaluated using Spearman’s rank correlation and the cutoff was set at 0.2. The correlation matrix was visualized using the R package ‘ggcorrplot’, in which positive correlations were colored blue and negative correlations were colored red.
Functional profile inference and analysis
The metabolic functional profile of the microbiota was predicted using the PICRUSt2 algorithm based on OTU-level read abundance. The predicted abundance of functional groups, including the KEGG Orthology (KO) and MetaCyc pathways was normalized by total read counts per sample for downstream analysis. The KO pathway (KOPath) abundance was also calculated as the sum of the involved KO abundances and normalized as mentioned for downstream analysis. Differential enrichment analysis was performed using the Kruskal–Wallis test. The results were first visualized by a volcano plot showing false discovery rate (FDR, in log 10) versus fold changes (in log 2) between groups. The functional pathways with significant differences were further explored for abundance distribution using a boxplot for those with >50% of samples with nonzero abundance in at least one compared group, and reported in descending order of group mean differences; that is, the greater the difference, the higher the priority.
Statistical analysis
Comparisons of different groups were performed using the two-tailed t-test and one-way ANOVA. Values of p less than p < 0.05, p < 0.01, and p < 0.001 were considered statistically significant. The specificity and sensitivity of the microbial markers were determined using the receiver operating characteristic curve (ROC curve) and the area under the curve (AUC) values. Correlations were calculated using the Pearson correlation coefficient. All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA)19.
Results
Difference in diversity and taxonomy of gut microbiota between control individuals and breast cancer patients with different menopausal statuses
In this study, we recruited 267 participants belonging to four groups: premenopausal female controls (Pre-C, N = 50), premenopausal breast cancer patients (Pre-BC, N = 100), postmenopausal female controls (Post-C, N = 17), and postmenopausal breast cancer patients (Post-BC, N = 100). The clinical characteristics of the individuals in the four groups are summarized in Table 1. Patients in the Pre-BC and Post-BC groups had similar body mass indices (BMIs) and grades; however, a significant difference was found in tumor size and progesterone receptor (PR) status.
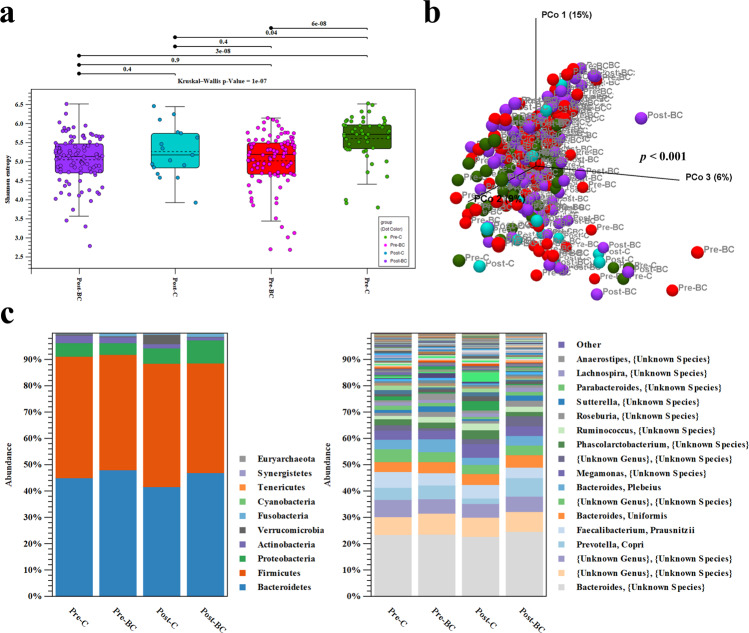
Initially, we analyzed the α- and β-diversities among the four groups. The α-diversity of Shannon entropy was significantly reduced in the Pre-BC group (red box) compared to in the Pre-C group (green box); however, no significant differences were found in α-diversity between the Post-C (blue box) and Post-BC groups (purple box) or the Pre-BC and Post-BC groups (Fig. 1a). In terms of β-diversity, principal coordinate analysis (PCoA) of D_0.5 UniFrac was performed to determine the total microbial composition of the different groups. The PERMANOVA test revealed a significant difference in the overall microbial composition among the different groups (p < 0.001, Fig. 1b). To further explore the microbial composition, the relative abundances of operational taxonomic units (OTUs) were evaluated among the different groups. At the phylum and species levels, Actinobacteria was enriched in Pre-C, whereas Verrucomicrobia was enriched in Post-C and Proteobacteria in Post-BC (Fig. 1c). These results indicate that α-diversity was specifically decreased in premenopausal breast cancer patients. In addition, the overall microbial composition was significantly different between control individuals and breast cancer patients.
Fig. 1. Microbial diversity among different groups of control individuals and breast cancer patients.
a The α-diversity of Shannon entropy in Pre-BC (red box) was significantly lower than that in Pre-C (green box); this phenomenon was not observed in Post-BC (purple box). b The β-diversity of PCoA demonstrated significant differences (p < 0.001) in the total microbial composition among the four groups. c The relative abundance of OTUs at the phylum and species levels among the four groups.
Identification of the microbial markers of the different groups
To further reveal the microbial markers of the different groups, LDA effect size (LEfSe), Venn diagram, and heatmap analyses were used to identify (1) critical microbial markers of premenopausal breast cancer without age effects; (2) microbial markers that fluctuated with age but were more obviously altered in breast cancer; and (3) universal microbial markers of breast cancer.
