Abstract
Background
Antimicrobial resistance (AMR) is an emerging public health crisis in Uganda. The World Health Organization (WHO) Global Action Plan recommends that countries should develop and implement National Action Plans for AMR. We describe the establishment of the national AMR program in Uganda and present the early microbial sensitivity results from the program.
Objective
The aim of this study is to describe a national surveillance program that was developed to perform the systematic and continuous collection, analysis, and interpretation of AMR data.
Methods
A systematic qualitative description of the process and progress made in the establishment of the national AMR program is provided, detailing the progress made from 2015 to 2020. This is followed by a report of the findings of the isolates that were collected from AMR surveillance sites. Identification and antimicrobial susceptibility testing (AST) of the bacterial isolates were performed using standard methods at both the surveillance sites and the reference laboratory.
Results
Remarkable progress has been achieved in the establishment of the national AMR program, which is guided by the WHO Global Laboratory AMR Surveillance System (GLASS) in Uganda. A functional national coordinating center for AMR has been established with a supporting designated reference laboratory. WHONET software for AMR data management has been installed in the surveillance sites and laboratory staff trained on data quality assurance. Uganda has progressively submitted data to the WHO GLASS reporting system. Of the 19,216 isolates from WHO GLASS priority specimens collected from October 2015 to June 2020, 22.95% (n=4411) had community-acquired infections, 9.46% (n=1818) had hospital-acquired infections, and 68.57% (n=12,987) had infections of unknown origin. The highest proportion of the specimens was blood (12,398/19,216, 64.52%), followed by urine (5278/19,216, 27.47%) and stool (1266/19,216, 6.59%), whereas the lowest proportion was urogenital swabs (274/19,216, 1.4%). The mean age was 19.1 (SD 19.8 years), whereas the median age was 13 years (IQR 28). Approximately 49.13% (9440/19,216) of the participants were female and 50.51% (9706/19,216) were male. Participants with community-acquired infections were older (mean age 28, SD 18.6 years; median age 26, IQR 20.5 years) than those with hospital-acquired infections (mean age 17.3, SD 20.9 years; median age 8, IQR 26 years). All gram-negative (Escherichia coli, Klebsiella pneumoniae, and Neisseria gonorrhoeae) and gram-positive (Staphylococcus aureus and Enterococcus sp) bacteria with AST showed resistance to each of the tested antibiotics.
Conclusions
Uganda is the first African country to implement a structured national AMR surveillance program in alignment with the WHO GLASS. The reported AST data indicate very high resistance to the recommended and prescribed antibiotics for treatment of infections. More effort is required regarding quality assurance of laboratory testing methodologies to ensure optimal adherence to WHO GLASS–recommended pathogen-antimicrobial combinations. The current AMR data will inform the development of treatment algorithms and clinical guidelines.
Keywords: antimicrobial resistance, surveillance, microbiology, laboratory, Uganda, implementation, WHO, collection, analysis, data, antimicrobial, progress, bacteria, feasibility, resistance, antibiotic
Introduction
Background
Antimicrobial resistance (AMR) is associated with increased morbidity and mortality and is recognized as an emerging global health threat. If left unchecked, by 2050, AMR may contribute up to 10 million deaths per year [1]. In low- and middle-income countries (LMICs), particularly in Africa, data on drug-resistant infections are extremely scarce [2]. A few available reports indicate that resistance to commonly prescribed antibiotics is prevalent, but the methodology for the generation and reporting of AMR data is suboptimal [3,4]. A systematic review targeting policy makers in East Africa found significant knowledge gaps in AMR and recommended strengthening antimicrobial stewardship and AMR surveillance in the region [5].
In Uganda, early efforts against AMR identified the critical gap as a lack of routine surveillance systems with limited data, making it difficult to track the AMR burden [6]. Moreover, a substantial proportion of methicillin-resistant Staphylococcus aureus and gram-negative organisms in different sample types has been reported [7]. Another study at a Ugandan regional referral hospital (RRH) on antimicrobial-resistant infections among postpartum mothers recommended increased microbiological testing [8]. This informs appropriate antibiotic use, development of antimicrobial stewardship programs, and strengthening of infection prevention and control practices as top priorities. Notably, through an ongoing sentinel surveillance program, microbiology capacity has been enhanced in selected RRHs that contribute significantly toward bacterial ID and antimicrobial susceptibility testing (AST) in Uganda [9]. In addition, Uganda is among the few African countries that have adopted and established a quality-assured World Health Organization (WHO) Enhanced Gonococcal Antimicrobial Surveillance Program and reported data locally and globally [10,11].
In 2015, the WHO launched the Global Laboratory AMR Surveillance System (GLASS) and initiated its implementation in the human health sector [12]. The GLASS program provides national guidance on AMR, focusing on different surveillance methods for adoption and priority specimens, pathogens, and pathogen-antibacterial combinations for use within national surveillance programs. GLASS enables monitoring of emerging AMR profiles at the country level and facilitates the development of hospital-based antibiograms to inform clinical treatment decisions.
In line with global calls to enhance support for AMR systems in LMICs, development partners are currently supporting Uganda’s national laboratory system and, more recently, the National Action Plan (NAP) for AMR [13] using a system-strengthening approach [14]. Since 2015, the United States with the help of the Centers for Disease Control and Prevention has been supporting the laboratory capacity at national and regional referral levels to enhance sample transportation systems for microbiology samples. In 2018, the Fleming Fund of the United Kingdom initiated support to the Government of Uganda to strengthen national coordination efforts for AMR using the One Health approach. The efforts are targeting to expand the microbiology testing capacity at the national level and selected RRHs and generate quality-assured AMR data. With the support of these and other partners, Uganda is implementing its NAP for AMR, which was formally launched in 2019.
Objective
In this paper, we highlight the progress on the implementation of GLASS in Uganda from October 2015 to June 2020 and describe laboratory-based AMR surveillance data obtained from selected surveillance sites in the same period. The data include corresponding participant characteristics (sex and age), source of bacterial infection for surveillance hospital-acquired infections (HAIs), bacterial recovery rates, and resistance profiles.
