Abstract
The oncogenic transcription factor signal transducer and activator of transcription 3 (STAT3) is activated constitutively in a wide array of human cancers. It is an appealing molecular target for novel therapy as it directly regulates expression of genes involved in cell proliferation, survival, angiogenesis, chemoresistance and immune responsiveness. In addition to these well-established oncogenic roles, STAT3 has also been found to mediate a wide array of functions in modulating cellular behavior. The transcriptional function of STAT3 is canonically regulated through tyrosine phosphorylation. However, STAT3 phosphorylated at a single serine residue can allow incorporation of this protein into the inner mitochondrial membrane to support oxidative phosphorylation (OXPHOS) and maximize the utility of glucose sources. Conflictingly, its canonical transcriptional activity suppresses OXPHOS and favors aerobic glycolysis to promote oncogenic behavior. Apart from mediating the energy metabolism and controversial effects on ATP production, STAT3 signaling modulates lipid metabolism of cancer cells. By mediating fatty acid synthesis and beta oxidation, STAT3 promotes employment of available resources and supports survival in the conditions of metabolic stress. Thus, the functions of STAT3 extend beyond regulation of oncogenic genes expression to pleiotropic effects on a spectrum of essential cellular processes. In this review, we dissect the current knowledge on activity and mechanisms of STAT3 involvement in transcriptional regulation, mitochondrial function, energy production and lipid metabolism of malignant cells, and its implications to cancer pathogenesis and therapy.
Keywords: STAT3 transcription factor; Protein processing, Post-translational; Adenosine triphosphate; Lipid metabolism; Metabolism; Neoplasms
Abbreviations: AML, acute myeloid leukemia; ATP, adenosine triphosphate; BCL, B-cell lymphoma; BCSC, breast cancer stem cells; CDK, cyclin-dependent kinase; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; CNTF, ciliary neurotrophic factor; CPT1B, carnitine palmitoyltransferase 1B; CREB, cyclic AMP response element-binding protein; EGF, epidermal growth factor; ERH, enhancer of rudimentary homolog; ETC, electron transport chain; FABP6, fatty acid binding protein 6; FAO, fatty acid oxidation; FASN, fatty acid synthase; FBP1, fructose-bisphosphatase 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GLUT1, glucose transporter 1; GP130, glycoprotein 130; GPI, glucose-6-phosphate isomerase; GRIM-19, gene associated with retinoid-IFN-induced mortality; HDL-C, high-density lipoprotein cholesterol; HIF1α, hypoxia inducible factor 1α; IAP, inhibitor of apoptosis; IFN, interferon; IL, interleukin; JAK, Janus kinase; LDHA, lactate dehydrogenase A; LDL-C, low-density lipoprotein cholesterol; LIF, leukemia inhibitory factor; MCL, myeloid cell leukemia; MEF2D, Myocyte Enhancer Factor 2D; MMP, matrix metalloproteinase; MPTP, mitochondrial permeability transition pore; NHL, non-Hodgkin's lymphoma; NFκB, nuclear factor kappa B; OSM, oncostatin M; OXPHOS, oxidative phosphorylation; p53, tumor protein 53; PBP, PPARγ binding protein; PC, phosphatidylcholine; PDGF, platelet-derived growth factor; PFK, phosphofructokinase; PGK1, phosphoglycerate kinase; PIAS, protein inhibitors of activated STATs; PKCδ, protein kinase C δ; PPARγ, peroxisome proliferator-activated receptor gamma; PTP, protein tyrosine phosphatases; RIME, rapid immunoprecipitation mass spectrometry of endogenous proteins; SM, sphingomyelin; SOCS, suppressors of cytokine signaling; SREBP1, sterol regulatory element-binding protein 1; STAT, signal transducer and activator of transcription; TAG, triacylglycerol; TBK1, TANK-binding kinase 1; TF, transcription factors; TGFβ, transforming growth factor beta; TNFα, tumor necrosis factor alpha; VEGF, vascular endothelial growth factor
Introduction
Transcription factors (TF) exquisitely regulate expression of genes, representing crucial mediators in defining fundamental cellular behavior and identity. Impairment of TF function can thus result in various pathological phenotypes, and has been recognized as an important mediator in malignant transformation and progression (1). The most evident and direct relation between transcription factors and cancer development comes from TFs that regulate genes involved in proliferation, invasion, immune response and self-renewal, as the hallmarks of malignant behavior. Additionally, due to insufficient vascularization, hypoxic environment and nutrient deficiency, cancer cells commonly employ compensatory mechanisms to achieve continued proliferation under metabolic stress (2). Thus, tumors are generally characterized by altered metabolism and utilization of glucose and lipids to satisfy their distinct energy needs and provide building blocks for rapid cell division (3,4).
The crucial interplay between metabolic and transcriptional cellular regulation has recently been recognized (5,6). For certain metabolic enzymes, such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (7) and fructose-bisphosphatase 1 (FBP1) (8), it is becoming clear that they not only regulate metabolism through their enzymatic function, but they can also serve as co-regulators of transcription factors on chromatin with clear roles in cancer development. Conversely, a number of TFs known to affect cell survival and proliferation through transcriptional mechanisms have also been found to utilize non-transcriptional mechanisms to shape the metabolic architecture of neoplastic cells. For some TFs, subcellular localization is not restricted to shuttling between cytoplasm and nucleus, but they are found to be present in mitochondria to directly modulate energy metabolism. These include TFs such as nuclear factor kappa B (NFκB), tumor protein 53 (p53), cAMP response element-binding protein (CREB) and myocyte enhancer factor 2D (MEF2D) (9, 10, 11, 12).
One of the transcription factors that displays both transcriptional and direct metabolic regulation, and which has been shown to play a critical role in tumorigenesis, is signal transducer and activator of transcription 3 (STAT3) (13). Increasing evidence suggests that STAT3 plays a direct role in modulating the biology of cancer cells, through its involvement in energy metabolism, and metabolism of glucose and lipids (13, 14, 15, 16). While the transcriptional regulation of cancer-promoting genes by STAT3 has been well established, a better understanding of its influence on the metabolic milieu of malignant cells may provide the basis for the development of novel therapeutic strategies.
The STAT3 signaling pathway
The STAT family of proteins consists of seven related transcription factors termed STAT1 to STAT6, including the closely related STAT5A and STAT5B (17). Under basal conditions in normal cells, STAT3 resides in the cytoplasm in a latent state and can become transcriptionally active in response to external signals. These include cytokines such as IL-6, IL-10, IL-23, IL-21, IL-11, oncostatin M (OSM), leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNTF) as well as growth factors such as epidermal growth factor (EGF), platelet-derived growth factor (PDGF) and others (6,18,19). Upon binding to their cognate cell surface receptor, these receptors oligomerize, thereby bringing their associated tyrosine kinases into close proximity to initiate their transphosphorylation and activation of kinase activity. In particular, cytokine receptors associate with the Janus kinases (JAK), JAK1, JAK2, JAK3, and TYK2 (20). These JAKs then phosphorylate the intracellular domain of receptors, forming a docking site for the SH2 domains of cytoplasmic STAT3. Following STAT3 recruitment, JAKs phosphorylate STAT3 on tyrosine 705 (Y705). In addition to membrane receptor kinases, non-receptor cytoplasmic kinases that signal through STATs include ABL and Src-related kinases. These can be activated in a variety of cancers, such as chronic myeloid leukemia (CML) characterized by the BCR-ABL fusion (19,21). STAT3 Y705 phosphorylation further initiates conformational changes and SH2 domain-dependent formation of an activated homodimer or STAT1-STAT3 heterodimer. This activating dimerization reveals a nuclear localization signal that facilitates its translocation to the nucleus and binding to the nine base pair sequence TTCN3GAA in the promoter of target genes to modulate their transcription (22). Given its role in essential cellular events, STAT3 function is tightly regulated by endogenous inhibitors including suppressors of cytokine signaling (SOCS), protein tyrosine phosphatases (PTPs) and protein inhibitors of activated STATs (PIAS) (18). As STATs are downstream of multiple kinases and cytokine receptors, they lay at the convergence point of various signaling pathways and can integrate signals from a variety of internal and external cellular stimuli (19,23).
Genes whose transcription is regulated by STAT3 include key mediators of cell cycle progression and promoters of oncogenic cell behavior. Such pro-tumorigenic genes include regulators of cell survival and suppressors of apoptosis, such as B-cell lymphoma (BCL)-2, BCL-6, BCL-xL, myeloid cell leukemia (MCL)-1, survivin (a member of the inhibitor of apoptosis (IAP) family); regulators of proliferation such as MYC and cyclin D1; promoters of migration and metastasis including matrix metalloproteinases (MMPs); mediators of angiogenesis such as vascular endothelial growth factor (VEGF); pro-inflammatory cytokines such as IL-6 and immunosuppressive cytokines including IL-10 (18,22,24). These genes are general STAT3 transcriptional targets, as they show responsiveness to STAT3 activity across multiple cell types (reviewed in 25).
The transcriptional effects of STAT3 vary among tissue types, suggesting that cell-type specific interactions with other proteins, such as other transcription factors and transcriptional co-regulators, modulate the potency and biological outcome of STAT3 activity (21,26). Chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) has been used to identify cell-type specific genomic binding sites of activated STAT3. These analyses have confirmed near universal STAT3 binding to the promoters of SOCS3, BCL3, JUNB and STAT3 itself in a spectrum of cell types (27, 28, 29, 30, 31, 32). They also revealed cell type-specific binding patterns, underlining the importance of the specific genetic and biological context. Activated STAT3 was associated with inflammatory gene signature expression patterns in Src-transformed MCF-10A mammary epithelial cells (29), Th17 polarization in naïve CD4+ cells (30), anti-inflammatory phenotype development in macrophages (31), and pluripotency markers in ESCs (32). Based on these findings, Hutchins et al (33,34) proposed two distinct modes of STAT3 target DNA-binding. The first represents universal, evolutionary conserved STAT3 DNA-binding sites comprising a set of 35 regions that mediate STAT3 auto-regulation and cell growth. The second binding mode is cell-type specific, mediating a diverse spectrum of functions depending on the cellular context.
