Abstract
Background
Immune-checkpoint inhibitors have propelled the field of therapeutics for small cell lung cancer (SCLC) treatment, but are only beneficial to some patients. The objective of this study was to identify valid biomarkers for good potential response to immunotherapy.
Material/Methods
We performed an integrated analysis of the available datasets from the Gene Expression Omnibus (GEO) projects, Cancer Cell Line Encyclopedia (CCLE), TISIDB database, and Lung Cancer Explorer (LCE) database. Six prognosis-related genes (MCM2, EZH2, CENPK, CHEK1, CDKN2A, and EXOSC2) were identified utilizing the meta workflow of data analysis methods. We performed subclass mapping to compare their expression profiles to other datasets of patients who responded to immunotherapy. A drug sensitivity predictive model was used to predict the chemotherapeutic response to cisplatin and etoposide.
Results
Our results showed that the expression of the 6 key genes was significantly associated with the overall survival of patients with SCLC. Lower expression of these 6 genes was correlated to the response to anti-PD-1 treatment. Additionally, low expression of MCM2, EZH2, CENPK, and CHEK1 was correlated with increased sensitivity to cisplatin, but not etoposide.
Conclusions
Overall, our data showed that MCM2, EZH2, CENPK, CHEK1, CDKN2A, and EXOSC2 are potential prognostic and predictive biomarkers for response to immune-checkpoint inhibitor treatment in patients with SCLC. Further studies with large sample sizes are required to validate our findings and to explore the detailed mechanisms underlying the role of these genes in SCLC.
Keywords: Antineoplastic Agents, Immunological; Antineoplastic Combined Chemotherapy Protocols; Small Cell Lung Carcinoma
Background
Small cell lung cancer (SCLC) is one of the deadliest and most aggressive lung cancers due to its rapid growth, early metastasis, and acquired therapeutic resistance [1]. Unlike non-small cell lung cancer (NSCLC), which can be treated with a plethora of drugs, few treatment options are available for patients with SCLC. The current standard of care, which was defined several decades ago [2], relies on platinum-based chemotherapy and has a median survival time of only 8–13 months [3].
However, the treatment landscape has recently changed with the introduction of immune-checkpoint inhibitors (ICIs). Incorporation of the anti-PDL1 antibody atezolizumab with platinum-doublet chemotherapy has become the first-line therapy for extensive SCLC, with an improvement in median overall survival (OS) from 10.3 to 12 months for patients with metastatic disease [4]. Ipilimumab (an anti-CTLA4 monoclonal antibody) also showed increased survival in a phase II trial [5].
Unfortunately, only a subset of patients with SCLC benefits from immunotherapy [4]. More recent studies have reported the use of drugs such as CheckMate-032 and CheckMate-331 for maintenance and second-line therapy, but these fail to improve survival for patients with SCLC [6,7]. Recent research suggest that high tumor mutational burden (TMB) may be correlated with efficacy of immunotherapy in SCLC [8]. Moreover, no study used genetic marker to select patients more likely to benefit from these treatments, which may explain their clinical failure. Therefore, there is an urgent need to identify biomarkers that can predict the responsiveness to immune-checkpoint blockade treatment.
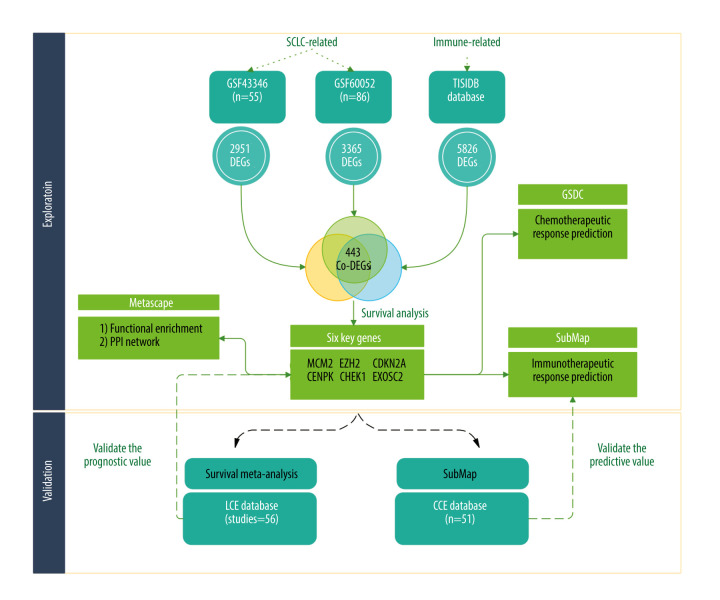
To address this, we analyzed the expression of different genes in healthy and tumor tissues from patients with SCLC in 2 independent cohorts. We then identified genes associated with immunotherapeutic response. Using subclass mapping and survival analysis, we found 6 prognostic genes predictive for response to immune-checkpoint inhibitors. These results were validated by meta-analysis and a third cohort (a summary of the statistical analysis is presented in Figure 1).
Figure 1.
The flowchart of our study (Biorender website, https://biorender.com/).
Material and Methods
Data and Resources
We used 3 different cohorts for this study. Expression data for the first 2 cohorts were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) [9]. The first cohort was obtained from the Affymetrix U133plus2 GEO SCLC Cohort (accession number GSE43346) and comprised 42 normal samples and 23 clinical SCLC samples. The second cohort downloaded from GEO (accession number GSE60052) consisted of 7 normal lung samples and 79 SCLC samples, of which only 48 had associated clinical data. Gene expression profiles were obtained using the Illumina HiSeq2000 platform. The third cohort was downloaded from the Cancer Cell Line Encyclopedia (CCLE) database (https://portals.broadinstitute.org/ccle) [10]. The CCLE datasets contain information from cell lines derived from different tumor types; we used data from SCLC tumors, resulting in 51 samples for this cohort.
