Abstract
The COVID-19 pandemic worldwide has resulted in over 176 million cases and roughly 3.8 million deaths so far. We could analyze mutation dynamics across the genome from countries such as the USA, Italy, the UK, France, Brazil, and India considering the rapid mutations of the SARS-CoV-2 genome. The analysis would help us to understand the genome diversity, the implications of the mutations in protein stability, and viral transmission. Among the 11 genes, surface glycoprotein (S) was singled out because of its crucial function associated with the entry of virion into the human cell upon binding with the hACE2 receptor. 749 S protein sequences from India were retrieved from the NCBI database for our study. The S protein is an important antigenic component responsible for inducing host immune responses, neutralizing antibodies, and providing protective immunity against viral infection. During an epitope prediction from a mutation-prone S-protein region, it is necessary to ascertain how new mutations significantly change the S protein, such that our vaccine is effective against all the mutated strains as well. The S1 region of the S protein had been our prime focus for identifying immune epitopes against SARS-COV-2. Antigenic B- cell epitopes were YYPDKVF from NTD and LFRKSNLKP from RBD. Cytotoxic T-cell epitopes WTAGAAAYY (within NTD) and CVADYSVLY (within RBD) exhibited binding with a maximum number of MHC I alleles. The T-cell epitopes which showed a maximum affinity for MHC II alleles were FLPFFSNVT within NTD and YFPLQSYGF within RBD. Furthermore, the best epitopes were characterized in terms of their physicochemical properties to establish their potentiality.
Keywords: SARS-CoV-2, COVID-19, Spike protein, Mutation profile, B and T-cell epitopes
Introduction
A novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), widely recognized as the COVID-19 has led to the eruption of the global pandemic. As of April 2021, 223 countries, areas, or territories have reported 176,484,692 laboratory-confirmed cases of COVID-19 that originated from Wuhan, China, and the death toll worldwide has surpassed 3.8 million [26] (World Health Organization-WHO). The most common symptoms of SARS-CoV-2 are fever, coughing, and breathlessness, while 2% of the infected people show severe symptoms like pneumonia, multi-organ failure, acute respiratory illness, and even death. Aged individuals (> 60–65 years of age) and people with underlying cardiovascular, immunological, metabolic, and respiratory comorbidities are more susceptible to the severe COVID-19 compared to children and young adults [1, 14]. The virus has been found to inhabit the bronchoalveolar-lavage, sputum, saliva, throat, and nasopharyngeal regions [22]. The SARS-CoV-2 transmission is predominantly airborne, attributed to contact via respiratory droplets or aerosols either during coughing, sneezing, talking, or through surface contact [8]; and it revolves around humans, animals, and the environment [1].
Based on the differences in protein sequence, the coronavirus sub-family has four genera -alpha, beta, gamma, and delta coronaviruses [24]. SARS-CoV-2 belongs to the Betacoronavirus genus which includes severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), bat SARS-related coronaviruses, as well as others identified in humans and diverse animal species. Bat coronavirus RaTG13 appears to be the closest relative of the SARS-CoV-2, sharing more than 93.1% sequence identity in the spike (S) gene. SARS-CoV is however, distinct from SARS-CoV-2 and shares less than 80% sequence identity [15]. SARS-CoV-2 is reported to have originated in bats and subsequently transmitted to humans via pangolins which serve as intermediate host species. To cross a species barrier and infect a new mammalian host, the virus undergoes mutations in the spike proteins/surface glycoproteins (S) [8].
Structural aspects of SARS-CoV-2 genome
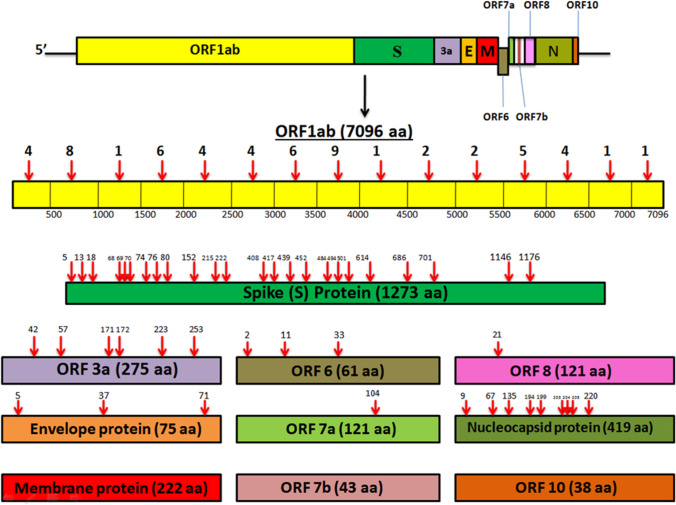
The SARS-CoV-2 virion has enveloped, positive-sense, single-stranded ribonucleic acid (RNA) harboring 26–32 kB nucleotides. The genome possesses 11 genes and 11 open reading frames (ORFs) encoding 27 proteins [14, 33]. More than two-thirds of the genome comprises two large overlapping ORF (ORF1a and ORF1b). A set of non-structural proteins (NSP1-16) are generated upon proteolytic cleavage of ORF1a and ORF1b polyprotein (pp1ab). The NSPs help in viral replication and transcription. The NSP12 functions as RNA-dependent RNA Polymerase (RdRp) that regulates the synthesis of viral RNA with the assistance of NSP7 and NSP8 as cofactors [18]. Other NSPs encoded by the ORFs include 3-chymotrypsin-like protease, papain-like protease, etc. [10]. The rest of the genome contains ORFs for the structural proteins namely Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) proteins [18]. The S protein possesses two subunits- S1 and S2 which mediates virus entry (receptor binding and entry of virion into host cells) and membrane fusion respectively. The M protein determines the shape of the virus envelope and stabilizes the nucleocapsids. The N protein regulates viral replication and mediates the response of host cells to viral infection. The E protein- the smallest structural protein plays a role in the assembly, production, and maturation of the virus [29]. ORF3a, ORF 6, ORF 7a, ORF 7b, ORF 8, and ORF 10 represent the accessory proteins that mediate viral replication and regulate virus-host interactions [33].
The S protein of SARS-CoV-2 is 180–200 kDa and consists of 1273 amino acid residues. Trimeric conformation of the S protein is extensively glycosylated, which is essential for proper folding, and for enhancing accessibility to host proteases and neutralizing antibodies (nAbs) [30]. The S protein encompasses a signal peptide (1–12) located at the N-terminus, the membrane distal S1 subunit (13–710), and the membrane-proximal S2 subunit (711–1273), which cumulatively occurs in the viral envelope as a homotrimer. The S1 and S2 subunits mediate receptor recognition & binding and membrane fusion respectively [5, 10]. The S1 subunit consists of an N-terminal domain (13–302) and a Receptor Binding Domain (RBD, 333–527). The S2 subunit consists of a fusion peptide (FP, 816–828), a heptad repeat region 1 (HR1, 912–983), an HR2 (1134–1213), a transmembrane domain (TM, 1214–1236), and a cytoplasmic domain (CT, 1237–1273) (Fig. 1). The S2 subunit also consists of some structural elements which include three long α helices, multiple α helical segments, extended twisted β sheets, membrane-spanning α helix, and an intracellular cysteine-rich segment [8]. The S protein usually exists in a metastable inactive prefusion state. Upon viral infection, the cellular proteases together with serine protease TMPRSS2 activate the S protein by splitting it into S1 and S2 subunits [10]. The PRRA sequence located between S1 and S2 serves as a furin cleavage site. In the S2 subunit, at the upstream of the fusion peptide, an additional proteolytic cleavage site is present. Both these cleavage sites regulate viral entry into host cells [8].
Fig. 1.
Monomeric and Trimeric state of the Spike protein. Each spike monomer/protomer consists of S1 and S2 subunits, connected at the furin cleavage site. The overview illustrates S1 and S2 regions displaying major domains. The spike protein comprises three spike monomers, which form a homotrimeric structure that is necessary for viral entry into the host cell. Viral fusion occurs via the fusion of the receptor-binding domain (RBD) with the host cell target receptor (ACE2). The fusion of the membrane is mediated by the S2 region. NTD- N terminal domain, and RBD- receptor binding domain of S1 region of spike protein; HR1- Heptad repeat region 1, HR2- Heptad repeat region 2, TM- Transmembrane domain, and CTD- the cytoplasmic tail domain of S2 region of the spike protein
The S1-NTD might be involved in virus attachment to host cells by recognizing specific carbohydrate molecules like sialic acid [8]. The NTD also plays an important role in the transition from prefusion to postfusion conformation [5]. The S1-NTD can be recognized as a potential target in the coronavirus (CoV) vaccine. The nAbs targeting NTD do not directly interfere with receptor binding, but they prevent conformational changes in the S protein that are necessary for the postfusion transformation of prefusion cells. Compared to RBD-specific nAbs, SARS-CoV-2 NTD-directed nAbs had lower neutralizing potency. It has been shown in a MERS-CoV mouse model that vaccination with NTD results in nAbs and NTD-specific T-cell responses [5]. Despite the significantly lower antigenicity of the NTD protein than the RBD protein, the NTD protein can still be regarded as a potential vaccine against COVID-19. COVID-19 vaccines based on NTD have not been reported yet. Angiotensin-converting enzyme (hACE2) receptor interacts with the RBD of the S1 subunit in the region where aminopeptidase N resides. The lungs, the intestine, the heart, the kidneys, and the alveolar epithelial type II cells display ACE2 on their surfaces. Viral entry is dependent upon RBD's binding to hACE2 [10]. The RBD can be modified to enhance its interactions with ACE2 which, in turn, could alter the virus' ability to infect [10]. The entire surface of S protein remains guarded by glycans to evade recognition by host cell antibodies except for RBD, which goes on to elucidate the predominance of RBD epitopes. nAbs targeted against RBD obstruct the RBD-hACE2 interaction, thus preventing virus attachment [5]. Thus, both NTD and RBD can be targeted to forestall infection.