Methods
Overview
A mixed methodology was used to obtain data presented in this study. Qualitative methods were used for the program setup, whereas quantitative methods were used to generate the AMR surveillance isolate data. A situational analysis report by the Uganda National Academy of Sciences was reviewed to understand the existing national AMR capacity and provide important information to guide program development [15]. To develop a sustainable national AMR surveillance program, the Ministry of Health (MoH) benchmarked on international guidance, using the approach first described in the WHO GLASS manual for the early implementation [12] and later interpreted according to the road map for participation in GLASS by Seale et al [16]. These recommendations were implemented under the cognizance of the local context of Uganda’s health systems. A systematic stepwise capacity-building approach [17] was used to set up and implement the program. The approach focused on setting up structures, systems, and roles at national and subnational levels; addressing staffing and infrastructure needs; and providing skills and tools to health workers. Stakeholders’ engagement was undertaken to ensure a supportive environment for the implementation of AMR surveillance. The partners supporting the national AMR surveillance program include the Centers for Disease Control and Prevention, the Fleming Fund, the World Bank, and academic institutions, including Makerere University and Mbarara University of Science and Technology. Using the outline by Seale et al [16], we describe the steps taken and the achievements of developing the national AMR surveillance program and later present the AMR data obtained from the program under the Results section.
Key Achievements
Enrollment of Uganda in GLASS
In 2015, Uganda responded to the WHO call for countries to enroll in the GLASS program. By enrolling in the GLASS program, Uganda committed to collecting and sharing national AMR surveillance data. As part of this process, the country also acquired the WHONET software [18] used to report AMR surveillance data. A WHO GLASS focal person was designated by the MoH to support the coordination of AMR data validation, quality assurance, and the reporting process. Uganda now participates in the annual AMR data submissions to GLASS, and the country AMR data are part of the WHO global AMR surveillance reports [19].
Establishment of the AMR National Coordinating Center
The National Coordinating Center (NCC) at the MoH has been set up to oversee the national AMR surveillance program in human health, including the collection and aggregation of data from surveillance sites. The NCC works in collaboration with the Uganda National AMR Sub-Committee (UNAMRsC) of the One Health approach to provide strategic oversights of the national AMR program. The UNAMRsC has also been established as part of the governance structure for AMR in the country. The membership of the UNAMRsC also includes representation from other relevant line ministries, such as animal health, wildlife, and the environment. The mandate of the UNAMRsC includes defining the national AMR surveillance objectives; developing and disseminating protocols; coordinating data collection, analysis, and reporting; and reviewing data before reporting to GLASS. To date, the NCC has supported the development of key AMR surveillance documents, including national AMR surveillance plans, protocols, guidelines, curricula, and microbiology standard operating procedures.
Finalization of the NAP for AMR
The WHO requested all member countries to develop multisectoral-wide NAPs that are aligned with the Global Action Plan for AMR to support the implementation of the national AMR programs. Working with partners, the UNAMRsC completed the development of the Uganda AMR NAP [13], which was launched in November 2018 and now supports the implementation of priority activities in the NAP for AMR.
Designation of the National Microbiology Reference Laboratory
Initially, the Department of Medical Microbiology Laboratory at Makerere University was designated as the AMR Surveillance Laboratory. However, the capacity of the Central Public Health Laboratories has been built gradually, and it is now the designated national microbiology reference laboratory for AMR surveillance. The capacity built at Central Public Health Laboratories includes human resource development, quality management systems toward accreditation, isolate transportation, enhanced biorepository, and enrollment of laboratories in an External Quality Assurance scheme. In addition, the state-of-the-art Becton and Dickson–manufactured equipment, including matrix assisted laser desorption/ionization time of flight [20] and Phoenix M50 [21], has been installed at the reference laboratories and BACTEC blood culture systems, including FX 200 and FX40 [22], at selected RRH laboratories. These results support bacterial ID and AST.
Selection and Capacity Building for AMR Surveillance Sites
In 2016, the AMR NCC designated different facilities as AMR surveillance sites. The sites were selected to ensure a balanced geographic, demographic, and socioeconomic distribution. They offer both outpatient and inpatient services, as per GLASS recommendations. However, the capacity of health facilities to conduct AMR surveillance varied between the different health facilities. As a result, selected sites have reported AMR surveillance data to the WHO and the capacity of the surveillance sites has been gradually developed. The Medicines and Therapeutics Committees oversee the implementation of the AMR surveillance program at the surveillance sites, which have been trained in collecting, analyzing, and reporting epidemiological, clinical, and laboratory data. The Medicines and Therapeutics Committee is usually headed by a senior consultant (Internal medicine, Gynecology, Surgery, and Pediatrics) who leads the stewardship of the AMR program at the site. On the basis of the clinician’s request for bacterial ID and AST as part of patient care, samples are collected according to the clinical protocols appropriate for the clinical presentation of patients and sent to the microbiology laboratory. The samples mainly include blood, urine, stool, and urogenital swabs and are accompanied by a microbiology laboratory request form that captures epidemiological information such as patient demographics and clinical presentation. The bacterial ID and AST data in this report were collected from 10 surveillance site microbiology laboratories between October 2015 and June 2020. The surveillance sites included Department of Medical Microbiology, Mbarara University of Science and Technology, Arua RRH, Kabale RRH, Mbarara RRH, Mubende RRH, Fort Portal RRH, Hoima RRH, Jinja RRH, Mbale RRH, and Soroti RRH (Figure 1). The surveillance sites have microbiology laboratories with the capacity to isolate, identify, and conduct microbial sensitivity testing for GLASS priority pathogens.
Figure 1.
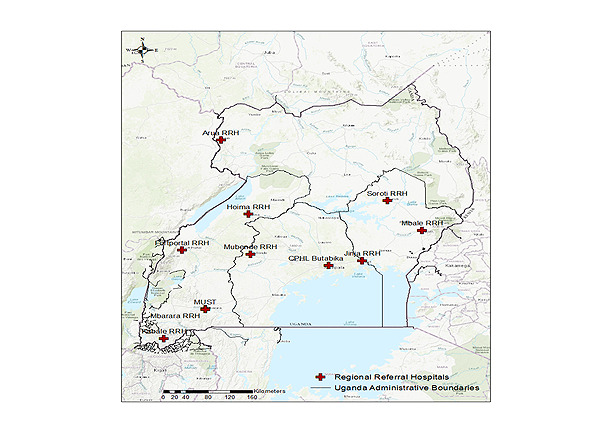
Geographic locations of the sentinel sites participating in the National Antimicrobial Resistance surveillance program.
