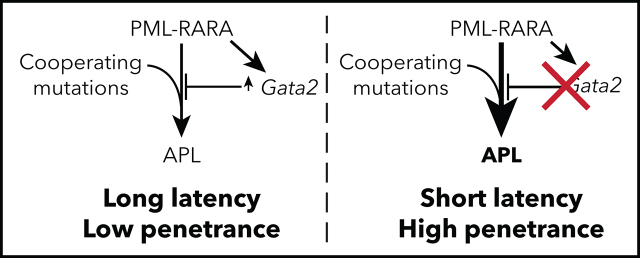
Katerndahl et al investigate genes dysregulated by PML-RARA in mouse and human cells and demonstrate that GATA2 is highly expressed in preleukemic cells expressing PML-RARA; furthermore, somatic GATA2 mutations occur during progression of acute promyelocytic leukemia (APL). They demonstrate that GATA2 knockout increases APL penetrance and decreases latency, while restoration of GATA2 expression suppresses penetrance and increases latency. These studies suggest a role for GATA2 as a tumor suppressor in AML development.
Key Points
Gata2 inactivation cooperates with PML-RARA, RUNX1-RUNX1T1, and Cebpa mutations.
Ectopic Gata2 expression suppresses PML-RARA–driven cell growth, even in fully transformed APL cells.
Visual Abstract
Abstract
Most patients with acute promyelocytic leukemia (APL) can be cured with combined all-trans retinoic acid (ATRA) and arsenic trioxide therapy, which induces the destruction of PML-RARA, the initiating fusion protein for this disease. However, the underlying mechanisms by which PML-RARA initiates and maintains APL cells are still not clear. Therefore, we identified genes that are dysregulated by PML-RARA in mouse and human APL cells and prioritized GATA2 for functional studies because it is highly expressed in preleukemic cells expressing PML-RARA, its high expression persists in transformed APL cells, and spontaneous somatic mutations of GATA2 occur during APL progression in mice and humans. These and other findings suggested that GATA2 may be upregulated to thwart the proliferative signal generated by PML-RARA and that its inactivation by mutation (and/or epigenetic silencing) may accelerate disease progression in APL and other forms of acute myeloid leukemia (AML). Indeed, biallelic knockout of Gata2 with CRISPR/Cas9-mediated gene editing increased the serial replating efficiency of PML-RARA–expressing myeloid progenitors (as well as progenitors expressing RUNX1-RUNX1T1, or deficient for Cebpa), increased mouse APL penetrance, and decreased latency. Restoration of Gata2 expression suppressed PML-RARA–driven aberrant self-renewal and leukemogenesis. Conversely, addback of a mutant GATA2R362G protein associated with APL and AML minimally suppressed PML-RARA–induced aberrant self-renewal, suggesting that it is a loss-of-function mutation. These studies reveal a potential role for Gata2 as a tumor suppressor in AML and suggest that restoration of its function (when inactivated) may provide benefit for AML patients.
Introduction
Inherited mutations in the transcription factor GATA2 cause monocytopenia and mycobacterial infection (MonoMAC) syndrome and Emberger syndrome, which strongly increase the risk of developing myelodysplastic syndrome or acute myeloid leukemia (AML; 80% probability by age 40).1-3 Somatic GATA2 mutations have been reported in ∼3.4% (135/4019 cases)4-10 of AML patients, especially in association with CEBPA mutations, where ∼21.1% (89/422 cases)4-6,8-12 have GATA2 mutations. Somatic GATA2 mutations have also been reported in AML cases initiated by mutations in DNMT3A and ASXL1 and several fusion oncogenes (eg, RUNX1-RUNX1T1, MLL fusions, and PML-RARA).5,9,13,14 Therefore, GATA2 mutations are found in many AML subtypes, suggesting that they can cooperate with several initiating mutations to promote AML.15,16 GATA2 is highly expressed in normal CD34+ cells, but not in T cells, B cells, or mature myelomonocytic cells (including promyelocytes).6 GATA2 is also expressed in virtually all AML subtypes (including acute promyelocytic leukemia [APL]) and in most AML samples tested to date.6 However, there is no consensus on the mechanism by which GATA2 contributes to AML or whether GATA2 acts as an oncogene or a tumor suppressor.
Some reports have suggested that GATA2 expression may promote AML by increasing cell proliferation17,18 and that high expression correlates with poor outcomes.19 However, other reports have suggested that GATA2 inhibits the proliferation of hematopoietic progenitors16,20,21 and that GATA2 function may be disrupted in many cases of AML.3,22-29 Finally, GATA2 mutations in AML are nearly always heterozygous,9,22,24 and the remaining GATA2WT allele is often epigenetically silenced,8,22,24,30,31 suggesting that abrogation of GATA2 function may contribute to AML progression in some patients.