The role of STAT3 in tumorigenesis
As noted, STAT3 regulates the expression of genes important for essential cellular processes including survival, proliferation, self-renewal, angiogenesis, and immune response (35). Given this, it is not surprising that constitutive STAT3 activation is a sufficient factor to cause malignant transformation and tumor development in model systems (36,37). While the activation of STAT3 is tightly controlled in a physiological setting, constitutive activation of this transcription factor has been reported to occur in more than 70% of all human malignancies and is associated with poor clinical prognosis (21). Aberrant STAT3 activity has been detected in most hematological malignancies, including acute myeloid leukemia (AML), multiple myeloma, non-Hodgkin lymphoma, and chronic lymphocytic leukemia (CLL), as well as solid tumors of the lung, breast, ovary, cervix, prostate, bladder, kidney, colon, liver, stomach, head and neck, and others (38, 39, 40, 41, 42, 43). Constitutive STAT3 activation can occur as a result of an activating mutation, most frequently found in the SH2 domain. However, much more common is constitutive phosphorylation of wildtype STAT3, driven by pathogenic mechanisms including paracrine or autocrine cytokine stimulation; hyper-activation of JAKs due to genetic or epigenetic alterations or overexpression; or, by downregulation of physiological inhibitors of STAT3 (18,21). In addition, therapeutic inhibition of several pathways in “oncogene addicted” tumors result in feedback activation of STAT3, as seen in non-small lung cancers with EGFR or KRAS activating mutation, ALK- and MET-addicted neuroblastomas, and HER2-driven breast cancer cells. In turn, activated STAT3 prevents apoptosis by promoting survival and chemoresistance patterns in tumor cells, and dual inhibition of these targets along with STAT3 has shown promise in inducing cancer apoptosis in preclinical models (44).
In conjunction with its tumor cell-intrinsic oncogenic roles, STAT3 is commonly overactivated in tumor-infiltrating immune cells, thereby affecting the tumor microenvironment in a dual manner (45). In tumors, STAT3 promotes tumor-promoting pro-inflammatory pathways, through both transcriptional upregulation of cytokines such as IL-6 (25), as well as by cooperating with other inflammatory mediators such as NFκB (46,47). In infiltrating immune cells, STAT3 induces strong immunosuppression, leading to an immune tolerant state and immune evasion of cancer cells (48). Such immunosuppressive effects of STAT3 transcriptional activity has been shown to impair both innate and adaptive immunity, reducing the functional activity of dendritic cells, neutrophils, natural killer cells, and T effector cells (49, 50, 51), while promoting differentiation of naïve T cells to T regulatory cells by upregulating FOXP3 expression (47). Another important aspect of STAT3 relates to its regulatory role in metabolism of energy, glucose, and lipids, which support cancer progression under conditions of metabolic stress. Thus, STAT3 acts as a pleiotropic protein, exerting both transcriptional and non-transcriptional functions in shaping cellular identity and behavior.
Posttranslational modifications of STAT3
In addition to canonical tyrosine phosphorylation, other posttranslational modification of cytoplasmic STAT3 include phosphorylation at serine 727, acetylation (at Lys685) (52), methylation (at Lys180) (53), S-glutathionylation (at Cys328 and Cys542) (54), ubiquitination (55), and SUMOylation (at Lys451) (56). Each of these diverse modifications modulate some part of the STAT3 activation process, including dimerization, nuclear translocation, nuclear retention and DNA binding ability, resulting in reduced or enhanced transcriptional activity. Although all of these modifications have been reported to mediate effects on STAT3 activity, current evidence supports the greatest role for serine phosphorylation, along with tyrosine phosphorylation, as contributing the most to STAT3 pathological cellular phenotypes. This likely reflects both transcriptional and non-transcriptional effects of serine-phosphorylated STAT3.
STAT3 serine phosphorylation
The potential importance of STAT3 phosphorylation on a serine residue within the C-terminal domain was recognized soon after the identification of the protein itself, however its function was less clear than that of pY-STAT3. So-called proline-directed serine/threonine kinases that are able to phosphorylate serine residues found in the P-M-S-P motif in which Ser727 is found include MAP kinases (including ERK (57), p38 (58) and JNK (59)), mTOR (60), cyclin-dependent kinase (CDK) 1, protein kinase C δ (PKCδ) (61) and TANK-binding kinase 1 (TBK1) (62). The interconnection with these crucial players in carcinogenesis underlies the importance of STAT3 as a convergence node for multiple oncogenic signaling pathways. A quarter of a century ago it was reported that STAT3 (as well as STAT1) require serine phosphorylation in addition to classical tyrosine phosphorylation to achieve the maximal transcriptional effect (63), although this finding may not be universally true. More recent evidence indicated that serine phosphorylation of STAT3 is an independent tumor driving posttranslational modification of this TF, capable of facilitating pathological growth, proliferation and invasiveness of several tumor types independently of tyrosine phosphorylation. Accordingly, constitutive pS-STAT3 levels were found to be major oncogenic factors in a number of malignancies, including glioma, melanoma, hematological malignancies, breast and prostate cancers (64, 65, 66, 67, 68, 69).
It has also become clear that serine phosphorylation may modulate the effects of other STAT family members. For example, STAT5, which is frequently activated in hematologic malignancies, is frequently phosphorylated at two C-terminal serine residues (Ser725 and Ser779) in human myeloid malignancies (70). In fact, serine phosphorylation was shown to be required for STAT5 nuclear translocation and consequent oncogenic potential in these tumors, as mice with substitution of alanine at either Ser725 or Ser779 showed impeded leukemogenesis (70,71). Analogously, interferon (IFN) gamma-induced transcriptional activity of STAT1 was shown to be reduced by 80% when Ser727 is mutated, indicating the importance of serine phosphorylation in diverse STAT family members (63).
Transcriptional activity of pS-STAT3
In certain cancer systems, pS-STAT3 was shown capable of driving oncogenesis independently of the canonical pY-STAT3, due to the ability to autonomously promote STAT3 transcriptional activity (68,72, 73, 74) . In a melanoma model, IFNβ-induced serine phosphorylation was essential for STAT3 translocation to the nucleus, which was abrogated by S727 mutation (75). Serine phosphorylation of STAT3 may have direct pathogenic effects in at least some human cancers. For example, it has been reported that the malignant lymphocytes in all samples tested from patients with chronic lymphocytic leukemia (CLL) display constitutive pS-STAT3, whereas phosphorylation at Y705 occurred in a cytokine-inducible manner (76). This study further showed nuclear localization and direct DNA-binding capacity of the pS-STAT3, while pY-STAT3 levels did not affect DNA binding ability of STAT3 in the tested CLL cells. On the other hand, in solid tumors such as gastric cancer, pS-STAT3 was shown not to directly bind to DNA, nor to promote the DNA-binding affinity of STAT3. Instead, pS-STAT3 facilitated the transcriptional elongation of STAT3 target genes by RNA polymerase (Pol) II, without affecting Pol II recruitment and transcription initiation (77). These findings might be explained by the cooperation of pS-STAT3 with other coregulatory proteins. Accordingly, rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) coupled with a CRISPR-based gene editing screen revealed the chromatin and transcription regulating proteins pontin and enhancer of rudimentary homolog (ERH) to form nuclear complexes with pS-STAT3. Reducing expression of these genes markedly reduced the expression of a STAT3 target gene SOCS3, whereas SOCS1 expression remained unaltered, indicating conserved STAT1 activity.
Additionally, it is plausible that pS-STAT3 contributes to transcriptional potency of nuclear STAT3 through interaction with other transcriptional regulators, including other members of the STAT family (78). Considering its importance for transcriptional efficacy, it was not surprising that abrogation of STAT3 serine phosphorylation markedly reduced tumor formation and decreased disease severity in mice with gastric tumors (77) and certain hematological malignancies (79,80). On the contrary, JNK1/2-mediated stimulation of S727 phosphorylation with reciprocal reduction of pY-STAT3 levels, suppressed cell survival and proliferation of oral cancer cells in a dose dependent manner (81). Overall, it seems that pS-STAT3 driven malignancies occur less commonly than ones initiated and facilitated by the canonically active pY-STAT3. Interestingly, cooperation between the two posttranslational modifications of this oncogenic transcription factor proved important for orchestrating metastatic dissemination of lung cancer cells to bone marrow. While LIFR-mediated pS-STAT3 led dissemination and early migration ability of the lung cancer cells, IL6/pY-STAT3 signaling prevailed in the later metastatic phase, promoting proliferation and tumor initiation at the secondary site while attenuating motility of the metastatic cells (82).
