Abstract
APOBEC enzymes are strong mutagenic factors. In breast cancer, expression of APOBEC3B is increased and associated with mutation load and poor outcome. Other APOBEC3s can also mutate DNA but their clinical significance in breast cancer and its underpinnings have not been comprehensively studied. In our examination of 1,091 breast carcinoma cases, high expression of APOBEC3A or APOBEC3B genes was associated with greater tumor burden of mutations and other genomic aberrations. Expression of none of the five APOBEC3C-H genes had any correlation with these features, including T[C-T/G]W mutations, but their high expression levels indicated a robust anti-cancer immune response within tumors, with elevated CD8+ T cell abundance, T cell receptor diversity, and immune cytolytic activity. Concordantly, survival analyses of this and two other cohorts with > 3,000 patients each showed favorable prognostic benefit of high APOBEC3C-H expression for both cancer progression and mortality. A detrimental prognostic value was observed for APOBEC3A and APOBEC3B. Single-cell data revealed cancer epithelial and stromal immune cells as major sources of APOBEC3B and APOBEC3C-H expression in tumors, respectively. These observations on opposing associations with breast cancer of different APOBEC3s highlight the contrasting roles of these enzymes, promoting cancer through mutagenesis while antagonizing it through immune response.
Keywords: APOBEC, breast cancer, mutation, prognosis, RNA editing, tumor immune microenvironment
Introduction
Analyses of the sequence context of mutations that arise in cancer show that single nucleotide C-to-T or -G mutations in the TCW trinucleotide context (W = A or T) comprise a significant fraction of mutations in a number of solid tumors, especially those of bladder, breast, cervix, head & neck, and lung [1,2]. These mutations are believed to arise from the activity of APOlipoprotein B mRNA Editing Catalytic polypeptide-like (APOBEC) proteins. APOBECs are zinc-coordinating enzymes with one or more cytidine deaminase (CD) domains that can convert cytosine (C) bases in single-stranded DNA or RNA to uracil (U). Within cells, DNA deamination by APOBEC3s generates C-to-T as well as C-to-G mutations, the latter arising consequent to cellular repair processes. Among the 11 human APOBEC proteins, expression of APOBEC1, APOBEC2, APOBEC4, and Activation-Induced Deaminase (AID) is tissue-specific, respectively restricted to liver, muscle, gonads, and lymphocytes [3], and APOBEC2 and APOBEC4 lack any known ability to deaminase C bases [4]. Thus, mutations with APOBEC signature occurring in solid tumors likely originate from the activity of the remaining seven APOBECs, APOBEC3A-D and APOBEC3F-H (there is no APOBEC3E), which are encoded by a cluster of seven APOBEC3A-H genes on chromosome 22. APOBEC3A, 3C, and 3H have a single CD domain, whereas APOBEC3B, 3D, 3F, and 3G have double CD domains. All seven APOBEC3s have the ability to convert C bases of DNA to U, especially in the TCW context [5-7]. However, C-to-U conversion in RNA (RNA editing) has been reported for only APOBEC3A, 3B, and 3G [8]. DNA mutagenesis by APOBEC3s is widespread in human cancers [1], and is believed to contribute to cancer progression and therapy resistance. Localized hyper-mutation in cancer cell genomes, termed kataegis, which is characterized by clusters of C-to-U or -G mutations in preferential TCW context is also supposed to be a result of APOBEC activity [1,9,10].
Among the seven APOBEC3s, APOBEC3B has been studied the most in human cancer, especially that of breast. APOBEC3B has been described as a strong driver of breast cancer [2]. Expression of the APOBEC3B gene is upregulated in most primary breast cancer tumors and is associated with kataegis, aggressive clinical and pathological features like high nuclear grade and Ki67 index [11,12], and poor prognosis [12-17]. Oddly, higher APOBEC3B gene expression in breast cancer was significantly associated with achievement of pathological complete response to neoadjuvant chemotherapy in a cohort of 274 patients who underwent tumor resection, though the gene expression level was not prognostic of relapse-free interval or disease-specific survival [12]. Unlike APOBEC3B, the significance of the five APOBEC3C-H proteins in breast cancer is unclear. APOBEC3C-H also possess the C-to-U DNA mutating ability, which is believed to underlie their known role in inhibiting infections by viruses such as human immune deficiency virus type 1 (HIV-1), hepatitis B, and Epstein-Barr virus through deamination of viral cDNAs [3,18-20]. Here, to investigate the relevance of APOBEC3C-H in breast cancer, we performed systematic cancer genomics and transcriptomics association studies.
Materials and methods
Patient cohorts
Clinical and APOBEC3 gene expression data of three breast cancer patient cohorts are examined in this study (cohort characteristics in Table S1). The study focuses on the cohort of 1,097 breast carcinoma cases (BRCA) of The Cancer Genome Atlas (TCGA) project that undertook molecular analyses of treatment-naive tumors diagnosed during 1978-2013 (median 2009) [21]. The Sweden Cancerome Analysis Network - Breast (SCAN-B) cohort of 3,273 patients who were diagnosed during 2010-2015 and had transcriptome profiling of resected tumors is from an on-going study. The latest publicly available clinical data of these patients was obtained from resources noted in a recent SCAN-B study [22]. The “Kaplan-Meier (KM) Plotter” set of 5,134 patients is a meta-cohort of subjects and normalized tumor gene expression data that was developed for breast cancer research [23] and currently encompasses 35 studies (2016.10.13 update).
Gene expression data
For 1,091 fresh-frozen tumor and 112 normal tissues of the 1,092 subjects of the TCGA-BRCA project for whom RNA sequencing data was available, gene-level read counts were obtained in April 2018 from National Cancer Institute (USA) Genome Data Commons portal with TCGAbiolinks Bioconductor package for R (version 2.5.9) [24], and used to generate transcripts per million (TPM) values, with transcript lengths as per release 92 of Ensembl human gene annotations. Count data was also processed with edgeR Bioconductor package (version 3.20.9) [25] for normalization with the trimmed mean of M-values (TMM) method to generate gene expression data in counts per million (CPM), excluding genes without count of ≥ 2 in > 56 tissues. Values for rowsum.filter and prior.df parameters in edgeR estimateCommonDisp and estimateTagwiseDisp functions were set at 224 and 0.25, respectively. Data without a Human Genome Organization (HUGO) gene symbol was removed from the final set of gene expression values. For log2-transformation, the expression values were padded with 0.25. For the 523 fresh-frozen tumors for which mRNA transcriptome had also been profiled with Agilent microarrays, a log2-transformed, normalized, microarray-based gene expression dataset was obtained from Broad Institute’s Firebrowse (version 1.1.40) [26]. Log2-transformed RNA sequencing-based gene expression measurements of SCAN-B tumors, in terms of fragments per kb per million reads (FPKM), were obtained in July 2020 from Gene Expression Omnibus (dataset GSE96058). RNA sequencing-based gene expression measurements in TPM units for 55 human breast cancer cell-lines of the Cancer Cell Line Encyclopedia [27], 16 human tissues of the Illumina Body Map [28], and 27 types of human peripheral blood cells of the BLUEPRINT Epigenome [29] projects were obtained from the European Bioinformatics Institute Expression Atlas [30] in June 2018. Normalized single-cell gene expression and cell identity data of dissociated cells of six human breast cancer tumors examined by Karaayvaz et al. [31] were obtained from Gene Expression Omnibus in June 2020 (GSE118389).