Bacterial ID and AST
In the laboratory, bacterial ID and AST for the different samples were collected, and subsequently isolates were performed in accordance with the standardized microbiology protocols and standard operating procedures. Blood culture vials were placed in a BACTEC 9050 or FX40 blood culture system (Becton-Dickinson) according to the manufacturer’s instructions. Samples from flagged positive vials were subjected to Gram staining and then cultured on blood agar, chocolate agar, and MacConkey agar culture plates. The culture plates were incubated for 18 to 24 hours at 35°C to 37°C, with recovered colonies undergoing conventional biochemical testing to confirm ID. For stool samples, a loop full of emulsified sample was inoculated on deoxycholate citrate agar, or Xylose Lysine Deoxycholate agar, and MacConkey agar and incubated at 35°C to 37°C in ambient air for 18 to 24 hours. Isolation and ID of growth was performed using conventional methods. Urine samples were gently mixed and inoculated on MacConkey agar and blood agar using an appropriate calibrated loop and incubated in ambient air overnight for 18 to 24 hours at 35°C to 37°C. Isolation, conventional ID, and colony counting were performed where applicable. Both urine and stool were examined macroscopically and microscopically (Gram staining). All urogenital swabs were inoculated on selective modified Thayer Martin and nonselective chocolate agar culture plates and then incubated at 35°C to 37°C in 5% CO2-enriched humid conditions. ID of gonococci colonies was based on the growth of the colonies with typical morphology in the modified Thayer Martin medium with a positive oxidase test [23]. AST was performed using the Kirby-Bauer disk diffusion method according to the Clinical and Laboratory Standards Institute [24]. We followed the WHO GLASS pathogen-antimicrobial combinations to set the antibiotics for susceptibility testing [12]. Preliminary culture results were immediately sent to the hospital wards to help clinicians optimize patient management, whereas the final results were shared later.
Submission of AMR Data for National and WHO GLASS Reporting
All laboratory results (bacterial ID and AST), patient demographics, and clinical data were entered into the microbiology register. These data were then entered into the WHONET software program [18] on a weekly basis for data analysis to generate facility-based AMR surveillance reports. The AMR surveillance subcommittee technical working committee representatives conducted data quality assessments on a quarterly basis to inform key performance indicators and reports submitted to NCCs. Uganda has been consistently submitting data to GLASS reporting since its enrollment in 2016 [19].
Statistical Analysis
Summary statistics were calculated for key demographic characteristics of participants from whom samples were collected and stratified by the origin of the samples. Age was summarized as a continuous variable and categorized with age groups defined. P values based on chi-square tests were calculated for each variable to provide a sense of differences in demographic characteristics among different origins. For each specimen type, the percentage recovery for all bacterial pathogens and GLASS priority pathogens was expressed as the proportion of samples with a positive culture result out of the total samples cultured. Similarly, for each pathogen, resistance was expressed as the proportion of isolates with resistant or intermediate results out of the total number of isolates tested for susceptibility to a specific antibiotic. The binomial 95% CIs for the proportions of recovery and resistance were calculated using the Wilson method. The frequency of infection with resistant pathogens could not be calculated because data on the population at risk were unavailable. In addition, because of potential sampling bias, no statistical analysis was performed to identify any associations or risk factors for the occurrence of resistant pathogens. The analysis was performed using Excel (Microsoft), R version 3.6 (R Foundation for Statistical Computing), and WHONET.
Results
Demographics of Participants
Of the 19,216 participants involved in the surveillance program, 22.95% (4411/19,216) had community-acquired infections, 9.46% (1818/19,216) had HAIs, and 68.57% (12,987/19,216) had infections of unknown origin (Table 1). The mean age was 19.1 years (SD 19.8 years), whereas the median was 13 years (IQR 28 years). Approximately 49.13% (9440/19,216) of the participants were female, and 50.51% (9706/19,216) were male. Participants with community-acquired infection were older (mean age 28 years, SD 18.6 years; median age 26 years, IQR 20.5 years) than those with HAIs (mean age 17.3 years, SD 20.9 years; median age 8 years, IQR 26 years).
Table 1.
Characteristics of participants in the antimicrobial resistance surveillance program from 10 surveillance sites from October 2015 to June 2020.
| Characteristics | Origin | Total (N=19,216) | P value | |||||||||||
|
|
Community acquired (n=4411) | Hospital acquired (n=1818) | Unknown (n=12,987) |
|
|
|||||||||
| Value, mean (SD) | 28.0 (18.6) | 17.3 (20.9) | 15.9 (19.0) | 19.1 (19.8) | N/Aa | |||||||||
| Value, median (IQR) | 26 (20.5) | 8 (26) | 6 (27) | 13 (28) | N/A | |||||||||
| Age (years), n (%) | <.001 | |||||||||||||
|
|
<1 | 230 (5.21) | 192 (10.56) | 1753 (13.49) | 2175 (11.32) |
|
||||||||
|
|
1-4 | 365 (8.27) | 485 (26.68) | 3003 (23.12) | 3853 (20.05) |
|
||||||||
|
|
05-14 | 372 (8.43) | 459 (25.25) | 1600 (12.32) | 2431 (12.65) |
|
||||||||
|
|
15-24 | 909 (20.61) | 149 (8.19) | 1042 (8.02) | 2100 (10.92) |
|
||||||||
|
|
25-34 | 1083 (24.55) | 168 (9.24) | 1267 (9.76) | 2518 (13.1) |
|
||||||||
|
|
35-44 | 572 (12.97) | 120 (6.6) | 748 (5.76) | 1440 (7.49) |
|
||||||||
|
|
45-54 | 350 (7.93) | 88 (4.84) | 463 (3.56) | 901 (4.69) |
|
||||||||
|
|
55-64 | 170 (3.85) | 37 (2.04) | 268 (2.06) | 475 (2.47) |
|
||||||||
|
|
65-80 | 149 (3.38) | 60 (3.3) | 229 (1.76) | 438 (2.28) |
|
||||||||
|
|
>81 | 52 (1.18) | 30 (1.65) | 51 (0.39) | 133 (0.69) |
|
||||||||
|
|
Unknown | 159 (3.6) | 30 (1.65) | 2563 (19.74) | 2752 (14.32) |
|
||||||||
| Sex, n (%) | <.001 | |||||||||||||
|
|
Female | 2413 (54.72) | 836 (45.98) | 6191 (47.67) | 9440 (49.13) |
|
||||||||
|
|
Male | 1974 (44.75) | 980 (53.91) | 6752 (51.99) | 9706 (50.51) |
|
||||||||
|
|
Unknown | 24 (0.54) | 2 (0.11) | 44 (0.34) | 70 (0.36) |
|
||||||||
| Facility, n (%) | <.001 | |||||||||||||
|
|
Arua RRHb | 1026 (23.26) | 384 (21.12) | 1049 (8.08) | 2459 (12.79) |
|
||||||||
|
|
DMM MUSTc | 480 (10.88) | 35 (1.92) | 2351 (18.13) | 2866 (14.91) |
|
||||||||
|
|
Fort Portal RRH | 390 (8.84) | 270 (14.85) | 99 (0.76) | 759 (3.95) |
|
||||||||
|
|
Hoima RRH | 7 (0.16) | 2 (0.11) | 164 (1.26) | 173 (0.9) |
|
||||||||
|
|
Jinja RRH | 88 (1.99) | 336 (18.48) | 4398 (33.86) | 4822 (25.09) |
|
||||||||
|
|
Kabale RRH | 699 (15.84) | 106 (5.83) | 2191 (16.87) | 2996 (15.59) |
|
||||||||
|
|
Mbale RRH | 653 (14.8) | 163 (8.97) | 692 (5.33) | 1508 (7.85) |
|
||||||||
|
|
Mbarara RRH | 392 (8.89) | 277 (15.24) | 132 (1.02) | 801 (4.17) |
|
||||||||
|
|
Mubende RRH | 312 (7.07) | 153 (8.42) | 1464 (11.27) | 1929 (10.04) |
|
||||||||
|
|
Soroti RRH | 364 (8.25) | 92 (5.06) | 447 (3.44) | 903 (4.69) |
|
||||||||
| Department, n (%) | <.001 | |||||||||||||
|
|
Inpatient | 1186 (26.89) | 1509 (83) | 4527 (34.86) | 7222 (37.58) |
|
||||||||
|
|
Outpatient | 2850 (64.61) | 181 (9.96) | 3289 (25.32) | 6320 (32.89) |
|
||||||||
|
|
Unknown | 375 (8.5) | 128 (7.04) | 5171 (39.82) | 5674 (29.53) |
|
||||||||
aN/A: not applicable.