During normal hematopoiesis, GATA2 promotes the endothelial to hematopoietic transition during embryogenesis.32,33 Therefore, Gata2-knockout mice are embryonically lethal as the result of a lack of hematopoietic stem/progenitor cells (HSPCs) and anemia.32,34 Conditional deletion of Gata2 revealed that it is also required for the maintenance and survival of HSPCs in adult bone marrow.35 Therefore, Gata2 is thought to promote the self-renewal of HSPCs during normal hematopoiesis.36
In this study, we examined the role of GATA2 in AML pathogenesis using the well-characterized Ctsg-PML-RARA mouse model, in which human PML-RARA is knocked into the 5′ untranslated region of the mouse Ctsg gene.37 This leads to the expression of PML-RARA in early myeloid progenitors,38 aberrant self-renewal in replating assays,39 and the development of APL with long latency (∼50% penetrance at 1 year in C57BL/6 mice).37 Using this mouse model, along with expression data from human APL samples, we were able to define genes that are canonically dysregulated in preleukemic and fully transformed APL cells. Among these genes, GATA2 has unique properties that strongly suggest that it plays a role in the pathogenesis of this disease, as well as other AML subtypes.
Methods
Study approval
All mouse experiments were done in accordance with institutional guidelines and approved by the Animal Studies Committee at Washington University.
Mice
Details about the mouse strains, husbandry, and analysis are provided in supplemental Methods, available on the Blood Web site.
Exome and bulk RNA sequencing and single-cell RNA sequencing
Details are provided in supplemental Methods.
CRISPR/Cas9 gene editing
Details about Gata2-directed gene editing and analysis are provided in supplemental Methods.
Methylcellulose colony assays
Methylcellulose assays were performed as previously described39 and are detailed in the supplemental Methods.
Retroviral transductions
Details about the retroviral transduction strategies are provided in supplemental Methods.
Western blotting
Western blots were performed using the Jess Western Blotting System (ProteinSimple; https://www.proteinsimple.com/jess.html), as described in supplemental Methods.
Flow cytometry and cellular purification
Flow studies were performed as previously described.40 Mouse promyelocytes were flow purified as previously described.38
ATRA treatment of APL mice
Details about all-trans retinoic acid (ATRA) pellet treatment of APL-engrafted mice are presented in supplemental Methods.
Details about gene set enrichment analysis are presented in supplemental Methods.
Sex differences
Sex differences were evaluated for the donor source of mice for all assays, including colony assays, APL penetrance studies, and ATRA sensitivity. None of the sex-based differences were statistically significant.
Results
GATA2/Gata2 is dysregulated and mutated in human and mouse APL
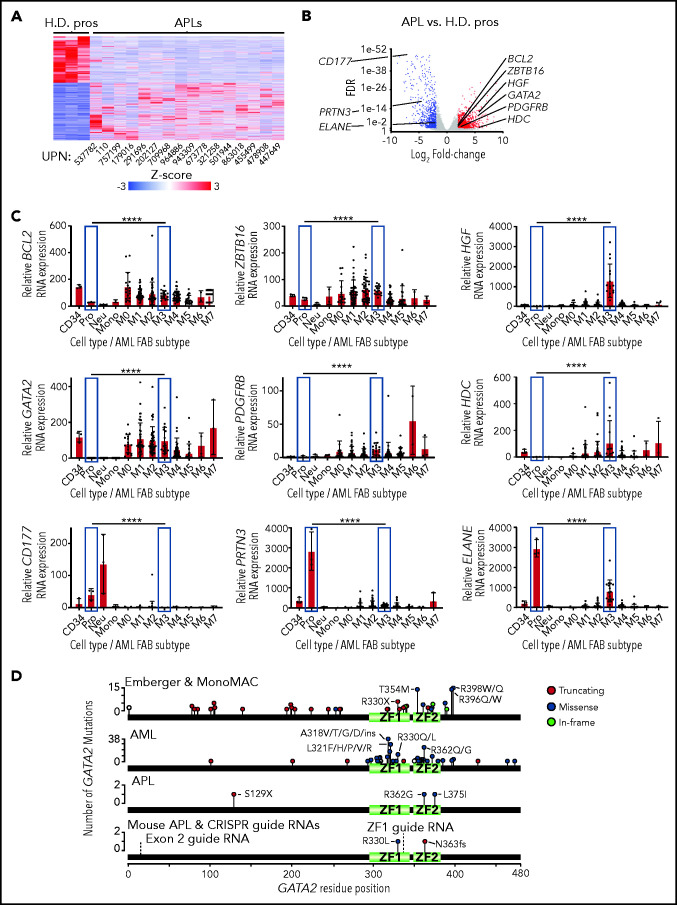
To identify genes that are dysregulated by PML-RARA in human APL patients, we evaluated bulk RNA-sequencing (RNA-seq) data from the AML TCGA study, derived from 16 primary human APL cases and 3 normal promyelocyte-enriched samples.6 Using this approach, we identified 3922 differentially expressed genes (DEGs), of which 2189 were upregulated in APL and 1733 were downregulated (fold change ≥2 and false discovery rate [FDR] <0.05 by analysis of variance [ANOVA]; Figure 1A-B; supplemental Table 1). Several of the genes that are differentially expressed in APL have previously been implicated in AML, APL, self-renewal, and/or myeloid development, including the upregulated genes BCL2 (2.9×; previously shown to cooperate with PML-RARA to decrease the latency of APL39), ZBTB16/PLZF1 (2.2×), GATA2 (63.9×), HGF (55.7×), PDGFRB (9.3×), and HDC (64.3×) and the downregulated genes PRTN3 (25.8×), ELANE (3.6×), and CD177 (157.8×). GATA2 is normally expressed in CD34+ cells, but its expression is essentially absent in normal promyelocytes, neutrophils, and monocytes. However, GATA2 is highly expressed in most AML French-American-British subtypes, (including M3/APL) (Figure 1C). In APL patients, somatic GATA2 mutations have been reported in ∼4.5% of cases that have been sequenced to date (6/134 cases)4,6-8,10,14; GATA2 mutations cooperate with PML-RARA,7 as well as with rare APL-initiating fusions14 (Figure 1D).
