Abstract
Morphological variation of the skull was examined in the northern treeshrew (Tupaia belangeri) from various localities across Southeast Asia. Through a multivariate analysis, the treeshrews from South Vietnam exhibited distinct morphological characteristics compared to other populations from Thailand and Laos, and Malaysia. The plots of the specimens of North Vietnam are not randomly mixed with Thailand plots segregation in the scatteregrams of canonical discriminant analysis. Since the skulls of the population from North Vietnam were morphologically similar to those form central Laos and northern and northeastern Thailand, the zoogeographical barrier effect of Mekong River was not clearly confirmed. The population of the Kanchanaburi in western Thailand is clearly smaller in size compared to the other populations. The southern border of the distribution of this species is determined by the Isthmus of Kra or Kangar-Pattani Line. In the northern treeshrew, which is distributed from southern China to Bangladesh and southern Thailand, we have detected osteometrical geographical variation driven by geography. These results indicate that the skull morphology in the Tupaia glis-belangeri complex distinctively differs in South Vietnam, western Thailand, and southern Thailand. The zoogeographical barrier and factor separating these districts are expected to clarify in the future.
Keywords: geographical variation, Isthmus of Kra, Mekong River, northern treeshrew, skull
The northern (Tupaia belangeri) and common treeshrew (Tupaia glis) are distributed across Southeast Asia, throughout the Indochinese region, Malayan Peninsula, and Indonesian Islands. The two species have been distinguished in external color characters [14], with T. belangeri exhibiting green-gray pelage and T. glis appearing red-brown in the dorsal and lateral aspects. Interestingly, the sympatric distribution of the two species has been osteometrically and karyologically investigated in southern Thailand regions including the Isthmus of Kra and Kangar-Pattani Line [12,13,14, 24].
The zoogeographical role of the Isthmus of Kra and the Kangar-Pattani Line on morphological variation has been strongly supported not only in mammals [15,16,17,18, 40], but also in birds [25], freshwater crustacean [10], insects [4, 8] and other various taxa including plants [3, 33]. As the relationships between sea level changes and the breaches of Peninsular Malaysia were geologically and biogeographically examined [33, 38, 40], it has been pointed out that the narrow and low land around the Isthmus of Kra and the Kangar-Pattani Line in Malayan Peninsula is the transitional zone from Sundaic to Indochinese fauna. With regard to the treeshrews, this line may function as a speciation barrier of T. belangeri and T. glis. Molecular data estimates that the species or populations of the glis-belangeri complex continuously split in the late Miocene and early Pliocene [35]. Geological data suggests that the large transgressions of about 25 meters may have occurred in coast area of Indomalayan Region in the late Miocene and early Pliocene [40]. The sea level was thought to have then risen in the Holocene and Pleistocene [33]. We predict that the glis-belangeri complex has continued to diversify due to various geographical factors in this region and throughout geological eras.
We also reported that the distinct size of specimens from Kanchanaburi in western Thiland may be mainly due to their generally smaller body size. A geographical discontinuation has been taxonomically indicated in mammals in western Thailand, and also between Myanmar and Thailand. Kanchanaburi is located at 14-degrees north latitude. In the case of primates, the hybrid zone between Macaca mulatta and Macaca fascicularis has been observed from 15 to 20-degrees north latitude in western Thailand [5, 6]. The western part of Thailand and the Kanchanaburi population of T. belangeri are noticeable on the unknown zoogeographical factor of the terrestrial mammals in the Indochinese Peninsula.
Previous researches have shown zoogeographical barriers to be of significant impact to morphological variation in mammals in the Indochinese Peninsular region [9, 26]; thus, T. belangeri is of particular interest due to its large distribution. This study sought to examine the oeteometrical characteristics of the Vietnam populations of the northern tree shrew and to compare the populations of the glis-belangeri complex from Vietnam, Thailand and Malaysia. It will contribute to reveal the zoogeographical factors that may have induced variation in skull morphology among northern and common treeshrews in the Southeast Asian region.
MATERIALS AND METHODS
Specimens
We examined totally 21 skulls of the northern treeshrew from North and South Vietnam. The measurement data of other 203 skulls published in our previous studies [12,13,14] were also used in this comparative analysis. These specimens are stored in Thailand Institute of Scientific and Technological Research (TISTR) (Bangkok, Thailand), Department of Wildlife and National Parks (Kuala Lumpur, Malaysia), Muséum National d’Histoire Naturelle (MNHN) (Paris, France), Department of Vertebrate Zoology, Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology (Hanoi, Vietnam), and the Department of Zoology, National Museum of Nature and Science, Tokyo (Tokyo, Japan) [12,13,14]. Although the specimens have been used also in our previous studies [12,13,14], we selected the localities with larger specimen numbers and added the specimens from Vietnam in this study. All specimens used in this study had completely erupted molars. The replacement of premolar and incisors has not been checked. The skulls used in this study were not considered as full-adult specimens, since the permanent premolars and incisors are erupted after the molars in the taxa [37]. However, we applied the collections to this analysis, since the number of treeshrew specimens in protected populations is not so large in the museum collections. Locality and composition of the specimens are shown (Table 1, Fig. 1, Supplementary Tables 1 and 2).
Table 1. Localities and compositions of specimens used in this study.
| Localities | Locality symbol | Male | Female |
|---|---|---|---|
| Chiang Rai, Mae Hong Son, Chiang Mai, Nan | 1 | 7 | 13 |
| Tak, Nakhon Sawan, U Thai Thani | 2 | 7 | 4 |
| Nakhon Phanom, Udon Thani, Loei, Khon Kaen | 3 | 5 | 5 |
| Ubon Ratchathani, Surin, Khorat | 4 | 22 | 20 |
| Kanchanaburi | 5 | 10 | 13 |
| Prachin Buri, Phetchaburi | 6 | 5 | 4 |
| Chanthaburi | 7 | 3 | 4 |
| Nakhon Si Thammarat, Krabi, Trang, Pattani, Yala, Narathiwat, Pangha | 8 | 7 | 6 |
| Xiengkhuang | 9 | 4 | 4 |
| Peninsular Malaysia | X | 22 | 38 |
| Hanoi, Bac Giang, Lao Cai | Y | 5 | 8 |
| Khanh Hoa, Dong Nai, | Z | 3 | 5 |
| Total | 100 | 124 | |
Fig. 1.
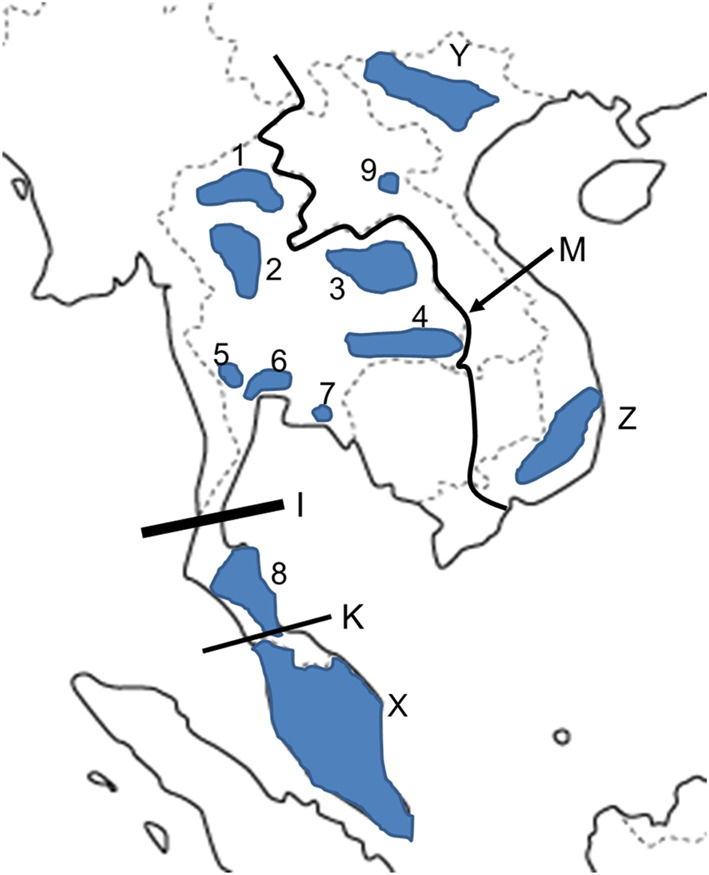
Distributions (grey areas) of the northern treeshrew populations in Indochinese and Malayan regions. I, Isthmus of Kra, K, Kangar-Pattani Line, M (arrow), present Mekong River. Locality symbols are explained in Table 1.
Measurements
We measured 17 characters of skull and mandible with a vernier caliper to the nearest 0.05 mm [11]. Measurements and abbreviations are defined in Table 2. From the raw measurement values, proportion indices were calculated by dividing each measurement by the geometric mean of all values.
Table 2. List of measurements and their abbreviations.
| 1. Cranium | |
| Profile length | PL |
| Condylobasal length | CL |
| Short lateral facial length | SL |
| Zygomatic width | ZW |
| Least breadth between the orbits | LBO |
| Greatest neurocranium breadth | GNB |
| Median palatal length | MPL |
| Length from Basion to Staphylion | LBS |
| Dental length | DL |
| Greatest palatal breadth | GPB |
| Greatest mastoid breadth | GMB |
| Height from Akrokranion to Basion | HAB |
| 2. Mandible | |
| Length from the condyle | LC |
| Length from the angle | LA |
| Length from Infradentale to aboral border of the alveolus | LIA |
| Height of the vertical ramus | HR |
| Height of the mandible at M1 | HM |
The measurement items were based on Driesch [11].
Statistical analysis
To visualize variation in skull morphology within our study sample, a principal component analysis and canonical discriminant analysis were carried out in both sexes using raw measurement values. The canonical discriminant analysis was also applied to the proportion indices calculated from the measurement values in each sex. In addition the morphological differences between populations of Vietnam (Y and Z) and the other populations were statistically examined by a t-test, Kruskal-Wallis test and Mann-Whitney U-test using the raw values and proportion indices. The multivariate analyses, Kruskal-Wallis test and Mann-Whitney U-test were carried out by using the software SPSS (IBM Japan, Ltd., Tokyo, Japan). The canonical discriminant analysis and Kruskal-Wallis test omitted the data of Locality 8 (Nakhon Si Thammarat, Krabi, Trang, Pattani, Yala, Narathiwat, Pangha) in which the two species are sympaptrically distributed. Since specimens originating here may contain both species, these data are inadequate to morphologically distinguish the populations [14].
RESULTS
Mean values and standard errors of the measurements in each locality are shown in Table 3, while proportion indices and their standard errors in each locality are shown in Table 4. The scattergrams and quantitative data are shown in the principal component analysis (Figs. 2 and 3, Table 5) and the canonical discriminant analysis (Figs. 4, 5, 6, 7, Table 6). The horizontal axis indicates the first principal component (PC1), whereas the vertical axis shows the second principal component (PC2) in Figs. 2 and 3. Scattergram shows the individual scores on the discriminant axes 1 (horizontal) and 2 (vertical) (Figs. 4, 5, 6, 7). The specimens and their measurement data of localities other than the populations of North Vietnam (Locality Y: Hanoi, Bac Giang, Lao Cai) and South Vietnam (Locality Z: Khanh Hoa, Dong Nai) were also used in our previous studies [12,13,14].