bRRH: regional referral hospital.
cDMM MUST: Department of Medical Microbiology, Mbarara University of Science and Technology.
A total of 19,216 WHO GLASS priority specimens were collected for microbiological testing from 10 surveillance sites over a period of 4 years and 6 months, from October 2015 to June 2020. The highest proportion of the specimens was blood (12,398/19,216, 64.52%), followed by urine (5278/19,216, 27.47%) and stool (1266/19,216, 6.59%), whereas the lowest proportion was that of urogenital swabs (274/19,216, 1.43%).
Recovery Rates and Distribution of Pathogens
The overall recovery rate of the GLASS priority pathogens from the GLASS priority specimens was 7.4% (1429/19,216; Table 2). The recovery rates from the different samples were as follows: urogenital swabs 17.9% (49/274), urine 12.1% (637/5278), stool 7.74% (98/1266), and blood 5.2% (645/12,398). The highest percentage of GLASS priority pathogens identified were Escherichia coli (652/1429, 45.62%), followed by S aureus (337/1429, 23.58%), with the lowest being Acinetobacter baumannii (6/1429, 0.42%).
Table 2.
Bacterial recovery rates from priority specimens collected from 10 surveillance sites, October 2015 to June 2020.
| Variable | Value, n (%) | Odds ratio (95% CI) | |||
| Samples cultured (n=19,216) | N/Aa | ||||
|
|
Blood | 12,398 (64.52) |
|
||
|
|
Urogenital swabs | 274 (1.43) |
|
||
|
|
Stool | 1266 (6.59) |
|
||
|
|
Urine | 5278 (27.47) |
|
||
| Samples with bacterial growth (n=4471) | 23.3 (22.7-23.9) | ||||
|
|
Blood | 1520 (33.99) | 12.3 (11.7-12.9) | ||
|
|
Urogenital swabs | 174 (3.89) | 63.5 (57.7-69.0) | ||
|
|
Stool | 491 (10.98) | 38.8 (36.1-41.5) | ||
|
|
Urine | 2286 (51.13) | 43.3 (42.0-44.6) | ||
| Samples yielding the GLASSb priority pathogens (n=1429) | 7.4 (7-7.8) | ||||
|
|
Blood | 645 (45.14) | 5.2 (4.8-5.6) | ||
|
|
Urogenital swabs | 49 (3.43) | 17.9 (13.8-22.9) | ||
|
|
Stool | 98 (6.86) | 7.7 (6.3-9.3) | ||
|
|
Urine | 637 (44.58) | 12.1 (11.3-13.0) | ||
| GLASS priority pathogens recovered (n=1429) | N/A | ||||
|
|
Escherichia coli | 652 (45.62) |
|
||
|
|
Staphylococcus aureus | 337 (23.58) |
|
||
|
|
Salmonella spp | 237 (16.58) |
|
||
|
|
Klebsiella pneumoniae | 109 (7.63) |
|
||
|
|
Neisseria gonorrhoeae | 49 (3.43) |
|
||
|
|
Shigella spp | 21 (1.47) |
|
||
|
|
S pneumoniae | 18 (1.26) |
|
||
|
|
Acinetobacter baumannii | 6 (0.42) |
|
||
aN/A: not applicable.
bGLASS: Global Laboratory Antimicrobial Resistance Surveillance System.
Pathogen Resistance
Resistance patterns for the most commonly isolated gram-negative bacteria, that is, E coli, Neisseria gonorrhoeae, Shigella sp, and Salmonella sp are shown in Table 3, with all gram-negative bacteria showing resistance to each of the tested antibiotics. High resistance of E coli was noted among commonly used antibiotics, with 52.7% (95% CI 46.8%-58.5%) resistance to ceftriaxone, 18.8% (95% CI 14.9%-23.4%) resistance to imipenem, and 52% (95% CI 47.5%-56.6%) resistance to ciprofloxacin. High resistance of Klebsiella pneumoniae was noted among the commonly used antibiotics. High resistance of N gonorrhoeae was noted among the commonly used antibiotics, with 10% (95% CI 1.8%-40.4%) resistance to ceftriaxone and 71.4% (95% CI 45.4%-88.3%) resistance to ciprofloxacin. High resistance of Salmonella and Shigella was noted among the commonly used antibiotics, including resistance to meropenem, ceftriaxone, and ciprofloxacin.
Table 3.
