Figure 1.
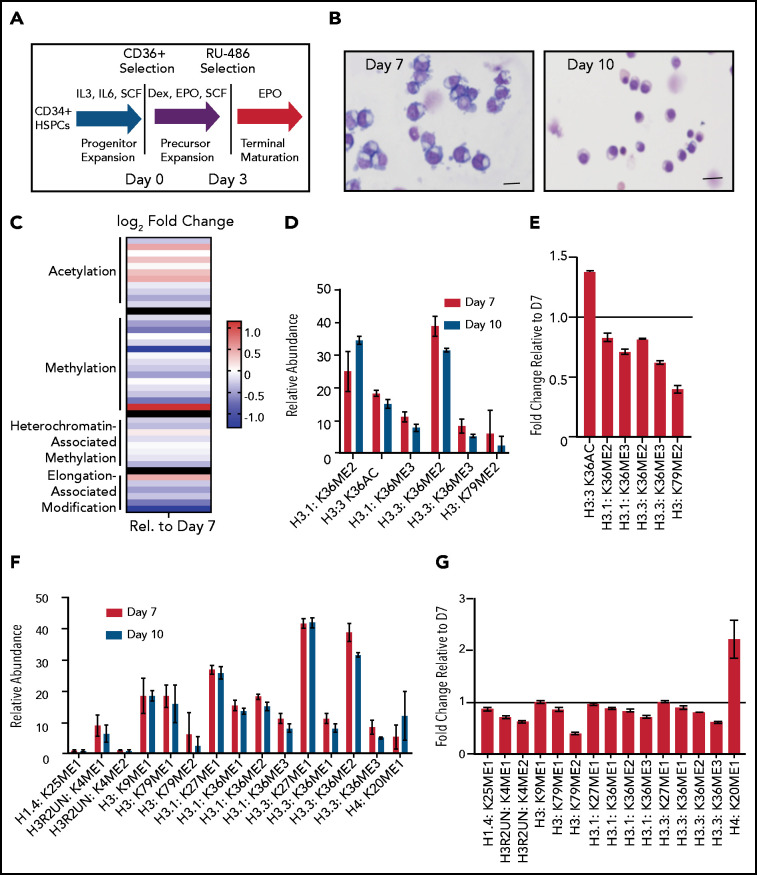
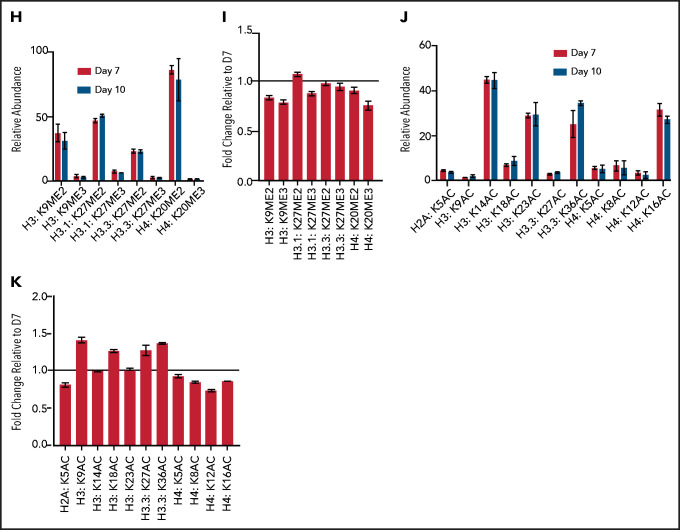
Terminal erythroid maturation is associated with loss of histone PTM associated with active transcriptional elongation. (A) Schematic demonstrating CD36 erythroid synchronization culture system. CD34-positive cells are expanded for 7 days before CD36 selection. CD36-positive cells are cultured for an additional 11 days allowing for terminal erythroid maturation. (B) Cytospins of cells at day 7 and day 10 of terminal erythroid maturation. (C) Heat map of changes in histone abundance during terminal erythroid maturation as determined by mass spectrometry. Values represent log2 fold change of day 10 compared with day 7 of terminal erythroid maturation. (D) Abundance of histone modifications associated with active transcription by RNA polymerase II on day 7 and day 10 of terminal maturation. (E) Fold change of transcription related histone marks at day 10 relative to day 7. (F) Abundance of histone methylation PTM at day 7 and day 10 of terminal maturation. Only histone PTM present at greater than 1% abundance are shown. (G) Fold change of histone methylation at day 10 relative to day 7. (H) Abundance of histone PTMs associated with heterochromatin at day 7 and day 10 of terminal maturation. Only histone PTMs with an abundance greater than 1% are shown. (I) Fold change of heterochromatin-associated PTMs at day 10 relative to day 7. (J) Abundance of histone acetylation at day 7 and day 10 of terminal maturation. Only histone PTMs with an abundance greater than 1% are shown. (K) Fold change of histone acetylation marks at day 10 relative to day 7. The data in this figure represent 2 biologic and 6 technical replicates. Error bars represent standard error of the mean.