Mitochondrial function of pS-STAT3
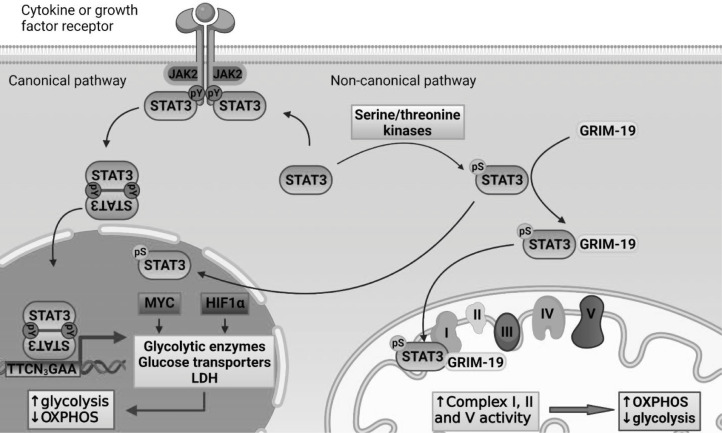
The role of STAT3, particularly pS-STAT3, extends beyond its nuclear function to the mitochondria, where it can exert significant effects on cellular energy metabolism and physiology (Fig. 1). The first indication of pS-STAT3 effects in mitochondria came from the observation that a subunit of electron transport chain (ETC) complex I, gene associated with retinoid-IFN-induced mortality (GRIM) 19, directly binds to STAT3 (83) and is likely responsible for its transport into mitochondria (84). This interaction is specific to STAT3, as GRIM-19 did not co-precipitate in cellular lysates with other family members, such as STAT1, STAT2 or STAT5. Inside the mitochondria, GRIM-19 integrates STAT3 into ETC complex I, where STAT3 associates with the inner mitochondrial membrane and is oriented towards the matrix (84). Besides direct incorporation into complex I, mitochondrial STAT3 enhances the activity of complexes I, II and ATP synthase (complex V) (85). Conversely, STAT3 functional inhibition (85,86), genetic deficiency (86) or serine mutation (84) attenuate their functional activities without affecting other ETC components, including Complexes III and IV. Of note, the STAT3-GRIM-19 interaction also reduces transcriptional effects of STAT3, possibly due to impairment of nuclear translocation and favoring mitochondrial localization (83).
Fig. 1.
Canonical and non-canonical pathway effects of STAT3 on energy metabolism.
Canonical STAT3 signaling is activated by its tyrosine phosphorylation, which enables STAT3 to regulate transcription of its target genes. In addition to regulating genes controlling proliferation, survival and self-renewal, STAT3 both directly and indirectly regulates the expression of genes important for energy metabolism. Consequently, canonical STAT3 transcriptional activation favors glycolysis over oxidative phosphorylation as the cellular mechanism of ATP production. Non-canonical STAT3 function occurs via STAT3 phosphorylation at Ser727. Serine phosphorylated STAT3 can regulate the transcription of target genes. In addition, pS-STAT3 can be transported into the inner mitochondrial membrane where it enhances the activity of ETC complexes, stimulating OXPHOS and increased ATP production.
ATP, adenosine triphosphate; CDK1, cyclin-dependent kinase 1; ETC, electron transport chain; GLUT1, glucose transporter 1; GRIM-19, gene associated with retinoid-IFN-induced mortality; HIF1α, hypoxia inducible factor 1α; LDHA, lactate dehydrogenase; OXPHOS, oxidative phosphorylation; PKC δ, protein kinase C δ; STAT3, signal transducer and activator of transcription 3; TBK1, TANK-binding kinase 1.
Contrary to the classical Warburg effect, mitochondrial STAT3 stimulates oxidative phosphorylation (OXPHOS) and respiration through the ETC (14,15). This results in increased production of ATP, providing additional energy needed for rapid oncogenic growth and cell division (85,87). Consequently, STAT3 mutation of Ser727 to alanine significantly reduced the activity of ETC and suppressed tumor growth in breast cancer-bearing mice (88). In addition, STAT3 activity may modulate Ras-dependent oncogenic transformation in divergent manners depending on the cellular context and subcellular localization of activated STAT3. Mitochondrial pS-STAT3 strongly supports Ras-mediated oncogenic transformation in mouse embryonic fibroblasts, breast, renal and myeloproliferative neoplasms, in which nuclear STAT3 was shown to be redundant for this process (80,89). While STAT3−/− cells with activated Ras showed survival dependency on high glucose media, reconstitution of STAT3 reversed this sensitivity, presumably due to the ability to provide sufficient ATP from restricted glucose resources (85). Mutational studies confirmed that mitochondrial STAT3 is responsible for this effect, as reconstitution of STAT3 with 727 with a serine to alanine substitution failed to rescue the survival in glucose deprived conditions, while mutations at Y705, the SH2 domain, or the DNA-binding site did not affect the cellular glucose dependence of Ras-transformed cells (85).
However, Ras-transformation and STAT3 signaling are found to be mutually exclusive in pancreatic cancers, in which KRAS mutation occurs at a 90% frequency (90). Analyses of primary human pancreatic adenocarcinomas and pancreatic cancer cellular systems showed that the LIF receptor is significantly downregulated in cancers with mutated KRAS, and this downregulation of LIFR was necessary for KRAS-mediated neoplastic transformation. Inhibition of the LIFR/pY-STAT3 axis led to upregulation of GLUT1 expression, resulting in increased levels of both glycolysis and OXPHOS. Thus, in addition to tumor promoting effects, STAT3 signaling may display tumor suppressing activity in certain cancer systems depending on the genetic background of neoplastic cells. In addition to supporting energy provision, it is plausible that mitochondrial STAT3 improves cell survival and apoptosis resistance by suppressing mitochondrial permeability transition pore (MPTP) opening and consequent cytochrome c release. While this effect has been described in cardiomyocytes, it needs to be further investigated in cancer models (91,92).
Effects of nuclear STAT3 on ATP production
In contrast to the mitochondrial effects on ATP production, STAT3 transcriptional activity in the nucleus has been shown to promote aerobic glycolysis to enhance energy production and proliferation of breast cancer cells (87). Consistent with this finding, inhibition of STAT3 tyrosine phosphorylation decreased glycolysis and lactate production, and consequently induced death of prostate and breast cancer cell lines (87). This glycolysis-promoting effect originates as a consequence of STAT3 transcriptional output, including positive regulation of HIF1α and MYC (93) expression, the two major promoters of the Warburg effect (94). MYC is well known for its transcriptional regulation of glycolytic enzymes, including glucose-6-phosphate isomerase (GPI), phosphofructokinase (PFK), GAPDH, phosphoglycerate kinase (PGK 1), and phosphopyruvate hydratase (95). In addition, MYC activity supports cellular glucose import through transcriptional upregulation of GLUT1 (96), but also increases metabolic flux from pyruvate to lactate via induction of lactate dehydrogenase A (LDHA) expression (97). Similar to MYC, HIF1α transcriptionally regulates the expression of numerous genes involved in glycolysis in hypoxic and also normoxic setting (98,99). In addition to STAT3-mediated HIF1α expression, STAT3 binds to the promoters of HIF1α target genes, contributing to formation of a transcriptional complex that supports HIFα target genes expression (100). Thus, in contrast to mitochondrial pS-STAT3, transcriptionally active pY-STAT3 favors glycolysis over the more efficient ATP production via OXPHOS.
Lipids in tumorigenesis
The crosstalk between lipid metabolism and tumorigenesis became intriguingly evident in the last decades, and can be observed through both paracrine signaling between adipose tissue and cancer cells, as well as intracellular alterations of cancer cells’ metabolic profile (4). Adipose tissue surrounding the tumor can influence its growth through secretion of adipokines, cytokines, hormones and growth factors (101). This adipocyte-tumor communication is particularly important in cancer types in which a significant portion of tumor environment comprises adipose tissue, such as in breast cancers (4). An intriguing implication of such an interplay comes from a clear link between obesity and cancer development (102). As estimated by the American Cancer Society, approximately 20% of all malignancies are associated with obesity (103), and more strikingly, one study reported that cancer-related death could be prevented in 14% of men and 20% of women if normal weight was maintained (104). The microenvironment of adipose tissue in obese people displays several similarities with the tumor microenvironment, such as a chronic low degree of inflammation and increased release of reactive oxidative species (101).
Additionally, adipose tissue functions as an endocrine organ, producing elevated levels of tumor-promoting hormones, growth factors and cytokines in obese people, such as leptin, estrogen, TNFα, VEGF, TGFβ and several members of interleukin family including IL-1β, IL-6, IL-8 and IL-17 (4, 105). As a response, different transcription factors, including STAT3 and NFκB, are continually activated, promoting oncogenic behavior (102). For instance, leptin, which circulates in elevated levels in obese people, has dual effects on STAT3 signaling. In addition to binding to its receptor (OBR) on the plasma membrane, which can itself mediate downstream signaling through STAT3 (106), leptin can bind to the IL-6 receptor glycoprotein 130 (GP130), initiating a cascade of events that lead to STAT3 Y705 phosphorylation (107,108). It is suggested that continuous leptin stimulation induces the STAT3-mediated expression of its major endogenous inhibitor and target gene SOCS3, which further decreases leptin-induced signal transduction and finally facilitates leptin resistance (109).
Furthermore, adipocyte-derived IL-6, which can trigger STAT3-mediated tumor promoting properties, was reported in a number of malignancies including multiple myeloma (110), hepatic (111), prostate (112) and breast cancer (113, 114, 115). A high-fat diet can induce STAT3 Y705 phosphorylation and prostate tumor progression in mice (116). Conversely, treatment of diet-induced obese and atherosclerotic rabbits with the JAK inhibitor ruxolitinib (which decreases STAT3 phosphorylation), significantly reduced the plasma levels of total cholesterol, triacylglycerol (TAG) and low-density lipoprotein cholesterol (LDL-C) and increased high-density lipoprotein cholesterol (HDL-C) levels (117). These findings indicate a potential feedback loop between IL-6-induced STAT3 signaling and lipid metabolism. Finally, STAT3 was shown to be an important factor in differentiation of preadipocytes into adipocytes and a promotor of mitotic clonal expansion and lipid accumulation in adipocytes (118), as well as fatty acid uptake in cancer cells (119).