Antimicrobial resistance profiles of selected gram-negative bacteria from 10 surveillance sites from October 2015 to June 2020.
| Bacteria name | Antibiotic name | Number | R+Ia (95% CI; %) |
| Escherichia coli | |||
|
|
Amikacin | 52 | 7.7 (3-18.2) |
|
|
Amoxicillin | 25 | 88 (70-95.8) |
|
|
Amoxicillin and clavulanic acid | 232 | 75 (69.1-80.1) |
|
|
Ampicillin | 359 | 91.6 (88.3-94.1) |
|
|
Cefoxitin | 27 | 25.9 (13.2-44.7) |
|
|
Ceftazidime | 77 | 45.5 (34.8-56.5) |
|
|
Ceftriaxone | 277 | 52.7 (46.8-58.5) |
|
|
Cefuroxime | 334 | 63.8 (58.5-68.7) |
|
|
Chloramphenicol | 366 | 42.1 (37.1-47.2) |
|
|
Ciprofloxacin | 455 | 52.1 (47.5-56.6) |
|
|
Clindamycin | 54 | 88.9 (77.8-94.8) |
|
|
Erythromycin | 100 | 92 (85-95.9) |
|
|
Gentamicin | 403 | 38.2 (33.4-42.8) |
|
|
Imipenem | 324 | 18.8 (14.9-23.4) |
|
|
Levofloxacin | 35 | 5.7 (1.6-18.6) |
|
|
Meropenem | 42 | 19 (10-33.3) |
|
|
Nalidixic acid | 181 | 76.8 (70.1-82.3) |
|
|
Nitrofurantoin | 266 | 30.8 (25.6-36.6) |
|
|
Penicillin G | 71 | 97.2 (90.3-99.2) |
|
|
Piperacillin or tazobactam | 33 | 36.4 (22.2-53.4) |
|
|
Tetracycline | 226 | 78.8 (73-83.6) |
|
|
Trimethoprim-sulfamethoxazole | 327 | 83.8 (79.4-87.4) |
|
|
Vancomycin | 94 | 76.6 (67.1-84) |
| Klebsiella pneumoniae | |||
|
|
Amoxicillin | 50 | 86 (73.8-93) |
|
|
Ampicillin | 79 | 97.5 (91.2-99.3) |
|
|
Ceftazidime | 20 | 65 (43.3-81.9) |
|
|
Ceftriaxone | 79 | 79.7 (69.6-87.1) |
|
|
Cefuroxime | 63 | 77.8 (66.1-86.3) |
|
|
Chloramphenicol | 78 | 53.8 (42.9-64.5) |
|
|
Ciprofloxacin | 86 | 53.5 (43-63.7) |
|
|
Gentamicin | 78 | 71.8 (61-80.6) |
|
|
Imipenem | 63 | 1.6 (0.3-8.5) |
|
|
Meropenem | 21 | 23.8 (10.6-45.1) |
|
|
Tetracycline | 42 | 54.8 (39.9-68.8) |
|
|
Trimethoprim-sulfamethoxazole | 67 | 82.1 (71.3-89.4) |
| Neisseria gonorrhoeae | |||
|
|
Ceftriaxone | 10 | 10 (1.8-40.4) |
|
|
Cefuroxime | 13 | 46.2 (23.2-70.9) |
|
|
Ciprofloxacin | 14 | 71.4 (45.4-88.3) |
|
|
Tetracycline | 16 | 100 (80.6-100) |
| Salmonella sp | |||
|
|
Amikacin | 37 | 5.4 (1.5-17.7) |
|
|
Amoxicillin | 33 | 100 (89.6-100) |
|
|
Ampicillin | 131 | 81.7 (74.2-87.4) |
|
|
Ceftazidime | 66 | 13.6 (7.3-23.9) |
|
|
Ceftriaxone | 98 | 17.3 (11.1-26) |
|
|
Cefuroxime | 108 | 20.4 (13.9-28.9) |
|
|
Chloramphenicol | 138 | 66.7 (58.4-74) |
|
|
Ciprofloxacin | 116 | 24.1 (17.3-32.7) |
|
|
Gentamicin | 52 | 17.3 (9.4-29.7) |
|
|
Imipenem | 83 | 3.6 (1.2-10.1) |
|
|
Levofloxacin | 60 | 1.7 (0.3-8.9) |
|
|
Nalidixic acid | 127 | 15.7 (10.4-23.1) |
|
|
Tetracycline | 96 | 87.5 (79.4-92.7) |
|
|
Trimethoprim-sulfamethoxazole | 114 | 69.3 (60.3-77) |
| Shigella sp | |||
|
|
Amikacin | 15 | 93.3 (70.2-98.8) |
|
|
Ceftriaxone | 19 | 15.8 (5.5-37.6) |
|
|
Cefuroxime | 10 | 50 (23.7-76.3) |
|
|
Chloramphenicol | 11 | 54.5 (28-78.7) |
|
|
Ciprofloxacin | 20 | 30 (14.5-51.9) |
|
|
Gentamicin | 13 | 23.1 (8.2-50.3) |
|
|
Nalidixic acid | 12 | 33.3 (13.8-60.9) |
|
|
Tetracycline | 12 | 50 (25.4-74.6) |
|
|
Trimethoprim-sulfamethoxazole | 13 | 38.5 (17.7-64.5) |
aR+I: Resistance + Intermediate.
Among gram-positive bacteria, high resistance of S aureus was noted among the commonly used antibiotics, with 42.9% (95% CI 28%-59.1%) resistance to cefoxitin, 30.9% (95% CI 21.2%-42.6%) resistance to oxacillin, 76.9% (95% CI 69%-83.2%) resistance to TMP-SMX, and 15.5% (95% CI 9.6%-24%) resistance to vancomycin (Table 4). High resistance of Enterococcus sp was noted among the commonly used antibiotics, with 81.8% (95% CI 61.5%-92.7%) resistance to ciprofloxacin and 50% (95% CI 33.6%-66.4%) resistance to vancomycin. A high resistance of Streptococcus sp was noted between vancomycin and ceftriaxone.
Table 4.