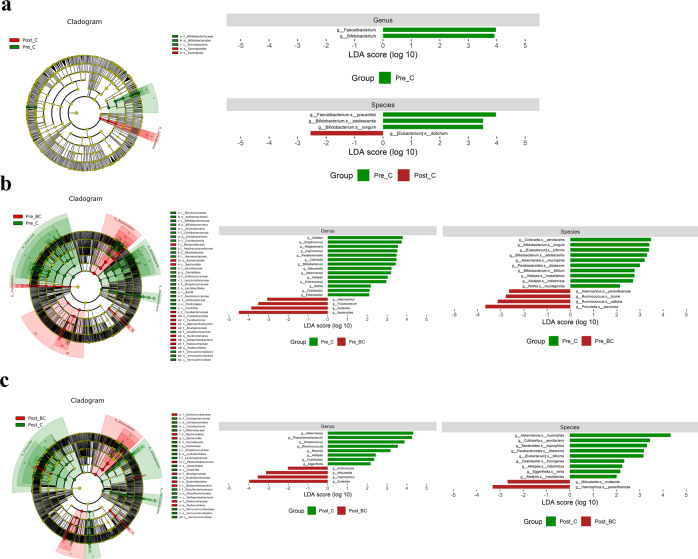
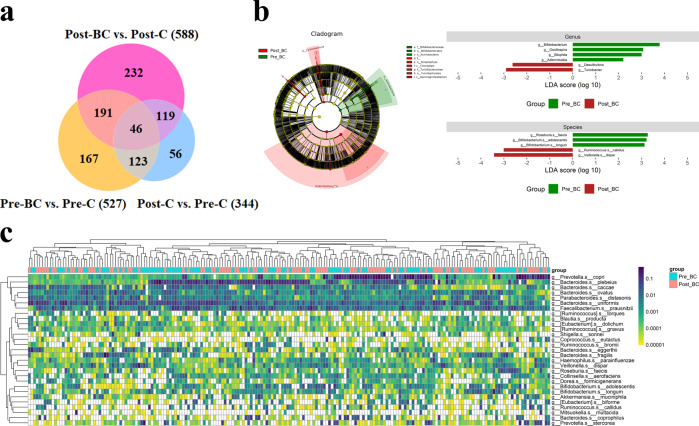
We specifically focused on the genus/species levels among the different groups. Based on LEfSe analysis, Faecalibacterium and Bifidobacterium were identified to be more enriched in Pre-C than in Post-C. When Pre-C was compared with Pre-BC and Post-C with Post-BC, Bifidobacterium and Akkermansia were enriched in control individuals, while Sutterella and Haemophilus were enriched in breast cancer patients (Fig. 2). A Venn diagram was subsequently generated to verify the intersection of OTUs among the different groups with the criteria of absolute fold change ≧ 5 and p < 0.05 (Fig. 3a). We found 167 OTUs belonging to Pre-BC versus Pre-C and 191 OTUs belonging to breast cancer (overlap of Pre-BC and Post-BC). We also compared Pre-BC with Post-BC with LEfSe and heatmap analyses to identify differential microbiomes (Fig. 3b, c). Accordingly, several potential microbial markers among the different groups based on LEfSe, Venn diagram, and heatmap analyses were selected as candidates for further analysis.
Fig. 2. Microbial markers at the genus/species levels between control individuals and breast cancer patients.
a The potential microbial markers between Pre-C and Post-C. b The potential microbial markers between Pre-C and Pre-BC. c The potential microbial markers between Post-C and Post-BC.
Fig. 3. The intersection of OTUs among different groups of control individuals and breast cancer patients.
a The Venn diagram analysis shows that 167 OTUs belonged to Pre-BC versus Pre-C and 191 OTUs belonged to breast cancer (overlap of Pre-BC and Post-BC). b, c The potential microbial markers between Pre-BC and Post-BC.
Unique gut microbial markers of different menopausal statuses of breast cancer
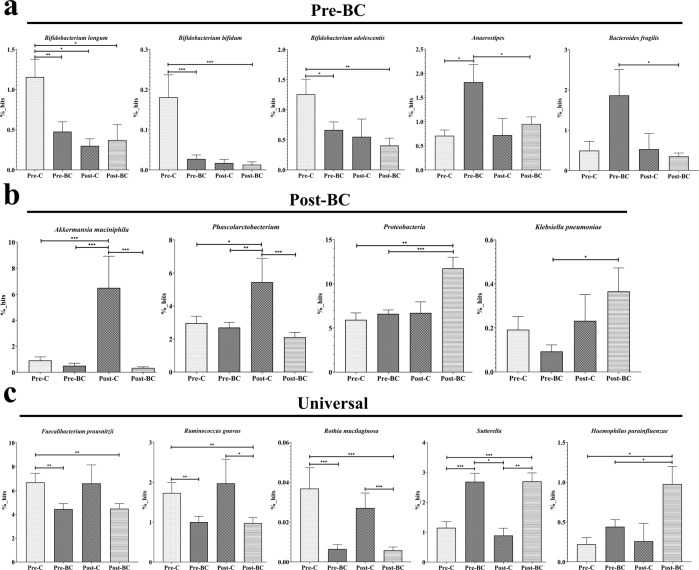
To further verify the gut microbial markers of different menopausal statuses, we utilized BaseSpace in the Ribosomal Database Project (RDP) classifier to reconfirm the OTUs at the genus/species levels. Using the RDP classifier, we screened all of the potential microbial markers of OTUs obtained via LEfSe, Venn diagram, and heatmap analyses at the genus/species levels and eliminated the microbiomes with low percentages or that were not present in statistically significant numbers in the four groups of Pre-C, Pre-BC, Post-C, and Post-BC. Eventually, we identified 14 bacterial taxa that were abundant and/or present in statistically significant quantities in the four groups.