Table 3. The mean value (mm) and standard deviation of measurements of skulls from various localities.
| Locality | Sex | PL | CL | SL | ZW | LBO | GNB | MPL | LBS | DL | GPB | GMB | HAB | LC | LA | LIA | HR | HM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 48.88 | 43.22 | 19.96 | 25.44 | 13.77 | 19.02 | 25.71 | 17.56 | 25.53 | 16.40 | 20.66 | 11.81 | 33.36 | 33.56 | 20.70 | 13.17 | 3.91 |
| 1.35 | 1.29 | 0.86 | 0.93 | 0.45 | 0.52 | 1.05 | 0.45 | 0.80 | 0.54 | 0.57 | 0.25 | 1.24 | 1.24 | 0.72 | 1.21 | 0.41 | ||
| Female | 47.89 | 41.92 | 19.29 | 24.26 | 12.91 | 18.99 | 25.24 | 16.76 | 24.76 | 15.92 | 20.09 | 11.74 | 32.53 | 32.68 | 20.41 | 13.14 | 3.56 | |
| 0.98 | 0.98 | 0.67 | 0.82 | 0.50 | 0.49 | 0.80 | 0.39 | 0.61 | 0.30 | 0.67 | 0.35 | 0.92 | 0.99 | 0.78 | 0.50 | 0.29 | ||
| 2 | Male | 50.26 | 43.69 | 20.04 | 26.18 | 13.59 | 19.38 | 26.06 | 18.01 | 25.86 | 16.49 | 21.49 | 12.22 | 34.19 | 34.64 | 21.34 | 13.76 | 4.06 |
| 1.10 | 0.98 | 0.74 | 1.05 | 0.51 | 0.31 | 0.68 | 0.72 | 0.73 | 0.42 | 0.57 | 0.35 | 0.76 | 1.06 | 0.25 | 0.73 | 0.38 | ||
| Female | 49.33 | 42.94 | 19.51 | 24.51 | 13.45 | 19.26 | 25.80 | 17.29 | 25.76 | 16.51 | 20.78 | 11.56 | 33.34 | 33.39 | 20.98 | 13.08 | 3.74 | |
| 1.12 | 1.16 | 0.88 | 0.50 | 0.33 | 0.31 | 1.00 | 0.29 | 0.71 | 0.20 | 0.33 | 0.33 | 0.72 | 0.71 | 0.60 | 0.49 | 0.15 | ||
| 3 | Male | 49.43 | 43.43 | 20.19 | 25.44 | 13.41 | 19.07 | 25.85 | 17.70 | 25.75 | 16.37 | 20.66 | 12.09 | 33.73 | 34.43 | 20.91 | 13.67 | 4.17 |
| 0.64 | 0.43 | 0.48 | 1.20 | 0.61 | 0.50 | 0.56 | 0.20 | 0.39 | 0.69 | 0.24 | 0.54 | 0.79 | 1.21 | 0.77 | 0.87 | 0.56 | ||
| Female | 49.80 | 44.00 | 20.20 | 25.11 | 13.66 | 19.15 | 26.39 | 17.75 | 26.39 | 16.88 | 21.24 | 12.00 | 33.91 | 33.97 | 21.38 | 13.30 | 3.67 | |
| 1.07 | 1.08 | 0.71 | 0.28 | 0.52 | 0.35 | 0.80 | 0.49 | 0.77 | 0.52 | 0.29 | 0.13 | 0.71 | 0.99 | 0.77 | 0.57 | 0.39 | ||
| 4 | Male | 50.16 | 44.04 | 19.95 | 25.43 | 13.93 | 19.29 | 26.10 | 17.96 | 25.76 | 16.51 | 20.60 | 12.03 | 33.95 | 34.40 | 21.27 | 13.64 | 4.08 |
| 1.14 | 1.24 | 0.66 | 1.10 | 0.64 | 0.56 | 0.64 | 0.80 | 0.77 | 0.55 | 0.58 | 0.37 | 1.16 | 1.42 | 0.79 | 0.77 | 0.48 | ||
| Female | 49.26 | 43.05 | 19.31 | 24.63 | 13.68 | 19.05 | 25.65 | 17.47 | 25.45 | 16.38 | 20.18 | 11.86 | 33.27 | 33.88 | 20.99 | 13.36 | 3.83 | |
| 1.34 | 1.16 | 0.84 | 1.10 | 0.55 | 0.43 | 0.79 | 0.55 | 0.86 | 0.56 | 0.85 | 0.28 | 1.01 | 1.25 | 0.71 | 0.83 | 0.33 | ||
| 5 | Male | 46.93 | 41.18 | 19.13 | 24.54 | 13.59 | 17.75 | 24.45 | 16.87 | 24.06 | 15.19 | 19.48 | 11.24 | 32.39 | 32.48 | 20.14 | 12.81 | 4.04 |
| 1.35 | 0.74 | 0.75 | 0.51 | 1.32 | 0.46 | 0.63 | 0.46 | 0.52 | 0.32 | 0.52 | 0.37 | 0.75 | 0.74 | 0.53 | 0.43 | 0.29 | ||
| Female | 45.62 | 39.75 | 18.37 | 23.07 | 12.53 | 17.37 | 23.89 | 15.98 | 23.65 | 14.73 | 18.92 | 11.09 | 31.37 | 31.33 | 19.61 | 12.41 | 3.72 | |
| 1.19 | 1.00 | 0.67 | 0.85 | 0.51 | 0.61 | 0.84 | 0.43 | 0.61 | 0.52 | 0.79 | 0.33 | 0.90 | 0.99 | 0.46 | 0.47 | 0.30 | ||
| 6 | Male | 49.58 | 42.97 | 19.61 | 25.04 | 13.85 | 19.06 | 25.66 | 17.50 | 25.42 | 15.97 | 20.53 | 12.00 | 33.24 | 33.69 | 20.68 | 13.64 | 3.77 |
| 1.52 | 1.08 | 0.74 | 1.24 | 0.62 | 0.50 | 1.09 | 0.41 | 0.73 | 0.41 | 0.92 | 0.53 | 1.19 | 1.19 | 0.53 | 1.07 | 0.33 | ||
| Female | 47.73 | 41.89 | 18.41 | 24.46 | 13.53 | 19.06 | 24.70 | 17.26 | 24.65 | 16.18 | 19.96 | 11.80 | 32.41 | 33.13 | 20.34 | 13.19 | 3.74 | |
| 1.31 | 1.23 | 0.74 | 0.28 | 0.35 | 0.49 | 0.63 | 0.77 | 0.43 | 0.20 | 0.52 | 0.27 | 0.31 | 0.25 | 0.51 | 0.63 | 0.28 | ||
| 7 | Male | 51.45 | 45.00 | 20.45 | 26.80 | 14.75 | 19.40 | 26.80 | 18.28 | 26.47 | 16.48 | 21.08 | 12.57 | 35.37 | 35.78 | 21.98 | 14.05 | 4.22 |
| 1.43 | 1.21 | 0.71 | 1.23 | 0.88 | 0.18 | 0.64 | 0.51 | 0.81 | 1.20 | 0.67 | 0.60 | 1.24 | 1.51 | 0.59 | 0.98 | 0.65 | ||
| Female | 52.23 | 45.84 | 22.54 | 25.93 | 14.16 | 19.51 | 28.38 | 18.38 | 27.63 | 17.14 | 21.11 | 12.41 | 35.50 | 35.86 | 22.11 | 14.09 | 4.03 | |
| 0.92 | 0.91 | 1.64 | 0.67 | 0.24 | 0.27 | 0.46 | 0.22 | 1.22 | 0.43 | 0.93 | 0.57 | 1.06 | 0.57 | 0.87 | 0.16 | 0.29 | ||
| 8 | Male | 51.65 | 44.72 | 22.95 | 25.28 | 14.21 | 19.25 | 27.67 | 17.23 | 27.47 | 16.54 | 20.37 | 12.00 | 35.19 | 35.14 | 22.36 | 13.59 | 3.76 |
| 1.44 | 1.43 | 0.76 | 0.56 | 0.86 | 0.32 | 0.94 | 0.56 | 0.90 | 0.35 | 0.45 | 0.41 | 1.07 | 1.16 | 0.70 | 0.72 | 0.23 | ||
| Female | 50.62 | 43.99 | 22.15 | 25.03 | 14.52 | 19.58 | 27.17 | 17.18 | 26.93 | 16.43 | 20.53 | 11.84 | 34.77 | 34.69 | 21.98 | 13.53 | 3.81 | |
| 1.06 | 1.30 | 0.68 | 0.46 | 0.69 | 0.70 | 1.12 | 0.28 | 0.89 | 0.22 | 0.68 | 0.34 | 1.26 | 1.20 | 0.53 | 0.65 | 0.46 | ||
| 9 | Male | 48.04 | 41.66 | 19.88 | 24.48 | 13.26 | 19.23 | 25.23 | 16.53 | 24.53 | 15.36 | 19.70 | 11.76 | 31.63 | 31.73 | 19.85 | 12.71 | 3.45 |
| 0.83 | 0.48 | 0.66 | 0.62 | 0.32 | 0.47 | 0.46 | 0.22 | 0.42 | 0.64 | 0.57 | 0.29 | 0.64 | 1.02 | 0.70 | 0.41 | 0.11 | ||
| Female | 48.23 | 42.08 | 19.94 | 24.28 | 13.35 | 19.09 | 25.34 | 16.93 | 24.88 | 15.89 | 19.79 | 11.94 | 31.89 | 32.01 | 20.29 | 12.99 | 3.39 | |
| 1.50 | 1.57 | 0.73 | 0.99 | 0.33 | 0.13 | 1.28 | 0.62 | 1.10 | 0.57 | 0.85 | 0.28 | 1.63 | 1.91 | 0.92 | 1.18 | 0.14 | ||
| X | Male | 52.57 | 45.59 | 23.02 | 26.34 | 14.85 | 19.67 | 28.18 | 17.72 | 27.64 | 16.39 | 21.01 | 12.51 | 36.02 | 36.14 | 22.61 | 14.33 | 3.91 |
| 1.07 | 1.06 | 0.96 | 0.87 | 0.67 | 0.30 | 0.81 | 0.54 | 0.62 | 0.53 | 0.58 | 0.42 | 1.03 | 1.12 | 0.68 | 0.84 | 0.39 | ||
| Female | 51.81 | 44.98 | 22.49 | 25.34 | 14.38 | 19.70 | 27.68 | 17.54 | 27.32 | 16.38 | 20.74 | 12.22 | 35.10 | 35.22 | 22.23 | 13.81 | 3.74 | |
| 1.14 | 1.04 | 0.73 | 0.89 | 0.54 | 0.45 | 0.72 | 0.54 | 0.79 | 0.50 | 0.47 | 0.35 | 0.86 | 0.89 | 0.64 | 0.52 | 0.25 | ||
| Y | Male | 50.53 | 44.08 | 20.88 | 24.77 | 13.95 | 19.72 | 26.62 | 17.71 | 25.76 | 16.21 | 20.67 | 12.15 | 33.55 | 33.96 | 20.94 | 13.61 | 3.74 |
| 0.84 | 1.18 | 0.93 | 0.55 | 0.25 | 0.42 | 0.77 | 0.62 | 0.96 | 0.62 | 0.25 | 0.41 | 0.91 | 0.86 | 0.55 | 0.50 | 0.34 | ||
| Female | 49.89 | 43.57 | 21.23 | 24.41 | 13.61 | 19.50 | 26.49 | 17.64 | 25.93 | 16.05 | 19.94 | 11.84 | 33.32 | 33.70 | 20.87 | 13.51 | 3.74 | |
| 0.96 | 1.14 | 0.80 | 0.69 | 0.28 | 0.42 | 0.66 | 0.40 | 0.61 | 0.50 | 0.78 | 0.35 | 0.79 | 0.78 | 0.36 | 0.39 | 0.26 | ||
| Z | Male | 53.87 | 47.58 | 23.55 | 27.23 | 15.13 | 20.30 | 28.85 | 19.05 | 28.58 | 18.02 | 20.82 | 12.50 | 36.85 | 36.90 | 22.97 | 15.15 | 4.48 |
| 1.49 | 1.00 | 0.48 | 0.88 | 0.12 | 0.59 | 0.93 | 0.54 | 0.91 | 0.36 | 0.20 | 0.57 | 0.61 | 0.69 | 0.71 | 0.66 | 0.33 | ||
| Female | 53.21 | 46.93 | 22.74 | 26.86 | 14.83 | 20.56 | 28.36 | 18.89 | 27.96 | 17.88 | 19.93 | 12.73 | 36.32 | 36.64 | 22.38 | 15.34 | 4.43 | |
| 1.18 | 1.15 | 0.80 | 0.74 | 0.39 | 0.22 | 1.09 | 0.20 | 0.96 | 0.48 | 0.63 | 0.16 | 0.80 | 0.72 | 0.68 | 0.90 | 0.14 | ||
The mean values are shown in the upper rows, and the standard errors in the lower rows. Abbreviations of the measurement are explained in Table 2.