DNA viruses have mutation rates on the order of 108 to 106 substitutions per nucleotide site per cell infection (s/n/c). On the contrary, RNA viruses have high mutation rates that range between 106 and 104 s/n/c [21]. High per-site mutation rates are attributed to RdRp, which governs the replication of RNA viruses. The RdRp lacks proofreading activity and thus fails to correct the replication errors, leading to mutations in the genome [21]. Thus, while some mutations in the SARS-CoV-2 virus lead to a novel RdRp variant, other amino acid changes enhance the transmissibility of the virus. This has a significant evolutionary advantage, as demonstrated in the SARS-CoV-2 variant exhibiting D614G mutation [19]. The continuous formation of mutants favors the adaptability of viruses in different environmental conditions. In a constant environment, the occurrence of mutations is improbable and in a perfectly adapted environment, the rate of beneficial mutation will be zero. On the contrary, if an organism is exposed to a new environment, the potential rate of beneficial mutation will be non-zero. From the above fact, it can be derived that the SARS-CoV-2 genome shows more deleterious and neutral mutations when compared to beneficial mutations as it has adapted to the environment in the past year [18]. Based upon computational studies, a strain with a high mutation rate can strengthen its adaptability in the short term, but eventually, it perishes in the due course of time owing to the accretion of deleterious mutations [21]. Thus, a sequence with a high number of deleterious mutations will get extinct easily owing to its increased mutation load, and as a result, it tends to lose out to competitors showing low mutations [21]. A missense mutation (change in single amino acid) within an antigenic determinant or epitope becomes resistant to nAbs. So, the ability to identify an epitope is critical for potential vaccine design and disease diagnosis [18].
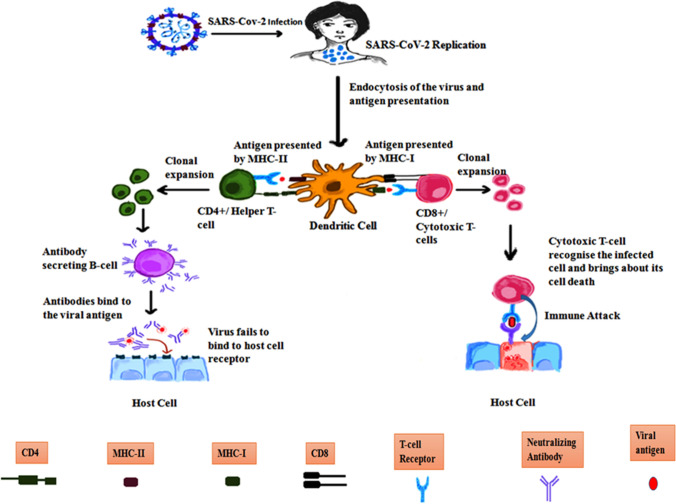
Identification of epitopes on antigens is vital for a better understanding of the disease etiology, disease prognosis, which eventually facilitates in developing diagnostic assays and epitope-based vaccines. The immune system is categorized into 2 major types- innate and adaptive. Innate immunity involves non-specific defense mechanisms that act spontaneously, or within hours after microbial entry into the body. On the contrary, adaptive immunity is highly specific and is mediated by lymphocytes, specifically by B- and T-cells, which are responsible for humoral and cell-mediated immunity. The B and T cells recognize molecular segments of the virus known as antigen employing receptors present on their cell surface [28]. The B cells derived from the bone marrow produce neutralizing antibodies that bind to virus particles stopping the spread of infection [9]. The T cells, that mature in the thymus, play a crucial role in the immune response to viral infections by eliminating virus-infected cells. The T-cell receptor recognizes the virus-derived peptides when they are presented on the surface of the infected cell by the Major Histocompatibility Complex (MHC) [9, 25]. The CD8 + T cells recognize viral antigens through their presentation by MHC I molecules, and upon recognition, the T cells become activated by clonal expansion and identical antigen recognition [25]. The CD8 + T cell clones, also called the cytotoxic T-cells, bring about lysis of the infected cell by the release of perforin/granzyme and induce apoptosis through tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or Fas ligand, and proinflammatory mediators such as interferon-γ (IFN-γ). If the CD4 + is capable of binding to the viral antigen presented by MHC II molecules, then it leads to activation of B-cells that identifies the antigen by causing them to clonally proliferate and secrete antibodies to target different parts of the SARS-CoV-2 virus [25]. Some memory B-cells procured from the plasma B-cells keep the invader's memory to exhibit faster recognition and quick response during subsequent encounters with the invader [2]. The T-cell mediated response in SARS-CoV-2 is shown in Fig. 2.
Fig. 2.
T-cell mediated response to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2).The virus gains entry into the human body upon binding with the hACE2 receptor present on the cell membrane of various cell types. As soon as the virus begins to replicate, the immune cells are alerted. This is followed by endocytosis of the virus into dendritic cells and antigen degradation. The antigen fragments are then presented on the cell surface by MHC class I and MHC class II molecules facilitating its binding with a T-cell. When CD8 + T-cells bind to antigens, MHCI molecules present the antigen—this causes CTLs to multiply and target host cells infected with the pathogen, ultimately leading to their death. If CD4 + T-cell binds, it eventually activates B-cells which secrete neutralizing antibodies that associate with different regions of the virus thereby preventing it from binding with the ACE2 receptor expressed on various cell surfaces
Traditional vaccine development is a long complex process, which involves growing pathogens, isolating, attenuating, and administering the disease-causing organism into the human body [32]. Thus, conventional vaccine design is a cumbersome and a time taking process. It is often associated with several drawbacks such as difficulties encountered during culture, isolation, attenuation of the virus, and the recognition of genetic variations that produce new strains [2]. Scientists have embraced the immunoinformatics approach for the design of multiepitope vaccine (MEV). One major benefit of the epitope vaccine is its high efficacy in both pre-exposure and post-exposure periods, which is crucial to combat repeated infection by SARS-CoV-2 in the same individual [2].
Our study is focused on intricately analyzing the mutation profile of the S protein of SARS-CoV-2 which is key for the viral attachment to human host cells. The study is further substantiated by the prediction of B-cell and T-cell (MHC class I and II) epitopes primarily within NTD and RBD of S protein, and their immunogenic properties. The S protein from the SARS-CoV-2 reference strain (YP_009724390.1) was undertaken to characterize its antigens as potential vaccine candidates.
Materials and methods
Distribution of mutations in whole-genome sequences from major countries affected by SARS-CoV-2
Genome analysis
Sequences of all proteins obtained from the SARS-CoV-2 genome (ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8, N, and ORF10) were collected from the NCBI database (https://www.ncbi.nlm.nih.gov/) in FASTA format. 6 countries were included in the study based on their high incidence rates—India, United States of America (USA), Brazil, United Kingdom (UK), France, and Italy. The sequences were studied thoroughly by comparing with the reference SARS-CoV-2 protein sequences from Wuhan-Hu-1, China. These countries recorded a high number of COVID-19 cases or showed a high rate of mortality.
Multiple sequence alignment
Clustal Omega was picked to align ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF 8, N, and ORF10 proteins of SARS-CoV-2 sequences originated from the above-mentioned countries relative to the reference protein sequences from Wuhan-Hu-1, China. Thus, multiple sequence alignment was performed to identify the non-synonymous mutation from all the protein sequences. The position of missense mutations and amino acid substitutions were noted.
Effect/outcome of amino acid alterations
The PROVEAN algorithm was used to study the functional impact of the non-synonymous mutations. It is an efficient method that computes the pairwise alignment scores between a protein sequence and numerous single-locus variations of a previously developed protein [3]. PROVEAN makes use of delta scores to scrutinize amino acid alterations. In our present study, the cut-off for PROVEAN scores was set to 2.5. Low delta scores indicate amino acid alterations which have a deleterious effect on protein function, while high delta scores indicate variations with a neutral impact on protein function.
Distribution of mutations in the spike glycoprotein (S) from India
Spike protein sequence retrieval
All the spike protein sequences from India contained in the NCBI database till 31st March 2021 were analyzed for our study. The sequences were retrieved in FASTA format.
Multiple sequence alignment
The multiple sequence alignment of the proteins was achieved using Clustal Omega. The human SARS-CoV-2 spike protein sequence from Wuhan-Hu-1, China (NCBI accession code: YP_009724390.1)1 was used as the reference sequence to study the mutation profile. The mutations within different regions of the S protein were analyzed thoroughly.
Effect of amino acid alterations
Again, the PROVEAN algorithm was used to detect neutral and deleterious mutations in the S protein sequence [3].
Epitope prediction for future vaccine construct from S protein
Detection of possible epitopes within S1 and S2 regions of the S protein
For identifying the antigenic epitopes within the S1 region (comprising the NTD and RBD) and S2 region proteins sequences, an in silico analysis was conducted. The antigenicity score was determined for the 3 sequences- NTD, RBD, and S2 subunit of S protein through VaxiJen v2.0 [6]. The threshold value was set at 0.4. VaxiJen v2.0 involves an alignment-free estimation of antigenicity based on physicochemical properties of amino acids. The immunogenicity was determined using IEDB Analysis Resource. Allergenicity was checked through the AllergenFP server to ensure the peptide does not elicit an allergic reaction when introduced into the human body.
B-cell epitope prediction within NTD and RBD of S protein
For B-Cell epitope prediction, Immune Epitope Database (IEDB) was used (https://www.iedb.org/). The reference sequence of SARS-CoV-2 from Wuhan, China was used. Five different methods were employed for this detection- Kolaskar & Tongaonkar antigenicity, Emini surface accessibility, BepiPred linear epitope prediction, Karplus & Schulz flexibility, and Chou & Fasman beta-turn method [4, 7, 12, 13, 16]. The B-cell epitopes were also predicted using ElliPro, which identifies linear and discontinuous epitopes based on a protein antigen's 3D structure [23, 27].
Cytotoxic T-cell (CTL) epitope prediction from NTD and RBD of S protein
CTL epitopes were predicted through NetCTL 1.2 server, the information of which is necessary for potential vaccine design. It is a web-based tool that integrates three approaches-predictions of proteasomal cleavage, TAP transport efficiency, and major histocompatibility complex (MHC) class I affinity to detect possible epitopes [17, 27]. The NetMHCI 4.0 server was used to predict the binding affinity of epitopes with 81 different HLA-A, HLA-B, HLA-C, and HLA-E human MHC-I alleles.
Helper T- cell epitope prediction from NTD and RBD of S protein
Understanding which peptides will be displayed by the MHC-II molecule is critical for the activation of Helper-T cells and can be used to identify T-cell epitopes. Helper T-cell epitopes were predicted by NetMHCII 2.3 server [11, 27]. The epitopes having a high affinity towards HLA-DR were taken into consideration. The formidable epitope length was set to 9-mer epitopes as HLA molecules exhibit a strong affinity for 9-mer epitopes.