Additionally, we analyzed a comprehensive list of 5826 immunotherapy-related genes available from the Tumor-Immune System Interactions Database (TISIDB) (http://cis.hku.hk/TISIDB/) [11]. These genes are known to differ in expression between patients that are responders vs those who are non-responders to immunotherapy.
Identification of Differentially Expressed Genes Related to Immunotherapy
We identified differentially expressed genes (DEGs) by comparing the normalized expression data of SCLC samples to that of normal samples from GSE43346 and GSE60052. Only DEGs with |log2FC| >2 and FDR <0.05 were extracted using the Limma package in R [12]. We calculated the difference between the 2 groups using the t test and adjusted the P value using the Benjamini-Hochberg (BH) method. To identify immunotherapy-related genes among the SCLC DEGs, Venn analysis was utilized to determine overlapping DEGs among the 2 GEO datasets and the TISIDB dataset above. To narrow the scope of candidate genes, we selected the top 300 DEGs, in ascending order of FDR, from each GEO dataset.
Functional Enrichment Analysis and PPI Network Construction
We conducted function and protein-protein interaction (PPI) enrichment analyses to predict the biological function of the aforementioned DEGs using Metascape online tools (http://metascape.org/gp/index.html#/main/step1) [13]. The DEGs were annotated according to gene ontology biological processes. The P value was set at <0.01, indicating a statistically significant enrichment score of the GO or pathway terms. The PPI network was also visualized by Metascape.
Survival Analysis
Fifty-five patients (48 SCLC samples and 7 normal lung samples) from GSE60052 were divided into high- and low-expression groups based on optimal cut-off values defined by R package “Survminer” [14] for candidate gene expression. We used Kaplan-Meier survival curves to show the differences in patients’ OS between the high and low expression groups. Genes with P values <0.05 were identified as prognosis-related genes.
Immunotherapeutic/Chemotherapeutic Response Prediction
We divided the expression profiles from GSE60052 (n=86) into high- and low-expression groups based on the median expression values of our key genes. We further utilized an unsupervised subclass mapping (https://cloud.genepattern.org/gp/) to compare these expression profiles with those of another dataset containing 47 patients with melanoma who responded to immunotherapy [15,16] to predict the response to ICI therapy in high- and low-subgroup. R package “pheatmap” was used for visualizing the results. We also analyzed the expression profiles of SCLC cell lines (n=51) from the CCLE database using the same method to validate the immunotherapeutic response prediction. Groups with Bonferroni-corrected P values <0.05 were identified as immunotherapy responders.
In addition to the immunotherapeutic response prediction, the chemotherapeutic response for 86 samples was predicted using the Genomics of Drug Sensitivity in Cancer database (GDSC) (https://www.cancerrxgene.org/) [17]. We predicted the responses to cisplatin and etoposide using the pRRophetic package in R [18]. Each sample’s half-maximal inhibitory concentration (IC50) was assessed by ridge regression, and the prediction accuracy was evaluated by 10-fold cross-validation relying on the GDSC training model. We summarized duplicate gene expression as the mean expression, and set the tissue type to “allSolidTumours” and the batch effect to “combat”. Meanwhile, the values of other parameters were set as default.
Survival Meta-Analysis of Key Henes
Using survival meta-analysis on data from the Lung Cancer Explorer (LCE) database (http://lce.biohpc.swmed.edu/lungcancer/) [19], we were able to validate whether our chosen key genes were prognosis-related in SCLC based on the meta-analysis module in LCE database. Survival meta-analysis was based on 56 studies, including over 6700 patients with lung cancer. Cox hazard models were fitted to evaluate the association between key genes expression and patient survival outcome in each dataset. After heterogeneity testing, the HR from individual datasets were then calculated to summary HR utilizing a random effects model.
Statistical and Bioinformatic Analysis
The t test was used to compare gene expression between tumor tissues and adjacent nontumorous tissues. The OS between different groups was compared by Kaplan-Meier analysis with the log-rank test. Bonferroni correction was used for adjust P value in subclass mapping. The Wilcox test was used to compare the IC50 between different subgroups. R software (Version 4.0.0, http://www.r-project.org), Metascape online tool, subclass mapping and LCE database were used to analyze data and plot graphs. If not specified above, a P value less than 0.05 was considered statistically significant.
Results
Identification of Candidate Genes Related to Immunotherapy
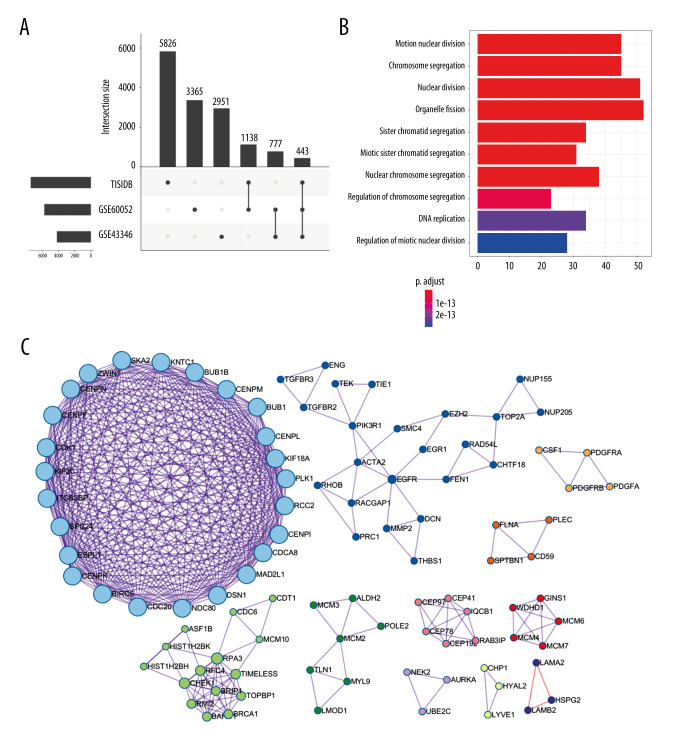
To select genes differentially expressed between SCLC and normal tissues, we conducted Limma analysis of data from GSE43346 and GSE60052. We identified 2951 and 3365 DEGs from each dataset, respectively, and the 2 datasets had 777 DEGs in common (co-DEGs). We then identified common genes between these co-DEGS and the immunotherapy-related genes from TISIDB, which included DEGs between immunotherapy responders and non-responders, resulting in 443 candidate genes retained for further analysis (Figure 2A).
