Abstract
Background
Panax notoginseng is a highly valued medicinal herb used widely in China and many Asian countries. Its root and rhizome have long been used for the treatment of cardiovascular and hematological diseases. Imaging the spatial distributions and dynamics of metabolites in heterogeneous plant tissues is significant for characterizing the metabolic networks of Panax notoginseng, and this will also provide a highly informative approach to understand the complex molecular changes in the processing of Panax notoginseng.
Methods
Here, a high-sensitive MALDI-MS imaging method was developed and adopted to visualize the spatial distributions and spatiotemporal changes of metabolites in different botanical parts of Panax notoginseng.
Results
A wide spectrum of metabolites including notoginsenosides, ginsenosides, amino acids, dencichine, gluconic acid, and low-molecular-weight organic acids were imaged in Panax notoginseng rhizome and root tissues for the first time. Moreover, the spatiotemporal alterations of metabolites during the steaming of Panax notoginseng root were also characterized in this study. And, a series of metabolites such as dencichine, arginine and glutamine that changed with the steaming of Panax notoginseng were successfully screened out and imaged.
Conclusion
These spatially-resolved metabolite data not only enhance our understanding of the Panax notoginseng metabolic networks, but also provide direct evidence that a serious of metabolic alterations occurred during the steaming of Panax notoginseng.
Keywords: Panax notoginseng, Metabolites, Spatial distribution, Steaming process, MALDI-MS imaging
Graphical abstract
1. Introduction
Panax notoginseng (P. notoginseng) has been widely used for the treatment of cardiovascular diseases and hematological diseases in traditional Chinese medicine [[1], [2], [3]]. It is also regarded as an important functional food in many countries [4]. The underground parts including rhizome and root are two historically utilized medicinal portions of P. notoginseng. Metabolites contained in the rhizome and root of P. notoginseng are the material basis for P. notoginseng to exert its pharmacological effects. Previous studies have demonstrated that in addition to notoginsenosides and ginsenosides, the rhizome and root of P. notoginseng also contains dencichine, flavonoids, amino acids, organic acids, sugars, etc. [[5], [6], [7]].
Liquid chromatography-mass spectrometry (LC-MS) is an effective technique to investigate the metabolite profiles of biological tissues [8,9], and it has made significant inroads into the qualitative and quantitative determination of metabolites in P. notoginseng rhizome and root. For example, Chan et al developed a LC-MS/MS method to confirm the structure of new saponins, and to identify artifactual and natural compounds in Panax japonicus var. major [1]. Jiang’s group developed a high sensitive UPLC-Q-TOF mass spectrometry method to quantitatively compare eight saponins in rhizome, main root, branch root, and fibrous root of P. notoginseng [10]. However, it is worth noting that P. notoginseng possesses a high degree of heterogeneity in its tissue structure. According to the position and shape, P. notoginseng can be divided into rhizome, main root, branch root and fibrous root. According to the cell type and components, rhizome and root tissue can be further divided into phloem, xylem, medulla, and cork tissues. Unfortunately, the complex sample pre-treatment carried out during LC-MS analysis severely destroy the spatial information regarding the locations of metabolites in heterogeneous P. notoginseng tissues.
The synthesis, transport and metabolism of metabolites in higher plants are precisely regulated by their metabolic networks. The region-specific distributions of metabolites in P. notoginseng also lead to the different medicinal effects of different P. notoginseng botanical parts. In addition, both raw P. notoginseng and steamed P. notoginseng have important pharmacological effects. Traditionally, characterizing the metabolic difference between raw and steamed P. notoginseng requires complex tissue homogenization and metabolite extraction, and the results are often obtained by averaging the metabolite information in different parts of many P. notoginseng samples. This makes it difficult to accurately characterize the spatiotemporal changes of metabolites in a specific part of a certain P. notoginseng tissue during the steaming process. Therefore, developing a spatially-resolved method to map the distribution of metabolites in heterogeneous P. notoginseng tissues is very imperative, and this will essentially facilitate our understanding of the complex metabolic alteration of P. notoginseng during steaming.
Matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) is a state-of-the-art tissue imaging technique, and it provides a label-free approach to localisation of different kinds of metabolites directed on the tissue sections [[11], [12], [13], [14], [15], [16], [17]]. Currently, MALDI-MSI technique has been successfully used to map the spatial distributions and dynamics of metabolites in different plant tissues [[18], [19], [20], [21]], to visualize the complex metabolic pathways of plant secondary metabolites [[22], [23], [24]], and to uncover the metabolic mechanism of plants in response to biotic and abiotic stresses [25,26]. However, there are still no reports on MALDI-MSI technique to investigate the spatial distributions of metabolites in P. notoginseng by now. More importantly, a precise characterization of metabolites in different P. notoginseng botanical parts and visualizing the spatiotemporal changes of metabolites during P. notoginseng steaming is of great significance for elucidating the pharmacological effects of raw and steamed P. notoginseng.
In this study, we develop a high-sensitive MALDI-MSI method to visualize the metabolites in P. notoginseng tissues. The spatial distributions of notoginsenosides, ginsenosides, amino acids, dencichine, gluconic acid, and low-molecular-weight organic acids in different locations of P. notoginseng (including rhizome, main root, branch root and fibrous root) and in different micro-regions of P. notoginseng root (including phloem, xylem, medulla, and cork) were imaged for the first time. In addition, the spatiotemporal alterations of metabolites during the steaming of P. notoginseng root were also imaged, and a series of metabolites that changed with the steaming of P. notoginseng root were successfully screened out. MALDI-MS imaging of metabolites in plant tissues not only expand our understanding of the spatial features of metabolites in plants, but also provides a new approach for more detailed exploration of the molecular changes in the processing of medicinal herb.
2. Material and methods
2.1. Chemicals and reagents
1,5-Naphthalenediamine (1,5-DAN), α-cyano-4-hydroxycinnamic acid (CHCA), and 9-aminoacridine (9-AA) were obtained from Aladdin Chemical Reagent Co. (Shanghai, China). Safranin-O stain and fast green stain were provided by Beijing Solarbio Co. (Beijing, China). Tissue-Tek O.C.T. compound was purchased from Sakura Finetek Japan Co., Ltd. (Tokyo, Japan). Acetonitrile (ACN) was obtained from Merck (Muskegon, USA). Ultrapure water was obtained from a Milli-Q Water System (Millipore Corporation, Bedford, MA, USA). Ethanol and xylene were purchased from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China).
2.2. Sample preparation
The root and rhizome of P. notoginseng were collected from P. notoginseng planting base in the Yunnan province of China. Fresh P. notoginseng samples were stored in −80 °C refrigerator. The rhizome, main root, branch root, and fibrous root of P. notoginseng were all cut into 20 μm tissue sections using a cryostat microtome (Thermo CryoStar NX50 NOVPD, Bremen, Germany). Then, the tissue sections were mounted onto conductive side of indium tin oxide (ITO)-coated glass slide. Three adjacent branch root sections were prepared for the optimization of MALDI matrix. Steamed P. notoginseng samples were prepared by steaming the raw P. notoginseng root and rhizome in an autoclave for 20 min, 1 h, and 2 h at 110°C.
2.3. Matrix coating
After drying in vacuum for 20 min, the P. notoginseng tissue sections were coated with MALDI matrix. The matrix coating was carried out on a HTX TM-SprayerTM (HTX Technologies, Carrboro, NC). 9-AA, CHCA, and 1,5-DAN were successively used as MALDI matrix for metabolites MS imaging. Since more metabolites can be detected, 1.5-DAN, 2.0 mg/mL in ACN/H2O (70:30, v/v), was finally selected as the MALDI matrix. The flow rate of matrix solution was set to 0.05 mL/min at 60 °C. Nozzle nitrogen gas pressure was set to 10 psi. Track speed, track spacing, and nozzle-to-target distance were set to 800 mm/min, 3 mm, and 4 cm, respectively. A total of six spray cycles were made over each tissue sections.
