Highlights
-
•
New knowledge about the potential use of the Pseudomonas spp. genus to bioremediate affluent contaminated with heavy metals.
-
•
The isolates of Pseudomonas aeruginosa, Pseudomonas nitroreducens, and Pseudomonas alcaligenes had the capacity to tolerate concentrations higher than 50 mg/ml of lead.
-
•
Statistically significant differences were detected (p < 0.05) in the production of the exopolysaccharide (EPS) among the isolates.
-
•
Pseudomonas aeruginosa and Pseudomonas nitroreducens have 80% capacity to biosorber lead using live mass (minimum range from 80.9% to 87%).
Keywords: Bioprospecting, Heavy metals, Toxic, Bioaccumulation, Live biomass, Dead biomass
Abstract
The mechanisms of tolerance to heavy metals used by some microorganisms identified by bioprospection processes are useful for the development and implementation of bioremediation strategies for contaminated environments with high toxic load caused by heavy metals. A total of seven native microbial isolates were obtained from wastewater bodies from an industrial zone in the municipality of Girardota, Antioquia, Colombia. Subsequently, they were selected to evaluate their lead tolerance capacity at different concentrations. In addition, some parameters were determined, such as the capacity to produce exopolysaccharides and their biosorption to understand potential mechanisms associated to lead tolerance. According to the biocehemical test (Vitek) and the molecular analysis of sequences of 16S rDNA, bacterial were identified as Pseudomonas aeruginosa, Pseudomonas nitroreducens, and Pseudomonas alcaligenes. We determined that the seven isolates had the capacity to tolerate concentrations higher than 50 mg/ml of lead, and that the concentration and exposure time (40 h) to this metal significantly affect the Pseudomonas spp. isolates. Statistically significant differences were detected (p < 0.05) in the production of the exopolysaccharide (EPS) among the isolates. P. aeruginosa (P16) was the strain with the maximum absorbance exopolysaccharide measured. We evidenced that P. aeruginosa (P14) and P. nitroreducens (P20) have 80% capacity to biosorber lead using live mass (minimum range from 80.9% to 87%). It is suggested that the tolerance to lead exhibited by the environmental isolates of Pseudomonas spp. can be attributed to the production of exopolysaccharides and biosorption, which are protection factors for its survival in contaminated places. Finally, it was determined that the adsorption measured from dead biomass was significant (p < 0.05) from 40 h of exposure to metal (Average 182.2 ± 7). We generated new knowledge about the potential use of the Pseudomonas spp. genus to bioremediate affluent contaminated with heavy metals.
This work was supported by the Institución Universitaria Colegio Mayor de Antioquia (IUCMA), convocatoria 2016–02
1. Introduction
Currently, heavy metals are still considered a global contamination problem due to their persistence, accumulation, and the toxic effects they could cause in the environment and on the health of exposed human populations [42]. Lead (Pb) is part of these metals and has been classified by the World Health Organization (WHO) as one of the 10 chemical products that cause health conditions and severe complications (WHO, 2014). Anemia, reproductive alterations, kidney failure, cancer, and neurodegenerative damage are among these conditions [10, 18, 24] . At molecular level, Pb can cause oxidative damage to the DNA, proteins, and lipids, and substitute essential metal ions (Zn, Ca, and Fe) present in some enzymes [24, 34, 44].
There is a wide variety of processes and treatments to reduce or recover heavy metals from contaminated environments, including physicochemical treatment, use of live microorganisms, and potentiation of the interaction mechanisms with metals through various bioremediation strategies [10, 36, 40]. Thus, some studies show that strains from microorganisms isolated from places contaminated with heavy metals are potential candidates for metal bioremediation, since they possess mechanisms that allow them to adapt to adverse conditions and tolerate these toxic elements. . Therefore, it is important to study native isolates from this kind of areas [27]. These mechanisms allow the cell to interact with the metal and avoid its toxicity. Some of them include biosorption, bioaccumulation, and the production of exopolysaccharides (EPS).
Progress in studies on these mechanisms has made it possible to consider their implementation as bioremediation strategies in areas contaminated with metals. Some studies have suggested the use of biomass produced from a microorganism (live or dead) could be effective as metal bio-adsorbent (at laboratory scale) of water matrices ( [11]; [19]). Likewise, the effectiveness of some bacterial ESP as bio-adsorbents of heavy metals has been evidenced. Such is the case of a strain of Paenibacillus jamilae CECT 5266, whose EPS was characterized and evaluated against metals like Pb, Cd, Co, Ni, Zn and Cu, where they found it had higher affinity for and absorption of Pb compared with other metals [26]. In addition, the capacity of Pb absorption has also been reported in association with the capacity of Pseudomonas strains of producing EPS [17].
Biological treatments based on metal tolerance mechanisms show some advantages compared with conventional treatments because they can be applied to systems with low metal concentrations, which implies a decrease in costs. Additionally, they can be carried out at the contamination site causing minimal alteration of the treated area [20, 23, 36].
In Colombia, studies associated to the identification of microorganisms present in waters or soils that are able to interact with heavy metals have been reported, identifying bacteria such as Corynebacterium spp., Bacillus spp., Pseudomonas spp. [38], along with a strain of Lysinibacillus sphaericus capable of interacting with Pb [32]. On the other hand, in Antioquia, rice fields contaminated with cadmium have been researched to determine the presence of microorganisms like Burkholderia sp. and Pseudomonas sp. [2]. Even though there are not many reports related to searching and bioprospection of microorganisms that could help in bioremediation of heavy metals at local level, we are interested in adding to the knowledge about the existence of microorganisms and the mechanisms with which they interact with heavy metals.
A previous study made it possible to carry out a preliminary identification of native isolates of the Pseudomonas spp. genus from the zone studied in this research, which showed viability to grow in the presence of up to 2.5 mg/ml of Pb solution [4]. However, the possible tolerance mechanisms were not explored. As such, this study aims at evaluating tolerance mechanisms to Pb like the production of EPS and biosorption in native isolates of Pseudomonas spp. obtained from a locality adjoining an industrial zone with potential risk of contamination. The purpose is to generate information for the planning of alternative or complementary strategies for the treatment and removal of heavy metals in the environments, as affluents contaminated.
2. Materials and methods
2.1. Isolation of Pseudomonas spp. potentially tolerant to lead
The isolates were obtained from wastewater samples collected from the Medellin river, north zone (Girardota), in an area adjoining factory that emit waste with Pb, located at 6°23′02.9′’N 75°27′09.9′’W. A total of four samples of water were taken for this study. Two samples of water were taken from the river, and the two remaining were taken in a basin near the river, both under the same sterile conditions. Each one had a volume of 500 mL and were deposited in a sterile container. Samples were transported to the LACMA Laboratory at the Institución Universitaria Colegio Mayor de Antioquia, under refrigeration conditions to be preserved. Subsequently, the samples were grown on selective medium (Cetrimide Agar, Merck®, USA) to obtain isolate colonies that were compatible with Pseudomonas spp. by means of serial cultures. Samples were incubated at 35 °C for 24–48 h.