Figure 2.
Identification of candidate genes related to immunotherapy. (A) Identification of common differentially expressed genes from 3 datasets. (B) The gene ontology annotation and pathway enrichment analysis of differentially expressed genes. (C) PPI networks and 11 subclusters of differentially expressed genes. R software Version 4.0.0 (http://www.r-project.org) and Metascape online tools http://metascape.org/gp/index.html#/main/step1.
To obtain further insight into their function, the candidate genes were uploaded to the Metascape database. GO and KEGG (Gene Ontology and Kyoto Encyclopedia of Genes and Genomes) analysis results showed that the candidate genes were mainly enriched through mitotic nuclear division, chromosome segregation, nuclear division, organelle fission, nuclear chromosome segregation, and regulation of chromosome segregation. Moreover, candidate genes for organelle fission were significantly enriched (Figure 2B).
Using the Metascape database, we also constructed PPI networks selecting samples that had a PPI minimum network size above 3. From these constructed PPI networks, we identified closely related proteins using molecular complex detection (MCODE). The candidate genes in the PPI networks were stratified into 11 subclusters through the MCODE algorithm. Figure 2C shows MCODE components.
Key genes as Prognostic Biomarkers
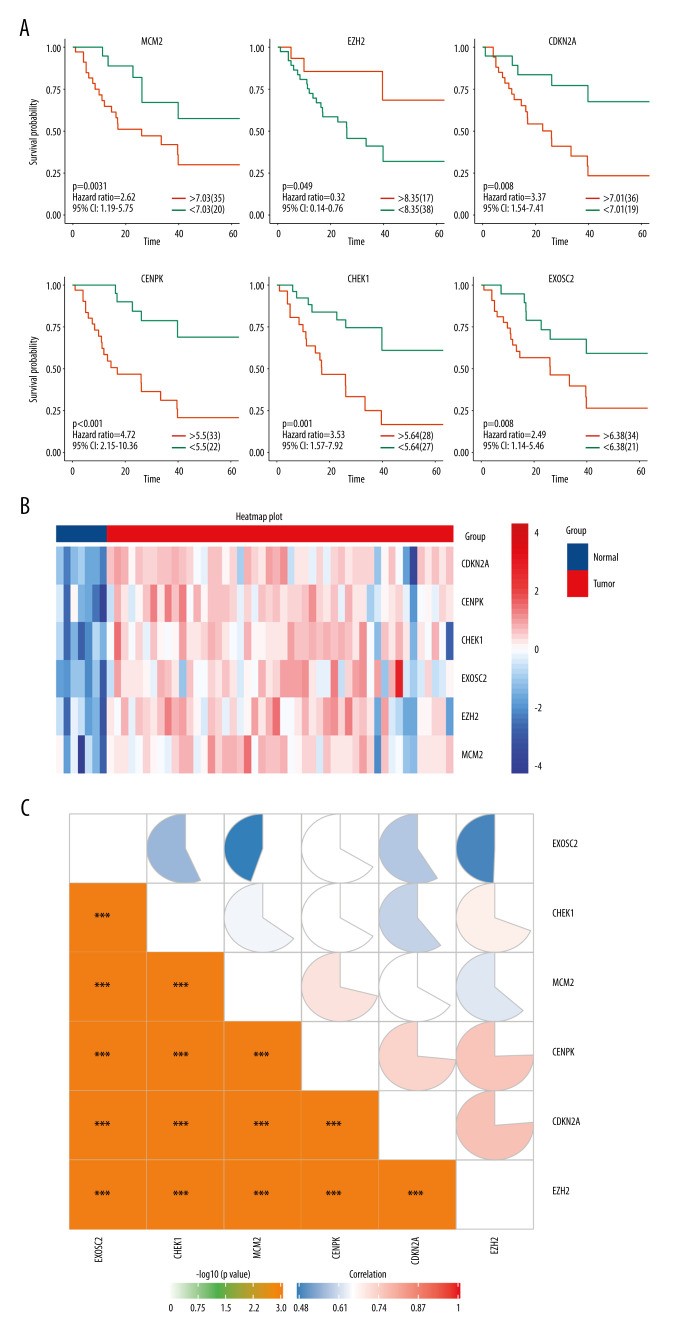
The prognostic importance of the 11 selected genes modulated in SCLC was evaluated using the GSE60052 clinical data, from which we retrieved 55 SCLC patient samples with clinical data. The gene expression values were categorized as high or low according to the optimal cut-off values set by the Survminer package for R. As a result, 6 genes were found to be independent prognostic factors for OS. Low expression of enhancer of zeste homolog 2 (EZH2) was associated with poor OS (P=0.049), whereas high expression of minichromosome maintenance complex component 2 (MCM2) (P=0.031), cyclin-dependent kinase inhibitor 2A (CDKN2A) (p=.008), centromere protein K (CENPK) (P<0.001), checkpoint kinase 1 (CHEK1) (P=0.001), and exosome component 2 (EXOSC2) (P=0.031) were associated with poor OS (Figure 3A). The heatmap showed that the expression level of 6 genes is lower in the normal tissue (Figure 3B). Moreover, the correlation heatmap showed a significant positive correlation among the 6 genes (Figure 3C).
