Abstract
SARS-CoV-2 exploits the host cellular machinery for virus replication leading to the acute syndrome of coronavirus disease 2019 (COVID-19). Growing evidence suggests SARS-CoV-2 also exacerbates many chronic diseases, including cancers. As mutations on the spike protein (S) emerged as dominant variants that reduce vaccine efficacy, little is known about the relation between SARS-CoV-2 virus variants and cancers. Compared to the SARS-CoV-2 wild-type, the Gamma variant contains two additional NXT/S glycosylation motifs on the S protein. The hyperglycosylated S of Gamma variant is more stable, resulting in more significant epithelial-mesenchymal transition (EMT) potential. SARS-CoV-2 infection promoted NF-κB signaling activation and p65 nuclear translocation, inducing Snail expression. Pharmacologic inhibition of NF-κB activity by nature food compound, I3C suppressed viral replication and Gamma variant-mediated breast cancer metastasis, indicating that NF-κB inhibition can reduce chronic disease in COVID-19 patients. Our study revealed that the Gamma variant of SARS-CoV-2 activates NF-κB and, in turn, triggers the pro-survival function for cancer progression.
Keywords: SARS-CoV-2 variant, glycosylation, epithelial-mesenchymal transition, spike, NF-κB
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the pathogen of the coronavirus disease 2019 (COVID-19), which leads to a total of 160,074,267 confirmed cases and 3,325,260 deaths globally (WHO situation report, 05/14/2021). SARS-CoV-2 is a single-stranded RNA virus that encodes 15 non-structural proteins (nsp1-10 and nsp12-16) as well as four structural proteins, nucleocapsid (N), membrane protein (M), envelope (E), and spike (S), for virus replication [1]. The viral spike (S) glycoprotein binds to the angiotensin-converting enzyme 2 (ACE2) receptor on the host cells, cleaved by serine protease TMPRSS2, resulting in SARS-CoV-2 virus entry [2]. Thus far, over a thousand SARS-CoV-2 variants were found worldwide, including SARS-CoV-2 Alpha (UK, B.1.1.7), SARS-CoV-2 Beta (South Africa, B.1.351), and SARS-CoV-2 Gamma (Brazil, P.1) variants. These variants have multiple changes in the S protein that influence virus entry. Mutations in the receptor recognition site on the S are of great concern for escaping immune surveillance [3]. S protein of Alpha variant contained eight amino acid changes, resulted in decreasing efficacy of the BNT162b2 vaccine [4]. Sera from 19 individuals received twice with Pfizer/BioNTech vaccine was less efficacious against Beta variant [5]. S protein of Beta variant appeared with mutations in K417N, E484K, N501Y, and D614G [6]. In those who received two standard doses of the ChAdOx1 nCoV-19 vaccine, the efficacy was 60% in the Alpha and 64% in Gamma strain against primary symptomatic COVID-19 [7].
Nuclear factor-kappaB (NF-κB) is a key regulator of inflammatory responses. The downstream targets of NF-κB include interleukin (IL)-1β, IL-6, and tumor necrosis factor-alpha (TNFα), which induce an innate immune response [8,9]. Excessive NF-κB activation contributes to inflammation and autoimmunity [10] and is a hallmark of SARS-CoV infection. Inhibition of NF-κB activity may suppress virus-mediated inflammation and reduce lung pathology [11]. Currently, NF-κB antagonists derived from natural food compounds are widely used in the treatment of various diseases, such as indole-3-carbinol (I3C) in cancers [12] and flavonoids in cardiovascular diseases [13]. I3C is an active natural anticancer compound found in cruciferous vegetables. It stabilizes IκBα and retains NF-κB in the cytoplasm [14]. Whether manipulation of NF-κB could be utilized as a strategy to combat the COVID-19 pandemic has not previously been explored in detail.
The role of SARS-CoV-2 in cancer progression remains largely unknown. Cancer patients are generally much more vulnerable to COVID-19 as the immune deficiency increases the SARS-CoV-2 infection rate [15]. The expression of viral receptor ACE2 or TMPRSS2 is positively correlated with EMT signature in lung cancer [16] and breast cancer [17] patient samples. The S of SARS-CoV-2 increases breast cancer metastasis through upregulation of Snail [17]. This study aimed to dissect the oncogenic potentials of SARS-CoV-2 variants in the EMT progression. While the 22 NXT/S motifs are highly conserved in SARS-CoV-2 variants, the S of Gamma variant contains two additional NXT/S motifs, resulting in S stabilization and NF-κB activation. We discovered that the extra glycosylation motifs in Gamma variant increase breast cancer metastatic potentials. Understanding the relation between SARS-CoV-2 and breast cancer metastasis may help reduce COVID-19-mediated chronic disease progression.
Materials and methods
Cell cultures and treatments
MCF7, MCF10A, Caco-2, and HEK293 cells were obtained from American Type Culture Collection. MCF10A cells were cultured in DMEM/F12 medium supplemented with 5% horse serum, 500 ng/ml hydrocortisone, 10 μg/ml insulin, and 20 ng/ml EGF.
Plasmids
The gene encoding amino acids 1-1273 and 1-218 of the SARS-CoV-2 S protein (Gene ID: MN908947) was subcloned into the pLAS2w-pPuro (RNAi core, Academia Sinica, Taiwan). The S variants of SARS-CoV-2 Alpha (Gene ID: VG40771-UT), Beta (Gene ID: VG40772-UT), and Gamma (Gene ID: VG40773-UT) were purchased from Sino Biological. All construct sequences were confirmed by enzyme digestion and DNA sequencing.