2.2. Identification of the isolates of Pseudomonas spp
We performed a differential study of the morphologies of the colonies observed from the growth obtained in culture media. We selected and isolated colonies in trypticase soy agar, TSA (Merck®, USA) for the subsequent microscopic characterization and biochemical analysis using the VITEK® 2 compact automatic identification system (bioMérieux, France). The isolates were preserved and stored in Brain Heart Infusion, BHI medium (Brain Heart Infusion medium, Merck®, USA) with glycerol (30%) at 4 °C until they were used.
Additionally, we extracted the total DNA of the selected isolates using the DNA extraction kit PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc., USA). The purity of the DNA was verified in 1% agarose gel. Gene RNAr 16S was amplified with primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′), and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [16], using the amplification conditions described by [16]. PCR products were analyzed, purified by means of Wizard ® PCR Preps (Promega, New England), and then their readings were obtained using an ABI-PRISM 3700 DNA automated sequencer (Applied Biosystems), through Macrogen Company Inc., in Korea.
To determine the identity of the sequences, they were initially edited with the MEGA 10 software (Kumar, 2004), and later submitted to BLASTN (National Center for Biotechnology Information; http://blast.ncbi.nlm.nih.gov/Blast.cgi) [1], comparing them with sequences published on GenBank, according to identity ranking (>97%) and E-values (0.0). In addition, Neighbor-Joining (NJ) clustering analysis was developed, based on the Kimura-2-parameters (K2P) as the molecular model of evolution for the data set (Saitou et al., 1987), using as ingroup several 16S rDNA sequences of nominal species matching with our isolates within a percent sequence similarity threshold of 97% [28]. Similarly, we evaluate the intra and interspecific p-distance threshold method (3%) to support the identification of isolates and to determine the most likely number of independents operational taxonomic unit – OTUs [28] among the previously NJ clusters. The nucleotide sequences generated during this study were deposited into GenBank (National Center for Biotechnology Information - NCBI) with the following access codes: MW661177 - MW661183.
2.3. Lead tolerance determination assay
2.3.1. Agar diffusion method
The assay was performed for all isolates identified as Pseudomonas spp., and the assay was done as previously described in the article by Bedoya et al. [4], with modifications. In brief, petri dishes with BHI agar (Merck®, USA) were inoculated with 100 µl suspension of each isolate in the exponential phase (1 × 106 cells/ml), and then filter paper disks was impregnated with an aliquot of a Pb(NO3)2 solution in increasing concentrations of 5, 10, 20, 50, 80, and 100 mg/ml concentrations. Each assay was performed in duplicate. They were incubated at 35 °C for 48 h. Tolerance was estimated by the growth capacity around the discs or the growth inhibition with halos greater than 2 mm [4]. As a negative control we used the medium supplemented with the solution without inoculation.
2.3.2. Lead minimum inhibitory concentration (MIC)
Considering the two previously identified biochemical groups (Vitek test) and genetic OTUs, we selected one isolate from each one, corresponding to P14 (P. aeruginosa), and P20 (P. nitroreducens). For the assay we prepared a series of tubes with 10 ml of BHI (Merck®, USA). The tests were done in duplicate and were supplemented with a Pb(NO3)2 solution in increasing concentrations of 0.1, 1, 10, and 100 mg/ml. Each tube was inoculated with 1 ml of each isolate in suspension that was previously prepared in the exponential phase (1 × 106 cells/ml). Samples were incubated at 35 °C, and after 0, 24, and 120 h, a sample was taken. The microbial growth were determined by turbidimetry measured by spectrophotometry (Mindray Microplate Reader® MR-96 A Spectrophotometer) at a wave length of 600 nm at the final time [4]. As negative control we used the Pseudomonas aeruginosa ATCC 27853 strain as reference.
2.4. Evaluation of exopolysaccharide production
Qualitaive EPS production was evaluated from the culture of each isolate using Congo red medium ( [8] with modifications), and the tube method [14]. For each of the tests, we used the Pseudomonas aeuriginosa ATCC 27853 strain as reference, and as negative control we used the supplemented medium without microorganism inoculation. For the Congo red test, black colonies with a dry and crystalline consistency would indicate a positive result for EPS production [8, 9, 14]. In regard to the tube test, EPS production was considered positive when observing violet halos o films adhered to the walls or bottom of the tube [14], when the test was reveled with cristal violet. Lastly, we carried out the assay by means of the microplate culture method, where we could quantify EPS production [7, 12, 14]. The results are interpreted based on the procedure described by Hassan et al. [14] in Table 1.
Table 1.
Interpretation of the microplate test for the production of biofilm in Pseudomonas spp. Isolates.
| Mean OD value | Biofilm production |
|---|---|
| ≤ ODc/DOc <∼≤ 2x ODc | Not produce / Weak produce |
| 2x ODc <∼≤ 4x ODc | Moderate |
| > 4x ODc | Strong |
ODc: Cut-off optical density value = Average value of the optical density of the negative control + 3x standard deviation (SD) of the negative control. Source: Hassan et al. [14].
2.5. Evaluation of lead biosorption capacity
Lead biosorption was evaluated using live and dead biomass. These assays were carried out on isolates P14 and P20, which were previously selected by the biochemical and genetic criteria explained above. For the biomass preparation, trypticase soy broth, TSB (TSB Merck®, USA) was used, starting with a suspension of each isolate previously prepared in the exponential phase (1 × 106 cells/ml). The media were supplemented with the Pb(NO3)2 solution at 0.5 mg/ml.
2.5.1. Live biomass lead biosorption determination
Under the previously described conditions, the preparation was incubated in agitation at 120 rpm, and samples were taken after 18 and 40 h. As negative control, samples with TSB medium (Merck®, USA) were used, and another one with lead solution. The samples were centrifugated at 5000 rpm during 10 min (Ballardo de la Cruz et al., 2015; with modifications). The supernatant were collected to determine the residual concentration of the metal in the samples using Atomic Absorption Spectroscopy (AAS) [11, 33].
2.5.2. Dead biomass lead biosorption determination
To increase performance when obtaining dead cells, this assay was carried out in a 5 liter reactor (BIOSTAT® A, Sartorius Stedim-Biotech) with 2.7 liters of TSB (Merck®, USA), which was inoculated with a fresh culture to an effective volume of 3 liters. Operation conditions were room temperature for 48 h, agitation at 120 rpm, and aeration of 2000 cc/m. The biomass produced by each isolate was subjected to 121 °C for 30 min (to obtain dead biomass) ( [19] with modifications). After the autoclave, the medium was centrifugated at 5000 rpm for 25 min. The sediment was washed twice with distilled water. The obtained precipitate was deposited in crucibles that were previously weighted and dried in the oven at 60 °C until we obtained a constant weigh.
Afterward, schott vials containing TSB (Merck®, USA), the lead solution and the dead biomass were prepared. We added 1 mg of dead biomass per each milliliter of metal solution [3]. The samples were incubated and subsequently treated under the same conditions as the live biomass samples. Likewise, the supernatant was collected to determine the residual concentration of the metal in the samples by means of AAS [11, 33].
2.5.3. Calculations to determine the percentage of removal and biosorption capacity
With the average results from the three assays, we calculated the percentage of metal removal with the following equation:
where R is the metal percentage bound to the biomass, and as such, removed from solution, C0 is the initial concentration of the metal in the solution (mg/l), and Cf is the final metal concentration in the solution (mg/l) [3, 31].