Figure 3.
(A) Overall survival according to the expression of the 6 selected genes modulated in SCLC. (B) Heatmap demonstrated the expression level of 6 selected genes between normal and tumor tissue. (C) Correlation heatmap of the 6 selected genes. R software Version 4.0.0 http://www.r-project.org.
Key Genes as Predictive Biomarkers of Immunotherapeutic/Chemotherapeutic Response
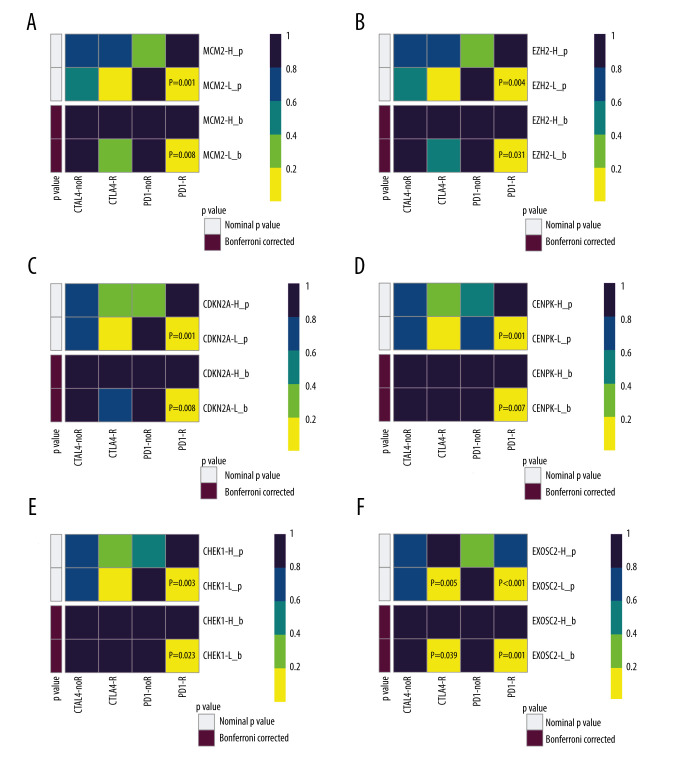
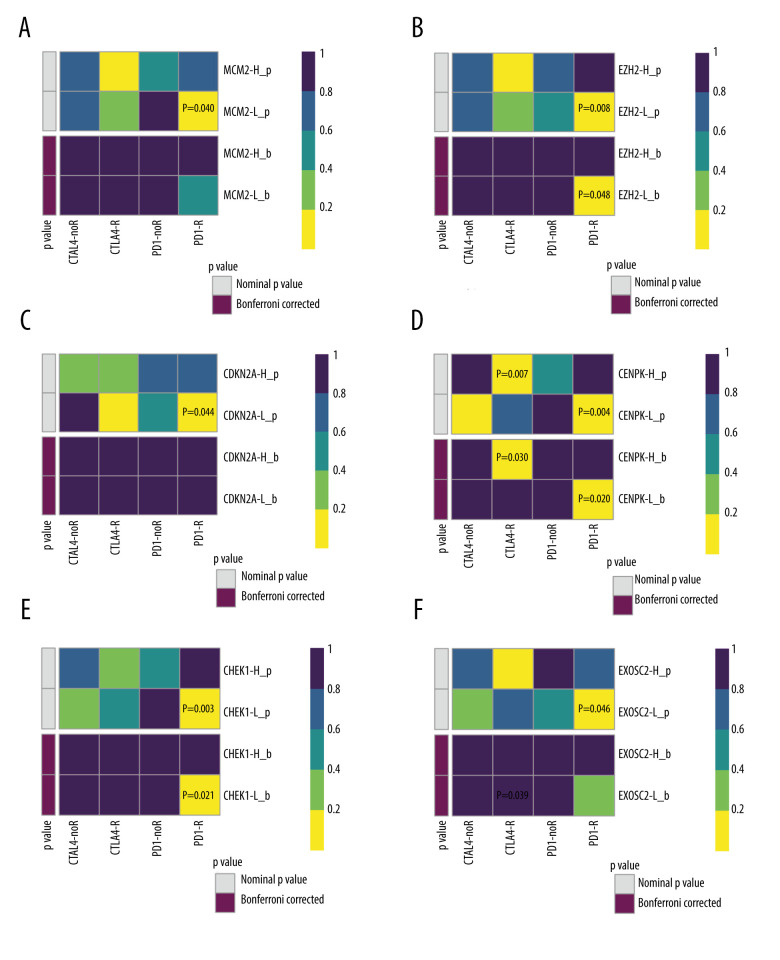
We subsequently performed a SubClass Mapping (SubMap) analysis to determine whether groups with high and low expression of these key genes were similar to other groups that clearly responded to immunotherapy [15]. SubMap results indeed showed that low expression of the 6 key genes was correlated with favorable response to anti-PD-1 treatment CDKN2A (Bonferroni-corrected P=0.008), CENPK (Bonferroni-corrected P=0.007), CHEK1 (Bonferroni-corrected P=0.023), EXOSC2 (Bonferroni-corrected P=0.001), EZH2 (Bonferroni-corrected P=0.031), and MCM2 (Bonferroni-corrected P=0.008). Moreover, lower expression of EXOSC2 was correlated with more favorable response to anti-CTLA4 treatment (Bonferroni-corrected p=0.039) (Figure 4).
