Abstract
Fusarium spp. have emerged as major opportunistic fungal agents. Since new antifungal agents exhibit variable activity against Fusarium isolates depending on the species, rapid identification at the species level is required. Conventional culture methods are difficult, fastidious, and sometimes inconclusive. In this work, we sequenced a 440-bp fragment encoding the 28S rRNA from 33 Fusarium isolates belonging to six Fusarium species associated with human infections. The data were then analyzed by the neighbor-joining method. By using distance matrix analysis and constructing the phylogram, we could easily distinguish the different species for all but one isolate. The method also allowed differentiation between the closely related genera Acremonium and Cylindrocarpon. In contrast to the case with conventional methods, the results could be obtained within 48 h from a 3-day culture and are independent of mycologist experience, making this method rapid and reliable for identification of Fusarium species isolated from patients.
The fungi belonging to the genus Fusarium are well-known plant pathogens and food contaminants that also cause superficial and subcutaneous infections, such as onychomycosis and keratomycosis, in humans (9). They have recently emerged as major opportunistic agents in immunocompromised hosts, especially in patients with hemopathy (2, 7). They are now considered the third most common fungal genus (after Candida and Aspergillus) isolated from systemic infections in bone marrow transplantation patients (8). Four species account for more than 95% of human infections: Fusarium solani, Fusarium moniliforme (Fusarium verticilloides), and Fusarium oxysporum are each responsible for about 30% of the cases, whereas Fusarium dimerum is involved in 5% of the cases (6, 7). Diagnosis of Fusarium at the species level is based on conventional methods, which include the description of colonies on appropriate media (texture, color, and pigment etc.) and microscopic description of conidiogenous cells and conidia. This can be best observed after 2 weeks of incubation, lengthening the time for a definitive diagnosis. Because of important variations of characters, such as pigmentation and growth rate, that are often seen within a given species, only well-trained mycologists are able to ensure the diagnosis (6). The results are thus frequently inconclusive, as seen in various reports where one-third to one-half of the isolates are not identified at the species level (2, 7). Identification at the species level is important for epidemiological purposes and may become absolutely necessary because some new antifungal agents exhibit variable activity against Fusarium depending on the species (1, 13). In this work, we report a rapid method for the identification of significant Fusarium species involved in human infections by using rRNA gene (rDNA) sequencing.
MATERIALS AND METHODS
Strains.
The fungal isolates used in this work are listed in Table 1. A total of 33 isolates belonging to six different Fusarium species were tested, including 18 reference strains and 15 clinical isolates collected from superficial and deep infections. Three clinical isolates and one reference strain belonging to the genus Acremonium and two Cylindrocarpon tonkinense reference strains were also studied due to the difficulties in distinguishing between these genera and Fusarium (6). In addition, nine control isolates from unrelated fungi were included to test the specificity of the designed primers (see below). The identification of Fusarium and Acremonium isolates was performed according to the colonial aspects and microscopic morphology after 7 and 14 days of culture at 25°C on potato dextrose agar (Difco Laboratories, Detroit, Mich.) plates in daylight. Examination of micro-, meso-, and macroconidia, chlamydospores, and phiallides allowed differentiation between the Fusarium species (4, 6). All of the isolates were stored in 10% glycerol at −80°C until tested.
TABLE 1.