Antibodies
The following antibodies were used: N protein (a gift from Dr. An-Suei Yang, Genomics Research Center, Academia Sinica, Taiwan), β-Tubulin (66240-1-Ig; Proteintech, Chicago, IL, USA), and IκBα (ab32518; Abcam, Cambridge, MA, USA). Antibodies for EMT markers, rabbit IgG-HRP, and mouse IgG-HRP were used as previously described [18].
Virus isolation and infection
The SARS-CoV-2 strain was isolated from a COVID-19 patient in Taiwan (TCDC#4) as described earlier [18]. Briefly, the virus was passaged on Vero E6 cells in MEM supplemented with 2% FBS. Target cells were infected with SARS-CoV-2 (MOI of 0.1) for 24 h. The infected cells were then fixed with 10% formaldehyde and permeabilized by 0.5% Triton X-100. All procedures were performed in the P3 lab in the IBMS, Academia Sinica. The experiments were strictly followed the Taiwan Centers for Disease Control’s laboratory biosafety guidelines. MCF10A-ACE2, Caco-2, and Vero E6 cells were treated with 10 μM I3C or JSH-23 (S2313; S7351, Selleckchem, Houston, TX, USA) at 37°C for 2 h and then infected with SARS-CoV-2 for 24 h.
Immunofluorescence staining
After infection, MCF10A-ACE2 and Vero E6 cells were fixed with 10% formaldehyde for 1 h, and then incubated with a primary antibody, anti-p65, or anti-N protein antibodies at 4°C overnight. The secondary antibody used was anti-rabbit 594 (1:1000, Z25307, Invitrogen, Waltham, MA) or anti-human 488 (1:1000, A-11013, Invitrogen, Waltham, MA) at RT for 1 h. Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI) for 15 mins in the dark. Images were collected using a Zeiss confocal microscope (LSM 700 Stage, Carl-Zeiss-Strasse 22, Oberkochen) and a high content analysis system (ImageXpress Micro XL, Molecular Devices, San Jose, CA).
Luciferase assay
Cells were collected after treating with pseudovirus (MOI of 1) or SARS-CoV-2 for two days. According to the manufacturer’s protocol, luciferase activities were measured by the luciferase reporter assay system (Promega, Madison, WI, USA). Luminescence measurement was detected via GloMax 96 Microplate Luminometer.
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated using Quick-RNA Miniprep Kit (R1055; ZYMO Research, Irvine, CA, USA) as described earlier [18]. 1 μg RNA were transcribed to cDNA using ToolsQuant II Fast RT Kit (KRT-BA06-2; Biotools, Taipei, Taiwan) according to the manufacture’s protocol. qPCR reactions were performed using SYBR® Green supermix (1708880; Bio-Rad, Hercules, CA, USA). Primers used are as follows: E-cadherin_F: 5’-TGCCCAGAAAATGAAA AAG-3’, E-cadherin_R: 5’-GTGTATGTGGCAATGCGTT-3’, N-cadherin_F: 5’-ACAGTGGCCACCTACAAAG-3’, N-cadherin_R: 5’-CCGAGATGGGGTTGATAAT-3’, SNAIL_F: 5’-CCTCCCTGTCAGATGAGG-3’, SNAIL_R: 5’-CCAGGCTGAGGTATTCCT-3’, FOXC2_F: 5’-TCACCTTGAACGGCATCTACCAG-3’, FOXC2_R: 5’-TGACGAAGCACTCGTTGAGCGA-3’, REL_F: 5’-AGTTGCGGAGACCTTCTGACCA-3’, REL_R: 5’-CGTGATCCTGGCACAGTTTCTG-3’, NFKB1_F: 5’-GCAGCACTACTTCTTGACCACC-3’, NFKB1_R: 5’-TCTGCTCCTGAGCATTGACGTC-3’, IRF7_F: 5’-CCACGCTATACCATCTACCTGG-3’, IRF7_R: 5’-GCTGCTATCCAGGGAAGACACA-3’, IL-6_F: 5’-AGACAGCCACTCACCTCTTCAG-3’, IL-6_R: 5’-TTCTGCCAGTGCCTCTTTGCTG-3’, IFITM1_F: 5’-TCAACATCCACAGCGAGACC-3’, IFITM1_R: 5’-TGTCACAGAGCCGAATACCAG-3’, E protein_F: 5’-ACAGGTACGTTAATAGTTAATAGCGT-3’, E protein_R: 5’-ATATTGCAGCAGTACGCACACA-3’.
Promoter assay
We used X-tremeGENE DNA Transfection Reagent (XTGHP-RO, Roche, Mannheim, Germany) for transient transfection according to the manufacture’s protocol. E-cadherin promoter [19] or NF-κB-Luc plasmid was co-transfected with S variants in MCF7 cells. Luciferase activity was assayed using a Dual-Luciferase Reporter Assay System.
Sphere formation assay
Cells were seeded in an ultra-low adherent plate and grown in serum-free conditions as described earlier [18]. Briefly, MCF7 cells expressing S variants were suspended in complete MammoCultTM Medium at the concentration of 1,000 cells/ml. The cell suspension was cultured in a 6-well plate for five days. Mammospheres over 100 µm were counted manually.
In vitro cell migration and invasion assays
We used transwell permeable supports (Corning-Costar, Cambridge, MA) to perform cell invasion assay as earlier described [18]. Briefly, MCF7 cells expressing S variants were serum-starved overnight before the experiment. Then, five thousand cells were seeded onto the upper chamber supplemented in serum-free DMEM. Two days later, cells that migrated to the bottom of the chamber were stained with crystal violet, and counted by ImageJ.