Figure 4.
SubClass Mapping analysis of the 6 key genes to predict the likelihood of response to immune therapy. (A) Low expression of MCM2 was correlated with favorable response to anti-PD-1 treatment. (B) Low expression of EZH2 was correlated with favorable response to anti-PD-1 treatment. (C) Low expression of CDKN2A was correlated with favorable response to anti-PD-1 treatment. (D) Low expression of CENPK was correlated with favorable response to anti-PD-1 treatment. (E) Low expression of CHEK1 was correlated with favorable response to anti-PD-1 treatment. (F) Low expression of EXOSC2 was correlated with favorable response to anti-CTLA4 treatment and anti-PD-1 treatment. R – response; noR – no response; -L – low-expression group of certain gene; -H – high-expression group of certain gene. (R software (Version 4.0.0, http://www.r-project.org)).
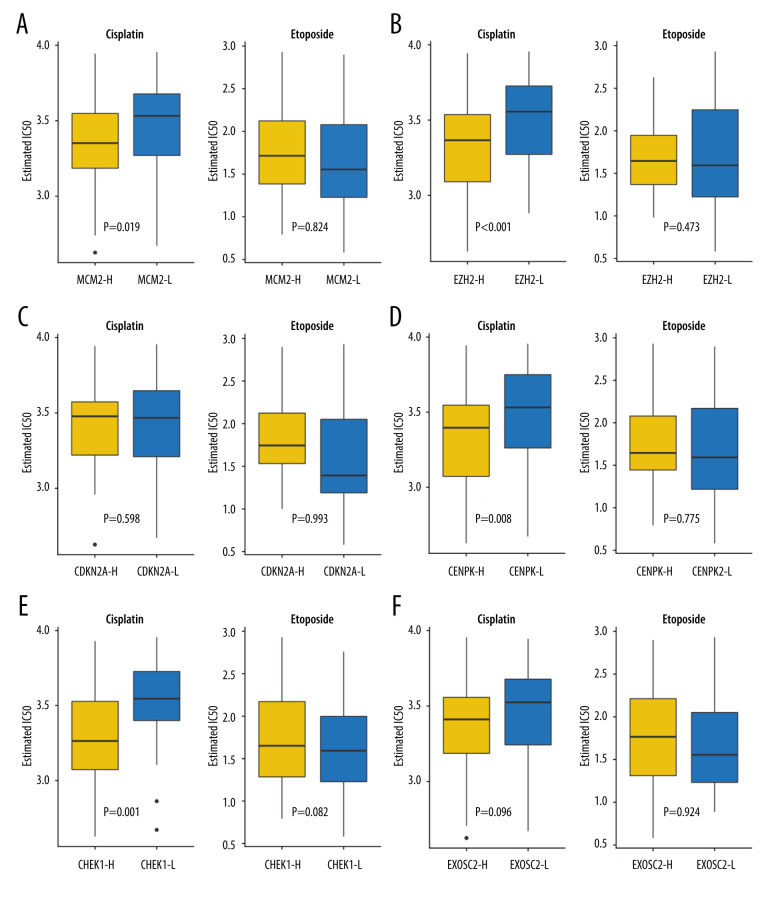
Because chemotherapy is the standard of care for SCLC, we assessed the GDSC data for chemotherapeutic responses to cisplatin and etoposide based on a predictive model. The estimated IC50 of cisplatin, but not of etoposide, significantly differed between the high- and low-expression groups. Low expression of MCM2 (P=0.019), EZH2 (p=0.0008), CENPK (P=0.008), and CHEK1 (P=0.0013) was correlated with favorable cisplatin response, whereas the expression of CDKN2A (p=0.598) and EXOSC2 (P=0.096) was not predictive of chemotherapeutic response. There were no statistically significant differences between the high and low expression groups in terms of predicting response to etoposide (MCM2, P=0.824; EZH2, P=0.473; CENPK, P=0.775; CHEK1, P=0.832; CDKN2A, P=0.993; and EXOSC2, P=0.924). Overall, our results suggest that MCM2, EZH2, CENPK, and CHEK1 are predictive biomarkers for chemotherapeutic response to cisplatin (Figure 5).
Figure 5.
Assessment for chemotherapeutic responses to cisplatin and etoposide. (A) The estimated IC50 of cisplatin and etoposide to MCM-2. (B) The estimated IC50 of cisplatin and etoposide to EZH2. (C) The estimated IC50 of cisplatin and etoposide to CDKN2A. (D) The estimated IC50 of cisplatin and etoposide to CENPK. (E) The estimated IC50 of cisplatin and etoposide to CHEK1. (F) The estimated IC50 of cisplatin and etoposide to EXOSC2. (R software [Version 4.0.0, http://www.r-project.org]).