Additionally, we calculated the values of biosorption capacity of the dead biomass from each isolate with the following equation:
where q is the milligrams of adsorbed metal ion per biomass gram (mg/g), C0 is the initial concentration of the metal in the solution (mg/l), Cf is the final concentration of the metal in the solution after treatment with biomass (mg/l), V is the volume of the solution (l), and m is the dry weigh of biomass [33].
2.6. Statistical analysis
To relate the Pb tolerance level of the obtained isolates, the study was done in two phases: a first phase was descriptive based on the disk diffusion assay, and a second phase for the analysis in MIC, with different lead concentrations, at two exposure times to determine the capacity of surviving and tolerating the metal. The data were studied through parametric and non-parametric tests, depending on the case.
For the evaluation of EPS production among the seven isolates, the experiment following a completely randomized design, considering each isolate as an independent treatment (n = 7). To determine the effects of the treatments on the response variable, we used a one-way ANOVA followed by a Tukey multiple comparison test.
For the biosorption assay, we did two factorial analysis designs. The first consisted in a triple interaction of the strain (P14 and P20), type of biomass (live or dead) and time of exposure (18 and 40 h) factors. Each one with two levels. The objective was to identify the individual (main) or combined (secondary) effects of those factors on the removal of lead in the medium. The second factorial analysis consisted in a double interaction of the strain (P14 and P20) and the time of exposure (18 and 40 h) factors, to identify the individual or combined effects of those factors on the amount of metal adsorbed (mg/g), using the dead biomass.
For all purposes, the response variables were expressed as the mean ± standard deviation (SD), and they were considered significant differences when p < 0.05 with 95% confidence interval. It is important to note that prior to any statistical analysis, a Levene test was done to determine variance homogeneity, and a Shapiro-Wilk test for normality of the obtained data. When the data did not fulfilled any of the previous criteria, they were analyzed with a non-parametric Kruskal- Wallis test. For the data analysis, a programming routine was carried out using statistical software SPSS version 25 (IBM® SPSS® Statistics GradPack).
3. Results
3.1. Isolation and identification of Pseudomonas spp. tolerant to lead
3.1.1. Morphological and biochemical characterization
We obtained 25 isolates with different morphologies (P01 and P25) from the growth in cetrimide agar (Merck®, USA), which were purified. Afterward, we carried out a gram test, where we observed that all isolates were Gram negative. None of the isolates were catalase positive, and only seven were oxidase positive (P07, P10, P14, P16, P18, P20, and P25). Including other characteristics, we could observe that four of the isolates (P07, P14, P18, and P25) produced pigment and fluorescence when exposed to the UV light lamp (Table 2).
Table 2.
Description of the microcroscopic and macroscopic characteristics of the selected isolates, results of catalase and oxidase tests and biochemical identification using VITEK®2 compact.
| Isolate code | Microscopic characteristics | Macroscopic characteristics | Oxidase | Catalase | Biochemical identification (Vitek) | Probability (%) | ||
|---|---|---|---|---|---|---|---|---|
| Form | Gram | Pigment | Fluorescencia | |||||
| P02 | Bacilli | Negative | No | No | Negative | Negative | Escherichia coli | 98 |
| P03 | Bacilli | Negative | No | No | Negative | Negative | Escherichia coli | 98 |
| P04 | Bacilli | Negative | No | No | Negative | Negative | Shiguella spp | 98 |
| P07 | Bacilli | Negative | Yes | Yes | Positive | Negative | Pseudomonas aeruginosa | 98 |
| P10 | Bacilli | Negative | No | No | Positive | Negative | Pseudomonas aeruginosa | 98 |
| P12 | Bacilli | Negative | No | No | Negative | Negative | Aeromonas hydrophila | 98 |
| P13 | Bacilli | Negative | No | No | Negative | Negative | Klebsiella oxytoca | 98 |
| P14 | Bacilli | Negative | Yes | Yes | Positive | Negative | Pseudomonas aeruginosa | 98 |
| P15 | Bacilli | Negative | No | No | Negative | Negative | Enterobacter cloacae | 98 |
| P16 | Bacilli | Negative | No | No | Positive | Negative | Pseudomonas aeruginosa | 98 |
| P18 | Bacilli | Negative | Yes | Yes | Positive | Negative | Pseudomonas fluorescens | 50 |
| P20 | Bacilli | Negative | No | No | Positive | Negative | Pseudomonas fluorescens | 50 |
| P23 | Bacilli | Negative | No | No | Negative | Negative | Serratia plymuthica | 98 |
| P24 | Bacilli | Negative | No | No | Negative | Negative | Escherichia coli | 98 |
| P25 | Bacilli | Negative | Yes | Yes | Positive | Negative | Pseudomonas aeruginosa | 98 |
Biochemical identification using VITEK® (biomerieux, France) showed that five of the seven oxidase positive isolates were consistent with Pseudomonas aeruginosa (P07, P10, P14, P16, and P25) with more than 95% probability. However, P18 and P20 classified as Pseudomonas, showed a probability of 50% without defining their species (Table 2).
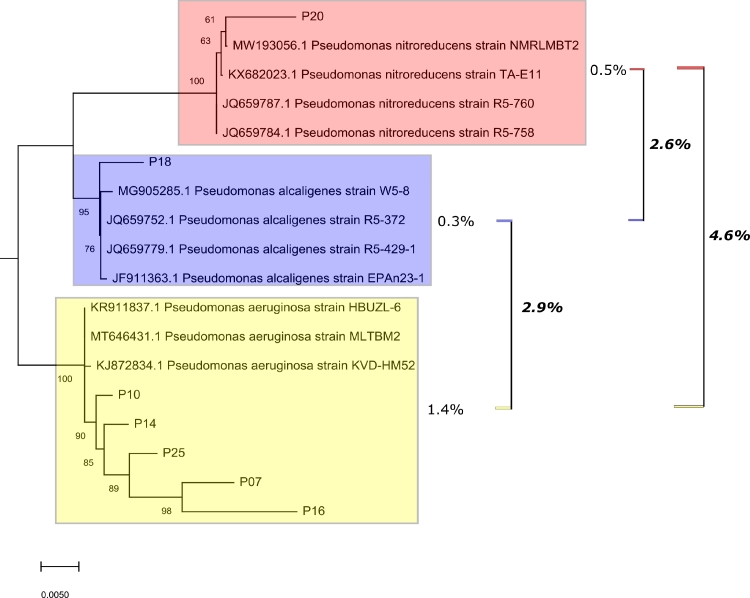
As for the molecular analyses of 16S, they attributed the seven isolates to the Pseudomonas genus, with five of them corroborated as Pseudomonas aeruginosa (P07, P10, P14, P16, P25) (% identity= average value from 96,49 to 99.57; E-value = 0,0), while the others were identified as Pseudomonas alcaligenes (P18) (98,85% identity; E-value = 0,0), and Pseudomonas nitroreducens (P20) (98,27% identity; E-value = 0,0). Regarding the NJ analysis, even though it shows a grouping pattern suggesting the same identification as BLASTN, some bootstraps show clusters with distant and varied ends, which could mean the existence of genetically different isolates to the strains used as reference or positive control (Fig. 1).