Profiling and mapping of B and T-cell epitopes
At first, the antigenicity of the peptide was estimated using the Immunomedicine Group server. If the antigenic score of a peptide is above 1.00, it is antigenic and competent to elicit an antibody response. For determining the physicochemical properties of the peptide-like molecular weight, charge, hydrophobicity, the AntiAngioPred web-server was used. For an epitope to be deemed effective and stable, it must be scrutinized for digestion by different enzymes. This was done through the PROTEIN ENZYME DIGEST server [27]. It examines digestion of the peptide with eleven different enzymes namely—Trypsin, Chymotrypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline Endopept, Staph Protease, Trypsin R, Trypsin K, AspN, and Elastase.
The NTD and RBD structures from the reference strain were predicted through the RaptorX structure prediction server. It is a distance-based protein folding server powered by deep learning [31]. The predicted B-cell, CTL, Helper T-cell epitopes were separately mapped on structures of NTD and RBD with the aid of BIOVIA Discovery Studio Visualizer.
Results
Spread of mutations in the SARS-CoV-2 genome from isolates of different countries
The SARS-CoV-2 genome has a total of 11 genes with 11 open reading frames (ORFs) namely—ORF1ab, ORF2 (Spike protein), ORF3a, ORF4 (E protein), ORF5 (M protein), ORF6, ORF7a, ORF7b, ORF8, ORF9 (N protein), and ORF10. We have studied the mutation profile of each of these proteins (Tables 1, 2). The mutation density was found to be relatively high in S, ORF3a, E, ORF6, and N, suggesting these are more prone to mutations as compared to others. M, ORF7b, and ORF10 did not show any mutations. ORF1ab which codes for replicase polyprotein that helps in viral replication and transcription spans across a region of 7096 amino acids and exhibits 60 missense mutations of which L3352F, S3884L, L5030F, H5614Y, V6237F are deleterious. At, 4715, an amino acid alteration from proline (P) to leucine (L) is found in 23 sequences (out of the 30 studied) distributed across countries- Brazil(5), Italy(4), USA(4), India(4), France(3), and UK(3). Another mutation site, T265I is found in 9 sequences—5 from the USA, 2 from France, and one each from Brazil and the UK. The Spike or Surface glycoprotein (S) comprises 1273 amino acids with 24 detected missense mutations all of which are neutral. The D614G mutation occurs in 26 sequences- USA(5), Italy(5), France(5), India(4), UK(4), and Brazil(3). This results in enhanced infectivity of the virion. Alongside D614G, V1176F is also associated with a severe outcome of COVID-19 disease [19]. The ORF3a is an ion channel protein, which exhibits 6 substitutions of which Q57H is deleterious and the most predominant, found in 3 sequences from the USA, 3 from France, 1 each from Brazil, UK, and India. The E protein, which is capable of oligomerization to create an ion channel [33], reveals 3 mutation points all of which are deleterious. At position 71, P is substituted to S (USA) and L (France). The M protein is a conserved protein considered to be associated with housekeeping functions and thus has a greater resistance to mutations. An accessory protein ORF6 responsible for viral pathogenesis shows three deleterious amino acid substitutions [33]. I33T found in QRN56062 (Brazil) is deleterious and poses a severe outcome of the disease [19]. The other accessory proteins—ORF7a, ORF7b, ORF8, and ORF10 do not show much variability. The N protein which directly binds to the viral RNA and confers stability [33], shows high mutability with 9 missense mutations of which S194L is deleterious. R203K and G204R mutations occur in sequences from countries like Brazil(1), Italy(3), India(2), and UK(1). These two mutations along with S194L are related to the severity of COVID 19 in patients. Figure 3 shows the mutation sites spread across the whole genome of SARS-CoV-2.
Table 1.
Distribution of mutations in the 11 protein sequences of the SARS-CoV-2 genome considering 6 major countries- USA, India, UK, France, Brazil, and Italy relative to the reference strain from Wuhan, China
| Sl no. | Protein sequence of SARS-CoV-2 genome | Length of the protein (amino acids) | Total no. of missense mutations | Percent mutation coverage/mutation density | No. of neutral mutations | No. of deleterious mutations | List of deleterious mutations |
|---|---|---|---|---|---|---|---|
| 1 | ORF1ab | 7096 | 60 | 0.817 | 55 | 5 | L3352F, S3884L, L5030F, H5614Y, V6237F |
| 2 | Spike/ Surface Glycoprotein | 1273 | 24 | 1.88 | 24 | – | – |
| 3 | ORF3a | 275 | 6 | 2.18 | 1 | 5 | P42L, Q57H, G172V, T223I, S253P |
| 4 | Envelope Protein | 75 | 4 | 4.00 | – | 4 | V5A, L37H, P71S, P71L |
| 5 | Membrane Protein | 222 | 0 | – | – | – | – |
| 6 | ORF6 | 61 | 3 | 4.91 | – | 3 | F2M, I11T, I33T |
| 7 | ORF7a | 121 | 1 | 0.82 | – | 1 | V104F |
| 8 | ORF7b | 43 | 0 | – | – | – | – |
| 9 | ORF8 | 121 | 1 | 0.82 | 1 | – | – |
| 10 | Nucleocapsid Protein | 419 | 9 | 2.14 | 8 | 1 | S194L |
| 11 | ORF10 | 38 | 0 | – | – | – | – |
The account of deleterious mutations was obtained from the PROVEAN algorithm. The mutation density was calculated as a function of the total number of mutations points divided by the length of the sequence corresponding to different protein regions of the SARS-CoV-2 genome
Table 2.
A detailed account of the missense mutations prevailing in the 11 protein sequences of the SARS-CoV-2 genome from isolates of six different countries- India, USA, UK, France, Italy and Brazil
| Position | Missense mutation | SARS-CoV-2 strain |
|---|---|---|
| ORF1ab protein | ||
| 28 | Valine (V)  Isoleucine (I) Isoleucine (I) |
USA_QQN00979.1 |
| 190 | Phenylalanine (F)  Leucine (L) Leucine (L) |
USA_QQG71397.1 |
| 265 | Threonine (T)  Isoleucine (I) Isoleucine (I) |
Brazil_QLD32026.1 |
| United Kingdom_QMX86939.1 | ||
| France_QLJ84623.1 | ||
| USA_ QQN00979.1 | ||
| France_QRI43205.1 | ||
| USA_QTP77835.1 | ||
| USA_QQM19139.1 | ||
| USA_QQG71397.1 | ||
| USA_QTP77978.1 | ||
| 496 | Tyrosine (Y)  Cysteine (C) Cysteine (C) |
Italy_QTK16400.1 |
| 671 | Isoleucine (I)  Threonine (T) Threonine (T) |
India_QHS34545.1 |
| 680 | Lysine (K)  Asparagine (N) Asparagine (N) |
USA_QQG71397.1 |
| 739 | Isoleucine (I)  Valine (V) Valine (V) |
France_QJT72084.1 |
| 757 | Valine (V)  Phenylalanine (F) Phenylalanine (F) |
Brazil_QRN46959.1 |
| 765 | Proline (P)  Serine (S) Serine (S) |
France_QJT72084.1 |
| 798 | Lysine (K)  Asparagine (N) Asparagine (N) |
USA_QTP77978.1 |
| 820 | Proline (P)  Glutamine (Q) Glutamine (Q) |
USA_ QQN00979.1 |
| 874 | Valine (V)  Alanine (A) Alanine (A) |
USA_QQG71397.1 |
| 1001 | Threonine (T)  Isoleucine (I) Isoleucine (I) |
Italy_QTK16400 |
| Italy_QQI07512.1 | ||
| 1515 | Serine (S)  Phenylalanine (F) Phenylalanine (F) |
India_QKM77226.1 |
| 1567 | Threonine (T)  Alanine (A) Alanine (A) |
Italy_QQI07512.1 |
| 1655 | Lysine (K)  Asparagine (N) Asparagine (N) |
France_QRI43205.1 |
| 1708 | Alanine(A)  Aspartic Acid (D) Aspartic Acid (D) |
Italy_QTK16400 |
| Italy_QQI07512.1 | ||
| 1812 | Alanine(A)  Aspartic Acid (D) Aspartic Acid (D) |
India_QPB40113.1 |
| 1946 | Glycine (G)  Serine(S) Serine(S) |
USA_QTP77978 |
| 2124 | Threonine (T)  Isoleucine (I) Isoleucine (I) |
Italy_QTK16400.1 |
| 2144 | Proline (P)  Serine (S) Serine (S) |
India_QHS34545.1 |
| 2228 | Asparagine (N)  Threonine (T) Threonine (T) |
United Kingdom_QOD07307 |
| 2230 | Isoleucine (I)  Threonine (T) Threonine (T) |
Italy_QTK16400 |
| Italy_QQI07512.1 | ||
| 2501 | Isoleucine (I)  Threonine (T) Threonine (T) |
Italy_QQI07488.1 |
| Italy_QQP27498.1 | ||
| 2596 | Asparagine (N)  Serine (S) Serine (S) |
France_QRI43205.1 |
| 2606 | Methionine (M)  Isoleucine (I) Isoleucine (I) |
USA_QQN00979.1 |
| 2667 | Glycine (G)  Cysteine (C) Cysteine (C) |
USA_QTP77835.1 |
| 3147 | Cysteine (C)  Phenylalanine (F) Phenylalanine (F) |
USA_QQG71397.1 |
| 3201 | Leucine (L)  Proline (P) Proline (P) |
USA_QTP77978.1 |
| 3324 | Lysine (K)  Arginine (R) Arginine (R) |
Italy_QQD89044.1 |
| 3352 | Leucine (L)  Phenylalanine (F) Phenylalanine (F) |