Strains and isolates tested for 28S rDNA sequences in this study
| Strain category | Species | Strain(s)a | 28S rDNA sequence class | Accession no. |
|---|---|---|---|---|
| Reference strains | Acremonium killiense | IP 1577 | Acremonium class 2 | AF130374 |
| Aspergillus flavus | IP 954 | |||
| Aspergillus fumigatus | IP 1079 | |||
| Candida albicans | ATCC 90028 | |||
| Candida glabrata | ATCC 90030 | |||
| Candida parapsilosis | ATCC 90018 | |||
| Cryptococcus neoformans | ATCC 90112 | |||
| Cylindrocarpon tonkinense | IP 1692 | C. tonkinense class 1 | AF130375 | |
| IP 2329 | C. tonkinense class 2 | AF130376 | ||
| Fusarium chlamydosporum | IP 1542 | F. chlamydosporum | AF130377 | |
| Fusarium dimerum | IP 1516, IP 2365 | F. dimerum class 1 | AF130378 | |
| IHEM 5322 | F. dimerum class 2 | AF130379 | ||
| Fusarium moniliforme | IHEM 3824, IHEM 7465, IP 1550 | F. moniliforme class 1 | AF130384 | |
| IHEM 4195, IP 1579 | F. moniliforme class 2 | AF130385 | ||
| Fusarium oxysporum | IHEM 13830, IHEM 3798, IHEM 9571, IP 625 | F. oxysporum | AF130373 | |
| Fusarium semitectum | IP 2239 | F. semitectum | AF130380 | |
| Fusarium solani | IP 1684 | F. solani class 1 | AF130381 | |
| IHEM 10154 | F. solani class 2 | AF130382 | ||
| IHEM 6092, IP 2330 | F. solani class 3 | AF130383 | ||
| Paecilomyces variotti | ATCC 22319 | |||
| Scedosporium apiospermum | IP 1698 | |||
| Scedosporium prolificans | IP 1974 | |||
| Clinical isolates | Acremonium sp. | N97110423 | Acremonium sp. class 1 | AF130386 |
| N97128916, N96114830 | Acremonium sp. class 2 | |||
| F. dimerum | HM94.1 | F. dimerum class 1 | ||
| F. oxysporum | GEMO9795,b GEMO9796,b GEMO9896, N95151312, N96148518, N98117566 | F. oxysporum | ||
| F. solani | C98.1 | F. solani class 1 | ||
| HM94.2, N96144236c | F. solani class 3 | |||
| F. moniliforme | GEMO9908,d N98119873, N96146918, N96149460, PS97.1 | F. moniliforme class 1 |
IP, Pasteur Institute collection; IHEM, Institute of Hygiene and Epidemiology Mycology; ATCC, American Type Culture Collection.
First identified as F. solani.
Not pigmented isolate.
First identified as a Fusarium sp.
DNA extraction.
After a 3-day incubation at 35°C on Sabouraud agar (Sanofi-Diagnostic Pasteur, Marnes la Coquette, France) plates, cultures were discharged in 300 μl of distilled water in a microcentrifuge tube. A volume of 100 μl of Chelex solution (10% [wt/vol] Chelex-100 [Bio-Rad, Hercules, Calif.] in an aqueous solution of 0.1% [wt/vol] sodium dodecyl sulfate sodium salt [Sigma, St. Louis, Mo.], 1% [vol/vol] Nonidet P-40 [Sigma], and 1% [vol/vol] Tween 80 [Sigma]) was added. The tubes were incubated at 95°C for 30 min and then on ice for 5 min. DNA was removed from the supernatant after 5 min of centrifugation (10,000 × g) and stored at −80°C until used.
Amplification and sequencing.
Two oligonucleotides (Fus1 [5′-TGAAATCTGGCTCTCGGG] and Fus2 [5′-CATGCGCGAACCTCAGTC]) were designed after comparison of Fusarium 28S rDNA sequences in the GenBank database. Amplification was performed in a volume of 50 μl with 10 mM Tris-HCl, 50 mM KCl, 2.5 mM MgCl2, 0.01% (wt/vol) gelatin, 5 μl of DNA, a 0.25 μM concentration of each oligonucleotide primer, 0.2 mM deoxynucleoside triphosphates (Pharmacia, Uppsala, Sweden), 0.3 U of Taq Gold (PE Applied Biosystems, Foster City, Calif.), and 5 μl of DNA. Thermal cycling parameters were as follows: 10 min at 94°C, followed by 35 cycles of 30 s at 94°C, 90 s at 64°C, and 90 s at 72°C and a final extension of 10 min at 72°C. The PCR products were separated by electrophoresis and visualized under UV. Amplification products were purified by using microspin S-400 HR purification columns (Pharmacia). The sequence reactions were made with 5 μl of purified amplified DNA, 4 pmol of primers, and 4 μl of Big Dye Terminator (PE Applied Biosystems) according to the manufacturer's instructions. The extension products were then precipitated, washed, and resuspended in Template Suppression Reagent (PE Applied Biosystems). After denaturation at 95°C for 2 min, the samples were loaded onto POP6 capillary columns in a ABI Prism 310 Genetic Analyzer (PE Applied Biosystems). Sequences of both strands were determined.
Sequence alignments and phylogenetic trees.
Sequences were edited with the Sequence Navigator software (PE Applied Biosystems). Multiple sequence alignments were performed with the Clustal X version 1.64 software (Higgins, Heidelberg, Germany). Trees and matrix distances were constructed with the Phylip package 3.572 by using the neighbor-joining method. The degree of confidence in phylogenetic branching was assessed by using 1,000 bootstrap resamplings.
Nucleotide sequence accession numbers.
The nucleotide sequences of the strains and isolates tested in this work appear in the GenBank nucleotide sequence database under the accession numbers shown in Table 1.