Table 4. The mean proportion indices and standard deviation of measurements of skulls from various localities.
| Locality | Sex | PL | CL | SL | ZW | LBO | GNB | MPL | LBS | DL | GPB | GMB | HAB | LC | LA | LIA | HR | HM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 2.422 | 2.141 | 0.988 | 1.260 | 0.682 | 0.943 | 1.274 | 0.870 | 1.265 | 0.813 | 1.024 | 0.585 | 1.653 | 1.662 | 1.025 | 0.651 | 0.194 |
| 0.032 | 0.039 | 0.020 | 0.021 | 0.006 | 0.023 | 0.024 | 0.033 | 0.016 | 0.024 | 0.033 | 0.012 | 0.024 | 0.011 | 0.015 | 0.040 | 0.015 | ||
| Female | 2.446 | 2.141 | 0.985 | 1.239 | 0.659 | 0.970 | 1.289 | 0.856 | 1.265 | 0.814 | 1.026 | 0.600 | 1.661 | 1.669 | 1.042 | 0.671 | 0.182 | |
| 0.030 | 0.032 | 0.023 | 0.029 | 0.022 | 0.019 | 0.028 | 0.020 | 0.021 | 0.022 | 0.027 | 0.014 | 0.028 | 0.027 | 0.032 | 0.024 | 0.013 | ||
| 2 | Male | 2.436 | 2.117 | 0.971 | 1.268 | 0.658 | 0.940 | 1.263 | 0.873 | 1.253 | 0.799 | 1.041 | 0.592 | 1.657 | 1.678 | 1.034 | 0.666 | 0.196 |
| 0.027 | 0.035 | 0.022 | 0.030 | 0.012 | 0.034 | 0.026 | 0.020 | 0.022 | 0.022 | 0.017 | 0.009 | 0.015 | 0.034 | 0.022 | 0.025 | 0.015 | ||
| Female | 2.461 | 2.142 | 0.973 | 1.223 | 0.671 | 0.961 | 1.287 | 0.863 | 1.285 | 0.824 | 1.037 | 0.577 | 1.663 | 1.666 | 1.046 | 0.652 | 0.187 | |
| 0.026 | 0.026 | 0.029 | 0.025 | 0.016 | 0.011 | 0.030 | 0.014 | 0.016 | 0.013 | 0.010 | 0.021 | 0.016 | 0.017 | 0.013 | 0.016 | 0.009 | ||
| 3 | Male | 2.422 | 2.128 | 0.989 | 1.246 | 0.657 | 0.934 | 1.266 | 0.868 | 1.262 | 0.802 | 1.012 | 0.592 | 1.652 | 1.686 | 1.024 | 0.669 | 0.204 |
| 0.042 | 0.043 | 0.016 | 0.031 | 0.019 | 0.027 | 0.011 | 0.030 | 0.021 | 0.020 | 0.020 | 0.016 | 0.018 | 0.039 | 0.017 | 0.028 | 0.024 | ||
| Female | 2.441 | 2.156 | 0.990 | 1.231 | 0.669 | 0.939 | 1.293 | 0.870 | 1.293 | 0.827 | 1.041 | 0.588 | 1.662 | 1.664 | 1.047 | 0.652 | 0.180 | |
| 0.054 | 0.031 | 0.019 | 0.017 | 0.011 | 0.020 | 0.024 | 0.018 | 0.017 | 0.016 | 0.027 | 0.015 | 0.007 | 0.022 | 0.022 | 0.027 | 0.016 | ||
| 4 | Male | 2.444 | 2.146 | 0.972 | 1.239 | 0.679 | 0.941 | 1.272 | 0.875 | 1.255 | 0.805 | 1.004 | 0.586 | 1.654 | 1.676 | 1.036 | 0.664 | 0.198 |
| 0.032 | 0.030 | 0.020 | 0.026 | 0.020 | 0.037 | 0.027 | 0.022 | 0.026 | 0.022 | 0.023 | 0.017 | 0.016 | 0.032 | 0.020 | 0.022 | 0.019 | ||
| Female | 2.453 | 2.143 | 0.961 | 1.226 | 0.681 | 0.949 | 1.277 | 0.870 | 1.267 | 0.815 | 1.004 | 0.591 | 1.656 | 1.686 | 1.045 | 0.665 | 0.190 | |
| 0.028 | 0.025 | 0.022 | 0.029 | 0.018 | 0.029 | 0.020 | 0.020 | 0.021 | 0.019 | 0.023 | 0.017 | 0.021 | 0.035 | 0.025 | 0.030 | 0.014 | ||
| 5 | Male | 2.418 | 2.123 | 0.985 | 1.265 | 0.700 | 0.915 | 1.260 | 0.869 | 1.240 | 0.783 | 1.004 | 0.579 | 1.669 | 1.674 | 1.038 | 0.660 | 0.208 |
| 0.038 | 0.026 | 0.024 | 0.019 | 0.066 | 0.024 | 0.023 | 0.025 | 0.018 | 0.011 | 0.026 | 0.015 | 0.025 | 0.027 | 0.016 | 0.016 | 0.013 | ||
| Female | 2.442 | 2.128 | 0.983 | 1.235 | 0.670 | 0.930 | 1.278 | 0.856 | 1.266 | 0.788 | 1.012 | 0.594 | 1.679 | 1.677 | 1.050 | 0.664 | 0.199 | |
| 0.030 | 0.026 | 0.024 | 0.030 | 0.020 | 0.033 | 0.030 | 0.024 | 0.025 | 0.022 | 0.032 | 0.010 | 0.025 | 0.025 | 0.019 | 0.016 | 0.014 | ||
| 6 | Male | 2.463 | 2.135 | 0.974 | 1.243 | 0.688 | 0.947 | 1.274 | 0.870 | 1.263 | 0.793 | 1.019 | 0.596 | 1.651 | 1.673 | 1.027 | 0.677 | 0.187 |
| 0.042 | 0.037 | 0.015 | 0.038 | 0.021 | 0.029 | 0.025 | 0.039 | 0.012 | 0.009 | 0.019 | 0.018 | 0.013 | 0.022 | 0.014 | 0.038 | 0.011 | ||
| Female | 2.425 | 2.128 | 0.936 | 1.243 | 0.687 | 0.969 | 1.255 | 0.877 | 1.253 | 0.822 | 1.014 | 0.600 | 1.647 | 1.683 | 1.034 | 0.670 | 0.190 | |
| 0.050 | 0.050 | 0.032 | 0.013 | 0.019 | 0.024 | 0.026 | 0.034 | 0.016 | 0.013 | 0.026 | 0.016 | 0.011 | 0.008 | 0.023 | 0.036 | 0.015 | ||
| 7 | Male | 2.435 | 2.130 | 0.968 | 1.268 | 0.697 | 0.919 | 1.268 | 0.865 | 1.252 | 0.779 | 0.998 | 0.594 | 1.673 | 1.693 | 1.040 | 0.664 | 0.199 |
| 0.040 | 0.037 | 0.013 | 0.003 | 0.011 | 0.033 | 0.031 | 0.014 | 0.020 | 0.026 | 0.012 | 0.012 | 0.017 | 0.023 | 0.021 | 0.018 | 0.022 | ||
| Female | 2.447 | 2.147 | 1.055 | 1.214 | 0.664 | 0.915 | 1.330 | 0.861 | 1.294 | 0.803 | 0.989 | 0.581 | 1.663 | 1.680 | 1.036 | 0.660 | 0.188 | |
| 0.015 | 0.008 | 0.055 | 0.015 | 0.009 | 0.031 | 0.053 | 0.029 | 0.029 | 0.014 | 0.023 | 0.023 | 0.013 | 0.014 | 0.017 | 0.018 | 0.010 | ||
| 8 | Male | 2.474 | 2.142 | 1.099 | 1.211 | 0.680 | 0.922 | 1.325 | 0.825 | 1.316 | 0.792 | 0.976 | 0.575 | 1.685 | 1.683 | 1.071 | 0.650 | 0.180 |
| 0.017 | 0.020 | 0.019 | 0.021 | 0.023 | 0.019 | 0.023 | 0.009 | 0.014 | 0.011 | 0.036 | 0.015 | 0.017 | 0.018 | 0.009 | 0.022 | 0.008 | ||
| Female | 2.444 | 2.123 | 1.069 | 1.209 | 0.701 | 0.945 | 1.311 | 0.829 | 1.300 | 0.793 | 0.991 | 0.572 | 1.678 | 1.674 | 1.061 | 0.653 | 0.184 | |
| 0.022 | 0.023 | 0.030 | 0.026 | 0.034 | 0.040 | 0.040 | 0.009 | 0.031 | 0.011 | 0.043 | 0.022 | 0.025 | 0.023 | 0.012 | 0.020 | 0.019 | ||
| 9 | Male | 2.475 | 2.147 | 1.024 | 1.261 | 0.683 | 0.991 | 1.300 | 0.852 | 1.264 | 0.792 | 1.015 | 0.606 | 1.629 | 1.634 | 1.023 | 0.655 | 0.178 |
| 0.022 | 0.018 | 0.024 | 0.023 | 0.013 | 0.025 | 0.014 | 0.020 | 0.020 | 0.031 | 0.033 | 0.015 | 0.023 | 0.041 | 0.038 | 0.017 | 0.006 | ||
| Female | 2.464 | 2.150 | 1.019 | 1.240 | 0.682 | 0.976 | 1.294 | 0.865 | 1.271 | 0.812 | 1.009 | 0.610 | 1.628 | 1.634 | 1.037 | 0.663 | 0.173 | |
| 0.024 | 0.016 | 0.014 | 0.022 | 0.013 | 0.032 | 0.024 | 0.025 | 0.012 | 0.005 | 0.018 | 0.011 | 0.023 | 0.037 | 0.034 | 0.046 | 0.001 | ||
| X | Male | 2.457 | 2.131 | 1.076 | 1.231 | 0.694 | 0.920 | 1.317 | 0.828 | 1.292 | 0.766 | 0.982 | 0.585 | 1.683 | 1.689 | 1.057 | 0.670 | 0.183 |
| 0.027 | 0.029 | 0.031 | 0.023 | 0.024 | 0.023 | 0.025 | 0.025 | 0.021 | 0.020 | 0.029 | 0.021 | 0.021 | 0.028 | 0.028 | 0.032 | 0.016 | ||
| Female | 2.471 | 2.145 | 1.072 | 1.209 | 0.686 | 0.940 | 1.320 | 0.837 | 1.303 | 0.781 | 0.989 | 0.583 | 1.674 | 1.679 | 1.060 | 0.659 | 0.178 | |
| 0.027 | 0.023 | 0.022 | 0.031 | 0.025 | 0.022 | 0.020 | 0.021 | 0.021 | 0.021 | 0.019 | 0.013 | 0.020 | 0.026 | 0.026 | 0.022 | 0.010 | ||
| Y | Male | 2.473 | 2.157 | 1.021 | 1.212 | 0.683 | 0.965 | 1.302 | 0.866 | 1.260 | 0.794 | 1.012 | 0.595 | 1.642 | 1.662 | 1.025 | 0.666 | 0.183 |
| 0.017 | 0.014 | 0.027 | 0.019 | 0.006 | 0.033 | 0.019 | 0.016 | 0.025 | 0.045 | 0.021 | 0.020 | 0.014 | 0.015 | 0.006 | 0.017 | 0.014 | ||
| Female | 2.464 | 2.151 | 1.048 | 1.205 | 0.672 | 0.963 | 1.308 | 0.871 | 1.281 | 0.793 | 0.985 | 0.585 | 1.645 | 1.664 | 1.031 | 0.667 | 0.185 | |
| 0.020 | 0.022 | 0.027 | 0.032 | 0.014 | 0.020 | 0.014 | 0.010 | 0.016 | 0.026 | 0.034 | 0.017 | 0.019 | 0.016 | 0.014 | 0.012 | 0.012 | ||
| Z | Male | 2.424 | 2.141 | 1.060 | 1.225 | 0.681 | 0.914 | 1.298 | 0.857 | 1.286 | 0.811 | 0.937 | 0.562 | 1.658 | 1.661 | 1.033 | 0.682 | 0.202 |
| 0.021 | 0.021 | 0.003 | 0.020 | 0.020 | 0.034 | 0.013 | 0.021 | 0.012 | 0.004 | 0.031 | 0.013 | 0.017 | 0.011 | 0.011 | 0.016 | 0.011 | ||
| Female | 2.424 | 2.138 | 1.036 | 1.224 | 0.676 | 0.937 | 1.292 | 0.861 | 1.274 | 0.815 | 0.908 | 0.580 | 1.655 | 1.669 | 1.020 | 0.698 | 0.202 | |
| 0.024 | 0.026 | 0.028 | 0.020 | 0.008 | 0.026 | 0.033 | 0.012 | 0.028 | 0.017 | 0.033 | 0.013 | 0.006 | 0.008 | 0.021 | 0.029 | 0.005 | ||
The mean values are shown in the upper rows, and the standard errors in the lower rows. Abbreviations of the measurement are explained in Table 2.