Lipid metabolism of malignant cells and the role of STAT3
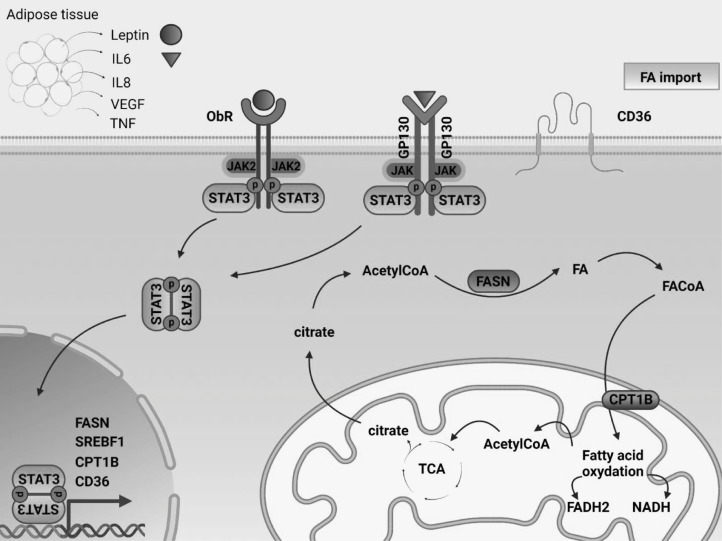
In addition to being influenced by adipose tissue through paracrine and endocrine mechanisms, cancer cells themselves change their intracellular lipid metabolism to maximize the utility of available resources. Interestingly, STAT3 may play a number of roles in this process (Fig. 2). Under stressful conditions of limited nutrient supply, malignant cells may employ lipids as a valuable alternative source of energy, membrane building units to support rapid cellular division and proliferation, buffers to resist excessive oxidative stress, and substrates to produce lipid-based hormones to promote their pathological growth (105,120). Endogenous lipids have been implicated in cellular events such as proliferation, differentiation, inflammation, autophagy, apoptosis, and membrane modeling (105). Thus, alterations in expression of proteins regulating lipid synthesis, catabolism, and storage have been extensively described in malignant cells (115,121). For instance, HER2/neu overexpression in breast cancers is commonly accompanied by amplification of the peroxisome proliferator-activated receptor gamma (PPARγ) binding protein (PBP) gene, possibly due to the co-localization of these genes on chromosome 17q12–21 (122). Interestingly, the STAT3 gene is also located in close proximity to these genetic loci, at chromosome 17q 21.2. Both PBP overexpression and STAT3 activity stimulate PPARγ transcriptional activity, which is one of the major regulators of lipid metabolism and a necessary mediator of adipogenesis and adipocyte differentiation (121,123,124).
Fig. 2.
STAT3 transcriptionally regulates lipid metabolism in malignant cells.
Adipose tissue can produce elevated levels of tumor-promoting hormones, growth factors and cytokines, some of which can directly initiate STAT3 tyrosine phosphorylation. Activated STAT3 can further regulate cellular lipid metabolism through transcriptional regulation of genes involved in this process. Alteration of these metabolic pathways supports proliferation and survival of malignant cells. The development of novel STAT3 inhibitors may be beneficial for both suppressing the transcription of oncogenic genes as well as blocking abnormal lipid metabolism.
CPT1B, carnitine palmitoyltransferase 1B; ETC, electron transport chain; FA, fatty acid; FAO, fatty acid oxidation; FASN, fatty acid synthase; GP130, glycoprotein 130; IL, interleukin; SREBP1, sterol regulatory element-binding protein 1; STAT3, signal transducer and activator of transcription 3; TCA, tricarboxylic acid cycle; TGFβ, transforming growth factor beta; TNFα, tumor necrosis factor alpha; VEGF, vascular endothelial growth factor.
Additionally, both fatty acid synthesis (mediated by fatty acid synthase (FASN)) and beta oxidation (FAO) are commonly upregulated in malignant cells (125,126). FASN-mediated de novo fatty acid synthesis and anabolic-driven proliferation, survival and migration can be triggered by upstream signaling pathways, such as HER/neu overexpression, and activation of PI3K/AKT/mTOR and MAPK (4,127). Importantly, STAT3 activity was shown to upregulate FASN expression (128,129). This relationship might hold therapeutic potential, as FASN inhibition reversed previously acquired resistance to trastuzumab in HER2 positive breast cancers (130,131). In addition, FA catabolism through FAO stimulates tumor growth by providing ATP to support cellular needs under the conditions of metabolic stress associated with glucose deprivation (132). Leptin, IL-6, or STAT3 activation through other means induces the expression of carnitine palmitoyltransferase 1B (CPT1B), a rate limiting enzyme in fatty acid oxidation (125,133). This pathway is crucial for self-renewal of breast cancer stem cells (BCSCs) and promotes their chemoresistance. FAO results in production of NADH and FADH2 which can both reduce oxidative stress and enhance ATP production by the electron transport system (134).
Another product of FAO is acetyl-CoA, which may be utilized for energy production through the Krebs cycle, or fatty acid synthesis and protein acetylation, all of which may promote tumor growth (125). Consistent with this relationship, acetyl-CoA supplementation partially reversed the effects of silencing STAT3 on proliferation and survival of BCSCs (125). Additionally, enhanced FAO is important for the development of chemoresistance in various malignant systems, including AML and breast, lung, and gastric cancers, and abrogating this pathway blocks aberrant growth of cancer cells and re-sensitizes them to chemotherapy (125,135, 136, 137)).
Another potential approach to therapeutically exploit STAT3-mediated lipid characteristics is to use it as a basis for distinguishing between cancer and normal cells, whose STAT3 activity is generally low. STAT3 activation and consequent malignant transformation of mammary epithelial cells results in a significant decline of cellular N-acyl taurine and arachidonic acid content (138). Since both of these molecules are important for plasma membrane modeling, it was considered whether this property can be therapeutically exploited by novel nanoparticle drug delivery systems. Nanoparticles whose surface layer was composed with poly-L-glutamic acid showed preferential cellular binding to cells characterized by abnormal STAT3 activity, and cytotoxic agents delivered via these particles selectively induced greater apoptosis in such cells.
In addition to effects on cell survival and proliferation, a recent report indicated the importance of lipid architecture on the metastatic predisposition of cancer cells. By creating a metastatic map of over 500 cell lines, this study found that cancer cells with high metastatic potential show significantly greater cellular content of cholesterol, phosphatidylcholine (PC), and sphingomyelin (SM), while having lower triacylglycerol levels (139). Interestingly, this was similar to trends identified independently in cellular TAG and SM levels following STAT3 activation in breast cancer cells (138). Jin et al. additionally found that several metabolic regulators strongly influenced the potential of breast cancer cells to form brain metastases, including the key mediator of FA and TAG synthesis, sterol regulatory element-binding protein 1 (SREBP1), the FA transporter CD36, and FA-binding protein FABP6 (139). Intriguingly, STAT3 activity was found to correlate with the mRNA expressions of SREBP1 and CD36 in different cellular and animal models. High glucose-induced SREBP1 expression displays dependency on STAT3 tyrosine phosphorylation (140), and this is strongly attenuated by STAT3 silencing (141). Similarly, overexpression of a constitutively active form of STAT3 significantly elevated CD36 expression (142) and JAK2/STAT3 signaling was required for IL13-induced CD36 expression (143). These data suggest that the metastatic potential of STAT3-driven malignancies may partially be explained by its pleiotropic effects on lipid metabolism, although further studies are needed to dissect the mechanistic aspects of these intriguing correlations.
Clinical development of STAT3 inhibitors
Given the evidence for a major role of STAT3 in cancer cell growth, and metabolic and immune regulation, there has been a growing interest in targeting this pathway therapeutically. Currently, 23 novel STAT3-directed therapies are being investigated in clinical trials (Table 1). A number of these treatments are directed towards upstream mediators of STAT3 activation, such as blocking antibodies to IL-6 or its receptor, and small molecule JAK kinase inhibitors. The most advanced stage JAK targeting cancer therapy is ruxolitinib, which is FDA approved for polycythemia vera and myelofibrosis, and has been tested for the treatment of pancreatic, colorectal, and triple-negative breast cancer (144). While these upstream-targeted therapies hold therapeutic potential, the effectiveness of this approach is often limited by the absence of a single driver cytokine or kinase in human cancers, and the development of resistance by kinase mutations or other upstream events that bypass the inhibition through collateral signaling pathways (26).
Table 1.
STAT3 inhibitors in clinical trials.