Single nucleotide mutation data
Counts of somatic single nucleotide substitutions in exomes of TCGA-BRCA tumors (n = 1,014) were estimated from publicly available mutation annotation format (MAF) files of the TCGA project [32]. Helmsman software (version 1.1.0) [33] was used to derive the trinucleotide contexts of the mutations, and deconstructSigs Bioconductor package (version 1.8.0) [34] was used for context-based mutational signature analysis. C-to-G or -T mutations in TCW trinucleotide context were considered to be APOBEC-mediated. For breast cancer cell-lines, APOBEC-mediated mutation counts were obtained from the Jarvis et al [35].
Other TCGA-BRCA data
Nottingham histological scores were manually collated from pathology reports by us for a recently published study [36]. Scores could be retrieved for 588 subjects. Relative abundances of 22 types of tumor-infiltrating immune cells were conjectured from tumor gene expression data using the CIBERSORT deconvolution method (online version 1.06) [37] with TPM gene expression values, the LM22 signature matrix, and 100 permutations. Samples (n = 176) with p value > 0.05 in the CIBERSORT examination were excluded from downstream analyses. Abundances for supersets of cell types were generated by adding abundance values of their constituents [38]. Immune cytolytic activity in tumors was quantified from TPM data of GZMB and PRF1 genes as the CYT score [39]. Quantifications of other tumor immune features such as T cell receptor (TCR) diversity and leukocyte infiltration were from Thorsson et al [38]. Clinical outcome data was obtained from the Pan-Cancer Clinical Data Resource [21], a standardized, curated, and filtered data-set for survival endpoints for TCGA cases. Prediction Analysis of Microarray (PAM50) subtype data was from Berger et al [40]. Information for other clinical and pathologic variables such as age, tumor stage, and status for estrogen receptor (ER) expression or human epidermal growth factor receptor 2 (HER2) overexpression was obtained through the cBio Cancer Genomics Portal [41]. Measurements of tumor genome characteristics like aneuploidy, copy number alterations (CNA), clonal heterogeneity quantified by ABSOLUTE method, and homologous recombination defects were from the study of Ellrott et al [38].
Comparisons of groups identified by APOBEC3 gene expression in tumors
Using within-cohort tertiles, patients were grouped by their tumor APOBEC3 gene expression into high (top tertile) and low (bottom tertile) expressors. Comparisons of these groups of patients for survival outcomes was performed with the survival package (version 3.1-12) in R; Kaplan-Meier method with log-rank test was used. Survival analyses of the KM Plotter cohort for similar comparison of within-cohort tertile groups using log-rank test was performed with the KM Plotter online tool at http://kmplot.com in August 2020. Enrichment of genesets in tumors of high relative to low expressors was analyzed with GSEA desktop software (version 4.0.2) using the 50-geneset Hallmark [42] collection of the Molecular Signatures Database (MSigDB; version 7.1). Genesets with normalized enrichment score > 1.5 and false discovery rate (FDR) q value < 0.05 were considered significant in the enrichment analyses.
Other
For the TCGA cohort, TPM values of APOBEC3 gene expression were used for inter-APOBEC3 comparisons, and in comparison of tumor and adjacent normal tissues; CPM values were used in other analyses. Statistical analyses and data plotting were performed using R (version 3.6.2) and Prism (version 8.4.3; GraphPad Software®, San Diego, USA). Unless noted otherwise, software options were default ones, and a threshold of 0.05 was used to deem significance from p values of statistical tests.
Results
All seven APOBEC3 genes are expressed in breast cancer tumors
Data of the Illumina Body Map project [28], in which transcriptomes of 16 healthy human tissues were profiled by RNA sequencing, shows that all seven APOBEC3 genes are expressed in breast, with APOBEC3C expressed the most compared to other APOBEC3s (Figure S1A). Expression of all APOBEC3 genes, and the highest expression for APOBEC3C, are also observed among the 1,091 treatment-naive breast carcinoma tumors of The Cancer Genome Atlas project (TCGA-BRCA) for whom RNA sequencing data was available (Figure 1A). There is good correlation among the six APOBEC3A and APOBEC3C-H genes for their expression levels in the tumors (Spearman ρ = 0.38-0.75), but their correlations with APOBEC3B levels are poor, with ρ of 0.09-0.29. Comparison of tumors with their matched normal adjacent tissues, for the 112 subjects with available data, shows that expression of APOBEC3A, 3B, and 3H is significantly higher in tumors compared to normal, by 1.5-, 3.9-, and 1.3-fold on average, with paired t test p values of 0.002, < 0.0001, and 0.005, respectively (Figure 1B). On the other hand, significantly reduced expression in tumor compared to normal tissue is observed for APOBEC3C, 3D, and 3G, with fold-changes of 0.5, 0.8, and 0.8 (P < 0.0001, 0.02, and 0.001), respectively. It should be noted, though, that the 112 patients varied widely for APOBEC3 gene expression differences between their tumor and normal tissues, and similar tumor-normal gene expression changes in > 75% of subjects were seen for only APOBEC3B and 3C (Figure 1B).
Figure 1.