A mouse model for lung metastasis
Six-week-old female BALB/c mice were purchased from the National Laboratory Animal Center (Taipei, Taiwan). The animals were housed at the Institute of Biomedical Sciences Animal Care Facility. The tumor metastasis assays were performed using a breast cancer mouse model through i.v. injection. 4T1 cells expressing S variants were injected into the lateral tail vein of mice (5 mice per group). Mice were treated with I3C 20 mg/kg via oral gavage. At the experimental endpoint, mice lungs were excised and stained with India ink. All animal procedures complied with the protocols approved by the Institutional Animal Care and Utilization Committee of Academia Sinica.
Statistical analysis
All data were analyzed and presented as mean ± SD (standard deviation). The data from individual experiments are examined by one-way or two-way ANOVA with Tukey’s post hoc test for multiple comparisons (GraphPad Prism Software Inc., San Diego, CA, USA). A probability value of P < 0.05 was considered to be statistically significant.
Results
Glycosylation of SARS-CoV-2 Gamma variant contains two additional NXT/S motifs
Glycosylation reduces immunogenicity that helps pathogens escape immune surveillance. To dissect the oncogenic function of COVID-19 variants, we focused on glycosylation-mediated cancer aggressiveness. To this end, PNGase F was used to remove all N-linked oligosaccharides from polypeptides. A glycostaining was employed to demonstrate that S protein was extensively modified by complex typed N-linked glycosylation (Figure 1A). The molecular weight of S protein was significantly reduced in Western blot analysis after treating N-linked but not O-linked glycosylation inhibitors (Figure 1B). Protein sequence alignment further demonstrated that the NXT/S motifs in the S protein are evolutionally conserved. Interestingly, SARS-CoV-2 Gamma variant contains two additional NXT/S motifs at N20 (give rise to an NRT motif) and N188 (give rise to an NLS motif), which are not present in the S of SARS-CoV-2 wild-type (WT), Alpha, or Beta (Figure 1C, 1D). To examine if N20 and N188 in Gamma variant are really modified by N-linked glycosylation, we subcloned S protein (1-218 amino acids) of WT, Alpha, Beta, and Gamma variants and then expressed in HEK293T cells. We noticed that the S of Gamma variant displayed a molecular weight shift compared to S of WT or other variants (Figure 1E). To better understand the function of the two NXT/S motifs on the Gamma variant, we analyzed protein stability using cycloheximide-chase assays (Figure 1F). The S of Gamma variant is more stable than that of WT, Alpha, or Beta variants, indicating glycosylation stabilizes S expression in host cells (Figure 1G). Similarly, the Gamma variant possessed stronger infectivity in HEK293-hACE2 cells in pseudovirus infection (Figure 1H). These results suggest hyper-glycosylated S of Gamma variant induces an aggressive phenotype than that of WT.
Figure 1.
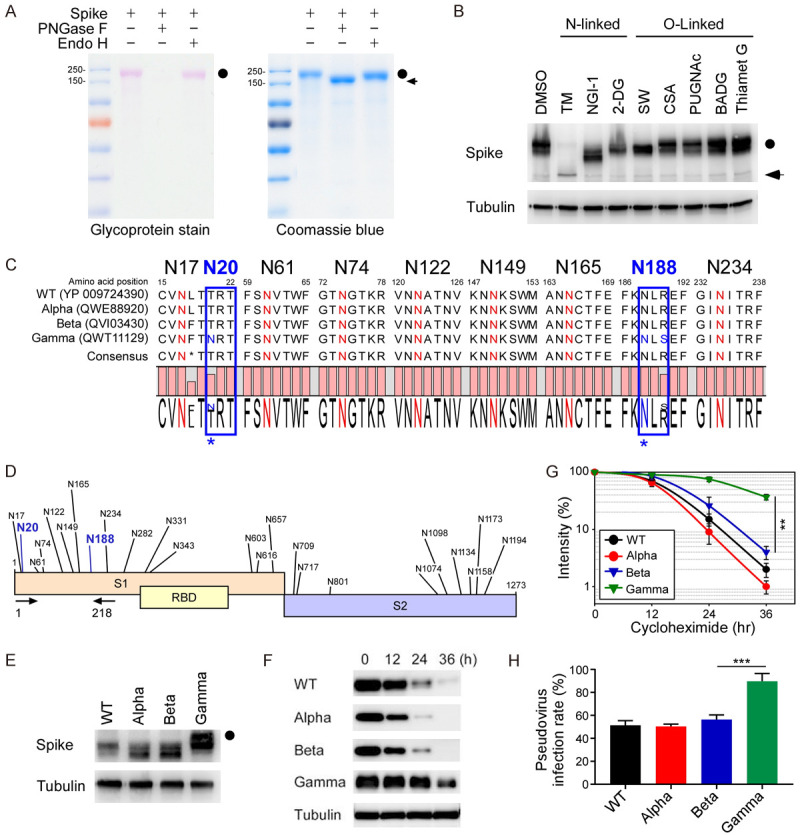
Hyperglycosylated S of SARS-CoV-2 Gamma variant is more stable in cells. A. The glycosylation pattern of the S protein. S protein was treated with PNGase F or Endo H and analyzed by glycoprotein staining (left panel) and Coomassie blue staining (right panel). Black circle, glycosylated S; arrowhead, non-glycosylated S. B. Western blot analysis of S protein expression pattern treated with N-linked and O-linked glycosyltransferase inhibitors. Circle, glycosylated S; arrow, non-glycosylated S. C. Amino acid sequences alignment of SARS-CoV-2 WT, Alpha, Beta, and Gamma variants for evolutionarily conserved NXT/S motifs. D. Diagram of NXT/S motifs in S protein of SARS-CoV-2. Conserved glycosylation sites on the S of Gamma variant are labeled in black. Two additional NTX motifs on the S of Gamma variant are marked in blue. Arrowhead indicates the PCR primers used to amplify the S of WT, Alpha, Beta, and Gamma variants. E. Western blot analysis of S protein (1-218 amino acids) of WT, Alpha, Beta, and Gamma variants. Circle, hyper-glycosylated Gamma variant. F. Western blot analysis of S protein expression in the cycloheximide-chase assay. G. The intensity of S protein in SARS-CoV-2 WT, Alpha, Beta, and Gamma variants was quantified using a densitometer. H. Luciferase activity was measured in HEK293T-ACE2 cells infected with SARS-CoV-2 pseudovirus generated from wild-type and mutant spike SARS-CoV-2.