USA_ QQN00979.1 |
| 3353 | Lysine (K)  Arginine (R) Arginine (R) |
France_QRI43205.1 |
| 3456 | Alanine (A)  Valine (V) Valine (V) |
France_QRI43205.1 |
| 3606 | Leucine (L)  Phenylalanine (F) Phenylalanine (F) |
USA_QQM19139.1 |
| France_QJT72084.1 | ||
| United Kingdom _QOD07307.1 | ||
| 3627 | Methionine (M)  Valine (V) Valine (V) |
USA_ QQN00979.1 |
| 3677 | Phenylalanine (F)  Leucine (L) Leucine (L) |
Italy_QTK16400.1 |
| Italy_QQI07512.1 | ||
| France_QRI43205.1 | ||
| USA_QTP77978.1 | ||
| 3718 | Valine (V)  Phenylalanine (F) Phenylalanine (F) |
USA_QTP77835.1 |
| 3752 | Methionine (M)  Isoleucine (I) Isoleucine (I) |
USA_QQG71397.1 |
| 3790 | Cysteine  Phenylalanine (F) Phenylalanine (F) |
USA_QQM19139.1 |
| 3884 | Serine (S)  Leucine (L) Leucine (L) |
Brazil_QLD32026.1 |
| 3930 | Leucine (L)  Phenylalanine (F) Phenylalanine (F) |
Brazil_QRN46959.1 |
| 3966 | Glutamine (Q)  Arginine (R) Arginine (R) |
Italy_QQI07512.1 |
| 4205 | Isoleucine (I)  Valine (V) Valine (V) |
USA_ QQM19139.1 |
| 4715 | Proline (P)  Phenylalanine (F) Phenylalanine (F) |
USA_QTP77835.1 |
| 4715 | Proline (P)  Leucine (L) Leucine (L) |
Italy_QTK16400.1 |
| Italy_QQI07512.1 | ||
| Italy_QQP27498.1 | ||
| Italy_QQD89044.1 | ||
| USA_QQN00979.1 | ||
| France_QRI43205.1 | ||
| France_QLJ84623.1 | ||
| France_QRG33471.1 | ||
| USA_QTP77978.1 | ||
| USA_QQG71397.1 | ||
| USA_QQM19139.1 | ||
| Brazil_QRN46959.1 | ||
| Brazil_QLD32026.1 | ||
| Brazil_QRN56056.1 | ||
| Brazil_QMT98138.1 | ||
| Brazil_QMB22609.1 | ||
| United Kingdom_QPC41142.1 | ||
| United Kingdom_QMX86939.1 | ||
| United Kingdom_QNC68216.1 | ||
| India_QJS39637.1 | ||
| India_QPB40113.1 | ||
| India_QKM77226.1 | ||
| India_QKM77274.1 | ||
| 4798 | Alanine (A)  Valine (V) Valine (V) |
India _QHS34545.1 |
| 5030 | Leucine (L)  Phenylalanine (F) Phenylalanine (F) |
USA_QQN00979.1 |
| 5412 | Glutamine (Q)  Histidine (H) Histidine (H) |
USA_QTP77978.1 |
| 5584 | Aspartic Acid (D) Tyrosine (Y) Tyrosine (Y) |
USA_QQM19139.1 |
| 5614 | Histidine (H)  Tyrosine (Y) Tyrosine (Y) |
Italy_QQI07488.1 |
| Italy_QQP27498.1 | ||
| 5716 | Arginine(R)  Cysteine (C) Cysteine (C) |
Italy_ QQI07488.1 |
| Italy_QQP27498.1 | ||
| 5784 | Lysine (K)  Arginine (R) Arginine (R) |
Italy_QQI07512.1 |
| 5805 | Threonine (T)  Methionine (M) Methionine (M) |
United Kingdom_QPC41142.1 |
| 6054 | Asparagine (N)  Aspartic Acid (D) Aspartic Acid (D) |
USA_QQN00979.1 |
| USA_QTP77835.1 | ||
| 6075 | Isoleucine (I)  Threonine (T) Threonine (T) |
United Kingdom_QOS14143.1 |
| 6237 | Valine (V)  Phenylalanine (F) Phenylalanine (F) |
USA_QQG71397.1 |
| 6245 | Alanine (A)  Valine (V) Valine (V) |
Brazil_QLD32026.1 |
| 6914 | Alanine (A)  Serine (S) Serine (S) |
Italy _QTK16400.1 |
| 6914 | Alanine (A)  Valine (V) Valine (V) |
United Kingdom_QOD07307.1 |
| 7014 | Arginine (R)  Cysteine (C) Cysteine (C) |
USA_QQN00979.1 |
| Spike/surface glycoprotein (S) | ||
| 5 | Leucine (L)  Phenylalanine (F) Phenylalanine (F) |
Italy_QQD89010.1 |
| 13 | Serine (S)  Isoleucine (I) Isoleucine (I) |
USA_QQM19141.1 |
| 18 | Leucine (L)  Phenylalanine (F) Phenylalanine (F) |
France_QRI43207.1 |
| 68 | Isoleucine (I)  Methionine (M) Methionine (M) |
Italy_QQI07490.1 |
| 69 | Histidine (H)  Proline (P) Proline (P) |
Italy_ QQI07490.1 |
| 70 | Valine (V)  Isoleucine (I) Isoleucine (I) |
Italy_ QQI07490.1 |
| 74 | Asparagine (N)  Lysine (K) Lysine (K) |
Brazil_QJA41641.1 |
| 76 | Threonine  Isoleucine (I) Isoleucine (I) |
Italy_QQD89046.1 |
| 80 | Aspartic Acid (D)  Alanine (A) Alanine (A) |
France_QRI43207.1 |
| 152 | Tryptophan (W)  Cysteine (C) Cysteine (C) |
USA_ QQM19141.1 |
| 215 | Aspartic Acid (D)  Glycine (G) Glycine (G) |
France_QRI43207.1 |
| 222 | Alanine (A)  Valine (V) Valine (V) |
Italy_QRZ59106.1 |
| 408 | Arginine (R)  Isoleucine (I) Isoleucine (I) |
India_ QHS34546.1 |
| 417 | Lysine (K)  Asparagine (N) Asparagine (N) |
France_QRI43207.1 |
| 439 | Asparagine (N)  Lysine (K) Lysine (K) |
United Kingdom_QNH88648.1 |
| United Kingdom_QNH88660.1 | ||
| Italy_ QQI07490.1 | ||
| 452 | Leucine (L)  Arginine (R) Arginine (R) |
USA_ QQM19141.1 |
| 484 | Glutamic Acid (E)  Lysine (K) Lysine (K) |
France_QRI43207.1 |
| 494 | Serine (S)  Proline (P) Proline (P) |
United Kingdom_QNH88648.1 |
| 501 | Asparagine (N)  Tyrosine (Y) Tyrosine (Y) |
France_QRI43207.1 |
| 614 | Aspartic Acid (D)  Glycine (G) Glycine (G) |
France_QRI43207.1 |
| France_QLJ84625.1 | ||
| France_QRG33473.1 | ||
| France_QPK91107.1 | ||
| France_QLJ57671.1 | ||
| United Kingdom_QNH88648.1 | ||
| United Kingdom_QNH88660.1 | ||
| United Kingdom_QNC68218.1 | ||
| United Kingdom_QMX86941.1 | ||
| USA_QPD96851.1 | ||
| USA_ QQM19141.1 | ||
| USA_QRJ38103.1 | ||
| USA_QPF81187.1 | ||
| USA_QQN00981.1 | ||
| Italy_QQI07490.1 | ||
| Italy_QQD89046.1 | ||
| Italy_QQD89010.1 | ||
| Italy_QRZ59106.1 | ||
| Italy_QQX94000.1 | ||
| Brazil_QRN46961 | ||
| Brazil_QRN56058.1 | ||
| Brazil_QMT98140.1 | ||
| India_QKM77228.1 | ||
| India_QKM77276.1 | ||
| India_QJS39639.1 | ||
| India_QPB40113.1 | ||
| 686 | Serine (S)  Glycine (G) Glycine (G) |
France_QLJ84625.1 |
| 701 | Alanine (A)  Valine (V) Valine (V) |
France_QRI43207.1 |
| 1146 | Aspartic Acid (D)  Tyrosine (Y) Tyrosine (Y) |
Italy_QQI07490.1 |
| 1176 | Valine (V)  Phenylalanine (F) Phenylalanine (F) |
Brazil_QRN46961.1 |
| ORF3a protein | ||
| 42 | Proline(P)  Leucine (L) Leucine (L) |
USA_QTP80668 |
| 57 | Glutamine (Q)  Histidine (H) Histidine (H) |
USA_QQN00982.1 |
| USA_QQM19142.1 | ||
| USA_QTP80668.1 | ||
| France_QRI43208.1 | ||
| France_QQV74459.1 | ||
| France_QLJ84626.1 | ||
| United Kingdom_QMX86942.1 | ||
| Brazil_QLD32029.1 | ||
| India _QJS39640.1 | ||
| 171 | Serine (S)  Leucine (L) Leucine (L) |
France_QRI43208.1 |
| 172 | Glycine (G)  Valine (V) Valine (V) |
USA_ QQN00982.1 |
| 223 | Threonine (T)  Isoleucine (I) Isoleucine (I) |
USA_QQM19142.1 |
| 253 | Serine (S)  Proline (P) Proline (P) |
Italy_QRX39426 |
| Envelope protein (E) | ||
| 5 | Valine (V)  Alanine (A) Alanine (A) |
Brazil _QMT98142 |
| 37 | Leucine (L)  Histidine (H) Histidine (H) |
United Kingdom_QPC41146.1 |
| 71 | Proline (P)  Serine (S) Serine (S) |
USA_QTI92374.1 |
| 71 | Proline (P)  Leucine (L) Leucine (L) |
France_QRI43209.1 |
| Membrane protein (M) | ||
| No mutations observed | ||
| ORF 6 protein | ||
| 2 | Phenylalanine (F)  Methionine (M) Methionine (M) |
Italy _QTK04753.1 |
| 11 | Isoleucine (I)  Threonine (T) Threonine (T) |
United Kingdom_QPC41148.1 |
| 33 | Isoleucine (I)  Threonine (T) Threonine (T) |
Brazil_QRN56062.1 |
| ORF 7a protein | ||
| 104 | Valine (V)  Phenylalanine (F) Phenylalanine (F) |
Italy_QQI07495.1 |
| ORF 7b protein | ||
| No mutations observed | ||
| ORF 8 protein | ||
| 24 | Serine (S)  Leucine (L) Leucine (L) |
USA_QQN00988.1 |
| Nucleocapsid Phosphoprotein (N) | ||
| 9 | Glutamine(Q)  Histidine (H) Histidine (H) |
Italy_QTD79359.1 |
| 67 | Proline  Serine (S) Serine (S) |
USA_ QQN00989.1 |
| 135 | Threonine (T)  Isoleucine (I) Isoleucine (I) |
Italy_ QTD79359.1 |
| 194 | Serine (S)  Leucine (L) Leucine (L) |
USA_QRJ35675.1 |
| 199 | Proline  Leucine (L) Leucine (L) |
USA_ QQN00989.1 |
| 203 | Arginine (R)  Lysine (K) Lysine (K) |
Brazil_QRN46969.1 |
| Italy_QRZ59100.1 | ||
| Italy_QQD89078.1 | ||
| Italy_ QQD89054.1 | ||
| India_ QKM77284.1 | ||
| India_ QPB40123.1 | ||
| United Kingdom_ QPC41152.1 | ||
| 204 | Glycine (G)  Arginine (R) Arginine (R) |
Brazil_QRN46969.1 |
| Italy_QRZ59100.1 | ||
| Italy_QQD89078.1 | ||
| Italy_ QQD89054.1 | ||
| India_ QKM77284.1 | ||
| India_ QPB40123.1 | ||
| United Kingdom_ QPC41152.1 | ||
| 205 | Threonine (T)  Isoleucine (I) Isoleucine (I) |
USA_ QQM19149.1 |
| 220 | Alanine (A)  Valine (V) Valine (V) |
Italy_QTD79359 |
| ORF 10 protein | ||
| No mutations observed | ||
The missense mutations are detected upon comparison with the reference strain of Wuhan, China. (ORF1ab—YP_009724389.1, S—YP_009724390.1, ORF3a—YP_009724391.1, E—YP_009724392.1, M—YP_009724393.1, ORF6—YP_009724394.1, ORF7a—YP_009724395.1, ORF7b—YP_009725318.1, ORF8—YP_009724396.1, N—YP_009724397.1, ORF10—YP_009725255.1)
Fig. 3.