2.4. MALDI-MSI experiment
MALDI-MSI experiment was performed on a RapifleX MALDI TissuetyperTM TOF/TOF MS (Bruker Daltonics, Billerica, MA) equipped with a smartbeamTM 3D laser. The laser was fired at a repetition rate of 5000 Hz and the spatial resolution was set to 100 μm. Number of shot was set to 200. Ion source voltage 1 and Ion source voltage 2 were set to 20 kV and 17.42 kV. Lens voltage was 11.6 kV. LasAtten was set to 30%. The raw mass spectral data were acquired in both positive and negative ion modes over the range of m/z 80-2000. For MALDI-MS/MS experiment, the ions of interest were listed as the targets in MS/MS scan mode. The fragment shot was set to 4000. The isolation window was selected according to the system recommendation. Default isolation window is 0.6% of precursor. The MS images were viewed and processed by using FlexImaging 5.0 software (Bruker Daltonics) and SCiLS Lab 2018b software (GmbH, Bremen, Germany). To directly compare the relative contents and distributions of metabolites in different P. notoginseng tissue sections, we import the MALDI-MSI data of different tissue sections into one file by SCiLS Lab 2018b software.
3. Results and discussion
3.1. Optimization of MALDI matrix for metabolite imaging in P. notoginseng
Choosing appropriate MALDI matrix is key for imaging more metabolites with high sensitivity. P. notoginseng contains a variety of natural products, mainly including primary metabolites such as fatty acids, amino acids and secondary metabolites such as notoginsenosides, ginsenosides, dencichine, etc. Amino acids and fatty acids are indispensable for cell energy metabolism and signal transduction. Dencichine, notoginsenosides, and ginsenosides are the material basis for P. notoginseng to treat cardiovascular and hematological diseases. Fatty acids, amino acids and dencichine contain carboxyl group, while notoginsenosides, ginsenosides contain multiple hydroxyl groups in their structure, which makes them easier to be detected in negative ion modes. 9-AA and 1,5-DAN are two most commonly used MALDI matrices in negative ion mode. Due to their low background interference and high sensitivity, 9-AA and 1,5-DAN have been successfully used for imaging the spatial locations of phenolic acids, carbohydrates, flavones, and flavone glycosides in different plant tissues [24]. CHCA was also reported to be an effective matrix for low-molecular-weight metabolites detection [27].
In this study, to acquire more abundant metabolites information, we compared the performance of MALDI-MSI on detecting metabolites in the root of P. notoginseng using 9-AA, CHCA, and 1,5-DAN as matrix. As shown in Fig. 1A, three adjacent sections from the same P. notoginseng root were subjected to MALDI-MSI analysis after spraying CHCA, 9-AA, and 1,5-DAN, respectively. The results suggest that all the detected primary and secondary metabolites exhibited stronger ion intensities when 1,5-DAN were used as MALDI matrix. For example, notoginsenoside-R1 (Fig. 1B, [M-H]-, m/z 931.5), ginsenoside-Re (Fig. 1C, [M-H]-, m/z 945.5), malic acid (Fig. 1D, [M-H]-, m/z 133.0), and palmitic acid (Fig. 1E, [M-H]-, m/z 255.2) cannot be detected when CHCA was used as matrix. Although the spatial locations of notoginsenoside-R1, ginsenoside-Re, malic acid, palmitic acid, dencichine (Fig. 1F, [M-H]-, m/z 175.0) and phenylalanine (Fig. 1G, [M-H]-, m/z 164.1) all can be imaged when 9-AA and 1,5-DAN were used as matrix, stronger ion intensities could be achieved for MALDI-MSI with 1,5-DAN as compared with 9-AA.
Fig. 1.
(A) Optimization of matrix for MALDI-MSI of P. notoginseng. (B-G) Comparison of 9-AA, CHCA, and 1,5-DAN as matrices for metabolites imaging in P. notoginseng root section.
Furthermore, we carried out in situ MS/MS experiment directly in the tissue regions with high metabolite levels on P. notoginseng root sections. Fig. 2 illustrates the typical in situ MALDI-MS/MS spectra and the fragmentation pathways of notoginsenoside-R1 and ginsenoside-Re. The results suggest that saponins tend to break glycoside bonds to generate characteristic product ions. For notoginsenoside-R1, the characteristic product ions include m/z 635.4 ([M-H-C11H20O9]-), m/z 617.4 ([M-H-C11H20O9-H2O]-), m/z 797.5 ([M-H-C5H10O4]-), m/z 767.5 ([M-H-C6H12O5]-), m/z 749.5 ([M-H-C6H12O5-H2O]-), etc. (Fig. 2A). For ginsenoside-Re, the characteristic product ions include m/z 635.4 ([M-H-C12H22O9]-), m/z 617.4 ([M-H-C12H22O9-H2O]-), m/z 781.5 ([M-H-C6H12O5]-), m/z 763.5 ([M-H-C6H12O5-H2O]-), m/z 797.5 ([M-H-C6H12O4]-), etc. (Fig. 2B).