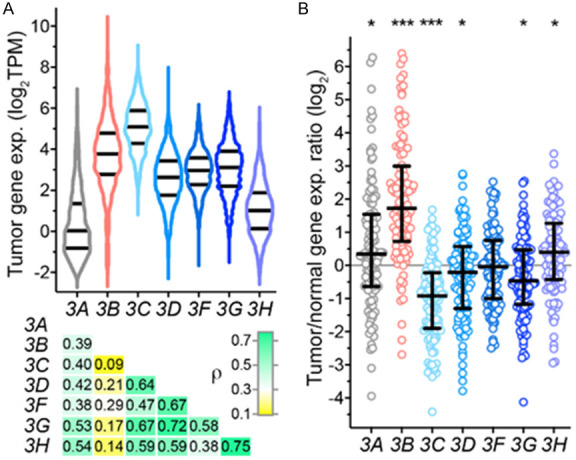
APOBEC3 gene expression in breast cancer tissues of TCGA. A. Violin plots of gene expression in 1,091 tumors with median and inter-quartile range (IQR) are shown for the seven APOBEC3 genes (3A-3H). Underneath is a heatmap of Spearman coefficients (ρ) of correlations between the various APOBEC3s. TPM, transcripts per million. B. Ratio of APOBEC3 gene expression of tumors and their matched adjacent normal breast tissue is plotted along with median and IQR values for 112 patients with data available for both tissues. Significant p values in paired tumor vs. normal comparison by Welch t test are indicated (*, < 0.05; ***, < 0.001).
Unlike APOBEC3A and 3B, APOBEC3C-H gene expression is not strongly associated with adverse cancer pathological features
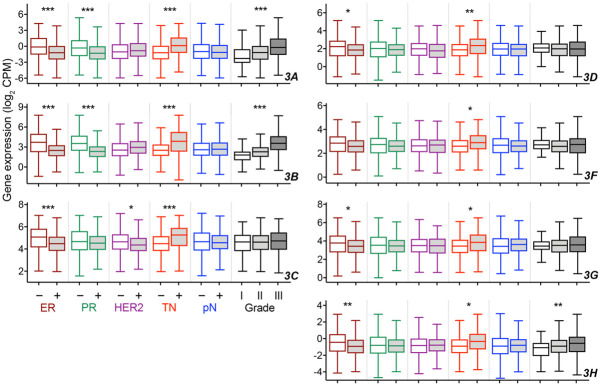
The positive association of high APOBEC3B gene expression in breast cancer with adverse pathological features has been demonstrated previously, with data of the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort of ~2,000 patients [11]. In the TCGA cohort too, APOBEC3B expression was higher among tumors that lack expression of ER or progesterone receptor (PR), are triple negative (TNBC) for ER, PR, and HER2 status, or are of elevated Nottingham histological grade (Welch t test p < 0.001, after adjustment for multiple testing with Benjamini-Hochberg method, for all comparisons; Figure 2). However, unlike for METABRIC, APOBEC3B gene expression did not differ by status for HER2 or lymph node metastasis in TCGA (Figure 2). Associations of APOBEC3A with tumor pathological features were like those of APOBEC3B. While each of the five APOBEC3C-H genes had higher expression in ER-negative (except for APOBEC3F) or TNBC tumors, similar to APOBEC3B, they had no association with histological grade (except for APOBEC3H) or node involvement. For APOBEC3C only, expression was significantly higher among HER2-negative tumors. These assessments of gene expression and tumor pathological features show a separation of APOBEC3C-H from APOBEC3A and 3B, with the former lacking a strong association with features that are prognostically adverse.
Figure 2.
Association of APOBEC3 gene expression with pathological features of TCGA breast cancer. Tukey boxplots of APOBEC3 gene expression are shown for tumors of different histological grade (data available for 77, 268, and 235 tumors for grades I, II, and III, respectively), and status for estrogen receptor (ER; 237 negative, 802 positive), progesterone receptor (PR; 342 negative, 694 positive), human epidermal growth factor receptor 2 (HER2; 764 negative, 185 positive), triple-negativity for the three receptors (TN; 858 false, 160 true), and pathologic lymph node metastasis (pN; 513 negative, 556 positive). Significant p values, adjusted for multiple testing with Benjamini-Hochberg method, are noted and were calculated with one-way Welch ANOVA test (*, < 0.05; **, < 0.005; ***, < 0.0005).
Expression of APOBEC3A and 3B but not APOBEC3C-H in breast cancer associates with genomic aberrations
To investigate the extent to which different APOBEC3s may reflect cancer-associated genomic changes in breast cancer, we examined the association of their gene expression with various genome features. APOBEC3A and 3B are believed to be the main drivers of APOBEC signature-bearing single nucleotide mutations (C-to-G or -T in the TCW trinucleotide context) [9,13,15,43,44]. Consistent with this, APOBEC3A and 3B gene expression of tumors correlated positively with their load of all exonic single nucleotide mutations, with Spearman ρ of 0.33 and 0.35 respectively (P < 0.001 for both; Figure 3A). Similarly, positive correlations were observed for the subset of APOBEC signature mutations (i.e., likely to be APOBEC-mediated), with ρ values of 0.33 and 0.31 respectively (both P < 0.01). On the other hand, Spearman coefficients were poor (0.10-0.21) for APOBEC3C-H in analyses of all and APOBEC-mediated mutations. In examination of the 55 human breast cancer cell-lines that have been characterized in the Cancer Cell Line Encyclopedia (CCLE) project, APOBEC3C-H expression levels of the cell-lines had similarly poor correlation with their load of APOBEC-mediated mutations (ρ = -0.28-0.03), whereas correlations for APOBEC3A and 3B were 0.52 and -0.11 respectively. As might be expected from their correlations with mutation burden, both APOBEC3A and 3B gene expression had good correlation with neoantigen load. Copy number alteration, aneuploidy, homologous recombination defects, and clonal heterogeneity of tumors too correlated positively with APOBEC3B level (ρ = 0.18-0.41), whereas such correlations were weak or absent for APOBEC3C-H (ρ = -0.15-0.12; Figure 3A).
Figure 3.
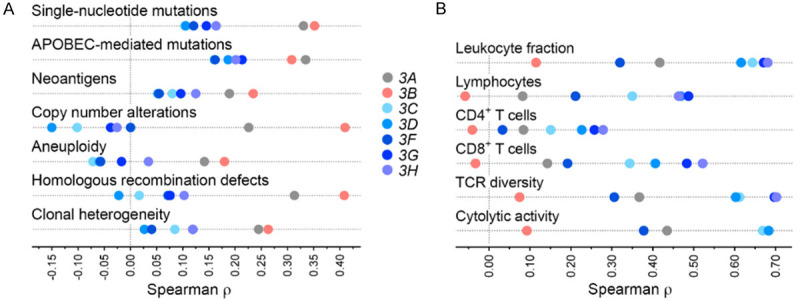
Association of APOBEC3 gene expression of breast tumors of The Cancer Genome Atlas with their cancer genome and immune characteristics. Shown are Spearman coefficient (ρ) values in analyses of correlation of gene expression of tumors with various features. A. For genome features, total and APOBEC-mediated single-nucleotide exonic mutation counts (n = 1,014 tumors with available data), counts of neoantigens predicted to arise from single-nucleotide mutations and indels (856), segments affected by copy number alteration (1,062), scorings for aneuploidy (1,034) and homologous recombination defects (1,035), and clonal heterogeneity (1,016) are analyzed. B. For immune features, coefficients are shown for infiltrating leukocyte fraction (n = 1,064 tumors with available data) and relative abundance of lymphocytes, CD4+, and CD8+ T cells among infiltrating immune cells (915), T cell receptor (TCR) Shannon diversity (1,038), and immune cytolytic activity measured as the CYT score (1,091).