The 14 bacterial taxa were divided into three groups: Pre-BC, Post-BC, and Universal. The abundances of Bifidobacterium longum, Bifidobacterium bifidum, and Bifidobacterium adolescentis were found to fluctuate with age but were more obviously reduced in premenopausal breast cancer patients. The abundance of Bifidobacterium spp. was higher in Pre-C but lower in Post-C due to the age effect; however, Bifidobacterium spp. was significantly reduced in Pre-BC compared with Pre-C individuals; this phenomenon was not observed in Post-BC patients. We also found that Anaerostipes and Bacteroides fragilis were not affected by age and were significantly higher in premenopausal breast cancer patients than in postmenopausal breast cancer patients (Fig. 4a). Thus, the above five bacterial taxa were unique to premenopausal breast cancer patients, with or without the age effect. In Post-BC, we found that the abundances of Akkermansia muciniphila and Phascolarctobacterium fluctuated with age but were more obviously reduced in postmenopausal breast cancer patients. Akkermansia muciniphila and Phascolarctobacterium were higher in Post-C individuals than in Pre-C individuals due to the age effect; however, they were significantly reduced in Post-BC patients compared with their levels in Post-C individuals; this phenomenon was not observed in Pre-BC patients. We also found that Proteobacteria and Klebsiella pneumoniae were not affected by age, with significantly higher levels in postmenopausal breast cancer patients than in premenopausal breast cancer patients (Fig. 4b). Thus, the above four bacterial taxa were unique to postmenopausal breast cancer patients with or without the age effect.
Fig. 4. The 14 microbial markers according to the different menopausal statuses of breast cancer.
a The percentage of Bifidobacterium spp. fluctuated with age but was significantly reduced in Pre-BC compared with Pre-C, whereas Anaerostipes and Bacteroides fragilis were specifically increased in Pre-BC. b The percentages of Akkermansia muciniphila and Phascolarctobacterium fluctuated with age but were significantly reduced in Post-BC compared with Post-C, whereas Proteobacteria and Klebsiella pneumoniae were specifically increased in Post-BC. c The percentages of Faecalibacterium prausnitzii, Ruminococcus gnavus, and Rothia mucilaginosa were simultaneously reduced in Pre-BC, and Post-BC, while Sutterella, and Haemophilus parainfluenzae were simultaneously increased in Pre-BC and Post-BC.
In the Universal group, the abundances of Faecalibacterium prausnitzii, Ruminococcus gnavus, and Rothia mucilaginosa were simultaneously reduced in Pre-BC and Post-BC patients, while Sutterella and Haemophilus parainfluenzae were simultaneously increased in Pre-BC and Post-BC patients (Fig. 4c). Thus, the above five bacterial taxa were universal microbiomes for breast cancer patients (for both premenopausal and postmenopausal breast cancer) without age effects. The above results indicate that the gut microbiome is unique according to different menopausal statuses of breast cancer. We also verified the above bacterial taxa at the genus/species level in individuals with different receptor statuses and stages. The 13 bacterial taxa did not show a significant trend among the different receptor statuses (luminal, HER2, basal) or stages (stages 1, 2), with only a small proportion of bacterial taxa reaching statistical significance (Supplementary Figs. 1, 2).
Certain microbial markers correlate with age in control individuals and breast cancer patients
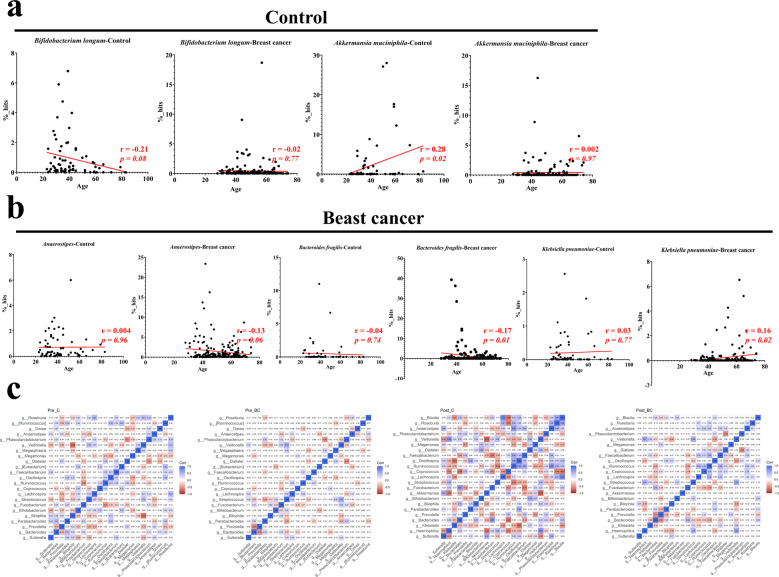
To further determine the microbial markers that are crucial in breast cancer with age fluctuation, especially in premenopausal of young breast cancer. We analyzed the correlation between 14 bacterial taxa and age in two separate groups: control (Pre-C + Post-C) and breast cancer (Pre-BC + Post-BC). Among the 14 bacterial taxa, Bifidobacterium longum was negatively correlated (r = −0.21, p = 0.08) and Akkermansia muciniphila was positively (r = 0.28, p = 0.02) correlated with age in the control group but not in the breast cancer group (Fig. 5a), indicating that Bifidobacterium longum and Akkermansia muciniphila fluctuated with age in female controls but were not involved in the different menopausal statuses of breast cancer. In contrast, we found that Anaerostipes (r = −0.13, p = 0.06) and Bacteroides fragilis (r = −0.17, p = 0.01) were negatively correlated and Klebsiella pneumoniae was positively correlated (r = 0.16, p = 0.02) with age in the breast cancer group but not in the control group (Fig. 5b), indicating that Anaerostipes, Bacteroides fragilis, and Klebsiella pneumoniae were microbiomes that were specifically involved in the different menopausal statuses of breast cancer.