Fig. 1.
Neighbor joining (NJ) analysis of the 16S rRNA gene partial sequences using 1000 bootstraps, for identification of seven Pseudomonas isolates (P) obtained from wastewater from the Aburrá Valley, Colombia. The figure is showing three NJ clusters/species obtained: red (Pseudomonas nitroreducens), violet (Pseudomonas alcaligenes) and yellow (Pseudomonas aeruginosa). Percentages of intraspecific and interespecific genetic distance (based on p-distance method) is showed in unbold and bold-italic, respectively. All nominal species used as ingroup of the analyses, were obtained from GenBank by BLAST (E-value = 0 and identity > 97%).
3.2. Lead tolerance determination
The microorganisms that were previously exposed to the metal allowed the identification of concentrations higher than 2.5 mg/ml (Table 3), for which they were tolerant. Consequently, exposure to higher concentrations in this study, evidenced that the most of the isolates have the capacity of grow up to 50 mg/ml in the disk diffusion test without showing inhibition halos. Additionally, in the MIC test, where four lead concentrations were evaluated for two of the identified biochemical variants, corresponding to P. aeruginosa (P14) and P. nitroreducens (P20), the non-parametric analysis identified a significant difference among the data analyzed, recognizing that from different concentrations of Pb exposure (>1 mg/ml) and longer exposure time, the biomass concentration is reduced, regardless of the strain used. This is confirmed by physical observation, where a precipitate of solid material is identified form the second concentrations of Pb supplementation. Nevertheless, cell viability occurs when the sample is grown in culture medium.
Table 3.
Disk diffusion test corresponding to the Pseudomonas isolates ability to tolerate lead.
| Pseudomonas spp. Isolate | Lead concentration | |||||
|---|---|---|---|---|---|---|
| 5 (mg/ml) | 10 (mg/ml) | 20 (mg/ml) | 50 (mg/ml) | 80 (mg/ml) | 100 (mg/ml) | |
| P07 | 0 | 0 | 0 | 0 | 14 | 19 |
| P10 | 0 | 0 | 0 | 0 | 14 | 17 |
| P14 | 0 | 0 | 0 | 0 | 9 | 10,5 |
| P16 | 0 | 0 | 0 | 0 | 16 | 12 |
| P18 | 0 | 0 | 0 | 0 | 11 | 14 |
| P20 | 0 | 0 | 0 | 0 | 9 | 14,5 |
| P25 | 0 | 0 | 0 | 0 | 14 | 21 |
The value of each cell represents the inhibition halo in mm.
3.3. EPS production evaluation
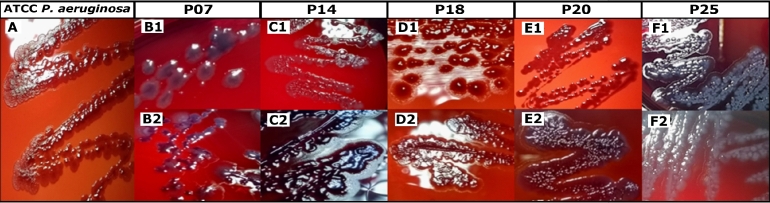
The results obtained through the Congo red agar method were not conclusive since we did not observe isolates that presented black colonies as described by some authors for the data interpretation. However, the identification of other characteristics as their coarse and dry aspect made it possible to correlate a positive result for EPS production, while the observation of smooth colonies was consistent with non-productive strains. Keeping this in mind, native isolates (without Pb exposure) presented two different colony morphologies.
On the one hand, six of the seven isolates (P07, P10, P16, P18, P20, and P25) showed humid, smooth aspect colonies; some of these strains of P. aeruginosa (P07, P10, and P25) presented a coloration that turned darker toward the center and lighter borders. On the other hand, only the P. aeruginosa (P14) strain showed a dry and coarse aspect. After the exposure to Pb, two of the seven isolates (P. alcanigenes y P. nitroreducens (P18, P20, respectively), consistent with the second biochemical group, changed their morphology, from humid and smooth to coarser and drier, while the other strains kept the initial colony characteristics (Fig. 2). Despite the fact that the isolate colonies did not present a black coloration per se, we could correlate the coarse and dry morphology of the colonies with possible EPS production, considering that the morphological characteristics presented by the colonies of the reference strain Pseudomonas aeruginosa ATCC 27853 has a dry and coarse aspect.
Fig. 2.
Morphology of the Pseudomonas spp. isolates colonies on Congo red agar. A: positive control strain ATCC 27853 Pseudomonas aeruginosa, showing dry and rough-looking colonies. B1-F1: morphology of isolates without exposure to lead. B2-F2: morphology of isolates exposed to 0.5 mg / ml of lead.
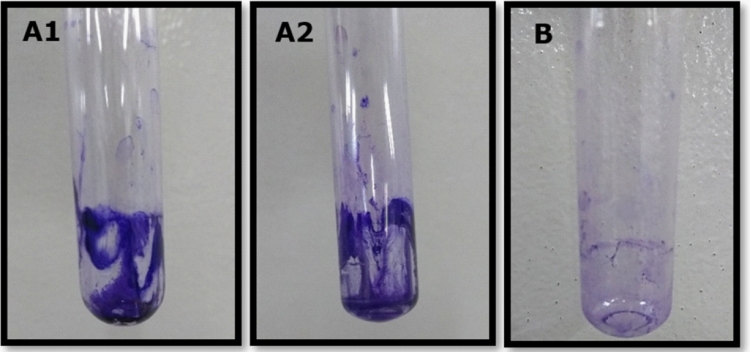
On the contrary, by means of the tube method, we could establish that six of the seven isolates were EPS producers, since we could observe a colored film that covered the walls and the bottom of the tube, as seen in Fig. 3.
Fig. 3.
Tube method to evaluate biofilm production, developing with crystal violet. A: positive biofilm production, with biofilm attached to the tubefor Pseudomonas aeruginosa (P16) (A1) and Pseudomonas alcaligenes (P18) (A2). B: negative production of biofilm (no presence of biofilm attached to the tube) for Pseudomonas aeruginosa (P25) .
From the results of the microplate method, we determined that from the seven evaluated strains, only P. aeruginosa (P25) was classified as a non-producer of EPS. The rest of the P. aeruginosa, P. alcalinegenes and P. nitroreducens strains were strong EPS producers (Table 4). This correlates to the results of the tube test, in which the same isolate had a negative result, while the other six had positive results.
Table 4.
Optical density metrics of the exopolysaccharide production test by the tube and microplate methods in Pseudomonas isolates.
| Methods | Pseudomonas spp. isolates | ||||||
|---|---|---|---|---|---|---|---|
| P07 | P10 | P14 | P16 | P18 | P20 | P25 | |
| Tube | 2,20 +/- 0,26 | 2,26 +/- 0,33 | 1,9 +/- 0,002 | 2,54 +/- 0,008 | 2,52 +/- 0,87 | 2,01 + /- 0,24 | 0,50 +/- 0,23 |
| Microplate | 0,304 +/- 0,35 | 0,05 +/- 0,39 | 0,281 +/- 0,28 | 1152 +/- 0,804 | 0,291 +/- 0,48 | 0,66 +/- 0,57 | 0,19 +/- 0,36 |
± mean standar desviation. Values in bold are values of absorbance high to production of exopolysaccharide.
