Abstract
The capacity of human induced pluripotent stem cells (hiPSCs) for indefinite self-renewal warrants their application in disease modeling, drug discovery, toxicity assays and efficacy screening. However, their poor proliferation ability, inability to adhere to surfaces without Matrigel coating and tendency to spontaneously differentiate in vitro hinder the application of hiPSCs in these fields. Here we study the ability to culture hiPSCs inside 200 nL droplets on the droplet microarray (DMA) platform. We demonstrate that (1) hiPSCs can attach to the Matrigel (MG)-free surface of DMA and show good viability after 24 h culture; (2) hiPSC do not spontaneously differentiate when cultured on the MG-free surface of DMAs; (3) culturing of hiPSCs in 200 nL as compared to 2 mL culture leads to higher expression of the Nanog pluripotency marker. Overall, the results demonstrate the possibility to culture undifferentiated hiPSCs in 200 nL droplets on DMA, thereby opening the possibility for high-throughput screenings of hiPSCs with various factors without compromising the results through the involvement of animal-derived materials, such as Matrigel.
Keywords: Human induced pluripotent stem cells, Droplet microarray, Xeno-free culture, Cell culture substrates, Stem cell pluripotency, High-throughput screening
Graphical abstract

1. Introduction
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) derived from blastocysts [1] and human induced pluripotent stem cells (hiPSCs) reprogrammed from somatic cells [2,3], hold tremendous promise for regenerative medicine due to their infinite capacity for self-renewal and potential to differentiate into all cell types in the human body. Organoids derived from hiPSC are attracting increasing interest due to their remarkable cell type complexity, architecture and functions that are similar to their in vivo analogs. These factors make hiPSCs attractive in the fields of tissue engineering [4], disease modeling [5], drug discovery, toxicity assays [6,7], and human developmental biology [8]. Since hiPSCs are reprogrammed from adult somatic cells, they overcome the ethical concerns and immunological rejection by the host, which have hindered the application of hESCs [9]. Therefore, hiPSCs have great potential for use in a wide range of clinical applications in various therapeutic approaches and personalized medicine.
In vitro culture and expansion of hiPSC is considered technically difficult since the self-renewal and differentiation of these cells is extremely sensitive and responsive to cell culture substrates [10]. Typically, hiPSCs are cultured on a layer of inactivated feeder cells such as irradiated or mitomycin-C-treated mouse embryonic fibroblasts, or an immortalized embryonic fibroblast line [11]. Alternatively, a layer of Matrigel (MG) is used to coat tissue culture-treated plastic labware [12]. MG is a mixture of proteins secreted by Engelbreth–Holm–Swarm mouse sarcoma cells and may contain xenogeneic contaminants, which restricts its use for humans [13]. In addition, the application of MG is limited due to its complexity as well as its ill-defined and variable composition, which results in batch-to-batch variations [14]. Several research groups have reported well-defined cell culture substrates that exhibit comparable functionality to MG in terms of the gene expression pattern and level of pluripotency of the cultured hiPSCs [[15], [16], [17], [18]]. These exciting findings have promoted great interest in the development of more advanced and well-defined feeder- and xeno-free substrates for hiPSC culture. Extracellular matrix proteins, such as laminin [19], fibronectin [20], and vitronectin [21], are recognized as well-defined, xeno-free cell culture substrates for hiPSCs. However, the application of these proteins is laborious and time-consuming, as well as very expensive due to the exorbitant costs of production of large amounts of high-purity functional proteins under conditions of good manufacturing practice (GMP). For example, human laminin and vitronectin are approximately 20-times and 80-times more expensive compared to MG, respectively [22]. Replacing feeder cells, MG or recombinant proteins with a synthetic substrate, which is well-defined in terms of chemical composition and surface cues, will avoid the exposure of cells to animal-derived products, increase the reproducibility of experiments and reduce the cost of cell culture.
Due to the unique ability of hiPSCs to differentiate into all cell types and their potential to be reprogrammed from any human individual, these cells represent an indispensable and physiologically relevant model of various diseases that can be used for high-throughput screening (HTS) in drug discovery. HTS applications usually require very large quantities of cells. For instance, approximately 1010 cells may be needed to screen a million-compound library [6]. However, it is difficult to obtain hiPSCs in sufficient quantities due to their low proliferation rate and tendency to differentiate spontaneously in vitro [23]. Thus, there is a need to devise efficient and cost-effective methods that can be used to obtain large quantities of pure pluripotent hiPSCs. Highly miniaturized and high-throughput platforms suitable for the maintenance of hiPSCs that can be used in HTS strategies are also required.
The droplet microarray (DMA) platform consists of hydrophilic areas on a superhydrophobic background. Due to the extreme difference in wettability of hydrophilic and superhydrophobic areas, arrays of stable immobilized and separated nanoliter droplets can be formed either by discontinuous dewetting or using a liquid dispenser. Culturing and screening of cells on DMAs reduces the consumption of compounds, reagents and valuable cells up to 10,000 times compared with the use of six-well plates. DMAs can be prepared on different surfaces with controllable well-defined chemical composition and surface cues [24,25]. It has been shown that surface properties, such as topography, have an impact on hiPSC pluripotency, self-renewal and differentiation [26,27]. Micro-nano scale roughness of the surface was shown to be the main factor that controls the retention of mouse embryonic stem cell (mESC) stemness [28]. Furthermore, nanoporous polymer-based DMAs inhibit the spontaneous differentiation of mESC [29], making HTS of embryonic bodies possible [30]. Thus, we hypothesized that both surface properties and small volume may have an influence on hiPSCs cultured on DMAs. However, DMAs have not yet been used to culture hiPSCs in vitro. Therefore, to determine the feasibility of using the miniaturized and high-throughput DMA technology to culture hiPSCs and assess its potential for their cultivation without MG coating (MG−), we investigated and compared two distinct types of commercially available droplet microarrays: type A (TA) and type B (TB). Both DMA types are used for miniaturized screening applications, such as screening of various adherent and suspension cells, as well as in three-dimensional (3D) cell culture models by means of spheroids and 3D scaffolds, such as hydrogels [[31], [32], [33], [34], [35], [36]]. Since the manufacturing of these substrates and patterns is not known, we performed a thorough physicochemical characterization of the surfaces in order to correlate their physical properties with biological performance. We compared the surface characteristics of TA and TB surfaces and investigated the topography, printing parameters, viability, and pluripotency of hiPSCs cultured on TA and TB DMAs. We demonstrated that hiPSCs cultured in 200 nL volumes on DMAs do not require MG for adhesion and do not differentiate for 24 h culture, which indicates the feasibility of HTS using hiPSCs without compromising the results through the involvement of animal-derived materials.
2. Materials and methods
2.1. Materials
Formaldehyde solution, propidium iodide (PI, 1.0 mg/mL in water), calcein AM (1 mg/mL in DMSO), 4′, 6-diamidino-2-phenylindole (DAPI) from Thermo Fisher Scientific Inc. (MA, USA) were used. Fetal bovine serum (FBS) and phosphate-buffered saline (PBS−/−) were purchased from Gibco, Life Technologies GmbH (Darmstadt, Germany). Serum-free hiPSCs culture medium mTeSR™ plus, Y-27632 and hiPSCs detaching reagent ReLeSR™ were purchased from STEMCELL technologies (Vancouver, Canada). Dulbecco's Modified Eagle Medium/F12 (DMEM/F12) was purchased from PAN Biotech (Aidenbach, Germany). Bovine serum albumin (BSA) was purchased from VWR international (Radnor, USA). Rabbit anti-Nanog and mouse anti-TRA-1-81 antibodies was purchased from Cell signaling Technology (CST, MA, USA). Alexa Fluor® 488 goat anti rabbit IgG H&L and Alexa Fluor® 594 goat anti mouse IgG H&L antibodies were bought from Abcam (San Francisco, USA). LDEV-free Corning®Matrigel® hESC qualified Matrix was purchased from Corning (MA, USA). Doxycycline hyclate and Triton™ X-100 were purchased from Sigma-Aldrich (Steinheim, Germany). Normocin was purchased from InvivoGen (CA, USA). The RNeasy Mini kit (QIAGEN GmbH, Germany), Superscript IV kit for reverse transcription (Life Technologies GmbH, Germany), Gotaq qPCR master mix (Promega GmbH, Germany) were used. TA surface (catalogue number: G-np-Custom-0001), TA DMA (catalogue number: G-np-Custom-0002), TB surface (catalogue number: G-np-602), and TB DMA (catalogue number: G-np-102) were purchased from Aquarray GmbH (Eggenstein-Leopoldshafen, Germany). Each DMA slide contains 672 individual hydrophilic spots with side length of 1 mm and 500 μm superhydrophobic distance between the side of square spot. The primers for qPCR were purchased from Integrated DNA Technologies (Heidelberg, Germany).