SARS-CoV-2 Gamma variant induces a stronger EMT in breast epithelial cells
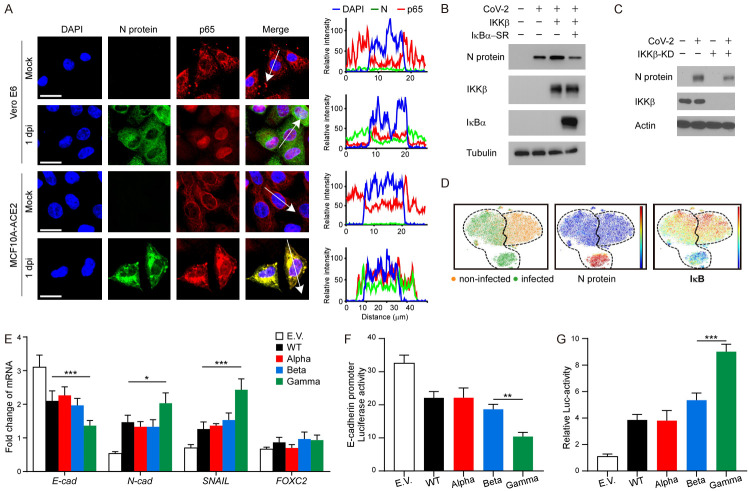
NF-κB activation is critical for SARS-CoV-2 infection. To further validate if SARS-CoV-2 transduction activates the NF-κB pathway, we performed an immunofluorescence assay (IFA) to examine the subcellular localization of p65, one of five components of the NF-κB pathway [20]. We observed p65 translocation from the cytoplasm into the nucleus upon SARS-CoV-2 infection in both Vero E6 and MCF10A-ACE2 cells (Figure 2A). As shown in Figure 2B, 2C, transient expression of IKKβ (another NF-κB pathway protein) increased viral N protein levels, whereas expression of dominant-negative IκBα super repressor (IκBα-SR) and IKKβ knockdown (IKKβ-KD) diminished them, indicative of alleviated SARS-CoV-2 infection. Consistent with the Western blot analysis, the CyTOF analysis indicated that SARS-CoV-2 significantly induced the expression levels of viral N protein, and the N protein levels were negatively correlated with levels of IκB (Figure 2D). Our earlier results showed that an S of SARS-CoV-2 induces EMT through NF-κB signaling [17]. To investigate the S-induced EMT of each virus strain, we transiently expressed the S proteins of SARS-CoV-2 WT, Alpha, Beta, or Gamma variants in MCF7 cells. Results from qPCR revealed that Gamma variant induces a stronger downregulation of E-cadherin and upregulation of N-cadherin and SNAIL (Figure 2E) than that of WT, Alpha, or Beta variants. Similarly, the Gamma variant exhibits more E-cadherin promoter repression (Figure 2F) and NF-κB activation (Figure 2G). These results suggest that the Gamma variant has more potential to induce breast cancer metastasis through NF-κB activation.
Figure 2.
S of SARS-CoV-2 Gamma variant induces a stronger EMT in breast cancer cells. A. Representative confocal microscopic images of p65 translocation in Vero E6 and MCF10A-ACE2 cells after SARS-CoV-2 infection (left panel). Scale bar, 20 μm. The plot of relative intensity through the white arrow in the left panel. B. Western blot analysis of N protein, IKKβ, and IκBα after SARS-CoV-2 infection. 293T-ACE2 cells were transiently transfected with pcDNA3 plasmid encoding p65 and IκBα-SR before SARS-CoV-2 infection. C. Western blot analysis of N protein and IKKβ after SARS-CoV-2 infection. 293T-ACE2 cells were transiently transfected with pcDNA3 plasmid encoding IKKβ-KD before SARS-CoV-2 infection. D. Immunophenotypic and signaling pathway analysis of the SARS-CoV-2 infected cell lines by CyTOF. ViSNE analysis showed non-infected (orange dots) and infected (green dots) cells (bottom left). The dashed lines indicated the ViSNE spaces for infected or non-infected cells, respectively. The expression levels of N-protein (bottom middle) and IκB (bottom right) for all analyzed cells. Red is high. Blue is low. E. qPCR analysis of E-cadherin, N-cadherin, SNAIL, and FOXC2 in MCF7 cells expressing S of SARS-CoV-2 WT, Alpha, Beta, or Gamma variants. Two-way ANOVA with Tukey’s post hoc test. *P < 0.05; ***P < 0.001. F. Luciferase reporter assay of E-cadherin. MCF7 cells were transfected with the E-cadherin reporter and S of SARS-CoV-2 WT, Alpha, Beta, or Gamma variants. One-way ANOVA with Tukey’s post hoc test. **P < 0.01. G. Luciferase reporter assay of NF-κB. One-way ANOVA with Tukey’s post hoc test. ***P < 0.001. E.V. means empty vector.