Fig. 2.
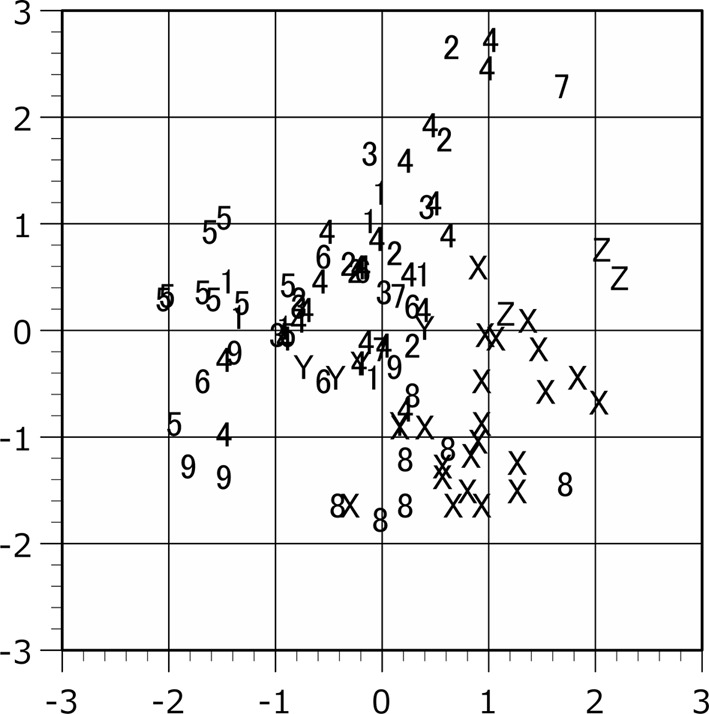
Scattergram of the plots in the principal component analysis. The raw measurement values of male are applied to the analysis. Plot symbols indicate the locality of the specimen as shown in Table 1. Horizontal axis, the first principal component. Vertical axis, the second principal component.
Fig. 3.
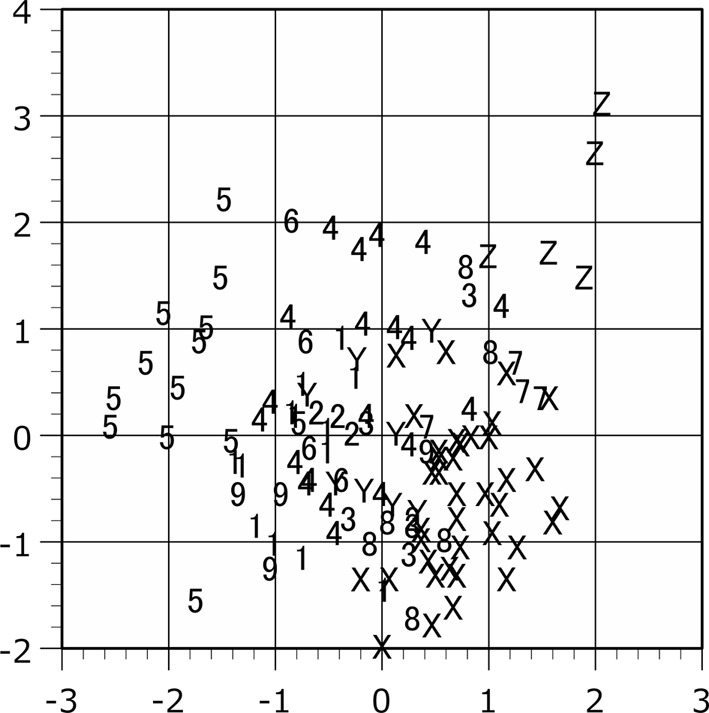
Scattergram of the plots in the principal component analysis. The raw measurement values of female are applied to the analysis. Plot symbols indicate the locality of the specimen as shown in Table 1. Horizontal axis, the first principal component. Vertical axis, the second principal component.
Table 5. Character loading factors observed from the principal component analysis.
| PL | CL | SL | ZW | LBO | GNB | MPL | LBS | DL | GPB | GMB | HAB | LC | LA | LIA | HR | HM | p.v.e. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male, measurement raw values | ||||||||||||||||||
| PC1 | 0.964 | 0.954 | 0.836 | 0.838 | 0.727 | 0.672 | 0.914 | 0.625 | 0.927 | 0.708 | 0.663 | 0.760 | 0.964 | 0.947 | 0.908 | 0.823 | 0.414 | 66.6 |
| PC2 | −0.137 | −0.052 | –0.410 | 0.315 | –0.024 | –0.222 | –0.333 | 0.518 | –0.244 | 0.314 | 0.316 | 0.029 | –0.057 | 0.010 | –0.184 | 0.124 | 0.702 | 9.0 |
| PC3 | 0.021 | –0.023 | –0.211 | –0.105 | –0.312 | 0.552 | –0.089 | 0.148 | –0.074 | 0.210 | 0.426 | 0.325 | –0.134 | –0.089 | –0.075 | –0.062 | –0.405 | 6.0 |
| Female, measurement raw values | ||||||||||||||||||
| PC1 | 0.971 | 0.975 | 0.882 | 0.859 | 0.831 | 0.800 | 0.936 | 0.805 | 0.951 | 0.790 | 0.699 | 0.796 | 0.961 | 0.953 | 0.886 | 0.778 | 0.429 | 72.5 |
| PC2 | –0.093 | –0.041 | –0.182 | 0.165 | 0.008 | –0.207 | –0.146 | 0.157 | –0.114 | 0.193 | –0.268 | 0.008 | –0.028 | 0.008 | –0.154 | 0.334 | 0.822 | 6.4 |
| PC3 | –0.095 | –0.058 | –0.328 | 0.250 | –0.008 | 0.316 | –0.202 | 0.251 | –0.165 | 0.408 | 0.346 | 0.062 | –0.134 | –0.083 | –0.233 | 0.010 | –0.199 | 4.8 |
p.v.e., percentage of the variation explained by each factor. Abbreviations of the measurement are explained in Table 2.
Fig. 4.
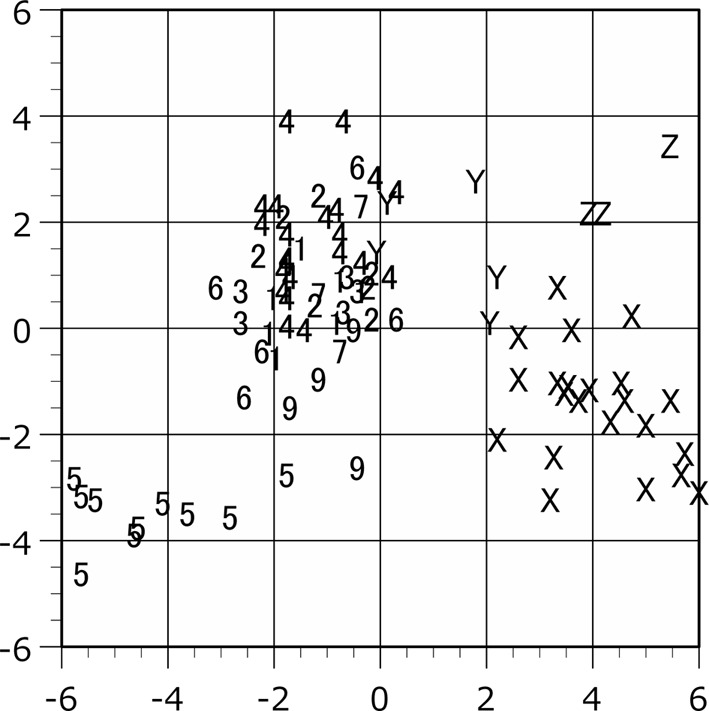
Scattergram of the plots in the canonical discriminant analysis. The raw measurement values of male are applied to the analysis. The individual scores are shown on the discriminant axes 1 (horizontal) and 2 (vertical). Plot symbols indicate the locality of the specimen as shown in Table 1.
Fig. 5.
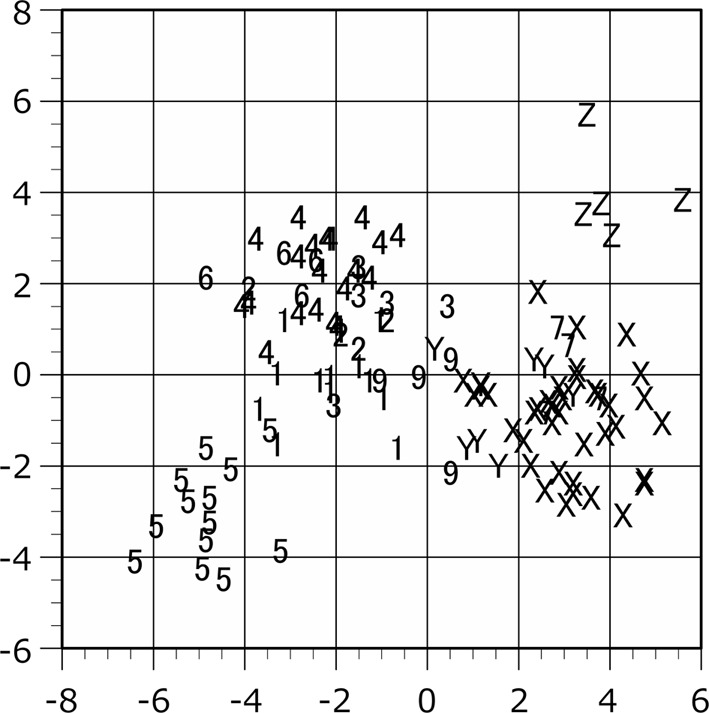
Scattergram of the plots in the canonical discriminant analysis. The raw measurement values of female are applied to the analysis. The individual scores are shown on the discriminant axes 1 (horizontal) and 2 (vertical). Plot symbols indicate the locality of the specimen as shown in Table 1.
Fig. 6.
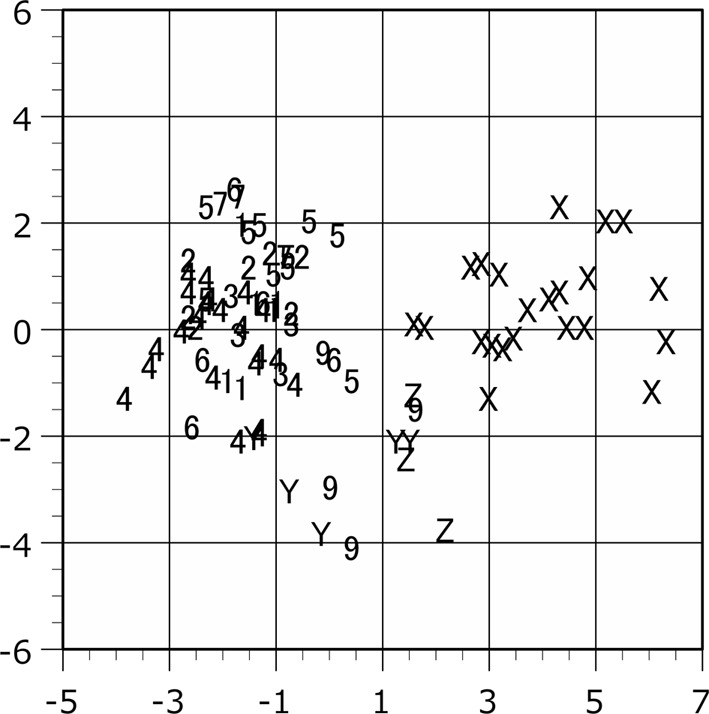
Scattergram of the plots in the canonical discriminant analysis. The proportion indices of male are applied to the analysis. The individual scores are shown on the discriminant axes 1 (horizontal) and 2 (vertical). Plot symbols indicate the locality of the specimen as shown in Table 1.