Fig. 2.
In situ MS/MS spectra of notoginsenoside-R1 (A) and ginsenoside-Re (B).
3.2. Spatial distributions of metabolites in different tissues and different tissue micro-regions of P. notoginseng
According to the Safranin O-Fast Green (SOFG) stain image, P. notoginseng root section can be further divided into phloem, xylem, medulla, and cork tissues (Fig. 3A). The spatial distributions of different classes of metabolites in P. notoginseng, including notoginsenosides, ginsenosides, amino acids, dencichine, gluconic acid, and low-molecular-weight organic acids were successfully imaged using the optimized MALDI-MSI method. For example, notoginsenoside-R1 showed stronger ion signal in the phloem and outer xylem (Fig. 3B). On the contrary, the level of ginsenoside-Re in the medulla and inner xylem is much higher than that in the phloem and outer xylem (Fig. 3C). Ginsenoside-20Rh1 was found to be mainly distributed in the phloem region (Fig. 3D). Amino acids such as phenylalanine, glutamine, and asparagine were mainly distributed in the medulla and phloem regions (Fig. 3E–G). Dencichine (Fig. 3H) and gluconic acid (Fig. 3I) presented the highest ion signal in the phloem, followed by the medulla, then the xylem. Low-molecular-weight organic acids, including malic acid (Fig. 3J) and citric acid (Fig. 3K) all exhibited stronger ion intensities in phloem and medulla than xylem. In addition, some unknown metabolite ions showed region-specific distributions in the root of P. notoginseng. For instance, the metabolite ions of m/z 274.2 and m/z 288.2 mainly distributed in the phloem region (Fig. 3L and M); the metabolite ion of m/z 594.3 exhibited stronger ion intensity in the xylem region (Fig. 3N); the metabolite ion of m/z 795.5 was more expressed in the cork layer region (Fig. 3O).
Fig. 3.
MS images of representative metabolites in P. notoginseng root section.
The rhizome and root of P. notoginseng have long been used for the treatment of hematological and cardiovascular diseases. Actually, the root of P. notoginseng includes main root, branch root and fibrous root. Fig. 4A and Fig. 4B illustrate the optical photo and SOFG stain image of P. notoginseng rhizome, main root, branch root and fibrous root. In this study, we carried out MALDI-MSI analysis on rhizome, main root, branch root and fibrous root from the same P. notoginseng plant. Furthermore, we conducted principal component analysis (PCA) based on the in situ MALDI-MS spectra of rhizome and different root tissue sections. A clear indication of separation was observed between rhizome and root tissues (Fig. 4C), which indicated that the metabolite profile of P. notoginseng rhizome was very different from that of P. notoginseng root. The MS images of representative metabolites in rhizome and different root tissue sections were shown in Fig. 4D–L. The levels of notoginsenoside-R1 in rhizome and different root tissues were similar (Fig. 4D). Other saponins, such as notoginsenoside-Fc and ginsenoside-Re, were more expressed in the rhizome tissue (Fig. 4E and F). The result was consistent with the report by Wang et al [10]. The levels of dencichine (Fig. 4G) and citric acid (Fig. 4H) in main root were much higher than that in rhizome, branch root and fibrous root. Glutamine (Fig. 4I) and asparagine (Fig. 4J) presented much stronger ion signals in rhizome than root tissues. The metabolite ion m/z 221.1 was mainly distributed in main root tissue (Fig. 4K). The metabolite ion m/z 624.5 is almost exclusively expressed in rhizome (Fig. 4L).
Fig. 4.
(A) Optical photo of P. notoginseng. (B) Safranin O-Fast Green (SOFG) stain image of P. notoginseng rhizome, main root, branch root, and fibrous root. (C) PCA score plot based on the MALDI-MS data of different P. notoginseng tissue sections. (D-L) MS images of representative metabolites in P. notoginseng rhizome, main root, branch root, and fibrous root.