| Compound | Mechanism of action | Indication | Phase | Clinical trial identifier |
|---|---|---|---|---|
| Inhibitors of STAT3 activation | ||||
| Tocilizumab | Anti-IL6 mAb | Solid (breast, lung, ovarian, pancreatic, prostate, liver etc.) and hematological malignancies (lymphomas, AML, ALL) | 2 & 3 | 66 clinical trials |
| Atovaquone | GP130 inhibitor | NSCLC | 1 | NCT04648033 |
| NSCLC | 1 | NCT02628080 | ||
| AML | 1 | NCT03568994 | ||
| Ruxolitinib / INCB018424 | JAK1/2 inhibitor | Head and neck cancer | 2 | NCT03153982 |
| TNBC | 2 | NCT02876302 | ||
| AML, ALL, CML, MDS | 2 | NCT00674479 | ||
| SAR302503 / TG101348 | JAK2 inhibitor | Hematopoietic malignancies | 2 | NCT01420783 |
| Pacritinib | JAK2 inhibitor | AML | 2 | NCT02532010 |
| WP1066 | JAK2 inhibitor | Recurrent glioma or metastatic melanoma in the brain | 1 | NCT01904123 |
| Pediatric progressive and refractory brain tumor | 1 | NCT04334863 | ||
| OPB-31121 | JAK2 inhibitor | Advanced solid tumors | 1 | NCT00955812 |
| AZD1480 | JAK2 inhibitor | Solid tumors | 1 | NCT01112397 |
| Icaritin | IL-6/JAK inhibitor | HCC | 2 | NCT01972672 |
| Imx-110 | NF-kB/Stat3/ pan-kinase inhibitor | Advanced solid tumors | 1 & 2 | NCT03382340 |
| Direct binding small molecule STAT3 inhibitors | ||||
| Napabucasin / BBI608 | Phosphorylation inhibitor | NSCLC | 3 | NCT02826161 |
| Metastatic Colorectal Cancer | 3 | NCT03522649 | ||
| Metastatic Pancreatic Ductal Adenocarcinoma | 3 | NCT02993731 | ||
| Colorectal metastatic cancer | 2 | NCT03647839 | ||
| OPB-51602 | Phosphorylation inhibitor | Advanced solid tumors | 2 | NCT01423903 |
| Nasopharyngeal Carcinoma | 1 | NCT02058017 | ||
| OPB-111077 | Phosphorylation inhibitor | Solid tumors | 1 | NCT01711034 |
| Disulfiram | Phosphorylation inhibitor | Refractory solid tumors and metastatic pancreatic cancer | 1 | NCT02671890 |
| WP1220 | Phosphorylation inhibitor | Cutaneous T-cell lymphoma | 1 | NCT04702503 |
| TTI-101 /C188-9 | Phosphorylation inhibitor | Advanced tumors (breast, HNC, NSCLC, HCC, colorectal, gastric, melanoma) | 1 | NCT03195699 |
| Oligonucleotide inhibitors | ||||
| IONIS-STAT3Rx / Danvatirsen | STAT3 Antisense Oligonucleotide | Advanced and refractory pancreatic, NSCLC, and colorectal cancer | 2 | NCT02983578 |
| DLBCL, Lymphoma | 1 & 2 | NCT01563302 | ||
| AZD9150 | STAT3 Antisense Oligonucleotide | Malignant ascites in ovarian and gastrointestinal cancers | 2 | NCT02417753 |
| NHL, DLBCL | 1 | NCT03527147 | ||
| Advanced solid tumors | 1 | NCT03421353 | ||
| STAT3 decoy | STAT3 decoy | Head and neck cancer | 1 | NCT00696176 |
| CpG-STAT3 siRNA CAS3/SS3 | STAT3 siRNA silencer | Relapsed/Refractory B-cell NHL | 1 | NCT04995536 |
| Indirect STAT3 inhibitors | ||||
| Pyrimethamine | STAT3 transcriptional activity inhibitor | CLL/Small Lymphocytic Lymphoma | 1 & 2 | NCT01066663 |
| SC-43 | SHP-1-mediated inhibitor of STAT3 phosphorylation | NSCLC and Biliary Tract Cancer | 1 & 2 | NCT04733521 |
| Bazedoxifene | ER modulator, GP130 inhibitor | Pancreatic Cancer | N/A | NCT04812808 |
ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; DLBCL, diffuse large B cell lymphoma; ER, estrogen receptor; HCC, hepatocellular carcinoma; HNC, head and neck cancer; N/A, non-applicable; NHL, non-Hodgkin lymphoma; NSCLC, non-small cell lung carcinoma; MDS, myelodysplastic syndrome; TNBC, triple-negative breast cancer.
Direct binding STAT3 inhibitors have been challenging to design due to structural similarities with other STAT family members and the steric limitations of the relatively flat interacting surfaces of transcription factors (145). However, a number of compounds have been successfully generated and have advanced to clinical investigations. For some of these molecules, such as for OPB-51602 and OPB‐111077, it has been shown that their anti-cancer effect depends on the mitochondrial STAT3-mediated modulation of oxidative phosphorylation (146,147). The other strategies most likely at least partially achieve their effectiveness through metabolic modulation, however, their specific effects are still largely unknown. Many STAT3-targeted molecules are designed to interact with the STAT3 SH2 domain, thereby blocking both STAT3 phosphorylation and its activating dimerization. Given the structural and binding similarities of many SH2 domains, it can be challenging to achieve specificity with this approach.
Napabucasin (148), is being studied in phase 3 trials for the treatment of metastatic pancreatic and colorectal cancer. Preclinical data indicated potent efficacy of this molecule in reducing glioma proliferation and spheroid formation in a STAT3-mediated manner (149). These therapeutic effects were further enhanced when this drug was administered together with gemcitabine and nab-paclitaxel in the clinical setting, perhaps reflecting the effect of STAT3 inhibition on decreasing expression of pro-survival proteins. This combination therapy achieved disease control in over 90% of patients with metastatic pancreatic carcinoma (150), encouraging further clinical studies.
An alternate approach is to identify drugs that specifically inhibit STAT3-dependent gene expression, though may do so by indirect means. One example is pyrimethamine, an FDA approved anti-parasitic drug that blocks STAT3 transcriptional activity, and shows anti-cancer effects in a number of systems (41,151). Of note, pyrimethamine seems to enhance the anti-cancer immune response. Given the role of STAT3 in mediating immune resistance of tumor cells and an immunosuppressive microenvironment, this effect of a STAT3 inhibitor is not unexpected. This finding also raises the possibility that combinations of STAT3 inhibitors with immune-mediated therapies, such as immune checkpoint inhibitors or engineered T cells or NK cells, may show synergistic benefit.
While most of the STAT3 inhibitors moving ahead with clinical development are small organic molecules, larger molecular approaches are being used as well. For example, heterobifunctional molecules to induce the targeted degradation of STAT3, such as PROTACs, are under development. In addition, nucleic acid-based approaches, such as, antisense oligonucleotides, decoy oligonucleotides, and RNA interference-based silencers of STAT3 mRNA expression are all under active development. While all of these approaches hold promise, the clinical benefit of these strategies remain to be demonstrated.
Conclusion
The oncogenic transcription factor STAT3 is aberrantly activated in a large proportion of human malignancies, driving the persistent expression of its target genes that underlie cancer initiation and progression. Cancers with hyperactivated STAT3 generally display a poor prognosis (24), thus STAT3-directed pharmacological approaches represent an appealing strategy in cancer therapy. In addition to mediating the hallmarks of tumorigenic cellular behavior, STAT3 activity leads to a number of metabolic alterations, rendering cancer cells resistant to the metabolic stress associated with a lack of nutrients and oxygen. The elevated production of ATP to support rapid growth and proliferation is strongly regulated by STAT3 activation, with dual and opposing roles mediated by its two key posttranslational modifications, Ser-727 and Tyr-705 phosphorylation. Aberrant growth of malignant cells is clearly supported by pathological adaptation of lipid metabolism, and cancers with high metastatic potential display a discrete lipid fingerprint. While acting as an important mediator of lipolysis, beta oxidation, and membrane lipid raft modeling, activated STAT3 promotes neoplastic development and progression (125,152, 153, 154). Therefore, STAT3 represents a key regulator of both transcriptional and metabolic identity of the cell, and the development of specific inhibitors of this pleiotropic oncogenic transcription factor holds great promise for the treatment of STAT3-driven malignancies.
CRediT authorship contribution statement
Isidora Tošić: Conceptualization, Writing – original draft. David A. Frank: Conceptualization, Writing – review & editing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Bhagwat AS, Vakoc CR. Targeting transcription factors in cancer. Trends Cancer. 2015;1(1):53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Purkayastha BP, Roy JK. Cancer cell metabolism and developmental homeodomain/POU domain transcription factors: a connecting link. Cancer Lett. 2015;356(2 Pt A):315–319. doi: 10.1016/j.canlet.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Baumann J, Sevinsky C, Conklin DS. Lipid biology of breast cancer. Biochimica et biophysica acta. 2013;1831:1509–1517. doi: 10.1016/j.bbalip.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camarda R, Zhou AY, Kohnz RA, Balakrishnan S, Mahieu C, Anderton B. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14(11):736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 7.Liu K, Tang Z, Huang A, Chen P, Liu P, Yang J. Glyceraldehyde-3-phosphate dehydrogenase promotes cancer growth and metastasis through upregulation of SNAIL expression. Int. J. Oncol. 2017;50:252–262. doi: 10.3892/ijo.2016.3774. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Qiu B, Lee DSM, Walton ZE, Ochocki JD, Mathew LK. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szczepanek K, Lesnefsky EJ, Larner AC. Multi-tasking: nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 2012;22(8):429–437. doi: 10.1016/j.tcb.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13(10):1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Kim CH, Simon DK, Aminova LR, Andreyev AY, Kushnareva YE. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J Biol Chem. 2005;280(49):40398–40401. doi: 10.1074/jbc.C500140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O. p53 regulates mitochondrial respiration. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 13.Valle-Mendiola A, Soto-Cruz I. Energy metabolism in cancer: the roles of STAT3 and STAT5 in the regulation of metabolism-related genes. Cancers (Basel) 2020;12(1):124. doi: 10.3390/cancers12010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang R, Rincon M. Mitochondrial Stat3, the need for design thinking. Int J Biol Sci. 2016;12(5):532–544. doi: 10.7150/ijbs.15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee M, Hirpara JL, Eu JQ, Sethi G, Wang L, Goh BC. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2019;25 doi: 10.1016/j.redox.2018.101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poli V, Camporeale A. STAT3-mediated metabolic reprograming in cellular transformation and implications for drug resistance. Front Oncol. 2015;5:121. doi: 10.3389/fonc.2015.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groner B, von Manstein V. Jak Stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol. 2017;451:1–14. doi: 10.1016/j.mce.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 18.Heppler LN, Frank DA. Targeting oncogenic transcription factors: therapeutic implications of endogenous STAT inhibitors. Trends Cancer. 2017;3(12):816–827. doi: 10.1016/j.trecan.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 20.Reddy EP, Korapati A, Chaturvedi P, Rane S. IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled. Oncogene. 2000;19(21):2532–2547. doi: 10.1038/sj.onc.1203594. [DOI] [PubMed] [Google Scholar]