2.2. Surface water contact angle measurement
The water contact angle on TA surface and TB surface were characterized using Drop Shape Analyzer DSA 25 goniometer (Krüss) under ambient conditions (25 °C). A water droplet of 8 μL was deposited on the substrate and the water contact angle was measured within 5 s. The measurements were repeated for three times and the standard derivation is less than 2°.
2.3. Scanning electron microscope (SEM) and energy-dispersive X-ray spectroscopy (EDX) characterization of the surfaces
The surface topography and elemental analysis of TA surface and TB surface were determined by SEM and EDX, respectively. Specimen were analyzed with a LEO 1530 scanning electron microscope from Leica (Hillsboro, USA) with an accelerating voltage of 5–10 kV. For SEM analysis the specimen were sputtered with a thin layer of gold. For EDX a NORAN System SIX from Thermo Scientific (Waltham, USA) was used.
2.4. Atomic force microscope (AFM) characterization of the surfaces
The roughness of TA surface and TB surface were investigated by atomic force microscope (AFM) using a Dimension Icon with ScanAsyst from Bruker (Billerica, USA). Cantilevers with a resonance frequency of 325 kHz from Olympus (Shinjuku, Japan) were used. The amplitude setpoint, the proportional gain and the integral gain were adjusted for an optimal overlap of the trace and retrace profile. The scan rate was held constant at 1 Hz. Data analysis was performed with the software Gwyddion V. 2.56 (GPL). The scanned surface dimensions were 10 × 10 μm and three different spots were examined for each surface. The Ra values were calculated over the entire surface areas. Values are given as an average with standard deviation (n=3).
2.5. X-ray photoelectron spectroscopy (XPS) characterization of the surfaces
The chemical composition (C1s, Si2p, O1s, F1s, and S2p) of two surfaces was measured by XPS. XPS spectra were recorded on an Axis Ultra DLD from Shimadzu (Kyoto, Japan) utilizing monochromatized Al Kα radiation. The survey scan and the high-resolution scans were operated at an analyzer pass energy of 160.0 eV and 40.0 eV, respectively. The binding energy (BE) scale was referenced by setting the peak maximum in the C1s spectrum to 284.6 eV.
2.6. Cell culture
The human induced pluripotent stem cells (hiPSCs, D1 cell line, kindly provide by Prof. Dr. Martin Bastmeyer) were maintained on Matrigel in mTeSR™ plus serum-free medium in a humidified air with 5% CO2 at 37 °C. The cell culture medium was changed every day and the cells were manually cleaned daily with a sterilized 20 μL pipette tip. Cells were then passaged every 5–7 days.
The glass slides were sterilized by immersing into 70% ethanol for 60 min and air-dried under the clean bench before using. Cells were detached by cell detaching reagent ReLeSR™ according to the manufacturer's instructions. Then cells were seeded on the surfaces or printed onto each spot on DMAs with I-DOT One dispenser (Dispendix GmbH) in the presence of Rock inhibitor (Y-27632). The experiments were repeated three times to decrease experimental error. The cells were grown for 24 h before live/dead staining, staining for Nanog and TRA-1-81 expression and pluripotency gene expression.
2.7. Live/dead staining of hiPSCs
The cell viability of hiPSCs were assessed by a live/dead staining. calcein AM is a fluorogenic esterase substrate that is hydrolyzed to a green-fluorescent product (calcein). Thus, green fluorescence is an indicator of cells with active metabolism to retain esterase products, which visualizes live cells in green fluorescence [37]. Propidium iodide (PI) cannot pass through intact cell membranes but readily passes through damaged membranes and binds to DNA, which indicates dead cells in red fluorescence [38]. The final concentration of 0.5 μg/mL was used for both calcein AM and PI. Fluorescent images were taken by Keyence BZ-9000 (KEYENCE, Osaka, Japan) and Olympus IX81 microscope (Olympus, Tokyo, Japan). The calcein AM- and PI-positive areas were analyzed by Image J (https://imagej.nih.gov/ij/). The cell viability was calculated as calcein AM-positive area divided by the sum of calcein AM- and PI-positive area.
2.8. Immunofluorescence (IF) staining of hiPSCs
The hiPSCs cultured on different surfaces were fixed for 15 min at RT with 3.7% formaldehyde in PBS (−/−) and permeabilized for another 15 min with 0.1% Trinton X-100 in PBS (−/−). Afterwards, the cells were incubated with 1% BSA at 37 °C for 1 h to block non-specific binding. Then, the rabbit anti-Nanog and mouse anti-TRA-1-81 antibodies (diluted in PBS −/− as 1:200) were introduced and incubated with cells overnight at 4 °C. On the second day, the cells were washed and incubated with Alexa Fluor® 488 goat anti rabbit IgG H&L and Alexa Fluor® 594 goat anti mouse IgG H&L antibodies (diluted in PBS −/− as 1:400) for 1 h at 37 °C. Finally, the cells were stained with DAPI (1.43 μM) to counterstain the cell nuclear. The expression of biomarkers was then visualized by a confocal laser scanning microscope (Leica TCS SPE, Mannheim, Germany).
2.9. Real-time PCR (qPCR)
Pluripotency gene expression was quantified using qPCR analysis. Total cellular RNA was isolated by RNeasy Mini kit, according to manufacturer's protocol. cDNA was synthesized according to the usual protocol by Superscript IV kit for reverse transcription. Real-time PCR was performed on a StepOne Real-time PCR system (Thermo Fischer Scientific GmbH, Germany), after processing the cDNA samples with Gotaq qPCR master mix. Real-time data was analyzed as described elsewhere [39]. Primer sequences have been provided in Table S1 (Supporting Information). Data was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression and statistical differences between groups were analyzed using unpaired t-test.
2.10. Statistical analysis
All experiments were conducted at least in three biological repeats. The data were presented as mean ± standard deviation (SD) or mean ± standard error of the mean (SEM). Comparisons were conducted via unpaired student's t-test, unless otherwise specified. A significant difference was defined as ∗p < 0.05.
3. Results and discussion
Droplet Microarray (DMA) platform has dimensions of standard microscope glass slide and consists of an array of hydrophilic spots (1 mm side length) on a superhydrophobic background (Fig. 1). In this study, we optimized and adapted the DMA platform for culturing and screening of hiPSC in 200-nL droplets. To fully characterize the phenotype and behavior of hiPSC cultured on the DMA platform, we investigated the morphology, viability and pluripotency of cells cultured on commercially available DMAs fabricated on two different types of surfaces, TA and TB (Fig. 1). To distinguish the influence of surface properties and small nanoliter volumes on these characteristics, we investigated and compared the phenotypes of hiPSCs cultured on a large area (2.5 cm × 7.5 cm) of hydrophilic surface covered with 2 mL of culture media (TA and TB surfaces) and in confined 200-nL droplets formed on these two surfaces (TA and TB DMAs). Since hiPSCs are known to be sensitive to surface cues, such as hydrophilicity, topography, and roughness [40,41], we first characterized the two types of surfaces.