3.3. Spatiotemporal changes of metabolites during P. notoginseng steaming
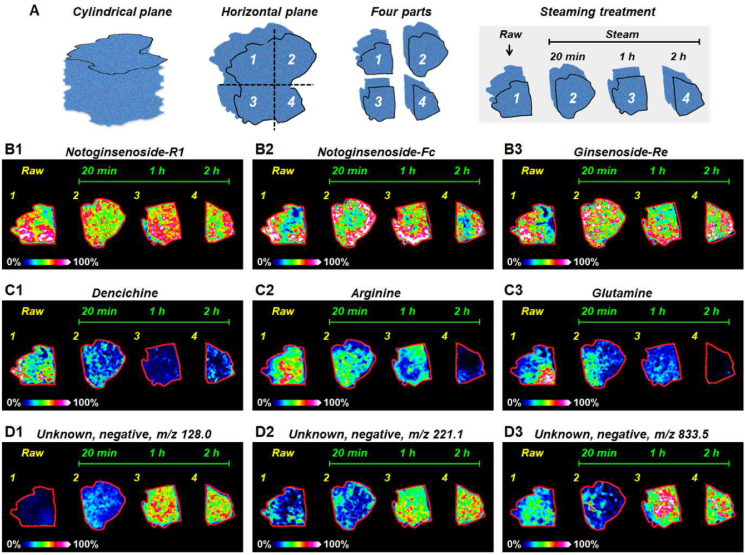
Both raw and steamed P. notoginseng were recognized as important medicine for the treatment of various diseases. The alteration of metabolites during the steaming process is regarded as an important reason for the difference of pharmacological effects between raw and steamed P. notoginseng. MALDI-MS imaging offers an efficient approach to explore the spatiotemporal alterations of metabolites during the steaming of P. notoginseng. In this study, we divided a P. notoginseng root tissue into four equal parts, one part was not steamed, and the other three parts were steamed in 110°C for 20 min, 1 h and 2 h, respectively (Fig. 5A). Then, one raw and three steamed P. notoginseng root tissues were cut into 20 μm tissue sections and subjected to MALDI-MS imaging analysis. The results are shown in Fig. 5B, C, and Fig. 5D. The levels of notoginsenoside-R1 (Fig. 5B1), notoginsenoside-Fc (Fig. 5B2), and ginsenoside-Re (Fig. 5B3) in raw and steamed P. notoginseng root differ slightly. Therefore, the high-abundance saponins such as notoginsenoside-R1, notoginsenoside-Fc, and ginsenoside-Re may not be the main reason for the biological difference between raw and steamed P. notoginseng. However, previous studies based on HPLC technique have found that notoginsenoside-R1 and ginsenoside-Re P. notoginseng were decreased gradually during the steaming process [3,28], which was different from our results. This may be due to the fact that the samples analyzed by MALDI-MSI were from the same cross-section of a certain P. notoginseng root, while the HPLC analysis was carried out on the mixed extracts of many P. notoginseng roots. Dencichine is the main hemostatic component in P. notoginseng. The MALDI-MSI results suggest that the content of dencichine in the roots of P. notoginseng decreased significantly after steaming (Fig. 5C1), which corresponds to the better hemostatic effect of raw P. notoginseng. Arginine and glutamine are two important amino acids in P. notoginseng. And the MALDI-MSI results suggest that their contents gradually increase with the extension of steaming time (Fig. 5C2 and Fig. 5C3). Some metabolite ions such as m/z 128.0, m/z 221.1, and m/z 833.5 showed very low intensities in raw P. notoginseng. However, as the steaming progresses, their signal intensities have increased significantly (Fig. 5D). These metabolites, which changed with steaming, may be the main cause of the different pharmacological effects of raw and steamed P. notoginseng.
Fig. 5.
(A) The process of obtaining raw and steamed P. notoginseng tissues from same P. notoginseng root. (B-D) MS images of representative metabolites in raw and steamed P. notoginseng root sections.