Validation of the Prognostic and Predictive Value of the 6 Key Genes
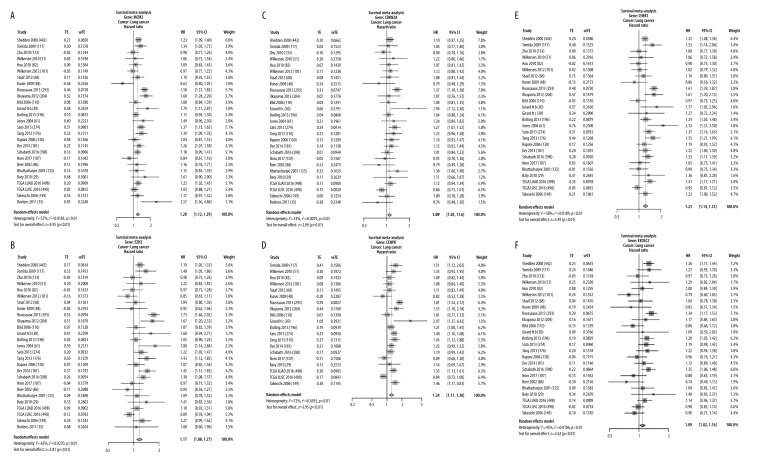
To validate the expression of the 6 potential biomarkers for the prognosis of SCLC, we examined their impact on the OS of patients with SCLC. In multiple selected lung cancer studies, we found that the survival meta-analyses for MCM2, EZH2, CENPK, CHEK1, CDKN2A, and EXOSC2 were significant in patients with SCLC (P<0.01). Our results indicate that these 6 genes can predict the prognosis of SCLC (Figure 6).
Figure 6.
Validation of the 6 key genes to predict immunotherapeutic response in the CCLE cohort. (A) There was no significant correlation between MCM2 and immune response. (B) Low expression of EZH2 was correlated with favorable response to anti-PD-1 treatment. (C) There was no significant correlation between CDKN2A and immune response (D) Low expression of CENPK was correlated with favorable response to anti-PD-1 treatment. (E) Low expression of CHEK1 was correlated with favorable response to anti-PD-1 treatment. (F) There was no significant correlation between EXOSC2 and immune response. R – response; noR – no response; -L – low-expression group of certain gene; -H – high-expression group of certain gene. (R software (Version 4.0.0, http://www.r-project.org)).
Finally, we selected 51 SCLC cell lines to validate the ability of our 6 key genes to predict immunotherapeutic response. Using SubMap analysis, we found that low expression of 4 of the 6 genes predicted response to anti-PD-1 therapy (CENPK (Bonferroni-corrected P=0.020), CHEK1 (Bonferroni-corrected P=0.021), and EZH2 (Bonferroni-corrected P=0.048)). Low expression of MCM2, CDKN2A, and EXOSC2 did not significantly predict response (MCM2 Bonferroni-corrected P=0.479, CDKN2A Bonferroni-corrected P=1.000, and EXOSC2 Bonferroni-corrected P= 0.315). Interestingly, the high expression of CENPK predicted response to anti-CTLA4 therapy (CENPK Bonferroni-corrected P=0.030) (Figure 7).
Figure 7.
Validation of expression of the 6 potential biomarkers for the prognosis of SCLC in the LCE database. The forest plot presented the results of survival meta-analyses. (A) MCM2 was a risk factor in lung cancer (HR=1.20, 95% CI: 1.12–1.29). (B) EZH2 was a risk factor in lung cancer (HR=1.17, 95% CI: 1.08–1.27). (C) CDKN2A was a risk factor in lung cancer (HR=1.09, 95% CI: 1.03–1.16). (D) CENPK was a risk factor in lung cancer (HR=1.23, 95% CI: 1.11–1.38). (E) CHEK1 was a risk factor in lung cancer (HR=1.23, 95% CI: 1.14–1.33). (F) EXOSC2 was a risk factor in lung cancer (HR=1.09, 95% CI: 1.02–1.16). TE – log (HR); seTE – SE (log (HR)). (LCE database (http://lce.biohpc.swmed.edu/lungcancer/), meta-analyses module).
Discussion
The addition of ICIs to chemotherapy regimens for first-line management is a significant advancement in the treatment of patients with SCLC. Predictive biomarkers are crucial to identify patients with SCLC who could benefit from ICIs. Recent studies have shown that TMB could serve as a predictive factor for immunotherapy in SCLC [20]. However, genetic markers that predict ICI response have not been well studied. To the best of our knowledge, this is the first study to assess potential genes that predict response to ICIs in SCLC. In this study, we also assessed chemotherapeutic response in patients with SCLC. Our results revealed that lower expression of 6 key genes (MCM2, EZH2, CDKN2A, CENPK, CHEK1, and EXOSC2) in patients with SCLC predicted responses to both anti-PD-1 therapy and cisplatin. We expect that low expression of these 6 key genes will be a significant predictor of clinical response to the combination of chemotherapy and ICIs in patients with SCLC, and the expression levels of 6 key genes were lower in the adjacent normal tissues. However, due to the lack of detailed clinical information, the expression level of the key genes was unclear in different stages. Thus, further studies are urgently needed to explore the correlation of these 6 key genes and tumor progression.
A limitation of this study is that we did not explore the potential mechanisms of our 6 key genes underlying the response of patients with SCLC to immunotherapy, as all the data analyzed in our study were retrieved from online databases. The prognostic significance of the 6 genes is validated in lung cancers, not specifically in small cell lung cancer. Due to the lack of valid verifying datasets of SCLC, further studies with larger sample sizes are required to validate our findings and to explore the detailed mechanisms of the 6 key genes in SCLC.
MCM2, which is essential for DNA replication and cell cycle, has been recently identified as a prognostic biomarker in oral, gastric, colon, and breast cancers [21–24]. Fujii et al reported that MCM2 could be a strong predictor of poor outcome in SCLC [25]. These studies suggest that high expression of MCM2 may play a role in the poor prognosis of cancers, which is consistent with our results [26]. However, the specific immunotherapeutic function of MCM2 in SCLC has not yet been explored. In our study, low MCM2 expression was correlated with increased response to anti-PD-1 treatment in the GEO datasets, but this was not statistically significant in the CCLE dataset. Further studies are needed to determine whether MCM2 plays a vital role in immunotherapeutic response of SCLC.