- 21.Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egusquiaguirre SP, Yeh JE, Walker SR, Liu S, Frank DA. The STAT3 target gene TNFRSF1A modulates the NF-κB pathway in breast cancer cells. Neoplasia. 2018;20(5):489–498. doi: 10.1016/j.neo.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudley AC, Thomas D, Best J, Jenkins A. The STATs in cell stress-type responses. Cell Commun Signal. 2004;2:8. doi: 10.1186/1478-811X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker SR, Xiang M, Frank DA. STAT3 activity and function in cancer: modulation by STAT5 and miR-146b. Cancers (Basel) 2014;6(2):958–968. doi: 10.3390/cancers6020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter RL, Lo HW. STAT3 target genes relevant to human cancers. Cancers (Basel) 2014;6(2):897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh J, Frank DA. STAT3-interacting proteins as modulators of transcription factor function: implications to targeted cancer therapy. ChemMedChem. 2016;11(8):795–801. doi: 10.1002/cmdc.201500482. [DOI] [PubMed] [Google Scholar]
- 27.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32(5):605–615. doi: 10.1016/j.immuni.2010.05.003. May 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47(1):38–49. doi: 10.1016/j.molcel.2012.04.021. Jul 13. [DOI] [PubMed] [Google Scholar]
- 29.Fleming JD, Giresi PG, Lindahl-Allen M, Krall EB, Lieb JD, Struhl K. STAT3 acts through pre-existing nucleosome-depleted regions bound by FOS during an epigenetic switch linking inflammation to cancer. Epigenetics Chromatin. 2015;8:7. doi: 10.1186/1756-8935-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi SK, Chen Z, Larjo A, Kanduri K, Nousiainen K, Äijo T. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 2017;19(9):1888–1901. doi: 10.1016/j.celrep.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Hutchins AP, Poulain S, Miranda-Saavedra D. Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Blood. 2012 Mar 29;119(13):e110–e119. doi: 10.1182/blood-2011-09-381483. [DOI] [PubMed] [Google Scholar]
- 32.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008 Jun 13;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Hutchins AP, Diez D, Miranda-Saavedra D. Genomic and computational approaches to dissect the mechanisms of STAT3’s universal and cell type-specific functions. JAKSTAT. 2013;2(4):e25097. doi: 10.4161/jkst.25097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML. Distinct transcriptional regulatory modules underlie STAT3’s cell type-independent and cell type-specific functions. Nucleic Acids Res. 2013;41(4):2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong A, Yang Z, Shen Y, Zhou J, Shen Q. Transcription factor STAT3 as a novel molecular target for cancer prevention. Cancers (Basel) 2014;6(2):926–957. doi: 10.3390/cancers6020926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. Aug 6. [DOI] [PubMed] [Google Scholar]
- 37.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251(2):199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 38.Bi S, Chen K, Feng L, Fu G, Yang Q, Deng M. Napabucasin (BBI608) eliminate AML cells in vitro and in vivo via inhibition of Stat3 pathway and induction of DNA damage. Eur J Pharmacol. 2019;855:252–261. doi: 10.1016/j.ejphar.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Severin F, Frezzato F, Visentin A, Martini V, Trimarco V, Carraro S. In chronic lymphocytic leukemia the JAK2/STAT3 pathway is constitutively activated and its inhibition leads to CLL cell death unaffected by the protective bone marrow microenvironment. Cancers (Basel) 2019;11(12):1939. doi: 10.3390/cancers11121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teoh PJ, Chung TH, Chng PYZ, Toh SHM, Chng WJ. IL6R-STAT3-ADAR1 (P150) interplay promotes oncogenicity in multiple myeloma with 1q21 amplification. Haematologica. 2020;105(5):1391–1404. doi: 10.3324/haematol.2019.221176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan MW, Saadalla A, Ewida AH, Al-Katranji K, Al-Saoudi G, Giaccone ZT. The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer. Cancer Immunol Immunother. 2018;67(1):13–23. doi: 10.1007/s00262-017-2057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takemoto S, Ushijima K, Kawano K, Yamaguchi T, Terada A, Fujiyoshi N. Expression of activated signal transducer and activator of transcription-3 predicts poor prognosis in cervical squamous-cell carcinoma. Br J Cancer. 2009;101:967–972. doi: 10.1038/sj.bjc.6605212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CL, Cen L, Kohout J, Hutzen B, Chan C, Hsieh FC. Signal transducer and activator of transcription 3 activation is associated with bladder cancer cell growth and survival. Mol Cancer. 2008;7:78. doi: 10.1186/1476-4598-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26(2):207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 45.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 46.Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15(4):283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117–128. doi: 10.1016/j.canlet.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10(1):48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 50.Iwata-Kajihara T, Sumimoto H, Kawamura N, Ueda R, Takahashi T, Mizuguchi H. Enhanced cancer immunotherapy using STAT3-depleted dendritic cells with high Th1-inducing ability and resistance to cancer cell-derived inhibitory factors. J Immunol. 2011;187:27–36. doi: 10.4049/jimmunol.1002067. [DOI] [PubMed] [Google Scholar]
- 51.Herrmann A, Kortylewski M, Kujawski M, Zhang C, Reckamp K, Armstrong B. Targeting Stat3 in the myeloid compartment drastically improves the in vivo antitumor functions of adoptively transferred T cells. Cancer Res. 2010;70(19):7455–7464. doi: 10.1158/0008-5472.CAN-10-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee H, Zhang P, Herrmann A, Yang C, Xin H, Wang Z. Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci U S A. 2012;109(20):7765–7769. doi: 10.1073/pnas.1205132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23(6):839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butturini E, Darra E, Chiavegato G, Cellini B, Cozzolino F, Monti M. S-Glutathionylation at Cys328 and Cys542 impairs STAT3 phosphorylation. ACS Chem Biol. 2014;9(8):1885–1893. doi: 10.1021/cb500407d. [DOI] [PubMed] [Google Scholar]
- 55.Yuan J, Zhang F, Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep. 2015;5:17663. doi: 10.1038/srep17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Z, Wang M, Li J, Xiao M, Chin YE, Cheng J. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene. 2016;35(45):5826–5838. doi: 10.1038/onc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gough DJ, Koetz L, Levy DE. The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS One. 2013;8(11):e83395. doi: 10.1371/journal.pone.0083395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gollob JA, Schnipper C, Murphy EA, Ritz J, Frank DA. The functional synergy between IL-12 and IL-2 involves p38 mitogen-activated protein kinase and is associated with the augmentation of STAT serine phosphorylation. J Immunol. 1999;162:4472–4481. [PubMed] [Google Scholar]
- 59.Chen B, Liu J, Chang Q, Beezhold K, Lu Y, Chen F. JNK and STAT3 signaling pathways converge on Akt-mediated phosphorylation of EZH2 in bronchial epithelial cells induced by arsenic. Cell Cycle. 2013;12(1):112–121. doi: 10.4161/cc.23030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dodd KM, Yang J, Shen MH, Sampson JR. Tee AR. mTORC1 drives HIF-1α and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene. 2015;34(17):2239–2250. doi: 10.1038/onc.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gartsbein M, Alt A, Hashimoto K, Nakajima K, Kuroki T, Tennenbaum T. The role of protein kinase C δ activation and STAT3 Ser727 phosphorylation in insulin-induced keratinocyte proliferation. J Cell Sci. 2006;119(Pt 3):470–481. doi: 10.1242/jcs.02744. [DOI] [PubMed] [Google Scholar]
- 62.Balic JJ, Albargy H, Luu K, Kirby FJ, Jayasekara WSN, Mansell F. STAT3 serine phosphorylation is required for TLR4 metabolic reprogramming and IL-1β expression. Nat Commun. 2020;11(1):3816. doi: 10.1038/s41467-020-17669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wen Z, Zhong Z, Darnell JE., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 64.Tkach M, Rosemblit C, Rivas MA, Proietti CJ, Díaz Flaqué MC, Mercogliano MF. p42/p44 MAPK-mediated Stat3Ser727 phosphorylation is required for progestin-induced full activation of Stat3 and breast cancer growth. Endocr Relat Cancer. 2013;20(2):197–212. doi: 10.1530/ERC-12-0194. [DOI] [PubMed] [Google Scholar]
- 65.Sakaguchi M, Oka M, Iwasaki T, Fukami Y, Nishigori C. Role and regulation of STAT3 phosphorylation at Ser727 in melanocytes and melanoma cells. J Invest Dermatol. 2012;132(7):1877–1885. doi: 10.1038/jid.2012.45. [DOI] [PubMed] [Google Scholar]