Immune activity is increased in tumors with high APOBEC3C-H expression
For each of APOBEC3C-H, tumor gene expression correlated positively with infiltration of the tumors with leukocytes (ρ = 0.32-0.68) and their lymphocyte subset (ρ = 0.21-0.49), whereas the correlations were poor for APOBEC3B (ρ = 0.11 and -0.06, respectively) (Figure 3B). Correlations of APOBEC3C-H levels with relative abundances of many major tumor-infiltrating immune cell subsets, such as B cells, macrophages, natural killer cells, and neutrophils were poor (Figure S2), positive associations of APOBEC3C-H expression, though modest, were observed for relative abundance of CD8+ T cells (ρ = 0.19-0.52). Concordantly, expression of each of these genes correlated positively with estimates of TCR diversity (ρ = 0.31-0.70) as well as immune cytolytic activity [39] within tumors (ρ = 0.38-0.78). For APOBEC3B, the correlations of gene expression with these features were much weaker (Figure 3B). Evidence of the positive association of APOBEC3C-H gene expression with increased anti-cancer immune activity within tumors was also noted in examination of the global tumor transcriptome with geneset enrichment analysis. For this, we scrutinized the 50 genesets of the mSigDB Hallmark collection [42] for enriched expression among tumors with high relative to tumors with low APOBEC3 gene expression, respectively identified as those in the top and bottom tertiles. With significance thresholds of 0.05 for FDR (q value) and 1.5 for normalized enrichment score, between 7 and 15 genesets were associated with each of the seven APOBEC3s. Six genesets were common to the five APOBEC3C-H genes as well as APOBEC3A. All of these genesets are related to immune activity (allograft rejection, complement, IL2-STAT5 and IL6-JAK-STAT3 signaling pathways, inflammatory response, and interferon gamma response). On the other hand, six of the 14 enriched genesets for APOBEC3B are associated with cell proliferation (E2F targets, G2M checkpoint, mitotic spindle, mTORC1 signaling, and Myc targets v1 and v2). None of those 14 genesets is for immune-related processes. Figure 4 depicts the functional enrichment of immune and proliferative pathways respectively in tumors with high APOBEC3C-H and APOBEC3B levels.
Figure 4.
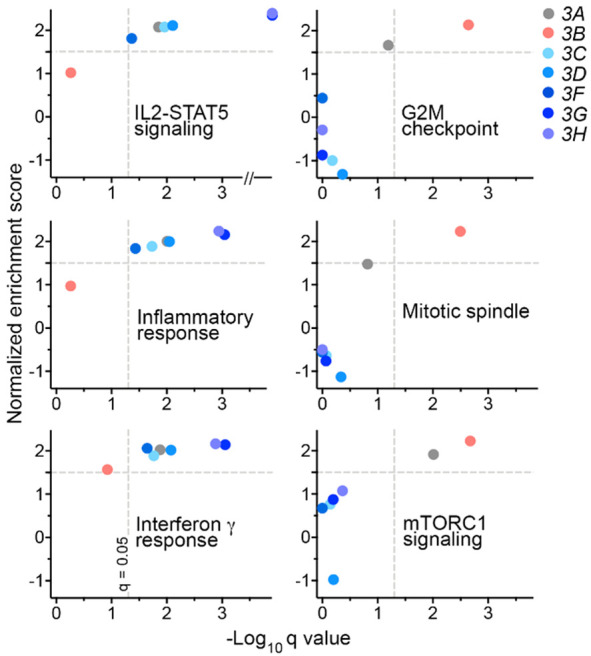
Breast cancer tumors with high APOBEC3C-H transcript levels have enriched expression of genesets for immune but not cell proliferation processes. Subjects (n = 1,091) were binned into high and low groups (top and bottom tertiles) by their tumor APOBEC3 gene expression to compare the two groups in geneset enrichment analysis. Normalized enrichment score and q values, generated from nominal p values after correction for false discovery, are plotted for each APOBEC3 gene for three each of immune- and cell proliferation-related mSigDB Hallmark genesets.
Breast cancer patients with high tumor APOBEC3C-3H expression have better survival
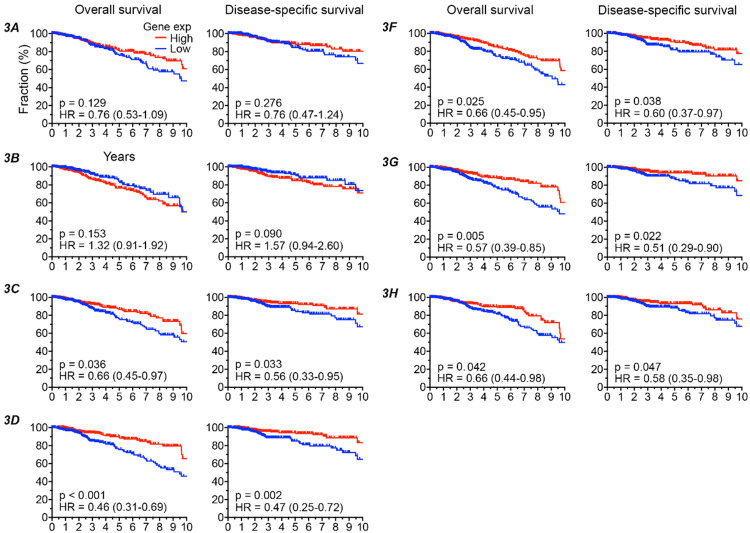
Consistent with its activity as a DNA mutator, high expression of APOBEC3B gene in breast cancer tumors has been associated with worse disease-free survival in the METABRIC cohort [11]. Because the other six APOBEC3s are capable of mutating DNA [3], we had hypothesized that their tumor gene expression too will have a negative impact on disease outcome. On the other hand, the positive associations of APOBEC3C-H with favorable tumor genome and immune features that we observed suggest a good prognosis for patients with high tumor APOBEC3C-H expression. To clarify this, we examined overall (OS) and disease-specific (DSS) survivals of the TCGA breast cancer patients, comparing the top and bottom one-thirds of patients defined by their tumor gene expression for each APOBEC3. Uniformly curated and filtered TCGA data for survival endpoints was used for this analysis [21]. In univariate analyses, though both OS and DSS were worse for high APOBEC3B expressors compared to low expressors, with hazard ratios (HR) of 1.32 and 1.57 respectively, the differences were not statistically significant (log-rank test p > 0.05) (Figure 5). Similarly, no significant association between APOBEC3A expression and survival was observed. However, for each of APOBEC3C-H, both OS and DSS were significantly better among high expressors, with hazard ratios of 0.43 to 0.66 (Figure 5).