Spread of missense mutations across the whole genome of SARS-CoV-2, taking into consideration sequences from 6 countries—India, United States of America (USA), Brazil, United Kingdom (UK), France, and Italy. ORF1ab (7096 amino acids) bears a total of 58 mutation sites; the total number of mutations occurring every 500 amino acids in ORF1ab is depicted in the figure. S protein (1273 amino acids) bears 24 mutation sites. ORF3a, E, ORF6, ORF7a, ORF8, and N protein shows 6, 3, 3, 1, 1, and 9 mutation points respectively. M, ORF7b, and ORF10 proteins do not exhibit any amino acid alterations. The red arrows indicate the mutation points. The bars are not drawn to scale
Distribution and implication of mutations in the spike protein from India
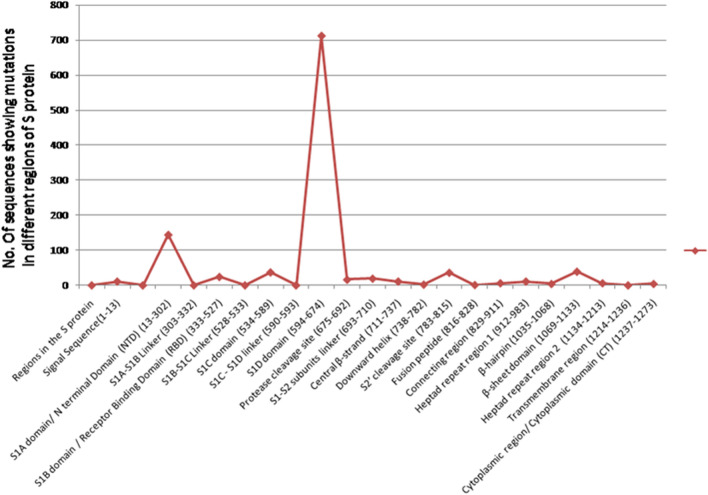
The NCBI database contained 749 complete human SARS-CoV-2 spike proteins from India, all of which were analyzed. The length of all the protein sequences was 1273 amino acid residues. One or more amino acid mutations were observed in 702 proteins. 47 spike protein sequences did not exhibit any mutations. A total of 98 mutations were observed corresponding to 55 distinct mutation sites. 42 mutation sites occurred in more than one sequence. The total number of sequences showing mutations distributed in various regions of S protein is given in Table 3. S is the major hotspot of amino acid alterations among all other proteins of SARS-CoV-2. A line graph illustrating the number of mutation sites distributed across different regions of the S protein is given in Fig. 4. Through its RBD domain, S protein mediates receptor recognition and binding with hACE2. Mutations in S might lead to alteration of the virion's virulence potential which contributes to rapid human-to-human transmission. The total number of neutral mutations in the S protein is higher than the deleterious mutations. Seven deleterious mutations are found in the S protein as predicted by the PROVEAN algorithm- C301F, T716F, A871V, A930V, D1153Y, Q1071L, and C1243F. Mutations observed in the RBD domain of the S protein are- A344T, R346T A348S, R408I, K444R, E471Q, E484Q, F490S, N501Y, Q506H, P507H, Y508N. The amino acid alterations written in italics occur in more than one sequence. N501Y impacts the association of RBD with the ACE2 receptor. This particular amino acid change affects the shape and charge of the protein near the protein-receptor interaction site. Therefore, alterations in the shape of the protein surface due to the mutations, especially near the RBD-ACE2 binding regions would be important for future considerations- in antibody, vaccine, and drug development [8]. Mutations are dispersed in almost all regions of the spike protein. The S1D domain with D614G mutation is the most predominant, and 684 out of 749 sequences possess this mutation. D614G mutation involves a change of large acidic residue D (aspartic acid) into small hydrophobic residue G (glycine) so this results in a large difference in both size and charge that might lower the binding affinity of antibodies against S protein, owing to electrostatic interactions in the tertiary structure of protein group. This might hinder the development of vaccines and make the virus prone to antigenic drift [29]. It was reported that D614G does not alter the binding affinity of S to the ACE2 receptor or its neutralization sensitivity, it increases infectivity by assembling more functional conformation of S protein into the virion, allowing an increased person to person transmission [34]. The second most dominant genomic alteration found in the S protein sequences from India is that of L54F found in at least 92 isolates. This alteration has significantly led to the difference in the size which might lead to the severity of the disease by lowering the binding affinity of antibodies. However, more studies need to be conducted to obtain clarity in this regard. Figure 5 is a representation of sequence alignment showing the points of missense mutations present across the S protein isolates from India with respect to the S protein sequence of Wuhan, China.
Table 3.
Spread of Mutations in different domains of S Protein from sequences of India
| Regions in the S protein | Total no. of sequences showing mutations | List of mutations |
|---|---|---|
| Signal Sequence(1–13) | 11 | L5F, L7S, S12F S13I, |
| S1A domain/ N terminal Domain (NTD) (13–302) | 144 | Q23R, A27V, Y28H, G35V, L54F, F59S, F65S, G75V, T76I, K77M, R78M, D88H, K97Q, Y144F, N148Y,W152L, M153I, S155I, E156D, S162I, S172F, Q173H, M177I, G181A, I233V, S255F, W258L G261S, A262S, Q271R, C301F |
| S1A-S1B Linker (303–332) | No mutations detected | |
| S1B domain / Receptor Binding Domain (RBD) (333–527) | 24 | A344T, R346T A348S, R408I, K444R, E471Q, E484D E484Q, E490S, N501Y, Q506H, P507H, Y508N |
| S1B-S1C Linker (528–533) | No mutations detected | – |
| S1C domain (534–589) | 37 | A570D, T572I, D574Y E583D, |
| S1C-S1D linker (590–593) | No mutations detected | – |
| S1D domain (594–674) | 712 | Q613H, D614G, D627P, H655Y |
| Protease cleavage site (675–692) | 16 | Q677H, P681H, A688V Q690H |
| S1-S2 subunits linker (693–710) | 19 | A701T, S704L, A706S |
| Central β-strand (711–737) | 11 | S711A, T716I, M731I |
| Downward helix (738–782) | 2 | E780D |
| S2’ cleavage site (783–815) | 36 | K786N, P809S, P812L |
| Fusion peptide (816–828) | 1 | T827I |
| Connecting region (829–911) | 6 | A871V, A879S, A892V |
| Heptad repeat region 1 (912–983) | 11 | A930V, S943P, S982A |
| β-hairpin (1035–1068) | 4 | K1045N, M1050I, L1063F |
| β-sheet domain (1069–1133) | 39 | Q1071L, T1077I, H1083Q, D1084Y, H1088R, F1089V, R1091L, V1104L, D1118H |
| Heptad repeat region 2 (1134–1213) | 6 | D1153Y, V1176F, K1181R, I1183V, N1187K, Q1201K, |
| Transmembrane region (1214–1236) | No mutations detected | – |
| Cytoplasmic region/ Cytoplasmic domain (CT) (1237–1273) | 4 | C1243F, D1259Y, P1263L, |
The ones highlighted indicate mutations that occurred in more than 1 sequence
Fig. 4.
A line graph illustrating the number of sequences showing mutations located at different domains of the spike protein. The S1D domain of spike protein contains the most predominant mutation- D614G found in 712 spike protein sequences from India. The N Terminal Domain exhibits the highest number of mutations
Fig. 5.
Sequence alignment of S protein isolates from India with the S protein sequence from China, Wuhan to detect the sites of mutations. 749 isolates from India, exhibited a total of 98 amino acid alterations. All these mutations were compiled in a single sequence for a better visual representation to study the missense mutations against the reference sequence. The missense mutations are indicated in red
Predicted epitopes within the s protein
The antigenicity, immunogenicity, and allergenicity of 3 regions of the S protein were determined (Table 4)
Table 4.
Initial Determination of Antigenicity, Immunogenicity, and Allergenicity from different regions of the S protein (S1 and S2) of reference strain (Wuhan) for prospective epitope interpretation
| Different regions of spike protein (strain- YP_009724390.1) | Antigenicity score | Immunogenicity score | Allergenicity score |
|---|---|---|---|
| N Terminal Domain (NTD) (13–302) of S1 subunit of S protein | 0.4250 (Probable Antigen) | 1.23938 | 0.79 (Probable Non- Allergen) |
| Receptor Binding Domain (RBD) 333 to 527 of S1 subunit of Spike Protein | 0.4947 (Probable Antigen) | 0.57391 | 0.79 (Probable Non- Allergen) |
| S2 Subunit of Spike protein (711–1273) | 0.3989 (Probable Non-Antigen) | − 1.41128 | 0.82 |
The antigenicity, immunogenicity, and allergenicity were estimated through the VaxiJen v 2.0 server, IEDB analysis resource, and Allergen FP server respectively
The antigenicity score of NTD and RBD was found to be higher than the threshold (0.4 was set for VaxiJen v 2.0), thus they were deemed as probable antigens. The antigenicity of RBD was found to be on a higher side (0.4947) when compared to NTD (0.4250). This makes the RBD a better potential site for vaccine construct. The NTD region can also be exploited as sites of possible epitopes. We went on to further predict the B and T-cell epitopes within the NTD and RBD regions of the S protein. However, the antigenicity score of the S2 region protein sequence was below par and thus, was regarded as a probable non-antigen. No further analyses for B and T-cell epitopes were carried out for the S2 region.