- 66.Aziz MH, Hafeez BB, Sand JM, Pierce DB, Aziz SW, Dreckschmidt NE. Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2) Oncogene. 2010;29(21):3100–3109. doi: 10.1038/onc.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villalva C, Martin-Lannerée S, Cortes U, Dkhissi F, Wager M, Le Corf A. STAT3 is essential for the maintenance of neurosphere-initiating tumor cells in patients with glioblastomas: a potential for targeted therapy? Int J Cancer. 2011;128(4):826–838. doi: 10.1002/ijc.25416. [DOI] [PubMed] [Google Scholar]
- 68.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100(12):3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu FN, Chen MC, Lin KC, Peng YT, Li PC, Lin E. Cyclin-dependent kinase 5 modulates STAT3 and androgen receptor activation through phosphorylation of Ser⁷²⁷ on STAT3 in prostate cancer cells. Am J Physiol Endocrinol Metab. 2013;305(8):E975–E986. doi: 10.1152/ajpendo.00615.2012. [DOI] [PubMed] [Google Scholar]
- 70.Friedbichler K, Kerenyi MA, Kovacic B, Li G, Hoelbl A, Yahiaoui S. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood. 2010;116(9):1548–1558. doi: 10.1182/blood-2009-12-258913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berger A, Hoelbl-Kovacic A, Bourgeais J, Hoefling L, Warsch W. PAK-dependent STAT5 serine phosphorylation is required for BCR-ABL-induced leukemogenesis. Leukemia. 2014;28(3):629–641. doi: 10.1038/leu.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qin HR, Kim HJ, Kim JY, Hurt EM, Klarmann GJ, Kawasaki BT. Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 2008;68(19):7736–7741. doi: 10.1158/0008-5472.CAN-08-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kondo K, Shaim H, Thompson PA, Burger JA, Keating M, Estrov Z. Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway. Leukemia. 2018;32(4):960–970. doi: 10.1038/leu.2017.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dimri S, Malhotra R, Shet T, Mokal S, Gupta S, De A. Noncanonical pS727 post translational modification dictates major STAT3 activation and downstream functions in breast cancer. Exp Cell Res. 2020;396(2) doi: 10.1016/j.yexcr.2020.112313. [DOI] [PubMed] [Google Scholar]
- 75.Liu Y, Lv J, Liu J, Liang X, Jin X, Xie J. STAT3/p53 pathway activation disrupts IFN-β-induced dormancy in tumor-repopulating cells. J Clin Invest. 2018;128(3):1057–1073. doi: 10.1172/JCI96329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood. 2010;115(14):2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balic JJ, Garama DJ, Saad MI, Yu L, West AC, West AJ. Serine-phosphorylated STAT3 promotes tumorigenesis via modulation of RNA polymerase transcriptional activity. Cancer Res. 2019;79(20):5272–5287. doi: 10.1158/0008-5472.CAN-19-0974. [DOI] [PubMed] [Google Scholar]
- 78.Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer — using tissue repair as a road map. Nat Rev Cancer. 2019;19(2):82–96. doi: 10.1038/s41568-018-0090-8. [DOI] [PubMed] [Google Scholar]
- 79.Balic JJ, White CL, Dawson R, Gough D, McCormack MP, Jenkins BJ. STAT3-driven hematopoiesis and lymphopoiesis abnormalities are dependent on serine phosphorylation. Cytokine. 2020;130 doi: 10.1016/j.cyto.2020.155059. [DOI] [PubMed] [Google Scholar]
- 80.Gough DJ, Marié IJ, Lobry C, Aifantis I, Levy DE. STAT3 supports experimental K-RasG12D-induced murine myeloproliferative neoplasms dependent on serine phosphorylation. Blood. 2014;124(14):2252–2261. doi: 10.1182/blood-2013-02-484196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gkouveris I, Nikitakis N, Karanikou M, Rassidakis G, Sklavounou A. JNK1/2 expression and modulation of STAT3 signaling in oral cancer. Oncol Lett. 2016;12(1):699–706. doi: 10.3892/ol.2016.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin WH, Chang YW, Hong MX, Hsu TC, Lee KC, Lin C. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT–MET switch and cancer metastasis. Oncogene. 2021;40(4):791–805. doi: 10.1038/s41388-020-01566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lufei C, Ma J, Huang G, Zhang T, Novotny-Diermayr V, Ong CT. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 2003;22(6):1325–1335. doi: 10.1093/emboj/cdg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NB. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J. Biol. Chem. 2013;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324(5935):1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323(5915):793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D'Angeli L, Bartoli A. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2010;2:823–842. doi: 10.18632/aging.100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Q, Raje V, Yakovlev VA, Yacoub A, Szczepanek K, Meier J. Mitochondrial localized stat3 promotes breast cancer growth via phosphorylation of serine 727. J Biol Chem. 2013;288(43):31280–31288. doi: 10.1074/jbc.M113.505057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu C, Huo X, Agoston AT, Zhang X, Theiss AL, Cheng E. Mitochondrial STAT3 contributes to transformation of Barrett's epithelial cells that express oncogenic Ras in a p53-independent fashion. Am J Physiol Gastrointest Liver Physiol. 2015;309(3):G146–G161. doi: 10.1152/ajpgi.00462.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu S, Gandler HI, Tošić I, Ye DQ, Giaccone ZT, Frank DA. Mutant KRAS downregulates the receptor for leukemia inhibitory factor (LIF) to enhance a signature of glycolysis in pancreatic cancer and lung cancer. Mol Cancer Res. 2021 doi: 10.1158/1541-7786.MCR-20-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harhous Z, Badawi S, Bona NG, Pillot B, Augeul L, Paillard M. Critical appraisal of STAT3 pattern in adult cardiomyocytes. J Mol Cell Cardiol. 2019;131:91–100. doi: 10.1016/j.yjmcc.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 92.Boengler K, Hilfiker-Kleiner D, Heusch G, Schulz R. Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res Cardiol. 2010;105(6):771–785. doi: 10.1007/s00395-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doherty JR, Yang C, Scott KE, Cameron MD, Fallahi M, Li W. Blocking lactate export by inhibiting the Myc target MCT1 disables glycolysis and glutathione synthesis. Cancer Res. 2014;74(3):908–920. doi: 10.1158/0008-5472.CAN-13-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiang S, Gu H, Jin L, Thorne RF, Zhang XD, Wu M. LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci U S A. 2018;115(7):E1465–E1474. doi: 10.1073/pnas.1711257115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275(29):21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 96.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14(12):1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107(5):2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kappler M, Pabst U, Weinholdt C, Taubert H, Rot S, Kaune T. Causes and Consequences of A Glutamine Induced Normoxic HIF1 Activity for the Tumor Metabolism. Int J Mol Sci. 2019;20(19):4742. doi: 10.3390/ijms20194742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nagao A, Kobayashi M, Koyasu S, Chow CCT, Harada H. HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int J Mol Sci. 2019;20(2):238. doi: 10.3390/ijms20020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pawlus MR, Wang L, Hu CJ. STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene. 2014;33(13):1670–1679. doi: 10.1038/onc.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cao Y. Adipocyte and lipid metabolism in cancer drug resistance. J Clin Invest. 2019;129(8):3006–3017. doi: 10.1172/JCI127201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zimta AA, Tigu AB, Muntean M, Cenariu D, Slaby O, Berindan-Neagoe I. Molecular Links between Central Obesity and Breast Cancer. Int J Mol Sci. 2019;20(21):5364. doi: 10.3390/ijms20215364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gallagher E, Novosyadlyy R, Yakar S, LeRoith D. In: Principles of Diabetes Mellitus. Poretsky L, editor. Springer; US: 2010. The increased risk of cancer in obesity and type 2 diabetes: potential mechanisms; pp. 579–599. editor. [Google Scholar]
- 104.Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 105.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279(15):2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 106.Banks AS, Davis SM, Bates SH, Myers MG., Jr Activation of downstream signals by the long form of the leptin receptor. J. Biol. Chem. 2000;275:14563–14572. doi: 10.1074/jbc.275.19.14563. [DOI] [PubMed] [Google Scholar]
- 107.Alshaker H, Wang Q, Frampton AE, Krell J, Waxman J, Winkler M. Sphingosine kinase 1 contributes to leptin-induced STAT3 phosphorylation through IL-6/gp130 transactivation in oestrogen receptor-negative breast cancer. Breast Cancer Res Treat. 2015;149(1):59–67. doi: 10.1007/s10549-014-3228-8. [DOI] [PubMed] [Google Scholar]
- 108.Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nat Rev Cancer. 2011;11(12):886–895. doi: 10.1038/nrc3174. [DOI] [PubMed] [Google Scholar]
- 109.Hu W, Lv J, Han M, Yang Z, Li T, Jiang S. STAT3: The art of multi-tasking of metabolic and immune functions in obesity. Prog Lipid Res. 2018;70:17–28. doi: 10.1016/j.plipres.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 110.Bullwinkle EM, Parker MD, Bonan NF, Falkenberg LG, Davison SP, DeCicco-Skinner KL. Adipocytes contribute to the growth and progression of multiple myeloma: Unraveling obesity related differences in adipocyte signaling. Cancer Lett. 2016;380(1):114–121. doi: 10.1016/j.canlet.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 111.Li Z, Zhang C, Du JX, Zhao J, Shi MT, Jin MW. Adipocytes promote tumor progression and induce PD-L1 expression via TNF-α/IL-6 signaling. Cancer Cell Int. 2020;20:179. doi: 10.1186/s12935-020-01269-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu L, Shen M, Chen X, Zhu R, Yang DR, Tsai Y. Adipocytes affect castration-resistant prostate cancer cells to develop the resistance to cytotoxic action of NK cells with alterations of PD-L1/NKG2D ligand levels in tumor cells. Prostate. 2018;78(5):353–364. doi: 10.1002/pros.23479. [DOI] [PubMed] [Google Scholar]