Figure 5.
High APOBEC3C-H gene expression in breast tumors of The Cancer Genome Atlas is associated with improved survival. The 1,091 subjects with available tumor gene expression data were binned into high and low groups (top and bottom tertiles) by their APOBEC3 gene expression. Survival plots comparing the high and low groups along with hazard ratio (HR) with its 95% confidence interval and p values in log-rank tests are shown for association of APOBEC3 gene expression with overall and disease-specific survival of patients. The axis for time since cancer diagnosis is truncated at 10 years. Group-sizes in these analyses vary from 346 to 362 because of unavailability of accurate survival data for some subjects.
We analyzed two independent and large breast cancer cohorts to validate the prognostic benefit of high APOBEC3C-H expression that was observed for TCGA. The SCAN-B cohort was comprised of 3,273 patients of the on-going Swedish initiative [45] for whom clinical and RNA sequencing-based gene expression data is currently publicly available. The other cohort, KM Plotter, is an integrated clinical and hybridization microarray-based tumor gene expression dataset of 5,134 patients of 35 studies [23]. These two cohorts significantly differ from TCGA for certain characteristics such as age at diagnosis and tumor ER status (Table S1). Nevertheless, comparison of OS between within-cohort APOBEC3 gene expression-based top and bottom tertiles of patients showed a survival benefit of high APOBEC3C-G expression in one or both of the KM Plotter and SCAN-B cohorts, with HR values for comparisons with log-rank test p values < 0.05 associated with high gene expression ranging from 0.64 to 0.96 (Figure 6). For APOBEC3H, there was no association with OS in SCAN-B (P = 0.97); measurements of this gene are unavailable in the KM Plotter dataset. Both datasets lack DSS information. Unlike for TCGA, a significantly (P < 0.05) unfavorable association of high expression was noted for APOBEC3A and 3B in OS analysis of both KM Plotter and SCAN-B data, with HR values of 1.35 to 2.35. In these survival analyses, we ignored patient treatment because detailed data for this aspect was unavailable for all three cohorts. Besides surgery, a majority of patients receive neoadjuvant and/or adjuvant treatments of various types and variable efficacies (chemotherapy, hormonal therapy, radiation, etc.; Table S1).
Figure 6.
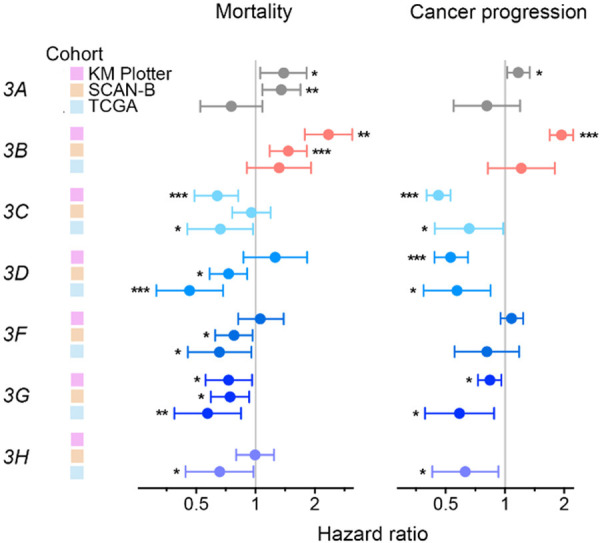
Favorable prognostic value of high tumor APOBEC3C-H gene expression in multiple breast cancer cohorts. For three independent cohorts, Kaplan-Meier (KM) Plotter (n = 5,134), Sweden Cancerome Analysis Network-Breast (SCAN-B; 3,273), and The Cancer Genome Atlas (TCGA; 1,091), subjects were binned into high and low groups by their APOBEC3 gene expression (within-cohort top and bottom tertiles). The groups were compared for mortality (overall survival) and cancer progression (disease recurrence or progression for KM Plotter and TCGA, respectively) in Kaplan-Meier analysis. Shown are hazard ratios and their 95% confidence intervals, with indicators of p values in log-rank tests (* < 0.05; ** < 0.01; *** < 0.001). The KM Plotter dataset lacks APOBEC3H values.
We also compared high and low APOBEC3C-H gene expressors for disease outcomes other than survival. For two of the three patient cohorts, time data was available for cancer progression - time to disease recurrence for KM Plotter, and disease progression-free interval for TCGA. For four of the five APOBEC3C-H genes, a protective benefit of high expression against cancer progression was observed in these cohorts, with HR values from 0.46 to 0.84 (Figure 6). In the KM Plotter but not the TCGA cohort, high APOBEC3B expression was had significant association with likelihood of disease progression. Taken together, these analyses demonstrate the favorable prognostic value of tumor APOBEC3C-H and the opposing detrimental value of APOBEC3A and 3B levels. In joint analyses of tumor APOBEC3 expression levels with various patient demographic and tumor pathological features using multivariable Cox proportional-hazards regression models, none of the seven APOBEC3s was associated with cancer mortality or progression independently of histological grade, and node, ER, and PR status, which are routinely evaluated in patients (all Wald test P > 0.05; Table S2). Thus, while the associations of tumor APOBEC3 gene expression with disease outcome are of biological interest, tumor APOBEC3 gene expression does not appear to be of practical translational value as prognostic biomarkers in breast cancer.