B-cell epitopes within the RBD and NTD
B-cell epitopes were determined using 5 methods from the Immune Epitope Database (IEDB) (https://www.iedb.org/)—First being, Kolaskar & Tongaonkar method helped us predict 12 epitopes from NTD and 10 epitopes from RBD [13]. Next, Emini's surface accessibility method predicts 7 epitopes from NTD and 4 epitopes within RBD which exhibited surface accessibility [7]. The third method by BepiPred predicts the residues with scores above the threshold value (0.35) to be a part of the peptide designated as an epitope [16]. This method revealed 14 and 9 epitopes from NTD and RBD respectively. The epitopes figured out from these methods are given in Tables 5 and 6. Chou & Fasman's beta-turn method was used to predict the antigenic regions, showing beta-turn conformations as such regions are highly accessible [4]. The last one, Karplus & Schulz's flexibility method is based on the flexibility of amino acids which depends on the mobility of the segments. The flexibility is correlated to antigenicity [12]. Linear and discontinuous epitopes were also found using ElliPro. It calculates the residues' protrusion index (PI) and then the residues are clustered based on their PI values [23]. By comparing and compiling the results of the above-mentioned methods, the predicted epitopes which can prompt B-cell responses are -
Within NTD- YYPDKVF, FDNPVLP, and LDSKTQSL
Within RBD- YGFQPT, YKLPDDFT, and LFRKSNLKP
Table 5.
Prediction of B-cell epitopes within the N Terminal Domain (NTD) of S protein through Kolaskar and Tongaonkar antigenicity, Emini Surface Accessibility Prediction, Bepipred Linear Epitope Prediction methods-all of which are a part of B-cell epitope prediction tools from Immune Epitope Database and Analysis Resource (IEDB)
| B cell epitope prediction for N terminal domain (13–302) | |||||
|---|---|---|---|---|---|
| Method of prediction | Sl no. | Start point | End point | Peptide sequence | Length of peptide |
| Kolaskar and Tangaonkar antigenicity | 1 | 23 | 29 | QLPPAYT | 7 |
| 2 | 34 | 71 | RGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVS | 38 | |
| 3 | 81 | 94 | NPVLPFNDGVYFAS | 14 | |
| 4 | 115 | 121 | QSLLIVN | 7 | |
| 5 | 123 | 146 | ATNVVIKVCEFQFCNDPFLGVYYH | 24 | |
| 6 | 157 | 163 | FRVYSSA | 7 | |
| 7 | 168 | 178 | FEYVSQPFLMD | 11 | |
| 8 | 200 | 207 | YFKIYSKH | 8 | |
| 9 | 209 | 233 | PINLVRDLPQGFSALEPLVDLPIGI | 25 | |
| 10 | 238 | 251 | FQTLLALHRSYLTP | 14 | |
| 11 | 262 | 278 | AAAYYVGYLQPRTFLLK | 17 | |
| 12 | 286 | 296 | TDAVDCALDPL | 11 | |
| Emini Surface Accessibility Prediction | 1 | 19 | 32 | TTRTQLPPAYTNSF | 14 |
| 2 | 35 | 43 | GVYYPDKVF | 9 | |
| 3 | 73 | 80 | TNGTKRFD | 8 | |
| 4 | 110 | 115 | LDSKTQ | 6 | |
| 5 | 144 | 153 | YYHKNNKSWM | 10 | |
| 6 | 179 | 185 | LEGKQGN | 7 | |
| 7 | 203 | 208 | IYSKHT | 6 | |
| Bepipred Linear Epitope Prediction | 1 | 21 | 31 | RTQLPPAYTNS | 11 |
| 2 | 37 | 38 | YY | 2 | |
| 3 | 71 | 81 | SGTNGTKRFDN | 11 | |
| 4 | 84 | 85 | LP | 2 | |
| 5 | 91 | 91 | Y | 1 | |
| 6 | 95 | 95 | T | 1 | |
| 7 | 97 | 98 | KS | 2 | |
| 8 | 110 | 113 | LDSK | 4 | |
| 9 | 150 | 152 | KSW | 3 | |
| 10 | 181 | 186 | GKQGNF | 6 | |
| 11 | 218 | 219 | QG | 2 | |
| 12 | 221 | 221 | S | 1 | |
| 13 | 249 | 261 | LTPGDSSSGWTAG | 13 | |
| 14 | 282 | 287 | NGTITD | 6 | |
Table 6.
Prediction of B-cell epitopes within the Receptor Binding Domain (RBD) of S protein through Kolaskar and Tongaonkar antigenicity, Emini Surface Accessibility Prediction, Bepipred Linear Epitope Prediction methods-all of which are a part of B-cell epitope prediction tools from Immune Epitope Database and Analysis Resource (IEDB)
| B cell epitope prediction for receptor binding domain (RBD)- 333 to 527 | |||||
|---|---|---|---|---|---|
| Method of prediction | Sl no. | Start point | End point | Peptide sequence | Length of peptide |
| Kolaskar and Tangaonkar antigenicity | 1 | 336 | 341 | CPFGEV | 6 |
| 2 | 347 | 353 | FASVYAW | 7 | |
| 3 | 358 | 372 | ISNCVADYSVLYNSA | 15 | |
| 4 | 374 | 385 | FSTFKCYGVSPT | 12 | |
| 5 | 387 | 404 | LNDLCFTNVYADSFVIRG | 18 | |
| 6 | 407 | 412 | VRQIAP | 6 | |
| 7 | 429 | 436 | FTGCVIAW | 8 | |
| 8 | 450 | 456 | NYLYRLF | 7 | |
| 9 | 470 | 478 | TEIYQAGST | 9 | |
| 10 | 485 | 497 | GFNCYFPLQSYGF | 13 | |
| Emini Surface Accessibility Prediction | 1 | 419 | 428 | ADYNYKLPDD | 10 |
| 2 | 437 | 442 | NSNNLD | 6 | |
| 3 | 455 | 468 | LFRKSNLKPFERDI | 14 | |
| 4 | 495 | 500 | YGFQPT | 6 | |
| Bepipred Linear Epitope Prediction | 1 | 382 | 385 | VSPT | 4 |
| 2 | 407 | 420 | VRQIAPGQTGKIAD | 14 | |
| 3 | 423 | 428 | YKLPDD | 6 | |
| 4 | 439 | 447 | NNLDSKVGG | 9 | |
| 5 | 461 | 463 | LKP | 3 | |
| 6 | 466 | 467 | RD | 2 | |
| 7 | 469 | 469 | S | 1 | |
| 8 | 473 | 483 | YQAGSTPCNGV | 11 | |
| 9 | 495 | 506 | YGFQPTNGVGYQ | 12 | |
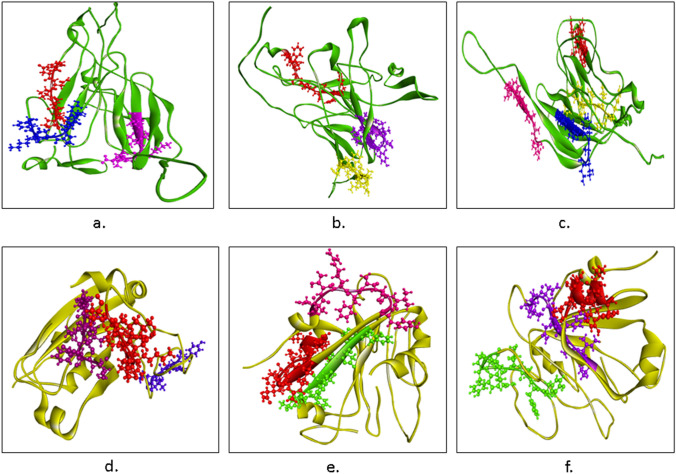
Profiling of these peptides was carried out (Tables 7, 8). YYPDKVF from NTD and LFRKSNLKP from RBD has the highest antigenic score. The epitopes were mapped on their respective 3D structures of NTD and RBD (Fig. 6a, d).
Table 7.
Physicochemical properties of all the predicted epitopes within the NTD of S protein
| Type of epitopes | Peptide Sequence | Antigenicity Score | Mutation | Hydroph-obicity | Charge | Molecular weight | Non- Digesting Enzymes |
|---|---|---|---|---|---|---|---|
| B-cell epitopes | YYPDKVF | 1.0937 | No Mutation | − 0.10 | 0.00 | 931.14 | Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Trypsin R |
| FDNPVLP | 1.0706 | No Mutation | 0.03 | − 1.00 | 801.00 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Trypsin R, Trypsin K | |
| LDSKTQSL | 1.0305 | No Mutation | − 0.27 | 0.00 | 891.10 | Chymotrypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin R | |
| Cytotoxic T-cell epitopes | YSSANNCTF | 1.0237 | No Mutation | − 0.12 | 0.00 | 1006.16 | Trypsin, Chymotrypsin, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin K, AspN |
| SANNCTFEY | 1.0058 | No Mutation | − 0.16 | − 1.00 | 1048.20 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin K, | |
| NIDGYFKIY | 1.0181 | No Mutation | -0.02 | 0.00 | 1132.41 | Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin R | |
| WTAGAAAYY | 1.0237 | No Mutation | 0.14 | 0.00 | 902.07 | Trypsin, Clostripain, Cyanogen Bromide, Proline endopept, Staph Protease, Trypsin K | |
| Helper T-cells epitopes | FQTLLALHR | 1.08 | No Mutation | − 0.06 | − 1.50 | 1098.45 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Proline endopept, Trypsin K, Trypsin R, AspN |
| FLPFFSNVT | 1.0741 | No Mutation | 0.19 | 0.00 | 1071.36 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Trypsin K, Trypsin R, AspN | |
| LLIVNNATN | 1.03 | No Mutation | 0.05 | 0.00 | 971.26 | Trypsin, Chymotrypsin, Clostripain, Cyanogen Bromide, Proline endopept, IodosoBenzoate, Staph Protease, Trypsin K, Trypsin R, AspN | |
| FRVYSSANN | 1.0164 | No Mutation | − 0.24 | 1.00 | 1057.24 | Cyanogen Bromide, Proline endopept, IodosoBenzoate, Staph Protease, Trypsin K, AspN |
Table 8.