Fig. 5. Critical microbial markers that correlate with age in control individuals and breast cancer patients.
a Bifidobacterium longum was negatively correlated (r = −0.21, p = 0.08) and Akkermansia muciniphila was positively correlated (r = 0.28, p = 0.02) with age in female controls but did not correlate with age in breast cancer patients. b Anaerostipes (r = −0.13, p = 0.06) and Bacteroides fragilis (r = −0.17, p = 0.01) were negatively correlated with age, while Klebsiella pneumoniae was positively correlated (r = 0.16, p = 0.02) with age in breast cancer patients but did not correlate with age in female controls. c Spearman correlation of Pre-C, Pre-BC, Post-C, and Post-BC at the genus level.
The correlations among the taxonomic groups was also verified by correlation matrix analysis to determine the effect of the dominant genus in female controls. In Pre-C, Bifidobacterium was negatively correlated with Sutterella (−0.35) and Anaerostipes (−0.11), thereby indicating the potential protective effect of Bifidobacterium in premenopausal female controls (Fig. 5c, left). On the other hand, in Post-C, Akkermansia and Phascolarctobacterium were negatively correlated with Haemophilus (Akkermansia: −0.64; Phascolarctobacterium: −0.22) and Klebsiella (Akkermansia: −0.12; Phascolarctobacterium: −0.17), indicating a potential protective effect of Akkermansia and Phascolarctobacterium in postmenopausal female controls (Fig. 5c, right).
Diagnostic value of the gut microbiota in the different menopausal statuses of breast cancer
In addition to elucidating the specific microbial markers in the different menopausal statuses of breast cancer, we sought to determine the potential diagnostic value of the gut microbiota for breast cancer. As specific microbial markers existed in premenopausal and postmenopausal breast cancer, the diagnostic value was verified by the menopausal status of three groups: premenopausal, postmenopausal, and all breast cancer patients (premenopausal and postmenopausal).
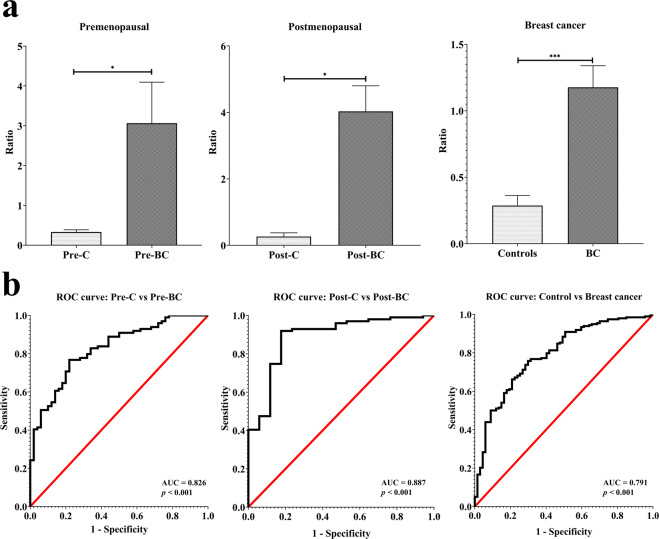
Regarding premenopausal status, ten bacterial taxa were calculated by dividing the increasing taxa by the decreasing taxa. The percentage of the sum (Bacteroides fragilis + Anaerostipes + Haemophilus parainfluenzae + Sutterella) divided by the sum (Faecalibacterium prausnitzii + Bifidobacterium adolescentis + Bifidobacterium longum + Bifidobacterium bifidum + Ruminococcus gnavus + Rothia mucilaginosa) yielded an average value (Pre-C: 0.33 versus Pre-BC: 3.06, p = 0.03). This average value was further evaluated using a receiver operating characteristic curve (ROC curve). The area under the curve (AUC) was 0.826 (p < 0.001), indicating excellent discrimination for distinguishing between Pre-BC and Pre-C (Fig. 6a, b left). For postmenopausal status, seven bacterial taxa were calculated by dividing the percentage of the sum (Klebsiella pneumoniae + Haemophilus parainfluenzae + Sutterella) by the sum (Akkermansia muciniphila + Phascolarctobacterium + Ruminococcus gnavus + Rothia mucilaginosa) to obtain an average value (Post-C: 0.27 versus Post-BC: 4.03, p = 0.02). For this average value, the AUC was 0.887 (p < 0.001), indicating excellent discrimination for distinguishing Post-BC from Post-C (Fig. 6a, b middle). In breast cancer patients regardless of menopausal status (premenopausal + postmenopausal), five bacterial taxa were calculated by dividing the percentage of the sum (Haemophilus parainfluenzae + Sutterella) by the sum (Faecalibacterium prausnitzii + Ruminococcus gnavus + Rothia mucilaginosa) to obtain an average value (Controls: 0.29 versus Breast cancer: 1.18, p < 0.001). For this average value, the AUC was 0.791 (p < 0.001), indicating acceptable discrimination for distinguishing breast cancer patients from control individuals (Fig. 6a, b right). The above results indicate that premenopausal and postmenopausal breast cancer differ in terms of the gut microbiota, which indicates diagnostic value for breast cancer.
Fig. 6. The potential of diagnosis using the microbial markers found in the different menopausal statuses of breast cancer.
a, b Microbial markers were determined for the different menopausal statuses of breast cancer or all breast cancer (Pre-BC + Post-BC) by dividing the increasing taxa by the decreasing taxa. The average values displayed excellent discrimination for use in distinguishing Pre-BC/Post-BC from Pre-C/Post-C.