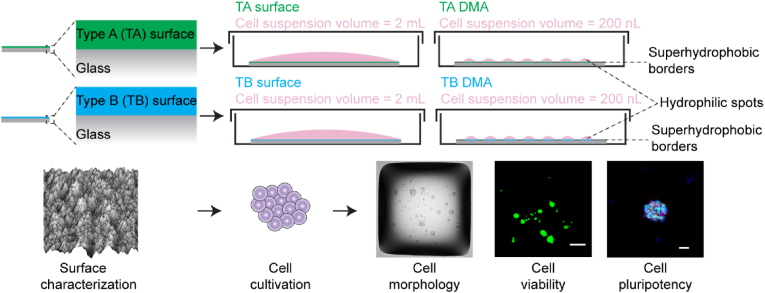
Fig. 1.
Overview of investigated surfaces and workflow of the study. (A) Two types of commercially available surfaces, type A (TA) and type B (TB), were used for the study. “TA and TB surfaces” refer to a large area (2.5 cm × 7.5 cm) of hydrophilic surfaces used for culturing hiPSC in 2 mL volumes. “TA and TB DMAs” refers to the DMA containing an array of hydrophilic spots with TA and TB coatings, respectively. DMA platform has dimensions of standard microscope glass slide and consists of an array of square hydrophilic spots with 1 mm side length on a superhydrophobic background. Cells were cultured in 200 nL volumes on the TA and TB DMAs. (B) The workflow of the conducted study. As a first step, the TA and TB surfaces were characterized using water contact angle (WCA) goniometry, energy-dispersive X-ray spectroscopy (EDX), scanning electron microscopy (SEM), atomic force microscopy (AFM), and X-ray photoelectron spectroscopy (XPS). As a second step, the hiPSCs were cultured in 2 mL media on TA and TB surfaces and in 200 nL media on TA and TB DMAs. As a third step, morphology, viability, and pluripotency of hiPSCs cultured on different surfaces were investigated.
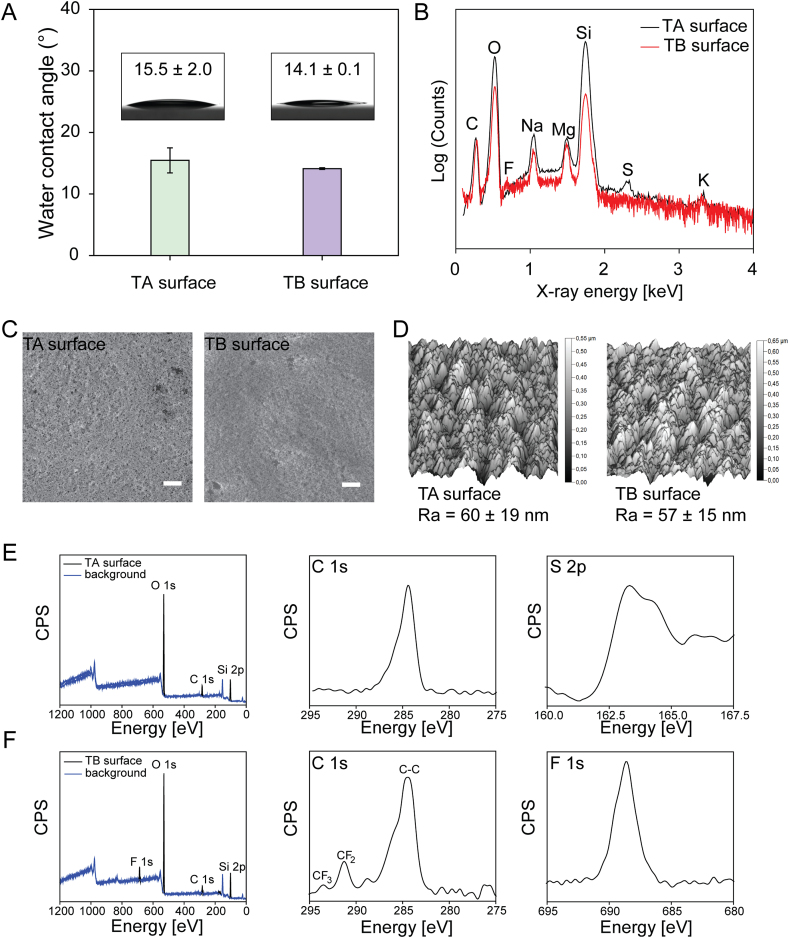
Two types of surfaces were characterized by water contact angle (WCA) goniometry, energy-dispersive X-ray spectroscopy (EDX), scanning electron microscopy (SEM), atomic force microscopy (AFM), and X-ray photoelectron spectroscopy (XPS). The hydrophilic areas of the TA and TB surfaces exhibited similarly low water contact angles of 15.5° ± 2.0° and 14.1° ± 0.1°, respectively, thus confirming the hydrophilic properties of the surfaces (Fig. 2A). EDX spectra showed the following elements present on the hydrophilic surfaces (Fig. 2B): a sulfur (S) peak was uniquely detected on the TA surface and a fluorine (F) peak was uniquely detected on the TB surface, whereas neither of the surfaces showed any additional characteristic elements apart from those of glass (e.g. Si, O, Na, Mg, and K). The surface topography and roughness were characterized by SEM and AFM (Fig. 2C and D). Both the TA and TB surfaces exhibited a homogenously rough morphology at the nanoscale. Surface roughness (Ra) determined from the AFM height profiles further confirmed the topological similarity of the TA and TB surfaces (60 nm ± 19 nm and 57 nm ± 15 nm, respectively: n = 3). XPS was employed to investigate the surface chemistry in more detail. The survey scan XPS spectra displayed the differences in the chemical elements of the TA and TB surfaces (Fig. 2E and F). For the survey scan of the TA surface, only C, Si, and O were detected, although the presence of sulfur was confirmed by the occurrence of a characteristic S 2p doublet at 163.3 eV in the narrow scan. A shoulder leaning to higher energies (approximately 286 eV) in the C 1s narrow scan indicated the presence of C–O species on the TA surface, as expected for adventitious carbon (Fig. 2E). A fluorine peak was observed in survey scan XPS spectra of the TB surface (Fig. 2F). A C 1s scan of the TB surface then revealed binding energies at 293.6 and 291.3 eV, which are indicative of –CF3 and –CH2 bonds, respectively. A shoulder toward higher energies (approximately 286 eV) might stem from oxidized carbon species or adventitious carbon. In the narrow scan of F 1s, a peak at 688.7 eV was detected, which is characteristic of organic fluoro-compounds. The Si 2p and O 1s narrow scans of both the TA and TB surfaces were practically identical and indicative of the silicon dioxide present in the coating (Fig. S1A, C). To ensure reproducibility and homogeneity of the surface functionalization, three high-resolution scans were conducted on different spots of the surfaces (Fig. S1B, D). To trace sulfur, 10 scans were conducted without scanning other energies (except for a C 1s scan as a reference) to avoid photochemical destruction of sulfur-carbon bonds (Fig. S1B). In conclusion, both the TA and TB surfaces possessed almost identical characteristics in terms of hydrophilicity and topography, but differed in their chemical environment.
Fig. 2.
Characterization of TA and TB surfaces. (A) Water contact angles (WCA) of the hydrophilic TA and TB surfaces measured with 8 μL water droplets under ambient conditions (25 °C). Data represent the mean ± SD (n = 3). (B) Energy-dispersive X-ray spectra (EDX) of the TA and TB surfaces. The surfaces were coated with carbon to ensure conductivity. A sulfur peak was detected on the TA surface and a fluorine peak was detected on the TB surface. (C) The surface morphology of TA and TB surfaces was characterized by scanning electron microscopy (SEM). Scale bar: 2 μm. (D) The surface topography was characterized by atomic force microscopy (AFM). Surface roughness (Ra) was determined from the AFM height profiles (n = 3). (E) Survey scan X-ray photoelectron spectroscopy (XPS) spectra of the TA surface, and XPS spectra of C 1s and S 2p on the TA surface. (F) Survey scan XPS spectra of the TB surface, and XPS spectra of C 1s and F 1s on the TB surface.