Figure 1.
GATA2 expression in normal and malignant hematopoietic cells. (A) Heat map of the 4094 DEGs between primary human APLs and healthy donor promyelocytes (H.D. pros) by RNA-seq fold change ≥2 and FDR <0.05. (B) Volcano plot of the samples from panel A. Expression changes of BCL2, ZBTB16, HGF, GATA2, PDGFRB, HDC, CD177, PRTN3, and ELANE are labeled. (C) BCL2, ZBTB16, HGF, GATA2, PDGFRB, HDC, CD177, PRTN3, and ELANE expression in flow purified healthy donor human CD34+ progenitors (CD34), promyelocytes (Pro), neutrophils (Neu), monocytes (Mono), and the AML French-American-British subtypes M0-M7 by RNA-seq using the AML TCGA data set.6 Promyelocytes and M3 (APL) subtype samples are highlighted in blue. Statistical significance was determined by edgeR, which includes correction for multiple testing. P values are corrected for multiple testing. (D) Distribution of GATA2 mutations from patients with Emberger syndrome, monocytopenia and mycobacterial infection (MonoMAC) syndrome, AML, or APL,1-5,7,11,30,34,59-64 as well as those from mouse APLs identified by exome sequencing. The locations of CRISPR guide RNAs used in this study (labeled exon 2 and ZF1) are annotated with dashed lines. ****P < .0001. UPN, unique patient number; ZF1, zinc finger domain 1; ZF2, zinc finger domain 2.
To determine whether Gata2 is mutated in APLs arising spontaneously in Ctsg-PML-RARA mice, we performed exome sequencing of 13 APL samples with paired normal (tail DNA) samples to define somatic mutations and copy number events (supplemental Figure 1; supplemental Table 2). Similar to previous studies, 2 of these APLs contained interstitial deletions of chromosome 2 that included the PU.1/Spi1 and Wt1 genes42,43; 7 contained amplifications of chromosome 15, where Myc is located42,44,45; and 9 had loss of 1 copy of the X chromosome, where Kdm6a is located.46 Mutations in the AML-associated genes Eppk1, Ptpn11, Kras, Jak1, and Kdm6a were identified in 1 APL sample each,6,46,47 and a novel somatic frameshift mutation that produces a premature in-frame stop codon was identified in Gata2 (N363fs) in 1 case. No deletions of the Gata2 locus were identified. To determine whether Gata2 was recurrently mutated in this model, we sequenced all exons of the Gata2 gene in 94 additional mouse APL samples and discovered an additional somatic R330L mutation in 1 case (data not shown). The R330L and N363fs mutations are located in zinc finger 1 and zinc finger 2 of Gata2, respectively.
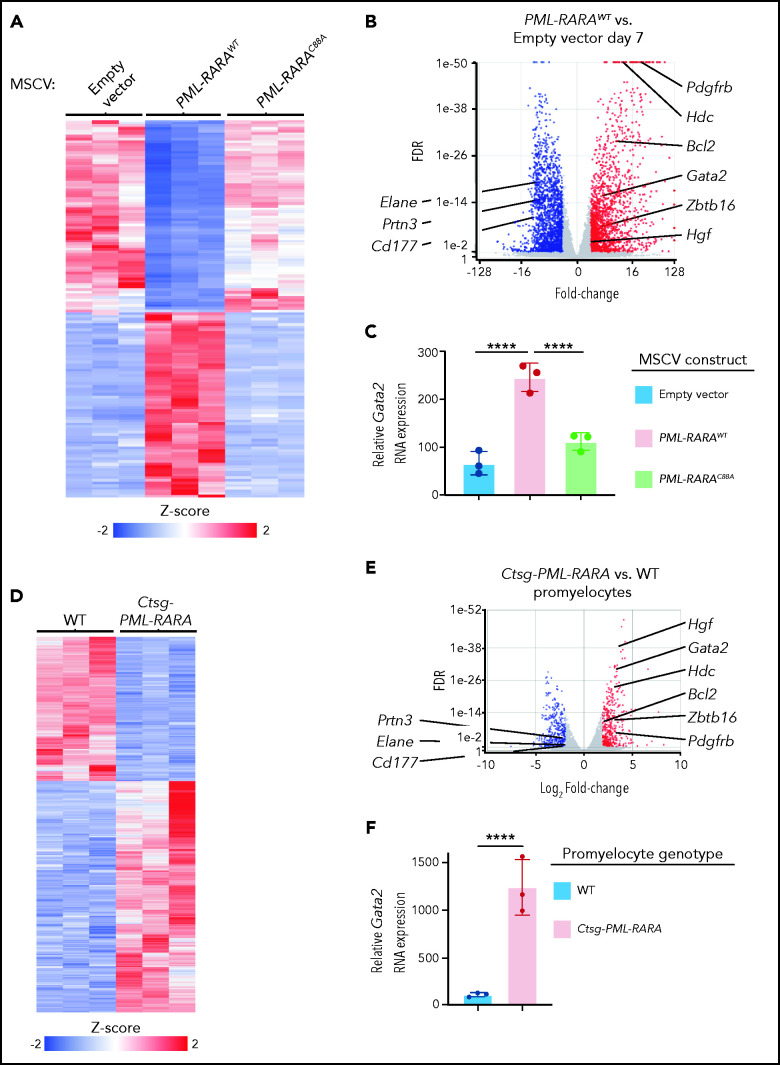
To more precisely identify genes that may be dysregulated by PML-RARA, we transduced lineage-depleted wild-type (WT) mouse bone marrow cells with murine stem cell virus (MSCV)–internal ribosome entry site (IRES)-GFP–based retroviruses containing no insert (empty vector), WT PML-RARA complementary DNA, or a C88A mutant of PML-RARA (PML-RARAC88A). The C88A mutation has previously been shown to abrogate the DNA-binding activity of PML-RARA,46 and we found that PML-RARAC88A was unable to induce aberrant self-renewal (supplemental Figure 2A-C), despite its robust expression 3 days following transduction (supplemental Figure 2B). Therefore, the PML-RARAC88A fusion serves as a control for genes regulated by PML-RARA. Seven days after transduction, we sorted on GFP+ cells and performed RNA-seq. This analysis