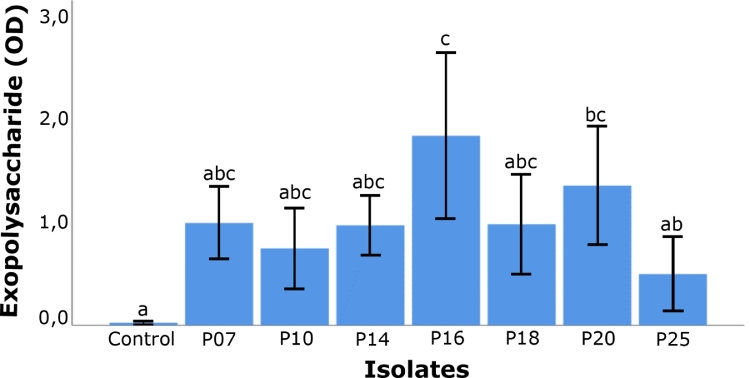
From the statistical analysis, we identified a significant difference (p < 0.05) in EPS production among the isolates by the microplate method. In addition, a Tukey test clearly identified a significant difference grouping the strains in two, where P16 (P. aeruginosa) was the strain that showed the greatest difference in the production of exopolisacaride, with reference to control (without production of exopolisarides), followed by strain P20 (P. nitroreducens), while the rest behaved very similar to control (Fig. 4).
Fig. 4.
Tukey's multiple comparison test for the average exopolysaccharide (EPS) production from different isolates of Pseudomonas spp. estimated by the crystal violet method. In the Y-axis the EPS production is expressed in arbitrary units of optical density. Same letters indicate non-significant statistical differences among treatments (P > 0.05). Values are represented as mean ± SD.
3.4. Lead biosorption capacity evaluation
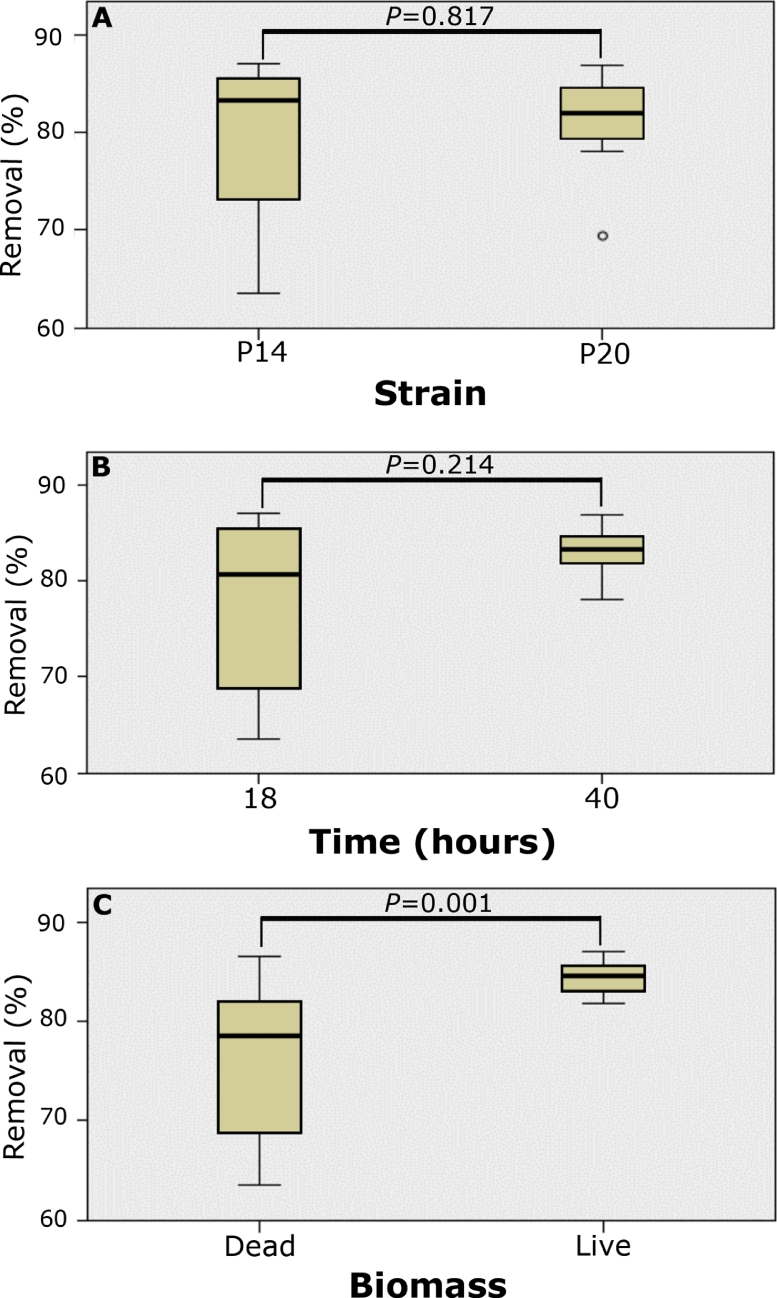
The results obtained from both the live and dead biomass of the studied isolates evidenced metal removal percentages above 80% in the maximum contact time of exposure (40 h). As seen in Fig. 5, the removal percentage of the P. aeruginosa (P14) strain at 40 h, was very similar with the live and dead biomass (83.43% and 82.62%, respectively); while the live biomass of P. nitroreducens (P20), showed a higher removal percentage (85.34%) than the dead biomass (81.04%) after 40 h of contact. Nevertheless, from the factorial analysis, we could identify there is a significant difference (p < 0.05) mediated by the type of biomass, concluding that the live biomass removes more lead in the medium independent of the time and strain (Fig. 5).
Fig. 5.
Kruskal-Wallis statistical analysis to test the effect of different factors on the percentage of removal of lead in the broth. A: effect of the strain, B: effect of the time of exposure of microorganisms to the metal, C: effect of the live or dead biomass exposure. Values are represented as mean ± SD.
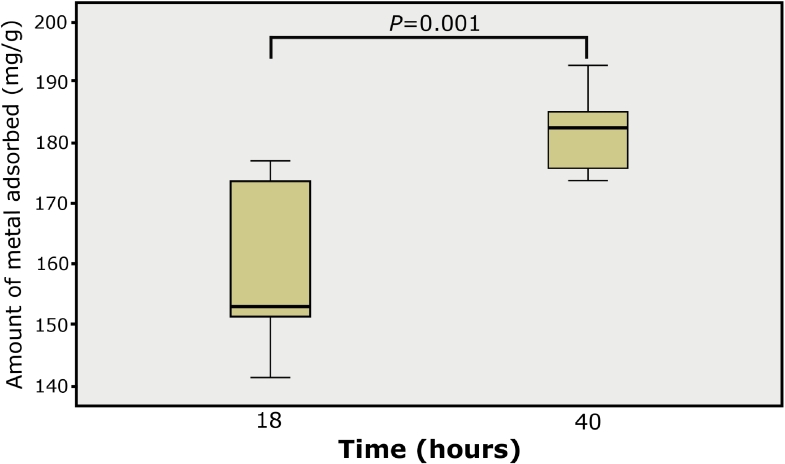
Concerning the metal adsorption factorial analysis mediated by the dead biomass of the organism, we demonstrated that there is a significant difference within the evaluated factors (p < 0.05), where we define that independently from the strain, the higher the exposure time, the higher the metal removal in the culture medium (Fig. 6). In addition, it was found that the interaction between strain and time did not have a significant effect on the adsorption of the metal.