HiPSCs are commonly maintained in different culturing vessels, including flasks, Petri dishes and multi-well plates, in volumes ranging from one to dozens of mL on a MG layer. MG promotes the attachment and proliferation of hiPSCs in vitro. In this study, we investigated the feasibility of culturing hiPSC in 200-nL droplets on DMAs coated with (MG+) and without (MG−) Matrigel (1% v/v) while preserving all important characteristics of these cells such as morphology, viability and the most important factor – pluripotency.
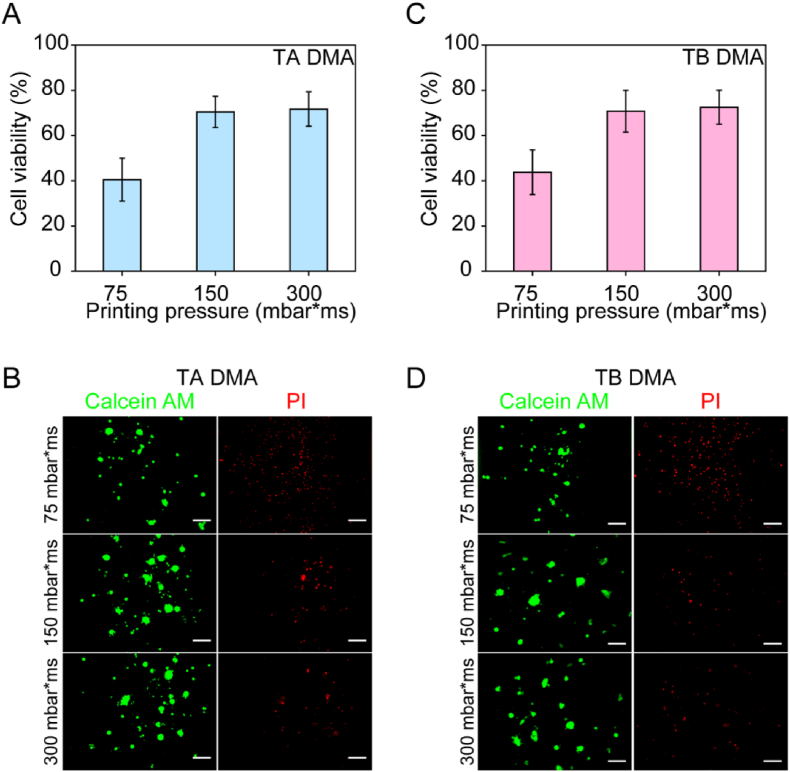
HiPSCs are very sensitive to environmental stresses, such as compression and shear, which can occur during dispensing of cells and cause dissociation-induced cell death [42,43]. The Rock inhibitor was used to block the dissociation associated apoptosis of hiPSCs. It could increase the cell survival and cloning efficiency of hiPSCs without influencing the pluripotency of hiPSCs. We firstly investigated the viability of cells with and without Rock inhibitor. As presented in Figure S2, the viability of hiPSCs cultured for 24 h without Rock inhibitor was 15.60 ± 1.42%, while cells cultured with Rock inhibitor for 24 h was 72.80 ± 6.87%. Thus, we selected culturing hiPSCs with Rock inhibitor on DMA slides. Then we compared the viability of hiPSCs dispensed onto DMAs using different printing settings, such as the pressure applied during dispensing of cells and reagents with the non-contact low volume dispenser used in this study (Fig. 3). A live/dead staining method was used to assess the viability of hiPSCs on MG− TA DMA and MG− TB DMA after dispensing with distinct printing pressures of 75, 150, and 300 mbar∙ms after 24 h of culture. For this, a solution containing calcein AM (0.5 μg/mL) and propidium iodide (PI, 0.5 μg/mL) was dispensed directly onto the droplets containing cells to stain live and dead cells, respectively. Cell viability was calculated as the ratio of the calcein AM-positive area to the sum of the calcein AM- and PI-positive areas. The viability of cells cultured for 24 h and dispensed under pressures of 150 and 300 mbar∙ms was comparable, while the viability of cells dispensed under 75 mbar∙ms was approximately 28% lower. This might be because single cells are more commonly dispensed than aggregates of cells under lower printing pressure, and single hiPSCs are more prone to cell death. Furthermore, we stochastically selected images from each printing pressure group and used ImageJ to calculate the mean area of clusters on the images. The results showed that the cluster area of cells printed by 75 mbar∙ms was smaller than that of 150 and 300 mbar∙ms (Figure S3). The viabilities of hiPSCs cultured on TA DMA for 24 h were 43.74% ± 9.90%, 70.73% ± 6.25% and 72.52% ± 7.51% for printing pressure of 75, 150 and 300 mbar∙ms, respectively (Fig. 3A and B). The viabilities of cells cultured on TB DMA under these three conditions were 40.50% ± 9.47%, 70.43% ± 6.91%, and 71.70% ± 7.63%, respectively (Fig. 3C and D). There were no significant differences in the viability of hiPSCs cultivated on MG− TA and MG− TB DMAs under the same conditions. These observations suggest that the DMA printing pressure has an impact on viability of hiPSCs. Therefore, we used a printing pressure of 150 mbar∙ms and cultivation time of 24 h for all further experiments.
Fig. 3.
The influence of different printing pressure on cell viability. HiPSCs were cultured in mTeSR plus medium in 6-well plates prior to being detached by ReLeSR™. The detached cells were then dispensed onto DMA spots with the volume of 200 nL/spot using pressures of 75, 150, and 300 mbar ms. The cells were cultured on DMA slides for 24 h, followed by adding calcein AM and PI for live/dead staining. The cell viability was calculated as the ratio of the calcein AM positive area to the sum of the calcein AM and PI positive areas. (A) Viability of hiPSCs printed on TA and cultured for 24 h (n = 3 biological replicates). (B) Representative fluorescence images of hiPSCs seeded onto TA DMA and cultured for 24 h. Scale bar: 100 μm. (C) Viability of hiPSCs printed onto TB DMA and cultured for 24 h (n = 3 biological replicates). (D) Representative fluorescence images of hiPSCs seeded onto TB DMA and cultured for 24 h. Data represent the mean ± SD. Scale bar: 100 μm.