led to the identification of 3299 DEGs when comparing PML-RARAWT–transduced cells with empty vector (or PML-RARAC88A)–transduced cells (1552 upregulated, 1747 downregulated with PML-RARAWT; fold change ≥2 and FDR <0.05 by DESeq2; Figure 2A-C; supplemental Figure 2C; supplemental Table 3). PML-RARAC88A led to the dysregulation of a smaller number of DEGs (353 upregulated, 202 downregulated compared with empty vector; fold change ≥2 and FDR <0.05 by DESeq2), perhaps via its ability to interact with RXRA.46 As noted in the corresponding volcano plot (Figure 2B), expression of PML-RARAWT for 7 days caused the dysregulation of many of the same genes that were differentially expressed in human APL samples, including Gata2 (Figure 2C).
Figure 2.
Increased Gata2 expression in preleukemic mouse myeloid cells expressing PML-RARA. (A) Heat map of the 3361 DEGs between lineage-depleted and flow-purified GFP+ WT mouse bone marrow cells 7 days following retroviral transduction with PML-RARAWT compared with those transduced with an empty MSCV-IRES-GFP vector control by bulk RNA-seq (fold change ≥2 and FDR <0.05). Also plotted is the expression of the same DEGs in cells transduced with PML-RARAC88A MSCV-IRES-GFP vector. (B) Volcano plot of the expression changes in (A). Upregulated (red) and downregulated (blue) DEGs (fold change ≥2 and FDR <0.05) are highlighted. Expression changes in Bcl2, Zbtb16 (Plzf1), Hgf, Pdgfrb, Hdc, Elane, Prtn3, and Cd177 are labeled. (C) Relative Gata2 RNA expression in lineage-depleted and flow-purified GFP+ WT mouse bone marrow cells 7 days following retroviral transduction with PML-RARAWT, PML-RARAC88A, or an empty MSCV-IRES-GFP vector. Data in panels A-C are from 3 independent biological replicates, each from a separate 6- to 8-week-old WT mouse. (D) Heat map of the 1838 DEGs between flow-purified promyelocytes from preleukemic Ctsg-PML-RARA mice compared with those from WT littermates by bulk RNA-seq (fold change ≥2 and FDR <0.05). (E) Volcano plot of the expression changes in (D). Upregulated (red) and downregulated (blue) DEGs (fold change ≥2 and FDR <0.05) are highlighted. Expression changes in Bcl2, Zbtb16 (Plzf1), Hgf, Pdgfrb, Hdc, Elane, Prtn3, and Cd177 are labeled. (F) Relative Gata2 RNA expression in flow-purified promyelocytes from preleukemic Ctsg-PML-RARA mice compared with those from WT mice by RNA-seq. Data in panels D-F are from 3 pairs of mice that were littermate matched and 6 to 8 weeks of age; each flow-purified promyelocyte sample was from a unique mouse. P values are corrected for multiple testing. ****P < .0001.
To extend these results, we performed RNA-seq on flow-sorted preleukemic promyelocytes from Ctsg-PML-RARA and littermate-matched WT mouse bone marrow samples. We identified 1826 DEGs comparing Ctsg-PML-RARA vs WT promyelocytes (959 upregulated and 867 downregulated with PML-RARA expression; fold-change ≥2 and FDR <0.05 by ANOVA; Figure 2D-F; supplemental Figure 2D; supplemental Table 4). Two hundred and forty-one of these DEGs were coordinately dysregulated in the mouse and human data sets (135 upregulated and 106 downregulated; Figure 2E), including Gata2 (Figure 2F). Among these DEGs, only 2, Gata2 (upregulated 10.1-fold) and Kit (upregulated 2.93-fold), were coordinately dysregulated in preleukemic mouse and human APL samples.