APOBEC3C-H expression primarily occurs in immune cells of tumors
To obtain an insight on the source of APOBEC3C-H gene expression associated with survival benefit in breast cancer patients, we evaluated data from a number of previous studies. APOBEC3 gene expression is well-known to be high in peripheral blood leukocytes [14]. Illumina Body Map data shows that expression of all seven APOBEC3s is higher in these cells compared to normal breast (Figure S1A). Data of the BLUEPRINT Epigenome project [29] shows that APOBEC3C-G are expressed in both myeloid and lymphoid leukocyte subsets in peripheral blood (Figure S1B). On the other hand, expression of APOBEC3B is stronger than APOBEC3C-H in the CCLE data for 55 breast cancer cell-lines (Figure S1C). These observations suggest that cancer epithelial and immune cells largely account for tumor APOBEC3B and APOBEC3C-H gene expression, respectively. To further explore the cellular origin of APOBEC3C-H expression within breast cancer tumors, we examined publicly available single-cell RNA sequencing data of breast cancer tumors. There is only one such study in which all cells were profiled without selection of specific cell-types [31]. Six triple-negative tumors were profiled in the study and cellular identities were established through their expression of canonical markers of cell-types, including VWF, EPCAM, LUM, and PTPRC (CD45) genes, respectively, for endothelial, epithelial, fibroblastic, or immune cells. As has been reported with immunohistochemical analysis of tumors [46], the study’s data also shows cancer epithelial cells to be the main source of APOBEC3B in breast cancer tumors (Figure 7). On the other hand, expression levels of APOBEC3C and 3G were much higher in immune compared to cancer cells. APOBEC3H was not detected in any type of cell and levels of APOBEC3D and 3F, though low, were higher in immune compared to cancer cells.
Figure 7.
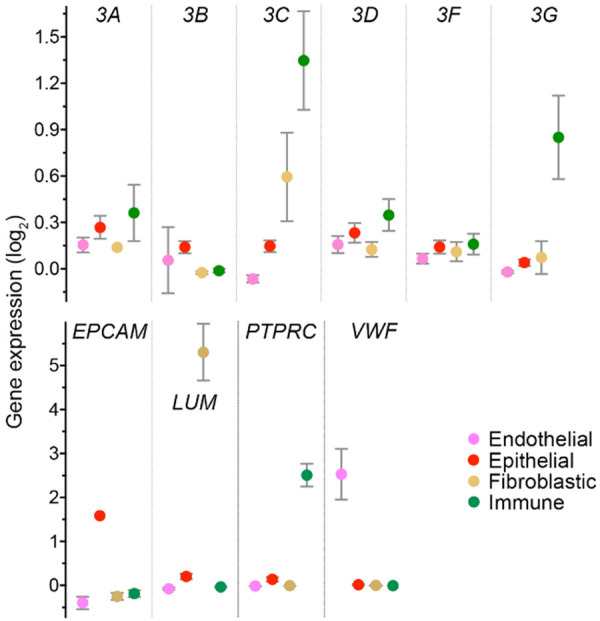
APOBEC3 gene expression in various components of breast cancer tumors. Gene expression in endothelial (n = 14), epithelial (868), fibroblastic (94), and immune (136) cells of six triple-negative breast cancer tumors are shown for the APOBEC3 genes, and for well-known gene markers of these types of cells. Mean and its 95% confidence interval are plotted for normalized gene expression data from the single-cell RNA sequencing study of Karaayvaz et al. APOBEC3H data was not captured for any of the cells. EPCAM, epithelial cell adhesion molecule; LUM, lumican; PTPRC, protein tyrosine phosphatase, receptor type, C (CD45); VWF, von Willebrand factor.
Discussion
In contrast to the other APOBECs, the seven human APOBEC3 proteins have demonstrable cytidine deamination activity and less restrictive tissue specificity of expression. These APOBEC3s are therefore likely to be the source of the APOBEC pattern of mutations that are prevalent in cancers of multiple organs [1,2]. A number of observations indicate that among the APOBEC3s, APOBEC3B is the cause of these mutations in breast cancer. Expression of APOBEC3B is higher in tumors relative to adjacent normal breast tissue, and in cancer relative to normal breast epithelial cell-lines, artificial overexpression of the gene in cell-lines generates mutations with APOBEC pattern, and there is a positive association between APOBEC3B gene expression of tumors and mutation and kataegis burdens [15,44]. In line with this, APOBEC3B gene expression in breast cancer is elevated in tumors with adverse pathological features and worse clinical outcome [11,16]. Unlike for APOBEC3B, the significance in breast cancer of the other APOBEC3s, which too are capable DNA mutators, has remained unclear, and we sought to address this with our study.
We find that in most breast cancer tumors of the TCGA cohort, and in many breast cancer cell-lines, all APOBEC3 genes are expressed, and among these genes, the RNA levels of APOBEC3C and not APOBEC3B are the highest (Figures 1A, S1C). However, unlike APOBEC3A and APOBEC3B, for which expression is on average > 3x higher in tumor compared to normal breast, APOBEC3C-H expression is either lower in tumors (APOBEC3C) or not significantly altered (Figure 1B). Similarity of APOBEC3A and APOBEC3B, and their dissimilarity from APOBEC3C-H are also seen in the associations of gene expression with tumor pathological features. We observed strong associations with prognostically adverse features like ER, PR, or triple negativity, and high tumor grade only for APOBEC3A and APOBEC3B and not APOBEC3C-H. However, contradicting what has been described for the METABRIC cohort [11], APOBEC3B expression was not associated with node involvement or positive HER2 status in the TCGA cohort. Consistent with their associations with pathological features, high APOBEC3C-H expression in tumors generally correlated with improved disease outcome for both progression and mortality while high expression of either APOBEC3A or APOBEC3B prognosticated worse outcomes (Figures 5, 6). We conclude so from survival analyses of three large and independent breast cancer patient cohorts. There was variation among the cohorts for some results of these analyses, such as the OS hazard reduction of high APOBEC3D or APOBEC3F expression reaching significance threshold (P < 0.05) for the SCAN-B and TCGA but not KM Plotter cohorts (Figure 6). This could be reflective of not only systemic differences between the cohorts, but also the platforms that were used for measuring gene expression (Table S1). Sequence similarity is high among APOBEC3 genes [47], which can reduce precision of measurement made using microarrays that rely on oligonucleotide probe hybridization, as is the case of KM Plotter and METABRIC cohorts. Indeed, an analysis of 523 TCGA-BRCA tumors whose transcriptomes were profiled using both RNA sequencing and microarray shows that correlation between the methods is either poor or modest for four of the seven APOBEC3s, with Spearman ρ of 0.80, 0.21, 0.46, and 0.81 for APOBEC3A, 3D, 3F, and 3H, respectively (Figure S3).