Fig. 7.
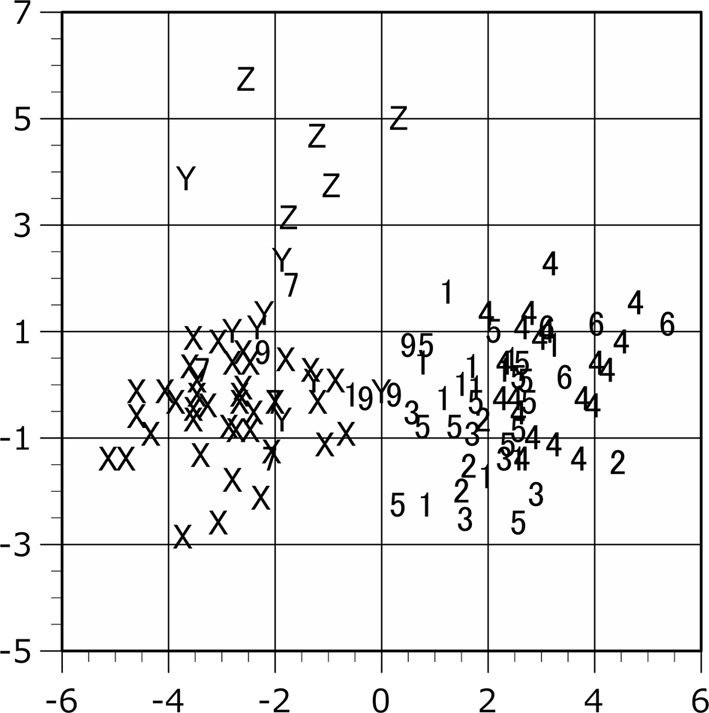
Scattergram of the plots in the canonical discriminant analysis. The proportion indices of female are applied to the analysis. The individual scores are shown on the discriminant axes 1 (horizontal) and 2 (vertical). Plot symbols indicate the locality of the specimen as shown in Table 1.
Table 6. Coefficients in each function of the canonical discriminant analysis.
| PL | CL | SL | ZW | LBO | GNB | MPL | LBS | DL | GPB | GMB | HAB | LC | LA | LIA | HR | HM | e.v. | p.v.e. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male, measurement raw values | |||||||||||||||||||
| CAV1 | 0.159 | –0.833 | 1.014 | –0.577 | 0.173 | 0.352 | 0.563 | 0.325 | 0.312 | 0.162 | 0.178 | 0.182 | –0.004 | –0.264 | 0.183 | –0.416 | –0.327 | 8.7 | 57.0 |
| CAV2 | 0.231 | 1.068 | –0.898 | –0.462 | –0.228 | 0.475 | 0.303 | 0.307 | –0.011 | 0.589 | 0.224 | –0.115 | –0.789 | 0.100 | –0.260 | 0.132 | 0.163 | 3.5 | 22.8 |
| CAV3 | –0.058 | –0.435 | 1.120 | –0.386 | –0.136 | 0.603 | 0.671 | 0.787 | –0.894 | 0.130 | –0.545 | 0.008 | –1.267 | –0.464 | 0.178 | 0.584 | 0.571 | 1.2 | 7.8 |
| Female, measurement raw values | |||||||||||||||||||
| CAV1 | –0.063 | –1.678 | 1.332 | –0.264 | 0.279 | 0.415 | 0.885 | 0.627 | –0.047 | 0.208 | –0.197 | 0.284 | –0.051 | –0.364 | 0.238 | 0.184 | –0.255 | 9.8 | 58.9 |
| CAV2 | 0.434 | 1.762 | –1.118 | –0.138 | 0.136 | 0.254 | –0.770 | –0.121 | –0.331 | 0.526 | –0.272 | 0.099 | –0.512 | 0.321 | 0.106 | 0.135 | 0.121 | 3.5 | 21.1 |
| CAV3 | 0.743 | –0.336 | –0.452 | –0.477 | 0.256 | –0.290 | 0.057 | –0.156 | 0.230 | 0.269 | 1.086 | –0.135 | –0.425 | –0.178 | 0.509 | –0.197 | –0.376 | 1.3 | 7.6 |
| Male, proportion indices | |||||||||||||||||||
| CAV1 | 0.344 | –0.398 | 1.558 | 0.439 | 1.225 | 0.843 | 0.731 | 0.870 | 0.639 | 0.597 | 0.640 | 0.971 | 0.513 | 0.330 | 0.750 | 0.941 | 2.105 | 6.4 | 63.6 |
| CAV2 | 0.151 | 0.379 | –0.179 | 0.670 | 0.780 | –0.420 | –0.192 | –0.239 | 0.639 | 0.110 | 0.742 | 0.478 | 0.774 | 0.338 | 0.307 | 0.177 | 0.867 | 1.4 | 14.1 |
| CAV3 | 0.493 | –0.034 | 0.366 | 0.649 | 0.124 | 0.362 | –0.139 | 0.041 | 0.103 | –0.241 | 0.610 | 0.442 | –0.533 | 0.439 | 0.276 | 0.186 | 0.345 | 0.71 | 7.1 |
| Female, proportion indices | |||||||||||||||||||
| CAV1 | 0.434 | 1.349 | –0.711 | 0.543 | 0.438 | 0.257 | –0.329 | –0.048 | 0.191 | 0.531 | 0.580 | 0.359 | 0.134 | 0.483 | 0.448 | 0.598 | 1.444 | 7.3 | 64.1 |
| CAV2 | 0.000 | 0.660 | 0.702 | 0.701 | 0.598 | 1.066 | 0.285 | 0.691 | 0.284 | 0.633 | –0.248 | 0.680 | 0.195 | 0.626 | 0.158 | 1.081 | 1.736 | 1.4 | 12.2 |
| CAV3 | 0.580 | –0.034 | 0.321 | 0.335 | 1.115 | 0.438 | 0.470 | 0.720 | 0.722 | 0.426 | 0.493 | 0.325 | 0.854 | 0.649 | 0.510 | 0.936 | 2.174 | 0.90 | 7.9 |
e.v., eigen values; p.v.e., percentage of the variation explained by each factor. Abbreviations of the measurement are explained in Table 2.
The population plots clearly indicated the function of zoogeographical barriers on skull morphology, particularly within the Isthmus of Kra and Kangar-Pattani Line as shown in our previous studies [12,13,14]. In both principal component and canonical discriminant analyses, the specimens from Peninsular Malaysia were distinguished from the northern population plots in almost cases (Figs. 2–7). The principal component analyses displayed a weak divide between Peninsular Malaysia specimens and those from other populations; however, morphological differences between populations were clearly discerned (Figs. 2 and 3). Particularly in males, specimens from Peninsular Malaysia plots were distinct in the canonical discriminant analysis (Figs. 4 and 6), whereas a few plots of Chanthaburi, Xiengkhuang of Laos and North Vietnam were exceptionally intermingled with those of Peninsular Malaysia in case of females (Figs. 5 and 7).
The tendency that Z population plots from South Vietnam were segregated and distinguished from the other plots is noticeable (Figs. 3, 4, 5, 6, 7), whereas the Y plots from North Vietnam could not be clearly separated from the other populations. The results of t-test in raw data (Table 7) and U-test in proportion indices (Table 8) were shown with Kruskal-Wallis test results. Based on the number of the measurements showing statistical differences, the Z population was strongly separated from the other populations using the raw data except for population of Locality 7 (Table 7).
Table 7. Significance probability of raw data in measurements between populations related to Vietnam localities.
| PL | CL | SL | ZW | LBO | GNB | MPL | LBS | DL | GPB | GMB | HAB | LC | LA | LIA | HR | HM | Total* | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | |||||||||||||||||||
| Kruskal- Wallis test | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.197 | 16 | |
| t-test | |||||||||||||||||||
| Y-1 | 0.037 | 0.266 | 0.106 | 0.186 | 0.444 | 0.033 | 0.133 | 0.631 | 0.658 | 0.585 | 0.984 | 0.106 | 0.783 | 0.548 | 0.546 | 0.465 | 0.453 | 2 | |
| Y-2 | 0.662 | 0.548 | 0.112 | 0.022 | 0.181 | 0.137 | 0.208 | 0.465 | 0.834 | 0.367 | 0.013 | 0.751 | 0.212 | 0.265 | 0.121 | 0.707 | 0.167 | 2 | |
| Y-3 | 0.048 | 0.281 | 0.179 | 0.289 | 0.103 | 0.057 | 0.107 | 0.974 | 0.983 | 0.711 | 0.950 | 0.848 | 0.748 | 0.499 | 0.945 | 0.897 | 0.182 | 1 | |
| Y-4 | 0.500 | 0.950 | 0.014 | 0.207 | 0.951 | 0.120 | 0.128 | 0.522 | 0.997 | 0.287 | 0.810 | 0.529 | 0.474 | 0.511 | 0.387 | 0.943 | 0.155 | 1 | |
| Y-5 | 0.000 | 0.000 | 0.002 | 0.425 | 0.556 | 0.000 | 0.000 | 0.011 | 0.001 | 0.001 | 0.000 | 0.001 | 0.020 | 0.004 | 0.016 | 0.007 | 0.100 | 14 | |
| Y-6 | 0.257 | 0.159 | 0.044 | 0.668 | 0.746 | 0.055 | 0.145 | 0.547 | 0.545 | 0.491 | 0.751 | 0.628 | 0.657 | 0.692 | 0.467 | 0.956 | 0.890 | 1 | |
| Y-7 | 0.285 | 0.331 | 0.520 | 0.016 | 0.092 | 0.269 | 0.746 | 0.231 | 0.329 | 0.678 | 0.243 | 0.280 | 0.053 | 0.068 | 0.044 | 0.423 | 0.213 | 2 | |
| Y-9 | 0.003 | 0.007 | 0.113 | 0.476 | 0.008 | 0.139 | 0.015 | 0.009 | 0.049 | 0.084 | 0.011 | 0.154 | 0.009 | 0.009 | 0.034 | 0.023 | 0.150 | 11 | |
| Y-X | 0.001 | 0.009 | 0.000 | 0.001 | 0.008 | 0.750 | 0.001 | 0.976 | 0.000 | 0.518 | 0.213 | 0.088 | 0.000 | 0.000 | 0.000 | 0.079 | 0.364 | 10 | |
| Y-Z | 0.006 | 0.005 | 0.004 | 0.003 | 0.000 | 0.152 | 0.010 | 0.022 | 0.006 | 0.004 | 0.421 | 0.345 | 0.002 | 0.003 | 0.004 | 0.009 | 0.024 | 14 | |
| Z-1 | 0.001 | 0.001 | 0.000 | 0.022 | 0.001 | 0.009 | 0.002 | 0.002 | 0.001 | 0.002 | 0.673 | 0.024 | 0.002 | 0.003 | 0.002 | 0.031 | 0.067 | 15 | |
| Z-2 | 0.003 | 0.000 | 0.000 | 0.170 | 0.001 | 0.010 | 0.001 | 0.059 | 0.001 | 0.001 | 0.087 | 0.359 | 0.001 | 0.010 | 0.000 | 0.022 | 0.130 | 12 | |
| Z-3 | 0.001 | 0.000 | 0.000 | 0.067 | 0.003 | 0.019 | 0.001 | 0.002 | 0.001 | 0.010 | 0.388 | 0.346 | 0.001 | 0.019 | 0.009 | 0.045 | 0.422 | 13 | |
| Z-4 | 0.000 | 0.000 | 0.000 | 0.013 | 0.004 | 0.008 | 0.000 | 0.033 | 0.000 | 0.000 | 0.546 | 0.063 | 0.000 | 0.007 | 0.002 | 0.004 | 0.171 | 14 | |
| Z-5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.074 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.001 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.041 | 16 | |
| Z-6 | 0.008 | 0.001 | 0.000 | 0.038 | 0.014 | 0.019 | 0.006 | 0.004 | 0.002 | 0.000 | 0.625 | 0.252 | 0.003 | 0.006 | 0.002 | 0.073 | 0.025 | 14 | |
| Z-7 | 0.112 | 0.047 | 0.003 | 0.646 | 0.496 | 0.065 | 0.035 | 0.149 | 0.040 | 0.102 | 0.546 | 0.896 | 0.137 | 0.309 | 0.137 | 0.183 | 0.563 | 4 | |
| Z-9 | 0.001 | 0.000 | 0.000 | 0.004 | 0.000 | 0.042 | 0.001 | 0.000 | 0.000 | 0.001 | 0.025 | 0.071 | 0.000 | 0.001 | 0.002 | 0.002 | 0.002 | 16 | |
| Z-X | 0.070 | 0.005 | 0.361 | 0.109 | 0.479 | 0.006 | 0.194 | 0.001 | 0.026 | 0.000 | 0.575 | 0.959 | 0.191 | 0.268 | 0.400 | 0.122 | 0.023 | 6 | |
| Female | |||||||||||||||||||
| Kruskal- Wallis test | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.030 | 17 | |
| t-test | |||||||||||||||||||
| Y-1 | 0.000 | 0.002 | 0.000 | 0.675 | 0.002 | 0.026 | 0.001 | 0.000 | 0.000 | 0.475 | 0.648 | 0.524 | 0.060 | 0.023 | 0.133 | 0.089 | 0.156 | 9 | |
| Y-2 | 0.384 | 0.389 | 0.007 | 0.792 | 0.407 | 0.344 | 0.178 | 0.156 | 0.676 | 0.114 | 0.072 | 0.210 | 0.969 | 0.516 | 0.704 | 0.120 | 0.966 | 1 | |