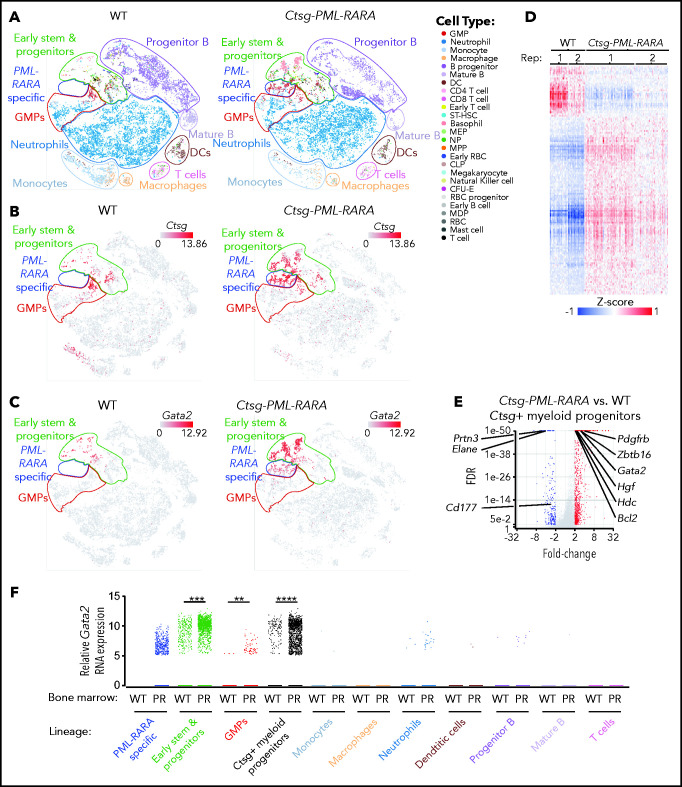
To better define the effects of PML-RARA on bone marrow populations and gene expression, we performed single-cell RNA-seq (scRNA-seq) on bone marrow cells from young littermate-matched nonleukemic Ctsg-PML-RARA and WT mice. This analysis revealed an expansion of myeloid progenitors in Ctsg-PML-RARA marrow (Figure 3A)37 that was restricted to the Ctsg-expressing (Ctsg+) myeloid progenitor cell population (Figure 3B) (PML-RARA expression is directed by the Ctsg locus in this model). Ctsg+ myeloid progenitors from WT and Ctsg-PML-RARA mice expressed several markers of myeloid precursors, including Cd34, Mpo, and Elane (supplemental Figure 3A). Therefore, Ctsg marks normal myeloid progenitors and precursors from WT and Ctsg-PML-RARA mice. We identified a unique cluster of myeloid progenitor cells in Ctsg-PML-RARA marrow and identified DEGs within this compartment: 850 genes were upregulated and 240 were downregulated in the PML-RARA–expressing cells compared with WT Ctsg+ myeloid progenitor cells (fold-change ≥ 2 and FDR < 0.05 by ANOVA; Figure 3C-E; supplemental Table 5). Gene set enrichment analysis revealed a positive correlation with the DEGs identified in this scRNA-seq data set, the human APL RNA-seq data set, the mouse promyelocyte RNA-seq data set, and a previously published data set of APL-specific DEGs47 (supplemental Figure 3B-D). More specifically, among these 4 data sets, 37 genes showed coordinate dysregulation (24 upregulated and 13 downregulated), including Gata2, which was upregulated 3.2-fold in PML-RARA–expressing myeloid progenitors vs WT progenitors (FDR <1E-307; P < 1E-303; Figure 3F).
Figure 3.
PML-RARA expression drives the formation of a unique population of preleukemic myeloid progenitors that express Gata2. (A) t-Distributed stochastic neighbor embedding (t-SNE) plots of scRNA-seq data from whole bone marrow cells from young nonleukemic WT mice (left panel) or Ctsg-PML-RARA mice (right panel) mice. Known hematopoietic cell types are labeled based on Haemopedia gene expression profiling40,58). Major cell populations (early stem and progenitors, GMPs, neutrophils, monocytes, macrophages, T cell, DCs, mature B, and progenitor B) are inferred, outlined, and labeled based on Haemopedia lineage assignment software and based on graph-based clustering analysis. A unique population of myeloid precursor cells that are only present in the bone marrow from Ctsg-PML-RARA mice is outlined in blue (“PML-RARA specific”). (B) t-SNE plots of the relative expression of Ctsg in whole bone marrow cells from WT mice (left panel) or Ctsg-PML-RARA mice (right panel) by scRNA-seq. (C) t-SNE plots of the relative expression of Gata2 in whole bone marrow cells from WT mice (left panel) or Ctsg-PML-RARA mice (right panel) by scRNA-seq. (D) Heat map of the 1090 DEGs between Ctsg+ myeloid precursors from Ctsg-PML-RARA mice compared with those from WT littermates by scRNA-seq. Replicate littermate pairs are labeled as Rep 1 and Rep 2 (fold change ≥2 and FDR <0.05 by ANOVA). (E) Volcano plot of expression changes between Ctsg+ myeloid progenitors from Ctsg-PML-RARA vs WT mouse bone marrow. Expression changes in Bcl2, Zbtb16 (Plzf1), Hgf, Pdgfrb, Hdc, Elane, Prtn3, and Cd177 are labeled. (F) Relative Gata2 expression by scRNA-seq in the various lineage populations from panel A in WT or Ctsg-PML-RARA (PR) bone marrow. Data in panels A-F are from 2 pairs of mice that were littermate matched and 8 to 12 weeks of age. Samples of the same genotype were concatenated for the analyses in panels A-C and E-F. **P = 1.37E-4, ***P = 1.73E-142, ****P < 1.00E-298; ANOVA. CFU-E, colony forming unit-erythroid; CLP, common lymphoid progenitor; DC, dendritic cell; GMPs, granulocyte-monocyte progenitors; MDP, monocyte dendritic cell progenitor; MEP, megakaryocyte erythroid progenitor; MPP, multipotent progenitor; NP, neutrophil progenitor; RBC, red blood cell; ST-HSC, short-term hematopoietic stem cell.