Metabolic functional pathway of the gut microbiota in breast cancer
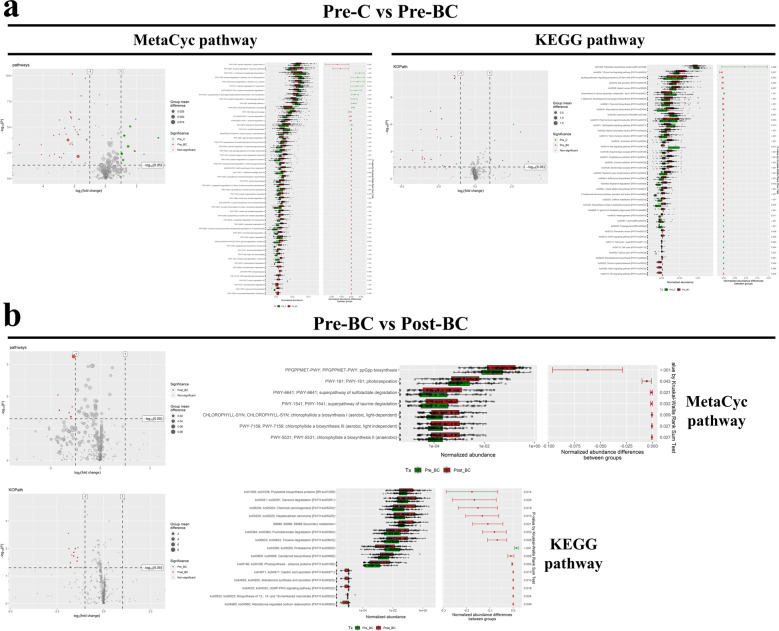
To further understand the function of the microbiome in breast cancer, we employed PICRUSt2 prediction of the abundance of different microbiomes against MetaCyc (pathways) and KEGG pathways (KOPath). We specifically focused on premenopausal breast cancer to elucidate functional pathways of the gut microbiota. Compared with the control (Pre-C vs. Pre-BC), premenopausal breast cancer was enriched with pathways contributing to the abundance of the microbiome against the steroid-related (MetaCyc pathway: meta cleavage pathway of aromatic compounds; aromatic biogenic amine degradation; androstenedione degradation, Fig. 7a left) and oncogenic-related pathways (KEGG pathway: Cell cycle, Tight junction, Notch/Wnt signaling pathway, Fig. 7a right). We also compared premenopausal patients with postmenopausal breast cancer patients (Pre-BC vs. Post-BC) to verify the functional pathways of the microbiome in different menopausal statuses of breast cancer. Interestingly, we found that postmenopausal breast cancer patients exhibited greater enrichment of steroid-related (KEGG pathway: aldosterone synthesis and secretion; aldosterone−regulated sodium reabsorption) and chemical carcinogenesis pathways than premenopausal breast cancer patients (Fig. 7b).
Fig. 7. The potential functional pathway involved in breast cancer.
a Compared with the control individuals, premenopausal breast cancer was enriched with pathways contributing to the abundance of the microbiome against the steroid-related (MetaCyc pathway) and oncogenic-related pathway (KEGG pathway). b Compared with premenopausal breast cancer patients, postmenopausal breast cancer patients exhibited greater enrichment in the steroid-related pathway (KEGG pathway).
Discussion
Increasing evidence has suggested that the gut microbiota plays a critical role in cancer, which is termed the “oncobiome” and can drive cancer initiation and progression. The mechanisms of the oncobiome that contribute to carcinogenesis are divided into three categories: (1) indirect effects through metabolites of microbiomes, (2) direct effects by impacting genomic stability of DNA damage and signaling related to host cell proliferation and death, and (3) direct effects by guiding immune system function20. In breast cancer, the metabolism of estrogen by the gut microbiota, which is termed the “estrobolome“, has set a foundation for the role of the gut microbiota. However, the concept of the estrobolome mainly exists in postmenopausal breast cancer patients. Little is known about the role of gut microbiota in premenopausal breast cancer patients.
The clinical and pathological features of premenopausal breast cancer differ from those of postmenopausal breast cancer. Premenopausal breast cancer is more aggressive and has a poorer prognosis than postmenopausal breast cancer5. In Asian countries, the high proportion of premenopausal breast cancer is a critical clinical issue that differs from the situation in Western countries. In addition, the luminal molecular subtype of premenopausal breast cancer in Asian countries, such as Taiwan, is more prevalent than in Western countries, where the basal-like subtype with a poor prognosis is dominant21. In our study of 200 breast cancer patients, the proportion of patients with premenopausal breast cancer was 50%, with greater tumor size and nearly the same distribution of grade and stage as that in patients with postmenopausal breast cancer. The molecular subtype of premenopausal breast cancer was true, with a high proportion of the luminal subtype, which accounted for more than 80% of cases.
In this study, we found that the α-diversity of the Shannon index was significantly reduced in premenopausal breast cancer patients compared with that in premenopausal controls. The phenomenon of lower α-diversity was unique to premenopausal breast cancer patients but not observed in postmenopausal breast cancer patients, whereas the α-diversity in postmenopausal controls was reduced compared with that in premenopausal controls. To date, only one study has investigated the gut microbiota in breast cancer patients and control individuals with different menopausal statuses. The researchers found that α-diversity was higher in breast cancer patients than in control individuals22. However, in our study, we provided a comprehensive analysis in which a lower α-diversity was specifically observed in premenopausal breast cancer.
The α-diversity is a general indicator used to evaluate the condition of the gut environment, including richness and evenness. Decreased diversity is generally correlated with aging, an unhealthy gut environment and disease status. This phenomenon of decreased diversity, termed loss of microbiota diversity (LOMD), is frequently associated with dysbiosis, which is the imbalance and abnormalities of the microbiota that result in negative effects on the host. LOMD is a general feature associated with many diseases and can even predict treatment response23. Herein, we also found a lower diversity in postmenopausal controls than in premenopausal controls due to the aging process. Several human studies have focused on the gut microbiota in terms of gender or menopausal status of healthy controls but not in breast cancer patients and demonstrated that α-diversity was lower in postmenopausal women24,25. However, there was no significant difference in α-diversity between premenopausal breast cancer patients and postmenopausal breast cancer patients, indicating that the LOMD was specific in premenopausal breast cancer without an age effect. There was a significant difference in the β-diversity of total microbial composition among the different menopausal statuses of breast cancer patients and controls. A study by Jia Zhu et al. also showed that the relative abundance of the gut microbiota did not differ significantly between premenopausal breast cancer patients and control individuals. Thus, the study by Jia Zhu et al. mainly focused on postmenopausal breast cancer22. However, our study provides the first evidence that gut microbiota profiles differ significantly according to menopausal status of breast cancer patients and control individuals.