Physicochemical properties of all the predicted epitopes within the RBD of S protein
| Type of epitopes | Peptide sequence | Antigenicity Score | Mutation | H ydroph-obicity | Charge | Molecular weight | Non- digesting enzymes |
|---|---|---|---|---|---|---|---|
| B-cell epitopes | YGFQPT | 1.0190 | No Mutation | − 0.02 | 0.00 | 711.85 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Trypsin K, Trypsin R, AspN, Elastase |
| YKLPDDFT | 1.0171 | No Mutation | − 0.20 | − 1.00 | 998.20 | Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Trypsin R, Elastase | |
| LFRKSNLKP | 1.0196 | No Mutation | − 0.36 | 3.00 | 1102.47 | Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, AspN | |
| Cytotoxic T-cell epitopes | NATRFASVY | 1.03 | No Mutation | − 0.13 | 1.0 | 1028.24 | Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin K, AspN |
| RISNCVADY | 1.07 | No Mutation | − 0.20 | 0.00 | 1040.27 | Chymotrypsin, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin K | |
| CVADYSVLY | 1.18 | No Mutation | 0.11 | 1.00 | 1032.29 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin K, Trypsin R | |
| FTNVYADSF | 1.03 | No Mutation | 0.03 | -1.00 | 10,632.24 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Proline endopept, Staph Protease, Trypsin K, Trypsin R | |
| Helper T-cells epitopes | YFPLQSYGF | 1.0799 | No Mutation | − 1.21 | 0.00 | 1121.38 | Trypsin, Clostripain, Cyanogen Bromide, IodosoBenzoate, Staph Protease, Trypsin K, Trypsin R, AspN |
| VIAWNSNNL | 1.0091 | No Mutation | − 0.90 | 0.00 | 1030.28 | Trypsin, Clostripain, Cyanogen Bromide, Proline endopept, IodosoBenzoate, Staph Protease, Trypsin K, Trypsin R, AspN | |
| YSVLYNSAS | 1.09 | No Mutation | − 0.81 | 0.00 | 1003.19 | Trypsin, Clostripain, Cyanogen Bromide, Proline endopept, IodosoBenzoate, Staph Protease, Trypsin K, Trypsin R, AspN |
Fig. 6.
Epitopes mapped on NTD (Green). a B-cell epitopes- YYPDKVF (Blue), FDNPVLP (Red), and LDSKTQSL (Pink). b Cytotoxic T-cell epitopes- YSSANNCTF (Yellow), SANNCTFEY (Yellow), NIDGYFKIY (Purple), and WTAGAAAYY (Red). c Helper T-cell- FQTLLALHR (Yellow), FLPFFSNVT (Red), LLIVNNATN (Blue), FRVYSSANN (Pink). d–f Epitopes mapped on RBD (yellow). g B-cell epitopes- YGFQPT (Blue), YKLPDDFT (Violet), and LFRKSNLKP (Red). h Cytotoxic T-cell epitopes- NATRFASVY (Magenta), RISNCVADY (Red), CVADYSVLY (Red), and FTNVYADSF (Green). i Helper T-cell- YFPLQSYGF (Green), VIAWNSNNL (Purple), and YSVLYNSAS (Red) (color figure online)
Cytotoxic T-cell within NTD and RBD
Five epitopes from each of NTD and RBD, which had the highest NetCTL score, were considered for further analysis. Then they were individually checked for their binding affinity with 81 different MHC-I alleles, HLA-A, B, C, and E. For NTD, the peptides were- WMESEFRVY (HLA-A0101, HLA-A2602, HLA-B1502, HLA-B1503), YSSANNCTF (HLA-A0101, HLA-B0803, HLA-B1501, HLA-B1502, HLA-B1503, HLA-B1517, HLA-B4601, HLA-B5801, HLA-C0303, HLA-C0501, HLA-C1203, HLA-C1402), SANNCTFEY (HLA-A2902, HLA-A3002, HLA-A8001, HLA-B4601), NIDGTFKIY (did not show strong binding with any of the MHC-I alleles), WTAGAAAYY (HLA-A0101, HLA-A2501, HLA-A2601, HLA-A2602, HLA-A2603, HLA-A2902, HLA-A3002, HLA-A6601, HLA-A6801, HLA-A6823, HLA-A8001, HLA-B1517, HLA-B3501, HLA-B5801, HLA-C0401). The peptide which showed maximum binding with MHC-I alleles was WTAGAAAYY. It was found to bind with 15 different MHC-I alleles. Of all these five peptides, the highest antigenic score was shown by YSSANNCTF and WTAGAAAYY, whereas, WMESEFRVY displayed an antigenic score below the threshold and thus it was not considered as a probable epitope to Cytotoxic T-cell response. The predicted Cytotoxic T-cell epitopes within the NTD are—YSSANNCTF, SANNCTFEY, and WTAGAAAYY.
For RBD, the 5 peptides shortlisted based on their NetCTL score were- NATRFASVY (HLA-A0101, HLA-B3501), RISNCVADY ( HLA-A0101, HLA-A0301, HLA-A3002), CVADYSVLY (HLA-A0101, HLA-A0301, HLA-1101, HLA-A2601, HLA-A3002, HLA-A6801, HLA-B1501, HLA-B3501, HLA-B5301, HLA-C0701), FTNVYADSF (HLA-A0101, HLA-A2501, HLA-A2601, HLA-B0803, HLA-B1501, HLA-B1517, HLA-B5801, HLA-C0303), ERIDISTEIY (HLA-A0101). Maximum binding with MHC-I alleles was shown by the CVADYSVLY peptide. Out of the 5 peptides, the CVADYSVLY peptide had the highest antigenic score. ERIDISTEIY was found to be non-antigenic. So, the predicted cytotoxic T-cell epitopes within the RBD are- NATRFASVY, RISNCVADY, CVADYSVLY, and FTNVYADSF. Physicochemical properties of all these epitopes were determined and then they were mapped on 3D NTD and RBD structures (Tables 7, 8) (Fig. 6b, e).
Helper T-cell epitope prediction
Through NetMHCII 2.3 server, 9-mer peptides which showed strong binding with a maximum number of MHC-II allele- HLA-DR were selected. Further, their antigenic scores were determined for them to be regarded as possible epitopes. The Helper T-cell epitopes obtained within NTD are FQTLLALHR (DRB1_0101, DRB1_0801, DRB1_1001, DRB1_1101, DRB1_1301, DRB1_1501, DRB4_0103, and DRB5_0101), FLPFFSNVT (DRB1_0101, DRB1_0401, DRB1_0404, DRB1_0405, DRB1_0701, DRB1_0802, DRB1_1501, DRB1_1602, DRB5_0101), LLIVNNATN (DRB1_0401, DRB1_0403, DRB1_0404, DRB3_0202, DRB3_0301) and FRVYSSANN (DRB1_0404, DRB1_0801, DRB1_0901, DRB1_1001, DRB1_1602 and DRB3_0202). Among these peptides, FLPFFSNVT from NTD reflected affinity with the maximum number of alleles.
Three Helper T-cell epitopes were found within the RBD- YFLPQSYGF (DRB1_1001, DRB1_1201, DRB1_1602, DRB3_0101, DRB5_0101), VIAWNSNNL (DRB1_0402, DRB1_0302, DRB1_1501, DRB3_0202, DRB3_0301), YSVLYNSAS (DRB1_0401, DRB1_0405, DRB1_1602, DRB3_0202). Among these, YFLPQSYGF and VIAWNSNNL showed binding with a maximum number of alleles.
Profiles of the peptide’s physicochemical properties were obtained to ascertain them as good epitopes for vaccine construction (Tables 7, 8). Then the epitopes were mapped on the respective 3D structures of NTD and RBD (Fig. 6c, f).
Discussion
The mutation profiling of the SARS-CoV-2 whole-genome across the globe revealed mutations in almost all the regions of the viral genome. The mutation density was relatively high in S, ORF3a, E, ORF6, and N regions, suggesting them to be more prone to mutations as compared to others such as M, ORF7b, and ORF10, which did not show any mutations. Out of the 11 genes studied, the surface glycoprotein (S) has significantly revealed that the S-protein is surface-exposed, and mediates entry into host cells through binding with the hACE2 receptor. As a result, it is the main target of nAbs upon viral infection and the focus of therapeutic and vaccine construction. Thus, from the next section, the study was confined to the mutation profile in S protein sequences from India where there is the rampant spread of the SARS-CoV-2. We also predicted multiple epitopes from the S protein which would further be conducive for vaccine design. 749 S protein sequences from India were retrieved from the NCBI database for our study. 98 mutations were identified in 55 distinct mutation sites, seven of which were regarded as deleterious according to the PROVEAN algorithm—C301F, T716F, A871V, A930V, D1153Y, Q1071L, and C1243F. In a study, it was found out that S protein mutations- L54F, D614G, and V1176F are the ones associated with a severe form of COVID-19 disease in patients [19]. There may occur more than one mutation type linked with a mutation site. When one or more mutations persist, instead of being evolutionarily cast off, they form new variants which are distinct from the existing ones. The B.1.617 variant carries two known mutations- E484Q and L452R mutations, which confers immune escape and increased infectivity. A recent study conducted at the Council of Scientific and Industrial Research's Institute of Genomics and Integrative Biology (CSIR-IGIB) in New Delhi also reported a triple mutant variant- B.1.618 found in sequences of West Bengal, Maharashtra, Delhi, and Chhattisgarh, all of which are presently hit by a second wave marked by a surge in the COVID-19 cases. The new strain reveals deletion of two amino acids-H146 and Y145 as well as exhibiting E484K and D614G variants in spike protein. This is associated with increased transmissibility and is competent in evading antibodies launched by the host immune system. All the S protein sequences present in the NCBI database up to the date of the study did not possess E484K mutation, instead, E484Q or E484D was found to be present in our studied sequences. Thus, there can be more than one mutation type associated with a mutation site. Thus at position 484, E has been altered to K, Q, and D. The role of S protein in receptor binding and membrane fusion renders it as an important antigenic component responsible for inducing host immune responses, neutralizing antibodies, and protective immunity against viral infection. The S protein of SARS-CoV-2 can mutate and it changes throughout the evolution of the virus. Most mutations will not be beneficial and will either render the S protein inactive or have no impact on its function. But some mutations may provide the virus with a selective advantage eventually making it more transmissible or infectious. This prevents protective antibodies from binding to it. Thus an antigenic determinant or epitope predicted from such a region would be ineffective. Hence it is necessary to ascertain how new mutations significantly change the S protein, and whether our current control measures remain effective. Here, the S protein has been our prime focus for identifying immune epitopes against SARS-COV-2. The antigenicity, immunogenicity, and allergenicity were evaluated by VaxiJen v2.0, IDEB Analysis resource, and AllergenFP respectively before proceeding to multiepitope prediction. The S2 region of S protein was found to be non-antigenic, and hence epitopes were predicted within the S1 region of S protein comprising of NTD and RBD. The B-cell, cytotoxic T-cell, and helper T-cell epitopes were determined for both the NTD and RBD. The B-lymphocytes are a key player in humoral immunity by eliciting antibody production [20]. 5 methods were used from the IEDB database to effectively predict the B-cell epitopes. Kolaskar and Tangaonkar method makes use of physicochemical properties of amino acid residues and experimental data, to predict antigenic determinants [13]; Emini's surface accessibility method predicts the surface accessibility of epitopes and the increased probability is indicative of an epitope being found on the surface which would further enhance its recognition by the immune system [7]; BepiPred method is based on the hidden Markov model and the propensity scale method predicts linear epitopes in protein [16]; Karplus-Schulz method helped us in figuring out the regions within the peptide which showed flexibility [12]; Chou & Fasman's beta-turn method predicts a stretch of amino acid residues which show flexible β turn conformations [4]. The sixth method predicted linear and discontinuous epitopes using ElliPro by providing the tertiary structure of the protein [23]. The consensus of all these methods revealed all plausible B-cell epitopes within the NTD and RBD. The B- cell epitopes YYPDKVF from NTD and LFRKSNLKP from RBD have the highest antigenic score. Cytotoxic T-lymphocytes bring about the death of cells that are infected during viral infection [20]. Here, CTL receptor-specific immunogenic epitopes were predicted using NetCTL 1.2 server. 9-mer epitopes with the highest NetCTL score were further scrutinized for their strong binding affinity with MHC-I alleles. The Cytotoxic T-cell epitopes WTAGAAAYY (within NTD) and CVADYSVLY (within RBD) have the highest antigenic score and exhibit binding with a maximum number of MHC I alleles. The Helper T-lymphocytes help mediate both humoral and cell-mediated immune responses, thus they form a crucial part of the construction of the immunotherapeutic vaccine [20]. Through NetMHCII 2.3 server, 9-mer T-cell epitopes manifesting affinity with a maximum number of MHC II alleles and high antigenic scores are FLPFFSNVT within NTD and YFPLQSYGF within RBD. The physicochemical features of the most suitable epitopes validated their stability. The epitopes were then mapped onto the 3-D structures of NTD and RBD for visual confirmation. The potential antigenic epitopes were thoroughly screened for multiple HLA, B-Cell, CTL, and helper T lymphocyte epitopes thus augmenting its competency in inducing both humoral and cellular immune responses. Also, all the epitopes showed high antigenic scores and these peptides were indigestible by a range of enzymes which accentuates their efficacy for vaccine construction. A schematic representation of our workflow is elucidated in the form of a flowchart in Fig. 7a, b. Researchers and scientists should contemplate not just the RBD but the entire S protein for therapeutic interventions for SARS-CoV-2. Antibody cocktails combining the NTD targeting antibodies and RBD targeting antibodies can also be used as potential candidates for treating COVID-19.