Antimicrobial resistance profiles of selected gram-positive bacteria from 10 surveillance sites, from October 2015 to June 2020.
| Bacteria name | Antibiotic name | Number | R+Ia (95% CI; %) |
| Staphylococcus aureus | |||
|
|
Amoxicillin and clavulanic acid | 47 | 51.1 (37.2-64.7) |
|
|
Ampicillin | 54 | 81.5 (69.2-89.6) |
|
|
Cefoxitin | 35 | 42.9 (28-59.1) |
|
|
Ceftazidime | 57 | 12.3 (6.1-23.2) |
|
|
Ceftriaxone | 74 | 41.9 (31.3-53.3) |
|
|
Cefuroxime | 93 | 20.4 (13.5-29.7) |
|
|
Chloramphenicol | 176 | 56.2 (48.9-63.4) |
|
|
Ciprofloxacin | 161 | 41 (33.7-48.7) |
|
|
Clindamycin | 120 | 16.7 (11.1-24.3) |
|
|
Erythromycin | 181 | 68 (60.8-74.3) |
|
|
Gentamicin | 158 | 31 (24.3-38.6) |
|
|
Imipenem | 101 | 13.9 (8.4-21.9) |
|
|
Levofloxacin | 61 | 0 (0-5.9) |
|
|
Moxifloxacin | 43 | 0 (0-8.2) |
|
|
Ofloxacin | 61 | 0 (0-5.9) |
|
|
Oxacillin | 68 | 30.9 (21.2-42.6) |
|
|
Penicillin G | 106 | 86.8 (79-92) |
|
|
Tetracycline | 162 | 72.2 (64.9-78.5) |
|
|
Trimethoprim-sulfamethoxazole | 134 | 76.9 (69-83.2) |
|
|
Vancomycin | 97 | 15.5 (9.6-24) |
| Enterococcus sp | |||
|
|
Ampicillin | 32 | 87.5 (71.9-95) |
|
|
Chloramphenicol | 21 | 61.9 (40.9-79.2) |
|
|
Ciprofloxacin | 22 | 81.8 (61.5-92.7) |
|
|
Erythromycin | 35 | 91.4 (77.6-97) |
|
|
Gentamicin | 14 | 57.1 (32.6-78.6) |
|
|
Tetracycline | 15 | 73.3 (48-89.1) |
|
|
Vancomycin | 32 | 50 (33.6-66.4) |
|
|
Gentamicin-high | 14 | 64.3 (38.8-83.7) |
| Streptococcus sp | |||
|
|
Ceftriaxone | 14 | 64.3 (38.8-83.7) |
|
|
Chloramphenicol | 45 | 35.6 (23.2-50.2) |
|
|
Clindamycin | 49 | 32.7 (21.2-46.6) |
|
|
Erythromycin | 52 | 59.6 (46.1-71.8) |
|
|
Penicillin G | 15 | 66.6 (41.7-84.8) |
|
|
Tetracycline | 29 | 51.7 (34.4-68.6) |
|
|
Trimethoprim-sulfamethoxazole | 20 | 75 (53.1-88.8) |
|
|
Vancomycin | 24 | 25 (12-44.9) |
aR+I: Resistance + Intermediate.
Discussion
Principal Findings
The findings of our surveillance program show the feasibility of setting up a national AMR surveillance program based on the WHO GLASS manual recommendation while building systems for quality assurance, data sharing, linking results to patient care, and building partnerships. AMR is a global health threat, and the establishment of national surveillance systems is necessary to identify the emerging drug-resistant infections [2]. The African region still has suboptimal microbiology laboratory capacity and surveillance systems for AMR [3]. However, in Africa and Uganda in particular, resistance to recommended antibiotics has been reported for the WHO GLASS priority pathogens [12]. This paper presents the processes undertaken to set up a national AMR surveillance system according to WHO GLASS standards in Uganda, which could be benchmarked for other LMICs. The surveillance sites were RRHs and an academic institution. The RRHs represented the majority of the geographical distribution as they received referrals from district hospitals and health centers IVs and IIIs.
The success of establishing the national AMR surveillance program in Uganda highlights the feasibility of implementing the WHO GLASS program in LMICs. AMR surveillance programs are fundamental in Sub-Saharan African countries such as Uganda for generating antibiograms that can inform the development of treatment guidelines and antibiotic procurement plans and contribute toward standardized reporting [25,26]. Clinicians at surveillance sites can also access bacterial ID and AST results to inform patient care because of the availability of strengthened quality microbiology services.
In Uganda, the AMR surveillance system has been established using a systematic capacity-building pyramid model [17] and in alignment with the London School of Hygiene and Tropical Medicine stepwise road map for participating in GLASS [16]. The rolling out of the NAP for AMR [13], AMR national and subnational structures with Terms of Reference, and supporting surveillance plans and protocols has strengthened antimicrobial stewardship. In addition, the establishment of data-sharing platforms, including software programs such as WHONET [18], has supported data collation, analysis, reporting, and electronic archival, supplementing the existing paper-based methods.
The Uganda surveillance program identified selected resistant priority pathogens, including E coli, S aureus, K pneumoniae, and N gonorrhoeae, which present a high diversity of pathogens seen in Central Africa, Gabon [27]. E coli and S aureus isolates were the most prevalent WHO priority pathogens isolated in Uganda. This is fundamental baseline information that could be used for pretesting the novel WHO protocol [28] for estimating mortality attributable to AMR bloodstream infections. Monitoring priority pathogens and analyzing their antimicrobial susceptibilities together with epidemiological information on sex, age, and surveillance site can inform early hospital-level interventions [29].
There was a high rate of resistance of E coli to ampicillin and cotrimoxazole, as recently reported [3]. However, proportion of ceftriaxone- and ciprofloxacin-resistant E coli was slightly lower than that observed in Equatorial Guinea [30]. More worryingly, there was a significant proportion 18.8% (61/324) of E coli resistance to imipenem, which is considerably higher than 3%, recently reported in other parts of the African continent [3]. For K pneumoniae, there was notable resistance to ceftriaxone at 79% (63/79) and cotrimoxazole at 82% (55/67) in the surveillance program, similar to findings in Uganda’s neighboring country Kenya [31].
To our knowledge, this is the first documentation of the implementation of a national AMR surveillance program on the African continent using the WHO-recommended methodology. Although all components of the WHO GLASS manual are implemented, the approaches were not sequential and were contextualized to Uganda cognizance of existing national policies and programs.
The main limitations included suboptimal recovery of the AMR GLASS priority pathogens and inconsistent setting of the recommended antibiotics against the pathogen in the laboratory. This was attributed to the fact that staff members were still undergoing comprehensive training on microbiology skills, stock-outs, and acquiring extensive knowledge on AMR surveillance [32]. The number of samples sent to the microbiology laboratory was relatively low, coupled with low rates of completion of the microbiology laboratory request forms. This was partially attributed to the lack of a laboratory-clinician interface to bridge these anomalies.