EZH2 is highly expressed in various cancer types and is more frequently overexpressed in SCLC than in NSCLC [27]. Hubaux et al demonstrated that EZH2 inhibits apoptosis and promotes the cell cycle in SCLC by promoting the oncogenic RB1/E2F pathway [28]. Gardner reported that inhibition of EZH2 enhances the effectiveness of current standard chemotherapy [29]. Toyokawa et al found that the expression of EZH2 was not significantly associated with postoperative survival [30]. In SCLC, EZH2, and other PRC2 components contribute to gene repression, which may be associated with worse prognosis in SCLC [31]. Intriguingly, we found a correlation between low EZH2 expression and poor OS in our SCLC cohorts.
EZH2 is further reported to inhibit immunity by repressing the production of Th1-type chemokines CXCL9 and CXCL10 in colon and ovarian carcinomas [32,33]. Additionally, Bugide et al reported that inhibition of EZH2 enhances hepatocellular carcinoma eradication by NK cells [34]. In our study, low EZH2 expression was correlated with a favorable response to anti-PD-1 treatment, validated by 2 independent SCLC cohorts. Nevertheless, the potential mechanism of EZH2 in the regulation of SCLC immune responses remains unclear.
Hans et al reported that CDKN2A is a major driver of pancreatic carcinogenesis in patients with pancreatic ductal adenocarcinoma (PDAC) [35]. However, the prognostic role of CDKN2A in SCLC is yet to be investigated. In this study, we demonstrated that lower CDKN2A expression was correlated with poor OS and response to anti-PD-1 therapy, but was not correlated with response to cisplatin.
To date, little is known about the expression and roles of CENPK, CHEK1, or EXOSC2 in SCLC. Lee et al reported that overexpression of CENPK in ovarian cancer is correlated with poor patient survival [36]. Liu et al discovered that high expression of CHEK1 in NSCLC is associated with poor OS [37]. Other studies have found that CHEK1 inhibitors may affect sensitivity to radiotherapy and chemotherapy [38–40]. In our study, low CENPK, CHEK1, and EXOSC2 expression was correlated with both poor OS and favorable response to anti-PD-1 therapy. Low CENPK and CHEK1 expression was correlated with response to cisplatin, but EXOSC2 expression was not significantly correlated with this response.
Conclusions
In conclusion, our results showed that expression of the 6 key genes was significantly associated with OS of patients with SCLC, and that lower expression of these 6 genes was correlated with response to anti-PD-1 therapy. We also showed that low expression of MCM2, EZH2, CENPK, and CHEK1 could predict response to cisplatin, but not etoposide, indicating that MCM2, EZH2, CENPK, CHEK1, CDKN2A, and EXOSC2 could be prognostic and predictive biomarkers for response to ICIs in patients with SCLC.
Abbreviations
- SCLC
small cell lung cancer
- GEO
Gene Expression Omnibus
- CCLE
Cancer Cell Line Encyclopedia
- LCE
Lung Cancer Explorer
- ICIs
immune-checkpoint inhibitors
- DEGs
differentially expressed genes
- OS
overall survival
- IC50
half-maximal inhibitory concentration
- MCODE
molecular complex detection
- GO
Gene Ontology
- HR
hazard ratio
- CI
confidence interval
- P
Pearson correlation coefficient
Footnotes
Conflict of interest: None declared
Declaration of Figures Authenticity
All figures submitted have been created by the authors, who confirm that the images are original with no duplication and have not been previously published in whole or in part.
Financial support: This work was supported by the Foundation Research Project of Jiangsu Province Natural Science Youth Fund (Grant no. BK20200395) and Jiangsu Provincial Health Commission (Grant no. H2019107)
References
- 1.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: What we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17(12):725–37. doi: 10.1038/nrc.2017.87. [Erratum in: Nat Rev Cancer. 2017;17(12):765] [DOI] [PubMed] [Google Scholar]
- 2.Cairns J. The interface between molecular biology and cancer research. Mutat Res. 2000;462(2–3):423–28. doi: 10.1016/s1383-5742(00)00030-2. [DOI] [PubMed] [Google Scholar]
- 3.Demedts IK, Vermaelen KY, van Meerbeeck JP. Treatment of extensive-stage small cell lung carcinoma: current status and future prospects. Eur Respir J. 2010;35:202–15. doi: 10.1183/09031936.00105009. [DOI] [PubMed] [Google Scholar]
- 4.Horn L, Mansfield AS, Szczęsna A, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 379(23):2220–29. doi: 10.1056/NEJMoa1809064. 20186. [DOI] [PubMed] [Google Scholar]
- 5.Ku GY, Yuan J, Page DB, et al. Single-institution experience with ipilimumab in advanced melanoma patients in the compassionate use setting: Lymphocyte count after 2 doses correlates with survival. Cancer. 2010;116(7):1767–75. doi: 10.1002/cncr.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ready N, Farago AF, de Braud F, et al. Third-line nivolumab monotherapy in recurrent SCLC: CheckMate 032. J Thorac Oncol. 2019;14(2):237–44. doi: 10.1016/j.jtho.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spigel DR, Vicente D, Ciuleanu TE, et al. Second-line nivolumab in relapsed small-cell lung cancer: CheckMate 331. Ann Oncol. 2021;32(5):631–41. doi: 10.1016/j.annonc.2021.01.071. [DOI] [PubMed] [Google Scholar]
- 8.Hellmann MD, Callahan MK, Awad MM, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33:853–61.e854. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: Archive for functional genomics data sets – update. Nucleic Acids Res. 2013;41(Database issue):D991–95. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. doi: 10.1038/nature11003. [Erratum in: Nature. 2012;492(7428):290, Erratum in: Nature. 2019;565(7738):E5–E6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ru B, Wong CN, Tong Y, et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–2. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kassambara A, Kosinski M, Biecek P, et al. Survminer: Drawing survival curves using ‘ggplot2’R package version 0.4.4. [Accessed on: June 27, 2019]. Available from: https://CRAN.R-project.org/package=survminer.