| Y-3 | 0.881 | 0.512 | 0.039 | 0.056 | 0.810 | 0.150 | 0.815 | 0.660 | 0.256 | 0.015 | 0.005 | 0.363 | 0.202 | 0.594 | 0.127 | 0.436 | 0.688 | 3 | |
| Y-4 | 0.243 | 0.288 | 0.000 | 0.599 | 0.723 | 0.018 | 0.014 | 0.452 | 0.156 | 0.169 | 0.511 | 0.897 | 0.899 | 0.717 | 0.664 | 0.614 | 0.523 | 3 | |
| Y-5 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.851 | 16 | |
| Y-6 | 0.008 | 0.041 | 0.000 | 0.882 | 0.670 | 0.136 | 0.001 | 0.282 | 0.004 | 0.650 | 0.966 | 0.832 | 0.056 | 0.189 | 0.059 | 0.290 | 0.970 | 5 | |
| Y-7 | 0.002 | 0.006 | 0.085 | 0.005 | 0.007 | 0.959 | 0.000 | 0.007 | 0.008 | 0.004 | 0.044 | 0.056 | 0.002 | 0.001 | 0.005 | 0.019 | 0.119 | 13 | |
| Y-9 | 0.040 | 0.087 | 0.022 | 0.793 | 0.186 | 0.090 | 0.061 | 0.035 | 0.053 | 0.625 | 0.756 | 0.653 | 0.062 | 0.050 | 0.137 | 0.262 | 0.031 | 5 | |
| Y-X | 0.000 | 0.001 | 0.000 | 0.008 | 0.000 | 0.259 | 0.000 | 0.643 | 0.000 | 0.092 | 0.000 | 0.008 | 0.000 | 0.000 | 0.000 | 0.133 | 0.987 | 12 | |
| Y-Z | 0.000 | 0.000 | 0.007 | 0.000 | 0.000 | 0.000 | 0.002 | 0.000 | 0.001 | 0.000 | 0.974 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 16 | |
| Z-1 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.647 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 16 | |
| Z-2 | 0.002 | 0.001 | 0.001 | 0.001 | 0.001 | 0.000 | 0.008 | 0.000 | 0.007 | 0.001 | 0.046 | 0.000 | 0.001 | 0.000 | 0.014 | 0.003 | 0.000 | 17 | |
| Z-3 | 0.001 | 0.003 | 0.001 | 0.001 | 0.004 | 0.000 | 0.011 | 0.001 | 0.021 | 0.013 | 0.003 | 0.000 | 0.001 | 0.001 | 0.061 | 0.003 | 0.003 | 16 | |
| Z-4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.553 | 0.000 | 0.000 | 0.000 | 0.001 | 0.000 | 0.001 | 16 | |
| Z-5 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.021 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 17 | |
| Z-6 | 0.000 | 0.000 | 0.000 | 0.001 | 0.001 | 0.000 | 0.001 | 0.002 | 0.000 | 0.000 | 0.936 | 0.000 | 0.000 | 0.000 | 0.002 | 0.005 | 0.002 | 16 | |
| Z-7 | 0.215 | 0.166 | 0.813 | 0.091 | 0.021 | 0.000 | 0.980 | 0.008 | 0.657 | 0.047 | 0.057 | 0.270 | 0.227 | 0.124 | 0.619 | 0.031 | 0.027 | 6 | |
| Z-9 | 0.001 | 0.001 | 0.001 | 0.003 | 0.001 | 0.000 | 0.006 | 0.000 | 0.003 | 0.001 | 0.780 | 0.001 | 0.001 | 0.001 | 0.006 | 0.011 | 0.000 | 16 | |
| Z-X | 0.014 | 0.000 | 0.481 | 0.001 | 0.079 | 0.000 | 0.070 | 0.000 | 0.104 | 0.000 | 0.001 | 0.003 | 0.005 | 0.001 | 0.632 | 0.000 | 0.000 | 12 | |
The values showing significant differences (P<0.05) are underlined. *Total number of signigficantly different measurements. Abbreviations of the measurement are explained in Table 2.
Table 8. Significance probability of proportion indices in measurements between populations related to Vietnam localities.
| PL | CL | SL | ZW | LBO | GNB | MPL | LBS | DL | GPB | GMB | HAB | LC | LA | LIA | HR | HM | Total* | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | |||||||||||||||||||
| Kruskal- Wallis test | 0.008 | 0.265 | 0.000 | 0.001 | 0.005 | 0.003 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.075 | 0.000 | 0.059 | 0.026 | 0.906 | 0.005 | 13 | |
| U-test | |||||||||||||||||||
| Y-1 | 0.010 | 0.530 | 0.730 | 0.005 | 1.000 | 0.202 | 0.048 | 0.876 | 0.755 | 0.876 | 0.755 | 0.432 | 0.202 | 0.755 | 0.876 | 0.755 | 0.202 | 3 | |
| Y-2 | 0.010 | 0.018 | 0.010 | 0.005 | 0.003 | 0.149 | 0.018 | 0.530 | 0.639 | 1.000 | 0.048 | 1.000 | 0.149 | 0.343 | 0.639 | 0.876 | 0.149 | 7 | |
| Y-3 | 0.056 | 0.310 | 0.095 | 0.095 | 0.008 | 0.095 | 0.008 | 1.000 | 1.000 | 0.841 | 1.000 | 0.841 | 0.548 | 0.310 | 0.841 | 0.222 | 0.222 | 2 | |
| Y-4 | 0.033 | 0.485 | 0.000 | 0.064 | 0.739 | 0.165 | 0.016 | 0.344 | 0.524 | 0.976 | 0.377 | 0.377 | 0.165 | 0.284 | 0.344 | 0.928 | 0.075 | 3 | |
| Y-5 | 0.008 | 0.013 | 0.040 | 0.001 | 0.859 | 0.013 | 0.005 | 0.953 | 0.075 | 0.679 | 0.594 | 0.099 | 0.055 | 0.165 | 0.099 | 0.679 | 0.005 | 7 | |
| Y-6 | 0.841 | 0.222 | 0.016 | 0.151 | 0.841 | 0.548 | 0.151 | 0.690 | 0.548 | 1.000 | 1.000 | 0.841 | 0.310 | 0.548 | 0.841 | 0.548 | 0.690 | 1 | |
| Y-7 | 0.143 | 0.250 | 0.036 | 0.036 | 0.071 | 0.143 | 0.250 | 1.000 | 0.786 | 1.000 | 0.250 | 1.000 | 0.071 | 0.071 | 0.393 | 1.000 | 0.393 | 2 | |
| Y-9 | 1.000 | 0.556 | 0.730 | 0.032 | 0.556 | 0.413 | 1.000 | 0.413 | 0.730 | 1.000 | 1.000 | 0.190 | 0.730 | 0.413 | 1.000 | 0.730 | 0.413 | 1 | |
| Y-X | 0.146 | 0.055 | 0.001 | 0.086 | 0.314 | 0.008 | 0.232 | 0.002 | 0.001 | 0.284 | 0.019 | 0.564 | 0.000 | 0.023 | 0.005 | 0.880 | 0.880 | 8 | |
| Y-Z | 0.036 | 0.393 | 0.036 | 0.393 | 0.786 | 0.143 | 1.000 | 0.786 | 0.036 | 0.786 | 0.036 | 0.036 | 0.250 | 0.786 | 0.393 | 0.393 | 0.143 | 5 | |
| Z-1 | 0.833 | 0.833 | 0.017 | 0.067 | 0.833 | 0.183 | 0.183 | 1.000 | 0.117 | 0.383 | 0.017 | 0.017 | 0.667 | 1.000 | 0.517 | 0.517 | 0.383 | 3 | |
| Z-2 | 0.517 | 0.383 | 0.017 | 0.067 | 0.067 | 0.267 | 0.033 | 0.383 | 0.033 | 0.517 | 0.017 | 0.017 | 1.000 | 0.267 | 0.833 | 0.267 | 0.267 | 5 | |
| Z-3 | 0.786 | 0.786 | 0.036 | 0.393 | 0.250 | 0.571 | 0.036 | 0.571 | 0.250 | 0.571 | 0.036 | 0.036 | 0.571 | 0.571 | 0.571 | 1.000 | 1.000 | 4 | |
| Z-4 | 0.398 | 0.606 | 0.001 | 0.398 | 0.844 | 0.206 | 0.072 | 0.206 | 0.046 | 0.783 | 0.006 | 0.058 | 0.783 | 0.311 | 1.000 | 0.238 | 0.723 | 3 | |
| Z-5 | 1.000 | 0.287 | 0.007 | 0.028 | 0.937 | 1.000 | 0.028 | 0.692 | 0.007 | 0.007 | 0.014 | 0.161 | 0.371 | 0.217 | 0.811 | 0.161 | 0.573 | 6 | |
| Z-6 | 0.143 | 0.571 | 0.036 | 0.571 | 0.786 | 0.393 | 0.250 | 1.000 | 0.071 | 0.036 | 0.036 | 0.143 | 0.393 | 0.571 | 0.786 | 1.000 | 0.143 | 3 | |
| Z-7 | 1.000 | 1.000 | 0.100 | 0.100 | 0.400 | 1.000 | 0.400 | 1.000 | 0.100 | 0.100 | 0.100 | 0.100 | 0.400 | 0.100 | 1.000 | 0.400 | 0.700 | 0 | |
| Z-9 | 0.057 | 0.857 | 0.057 | 0.114 | 0.857 | 0.057 | 0.857 | 1.000 | 0.229 | 0.400 | 0.057 | 0.057 | 0.229 | 0.629 | 1.000 | 0.229 | 0.057 | 0 | |
| Z-X | 0.058 | 0.723 | 0.311 | 0.906 | 0.353 | 0.969 | 0.206 | 0.058 | 0.844 | 0.002 | 0.046 | 0.058 | 0.027 | 0.058 | 0.089 | 0.497 | 0.046 | 4 | |
| Female | |||||||||||||||||||
| Kruskal- Wallis test | 0.005 | 0.455 | 0.000 | 0.023 | 0.008 | 0.001 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.003 | 0.001 | 0.144 | 0.013 | 0.212 | 0.000 | 14 | |
| U-test | |||||||||||||||||||
| Y-1 | 0.140 | 0.268 | 0.000 | 0.089 | 0.210 | 0.336 | 0.185 | 0.076 | 0.064 | 0.140 | 0.010 | 0.121 | 0.210 | 0.697 | 0.750 | 0.750 | 0.414 | 2 | |
| Y-2 | 1.000 | 0.683 | 0.008 | 0.368 | 0.808 | 1.000 | 0.154 | 0.283 | 0.808 | 0.028 | 0.004 | 0.683 | 0.214 | 0.808 | 0.109 | 0.154 | 0.933 | 2 | |
| Y-3 | 0.524 | 0.833 | 0.011 | 0.354 | 0.622 | 0.093 | 0.127 | 0.524 | 0.127 | 0.030 | 0.011 | 0.833 | 0.065 | 1.000 | 0.284 | 0.284 | 0.435 | 3 | |
| Y-4 | 0.354 | 0.438 | 0.000 | 0.281 | 0.281 | 0.182 | 0.001 | 0.709 | 0.070 | 0.063 | 0.150 | 0.409 | 0.218 | 0.049 | 0.099 | 0.823 | 0.354 | 3 | |
| Y-5 | 0.037 | 0.025 | 0.000 | 0.064 | 0.804 | 0.020 | 0.025 | 0.104 | 0.121 | 0.500 | 0.104 | 0.301 | 0.003 | 0.374 | 0.020 | 0.595 | 0.030 | 8 | |
| Y-6 | 0.214 | 0.461 | 0.004 | 0.073 | 0.154 | 0.683 | 0.004 | 0.933 | 0.028 | 0.073 | 0.283 | 0.214 | 0.683 | 0.048 | 0.461 | 0.368 | 0.683 | 4 | |
| Y-7 | 0.214 | 0.808 | 0.461 | 0.933 | 0.214 | 0.048 | 0.808 | 0.214 | 0.368 | 0.808 | 1.000 | 1.000 | 0.109 | 0.214 | 0.461 | 0.461 | 0.461 | 1 | |
| Y-9 | 0.933 | 0.808 | 0.073 | 0.109 | 0.283 | 0.368 | 0.368 | 0.461 | 0.283 | 0.368 | 0.376 | 0.028 | 0.283 | 0.154 | 0.808 | 1.000 | 0.073 | 1 | |
| Y-X | 0.368 | 0.485 | 0.013 | 0.943 | 0.046 | 0.008 | 0.090 | 0.000 | 0.004 | 0.146 | 0.989 | 0.744 | 0.002 | 0.138 | 0.001 | 0.172 | 0.191 | 7 | |
| Y-Z | 0.019 | 0.284 | 0.435 | 0.524 | 0.622 | 0.065 | 0.354 | 0.093 | 0.724 | 0.093 | 0.011 | 0.524 | 0.284 | 0.724 | 0.435 | 0.045 | 0.019 | 4 | |
| Z-1 | 0.117 | 1.000 | 0.010 | 0.387 | 0.143 | 0.026 | 0.849 | 0.503 | 0.566 | 0.924 | 0.000 | 0.010 | 0.443 | 1.000 | 0.289 | 0.095 | 0.004 | 5 | |
| Z-2 | 0.111 | 0.905 | 0.032 | 1.000 | 0.730 | 0.190 | 0.730 | 0.905 | 0.413 | 0.556 | 0.016 | 1.000 | 0.286 | 0.556 | 0.063 | 0.032 | 0.032 | 4 | |
| Z-3 | 0.421 | 0.421 | 0.056 | 0.690 | 0.222 | 0.841 | 0.841 | 0.548 | 0.222 | 0.310 | 0.008 | 0.421 | 0.222 | 0.421 | 0.095 | 0.032 | 0.095 | 2 | |