Conclusions
Using the WHO guidance, Uganda has successfully completed key foundational building activities for the successful implementation of a national AMR surveillance program. The emerging antibiotic resistance data can be refined, appraised, and used for further improvement of the current methodological approaches being used to implement AMR programs in Uganda and other LMICs. Uganda successfully enrolled in the WHO GLASS system and has consistently reported annual program progress to the WHO since 2016. There is an extremely high prevalence of AMR to the most commonly used antibiotics, similar to what has been found in other studies conducted in the research context.
Recommendations
The current Uganda Clinical Guidelines need to be reviewed in response to the AMR burden in Uganda. In addition, strengthening the capacity of the microbiology laboratory is fundamental for the successful implementation of surveillance protocols in hospital wards to further profile the emerging global health threat of AMR.
Acknowledgments
The authors would like to thank the members of the National Antimicrobial Resistance subcommittee of One Health Platform, staff at the National Microbiology Reference Laboratory at the Central Public Health Laboratories, Regional Referral Hospitals, and the Ministry of Health Senior Management Leadership. Funding obtained from the Global Health Security Partner Engagement Project (1U2GGH001744) through the US Centers for Disease Control and Prevention and the Fleming Fund Country Grant (RFP/CG1/Uganda-FF5/53) UK Aid through Mott Mac Donald management agent.
Abbreviations
- AMR
antimicrobial resistance
- AST
antimicrobial susceptibility testing
- GLASS
Global Laboratory Antimicrobial Resistance Surveillance System
- HAI
hospital-acquired infection
- LMIC
low- and middle-income country
- MoH
Ministry of Health
- NAP
National Action Plan
- NCC
National Coordinating Center
- RRH
regional referral hospital
- UNAMRsC
Uganda National Antimicrobial Resistance Sub-Committee
- WHO
World Health Organization
Footnotes
Authors' Contributions: HM and SN provided strategic national leadership for the surveillance program. FK and RK wrote the first draft of the manuscript. AK, ML, RW, and HK provided technical oversights on the implementation of surveillance activities. RK, JM, and JB developed the surveillance documents. MS and IM coordinated the operational requirements for implementation. All authors reviewed the final manuscript.
Conflicts of Interest: None declared.
References
- 1.O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance. 2016. [2021-10-06]. https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf .
- 2.World Health Organization . Global Action Plan on Antimicrobial Resistance. Geneva: World Health Organization; 2016. pp. 1–45. [Google Scholar]
- 3.Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, González IJ, Dittrich S. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017 Sep 11;17(616):1–17. doi: 10.1186/s12879-017-2713-1. https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-017-2713-1 .10.1186/s12879-017-2713-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ampaire L, Muhindo A, Orikiriza P, Mwanga-Amumpaire J, Bebell L, Boum Y. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016 Feb 01;5(1):432. doi: 10.4102/ajlm.v5i1.432. http://europepmc.org/abstract/MED/28879114 .AJLM-5-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wangai FK, Masika MM, Lule GN, Karari EM, Maritim MC, Jaoko WG, Museve B, Kuria A. Bridging antimicrobial resistance knowledge gaps: the East African perspective on a global problem. PLoS One. 2019 Feb 11;14(2):e0212131. doi: 10.1371/journal.pone.0212131. https://dx.plos.org/10.1371/journal.pone.0212131 .PONE-D-18-28028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kajumbula H, Fujita AW, Mbabazi O, Najjuka C, Izale C, Akampurira A, Aisu S, Lamorde M, Walwema R, Bahr NC, Meya DB, Boulware DR, Manabe YC. Antimicrobial drug resistance in blood culture isolates at a tertiary hospital, Uganda. Emerg Infect Dis. 2018 Jan;24(1):174–5. doi: 10.3201/eid2401.171112. doi: 10.3201/eid2401.171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kateete DP, Namazzi S, Okee M, Okeng A, Baluku H, Musisi NL, Katabazi FA, Joloba ML, Ssentongo R, Najjuka FC. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res Notes. 2011 Sep 07;4(1):326. doi: 10.1186/1756-0500-4-326. https://bmcresnotes.biomedcentral.com/articles/10.1186/1756-0500-4-326 .1756-0500-4-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bebell LM, Ngonzi J, Bazira J, Fajardo Y, Boatin AA, Siedner MJ, Bassett IV, Nyehangane D, Nanjebe D, Jacquemyn Y, van Geertruyden J, Mwanga-Amumpaire J, Bangsberg DR, Riley LE, Boum Y. Antimicrobial-resistant infections among postpartum women at a Ugandan referral hospital. PLoS One. 2017 Apr 13;12(4):e0175456–69. doi: 10.1371/journal.pone.0175456. https://dx.plos.org/10.1371/journal.pone.0175456 .PONE-D-16-46066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamorde M, Mpimbaza A, Walwema R, Kamya M, Kapisi J, Kajumbula H, Sserwanga A, Namuganga JF, Kusemererwa A, Tasimwa H, Makumbi I, Kayiwa J, Lutwama J, Behumbiize P, Tagoola A, Nanteza JF, Aniku G, Workneh M, Manabe Y, Borchert JN, Brown V, Appiah GD, Mintz ED, Homsy J, Odongo GS, Ransom RL, Freeman MM, Stoddard RA, Galloway R, Mikoleit M, Kato C, Rosenberg R, Mossel EC, Mead PS, Kugeler KJ. A cross-cutting approach to surveillance and laboratory capacity as a platform to improve health security in Uganda. Health Secur. 2018 Dec 01;16(S1):76–86. doi: 10.1089/hs.2018.0051. [DOI] [PubMed] [Google Scholar]
- 10.Workneh M, Hamill MM, Kakooza F, Mande E, Wagner J, Mbabazi O, Mugasha R, Kajumbula H, Walwema R, Zenilman J, Musinguzi P, Kyambadde P, Lamorde M, Manabe YC. Antimicrobial resistance of neisseria gonorrhoeae in a newly implemented surveillance program in Uganda: surveillance report. JMIR Public Health Surveill. 2020 Jun 10;6(2):e17009. doi: 10.2196/17009. https://publichealth.jmir.org/2020/2/e17009/ v6i2e17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kakooza F, Musinguzi P, Workneh M, Walwema R, Kyambadde P, Mande E, Lubega C, Nakasi JM, Kiggundu R, Hamill MM, Bagaya BS, Lamorde M, Unemo M, Manabe YC. Implementation of a standardised and quality-assured enhanced gonococcal antimicrobial surveillance programme in accordance with WHO protocols in Kampala, Uganda. Sex Transm Infect. 2021 Jun 20;97(4):312–6. doi: 10.1136/sextrans-2020-054581.sextrans-2020-054581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization . Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation. Geneva: World Health Organization; 2015. pp. 1–36. [Google Scholar]
- 13.Antimicrobial resistance: national action plan. Government of Uganda. 2018. [2021-10-06]. http://cphl.go.ug/sites/default/files/2020-02/Uganda%20National%20Action%20Plan%20for%20Antimicrobial%20Resistance%202018-%202023-compressed_0.pdf .