The clinical benefit that we observe for high tumor APOBEC3C-H expression is supported from its association with markers of heightened anti-cancer immune response within tumors, especially elevated abundance of CD8+ T cells, TCR diversity, immune cytolytic activity, and expression of genesets of immune processes (Figures 3B, 4). For APOBEC3B, these associations were absent or weak, and enriched expression of genesets for cell proliferation processes was observed, as has been reported for METABRIC [11] and in concordance with association of high APOBEC3B expression with tumor grade (Figure 2) and Ki67 level [17].
As expected, in both breast cancer cell-lines and TCGA tumors, correlations of gene expression of tumors with their mutation burden and other facets of cancer genome aberrations, including neoantigenicity, were strongest for APOBEC3B, and 3A, and weak or absent for APOBEC3C-H (Figure 3A). The same was seen for the tumor load of mutations of APOBEC pattern. Although APOBEC3C-H have the ability to act as DNA mutators [5,6], association of APOBEC3C-H gene expression with mutations of APOBEC pattern was not observed in either the tumors (Figure 3A) or cell-lines. This does not necessarily indicate that APOBEC3B is the major cause of APOBEC signature mutations in breast cancer because the mutations arose in the past, when gene expression levels may have been different. Mutations can drive cancer forward, for instance, by dysregulating checkpoints for cell proliferation, but they can also generate antigens that stimulate anti-cancer immune responses. Cancer mutagenesis occurs over long periods of time, and while it is accumulative, clonally generated mutations are also lost to selection pressure, including that from the anti-cancer immune response. This and the fact that immune cells also express APOBEC3s can complicate the examination of the biological and clinical relevance of tumor APOBEC3 activity. Indeed, our examination of gene expression of individual cells of breast cancer tumors suggests that epithelial cancer and immune cells are respectively the major sources of APOBEC3B and APOBEC3C-H expression in these tumors (Figure 7).
A comprehensive examination of all APOBEC3s covering multiple aspects of cancer biology such as immune features and patient outcome has not been previously described. Though we did not generate new data with experiment work, we utilized multiple large patient cohorts in our examination to strengthen the conclusions that may be drawn from its observations. Our study did not examine the APOBEC3 members at a finer level. For instance, haplotype I of APOBEC3H protein has increased nuclear localization and may therefore be more mutagenic compared to haplotype II [48], and a germline variation with deletion of the APOBEC3B coding region generates an APOBEC3A-3B hybrid that may be hypermutagenic [49]. It also remains possible that levels of APOBEC3s at the protein or enzyme activity level do not correlate with that at the RNA level, which is used for all analyses of this study. Clearly, additional experimental work is necessary to confirm some of the observations that we have presented here (such as immunohistochemistry or RNA in situ hybridization to identify cellular sources of APOBEC3s). We hope that our analyses of existing data guides such future work. Observations similar to those that we have made about APOBEC3s for breast cancer have been noted for other cancers. For instance, a survival benefit of high tumor APOBEC3G expression, associating with high levels of tumor-infiltrating T cells, has been made for ovarian cancer [50].
In conclusion, we show that while the associations of each of the seven APOBEC3s in breast cancer tumors with their genomic, immune, clinical features are concordant, the associations for APOBEC3C-H are favorably prognostic and opposite to the detrimental values of APOBEC3A and APOBEC3B. The underlying basis for the prognostic benefit of high APOBEC3C-H levels in tumors remains unknown. It is possible that this merely reflects the infiltration of tumors by immune cells that appear to be the major tumor source of expression for these genes. On the other hand, functional activities of the APOBEC3C-H proteins within immune cells may be important for anti-cancer immune response. Besides mutating DNA, many APOBEC3 enzymes can also cause C-to-U RNA editing to affect activity and level of proteins, and we have shown that APOBEC3-mediated C-to-U RNA editing occurs in breast cancer tumors and is associated with improved disease outcome [8]. APOBEC3s may also have molecular activities other than cytidine deamination [51]. Identification of these activities will help us understand the sharply contrasting roles of these enzymes, promoting cancer through mutagenesis while antagonizing it through immune response.
Acknowledgements
This research was supported by grants R01CA160688 to K.T. and P30-CA016056 to Roswell Park Comprehensive Cancer Center from National Institutes of Health, USA.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 5.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, Matthews N, Rowan A, Chambers T, Moore DA, Turajlic S, Xu H, Lee SM, Forster MD, Ahmad T, Hiley CT, Abbosh C, Falzon M, Borg E, Marafioti T, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Shah R, Joseph L, Quinn AM, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Dentro S, Taniere P, O’Sullivan B, Lowe HL, Hartley JA, Iles N, Bell H, Ngai Y, Shaw JA, Herrero J, Szallasi Z, Schwarz RF, Stewart A, Quezada SA, Le Quesne J, Van Loo P, Dive C, Hackshaw A, Swanton C TRACERx Consortium. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 6.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 7.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asaoka M, Ishikawa T, Takabe K, Patnaik SK. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int J Mol Sci. 2019;20:5621. doi: 10.3390/ijms20225621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 11.Cescon DW, Haibe-Kains B, Mak TW. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A. 2015;112:2841–2846. doi: 10.1073/pnas.1424869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiki Y, Yamamoto Y, Sueta A, Yamamoto-Ibusuki M, Goto-Yamaguchi L, Tomiguchi M, Takeshita T, Iwase H. APOBEC3B gene expression as a novel predictive factor for pathological complete response to neoadjuvant chemotherapy in breast cancer. Oncotarget. 2018;9:30513–30526. doi: 10.18632/oncotarget.25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikkila J, Kumar R, Campbell J, Brandsma I, Pemberton HN, Wallberg F, Nagy K, Scheer I, Vertessy BG, Serebrenik AA, Monni V, Harris RS, Pettitt SJ, Ashworth A, Lord CJ. Elevated APOBEC3B expression drives a kataegic-like mutation signature and replication stress-related therapeutic vulnerabilities in p53-defective cells. Br J Cancer. 2017;117:113–123. doi: 10.1038/bjc.