| Z-4 | 0.060 | 0.869 | 0.000 | 0.974 | 0.668 | 0.408 | 0.447 | 0.272 | 0.575 | 0.974 | 0.000 | 0.216 | 1.000 | 0.083 | 0.051 | 0.051 | 0.060 | 2 | |
| Z-5 | 0.387 | 0.566 | 0.004 | 0.289 | 0.443 | 0.703 | 0.633 | 0.924 | 0.633 | 0.035 | 0.000 | 0.075 | 0.046 | 0.633 | 0.019 | 0.019 | 0.703 | 6 | |
| Z-6 | 0.556 | 0.730 | 0.016 | 0.190 | 0.286 | 0.190 | 0.190 | 0.730 | 0.286 | 0.730 | 0.016 | 0.190 | 0.286 | 0.032 | 0.190 | 0.413 | 0.286 | 3 | |
| Z-7 | 0.286 | 0.730 | 0.556 | 0.556 | 0.063 | 0.286 | 0.286 | 0.413 | 0.413 | 0.286 | 0.016 | 0.730 | 0.190 | 0.413 | 0.286 | 0.032 | 0.032 | 3 | |
| Z-9 | 0.063 | 0.556 | 0.413 | 0.286 | 0.556 | 0.111 | 1.000 | 1.000 | 1.000 | 0.413 | 0.036 | 0.032 | 0.286 | 0.286 | 0.556 | 0.286 | 0.016 | 3 | |
| Z-X | 0.001 | 0.543 | 0.006 | 0.211 | 0.128 | 0.672 | 0.059 | 0.014 | 0.036 | 0.004 | 0.000 | 0.811 | 0.008 | 0.472 | 0.001 | 0.005 | 0.000 | 10 | |
The values showing significant differences (P<0.05) are underlined. *Total number of signigficantly different measurements. Abbreviations of the measurement are explained in Table 2.
The raw data indicated the significantly larger size of Z population than the other populations (Tables 3 and 7). The mean values of the raw data were largest in the Z population compared to those in the other populations except for GMB and HAM in male and GMB and MPL in female. In the U-test using the proportion index, the measurements showing significant differences decreased (Table 8). However, the Z plots were clearly separated at least in female in both raw value and proportion index by canonical discriminant analyses (Figs. 5 and 7).
The skull size of Y population were obviously smaller than that of X and Z populations and larger than 5 population (Tables 3 and 7). The significantly different measurements were not few between Y and 5, and Y and X populations in proportional analysis, however, we suggest that the morphological characteristics of Y skulls are not distinguishable from those of the other populations using the proportion index (Tables 4 and 8). In case of PL, SL, GNB and MPL, only Z, X, 7 or 8 population could show larger mean value of raw data than Y population. With or without statistically significant differences, these measurements showed the relatively larger mean values in the Y population than in the 1–6 populations from Thailand (Tables 3 and 7). In contrast, ZW and GPB related to the width of the mouth were relatively smaller in the Y population compared to those in the other populations (Tables 3 and 7).
DISCUSSION
The genus Tupaia consists of many species, subspecies and local populations with morphological and genetic variations affected by various zoogeographical factors such as mountains, rivers, isthmus, and islands [21, 22]. The glis-belangeri complex is especially noteworthy. These two species known to be the most closely-related based on both morphological and molecular data [29, 34, 36], one report even noted that T. belangeri population may be paraphyletic to T. glis [35].
South Vietnam (Locality Z: Khanh Hoa, Dong Nai) is the most noteworthy locality in these analyses. The plots from South Vietnam are completely distinguished from the other T. belangeri plots of Indochinese region in both size-included analyses and size-omitted analysis in females (Figs. 2–5 and 7). Since even the principal component analysis can separate the plots of South Vietnam from the other population, the osteometrical characteristics of the skull from South Vietnam locality can be clearly identified.
The Z plots are located in the positive area of both x- and y-axes in the scattergrams of the principal component analyses as shown in Figs. 2 and 3. From the character loading factor (Table 5), all of the PC1 are positive, the x-axis is influenced by body size. From these data, we observed that the skulls of Z population are larger in size. The absolute values of the character loading factor of the PC2 are larger in HM and ZW. The measurements of the HN and ZW may be morphologically related to the function of the mastication [23]. The female skulls of the Z population seem relatively larger in PC2 (Fig. 3). At least in female, we suggest that the chewing movement may induce the development of the mastication-related osteometrical characteristics and have influence on the morphological identification of skull in the Z population. The larger sample size will be needed to detail the differences of PC2 scores between sexes in Z population. In the Y population, the larger SL and MPL and smaller ZW and GPB suggest the rostro-caudally longer and bilaterally narrower mouth in facial region. We suggest that it may reflect the characteristics of mastication and diet in the Y population.
It has been reported that South Vietnam including the Lower Mekong District may contain a complicated transitional area of various taxa and serve as a zoogeographical barrier at least in amphibians [20]. A latitude of from 14 to 17-degrees north in central Vietnam was considered as a barrier on the primate species [19]. Futhermore, morphological discontinuities at the species or population level in various taxa have been indicated between the northern Plateau area and southern Lower Mekong [1, 2, 7, 28]. From these present data, it is important to note the taxonomical relationships between North Vietnam and South Vietnam populations in the case of the northern treeshrew.
The principal component scores of Y population from North Vietnam were not separated from the other populations in the principal component analysis (Figs. 2 and 3). The Y population plots are not randomly mixed with Thailand plots segregation in the canonical discriminant analysis (Figs. 4, 5, 6, 7). The non-parametric U-test demonstrated that the measurements showing statistical differences of skull proportion between Y and 7, Y and 9, Z and 7, Z and 9 are few at least in male (Table 8). The Y and Z localities of Vietnam and Site 9 of Xiengkhuang in Laos are on the same side of the present Mekong River. It was suggested that the Mekong River has acted as a zoogeographical barrier at least in small mammals during the Pliocene and Pleistocene [27]. The cases of Callosciurus squirrels [30,31,32], genetic differentiation or speciation has been demonstrated between both sides of this river. In northern treeshrew of North Vietnam population, however, the t-test using raw data demonstrated that the total number of the siginificantly different measurements were few between Y and 2, Y and 3, and Y and 4 in both sexes and Y and 7 in male (Table 7). Since these sites are located on both sides across the Mekong River, the zoogeographical barrier of the Mekong River is not consistently confirmed. The evolutionary effect of the Mekong River should be examined in more detailed in the future.
A previous study described the evaluation of the treeshrew manus using X –ray imaging [39]. This report demonstrated that non-adult specimens show lower values of adult range in osteological measurements, and suggested that subadults should be excluded from the analyses or else used with appropriate caution. This study included subadult skulls with complete erupted molars, as our study sample would have been more restricted without these subadult data. We suggest further osteometrical examinations of skulls to determine whether age is a hinderance in continuous morphological data.
POTENTIAL CONFLICTS OF INTEREST
The authors have nothing to disclose.
Supplemental Material
Acknowledgments
This study was financially supported by JSPS KAKENHI Grant Numbers 18H03602, 19H00534, 20H01381, 20H01979 and 21K00985, and JSPS Bilateral Programs (QTJP01.02/18-20) entitled “Advanced establishment of basis of biodiversity informatics in Vietnam”. We were also financially supported by the NAGAO Natural Environmental Foundation’s Bio-ecological Nature Conservation Project in Mountainous Regions of Northern Vietnam. We also thank to the financial support from The program of developing basic sciences in Chemistry, Life sciences, Earth sciences and Marine sciences, Vietnam Academy of Science and Technology (KHCBSS.01/20-22).