To assess the differences in the total microbial composition, we conducted multiple analyses, including relative abundance of OTUs, LEfSe, Venn diagram, and heatmap assessments. The OTUs from the above analysis were reconfirmed by the RDP classifier to determine the critical microbial markers at the genus/species levels. Age is one of the crucial factors that need to be considered with regard to gut microbiota profiles, and gut microbiota patterns will change with age (i.e., from infants to elderly patients)9,26. Age differs between premenopausal and postmenopausal women, which is accompanied by different patterns of gut microbiota24,25. Thus, we carried out a comprehensive analysis to identify microbial markers that met the following criteria: (1) critical microbial markers that are not affected by age in premenopausal breast cancer patients, (2) microbial markers that fluctuate with age but are more obviously altered in breast cancer patients, and (3) universal microbial markers in breast cancer patients.
Five microbial markers were identified in premenopausal breast cancer patients. Bifidobacterium spp. are typical probiotics related to the maintenance of human health and exhibit reduced abundance with aging. Thus, the proportion of Bifidobacterium spp. was lower in postmenopausal controls. However, Bifidobacterium spp. were specifically reduced in premenopausal breast cancer patients, which was not observed in postmenopausal breast cancer patients. Bifidobacterium spp. are well known for their beneficial effects and disease prevention27. In cancer, Bifidobacterium spp. also play a critical role in tumor suppression through the immunomodulation and inhibition of DNA damage. The antitumor/proliferative effect of Bifidobacterium spp. against breast cancer was also observed in vitro and in vivo28,29, indicating the loss of tumor-suppressor-like Bifidobacterium spp. in premenopausal breast cancer patients. Anaerostipes has been reported to increase in endometrial cancer, hepatocellular carcinoma, and thyroid cancer30–32. In addition, Bacteroides fragilis is an oncobiome that contributes to colorectal cancer formation by inducing inflammation and DNA damage33. In breast cancer, a critical study demonstrated that Bacteroides fragilis exists in breast tissues and colonizes the gut to promote breast tumorigenesis and metastatic progression through the axes of the Notch and β-catenin pathways34. In postmenopausal breast cancer patients, Akkermansia muciniphila functions as a probiotic in individuals with obesity and diabetes. Several human studies have shown that the abundance of Akkermansia muciniphila decreases in individuals with metabolic diseases, such as obesity, diabetes, and hypertension35,36. The increasing abundance of Akkermansia muciniphila with aging in postmenopausal controls needs further investigation; however, Akkermansia muciniphila was found to be more abundant in elderly people than in young adults37. Furthermore, a slightly lower abundance of A. muciniphila was observed in Chinese centenarians than in Chinese elderly subjects38. In breast cancer patients, a high abundance of Akkermansia muciniphila was correlated with lower fat mass and higher diversity39. Phascolarctobacterium can produce short-chain fatty acids (SCFAs) and is decreased in individuals with many cancer types31,40. Regarding specific microbes in postmenopausal breast cancer patients, Proteobacteria is a phylum enriched with pathogenic bacteria and is a marker of dysbiosis/disease41. Klebsiella pneumoniae is also a pathogenic bacterium that elevates risk and promotes colorectal cancer development through the bacterial toxin colibactin42. Of the universal microbial markers, Faecalibacterium prausnitzii is a critical microbial marker for the generation of SCFAs and shows an antitumor effect in breast cancer43. In addition, Faecalibacterium prausnitzii is an index of dysbiosis, which indicates lower diversity and loss of tumor-suppressor-like microbes in premenopausal breast cancer patients44. Ruminococcus gnavus is a Crohn’s disease-related microbe that is reduced in gastrointestinal cancer45,46. Rothia mucilaginosa mainly exists in the oral cavity and is reduced in several cancer types47,48. The upregulation of Sutterella and Haemophilus parainfluenzae, which are pathogenic bacteria, is associated with autism, ulcerative colitis, and oropharyngeal cancer47,49,50.
To further clarify which microbial marker is critical in breast cancer according to age fluctuation, especially in premenopausal young breast cancer patients. Two populations, control and breast cancer populations, were divided according to the same age parameters. Correlation analysis provided critical evidence of two species of Bacteroides fragilis in young women with premenopausal breast cancer, which provides the first clinical evidence and is supported by the findings of Parida et al.34. Another species of Klebsiella pneumoniae in postmenopausal women with older breast cancer requires further investigation. In addition to microbial markers of the different menopausal statuses of breast cancer patients, we provided a novel noninvasive approach with a combination of multiple microbial markers for early detection of the different menopausal statuses of breast cancer patients. The concept of screening microbiomes of noninvasive specimens could be a future strategy for the diagnosis or prevention of the disease and has been indicated for many cancer types22,31,51.
In functional pathway analysis, microbes of premenopausal breast cancer patients were found to be involved in steroid-related aromatic and androstenedione degradation. Aromatic amines are byproducts of the manufacturing of rubber, industrial chemicals, dyes, etc. Females exposed to aromatic amines, especially during puberty and childbirth, have an increased risk of DNA damage and the development of breast cancer52,53. In the pathway of androstenedione degradation, which is converted to estrogen via the aromatase enzyme in the ovaries of premenopausal women. In addition to steroid-related pathways, the oncogenic-related pathway may be partially related to the activation of the Notch and β-catenin pathways of Bacteroides fragilis34. The microbiome of postmenopausal breast cancer patients is involved in chemical carcinogenesis and aldosterone-related pathways. In postmenopausal women, aldosterone levels will increase due to lower estrogen levels and may be involved in breast cancer54.