- 14.Seale A, Hutchison C, Fernandes S, Stoesser N, Kelly H, Lowe B, Turner P, Hanson K, Chandler CI, Goodman C, Stabler RA, Scott JG. Supporting surveillance capacity for antimicrobial resistance: laboratory capacity strengthening for drug resistant infections in low and middle income countries. Wellcome Open Res. 2017;2:91–2. doi: 10.12688/wellcomeopenres.12523.1. http://europepmc.org/abstract/MED/29181453 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mpairwe Y, Wamala S, UNAS, CDDEP, and GARP-Uganda . Antibiotic Resistance in Uganda: Situation Analysis and Recommendations. Kampala, Uganda: Uganda National Academy of Sciences; Center for Disease Dynamics, Economics and Policy; 2015. p. 107. [Google Scholar]
- 16.Seale AC, Gordon NC, Islam J, Peacock SJ, Scott JA. AMR Surveillance in low and middle-income settings - A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS) Wellcome Open Res. 2017 Sep 26;2:92. doi: 10.12688/wellcomeopenres.12527.1. http://europepmc.org/abstract/MED/29062918 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potter C, Brough R. Systemic capacity building: a hierarchy of needs. Health Policy Plan. 2004 Sep 01;19(5):336–45. doi: 10.1093/heapol/czh038. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh A. Application of WHONET in the antimicrobial resistance surveillance of uropathogens: a first user experience from Nepal. J Clin Diagnostic Res. 2013 May 01;:3–6. doi: 10.7860/jcdr/2013/5193.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Global Antimicrobial Resistance Use Surveillance System (GLASS) World Health Organization. 2020. [2021-10-06]. https://www.who.int/initiatives/glass .
- 20.Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015 Aug 05;6:791–3. doi: 10.3389/fmicb.2015.00791. doi: 10.3389/fmicb.2015.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.BD Phoenix™ M50 System by BD Life Sciences: diagnostics. Selectscience. [2021-01-01]. https://www.selectscience.net/products/bd-phoenix-m50-system/?prodID=208749 .
- 22.BD BACTECTM FX blood culture system. BD. [2021-07-10]. https://www.bd.com/en-us/offerings/capabilities/microbiology-solutions/blood-culture/blood-culture-instrumentation/bd-bactec-fx-blood-culture-system .
- 23.Weston EJ, Wi T, Papp J. Strengthening global surveillance for antimicrobial drug-resistant neisseria gonorrhoeae through the enhanced gonococcal antimicrobial surveillance program. Emerg Infect Dis. 2017 Oct;23(13):4–5. doi: 10.3201/eid2313.170443. doi: 10.3201/eid2313.170443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolinsky AL. M100-S11, Performance standards for antimicrobial susceptibility testing. Clin Microbiol Newslett. 2001 Mar;23(6):49. doi: 10.1016/s0196-4399(01)88009-0. [DOI] [Google Scholar]
- 25.Leopold S, van Leth F, Tarekegn H, Schultsz C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: a systematic review. J Antimicrob Chemother. 2014 Sep 01;69(9):2337–53. doi: 10.1093/jac/dku176.dku176 [DOI] [PubMed] [Google Scholar]
- 26.Mboowa G, Aruhomukama D, Sserwadda I, Kitutu FE, Davtyan H, Owiti P, Kamau EM, Enbiale W, Reid A, Bulafu D, Kisukye J, Lubwama M, Kajumbula H. Increasing antimicrobial resistance in surgical wards at Mulago National Referral Hospital, Uganda, from 2014 to 2018-cause for concern? Trop Med Infect Dis. 2021 May 19;6(2):82–6. doi: 10.3390/tropicalmed6020082. https://www.mdpi.com/resolver?pii=tropicalmed6020082 .tropicalmed6020082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alabi AS, Frielinghaus L, Kaba H, Kösters K, Huson MA, Kahl BC, Peters G, Grobusch MP, Issifou S, Kremsner PG, Schaumburg F. Retrospective analysis of antimicrobial resistance and bacterial spectrum of infection in Gabon, Central Africa. BMC Infect Dis. 2013 Oct 02;13(1):455. doi: 10.1186/1471-2334-13-455. https://bmcinfectdis.biomedcentral.com/articles/10.1186/1471-2334-13-455 .1471-2334-13-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization . Glass Method for Estimating Attributable Mortality of Antimicrobial Resistant Bloodstream Infections. Geneva: World Health Organization; 2020. pp. 1–65. [Google Scholar]
- 29.Abera B, Kibret M, Mulu W. Knowledge and beliefs on antimicrobial resistance among physicians and nurses in hospitals in Amhara Region, Ethiopia. BMC Pharmacol Toxicol. 2014 May 19;15(1):26. doi: 10.1186/2050-6511-15-26. https://bmcpharmacoltoxicol.biomedcentral.com/articles/10.1186/2050-6511-15-26 .2050-6511-15-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shatalov A. Prevalence and antibiotic resistance pattern of Escherichia coli and Klebsiella pneumoniae in urine tract infections at the La Paz Medical Center, Malabo, Equatorial Guinea. Open J Med Microbiol. 2015;5(4):177–83. doi: 10.4236/ojmm.2015.54022. [DOI] [Google Scholar]
- 31.Maina D, Makau P, Nyerere A, Revathi G. Antimicrobial resistance patterns in extended-spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae isolates in a private tertiary hospital, Kenya. Microbiol Discov. 2013 Jan 01;1(1):5–7. doi: 10.7243/2052-6180-1-5. [DOI] [Google Scholar]
- 32.Holloway K, Mathai E, Gray A, Community-Based Surveillance of Antimicrobial Use and Resistance in Resource-Constrained Settings Project Group Surveillance of antimicrobial resistance in resource-constrained settings - experience from five pilot projects. Trop Med Int Health. 2011 Mar;16(3):368–74. doi: 10.1111/j.1365-3156.2010.02696.x. doi: 10.1111/j.1365-3156.2010.02696.x. [DOI] [PubMed] [Google Scholar]