- 113.He JY, Wei XH, Li SJ, Liu Y, Hu HL, Li ZZ. Adipocyte-derived IL-6 and leptin promote breast Cancer metastasis via upregulation of Lysyl Hydroxylase-2 expression. Cell Commun Signal. 2018;16:100. doi: 10.1186/s12964-018-0309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gyamfi J, Lee YH, Min BS, Choi J. Niclosamide reverses adipocyte induced epithelial-mesenchymal transition in breast cancer cells via suppression of the interleukin-6/STAT3 signalling axis. Sci Rep. 2019;9:11336. doi: 10.1038/s41598-019-47707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gyamfi J, Lee YH, Eom M, Choi J. Interleukin-6/STAT3 signalling regulates adipocyte induced epithelial-mesenchymal transition in breast cancer cells. Sci Rep. 2018;8:8859. doi: 10.1038/s41598-018-27184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hayashi T, Fujita K, Nojima S, Hayashi Y, Nakano K, Ishizuya Y. High-fat diet-induced inflammation accelerates prostate cancer growth via IL6 signaling. Clin Cancer Res. 2018;24(17):4309–4318. doi: 10.1158/1078-0432.CCR-18-0106. [DOI] [PubMed] [Google Scholar]
- 117.Yang X, Jia J, Yu Z, Duanmu Z, He H, Chen S. Inhibition of JAK2/STAT3/SOCS3 signaling attenuates atherosclerosis in rabbit. BMC Cardiovasc Disord. 2020;20:133. doi: 10.1186/s12872-020-01391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang X, Liu M, Cai GH, Chen Y, Shi XC, Zhang CC. A potential nutraceutical candidate lactucin inhibits adipogenesis through downregulation of JAK2/STAT3 signaling pathway-mediated mitotic clonal expansion. Cells. 2020;9(2):331. doi: 10.3390/cells9020331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu C, Niu X, Du Y, Chen Y, Liu X, Xu L. IL-17A promotes fatty acid uptake through the IL-17A/IL-17RA/p-STAT3/FABP4 axis to fuel ovarian cancer growth in an adipocyte-rich microenvironment. Cancer Immunol Immunother. 2020;69(1):115–126. doi: 10.1007/s00262-019-02445-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vidavsky N, Kunitake JAMR, Diaz-Rubio ME, Chiou AE, Loh HC, Zhang S. Mapping and profiling lipid distribution in a 3D model of breast cancer progression. ACS Cent Sci. 2019;5(5):768–780. doi: 10.1021/acscentsci.8b00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19(3):380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kauraniemi P, Bärlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61(22):8235–8240. [PubMed] [Google Scholar]
- 123.Kourtidis A, Srinivasaiah R, Carkner RD, Brosnan MJ, Conklin DS. Peroxisome proliferator-activated receptor-gamma protects ERBB2-positive breast cancer cells from palmitate toxicity. Breast Cancer Res. 2009;11(2):R16. doi: 10.1186/bcr2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang D, Zhou Y, Lei W, Zhang K, Shi J, Hu Y. Signal transducer and activator of transcription 3 (STAT3) regulates adipocyte differentiation via peroxisome-proliferator-activated receptor γ (PPARγ) Biol Cell. 2009;102(1):1–12. doi: 10.1042/BC20090070. [DOI] [PubMed] [Google Scholar]
- 125.Wang T, Fahrmann JF, Lee H, Li YJ, Tripathi SC, Yue C. JAK/STAT3-regulated fatty acid β-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 2018;27(1):136–150. doi: 10.1016/j.cmet.2017.11.001. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yoshii Y, Furukawa T, Saga T, Fujibayashi Y. Acetate/acetyl-CoA metabolism associated with cancer fatty acid synthesis: overview and application. Cancer Lett. 2015;356(2 Pt A):211–216. doi: 10.1016/j.canlet.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 127.Menendez JA, Lupu R. Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin Ther Targets. 2017;21(11):1001–1016. doi: 10.1080/14728222.2017.1381087. [DOI] [PubMed] [Google Scholar]
- 128.Wu J, Du J, Fu X, Liu B, Cao H, Li T. Iciartin, a Novel FASN inhibitor, exerts anti-melanoma activities through IGF-1R/STAT3 signaling. Oncotarget. 2016;7(32):51251–51269. doi: 10.18632/oncotarget.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gao P, Wang LL, Liu J, Dong F, Song W, Liao L. Dihydroartemisinin inhibits endothelial cell tube formation by suppression of the STAT3 signaling pathway. Life Sci. 2020;242 doi: 10.1016/j.lfs.2019.117221. [DOI] [PubMed] [Google Scholar]
- 130.Menendezab JA, Vellon L, Lupu R. Targeting fatty acid synthase-driven lipid rafts: a novel strategy to overcome trastuzumab resistance in breast cancer cells. Med Hypotheses. 2005;64(5):997–1001. doi: 10.1016/j.mehy.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 131.Vazquez-Martin A, Colomer R, Brunet J, Menendez JA. Inhibition of fatty acid synthase reverses autoresistance to trastuzumab in breast cancer: Pharmacological blockade of fatty acid synthase (FASN) reverses acquired autoresistance to trastuzumab (Herceptin by transcriptionally inhibiting 'HER2 super-expression' occurring in high-dose trastuzumab-conditioned SKBR3/Tzb100 breast cancer cells. Int J Oncol. 2007;31(4):769–776. [PubMed] [Google Scholar]
- 132.Qu Q, Zeng F, Liu X, Wang QJ, Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis. 2016;7(5):e2226. doi: 10.1038/cddis.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Koo YD, Lee JS, Lee SA, Quaresma PGF, Bhat R, Haynes WG. SUMO-specific protease 2 mediates leptin-induced fatty acid oxidation in skeletal muscle. Metabolism. 2019;95:27–35. doi: 10.1016/j.metabol.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li J, Zhao S, Zhou X, Zhang T, Zhao L, Miao P. Inhibition of lipolysis by mercaptoacetate and etomoxir specifically sensitize drug-resistant lung adenocarcinoma cell to paclitaxel. PLoS One. 2013;8:e74623. doi: 10.1371/journal.pone.0074623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.He W, Liang B, Wang C, Li S, Zhao Y, Huang Q. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38(23):4637–4654. doi: 10.1038/s41388-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tošić I, Heppler LN, Egusquiaguirre SP, Boehnke N, Correa S, Costa DF. Lipidome-based targeting of STAT3-driven breast cancer cells using poly-L-glutamic acid-coated layer-by-layer nanoparticles. Mol. Cancer Ther. 2021;20(4):726–738. doi: 10.1158/1535-7163.MCT-20-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jin X, Demere Z, Nair K, Ali A, Ferraro GB, Natoli T. A metastasis map of human cancer cell lines. Nature. 2020;588(7837):331–336. doi: 10.1038/s41586-020-2969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guillet-Deniau I, Pichard AL, Koné A, Esnous C, Nieruchalski M, Girard J. Glucose induces de novo lipogenesis in rat muscle satellite cells through a sterol-regulatory-element-binding-protein-1c-dependent pathway. J Cell Sci. 2004;117(Pt 10):1937–1944. doi: 10.1242/jcs.01069. [DOI] [PubMed] [Google Scholar]
- 141.Chen J, Yue J, Liu Y, Liu J, Jiao K, Teng M. Blocking of STAT-3/SREBP1-mediated glucose-lipid metabolism is involved in dietary phytoestrogen-inhibited ovariectomized-induced body weight gain in rats. J Nutr Biochem. 2018;61:17–23. doi: 10.1016/j.jnutbio.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 142.Su T, Huang C, Yang C, Jiang T, Su J, Chen M. Apigenin inhibits STAT3/CD36 signaling axis and reduces visceral obesity. Pharmacol Res. 2020;152 doi: 10.1016/j.phrs.2019.104586. [DOI] [PubMed] [Google Scholar]
- 143.Yakubenko VP, Hsi LC, Cathcart MK, Bhattacharjee A. From macrophage interleukin-13 receptor to foam cell formation: mechanisms for αMβ2 integrin interference. J Biol Chem. 2013;288(4):2778–2788. doi: 10.1074/jbc.M112.381343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stover DG, Gil Del Alcazar CR, Brock J, Guo H, Overmoyer B, Balko J. Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast Cancer. 2018;4:10. doi: 10.1038/s41523-018-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Orlova A, Wagner C, de Araujo ED, Bajusz D, Neubauer HA, Herling M. Direct targeting options for STAT3 and STAT5 in cancer. Cancers (Basel) 2019;11(12):1930. doi: 10.3390/cancers11121930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brambilla L, Lahiri T, Cammer M, Levy DE. STAT3 inhibitor OPB-51602 is cytotoxic to tumor cells through inhibition of complex I and ROS induction. iScience. 2020;23(12) doi: 10.1016/j.isci.2020.101822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tolcher A, Flaherty K, Shapiro GI, Berlin J, Witzig T, Habermann T. A first-in-human phase I study of OPB-111077, a small-molecule STAT3 and oxidative phosphorylation inhibitor, in patients with advanced cancers. Oncologist. 2018;23(6):658–e72. doi: 10.1634/theoncologist.2017-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kawazoe A, Kuboki Y, Bando H, Fukuoka S, Kojima T, Naito Y. Phase 1 study of napabucasin, a cancer stemness inhibitor, in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2020;85(5):855–862. doi: 10.1007/s00280-020-04059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Han D, Yu T, Dong N, Wang B, Sun F, Jiang D. Napabucasin, a novel STAT3 inhibitor suppresses proliferation, invasion and stemness of glioblastoma cells. J Exp Clin Cancer Res. 2019;38(1):289. doi: 10.1186/s13046-019-1289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bekaii-Saab T, Starodub A, El-Rayes B, O'Neil B, Shahda S, Ciombor K. A phase 1b/II study of cancer stemness inhibitor napabucasin in combination with gemcitabine (gem) & nab-paclitaxel (nabptx) in metastatic pancreatic adenocarcinoma (mpdac) patients (pts) Ann Oncol. 2017;28(3):150–153. [Google Scholar]
- 151.Lapidot M, Case AE, Larios D, Gandler HI, Meng C, Tošić I. Inhibitors of the transcription factor STAT3 decrease growth and induce immune response genes in models of malignant pleural mesothelioma (MPM) Cancers (Basel) 2020;13(1):7. doi: 10.3390/cancers13010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Babaei R, Schuster M, Meln I, Lerch S, Ghandour RA, Pisani DF. Jak-TGFβ cross-talk links transient adipose tissue inflammation to beige adipogenesis. Sci Signal. 2018;11(527) doi: 10.1126/scisignal.aai7838. pii: eaai7838. [DOI] [PubMed] [Google Scholar]
- 153.Dambal S, Alfaqih M, Sanders S, Maravilla E, Ramirez-Torres A, Galvan GC. 27-hydroxycholesterol impairs plasma membrane lipid raft signaling as evidenced by inhibition of IL6-JAK-STAT3 signaling in prostate cancer cells. Mol Cancer Res. 2020;18(5):671–684. doi: 10.1158/1541-7786.MCR-19-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wu Y, Zhang S, Gong X, Tam S, Xiao D, Liu S. The epigenetic regulators and metabolic changes in ferroptosis-associated cancer progression. Mol Cancer. 2020;19:39. doi: 10.1186/s12943-020-01157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]