NF-κB inhibitors limit SARS-CoV-2 infection and reduce IL-6 levels
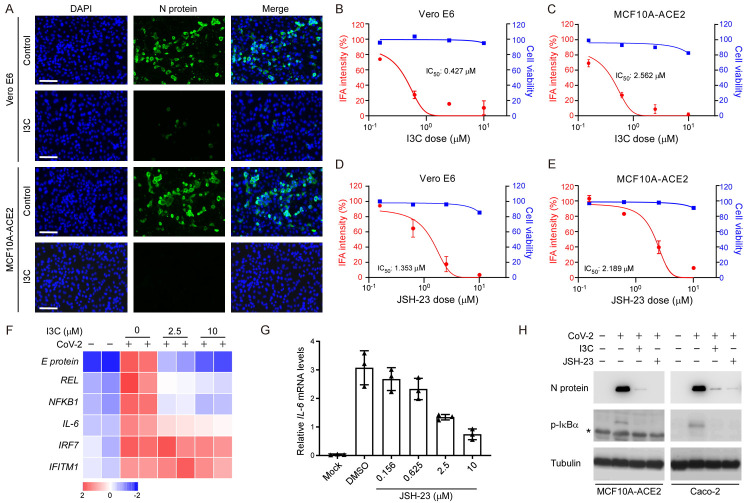
To investigate the role of NF-κB in SARS-CoV-2 infection, we utilized MCF10A-ACE2 and Vero E6 as models to examine if the NF-κB inhibitors I3C and JSH-23 could reduce SARS-CoV-2 infection. As shown in Figure 3A, IFA revealed that both I3C and JSH-23 (data not shown) dramatically reduced viral infection in both cell types. I3C presented a half-maximal inhibitory concentration (IC50) of 0.427 μM in Vero E6 and 2.562 μM in MCF10A-ACE2 cells upon four-fold serial dilution (Figure 3B, 3C), whereas the corresponding IC50 values for JSH-23 were 1.353 μM in Vero E6 cells (Figure 3D) and 2.189 μM in MCF10A-ACE2 cells (Figure 3E). We also found that mRNA expression levels of viral E protein, REL, NF-κB, and IL-6 were significantly reduced in SARS-CoV-2-infected MCF10A-ACE2 cells upon I3C treatment, but not interferon signaling, IRF7 and IFITM1 (Figure 3F), indicating that I3C specifically reduces NF-κB signaling without affecting the type I IFN response. Moreover, mRNA expression levels of IL-6 were significantly reduced in SARS-CoV-2-infected MCF10A-ACE2 cells upon JSH-23 treatment (Figure 3G). Administration of I3C and JSH-23 impeded SARS-CoV-2 infection by suppressing N protein and p-IκBα expression (Figure 3H). Thus, targeting NF-κB activity by administering NF-κB inhibitors such as I3C may protect against SARS-CoV-2 infection at the cellular level.
Figure 3.
Blocking NF-κB prevents SARS-CoV-2 infection. (A) Representative fluorescent microscopic images of N protein expression in Vero E6 and MCF10A-ACE2 cells after SARS-CoV-2 infection. Scale bar, 20 μm. (B) IC50 and cell viability of Vero E6 and (C) MCF10A-ACE2 cells pre-treated with I3C at the indicated doses prior to SARS-CoV-2 infection. (D) IC50 and cell viability of Vero E6 cells and (E) MCF10A-ACE2 pre-treated with JSH-23 and at the indicated doses before SARS-CoV-2 infection. (F) Heatmap of representative mRNA expression of MCF10A-ACE2 cells pre-treated with I3C at the indicated doses prior to SARS-CoV-2 infection. (G) Cellular IL-6 expression upon JSH-23 treatment. (H) Western blot analysis of I3C and JSH-23 on N protein and p-IκBα expression after SARS-CoV-2 infected MCF10A-ACE2 and Caco-2 cells. *Non-specific.