4. Conclusion
In this study, a high-coverage MALDI-MS imaging was developed to image the spatial distributions of notoginsenosides, ginsenosides, amino acids, dencichine, gluconic acid, and low-molecular-weight organic acids in P. notoginseng root and rhizome. The underlying metabolite information contained in different locations of P. notoginseng (including rhizome, main root, branch root and fibrous root) and in different micro-regions of P. notoginseng root (including phloem, xylem, medulla, and cork) were successfully imaged for the first time. Moreover, we investigated the spatiotemporal changes of metabolites during the steaming of P. notoginseng root, and screened out a series of metabolites that changed with the steaming process. Imaging the spatial locations of metabolites in different tissue regions of P. notoginseng not only offers a new approach to understand the complex plant metabolic pathways, but also helps to visualize the spatiotemporal changes of metabolites during P. notoginseng steaming.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the Taishan Scholars Program of Shandong Province, China; the financial support of the key project at the central government level: The Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources, China (2060302); the National Natural Science Foundation of China, China (21904080); Science, Education and Industry Integration Innovation Pilot Project (International Cooperation Project) from Qilu University of Technology (Shandong Academy of Sciences), China (2020KJC-GH08); and the Major Innovation Project of Shandong Biotechnology Technology Innovation Center, China (2019JSWGCCXZX001).
References
- 1.Chan H.H., Hwang T.L., Reddy M.V., Li D.T., Qian K., Bastow K.F. Bioactive constituents from the roots of Panax japonicus var. major and development of a LC-MS/MS method for distinguishing between natural and artifactual compounds. J Nat Prod. 2011;74:796–802. doi: 10.1021/np100851s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F., Ma N., He C., Hu Y., Li P., Chen M. Qualitative and quantitative analysis of the saponins in Panax notoginseng leaves using ultra-performance liquid chromatography coupled with time-of-flight tandem mass spectrometry and high performance liquid chromatography coupled with UV detector. J Ginseng Res. 2018;42:149–157. doi: 10.1016/j.jgr.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong Y., Chen L., Man J., Hu Y., Cui X. Chemical and bioactive comparison of Panax notoginseng root and rhizome in raw and steamed forms. J Ginseng Res. 2019;43:385–393. doi: 10.1016/j.jgr.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu D., Zhou Q., Li H., Li S., Dong Z., Li D. Pharmacokinetic characteristics of steamed notoginseng by an efficient LC-MS/MS method for simultaneously quantifying twenty-three triterpenoids. J Agric Food Chem. 2018;66:8187–8198. doi: 10.1021/acs.jafc.8b03169. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y.Y., Song J.Y., Li Y., Meng Y.Q., Zhao M.B., Jiang Y. Comparative study on excretive characterization of main components in herb pair notoginseng-safflower and single herbs by LC⁻MS/MS. Pharmaceutics. 2018;10 doi: 10.3390/pharmaceutics10040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu C., Wang W., Wang B., Zhang T., Cui X., Pu Y. Analytical methods and biological activities of Panax notoginseng saponins: recent trends. J Ethnopharmacol. 2019;236:443–465. doi: 10.1016/j.jep.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z., Zhu J., Zhang H., Fan X. Investigating chemical features of Panax notoginseng based on integrating HPLC fingerprinting and determination of multiconstituents by single reference standard. J Ginseng Res. 2018;42:334–342. doi: 10.1016/j.jgr.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimizu T., Watanabe M., Fernie A.R., Tohge T. Targeted LC-MS analysis for plant secondary metabolites. Methods Mol Biol. 2018;1778:171–181. doi: 10.1007/978-1-4939-7819-9_12. [DOI] [PubMed] [Google Scholar]
- 9.Wolfender J.L., Marti G., Thomas A., Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J Chromatogr A. 2015;1382:136–164. doi: 10.1016/j.chroma.2014.10.091. [DOI] [PubMed] [Google Scholar]