REFERENCES
- 1.Averyanov L. V., Loc P. K., Hiep N. T., Harder D. K.2003. Phytogeographic review of Vietnam and adjacent areas of eastern Indochina. Komarovia 3: 1–83. [Google Scholar]
- 2.Bain R. H., Hurley M. M.2011. A biogeographic synthesis of the amphibians and reptiles of Indochina. Bull. Am. Mus. Nat. Hist. 360: 1–138. doi: 10.1206/360.1 [DOI] [Google Scholar]
- 3.Baltzer J. L., Davies S. J., Noor N. S. M., Kassim A. R., Lafrankie J. V.2007. Geographical distributions in tropical trees: can geographical range predict performance and habitat association in co-occurring tree species? J. Biogeogr. 34: 1916–1926. doi: 10.1111/j.1365-2699.2007.01739.x [DOI] [Google Scholar]
- 4.Boontop Y., Kumaran N., Schutze M. K., Clarke A. R., Cameron S. L., Krosch M. N.2017. Population structure in Zeugodacus cucurbitae (Diptera: Tephritidae) across Thailand and the Thai–Malay peninsula: natural barriers to a great disperser. Biol. J. Linn. Soc. Lond. 121: 540–555. doi: 10.1093/biolinnean/blx009 [DOI] [Google Scholar]
- 5.Bunlungsup S., Imai H., Hamada Y., Matsudaira K., Malaivijitnond S.2017a. Mitochondrial DNA and two Y-chromosome genes of common long-tailed macaques (Macaca fascicularis fascicularis) throughout Thailand and vicinity. Am. J. Primatol. 79: 1–13. doi: 10.1002/ajp.22596 [DOI] [PubMed] [Google Scholar]
- 6.Bunlungsup S., Kanthaswamy S., Oldt R. F., Smith D. G., Houghton P., Hamada Y., Malaivijitnond S.2017b. Genetic analysis of samples from wild populations opens new perspectives on hybridization between long-tailed (Macaca fascicularis) and rhesus macaques (Macaca mulatta). Am. J. Primatol. 79: e22726. doi: 10.1002/ajp.22726 [DOI] [PubMed] [Google Scholar]
- 7.Cang D. N., Endo H., Nguyen T. S., Oshida T., Canh L. X., Phuong D. H., Lunde D., Kawada S., Hayashida A., Sasaki M.2008. Checklist of Wild Mammal Species of Vietnam. Shoukadoh, Kyoto. [Google Scholar]
- 8.Corbet A. S., Pendlebury H. M.1992. The Butterflies of the Malay Peninsula. 4th ed., Malayan Nature Society, Kuala Lumpur. [Google Scholar]
- 9.Corbet G. B., Hill J. E.1992. The Mammals of Indomalayan Region. A Systematic Review. Oxford University Press, London. [Google Scholar]
- 10.de Bruyn M., Nugroho E., Hossain M. M., Wilson J. C., Mather P. B.2005. Phylogeographic evidence for the existence of an ancient biogeographic barrier: the Isthmus of Kra Seaway. Heredity 94: 370–378. doi: 10.1038/sj.hdy.6800613 [DOI] [PubMed] [Google Scholar]
- 11.Driesch A.1976. A Guide to the Measurement of Animal Bones from Archaeological Sites, Harvard University Press, Cambridge. [Google Scholar]
- 12.Endo H., Cuisin J., Nadee N., Nabhitabhata J., Suyanto A., Kawamoto Y., Nishida T., Yamada J.1999. Geographical variation of the skull morphology of the common tree shrew (Tupaia glis). J. Vet. Med. Sci. 61: 1027–1031. doi: 10.1292/jvms.61.1027 [DOI] [PubMed] [Google Scholar]
- 13.Endo H., Nishiumi I., Hayashi Y., Rashdi A. B. M., Nadee N., Nabhitabhata J., Kawamoto Y., Kimura J., Nishida T., Yamada J.2000a. Multivariate analysis in skull osteometry of the common tree shrew from both sides of the Isthmus of Kra in Southern Thailand. J. Vet. Med. Sci. 62: 375–378. doi: 10.1292/jvms.62.375 [DOI] [PubMed] [Google Scholar]
- 14.Endo H., Hayashi Y., Rerkamnuaychoke W., Nadee N., Nabhitabhata J., Kawamoto Y., Hirai H., Kimura J., Nishida T., Yamada J.2000b. Sympatric distribution of the two morphological types of the common tree shrew in Hat-Yai Districts (South Thailand). J. Vet. Med. Sci. 62: 759–761. doi: 10.1292/jvms.62.759 [DOI] [PubMed] [Google Scholar]
- 15.Endo H., Kimura J., Oshida T., Stafford B. J., Rerkamuaychoke W., Nishida T., Sasaki M., Hayashida A., Hayashi Y.2003. Geographical variation and its functional significances of skull morphology in the red-cheeked squirrel. J. Vet. Med. Sci. 65: 1179–1183. doi: 10.1292/jvms.65.1179 [DOI] [PubMed] [Google Scholar]
- 16.Endo H., Kimura J., Oshida T., Stafford B. J., Rerkamnuaychoke W., Nishida T., Sasaki M., Hayashida A., Hayashi Y.2004a. Geographical and functional-morphological variations of the skull in the gray-bellied squirrel. J. Vet. Med. Sci. 66: 277–282. doi: 10.1292/jvms.66.277 [DOI] [PubMed] [Google Scholar]
- 17.Endo H., Kimura J., Oshida T., Stafford B. J., Rerkamnuaychoke W., Nishida T., Sasaki M., Hayashida A., Hayashi Y.2004b. Geographical variation of skull size and shape in various populations in the black giant squirrel. J. Vet. Med. Sci. 66: 1213–1218. doi: 10.1292/jvms.66.1213 [DOI] [PubMed] [Google Scholar]
- 18.Endo H., Hayashida A., Fukuta K.2007. Multivariate analyses of the skull size and shape in the five geographical populations of the lesser false vampire. Mammal Study 32: 23–31. doi: 10.3106/1348-6160(2007)32[23:MAOTSS]2.0.CO;2 [DOI] [Google Scholar]
- 19.Fooden J.1996. Zoogeography of Vietnamese primates. Int. J. Primatol. 17: 845–899. doi: 10.1007/BF02735268 [DOI] [Google Scholar]
- 20.Geissler P., Hartmann T., Ihlow F., Rödder D., Poyarkov N. A., Jr, Nguyen T. Q., Ziegler T., Böhme W.2015. The Lower Mekong: an insurmountable barrier to amphibians in southern Indochina? Biol. J. Linn. Soc. Lond. 114: 905–914. doi: 10.1111/bij.12444 [DOI] [Google Scholar]
- 21.Hawkins M. T.2018. Family tupaiidae (Treeshrews). pp. 242–269. In: Handbook of the Mammals of the World, vol. 8. Insectivores, Sloths and Colugos (Wilson, D. E. and Mittermeier, R. A. eds.), Lynx Edicions, Barcelona. [Google Scholar]
- 22.Helgen K. M.2005. Order Scandentia. pp. 104–109. In: Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed. (Wilson, D. E. and Reeder, D. M. eds.), Johns Hopkins University Press, Baltimore. [Google Scholar]
- 23.Hermanson J. W., Evans H. E.1993. The muscular system. pp. 258–384. In: Miller’s Anatomy of the Dog, 3rd ed. (Evans, H. E. ed.), Saunders, Philadelphia. [Google Scholar]
- 24.Hirai H., Hirai Y., Kawamoto Y., Endo H., Kimura J., Rerkamnuaychoke W.2002. Cytogenetic differentiation of two sympatric tree shrew taxa found in the southern part of the Isthmus of Kra. Chromosome Res. 10: 313–327. doi: 10.1023/A:1016523909096 [DOI] [PubMed] [Google Scholar]
- 25.Hughes J. B., Round P. D., Woodruff D. S.2003. The Indochinese-Sundaic faunal transition at the Isthmus of Kra: an analysis of resident forest bird species distributions. J. Biogeogr. 30: 569–580. doi: 10.1046/j.1365-2699.2003.00847.x [DOI] [Google Scholar]
- 26.Lekagul B., Mcneely J. A.1988. Mammals of Thailand, 2nd ed., Saha Karn Bhaet, Co., Bangkok. [Google Scholar]
- 27.Meijaard E., Groves C. P.2006. The geography of mammals and rivers in mainland Southeast Asia. pp. 305–329. In: Primate Biogeography: Progress and Prospects (Lehman, S. M. and Fleagle, J. G. eds.), Springer Science and Business Media, LLC., New York. [Google Scholar]
- 28.Nguyen T. S., Motokawa M., Oshida T., Thong V. D., Csorba G., Endo H.2015. Multivariate analysis of the skull and shape in tube-nosed bats of the genus Murina (Chiroptera: Vespertilionidae) from Vietnam. Mammal Study 40: 79–94. doi: 10.3106/041.040.0203 [DOI] [Google Scholar]
- 29.Olson L. E., Sargis E. J., Martin R. D.2005. Intraordinal phylogenetics of treeshrews (Mammalia: Scandentia) based on evidence from the mitochondrial 12S rRNA gene. Mol. Phylogenet. Evol. 35: 656–673. doi: 10.1016/j.ympev.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 30.Oshida T., Yasuda M., Endo H., Hussein N. A., Masuda R.2001. Molecular phylogeny of five squirrel species of the genus Callosciurus (Mammalia, Rodentia) inferred from cytochrome b gene sequences. Mammalia 65: 473–482. doi: 10.1515/mamm.2001.65.4.473 [DOI] [Google Scholar]
- 31.Oshida T., Dang C. N., Nguyen S. T., Nguyen N. X., Endo H., Kimura J., Sasaki M., Hayashida A., Takano A., Hayashi Y.2010. Phylogenetic relationship between Callosciurus caniceps and C. inornatus (Rodentia, Sciuridae): implication for zoogeographical isolation by the Mekong River. Ital. J. Zool. (Modena) 78: 328–335. doi: 10.1080/11250003.2010.490566 [DOI] [Google Scholar]
- 32.Oshida T., Can D. N., Nguyen T. S., Nghia N. X., Endo H., Kimura J., Sasaki M., Hayashida A., Takano A., Koyabu D., Hayashi Y.2013. Phylogenetic position of Callosciurus erythraeus griseimanus from Vietnam in the genus Callosciurus. Mammal Study 38: 41–47. doi: 10.3106/041.038.0105 [DOI] [Google Scholar]
- 33.Parnell J.2013. The biogeography of the Isthmus of Kra region: a review. Nord. J. Bot. 31: 1–15. doi: 10.1111/j.1756-1051.2012.00121.x [DOI] [Google Scholar]
- 34.Roberts T. E., Sargis E. J., Olson L. E.2009. Networks, trees, and treeshrews: assessing support and identifying conflict with multiple loci and a problematic root. Syst. Biol. 58: 257–270. doi: 10.1093/sysbio/syp025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts T. E., Lanier H. C., Sargis E. J., Olson L. E.2011. Molecular phylogeny of treeshrews (Mammalia: Scandentia) and the timescale of diversification in Southeast Asia. Mol. Phylogenet. Evol. 60: 358–372. doi: 10.1016/j.ympev.2011.04.021 [DOI] [PubMed] [Google Scholar]
- 36.Sargis E. J., Woodman N., Reese A. T., Olson L. E.2013. Using hand proportions to test taxonomic boundaries within the Tupaia glis species complex (Scandentia, Tupaiidae). J. Mammal. 94: 183–201. doi: 10.1644/11-MAMM-A-343.1 [DOI] [Google Scholar]
- 37.Slaughter B. H., Pine R. H., Pine N. E.1974. Eruption of cheek teeth in insectivora and carnivora. J. Mammal. 55: 115–125. doi: 10.2307/1379261 [DOI] [PubMed] [Google Scholar]
- 38.Voris H. K.2000. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J. Biogeogr. 27: 1153–1167. doi: 10.1046/j.1365-2699.2000.00489.x [DOI] [Google Scholar]
- 39.Woodman N., Miller-Murthy A., Olson L. E., Sargis E. J.2020. Coming of age: morphometric variation in the hand skeletons of juvenile and adult lesser treeshrews (Scandentia: Tupaiidae: Tupaia minor Günther, 1876). J. Mammal. 101: 1151–1164. doi: 10.1093/jmammal/gyaa056 [DOI] [Google Scholar]
- 40.Woodruff D. S., Turner L. M.2009. The Indochinese–Sundaic zoogeographic transition: a description and analysis of terrestrial mammal species distributions. J. Biogeogr. 36: 803–821. doi: 10.1111/j.1365-2699.2008.02071.x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