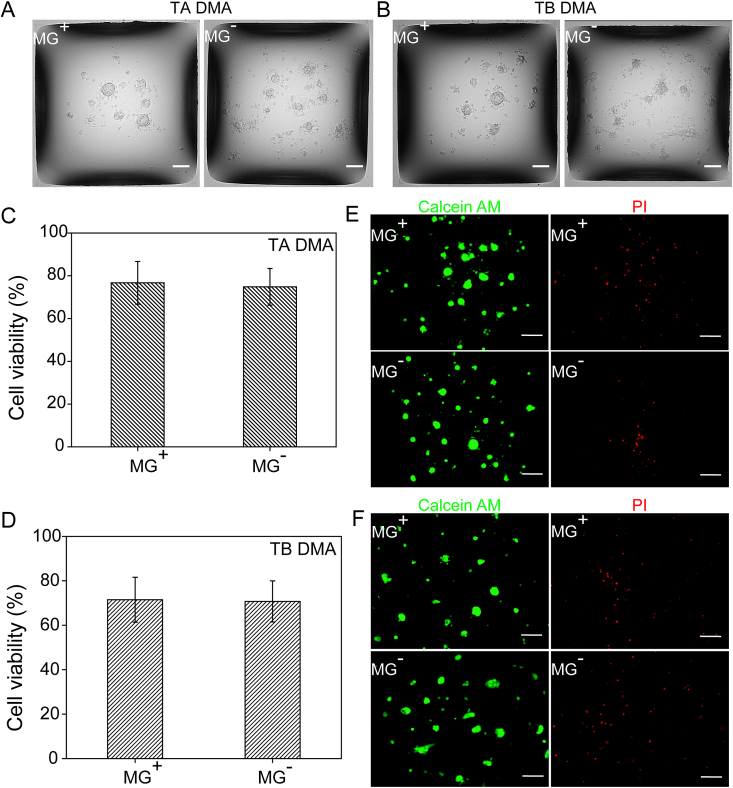
HiPSC cultured in vitro typically grow in tightly packed colonies, which distinguishes them from somatic cells [44,45]. Therefore, the morphology of hiPSC colonies is considered to be an important factor that indicates the pluripotency hiPSCs in vitro. HiPSCs cultured in vitro are usually passaged as multi-cellular clusters since single cells are more prone to cell death whereas colonies are quickly re-established by cell clusters [46]. We investigated the morphology of hiPSCs cultivated on MG+ and MG− TA DMA and TB DMAs (Fig. 4A and B, S4). HiPSCs exhibited typical morphology of tightly compacted, well-defined colonies consisting of round cells with large nuclei and a high nucleo-cytoplasmic ratio on both MG+ and MG− DMAs (Fig. 4A and B) [44,47]. These observations indicated the feasibility of utilizing the DMA platform for hiPSC culture. The morphology of hiPSCs grown on surfaces (MG+ 2 mL, MG− TA 2 mL, and MG− TB 2 mL) were also investigated (Fig. S5). Bright field images of 10 spots were acquired in three independent experiments (Fig. S6, S7). We compared the viability of hiPSCs on MG+ and MG− TA DMA and TB DMA after 24 h of culture. The viability of hiPSCs ranged from 70% to 76% (Fig. 3C, E) and live hiPSCs were abundant on DMAs (Fig. 3D, F). In order to further confirm the adhesion of hiPSCs to the surface of DMA slides without Matrigel coating, we quantified the number of hiPSCs present on the surface after washing the DMA slides with PBS (Figure S8). Our results showed that there were no significant differences in the viability of hiPSCs cultured on both MG+ and MG− DMAs, which indicated that MG coating is not crucial for culturing hiPSCs on DMAs for 24 h.
Fig. 4.
Morphology and viability of hiPSCs cultured on TA and TB DMAs with Matrigel coating (MG+) and without Matrigel coating (MG−). (A) Morphology of hiPSCs cultivated on TA DMA with Matrigel coating (MG+) and without Matrigel coating (MG−). (B) Morphology of hiPSCs cultivated on TB DMA in MG+ and MG-conditions. (C) Cell viability comparison of hiPSCs cultured 24 h on TA DMA. Data represent the mean ± SD. (D) Cell viability comparison of hiPSCs cultured 24 h on TB DMA. Data represent the mean ± SD. (E) Representative fluorescence images of live (green, calcein AM positive) and dead (red, PI positive) hiPSCs cultured on TA DMA. Scale bar: 100 μm. (F) Representative fluorescence images of live and dead hiPSCs cultured on TB DMA. Scale bar: 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The pluripotency of hiPSCs cultured in vitro is the most crucial and defining characteristic of these cells since it represents the ability of a cell to differentiate into any cell type. To maintain hiPSCs in the pluripotent state, several research groups have reported new cell culture substrates with the potential for use as MG substitutes. HiPSCs cultured on these substrates have similar gene expression patterns and a comparable level of pluripotency to cells grown on MG [[15], [16], [17]]. However, spontaneous differentiation of hiPSCs into random/multiple lineages during in vitro culture is still common. Thus, the search for a coating with well-defined composition that is xeno-free to replace the commonly used MG and facilitate the generation of a reproducible culturing environment for hiPSCs is still ongoing.
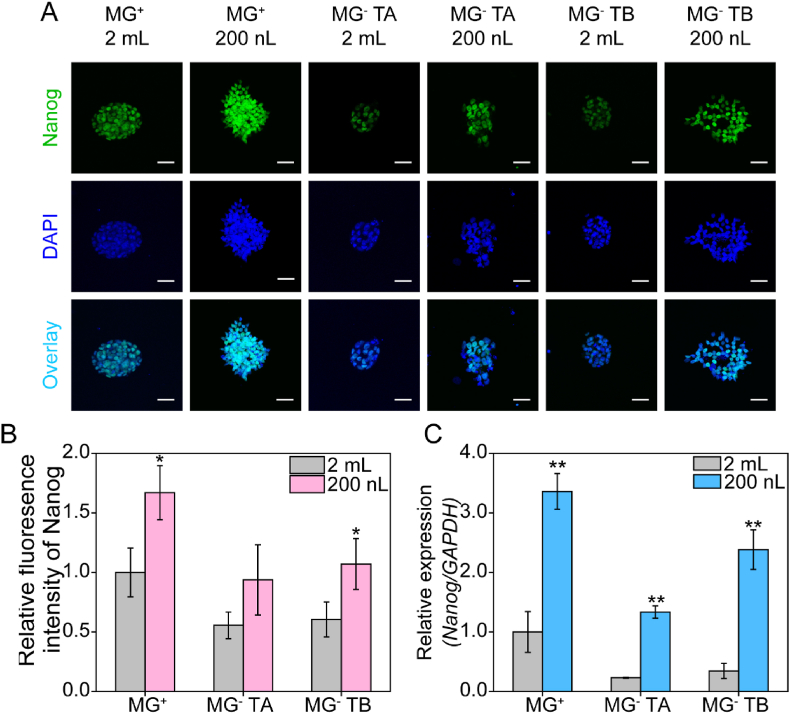
Therefore, as a next step, we characterized and compared the pluripotency of hiPSC cultured on MG+ and MG− TA and TB DMAs, as well as on MG+ and MG− TA and TB surfaces (Fig. 1). HiPSC pluripotency is precisely regulated by a core set of transcription factors, including Nanog, Oct4, and Sox2 [48,49]. However, Nanog is at the heart of the gene regulatory network and fluctuations in its expression have been linked to cell fate decisions such as self-renewal (Nanog high) and differentiation (Nanog low), making it a critical factor for maintaining pluripotency [50,51]. We therefore assessed the impact of cultivation environment on pluripotency of hiPSCs by immunofluorescence (IF) staining and qPCR analysis of Nanog protein and gene expression, respectively. We compared Nanog expression in hiPSCs cultured on MG− TA and TB DMAs (200 nL cell culture medium), as well as MG− TA and TB surfaces (2 mL cell culture medium) (Fig. 5A). Nanog expressions in hiPSCs cultured on a MG+ standard tissue culture plate (“MG+ 2 mL”) and a MG+ DMA (“MG+ 200 nL”) were taken as controls (Fig. 5A). Relative Nanog protein expression, calculated as the mean fluorescence intensity of Nanog IF staining and further normalized with the MG+ 2 mL group varied in hiPSCs cultured on different substrates (Fig. 5B). Generally, higher Nanog expression was observed in hiPSCs cultivated on DMAs (MG+ 200 nL, MG− TA 200 nL, and MG− TB 200 nL) compared with that in hiPSCs cultivated on surfaces (MG+ 2 mL, MG− TA 2 mL, and MG− TB 2 mL) (Fig. 5A and B). Nanog expression in hiPSCs cultivated on MG+ 200 nL was 1.67 ± 0.23-fold higher than that in MG+ 2 mL. Furthermore, Nanog expression levels in hiPSCs cultivated on MG− TA 200 nL and MG− TB 200 nL were 1.77 ± 0.35 and 1.69 ± 0.53 times higher than that on MG− TA 2 mL and MG− TB 2 mL, respectively. Compared to the Nanog expression on MG− TA 200 nL and MG- TB 200 nL, the Nanog expression levels on MG+ 200 nL were 1.78 ± 0.24-fold and 1.56 ± 0.21-fold higher, respectively. The same trend was observed for MG− TA 2 mL and MG− TB 2 mL, for which Nanog expression levels were 45% and 40% lower than for MG+ 2 mL, respectively. This trend was also observed in the qPCR analysis. HiPSCs grown on DMAs in 200-nL droplets (MG+ 200 nL, MG− TA 200 nL, and MG− TB 200 nL) generally displayed higher Nanog gene expression than that in cells grown on surfaces in a 2-mL volume (MG+ 2 mL, MG− TA 2 mL, and MG− TB 2 mL) (Fig. 5C). The Nanog expression of hiPSCs grown on MG+ 200 nL was 3.36 ± 0.3 times higher than that of hiPSCs grown on MG+ 2 mL. Nanog expression levels of hiPSCs grown on MG− TA 200 nL and MG− TB 200 nL were 5.74 ± 0.45-fold and 6.87 ± 0.96-fold higher than those of hiPSCs grown on MG− TA 2 mL and MG− TB 2 mL, respectively. However, similar Nanog expression levels were observed in hiPSCs grown on MG− TA 2 mL and MG− TB 2 mL, which were 76% and 66% lower than that in cells grown on MG+ 2 mL, respectively. We also analyzed the expression levels of the pluripotency markers genes, Oct4 and Sox2 (Fig. S9). The results showed a similar trend in the expression of these genes compared to that of Nanog, with higher levels of pluripotency gene expression in cells cultured on DMAs (200 nL) compared with that in cells cultured on surfaces (2 mL). Our results also showed that hiPSCs grown on different surfaces express different levels of Nanog. The XPS analysis of investigated surfaces showed the presence of sulfur and fluorine on TA and TB surface, respectively (Fig. 2), indicating the difference in chemical nature of the coatings, while the SEM images (Fig. 2) and roughness measurements show that both substrates have very similar topography. In our study we observed that cells cultured on TB surface (containing fluorine) displayed higher Nanog expression than that on TA (sulfur containing) surface, which shows that chemical modification plays an important role for the maintenance of the hiPSC pluripotency. In general, the pluripotency of hiPSCs cultured on DMAs in 200 nL droplets was higher than that of cells cultured on the same surfaces but in larger 2 mL volumes. Although Nanog expression was higher in cells cultured on MG+ 200 nL and MG+ 2 mL compared to MG− conditions, the pluripotency of hiPSCs cultured on MG− DMAs was maintained for 24 h, indicating that it is feasible to use DMAs without MG coating to successfully culture and screen hiPSCs in vitro.