I3C blocks S-induced EMT and breast cancer metastasis
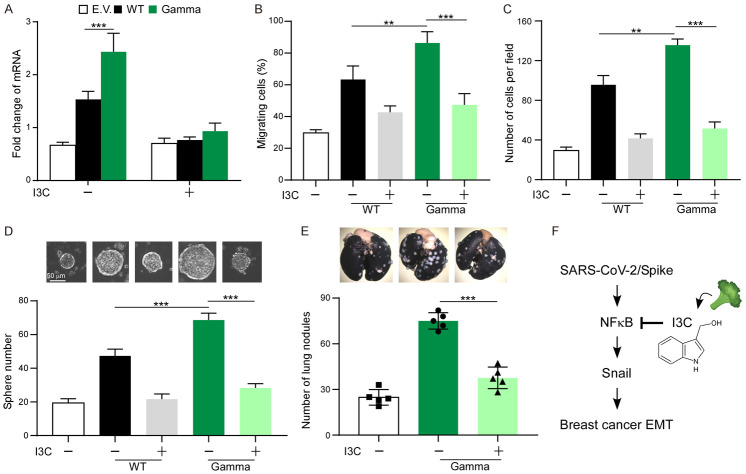
Inhibition of SARS-CoV-2 infection by I3C prompts us to investigate if I3C reduces SARS-CoV-2 Gamma variant-induced EMT. To do this, MCF7, MCF7-S (WT), and MCF7-S (Gamma) cells were treated with I3C and then examined the functional changes in cell-based assays. We found that NF-κB inhibition indeed impairs S of Gamma variant-induced EMT (Figure 4A). To explore the functional significance, we expressed the S of SARS-CoV-2 WT and Gamma variant in MCF7 cells. We found that downregulation of NF-κB by I3C reduced cell migration (Figure 4B) and invasion (Figure 4C), suggesting that NF-κB activation is required for S (Gamma)-induced EMT phenotype. Because EMT also increases cancer cell stemness, we further investigate if I3C reduces the tumorsphere formation ability of MCF7-S (WT) and MCF7-S (Gamma) cells (Figure 4D). While S protein increases tumorsphere by two-fold in S of WT and three-fold in S of Gamma variant, downregulation of NF-κB compromised S-mediated cancer stemness (Figure 4D). We next used a xenograft metastasis model to validate the abovementioned phenomenon. Consistently, I3C treatment antagonized S of Gamma variant-induced metastasis based on the number of lung nodules formed in mice (Figure 4E). Together, inhibition of NF-κB by I3C can effectively reduce Gamma variant-mediated breast cancer metastasis (Figure 4F).
Figure 4.
Blocking NF-κB suppresses S of Gamma variant-induced EMT. A. qPCR analysis of SNAIL expression in MCF7 expressing S of WT or Gamma variant. B. Quantification of migration activity of MCF7-S cells in the presence of I3C. C. Quantification of invasion activity of MCF7-S cells in the presence of I3C. D. Tumor initiation ability of MCF7 cells expressing S of WT or Gamma variant. E. 4T1-S cells were injected into female BALB/c mice via tail vein. Mice were treated with I3C by oral gavage. Lung nodules were stained with India ink at the experimental endpoint. Error bars represent the mean ± SD of five mice. F. Proposed working model indicating the mechanism of action of I3C. Statistic method: One-way ANOVA with Tukey’s post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Chronic inflammation is associated with a higher risk of infection and poor disease outcomes among COVID-19 patients. Previous studies have indicated that patients with inflammatory diseases such as cancers, chronic obstructive pulmonary diseases, and diabetes are more susceptible to SARS-CoV-2 infection and may develop more severe symptoms [21-23]. In this study, our infectious experiment has demonstrated that SARS-CoV-2 activates the NF-κB pathway, leading to up-regulation of inflammatory mediators such as TNFα and IL-6. Although NF-κB triggers both innate and adaptive immune reactions in response to pathogens, NF-κB may be hijacked by the virus for its propagation. Excessive activation of the NF-κB pathway upon MERS or SARS infection induces a cytokine storm, causing acute lung damage and acute respiratory distress syndrome, being a leading cause of death [24,25]. Therefore, targeting NF-κB may represent a promising therapeutic strategy for inhibiting stage-specific, inflammation-associated SARS-CoV-2 infection.
Activation of NF-κB is a hallmark of viral infections. NF-κB signaling is known to promote immune cell infiltration to eliminate viruses, and it is an important signal transduction pathway in the inflammatory response triggered by host cells at the early stages of viral infection [26,27]. However, viruses have evolved unique and sophisticated ability to modulate NF-κB signaling. Certain viruses, such as influenza A, HIV, or HSV-1, hijack the NF-κB signal to promote viral infection by enhancing viral replication [26,28]. Moreover, some viruses harness the NF-κB pathway to aid viral transmission by inducing apoptosis [29]. Previous studies also showed that inhibition of NF-κB can decrease the viral load by blocking viral replication [30,31]. In fact, viral entry, replication, and egression are all mediated by NF-κB [26,30]. The hepatitis C virus, an RNA virus, can utilize NF-κB for viral entry [32]. NF-κB can also stabilize membrane receptor expression, enabling continuous ACE2 expression to support viral entry [33]. Compared to the SARS-CoV-2 WT, the Gamma variant induced a stronger NF-κB activation, leading to a more aggressive cancerous phenotype. These results suggest cancer patients with COVID-19 Gamma variant infection may experience a strong potential of cancer metastasis or recurrence.
Our research links breast cancer metastasis to SARS-CoV-2 infection, and we further identify I3C as one of the potent antiviral agents that can reduce S-mediated metastasis. These potential therapeutic agents inhibit NF-κB activity by preventing its translocation into the nucleus [14,34]. Many NF-κB inhibitors, both clinically utilized and Food and Drug Administration (FDA)-approved, have been developed to block TNFR1, IKKβ, or the proteasome to treat diseases such as diabetes or cancers [35,36]. I3C is a relatively non-toxic natural compound [37] and effectively prevents transmission of SARS-CoV-2 by reducing NF-κB activity. Therefore, our results support that I3C may represent a promising alternative therapy for controlling SARS-CoV-2 infection. Given that NF-κB is ubiquitously expressed [20], administration of NF-κB inhibitors may significantly impede SARS-CoV-2 infection-mediated cancer progression.