- 15.Hoshida Y, Brunet JP, Tamayo P, et al. Subclass mapping: Identifying common subtypes in independent disease data sets. PLoS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh W, Chen PL, Reuben A, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379) doi: 10.1126/scitranslmed.aah3560. eaah3560. [Erratum in: Sci Transl Med. 2017;9(385):28251903] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–61. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geeleher P, Cox N, Huang RS. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai L, Lin S, Girard L, et al. LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38(14):2551–64. doi: 10.1038/s41388-018-0588-2. [Erratum in: Oncogene. 2020;39(3):718–19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricciuti B, Kravets S, Dahlberg SE, et al. Use of targeted next generation sequencing to characterize tumor mutational burden and efficacy of immune checkpoint inhibition in small cell lung cancer. J Immunother Cancer. 2019;7(1):87. doi: 10.1186/s40425-019-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Andrade BA, León JE, Carlos R, et al. Expression of minichromosome maintenance 2, Ki-67, and geminin in oral nevi and melanoma. Ann Diagn Pathol. 2013;17(1):32–36. doi: 10.1016/j.anndiagpath.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Czyzewska J, Guzińska-Ustymowicz K, Pryczynicz A, et al. Immunohistochemical evaluation of Ki-67, PCNA and MCM2 proteins proliferation index (PI) in advanced gastric cancer. Folia Histochem Cytobiol. 2009;47(2):289–96. doi: 10.2478/v10042-009-0042-y. [DOI] [PubMed] [Google Scholar]
- 23.Guzińska-Ustymowicz K, Pryczynicz A, Kemona A, Czyzewska J. Correlation between proliferation markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer. Anticancer Res. 2009;29(8):3049–52. [PubMed] [Google Scholar]
- 24.Yousef EM, Furrer D, Laperriere DL, Tahir MR, et al. MCM2: An alternative to Ki-67 for measuring breast cancer cell proliferation. Mod Pathol. 2017;30(5):682–97. doi: 10.1038/modpathol.2016.231. [DOI] [PubMed] [Google Scholar]
- 25.Fujii K, Miyata Y, Takahashi I, et al. Differential proteomic analysis between Small Cell Lung Carcinoma (SCLC) and pulmonary carcinoid tumors reveals molecular signatures for malignancy in lung cancer. Proteomics Clin Appl. 2018;12(6):e1800015. doi: 10.1002/prca.201800015. [DOI] [PubMed] [Google Scholar]
- 26.Gou K, Liu J, Feng X, et al. Expression of Minichromosome Maintenance Proteins (MCM) and cancer prognosis: A meta-analysis. J Cancer. 2018;9(8):1518–26. doi: 10.7150/jca.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012;2(9):798–811. doi: 10.1158/2159-8290.CD-12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubaux R, Thu KL, Coe BP, et al. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol. 2013;8(8):1102–6. doi: 10.1097/JTO.0b013e318298762f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31(2):286–99. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toyokawa G, Takada K, Tagawa T, et al. Prevalence of enhancer of zeste homolog 2 in patients with resected small cell lung cancer. Anticancer Res. 2018;38(6):3707–11. doi: 10.21873/anticanres.12649. [DOI] [PubMed] [Google Scholar]
- 31.Sato T, Kaneda A, Tsuji S, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagarsheth N, Peng D, Kryczek I, et al. PRC2 epigenetically silences Th1-type chemokines to suppress effector T-cell trafficking in colon cancer. Cancer Res. 2016;76(2):275–82. doi: 10.1158/0008-5472.CAN-15-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng D, Kryczek I, Nagarsheth N, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527(7577):249–53. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bugide S, Green MR, Wajapeyee N. Inhibition of enhancer of zeste homolog 2 (EZH2) induces natural killer cell-mediated eradication of hepatocellular carcinoma cells. Proc Natl Acad Sci USA. 2018;115(15):E3509–18. doi: 10.1073/pnas.1802691115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasen H, Ibrahim I, Ponce CG, et al. Benefit of surveillance for pancreatic cancer in high-risk individuals: Outcome of long-term prospective follow-up studies from three European expert xenters. J Clin Oncol. 2016;34(17):2010–19. doi: 10.1200/JCO.2015.64.0730. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y-C, Huang C-C, Lin D-Y, et al. Overexpression of centromere protein K (CENPK) in ovarian cancer is correlated with poor patient survival and associated with predictive and prognostic relevance. Peer J. 2015;3:e1386. doi: 10.7717/peerj.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6(11):9445–56. doi: 10.18632/oncotarget.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ERCC1 deficiency. Cancer Res. 2014;74(10):2835–45. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yashiro T, Koyama-Saegusa K, Imai T, et al. Inhibition of potential lethal damage repair and related gene expression after carbon-ion beam irradiation to human lung cancer grown in nude mice. J Radiat Res. 2007;48(5):377–83. doi: 10.1269/jrr.07029. [DOI] [PubMed] [Google Scholar]
- 40.Höglund A, Nilsson LM, Muralidharan SV, et al. Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells. Clin Cancer Res. 2011;17(22):7067–79. doi: 10.1158/1078-0432.CCR-11-1198. [DOI] [PubMed] [Google Scholar]