Fig. 6.
Lead biosorption capacity. Figure is showing the significant individual main effect of the time factor (18 and 40 h) on the amount of lead adsorbed from dead biomass of P14 and P20 isolates of Pseudomonas spp. The analysis is derived from a previously 2 × 2 factorial design [time (18/40 h) vs. Isolate dead biomass (P14/P20)] with non-significants effect for the interaction (P > 0.05) nor for isolate dead biomass main effect (P > 0.05). Values are represented as mean ± SD.
4. Discussion
The microbiological analysis of wastewater exposed to heavy metal potential contamination in a sector of the Aburrá Valley, indicates the presence of several isolates of the Pseudomonas genus, with high potential to bioremediate Pb, without having been previously reported for the area. By means of biochemical tests, we demonstrated some potential mechanisms of bioactivity. One of the identified isolates in these water sources corresponds to Pseudomonas aeruginosa, a species that has been widely associated with natural intrahospital environment, which indicates it is also considered an opportunistic pathogen [30], with easy adaptation to complex environments [6].
On the other hand, Pseudomonas nitroreducens has been reported for its presence in contaminated sites, and its capacity of degrading complex molecules, mainly xenobiotic aromatic substances [15]. However, this species does not have records where its capacity to interact with heavy metals has been described. Regarding Pseudomonas alcaligenes, it has been reported as a good biodegrader for toxic polycyclic aromatic hydrocarbons [39].
This study identified three species of Pseudomonas spp. isolated from water that receives industrial waste where the presence of lead had been previously reported in a concentration of 0.5 mg/l [4]. Published studies on residual water contaminated with heavy metals report the presence of a wide variety of microorganisms, and even studies like Wang et al. [41] report four types of microorganisms from their metagenomic research (Betaproteobacteria, Gammaproeobacteria, Alphaproteobacteria, and Actibacteria). The Pseudomonas genus is the most dominant in the Gammaproteobacteria class, both in residual and sediment waters [41]. It is important to highlight that according to Wang et al. [41], the variety of microorganisms is strictly related to redox activity and the presence of heavy metals, with respect to elements like phosphorus or nitrogen in the environment.
In our research, we evidenced that in concentrations above 50 mg/ml, Pseudomonas isolates remain active. There are other reports that describe the capacity of Pseudomonas spp. to tolerate Pb up to 0.05 mg/ml [21]. Gabr et al. [11] determined that the P. aeruginosa ASU 6a strain, was able to tolerate concentrations of 0.5 mg/ml of this metal [11]. Likewise, Li et al. [19] evidenced that the Pseudomonas sp. I3 strain could tolerate Pb concentrations of up to hasta 7.5 mM, and the Pseudomonas sp. 375 strain, studied by Xu et al. [43] showed a tolerance of up to 6 mM of Pb [43]. The tolerance of these bacterial species to other metals has also been reported: mercury (0.2 mg/ml), zinc (3.2 mg/ml) or copper 3 mg/ml [21, 22, 37].
It is necessary to precise that the species Pseudomonas aeruginosa and Pseudomonas nitroreducens did not present significant differences in the MIC study, and that in concentrations above 1 mg/ml after a 120-hour exposure, there was no evidence of cellular density, which suggests high toxicity of the compound for the cells. This could be explained as stated by Chien et al. [6], who say microorganisms in extreme conditions have the capacity of producing polysaccharides to protect their cell membrane, and they also allow for cell aggregation to search for an isolation mechanism, where the interaction with said toxic agent occurs with this extrenal component but not with the cell.
Thus, this could suggest the inability to register cellular density when the microorganism is exposed to concentrations equal or higher than 10 mg/ml. These data are supported by the experimental conditions observed in this study, since the culture medium where the cells were suspended turned thicker and had the tendency to precipitate or separate the solids in the experimental units. Additionally, when correlating the results with the disk diffusion test, they showed tolerance results in higher concentrations of metal exposure because their inhibition was recorded above 50 mg/ml in this last assay.
The composition of EPS produced by microorganisms like Pseudomonas are based on the presence of heteropolysaccharides that include compounds such as alginates, biosurfactants, and rhamnolipids, mainly reported in P. aeruginosa [25]. On the other hand, some studies suggest that the production of this type of compounds is promising for the removal of heavy metals in contaminated water from the interaction with this matrix of exopolysaccharides with aerobic mud and granular anaerobics [29].
Finally, regarding the biosorption assay where we calculated the percentage of Pb removal, we could not identify a marked difference among Pseudomonas species even with different exposure times. The difference was a more effective tendency with live biomass during the process. On the contrary, some authors have evidenced the effectiveness of using dead bacterial biomass for lead removal. [13]) achieved a maximum removal of 85.38% of Pb+2 from an aqueous solution and a biosorption capacity of 123 mg/g of dead cells, using Aeromonas hydrophila MTCC 646 biomass [13]. Removal percentages between 90 and 95% have even been obtained with other metals like Al+3 using dead Pseudomonas putida A (ATCC 12633) biomass, determining a biosorption capacity of up to 0.55 mg/g of biomass [5], which shows more efficiency. Some studies have also suggested that live cells are metabolically active and can produce H+ ions, which could generate competence with metal ions, thus reducing the biosorption capacity of the toxic metal [5, 11].
Other reports agree with the findings in this study. Li et al. [19] identified a Pseudomonas sp. strain (I3) that showed better results in Pb capture when they used live cells (49.8 mg/g of biomass), instead of dead biomass (42.37 mg/g of biomass). Furthermore, using Pseudomonas sp. strain 375 biomass it has been proven for other metals like Cd, for instance, Xu et al. [43] determined a maximum biosoprtion capacity of 92.59 mg/g of live biomass compared to the results with dead biomass (63.29 mg/g of biomass). It is worthy of note, that the main difference between both mechanims is the dependence on cell metabolism, i.e., the use of live biomass implies the cell is metabolically active and requires the transference of the metal through the cell wall to the cytoplasm [11].
It is possible to state that live cells carry out two processes that allow them to mediate the toxicity generated by toxic metals: they initially perform a biosorption process that, as explained above, is a passive process that does not require metabolic activity, so the bacterial cell wall acts as a first barrier that prevents the toxicity the metals present in the medium could cause. Subsequently, the metal ions could be transported to the cell interior through complex permeation, endocytosis, and/or an ion bomb mediated by carriers to be bioaccumulated [33, 35].
In addition, the calculated data to determine the absorption capacity of dead biomass indicated a significant difference mediated by the exposure time to the metal. This could be related to the available places where the metal joins and the interactions with the dead material [5], which could even be variable, but independent of the strain used, as evidenced in this study.