Acknowledgements
This work was funded by the following agency: Ministry of Science and Technology (MOST 109-2314-B-001-002 and MOST 109-2314-B-001-008 to C.-W.Li; MOST-108-3114-Y-001-002 to M.-H.Tao; MOST 108-2320-B-001-034-MY2 to S.-Y.Chen). Academia Sinica (AS-SUMMIT-109, AS-KPQ-109-BioMed to M.-H.Tao; AS-CDA-110-L09, AS-GC-110-05, and AS-KPQ-110-EIMD to S.-Y.Chen). University of Massachusetts Lowell (Faculty start-up D50210000000022 to Y.-J.Lai). Additionally, we thank Taiwan CDC for providing SARS-CoV-2 TCDC#4 (hCoV-19/Taiwan/4/2020) and funding support for the IBMS P3 facility (AS-CFII-108-102) and the Ministry of Science and Technology (MOST 109-3114-Y-001-001) for the COVID-19 study. We also thank the DNA Sequencing Core Facility (AS-CFII-108-115) and Light Microscopy Core Facility of Academia Sinica for the service.
Disclosure of conflict of interest
None.
References
- 1.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier DA, De Marco A, Ferreira IATM, Meng B, Datir RP, Walls AC, Kemp SA, Bassi J, Pinto D, Silacci-Fregni C, Bianchi S, Tortorici MA, Bowen J, Culap K, Jaconi S, Cameroni E, Snell G, Pizzuto MS, Pellanda AF, Garzoni C, Riva A CITIID-NIHR BioResource COVID-19 Collaboration; Elmer A, Kingston N, Graves B, McCoy LE, Smith KGC, Bradley JR, Temperton N, Ceron-Gutierrez L, Barcenas-Morales G COVID-19 Genomics UK (COG-UK) Consortium. Harvey W, Virgin HW, Lanzavecchia A, Piccoli L, Doffinger R, Wills M, Veesler D, Corti D, Gupta RK. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, Albert M, Donati F, Prot M, Behillil S, Enouf V, Maquart M, Smati-Lafarge M, Varon E, Schortgen F, Yahyaoui L, Gonzalez M, De Sèze J, Péré H, Veyer D, Sève A, Simon-Lorière E, Fafi-Kremer S, Stefic K, Mouquet H, Hocqueloux L, van der Werf S, Prazuck T, Schwartz O. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;25:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 6.Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, Xia H, Swanson KA, Cutler M, Cooper D, Menachery VD, Weaver SC, Dormitzer PR, Shi PY. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 7.Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, McGregor A, Morrison H, Mujadidi YF, Nana A, O’Reilly PJ, Padayachee SD, Pittella A, Plested E, Pollock KM, Ramasamy MN, Rhead S, Schwarzbold AV, Singh N, Smith A, Song R, Snape MD, Sprinz E, Sutherland RK, Tarrant R, Thomson EC, Török ME, Toshner M, Turner DPJ, Vekemans J, Villafana TL, Watson MEE, Williams CJ, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ Oxford COVID Vaccine Trial Group. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu TJ, Chang SS, Li CW, Hsu YH, Chen TC, Lee WC, Yeh CT, Hung MC. Severe hepatitis promotes hepatocellular carcinoma recurrence via NF-κB pathway-mediated epithelial-mesenchymal transition after resection. Clin Cancer Res. 2016;22:1800–1812. doi: 10.1158/1078-0432.CCR-15-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun SC, Chang JH, Jin J. Regulation of nuclear factor-κB in autoimmunity. Trends Immunol. 2013;34:282–289. doi: 10.1016/j.it.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, Castaño-Rodriguez C, Perlman S, Enjuanes L. Inhibition of NF-κB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. 2014;88:913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takada Y, Andreeff M, Aggarwal BB. Indole-3-carbinol suppresses NF-kappaB and IkappaBalpha kinase activation, causing inhibition of expression of NF-kappaB-regulated antiapoptotic and metastatic gene products and enhancement of apoptosis in myeloid and leukemia cells. Blood. 2005;106:641–649. doi: 10.1182/blood-2004-12-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy KW, Murugan D, Leong XF, Abas R, Alias A, Mustafa MR. Flavonoids as natural anti-inflammatory agents targeting Nuclear Factor-Kappa B (NFkappaB) signaling in cardiovascular diseases: a mini review. Front Pharmacol. 2019;10:1295. doi: 10.3389/fphar.2019.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad A, Biersack B, Li Y, Kong D, Bao B, Schobert R, Padhye SB, Sarkar FH. Targeted regulation of PI3K/Akt/mTOR/NF-κB signaling by indole compounds and their derivatives: mechanistic details and biological implications for cancer therapy. Anticancer Agents Med Chem. 2013;13:1002–1013. doi: 10.2174/18715206113139990078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z, Zhang Z, You H, Wu M, Zheng Q, Xiong Y, Xiong H, Wang C, Chen C, Xiong F, Zhang Y, Peng Y, Ge S, Zhen B, Yu T, Wang L, Wang H, Liu Y, Chen Y, Mei J, Gao X, Li Z, Gan L, He C, Li Z, Shi Y, Qi Y, Yang J, Tenen DG, Chai L, Mucci LA, Santillana M, Cai H. Patients with cancer appear more vulnerable to SARS-CoV-2: a multicenter study during the COVID-19 outbreak. Cancer Discov. 2020;10:783–791. doi: 10.1158/2159-8290.CD-20-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart CA, Gay CM, Ramkumar K, Cargill KR, Cardnell RJ, Nilsson MB, Heeke S, Park EM, Kundu ST, Diao L, Wang Q, Shen L, Xi Y, Maria Della Corte C, Fan Y, Kundu K, Pickering CR, Johnson FM, Zhang J, Kadara H, Minna JD, Gibbons DL, Wang J, Heymach JV, Byers LA. SARS-CoV-2 infection induces EMT-like molecular changes, including ZEB1-mediated repression of the viral receptor ACE2, in lung cancer models. bioRxiv. 2020 [Google Scholar]