RESULTS AND DISCUSSION
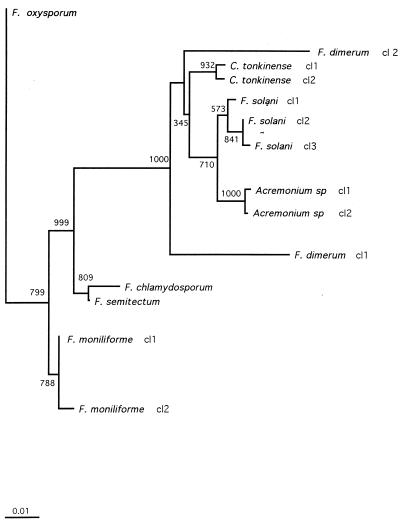
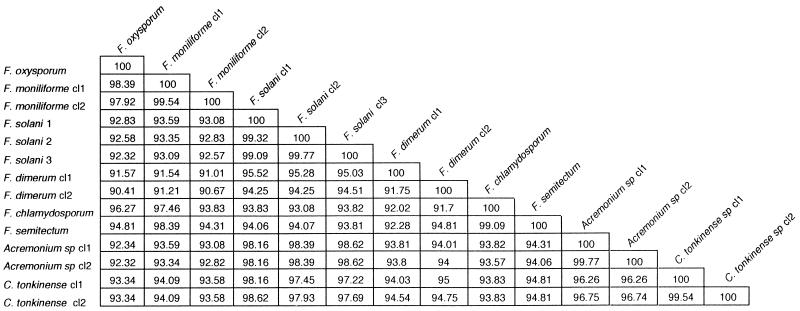
In this work, we compared the conventional methods for identification of Fusarium species with a 28S rDNA sequence method. Using the newly designed primers Fus1 and Fus2, we were able to amplify a 480-bp fragment from 33 Fusarium strains and isolates and from strains and isolates from the closely related genera Acremonium and Cylindrocarpon. In contrast, we failed to amplify the DNAs from nine reference strains belonging to the unrelated species listed in Table 1. Sequencing of the PCR products led to 10 sequence classes for Fusarium strains and isolates, each containing 1 to 10 isolates. Comparison with Fusarium sequences from the GenBank database demonstrates complete interspecies identity with those of Fusarium semitectum (accession no. X80813), F. oxysporum (U34537 and U34542), and F. moniliforme (U34528). The sequence of F. dimerum (U88105) exhibits close similarity with that of F. dimerum sequence class 2, with a difference of only 6 nucleotides (98.6% homology). Comparison of sequences for F. solani is possible only with sequences obtained from F. solani forma speciales phaseoli isolates (L36629, L36630, L36632, and L36634), which are involved in the soybean sudden death syndrome. They exhibit a closer homology with C. tonkinense sequence class 2 (98.16%) than with F. solani sequence class 1 (96.74%). Multiple alignment of our sequence classes was carried out by using Clustal X software (15). These data were used to construct an unrooted phylogenetic tree by the neighbor-joining method (12). As illustrated in Fig. 1, each Fusarium species as determined by conventional methods formed a distinct clade, with a minimum bootstrap confidence value of 71% for F. solani (Fig. 1). Interspecies homologies for Fusarium never reached higher than 99.01% (i.e., for Fusarium chlamydosporum and F. semitectum) (Fig. 2). In contrast, with the exception of F. dimerum (discussed below), intraspecies homologies ranged from 99.09% for F. solani to 100% for F. oxysporum. An isolate thus can be readily assigned to one species when its homology with one of the sequence classes is higher than 99.1%.
FIG. 1.
Unrooted phylogenetic tree derived from 28S rDNA sequences of Fusarium spp., C. tonkinense, and Acremonium spp. The tree was constructed by using the neighbor-joining method. Bar, 1% estimated sequence divergence. cl, class.
FIG. 2.
Homology between Fusarium spp., Acremonium spp., and C. tonkinense, derived from 28S rDNA sequences. The sequence classes (cl) correspond to those in Table 1.
The isolate IHEM 5322, considered F. dimerum, exhibited a unique sequence, with 8.25% divergence from the other sequence class containing three strains of F. dimerum. Interestingly, this isolate had deletions at positions 398 and 403, a characteristic shared only with the other sequence class corresponding to F. dimerum. We have no explanation for this discrepancy. (It should be mentioned that this strain had been collected not from a patient but from the recycled water of a spray humidifier.) Such a discrepancy between a molecular method (in this case using species-specific probes) and the conventional method has already been described for a Fusarium reference strain (3), suggesting that conventional methods may not be accurate for some strains with atypical phenotypic characteristics.