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sieuwerts AM, Willis S, Burns MB, Look MP, Meijer-Van Gelder ME, Schlicker A, Heideman MR, Jacobs H, Wessels L, Leyland-Jones B, Gray KP, Foekens JA, Harris RS, Martens JW. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer. 2014;5:405–413. doi: 10.1007/s12672-014-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga E, Yamashita N, Tanaka K, Inoue Y, Akiyoshi S, Saeki H, Oki E, Kitao H, Maehara Y. Expression of APOBEC3B mRNA in primary breast cancer of Japanese women. PLoS One. 2016;11:e0168090. doi: 10.1371/journal.pone.0168090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng AZ, Yockteng-Melgar J, Jarvis MC, Malik-Soni N, Borozan I, Carpenter MA, McCann JL, Ebrahimi D, Shaban NM, Marcon E, Greenblatt J, Brown WL, Frappier L, Harris RS. Epstein-Barr virus BORF2 inhibits cellular APOBEC3B to preserve viral genome integrity. Nat Microbiol. 2019;4:78–88. doi: 10.1038/s41564-018-0284-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suspene R, Guetard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome Analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brueffer C, Gladchuk S, Winter C, Vallon-Christersson J, Hegardt C, Häkkinen J, George AM, Chen Y, Ehinger A, Larsson C, Loman N, Malmberg M, Rydén L, Borg Å, Saal LH. The mutational landscape of the SCAN-B real-world primary breast cancer transcriptome. EMBO Mol Med. 2020;12:e12118. doi: 10.15252/emmm.202012118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 24.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JS. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep. 2016;49:607–611. doi: 10.5483/BMBRep.2016.49.11.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, Hu K, Andreev-Drakhlin AY, Kim J, Hess JM, Haas BJ, Aguet F, Weir BA, Rothberg MV, Paolella BR, Lawrence MS, Akbani R, Lu Y, Tiv HL, Gokhale PC, de Weck A, Mansour AA, Oh C, Shih J, Hadi K, Rosen Y, Bistline J, Venkatesan K, Reddy A, Sonkin D, Liu M, Lehar J, Korn JM, Porter DA, Jones MD, Golji J, Caponigro G, Taylor JE, Dunning CM, Creech AL, Warren AC, McFarland JM, Zamanighomi M, Kauffmann A, Stransky N, Imielinski M, Maruvka YE, Cherniack AD, Tsherniak A, Vazquez F, Jaffe JD, Lane AA, Weinstock DM, Johannessen CM, Morrissey MP, Stegmeier F, Schlegel R, Hahn WC, Getz G, Mills GB, Boehm JS, Golub TR, Garraway LA, Sellers WR. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nellore A, Jaffe AE, Fortin JP, Alquicira-Hernandez J, Collado-Torres L, Wang S, Phillips RA III, Karbhari N, Hansen KD, Langmead B, Leek JT. Human splicing diversity and the extent of unannotated splice junctions across human RNA-seq samples on the Sequence Read Archive. Genome Biol. 2016;17:266. doi: 10.1186/s13059-016-1118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens JH, Stunnenberg HG. BLUEPRINT: mapping human blood cell epigenomes. Haematologica. 2013;98:1487–1489. doi: 10.3324/haematol.2013.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, McMurry J, Marioni JC, Malone J, Megy K, Rustici G, Tang AY, Taubert J, Williams E, Mannion O, Parkinson HE, Brazma A. Expression Atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, Specht MC, Bernstein BE, Michor F, Ellisen LW. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun. 2018;9:3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, Sofia HJ, Hutter C, Getz G, Wheeler D, Ding L MC3 Working Group; Cancer Genome Atlas Research Network. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6:271–281. e277. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson J, Li JZ, Zollner S. Helmsman: fast and efficient mutation signature analysis for massive sequencing datasets. BMC Genomics. 2018;19:845. doi: 10.1186/s12864-018-5264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarvis MC, Ebrahimi D, Temiz NA, Harris RS. Mutation signatures including APOBEC in cancer cell lines. JNCI Cancer Spectr. 2018;2:pky002. doi: 10.1093/jncics/pky002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asaoka M, Patnaik SK, Zhang F, Ishikawa T, Takabe K. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res Treat. 2020;181:309–322. doi: 10.1007/s10549-020-05630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CE, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich L. The immune landscape of cancer. Immunity. 2018;48:812–830. e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, Rao A, Schultz A, Li X, Sumazin P, Williams C, Mestdagh P, Gunaratne PH, Yau C, Bowlby R, Robertson AG, Tiezzi DG, Wang C, Cherniack AD, Godwin AK, Kuderer NM, Rader JS, Zuna RE, Sood AK, Lazar AJ, Ojesina AI, Adebamowo C, Adebamowo SN, Baggerly KA, Chen TW, Chiu HS, Lefever S, Liu L, MacKenzie K, Orsulic S, Roszik J, Shelley CS, Song Q, Vellano CP, Wentzensen N, Weinstein JN, Mills GB, Levine DA, Akbani R. A comprehensive Pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33:690–705. e699. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris RS. Molecular mechanism and clinical impact of APOBEC3B-catalyzed mutagenesis in breast cancer. Breast Cancer Res. 2015;17:8. doi: 10.1186/s13058-014-0498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saal LH, Vallon-Christersson J, Hakkinen J, Hegardt C, Grabau D, Winter C, Brueffer C, Tang MH, Reutersward C, Schulz R, Karlsson A, Ehinger A, Malina J, Manjer J, Malmberg M, Larsson C, Ryden L, Loman N, Borg A. The Sweden Cancerome Analysis Network - Breast (SCAN-B) initiative: a large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015;7:20. doi: 10.1186/s13073-015-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown WL, Law EK, Argyris PP, Carpenter MA, Levin-Klein R, Ranum AN, Molan AM, Forster CL, Anderson BD, Lackey L, Harris RS. A rabbit monoclonal antibody against the antiviral and cancer genomic DNA mutating enzyme APOBEC3B. Antibodies (Basel) 2019;8:47. doi: 10.3390/antib8030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 48.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, Feng Y, Adolph MB, Chelico L, Law EK, Carpenter MA, Harris RS. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan JW, Zabidi MMA, Chong BK, Meng MY, Ng PS, Hasan SN, Sandey B, Bahnu S, Rajadurai P, Yip CH, Rueda OM, Caldas C, Chin SF, Teo SH. Germline APOBEC3B deletion increases somatic hypermutation in Asian breast cancer that is associated with Her2 subtype, PIK3CA mutations, and immune activation. Int J Cancer. 2021 doi: 10.1002/ijc.33463. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 50.Leonard B, Starrett GJ, Maurer MJ, Oberg AL, Van Bockstal M, Van Dorpe J, De Wever O, Helleman J, Sieuwerts AM, Berns EM, Martens JW, Anderson BD, Brown WL, Kalli KR, Kaufmann SH, Harris RS. APOBEC3G expression correlates with T-cell infiltration and improved clinical outcomes in high-grade serous ovarian carcinoma. Clin Cancer Res. 2016;22:4746–4755. doi: 10.1158/1078-0432.CCR-15-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willems L, Gillet NA. APOBEC3 interference during replication of viral genomes. Viruses. 2015;7:2999–3018. doi: 10.3390/v7062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.