In fact, the involvement of the gut microbiota in breast cancer was proposed in 1981, when constipation was found to increase the risk of breast cancer55. The gut microbiota is dynamic and an adjustable target in which diet, probiotics, and prebiotics could reduce the risk of breast cancer56. The therapeutic goal is termed “rebiosis”, which means restoring a healthy and highly diverse microbial environment23. A meta-analysis of a prospective study showed that total fiber (especially soluble fiber) consumption was associated with an 8% lower risk of breast cancer57. Moreover, in a study of an Asian population, regular consumption of Lactobacillus casei Shirota (BLS), soy isoflavone, and vitamin D reduced breast cancer risk in premenopausal women, whereas alcohol consumption was significantly associated with a higher risk of breast cancer, specifically in premenopausal women58–60.
In conclusion, we comprehensively analyzed the microbial profiles, diagnostic values, and functional pathways in breast cancer patients with different menopausal statuses. In addition, we provided the first evidence that the gut microbiota in premenopausal breast cancer patients differs from that in postmenopausal breast cancer patients and revealed menopausal-specific microbial markers for diagnosis and investigation. Premenopausal breast cancer patients were characterized by a lower diversity of dysbiosis, a lower abundance of probiotics with tumor suppressors, DNA damage, and SCFA production, whereas postmenopausal breast cancer patients were characterized by dysbiosis of increasing pathogenic bacteria, providing a noninvasive approach for breast cancer detection and a novel strategy for preventing premenopausal breast cancer in the future.
Supplementary information
Acknowledgements
The authors would like to thank the YongLin Healthcare Foundation for their sponsorship and Germark Biotechnology Co., Ltd. for performing the bioinformatics analyses. This study was supported by the YongLin Healthcare Foundation under study protocol No. QCR18001 and supported by grant 109-2314-B-037-142-MY2 from the Ministry of Science and Technology and grants KMUH104-4M26, and KMUH-DK(C)110005 from the Kaohsiung Medical University Hospital.
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study; the collection, analyses, or interpretation of data; the writing of the manuscript, or the decision to publish the results.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ming-Feng Hou, Fu Ou-Yang, Chung-Liang Li.
Supplementary information
The online version contains supplementary material available at 10.1038/s12276-021-00686-9.
References
- 1.Sung H, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Huang CS, Lin CH, Lu YS, Shen CY. Unique features of breast cancer in Asian women–breast cancer in Taiwan as an example. J. Steroid Biochem. Mol. Biol. 2010;118:300–303. doi: 10.1016/j.jsbmb.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 3.Lin CH, et al. Contrasting epidemiology and clinicopathology of female breast cancer in Asians vs the US population. J. Natl Cancer Inst. 2019;111:1298–1306. doi: 10.1093/jnci/djz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narod SA. Breast cancer in young women. Nat. Rev. Clin. Oncol. 2012;9:460–470. doi: 10.1038/nrclinonc.2012.102. [DOI] [PubMed] [Google Scholar]
- 5.Cheng SH, et al. Unique features of breast cancer in Taiwan. Breast Cancer Res. Treat. 2000;63:213–223. doi: 10.1023/A:1006468514396. [DOI] [PubMed] [Google Scholar]
- 6.Lin PH, et al. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget. 2016;7:8310–8320. doi: 10.18632/oncotarget.7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang J, et al. Impact of air pollution on breast cancer incidence and mortality: a nationwide analysis in South Korea. Sci. Rep. 2020;10:5392. doi: 10.1038/s41598-020-62200-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang S, El-Zaemey S, Heyworth J, Tang MC. DDT exposure in early childhood and female breast cancer: evidence from an ecological study in Taiwan. Environ. Int. 2018;121:1106–1112. doi: 10.1016/j.envint.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Blandino G, Inturri R, Lazzara F, Di Rosa M, Malaguarnera L. Impact of gut microbiota on diabetes mellitus. Diabetes Metab. 2016;42:303–315. doi: 10.1016/j.diabet.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 11.Festi D, et al. Gut microbiota and metabolic syndrome. World J. Gastroenterol. 2014;20:16079–16094. doi: 10.3748/wjg.v20.i43.16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchesi JR, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 13.Sampson TR, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Martel C, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 15.Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host. Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen KL, Madak-Erdogan Z. Estrogen and microbiota crosstalk: should we pay attention? Trends Endocrinol. Metab. 2016;27:752–755. doi: 10.1016/j.tem.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Flores R, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J. Transl. Med. 2012;10:253. doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miko E, et al. Microbiome-microbial metabolome-cancer cell interactions in breast cancer-familiar, but unexplored. Cells. 2019;8:293. doi: 10.3390/cells8040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih CT, Yeh YT, Lin CC, Yang LY, Chiang CP. Akkermansia muciniphila is negatively correlated with hemoglobin A1c in refractory diabetes. Microorganisms. 2020;8:1360. doi: 10.3390/microorganisms8091360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CH, et al. Molecular subtypes of breast cancer emerging in young women in Taiwan: evidence for more than just westernization as a reason for the disease in Asia. Cancer Epidemiol. Biomark. Prev. 2009;18:1807–1814. doi: 10.1158/1055-9965.EPI-09-0096. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6:136. doi: 10.1186/s40168-018-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosca A, Leclerc M, Hugot JP. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front. Microbiol. 2016;7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, et al. Compositional and functional features of the female premenopausal and postmenopausal gut microbiota. Febs. Lett. 2019;593:2655–2664. doi: 10.1002/1873-3468.13527. [DOI] [PubMed] [Google Scholar]