While the aim of this work was not a phylogenic reevaluation, we noted that there are closer relationships between some genera tested in this study than between some species within the Fusarium genus. This is illustrated by the grouping of F. solani, Acremonium spp., and C. tonkinense (intersequence class divergences lower than 6.24%), whereas F. moniliforme and F. oxysporum exhibited mean divergences from F. solani between 6.98 and 7.42%, respectively. Interestingly, F. solani, Acremonium spp., and Cylindrocarpon are characterized by teleomorphs belonging to the genus Nectria, whereas F. moniliforme and F. oxysporum have teleomorphs belonging to the genus Giberella, reflecting the heterogeneity of the genus Fusarium (5, 10, 11).
For diagnosis purposes, the sequencing method showed its objective value by easily and correctly identifying a nonpigmented F. solani isolate (N96144236), independent of the mycologist's experience. Also, isolates with uncertain or wrong conventional identifications were identified in agreement with the Institute of Hygiene and Epidemiology Mycology reference center identification. Finally, the sequencing method allows identification within 48 to 72 h following a 3-day culture of the isolate, as opposed to the time-consuming conventional methods. Recently, an exoantigen test was proposed for the species identification of Fusarium, which requires the production of antiserum against specific antigens (6 weeks of incubation) and the extraction of exoantigens from 10-day-old cultures (14). Our results demonstrate that 28S rDNA sequencing is a valuable method for the identification of significant Fusarium species involved in human infections.
ACKNOWLEDGMENTS
We thank C. de Bièvre for providing us the Pasteur Institute collection strains and E. Gueho for confirmation of the identification of some isolates. We are indebted to F. Botterel, S. Bretagne, J. Carrière, and A. Paugam for sending us some clinical isolates.
REFERENCES
- 1.Arikan S, Lozano-Chiu M, Paetznick V, Nagia S, Rex J. Program and abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Activity of amphotericin, itraconazole and voriconazole against Aspergillus and Fusarium, abstr. J-19. [Google Scholar]
- 2.Boutati E I, Anaissie E J. Fusarium, a significant emerging pathogen in patients with hematologic malignancy: ten years' experience at a cancer center and implications for management. Blood. 1997;90:999–1008. [PubMed] [Google Scholar]
- 3.Choi I, Westerman J, Morrison C. Program and abstracts of 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Rapid differenciation of filamentous fungi using species-specific DNA probes, abstr. C-288; p. 179. [Google Scholar]
- 4.de Hoog G, Guarro J. Atlas of clinical fungi. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures, Universitat Rivora i Virgili; 1995. p. 720. [Google Scholar]
- 5.Guadet J, Julien J, Lafay J F, Brygoo Y. Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Mol Biol Evol. 1989;6:227–242. doi: 10.1093/oxfordjournals.molbev.a040548. [DOI] [PubMed] [Google Scholar]
- 6.Guarro J, Gene J. Fusarium infections. Criteria for the identification of the responsible species. Mycoses. 1992;35:109–114. doi: 10.1111/j.1439-0507.1992.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 7.Hennequin C, Lavarde V, Poirot J L, Rabodonirina M, Datry A, Aractingi S, Dupouy C J, Caillot D, Grange F, Kures L, Morin O, Lebeau B, Bretagne S, Guigen C, Basset D, Grillot R. Invasive Fusarium infections: a retrospective survey of 31 cases. The French Groupe d'Etudes des Mycoses Opportunistes. J Med Vet Mycol. 1997;35:107–114. [PubMed] [Google Scholar]
- 8.Morrison V A, Haake R J, Weisdorf D J. The spectrum of non-Candida fungal infections following bone marrow transplantation. Medicine. 1993;72:78–89. doi: 10.1097/00005792-199303000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Nelson P E, Dignami M C, Anaissie E J. Taxonomy, biology, and clinical aspects of Fusarium species. Clin Microbiol Rev. 1994;7:479–504. doi: 10.1128/cmr.7.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell K. Fusarium and its near relatives. In: Reynolds D, Taylor J, editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, United Kingdom: CAB International; 1993. pp. 225–233. [Google Scholar]
- 11.O'Donnell K, Cigelnik E, Nirenberg H I. Molecular systematics and phylegeography of the Gibberella fujikoroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:437–443. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Sanche S, Fothergill A, Rinaldi M. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Interspecies variation of the susceptibility of Fusarium species to Schering 56592 in vitro, abstr. E66; p. 125. [Google Scholar]
- 14.Sekhon A S, Kaufman L, Moledina N, Summerbell R C, Padhye A A, Ambrosie E A, Panter T. An exoantigen test for the rapid identification of medically significant Fusarium species. J Med Vet Mycol. 1995;33:287–289. [PubMed] [Google Scholar]
- 15.Thompson J, Higgins D, Gibson T. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positio-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]