Gata2 suppresses serial replating induced by PML-RARA, RUNX1-RUNX1T1, and Cebpa inactivation
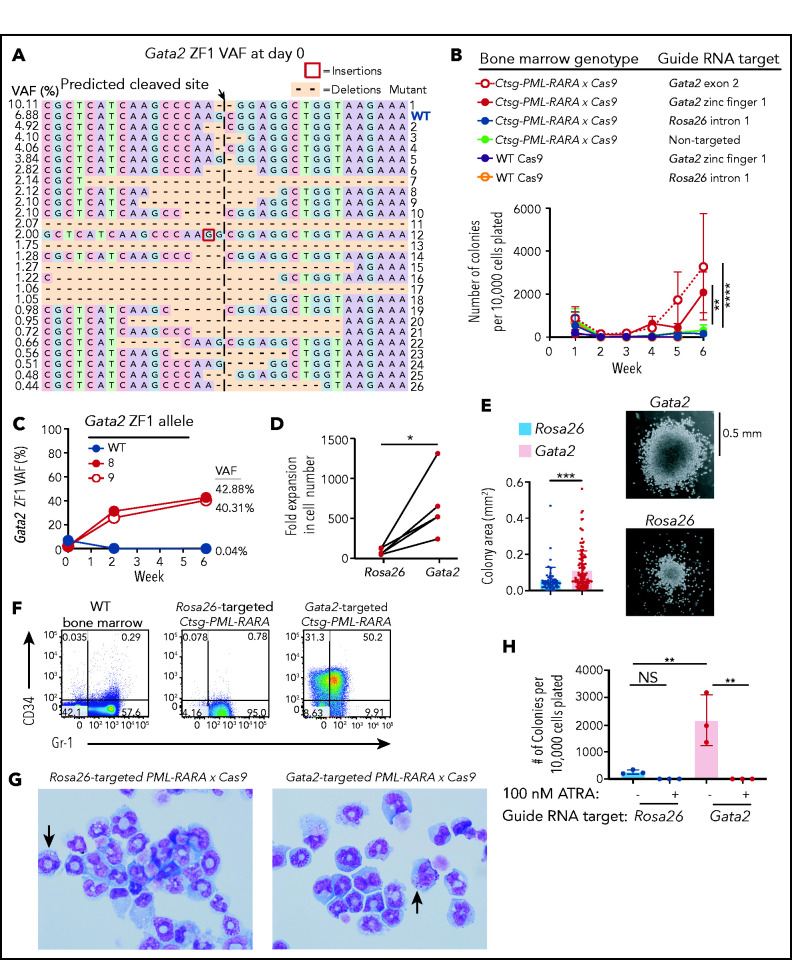
To determine whether Gata2 upregulation associated with PML-RARA expression was acting as a tumor promoter or a tumor suppressor, we inactivated it and assessed the consequences. We first generated PML-RARA × Cas9-GFP mice (hereafter referred to as PML-RARA × Cas9 mice), and then electroporated bone marrow from these mice with CRISPR guide RNAs that targeted the 5′ end (exon 2) of Gata2, the zinc finger 1 domain of Gata2, or intron 1 of Rosa2648 (as a neutral locus control; Figure 1D). Digital sequencing of polymerase chain reaction products centered around the targeted sites revealed the production of a wide “library” of insertion and deletion mutations at the appropriate target sites for each guide RNA (Figure 4A; supplemental Figure 4A,C), with an average targeting efficiency of 76.2% at the 3 target sites. Cells were then serially replated in MethoCult M3534 for 6 weeks. Targeting Rosa26 with CRISPR-Cas9 guide RNAs did not affect the ability of PML-RARA × Cas9 bone marrow cells to replate, as expected (Figure 4B). Gata2 inactivation caused a 17.0-fold increase in the serial replating efficiency of PML-RARA × Cas9 bone marrow compared with Rosa26-targeted cells or untransfected cells (nontargeted; Figure 4B). In addition, digital sequencing revealed that biallelic loss-of-function mutations of Gata2 were selected for with serial replating (Figure 4C; supplemental Figure 4A-B). However, Gata2 targeting with guide RNAs did not cause an increase in the replating efficiency within the first 3 weeks, even though Gata2 loss-of-function mutations were selected for within 2 weeks. This suggests that myeloid progenitor cells require several weeks to overcome the transcriptional program dictated by Gata2. Gata2 targeting in WT Cas9-GFP bone marrow cells did not induce serial replating, nor did targeting the Rosa26 locus (Figure 4B; supplemental Figure 4C-D). Gata2 deficiency in PML-RARA–expressing cells caused 2 effects: a 7.7-fold expansion in colonies (Figure 4D) and the production of larger colonies (1.43-fold increase in colony diameter) (Figure 4E). Phenotypically, the Rosa26- and Gata2-targeted PML-RARA × Cas9 cells maintained myeloid lineage markers by flow cytometry (Figure 4F; supplemental Figure 4E). However, a greater proportion of Gata2-targeted cells expressed CD34 and coexpressed CD34 and Gr-1 (32.6- and 30.7-fold increase in these populations respectively), a distinct feature of APL in this model system.36 Wright-Giemsa staining confirmed that the Gata2-targeted cells had early myeloid features with azurophilic granules (Figure 4G). Finally, Gata2-and Rosa26-targeted PML-RARA × Cas9 cells were equally sensitive to ATRA, indicating that Gata2-deficient cells are still dependent upon PML-RARA expression to cause the replating phenotype (Figure 4H).