Fig. 5.
Comparison of hiPSCs pluripotency for cells cultivated under diverse conditions. (A) Immunofluorescence (IF) staining for Nanog expression of hiPSCs cultivated on MG+ surface (MG+, 2 mL cell culture medium), MG+ DMA (MG+, 200 nL cell culture medium), MG− TA surface (MG− TA, 2 mL cell culture medium), MG− TB surface (MG− TB, 2 mL cell culture medium), MG− TA DMA (MG- TA, 200 nL cell culture medium), MG− TB DMA (MG- TA, 200 nL cell culture medium). Cell nuclei were counterstained with DAPI (blue) (n = 3 biological replicates). Scale bar: 20 μm. (B) Mean fluorescence intensity of Nanog IF staining for each experimental group was measured by ImageJ. Three images of each experimental group were randomly selected and analyzed. Data represent the mean ± SD. ∗P < 0.05, significant differences between the 2 mL and 200 nL groups. (C) Expression of the pluripotency specific gene Nanog was investigated by qPCR analysis of RNA isolated from cells cultured on different surfaces and volumes (n = 3 biological replicates). All gene expression were normalized to the reference gene GAPDH and represented as mean ± SEM. ∗∗P < 0.01, significant differences between the 2 mL and 200 nL groups. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In this study, we evaluated the DMA platform for culturing of hiPSC in 200-nL droplets for the first time. We investigated the impact of the printing process on the survival of hiPSCs on DMAs. We analyzed the viability, morphology and pluripotency of hiPSC cultured on MG− TA and MG− TB DMAs, and compared these with the properties of cells cultured on MG− TA and MG− TB surfaces and on MG. We demonstrated that hiPSCs cultured on TA and TB surfaces and TA and TB DMAs had typical colony morphology and cell survival in the presence and absence of MG. Based on our results, we conclude that hiPSCs exhibit high viability as well as expected morphology and pluripotency when cultured for 24 h in 200-nL droplets on both TA and TB DMAs without MG coating.
We observed that hiPSCs cultured in nanoliter droplets exhibited higher Nanog protein and gene expression and better maintained pluripotency compared to hiPSCs cultured on the same surfaces in 2 mL volumes. This observation may be accounted for by surface topography, which might contribute to the maintenance of pluripotency in a small volume (200 nL). Nanoscale surface topography possesses features with sizes comparable to the size of cellular filopodia, focal adhesions, lipid rafts, endocytic vesicles and extracellular matrix fibers. Such structured surfaces were shown to affect the adhesion and proliferation of hiPSCs [52]. It is reported that nanotopography affects downstream signaling such as activation of the integrin-linked kinase/β-catenin pathway [53]. Compared to the smooth surface of cell culture Petri dish or cell culture plates (Ra ≈ 3 nm), the TA and TB surfaces are significantly more rough (Ra ≈ 60 nm). The higher roughness, specific surface topography, and nanostructure that is visible on AFM and SEM images might have different effects on the cells. For example, it might induce the rearrangement of transmembrane adhesion proteins such as integrins, followed by the initiation of signal transduction and alteration of the cell behavior. This might explain why hiPSCs could attach on the TA and TB surfaces while could not attach on the non-coated smooth polystyrene cell culture well plates. However, more research needs to be done to shed light on the mechanistic aspects behind the observed effect of the nanorough surfaces on hiPSCs.
There is an urgent need for well-defined, xeno-free in vitro systems for culturing of hiPSCs. In this study, we demonstrated that it is possible to maintain hiPSCs in their pluripotent state during culture on DMAs for 24 h without any additional coating. DMAs can be precisely adjusted in terms of surface topography and chemical modification, as well as cell culture volumes, making it a well-defined and also animal source-free platform that is suitable for culturing hiPSCs in vitro.
HiPSCs have become a focus of research because of their potential in regenerative medicine, drug discovery and toxicology. There is still a high demand for cost- and labor-effective platforms for culturing and screening of hiPSCs. Existing screening methods usually require large number of cells and expensive reagents. Performing a screening of 300 substances on hiPSCs on DMA platform requires 600 times less reagents compared to the same screening performed in 96-well plates. In addition to well-defined culturing conditions, DMAs enable cultivation of hiPSC in nanoliter volumes in hundreds of parallel wells, making it a platform of choice for HTS applications.
4. Conclusion
In this study, we demonstrated that hiPSCs can be cultured for 24 h in 200-nL droplets on droplet microarray (DMA) slides with pluripotency maintained without a need for Matrigel (MG) coating, which is commonly used for hiPSC culture. In addition, compared to hiPSCs cultured in 2 mL cell culture medium on surfaces, cells grown in a small volume maintained better pluripotency. In this study, we used two types of commercial droplet microarray substrates (TA and TB), which were thoroughly characterized using various physico-chemical methods in order to understand whether surface topography, roughness, wettability and other properties can affect (positively or negatively) culture of hiPSCs on these substrates. This analysis is crucial for the use of droplet microarrays in high-throughput screenings of hiPSCs. TA and TB surfaces showed similar surface topography and roughness, and different chemical composition. HiPSCs cultured on both TA and TB DMAs exhibited high viability and typical morphology. The differences between the pluripotency of hiPSCs cultured on TA and TB surfaces (or TA and TB DMAs) demonstrated the influence of surface properties on the fate of hiPSCs. These results indicate that both surface physico-chemical properties and small culture volumes of influence the maintenance of hiPSCs pluripotency. Thus, miniaturized nanoliter volumes and the compatibility with high-throughput screenings (672 spots per microscope glass slide) make DMA a versatile and useful platform for short-term culture and xeno-free high-throughput screenings of hiPSCs.