Fig. 7.
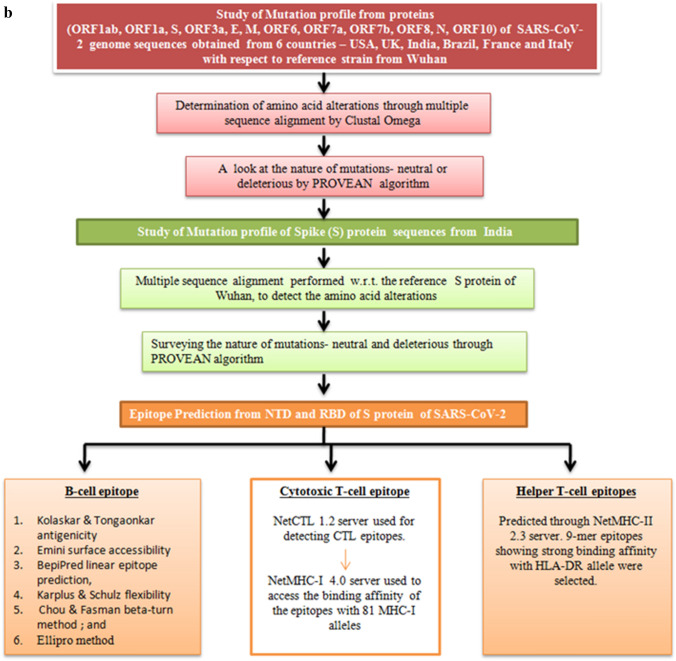
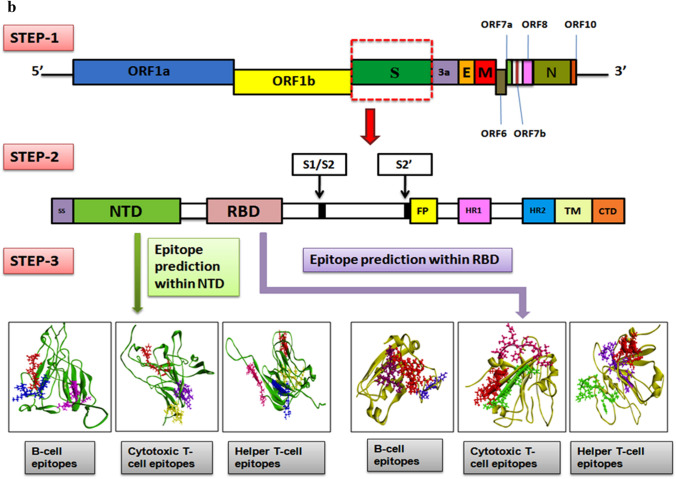
a, b Schematic workflow for studying mutation profile of SARS-CoV-2 genome throwing light on S protein isolates of India and intricate analysis of NTD and RBD regions of S protein for recognition of epitopes
Acknowledgements
The authors would like to thank the UGC-CAS program of the Department of Botany, the University of Calcutta for their support during the study. The authors also acknowledge the financial support provided by the Council of Scientific and Industrial Research.
Abbreviations
- COVID-19
Coronavirus disease 2019
- CT
Cytoplasmic domain
- CTL
Cytotoxic T-cell
- E
Envelope protein
- FP
Fusion peptide
- hACE2
Human angiotensin-converting enzyme 2
- HR
Heptad repeat region
- IEDB
Immune epitope database
- MEV
Muti-epitope vaccine
- MHC
Major histocompatibility complex
- M
Membrane protein
- MERS-CoV
Middle East respiratory syndrome coronavirus
- nAb
Neutralizing antibody
- NSP
Non-structural protein
- NTD
N terminal domain
- N
Nucleocapsid protein
- ORF
Open reading frame
- RBD
Receptor binding domain
- RdRp
RNA dependent RNA polymerase
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- S
Spike protein/surface glycoprotein
- TM
Transmembrane domain
- WHO
World Health Organization
Funding
The funds utilized during the study were provided by the Council of Scientific and Industrial Research in the form of a CSIR-JRF fellowship Grant.
Availability of data and material
All the data obtained during the study are included in the manuscript.
Declarations
Conflict of interest
The authors declare no conflict of interest associated with the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abdullahi IN, Emeribe AU, Ajayi OA, et al. Implications of SARS-CoV-2 genetic diversity and mutations on pathogenicity of the COVID-19 and biomedical interventions. J Taibah Univ Med Sci. 2020;15:258–264. doi: 10.1016/j.jtumed.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashik AI, Hasan M, Tasnim AT, et al. An immunoinformatics study on the spike protein of SARS-CoV-2 revealing potential epitopes as vaccine candidates. Heliyon. 2020;6:e04865. doi: 10.1016/j.heliyon.2020.e04865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 2006;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins Struct Funct Bioinform. 2021;89:569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez L, Beckford J, Alachkar H. Deciphering the TCR repertoire to solve the COVID-19 mystery. Trends Pharmacol Sci. 2020;41:518–530. doi: 10.1016/j.tips.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Yang C, Feng XX, et al. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen KK, Andreatta M, Marcatili P, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karplus PA, Schulz GE. Prediction of chain flexibility in proteins: a tool for the selection of peptide antigens. Naturwissenschaften. 1985;72:212–213. doi: 10.1007/BF01195768. [DOI] [Google Scholar]
- 13.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-Q. [DOI] [PubMed] [Google Scholar]
- 14.Kumar M, Al Khodor S. Pathophysiology and treatment strategies for COVID-19. J Transl Med. 2020;18:1–9. doi: 10.1186/s12967-020-02520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 16.Larsen JEP, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen MV, Lundegaard C, Lamberth K, et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007;8:1–12. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallick A, Chakrabarti J, Mandal S. Non-synonymous mutations of SARS-CoV-2 leads epitope loss and segregates its variants. Microbes Infect. 2020;22(2020):598–607. doi: 10.1016/j.micinf.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagy Á, Pongor S, Győrffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int J Antimicrob Agents. 2021;57:4. doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey RK, Bhatt TK, Prajapati VK. Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-19456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peck KM, Lauring AS. Complexities of viral mutation rates. J Virol. 2018;92:1–8. doi: 10.1128/jvi.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol. 2020 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponomarenko J, Bui HH, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008;9:1–8. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabaan AA, Al-Ahmed SH, Haque S, et al. SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 25.Ranasinghe S, Lamothe PA, Soghoian DZ, et al. Antiviral CD8+ T cells restricted by human leukocyte antigen class II exist during natural HIV infection and exhibit clonal expansion. Immunity. 2016;45:917–930. doi: 10.1016/j.immuni.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen SA. A novel coronavirus outbreak of global health concern. Ann Oncol. 2020 doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rehman Z, Fahim A, Bhatti MF. Scouting the receptor-binding domain of SARS coronavirus 2: a comprehensive immunoinformatics inquisition. Future Virol. 2021;16:117–132. doi: 10.2217/fvl-2020-0269. [DOI] [Google Scholar]
- 28.Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017 doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh PK, Kulsum U, Rufai SB, et al. Mutations in SARS-CoV-2 leading to antigenic variations in spike protein: a challenge in vaccine development. J Lab Phys. 2020;12:154–160. doi: 10.1055/s-0040-1715790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, McPartlon M, Li J. Improved protein structure prediction by deep learning irrespective of co-evolution information. bioRxiv. 2020 doi: 10.1101/2020.10.12.336859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep. 2021;11:1–21. doi: 10.1038/s41598-021-81749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Jackson CB, Mou H, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data obtained during the study are included in the manuscript.