Figure 4.
Effects of biallelic Gata2 mutations on PML-RARA–induced serial replating. Serial replating assays using preleukemic lineage-depleted PML-RARA × Cas9 or WT Cas9 bone marrow cells that were electroporated with CRISPR guide RNAs targeting Gata2 zinc finger domain 1 (ZF1), Gata2 exon 2, or Rosa26 intron 1 (neutral mutation control) prior to initial plating in MethoCult M3534. Cells were serially replated for 6 weeks. (A) The initial Gata2 variant allele frequencies (VAF) at day 0 (day of initial plating in MethoCult) by digital sequencing following CRISPR targeting with a Gata2 ZF1-specific guide RNA. Data are representative of 3 independent biological replicates. (B) Colony counts from serial replating assays in which preleukemic PML-RARA × Cas9 or WT Cas9 bone marrow cells were electroporated with CRISPR guide RNAs targeting Gata2 ZF1, Gata2 exon 2, or Rosa26 or were left untransfected (nontargeted) prior to initial plating in MethoCult. Three or 4 independent biological replicates are plotted for each guide RNA target. **P < .01, ****P < .0001; 2-way ANOVA at week 6. (C) VAFs of the Gata2 ZF1 alleles WT, 8, and 9 from panel A over time. Data are representative of 3 independent biological replicates. (D) The fold expansion in the total number of live cells from the number that was plated at the beginning of week 6 compared with those at the end of week 6 for Rosa26- and Gata2-targeted PML-RARA × Cas9 cells from panel B. Gata2-targeted samples include samples electroporated with ZF1 or exon 2 Gata2 guide RNAs. Five independent biological replicates are plotted for each target (Rosa26 or Gata2). *P < .05; paired, 2-tailed Student t test. (E) The colony area of Rosa26- and Gata2-targeted PML-RARA × Cas9 cells at week 6 (left panel). Data from 2 independent biological replicates are plotted for each target (Rosa26 or Gata2). Gata2-targeted samples include those electroporated with a ZF1 or an exon 2 Gata2 guide RNA. ***P < .001; unpaired, 2-tailed Student t test. Representative images of colony morphologies and sizes from PML-RARA × Cas9 cells electroporated with Gata2 or Rosa26 guide RNAs (right panels). Images were captured using an Olympus IX-51 microscope and a QImaging QIClick camera (original magnification ×4). (F) CD34 and Gr-1 expression by flow cytometry on WT bone marrow (not replated in MethoCult) or on Rosa26- or Gata2-targeted PML-RARA × Cas9 cells from panel A replated in MethoCult for 8 weeks. Data are representative of 3 or 4 independent biological replicates. (G) Wright-Giemsa staining of Rosa26 or Gata2 exon 2–targeted PML-RARA × Cas9 cells from panel A replated in MethoCult for 8 weeks and then cultured in suspension media containing SCF, FLT3L, interleukin-3, and TPO for 2 days. Arrows point to cells containing prominent cytoplasmic azurophilic granules. Data are representative of 3 independent biological replicates. Images were captured using an Olympus BX-53 microscope and an Olympus DP-72 camera (original magnification ×100). (H) The number of colonies per 10 000 cells plated at week 6 for Rosa26-targeted or Gata2-targeted PML-RARA × Cas9 cells that were cultured with 100 nM ATRA (+) or DMSO vehicle control (-). Data from 3 independent biological replicates are plotted for each target. **P < .01; 1-way ANOVA. NS, not significant.