It is important to highlight that these studies have shown that biosorption processes can be affected by factors such as pH, temperature, the initial metal or biomass concentration, and even the presence of other metal ions. Therefore, they are factors that should be evaluated and kept in mind when using microbial biomass as a heavy metal decontamination strategy.
5. Conclusions
This study evidenced the presence of several native isolates of the Pseudomonas genus with high biotechnological potential for lead removal and adsorption, from water sources that receive industrial waste and potentially contaminated with heavy metals, in regions in Aburrá Valley (Antioquia, Colombia). It was proven that there is a potential of the Pseudomonas genus, be it for EPS production or because the cell remains viable in high metal concentrations.
6. Funding information
This research was funded by the Research Direction of Institución Universitaria Colegio Mayor de Antioquia (internal call for project bank 2016–02), Colombia.
7. Author`s contributions
S.O.A. conceived and designed the study, performed field and laboratory work, analyzed and interpreted the data, prepared figures and/or tables, edited and wrote the manuscript. J.G.M. analyzed and interpreted the data, prepared figures and/or tables, edited and wrote the manuscript. J.B.V. performed field and laboratory work, analyzed and interpreted the data, edited and wrote the manuscript. J.T.O edited and wrote the manuscript. All authors approved the final manuscript.
Declaration of Competing Interest
Authors have no conflict of interest.
Acknowledgements
We would like to acknowledge the BIOCIENCIAS group from the School of Health Sciences, and the Research Direction in the internal call for project bank 2016–02, for financing the project: “Lead tolerance mechanisms of Pseudomonas strains, isolated from wastewater from the Aburrá Valley”. We would like to acknowledge Rafael Vivero Gomez and Manuela Tejada, for their valuable contributions in the development of this article.
References
- 1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. 10.1016/S0022-2836 (05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ayubb T.N., Cerra G.A., Chamorro A.L., Pérez C.A. Resistencia a cadmio (Cd) de bacterias endófitas y bacterias rizosféricas aisladas a partir de Oriza sativa en Colombia. Rev. Colomb. Cienc. Anim. - RECIA. 2017;9:281. doi: 10.24188/recia.v9.n2.2017.610. [DOI] [Google Scholar]
- 3.Ballardo de la Cruz C.E., Merino Rafael F.A., Gutiérrez Moreno S.M. Evaluación de la capacidad de bioadsorción de Cadmio (II) y Plomo (II) mediante el uso de biomasa bacteriana muerta en soluciones acuosas. Theorema, segunda época 2. 2016:95–106. https://revistasinvestigacion.unmsm.edu.pe/index.php/Theo/article/view/11967 Available in: [Google Scholar]
- 4.Bedoya Vélez J.M., Castaño G., Ochoa Agudelo S. Tolerancia al plomo de aislamientos nativos de Pseudomonas spp. de aguas residuales del Valle de Aburrá. Rev. Colomb. Biotecnol. 2019;21:135–143. 10.15446/rev.colomb.biote.v21n1.65146 [Google Scholar]
- 5.Boeris P.S., Agustín M.del R., Acevedo D.F., Lucchesi G.I. Biosorption of aluminum through the use of non-viable biomass of Pseudomonas putida. J. Biotechnol. 2016;236:57–63. doi: 10.1016/j.jbiotec.2016.07.026. [DOI] [PubMed] [Google Scholar]
- 6.Chien C.C., Lin B.C., Wu C.H. Biofilm formation and heavy metal resistance by an environmental Pseudomonas sp. Biochem. Eng. J. 2013;78:132–137. doi: 10.1016/j.bej.2013.01.014. [DOI] [Google Scholar]
- 7.Christensen G.D., Simpson W.A., Younger J.J., Baddour L.M., Barrett F.F., Melton D.M., Beachey E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fariñas Bravo L., Salazar Noblet D., Arce A.M., García H., Ramírez M., Cabrera L.E., Fernández A., Castañeda N. Estudio de factores de virulencia en cepas de Plesiomonas shigelloides aisladas de animales domésticos y afectivos (Study of virulence factors in Plesiomonas shigelloides isolated from domestic and affective animals) Rev. Electrónica Vet. REDVET. 2005;5:1–12. https://scielo.conicyt.cl/pdf/rci/v26n3/art05.pdf Available in: [Google Scholar]
- 9.Freeman D.J., Falkiner F.R., Keane C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu F., Wang Q. Removal of heavy metal ions from wastewaters: a review. J. Environ. Manage. 2011;92:407–418. doi: 10.1016/j.jenvman.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Gabr R.M., Hassan S.H.A., Shoreit A.A.M. Biosorption of lead and nickel by living and non-living cells of Pseudomonas aeruginosa ASU 6a. Int. Biodeterior. Biodegradation. 2008;62:195–203. doi: 10.1016/j.ibiod.2008.01.008. [DOI] [Google Scholar]
- 12.Gómez J., Gómez-Lus M.L., Bas P., Ramos C., Cafini F., Maestre J.R., Prieto J. ¿Es la cuantificación del biofilm un elemento diferenciador en la patogenia de bacilos gramnegativos? Rev. Esp. Quimioter. 2013;26:97–102. [PubMed] [Google Scholar]
- 13.Hasan S.H., Srivastava P., Talat M. Biosorption of lead using immobilized Aeromonas hydrophila biomass in up flow column system: factorial design for process optimization. J. Hazard. Mater. 2010;177:312–322. doi: 10.1016/j.jhazmat.2009.12.034. [DOI] [PubMed] [Google Scholar]
- 14.Hassan A., Usman J., Kaleem F., Omair M., Khalid A., Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Brazilian J. Infect. Dis. 2011;15:305–311. doi: 10.1016/s1413-8670(11)70197-0. [DOI] [PubMed] [Google Scholar]
- 15.Iyer R., Iken B., Damania A. Genome of Pseudomonas nitroreducens DF05 from dioxin contaminated sediment downstream of the San Jacinto River waste pits reveals a broad array of aromatic degradation gene determinants. Genom. Data. 2017;14:40–43. doi: 10.1016/j.gdata.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalita D., Joshi S.R. Study on bioremediation of Lead by exopolysaccharide producing metallophilic bacterium isolated from extreme habitat. Biotechnol. Reports. 2017;16:48–57. doi: 10.1016/j.btre.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang C.H., Oh S.J., Shin Y., Han S.H., Nam I.H., So J.S. Bioremediation of lead by ureolytic bacteria isolated from soil at abandoned metal mines in South Korea. Ecol. Eng. 2015;74:402–407. doi: 10.1016/j.ecoleng.2014.10.009. [DOI] [Google Scholar]
- 19.Li D., Xu X., Yu H., Han X. Characterization of Pb2+ biosorption by psychrotrophic strain Pseudomonas sp. I3 isolated from permafrost soil of Mohe wetland in Northeast China. J. Environ. Manage. 2017;196:8–15. doi: 10.1016/j.jenvman.2017.02.076. [DOI] [PubMed] [Google Scholar]