To exploit the benefits of hiPSCs fully, further studies utilizing the advantages of DMAs are required. Examples are: i. the generation of well-defined substrates for xeno-free hiPSC culture with maintained pluripotency; ii. analysis of the effects of small molecules or their combinations on signaling pathways, such as the Wnt signaling pathway, to induce or hinder hiPSCs differentiation; and iii. screening of small molecules favoring pluripotency of naïve hiPSCs.
Author contributions
Yanxi Liu, Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft and Writing – review & editing. Shraddha Chakraborty, Investigation, Visualization. Chatrawee Direksilp, Investigation. Johannes M. Scheiger, Investigation. Anna A. Popova, Conceptualization, Supervision, Funding acquisition, Writing – review & editing. Pavel A. Levkin, Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Both A.A.P. and P.A.L. in addition to being employed by the KIT are also shareholders of Aquarray GmbH. Other co-authors declare no conflict of interests.
Acknowledgement
The research was supported by DFG (Heisenbergprofessur Projektnummer: 406232485, LE2936/9–1). The authors would like to thank Prof. Dr. Martin Bastmeyer and Dr. Sarah Bertels at Zoological Institute of Cell and Neurobiology, Karlsruhe Institute of Technology (KIT) for the donation of hiPSCs and help organize the culture of hiPSCs. The work was further supported by the Helmholtz program “Materials Systems Engineering”. The Chinese Scholarship Council (fellowship to Y.L.) is also gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2021.100153.
Contributor Information
Anna A. Popova, Email: anna.popova@kit.edu.
Pavel A. Levkin, Email: levkin@kit.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Kwon S.G., Kwon Y., Lee T., Park G., Kim J. Recent advances in stem cell therapeutics and tissue engineering strategies. Biomater. Res. 2018;22:1–8. doi: 10.1186/s40824-018-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savoji H., Mohammadi M.H., Rafatian N., Toroghi M.K., Wang E.Y., Zhao Y., Korolj A., Ahadian S., Radisic M. Cardiovascular disease models: a game changing paradigm in drug discovery and screening. Biomaterials. 2019;198:3–26. doi: 10.1016/j.biomaterials.2018.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desbordes S.C., Studer L. Adapting human pluripotent stem cells to high-throughput and high-content screening. Nat. Protoc. 2013;8(1):111–130. doi: 10.1038/nprot.2012.139. [DOI] [PubMed] [Google Scholar]
- 7.Engle S.J., Puppala D. Integrating human pluripotent stem cells into drug development. Cell Stem Cell. 2013;12(6):669–677. doi: 10.1016/j.stem.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Zhu Z., Huangfu D. Human pluripotent stem cells: an emerging model in developmental biology. Development. 2013;140(4):705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamanaka S. Patient-specific pluripotent stem cells become even more accessible. Cell Stem Cell. 2010;7(1):1–2. doi: 10.1016/j.stem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi Y., Furue M.K. Biological effects of culture substrates on human pluripotent stem cells. Stem Cell. Int. 2016;2016:5380560. doi: 10.1155/2016/5380560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hovatta O., Mikkola M., Gertow K., Stromberg A.M., Inzunza J., Hreinsson J., Rozell B., Blennow E., Andäng M., Ährlund-Richter L. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum. Reprod. 2003;18(7):1404–1409. doi: 10.1093/humrep/deg290. [DOI] [PubMed] [Google Scholar]
- 12.Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., Thomson J.A. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 2006;3(10):637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 13.Park H., Yang K., Kim M., Jang J., Lee M., Kim D., Lee H., Cho S. Bio-inspired oligovitronectin-grafted surface for enhanced self-renewal and long-term maintenance of human pluripotent stem cells under feeder-free conditions. Biomaterials. 2015;50:127–139. doi: 10.1016/j.biomaterials.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Aisenbrey E.A., Murphy W.L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020;5(7):539–551. doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brafman D.A., Chang C.W., Fernandez A., Willert K., Varghese S., Chien S. Long-term human pluripotent stem cell self-renewal on synthetic polymer surfaces. Biomaterials. 2010;31(34):9135–9144. doi: 10.1016/j.biomaterials.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musah S., Morin S.A., Wrighton P.J., Zwick D.B., Jin S., Kiessling L.L. Glycosaminoglycan-binding hydrogels enable mechanical control of human pluripotent stem cell self-renewal. ACS Nano. 2012;6(11):10168–10177. doi: 10.1021/nn3039148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caiazzo M., Okawa Y., Ranga A., Piersigilli A., Tabata Y., Lutolf M.P. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater. 2016;15(3):344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 18.Chang P.H., Chao H.M., Chern E., Hsu S.H. Chitosan 3D cell culture system promotes naïve-like features of human induced pluripotent stem cells: a novel tool to sustain pluripotency and facilitate differentiation. Biomaterials. 2021;268:120575. doi: 10.1016/j.biomaterials.2020.120575. [DOI] [PubMed] [Google Scholar]
- 19.Mesquita F.C.P., Hochman-Mendez C., Morrissey J., Sampaio L.C., Taylor D.A. Laminin as a potent substrate for large-scale expansion of human induced pluripotent stem cells in a closed cell expansion system. Stem Cell. Int. 2019;2019:9704945. doi: 10.1155/2019/9704945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi Y., Chan T., Warashina M., Fukuda M., Ariizumi T., Okabayashi K., Takayama N., Otsu M., Eto K., Furue M.K., Michiue T., Ohnuma K., Nakauchi H., Asashima M. Reduction of N-glycolylneuraminic acid in human induced pluripotent stem cells generated or cultured under feeder- and serum-free defined conditions. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0014099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Badenes S.M., Fernandes T.G., Cordeiro C.S.M., Boucher S., Kuninger D., Vemuri M.C., Diogo M.M., Cabral J.M.S. Defined essential 8 (TM) medium and vitronectin efficiently support scalable xeno-free expansion of human induced pluripotent stem cells in stirred microcarrier culture systems. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0155296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam M.T., Longaker M.T. Comparison of several attachment methods for human iPS, embryonic and adipose-derived stem cells for tissue engineering. J. Tissue Eng. Regener. Med. 2012;6(S3):s80–s86. doi: 10.1002/term.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Jiang X., Li L., Chen Z., Gao G., Yao R., Sun W. 3D printing human induced pluripotent stem cells with novel hydroxypropyl chitin bioink: scalable expansion and uniform aggregation. Biofabrication. 2018;10(4) doi: 10.1088/1758-5090/aacfc3. [DOI] [PubMed] [Google Scholar]
- 24.Feng W., Ueda E., Levkin P.A. Droplet microarrays: from surface patterning to high-throughput applications. Adv. Mater. 2018;30(20) doi: 10.1002/adma.201706111. [DOI] [PubMed] [Google Scholar]
- 25.Feng W., Li L., Ueda E., Li J., Heissler S., Welle A., Trapp O., Levkin P.A. Surface patterning via thiol-yne click chemistry: an extremely fast and versatile approach to superhydrophilic-superhydrophobic micropatterns. Adv. Mater. Interfaces. 2014;1(7):1400269. [Google Scholar]