- 25.Mayneris-Perxachs J, et al. Gut microbiota steroid sexual dimorphism and its impact on gonadal steroids: influences of obesity and menopausal status. Microbiome. 2020;8:136. doi: 10.1186/s40168-020-00913-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Toole PW, Claesson MJ. Gut microbiota: changes throughout the lifespan from infancy to elderly. Int. Dairy J. 2010;20:281–291. doi: 10.1016/j.idairyj.2009.11.010. [DOI] [Google Scholar]
- 27.Arboleya S, Watkins C, Stanton C, Ross RP. Gut Bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik SS, et al. Anticarcinogenecity of microbiota and probiotics in breast cancer. Int. J. Food Prop. 2018;21:655–666. doi: 10.1080/10942912.2018.1448994. [DOI] [Google Scholar]
- 29.Wei H, et al. Antitumor mechanisms of bifidobacteria. Oncol. Lett. 2018;16:3–8. doi: 10.3892/ol.2018.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walther-Antonio MR, et al. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 2016;8:122. doi: 10.1186/s13073-016-0368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren Z, et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014–1023. doi: 10.1136/gutjnl-2017-315084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, et al. Dysbiosis of the gut microbiome is associated with thyroid cancer and thyroid nodules and correlated with clinical index of thyroid function. Endocrine. 2019;64:564–574. doi: 10.1007/s12020-018-1831-x. [DOI] [PubMed] [Google Scholar]
- 33.Cheng WT, Kantilal HK, Davamani F. The mechanism of Bacteroides fragilis toxin contributes to colon cancer formation. Malays. J. Med. Sci. 2020;27:9–21. doi: 10.21315/mjms2020.27.4.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parida S, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and beta-catenin axes. Cancer Discov. 2021;11:1138–1157. doi: 10.1158/2159-8290.CD-20-0537. [DOI] [PubMed] [Google Scholar]
- 35.Cani PD, de Vos WM. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front. Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 37.Biagi E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PloS ONE. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F, et al. Gut microbiota community and its assembly associated with age and diet in Chinese centenarians. J. Microbiol. Biotechnol. 2015;25:1195–1204. doi: 10.4014/jmb.1410.10014. [DOI] [PubMed] [Google Scholar]
- 39.Fruge AD, et al. Fecal Akkermansia muciniphila is associated with body composition and microbiota diversity in overweight and obese women with breast cancer participating in a presurgical weight loss trial. J. Acad. Nutr. Diet. 2020;120:650–659. doi: 10.1016/j.jand.2018.08.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, et al. Altered diversity and composition of the gut microbiome in patients with cervical cancer. Amb. Express. 2019;9:40. doi: 10.1186/s13568-019-0763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Strakova N, Korena K, Karpiskova R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development—a systematic review. Toxicon. 2021;197:126–135. doi: 10.1016/j.toxicon.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 43.Ma J, et al. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 2020;20:82. doi: 10.1186/s12866-020-01739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iebba V, et al. Eubiosis and dysbiosis: the two sides of the microbiota. N. Microbiol. 2016;39:1–12. [PubMed] [Google Scholar]
- 45.Henke MT, et al. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl Acad. Sci. USA. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, N. et al. Characterization of the fecal microbiota in gastrointestinal cancer patients and healthy controls. Preprint at https://www.researchsquare.com/article/rs-37119/v1 (2020). [DOI] [PubMed]
- 47.Panda M, et al. Alterations of salivary microbial community associated with oropharyngeal and hypopharyngeal squamous cell carcinoma patients. Arch. Microbiol. 2020;202:785–805. doi: 10.1007/s00203-019-01790-1. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Y, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. 2020;11:1030–1042. doi: 10.1080/19490976.2020.1737487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, et al. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun M, et al. Combined signature of the fecal microbiome and plasma metabolome in patients with ulcerative colitis. Med. Sci. Monit. 2019;25:3303–3315. doi: 10.12659/MSM.916009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mager DL, et al. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J. Transl. Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Vocht F, et al. Cancer mortality and occupational exposure to aromatic amines and inhalable aerosols in rubber tire manufacturing in Poland. Cancer Epidemiol. 2009;33:94–102. doi: 10.1016/j.canep.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 53.Heikkinen S, Pitkaniemi J, Sarkeala T, Malila N, Koskenvuo M. Does hair dye use increase the risk of breast cancer? a population-based case-control study of finnish women. PLoS ONE. 2015;10:e0135190. doi: 10.1371/journal.pone.0135190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rigiracciolo DC, et al. GPER is involved in the stimulatory effects of aldosterone in breast cancer cells and breast tumor-derived endothelial cells. Oncotarget. 2016;7:94–111. doi: 10.18632/oncotarget.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrakis NL, King EB. Cytological abnormalities in nipple aspirates of breast fluid from women with severe constipation. Lancet. 1981;2:1203–1204. doi: 10.1016/S0140-6736(81)91441-0. [DOI] [PubMed] [Google Scholar]
- 56.Laborda-Illanes A, et al. Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers (Basel) 2020;12:2465. doi: 10.3390/cancers12092465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farvid MS, Spence ND, Holmes MD, Barnett JB. Fiber consumption and breast cancer incidence: A systematic review and meta-analysis of prospective studies. Cancer. 2020;126:3061–3075. doi: 10.1002/cncr.32816. [DOI] [PubMed] [Google Scholar]
- 58.Toi M, et al. Probiotic beverage with soy isoflavone consumption for breast cancer prevention: a case-control study. Curr. Nutr. Food Sci. 2013;9:194–200. doi: 10.2174/15734013113099990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee MS, et al. Vitamin D decreases risk of breast cancer in premenopausal women of normal weight in subtropical taiwan. J. Epidemiol. 2011;21:87–94. doi: 10.2188/jea.JE20100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iwase M, et al. Alcohol consumption and breast cancer risk in Japan: a pooled analysis of eight population-based cohort studies. Int. J. Cancer. 2021;148:2736–2747. doi: 10.1002/ijc.33478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.