- 20.Majumder S., Gupta S., Raghuvanshi S., Gupta S., Majumder S. In: Heavy Metals in Water: Presence, Removal and Safety. Sharma S.K., editor. The Royal Society of Chemistry; Cambridge: 2014. Removal of dissolved metals by bioremediation; pp. 44–56. in: (Ed.) [Google Scholar]
- 21.Malik A., Aleem A. Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environ. Monit. Assess. 2011;178:293–308. doi: 10.1007/s10661-010-1690-2. [DOI] [PubMed] [Google Scholar]
- 22.Malik A., Jaiswal R. Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J. Microbiol. Biotechnol. 2000;16:177–182. doi: 10.1023/A:1008905902282. [DOI] [Google Scholar]
- 23.Mani D., Kumar C. Biotechnological advances in bioremediation of heavy metals contaminated ecosystems: an overview with special reference to phytoremediation. Int. J. Environ. Sci. Technol. 2014;11:843–872. doi: 10.1007/s13762-013-0299-8. [DOI] [Google Scholar]
- 24.Mohan M., Dubey S.K. Ecotoxicology and Environmental Safety Lead resistant bacteria : lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol. Environ. Saf. 2013;98:1–7. doi: 10.1016/j.ecoenv.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 25.More T.T., Yadav J.S.S., Yan S., Tyagi R.D., Surampalli R.Y. Extracellular polymeric substances of bacteria and their potential environmental applications. J. Environ. Manage. 2014;144:1–25. doi: 10.1016/j.jenvman.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 26.Morillo Pérez J.A., García-Ribera R., Quesada T., Aguilera M., Ramos-Cormenzana A., Monteoliva-Sánchez M. Biosorption of heavy metals by the exopolysaccharide produced by Paenibacillus jamilae. World J. Microbiol. Biotechnol. 2008;24:2699–2704. doi: 10.1007/s11274-008-9800-9. [DOI] [Google Scholar]
- 27.Muñoz A.J., Ruiz E., Abriouel H., Gálvez A., Ezzouhri L., Lairini K., Espínola F. Heavy metal tolerance of microorganisms isolated from wastewaters: identification and evaluation of its potential for biosorption. Chem. Eng. J. 2012;210:325–332. doi: 10.1016/j.cej.2012.09.007. [DOI] [Google Scholar]
- 28.Nguyen N.P., Warnow T., Pop M., White B. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. npj Biofilms Microbiomes. 2016 doi: 10.1038/npjbiofilms.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nouha K., Kumar R.S., Tyagi R.D. Heavy metals removal from wastewater using extracellular polymeric substances produced by Cloacibacterium normanense in wastewater sludge supplemented with crude glycerol and study of extracellular polymeric substances extraction by different methods. Bioresour. Technol. 2016;212:120–129. doi: 10.1016/j.biortech.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Ochoa S.A., López-montiel F., Escalona G., Cruz-córdova A., Dávila L.B., López-martínez B., Jiménez-tapia Y., Giono S., Eslava C., Hernández-castro R., Xicohtencatl-cortes J. Características patogénicas de cepas de Pseudomonas aeruginosa resistentes a carbapenémicos. asociadas con la formación de biopelículas. 2013;70:138–150. [Google Scholar]
- 31.Olukanni D.O., Agunwamba J.C., Ugwu E.I. Biosorption of heavy metals in industrial wastewater using micro- organisms (Pseudomonas aeruginosa) Am. J. Sci. Ind. Res. 2014;5:81–87. Available in: http://eprints.covenantuniversity.edu.ng/8944/1/AJSIR-5-2-81-87.pdf. [Google Scholar]
- 32.Osorio Herrera, L.N., Andes, T. (Ingeniero Q.–U. de los, 2015. Evaluación de las capacidades de absorción y desorción de plomo de Lysinibacillus sphaericus. instnameUniversidad los Andes 1–20. Available in: https://repositorio.uniandes.edu.co/bitstream/handle/1992/17783/u714283.pdf?sequence=1.
- 33.Oves, M., Saghir, M., Qari, H.A., 2017. Journal of the taiwan institute of chemical engineers ensifer adhaerens for heavy metal bioaccumulation, biosorption, and phosphate solubilization under metal stress condition 80, 540–552. 10.1016/j.jtice.2017.08.026.
- 34.Patrick L. Lead toxicity Part II: the role of free radical damage and the use of antioxidants in the pathology and treatment of lead toxicity. Altern. Med. Rev. 2006;11:114–123. [PubMed] [Google Scholar]
- 35.Priyadarshanee M., Das S. Biosorption and removal of toxic heavy metals by metal tolerating bacteria for bioremediation of metal contamination: a comprehensive review. J. Environ. Chem. Eng. 2020 doi: 10.1016/j.jece.2020.104686. [DOI] [Google Scholar]
- 36.Rajendran P., Muthukrishnan J., Gunasekaran P. Microbes in heavy metal remediation. Indian J. Exp. Biol. 2003;41:935–944. [PubMed] [Google Scholar]
- 37.Singh V., Chauhan P.K., Kanta R., Dhewa T., Kumar V. Isolation and characterization of Pseudomonas resistant to heavy metals contaminants. Int. J. Pharm. Sci. Rev. Res. 2010;3:164–167. [Google Scholar]
- 38.Soto, C., González, E., Gutiérrez, S., Leon, A., 2010. Biotransformation of heavy metals present in riverside sludge from the Bogotá and Tunjuelo rivers. Nova 8, 195. Available in: https://repository.unad.edu.co/handle/10596/30075.
- 39.Suzuki, M., Suzuki, S., Matsui, M., Hiraki, Y., Kawano, F., 2013. Genome sequence of a strain of the human pathogenic bacterium pseudomonas alcaligenes that caused bloodstream infection 1, 1–2. 10.1186/1471-2164-9-75.5. [DOI] [PMC free article] [PubMed]
- 40.Vidali M. Bioremediation: an overview. Pure Appl. Chem. 2001;73:1163–1172. doi: 10.1351/pac200173071163. [DOI] [Google Scholar]
- 41.Wang J., Yuan S., Tang L., Pan X., Pu X., Li R., Shen C. Science of the Total Environment Contribution of heavy metal in driving microbial distribution in a eutrophic river. Sci. Total Environ. 2020;712 doi: 10.1016/j.scitotenv.2019.136295. [DOI] [PubMed] [Google Scholar]
- 42.Wasi S., Tabrez S., Ahmad M. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: a review. Environ. Monit. Assess. 2013;185:8147–8155. doi: 10.1007/s10661-013-3163-x. [DOI] [PubMed] [Google Scholar]
- 43.Xu S., Xing Y., Liu S., Hao X., Chen W., Huang Q. Characterization of Cd2+ biosorption by Pseudomonas sp. strain 375, a novel biosorbent isolated from soil polluted with heavy metals in Southern China. Chemosphere. 2020;240 doi: 10.1016/j.chemosphere.2019.124893. [DOI] [PubMed] [Google Scholar]
- 44.Xu X., Liao W., Lin Y., Dai Y., Shi Z., Huo X. Blood concentrations of lead, cadmium, mercury and their association with biomarkers of DNA oxidative damage in preschool children living in an e-waste recycling area. Environ. Geochem. Health. 2018;40:1481–1494. doi: 10.1007/s10653-017-9997-3. [DOI] [PubMed] [Google Scholar]