- 26.Reimer A., Vasilevich A., Hulshof F., Viswanathan P., van Blitterswijk C.A., de Boer J., Watt F.M. Scalable topographies to support proliferation and Oct4 expression by human induced pluripotent stem cells. Sci. Rep. 2016;6:18948. doi: 10.1038/srep18948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan F., Zhang M., Wu G., Lai Y., Greber B., Scholer H.R., Chi L. Topographic effect on human induced pluripotent stem cells differentiation towards neuronal lineage. Biomaterials. 2013;34(33):8131–8139. doi: 10.1016/j.biomaterials.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 28.Jaggy M., Zhang P., Greiner A.M., Autenrieth T.J., Nedashkivska V., Efremov A.N., Blattner C., Bastmeyer M., Levkin P.A. Hierarchical micro-nano surface topography promotes long-term maintenance of undifferentiated mouse embryonic stem cells. Nano Lett. 2015;15(10):7146–7154. doi: 10.1021/acs.nanolett.5b03359. [DOI] [PubMed] [Google Scholar]
- 29.Tronser T., Popova A.A., Jaggy M., Bastmeyer M., Levkin P.A. Droplet microarray based on patterned superhydrophobic surfaces prevents stem cell differentiation and enables high-throughput stem cell screening. Adv. Healthcare Mater. 2017;6(23):1700622. doi: 10.1002/adhm.201700622. [DOI] [PubMed] [Google Scholar]
- 30.Tronser T., Demir K., Reischl M., Bastmeyer M., Levkin P.A. Droplet microarray: miniaturized platform for rapid formation and high-throughput screening of embryoid bodies. Lab Chip. 2018;18(15):2257–2269. doi: 10.1039/c8lc00450a. [DOI] [PubMed] [Google Scholar]
- 31.Rosenfeld A., Oelschlaeger C., Thelen R., Heissler S., Levkin P.A. Miniaturized high-throughput synthesis and screening of responsive hydrogels using nanoliter compartments. Mater. Today Bio. 2020;6:100053. doi: 10.1016/j.mtbio.2020.100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popova A.A., Tronser T., Demir K., Haitz P., Kuodyte K., Starkuviene V., Wajda P., Levkin P.A. Facile one step formation and screening of tumor spheroids using droplet-microarray platform. Small. 2019;15(25) doi: 10.1002/smll.201901299. [DOI] [PubMed] [Google Scholar]
- 33.Popova A.A., Schillo S.M., Demir K., Ueda E., Nesterov-Mueller A., Levkin P.A. Droplet-array (DA) sandwich chip: a versatile platform for high-throughput cell screening based on superhydrophobic-superhydrophilic micropatterning. Adv. Mater. 2015;27(35):5217–5222. doi: 10.1002/adma.201502115. [DOI] [PubMed] [Google Scholar]
- 34.Jogia G.E., Tronser T., Popova A.A., Levkin P.A. Droplet microarray based on superhydrophobic-superhydrophilic patterns for single cell analysis. Microarrays. 2016;5(4):5040028. doi: 10.3390/microarrays5040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui H., Wang X., Wesslowski J., Tronser T., Rosenbauer J., Schug A., Davidson G., Popova A.A., Levkin P.A. Assembly of multi-spheroid cellular architectures by programmable droplet merging. Adv. Mater. 2021;33(4) doi: 10.1002/adma.202006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Tronser T., Peravali R., Reischl M., Levkin P.A. High-throughput screening of cell transfection enhancers using miniaturized droplet microarrays. Adv. Biosyst. 2020;4(3):1900257. doi: 10.1002/adbi.201900257. [DOI] [PubMed] [Google Scholar]
- 37.Alisson-Silva F., Rodrigues D.de C., Vairo L., Asensi K.D., Vasconcelos-dos-Santos A., Mantuano N.R., Dias W.B., Rondinelli E., dos R.C., Goldenberg S.D., Urmenyi T.P., Todeschini A.R. Evidences for the involvement of cell surface glycans in stem cell pluripotency and differentiation. Glycobiology. 2014;24(5):458–468. doi: 10.1093/glycob/cwu012. [DOI] [PubMed] [Google Scholar]
- 38.Chen L., Liu X., Su B., Li J., Jiang L., Han D., Wang S. Aptamer-mediated efficient capture and release of T lymphocytes on nanostructured surfaces. Adv. Mater. 2011;23(38):4376–4380. doi: 10.1002/adma.201102435. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui H., Wang W., Shi L., Song W., Wang S. Superwettable surface engineering in controlling cell adhesion for emerging bioapplications. Small Methods. 2020;4(12):2000573. [Google Scholar]
- 41.Ross A.M., Jiang Z., Bastmeyer M., Lahann J. Physical aspects of cell culture substrates: topography, roughness, and elasticity. Small. 2012;8(3):336–355. doi: 10.1002/smll.201100934. [DOI] [PubMed] [Google Scholar]
- 42.Ohgushi M., Matsumura M., Eiraku M., Murakami K., Aramaki T., Nishiyama A., Muguruma K., Nakano T., Suga H., Ueno M., Ishizaki T., Suemori H., Narumiya S., Niwa H., Sasai Y. Molecular pathway and cell State responsible for dissociation-induced apoptosis in Human pluripotent stem cells. Cell Stem Cell. 2010;7(2):225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Goetzke R., Sechi A., De Laporte L., Neuss S., Wagner W. Why the impact of mechanical stimuli on stem cells remains a challenge. Cell. Mol. Life Sci. 2018;75(18):3297–3312. doi: 10.1007/s00018-018-2830-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kato R., Matsumoto M., Sasaki H., Joto R., Okada M., Ikeda Y., Kanie K., Suga M., Kinehara M., Yanagihara K., Liu Y., Uchio-Yamada K., Fukuda T., Kii H., Uozumi T., Honda H., Kiyota Y., Furue M.K. Parametric analysis of colony morphology of non-labelled live human pluripotent stem cells for cell quality control. Sci. Rep. 2016;6:34009. doi: 10.1038/srep34009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A., Suri S., Lee T., Chilton J.M., Cooke M.T., Chen W., Fu J., Stice S.L., Lu H., McDevitt T.C., Garcia A.J. Adhesion strength-based, label-free isolation of human pluripotent stem cells. Nat. Methods. 2013;10(5):438–444. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W., Deng C., Godoy-Parejo C., Zhang Y., Chen G. Developments in cell culture systems for human pluripotent stem cells, World. J. Stem Cell. 2019;11(11):968–981. doi: 10.4252/wjsc.v11.i11.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Courtot A.M., Magniez A., Oudrhiri N., Feraud O., Bacci J., Gobbo E., Proust S., Turhan A.G., Bennaceur-Griscelli A. Morphological analysis of human induced pluripotent stem cells during induced differentiation and reverse programming. Biores. Open Access. 2014;3(5):206–216. doi: 10.1089/biores.2014.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyer L.A., Lee T., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.R., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Gifford D.K., Melton D.A., Jaenisch R., Young R.A. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444(7117):364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 50.Blinka S., Rao S. Nanog expression in embryonic stem cells - an ideal model system to dissect enhancer function. Bioessays. 2017;39(12):1700086. doi: 10.1002/bies.201700086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarro P., Festuccia N., Colby D., Gagliardi A., Mullin N.P., Zhang W., Karwacki-Neisius V., Osorno R., Kelly D., Robertson M., Chambers I. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31(24):4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hulshof F.F., Zhao Y., Vasilevich A., Beijer N.R., de Boer M., Papenburg B.J., van Clemens B., Dimitrios S., de Boer J. NanoTopoChip: high-throughput nanotopographical cell instruction. Acta Biomater. 2017;62:188–198. doi: 10.1016/j.actbio.2017.08.023. [DOI] [PubMed] [Google Scholar]
- 53.Wang W., Zhao L., Wu K., Ma Q., Mei S., Chu P.K., Wang Q., Zhang Y. The role of integrin-linked kinase/β-catenin pathway in the enhanced MG63 differentiation by micro/nano-textured topography. Biomaterials. 2013;34(3):631–640. doi: 10.1016/j.biomaterials.2012.10.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.