- 10.Wang J.-R., Yau L.-F., Gao W.-N., Liu Y., Yick P.-W., Liu L. Quantitative comparison and metabolite profiling of saponins in different parts of the root of Panax notoginseng. J Agric Food Chem. 2014;62:9024–9034. doi: 10.1021/jf502214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boughton B.A., Thinagaran D., Sarabia D., Bacic A., Roessner U. Mass spectrometry imaging for plant biology: a review. Phytochem Rev. 2016;15:445–488. doi: 10.1007/s11101-015-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feenstra A.D., Alexander L.E., Song Z., Korte A.R., Yandeau-Nelson M.D., Nikolau B.J. Spatial mapping and profiling of metabolite distributions during germination. Plant Physiol. 2017;174:2532–2548. doi: 10.1104/pp.17.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norris J.L., Caprioli R.M. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C., Li Z., Ma C., Zang Q., Li J., Liu W. Acetone immersion enhanced MALDI-MS imaging of small molecule metabolites in biological tissues. J Pharm Biomed Anal. 2019;176:112797. doi: 10.1016/j.jpba.2019.112797. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y.J., Perdian D.C., Song Z., Yeung E.S., Nikolau B.J. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012;70:81–95. doi: 10.1111/j.1365-313X.2012.04899.x. [DOI] [PubMed] [Google Scholar]
- 16.Dong Y., Li B., Aharoni A. More than pictures: when MS imaging meets histology. Trends Plant Sci. 2016;21:686–698. doi: 10.1016/j.tplants.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Bhandari D.R., Wang Q., Friedt W., Spengler B., Gottwald S., Rompp A. High resolution mass spectrometry imaging of plant tissues: towards a plant metabolite atlas. Analyst. 2015;140:7696–7709. doi: 10.1039/c5an01065a. [DOI] [PubMed] [Google Scholar]
- 18.Li B., Neumann E.K., Ge J., Gao W., Yang H., Li P. Interrogation of spatial metabolome of Ginkgo biloba with high-resolution matrix-assisted laser desorption/ionization and laser desorption/ionization mass spectrometry imaging. Plant, Cell Environ. 2018;41:2693–2703. doi: 10.1111/pce.13395. [DOI] [PubMed] [Google Scholar]
- 19.Horn P.J., Korte A.R., Neogi P.B., Ebony L., Johannes F., Kerstin S. Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell. 2012;24:622–636. doi: 10.1105/tpc.111.094581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodfield H.K., Sturtevant D., Borisjuk L., Munz E., Guschina I.A., Chapman K. Spatial and temporal mapping of key lipid species in Brassica napus seeds. Plant Physiol. 2017;173:1998–2009. doi: 10.1104/pp.16.01705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun C., Liu W., Ma S., Zhang M., Geng Y., Wang X. Development of a high-coverage matrix-assisted laser desorption/ionization mass spectrometry imaging method for visualizing the spatial dynamics of functional metabolites in Salvia miltiorrhiza Bge. J Chromatogr A. 2020;1614:460704. doi: 10.1016/j.chroma.2019.460704. [DOI] [PubMed] [Google Scholar]
- 22.Bin L., Camilla K., Natascha Krahl H., Kirsten J.R., Rubini K., SRen B. Visualizing metabolite distribution and enzymatic conversion in plant tissues by desorption electrospray ionization mass spectrometry imaging. Plant J. 2013;74:1059–1071. doi: 10.1111/tpj.12183. [DOI] [PubMed] [Google Scholar]
- 23.Tocci N., Gaid M., Kaftan F., Belkheir A.K., Belhadj I., Liu B. Exodermis and endodermis are the sites of xanthone biosynthesis in Hypericum perforatum roots. New Phytol. 2018;217:1099–1112. doi: 10.1111/nph.14929. [DOI] [PubMed] [Google Scholar]
- 24.Sun C., Zhang M., Dong H., Liu W., Guo L., Wang X. A spatially-resolved approach to visualize the distribution and biosynthesis of flavones in Scutellaria baicalensis Georgi. J Pharm Biomed Anal. 2020;179:113014. doi: 10.1016/j.jpba.2019.113014. [DOI] [PubMed] [Google Scholar]
- 25.Shroff R., Vergara F., Muck A., Svatos A., Gershenzon J. Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc Natl Acad Sci U S A. 2008;105:6196–6201. doi: 10.1073/pnas.0711730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manuela P., Johannes T., Darin P., Winfriede W., Wim V.D.E., Hans-Peter M. Spatio-temporal dynamics of fructan metabolism in developing barley grains. Plant Cell. 2014;26:3728. doi: 10.1105/tpc.114.130211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porta T., Grivet C., Knochenmuss R., Varesio E., Hopfgartner G. Alternative CHCA-based matrices for the analysis of low molecular weight compounds by UV-MALDI-tandem mass spectrometry. J Mass Spectrom. 2011;46:144–152. doi: 10.1002/jms.1875. [DOI] [PubMed] [Google Scholar]
- 28.Wang D., Liao P.-Y., Zhu H.-T., Chen K.-K., Xu M., Zhang Y.-J. The processing of Panax notoginseng and the transformation of its saponin components. Food Chem. 2012;132:1808–1813. [Google Scholar]