- 17.Lai YJ, Chao CH, Liao CC, Lee TA, Hsu JM, Chou WC, Wang J, Huang HC, Chang SJ, Lin YL, Li CW. Epithelial-mesenchymal transition induced by SARS-CoV-2 required transcriptional upregulation of snail. Am J Cancer Res. 2021;11:2278–2290. [PMC free article] [PubMed] [Google Scholar]
- 18.Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, Chao CH, Yamaguchi H, Yang NK, Ding Q, Wang Y, Lai YJ, Labaff AM, Wu TJ, Lin BR, Yang MH, Hortobagyi GN, Hung MC. Epithelial-mesenchyme transition induced by TNF-α requires NF-κB mediated transcriptional upregulation of Twist1. Cancer Res. 2012;72:1290–300. doi: 10.1158/0008-5472.CAN-11-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno-Bueno G, Peinado H, Molina P, Olmeda D, Cubillo E, Santos V, Palacios J, Portillo F, Cano A. The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc. 2009;4:1591–1613. doi: 10.1038/nprot.2009.152. [DOI] [PubMed] [Google Scholar]
- 20.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonnet A, Chetboun M, Poissy J, Raverdy V, Noulette J, Duhamel A, Labreuche J, Mathieu D, Pattou F, Jourdain M LICORN and the lille COVID-19 and Obesity study group. High prevalence of obesity in Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity (Silver Spring) 2020;28:1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang K, Sheng Y, Huang C, Jin Y, Xiong N, Jiang K, Lu H, Liu J, Yang J, Dong Y, Pan D, Shu C, Li J, Wei J, Huang Y, Peng L, Wu M, Zhang R, Wu B, Li Y, Cai L, Li G, Zhang T, Wu G. Clinical characteristics, outcomes, and risk factors for mortality in patients with cancer and COVID-19 in Hubei, China: a multicentre, retrospective, cohort study. Lancet Oncol. 2020;21:904–913. doi: 10.1016/S1470-2045(20)30310-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alqahtani JS, Oyelade T, Aldhahir AM, Alghamdi SM, Almehmadi M, Alqahtani AS, Quaderi S, Mandal S, Hurst JR. Prevalence, severity and mortality associated with COPD and smoking in patients with COVID-19: a rapid systematic review and meta-analysis. PLoS One. 2020;15:e0233147. doi: 10.1371/journal.pone.0233147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar N, Xin ZT, Liang Y, Ly H. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J Virol. 2008;82:9880–9889. doi: 10.1128/JVI.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker RG, Hayden MS, Ghosh S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, He S, Minassian A, Li J, Feng P. Recent advances on viral manipulation of NF-κB signaling pathway. Curr Opin Virol. 2015;15:103–111. doi: 10.1016/j.coviro.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khandelwal N, Simpson J, Taylor G, Rafique S, Whitehouse A, Hiscox J, Stark LA. Nucleolar NF-kappaB/RelA mediates apoptosis by causing cytoplasmic relocalization of nucleophosmin. Cell Death Differ. 2011;18:1889–1903. doi: 10.1038/cdd.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang CW, Lee YZ, Hsu HY, Shih C, Chao YS, Chang HY, Lee SJ. Targeting coronaviral replication and cellular JAK2 mediated dominant NF-κB activation for comprehensive and ultimate inhibition of coronaviral activity. Sci Rep. 2017;7:4105. doi: 10.1038/s41598-017-04203-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palamara AT, Nencioni L, Aquilano K, De Chiara G, Hernandez L, Cozzolino F, Ciriolo MR, Garaci E. Inhibition of influenza A virus replication by resveratrol. J Infect Dis. 2005;191:1719–1729. doi: 10.1086/429694. [DOI] [PubMed] [Google Scholar]
- 32.Fletcher NF, Clark AR, Balfe P, McKeating JA. TNF superfamily members promote hepatitis C virus entry via an NF-κB and myosin light chain kinase dependent pathway. J Gen Virol. 2017;98:405–412. doi: 10.1099/jgv.0.000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takase O, Marumo T, Imai N, Hirahashi J, Takayanagi A, Hishikawa K, Hayashi M, Shimizu N, Fujita T, Saruta T. NF-kappaB-dependent increase in intrarenal angiotensin II induced by proteinuria. Kidney Int. 2005;68:464–473. doi: 10.1111/j.1523-1755.2005.00424.x. [DOI] [PubMed] [Google Scholar]
- 34.Shin HM, Kim MH, Kim BH, Jung SH, Kim YS, Park HJ, Hong JT, Min KR, Kim Y. Inhibitory action of novel aromatic diamine compound on lipopolysaccharide-induced nuclear translocation of NF-kappaB without affecting IkappaB degradation. FEBS Lett. 2004;571:50–54. doi: 10.1016/j.febslet.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 35.Suryavanshi SV, Kulkarni YA. NF-κβ: a potential target in the management of vascular complications of diabetes. Front Pharmacol. 2017;8:798. doi: 10.3389/fphar.2017.00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 37.Reed GA, Arneson DW, Putnam WC, Smith HJ, Gray JC, Sullivan DK, Mayo MS, Crowell JA, Hurwitz A. Single-dose and multiple-dose administration of indole-3-carbinol to women: pharmacokinetics based on 3,3’-diindolylmethane. Cancer Epidemiol Biomarkers Prev. 2006;15:2477–2481. doi: 10.1158/1055-9965.EPI-06-0396. [DOI] [PubMed] [Google Scholar]