We next tested whether Gata2 suppresses the aberrant self-renewal known to be caused by RUNX1-RUNX1T1 expression49,50 or Cebpa inactivation.51 To study RUNX1-RUNX1T1–induced self-renewal, we transduced WT Cas9-GFP bone marrow cells with RUNX1-RUNX1T1 or empty vector MSCV retroviruses. Cells were then electroporated with CRISPR guide RNAs targeting Gata2 or Rosa26 and serially replated in MethoCult. We found that biallelic loss-of-function mutations in Gata2 caused a 2.2- to 5.7-fold increase in the serial replating of RUNX1-RUNX1T1–expressing myeloid progenitors compared with those targeted at Rosa26 (supplemental Figure 5). To determine whether Gata2 inactivation affected the enhanced serial replating caused by biallelic Cebpa mutations, we used CRISPR guide RNAs to target the p42 isoform of Cebpa, which was previously shown to act as a tumor suppressor in AML.51 The resulting biallelic Cebpa mutations caused aberrant serial replating and were positively selected for over time (supplemental Figure 6). The subsequent targeting of Gata2 caused an additional 4.1-fold increase in the serial replating efficiency of Cebpa-deficient progenitors (compared with Cebpa-only or Cebpa- and Rosa26-targeted cells), whereas targeting the Rosa26 locus had no effect.
Gata2 deficiency affects APL penetrance and latency in Ctsg-PML-RARA mice
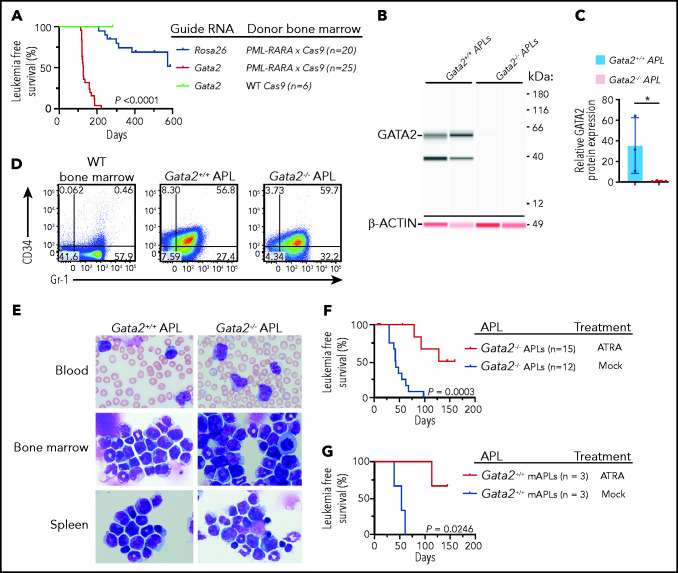
Based on the replating phenotype, we predicted that Gata2 deficiency would increase APL penetrance and decrease latency in the Ctsg-PML-RARA mouse model. Therefore, we electroporated PML-RARA × Cas9 bone marrow cells with CRISPR guide RNAs targeting Gata2 or Rosa26 and transplanted the cells into sublethally irradiated mice. Mice that received Gata2-targeted PML-RARA × Cas9 cells developed APL with 100% penetrance at 7.5 months posttransplantation compared with a concurrent cohort that received Rosa26-targeted PML-RARA × Cas9 cells and developed APL with 20% penetrance at 15 months posttransplantation (P < .0001, log-rank [Mantel-Cox] test). The median leukemia-free survival of mice transplanted with Gata2-targeted cells was 141 days vs >361 days for Rosa26-targeted cells (P < .01; Figure 5A). Targeting of Gata2 with guide RNAs in WT Cas9-GFP bone marrow cells did not lead to any leukemic deaths (Figure 5A). Digital sequencing and western blot analysis of APL samples showed that biallelic loss-of-function mutations were selected for in the APL samples in vivo, resulting in a complete absence of GATA2 protein in these tumors (Figure 5B-C; supplemental Figure 7A-C). Moreover, digital sequencing showed that 28 of 28 single-cell clones (obtained by single-cell deposition from a flow cytometer) from Gata2-targeted APLs contained the same biallelic loss-of-function mutations as did the bulk tumor (ie, both mutations were present in the same cells; supplemental Figure 7D). Exome sequencing of the Gata2-deficient APLs revealed that CRISPR/Cas9 specifically targeted Gata2 in all of these APLs, resulting in biallelic frameshift mutations between aa 9 and 16 that caused premature stop codons in all 5 samples (supplemental Figure 8A; supplemental Table 6). In addition to the biallelic Gata2 mutations, 1 Gata2-deficient APL harbored a missense mutation (F837V) in the kinase domain of Jak1, similar to that previously described.44,52 Gata2-deficient APLs were also found to have interstitial deletions of chromosomes 2 and X (involving the Kdm6a gene), as well as an amplification of all or part of chromosome 15 (which contains Myc); all were described in APLs arising in Gata2-sufficient Ctsg-PML-RARA mice6,40-45 (supplemental Figure 8A-B). Therefore, for 4 of 5 Gata2-deficient APLs sequenced, additional cooperating events probably contributed to APL development. Flow cytometry revealed that the leukemic cells derived from the Gata2-deficient PML-RARA × Cas9 cells coexpressed CD34 and Gr-1, as described above (Figure 5D; supplemental Figure 8C-D). Morphologically, Gata2-deficient APLs closely resembled Gata2-sufficient APLs; both had abundant azurophilic granules characteristic of promyelocytes (Figure 5E). Gata2-deficient or -sufficient APLs were similarly sensitive to ATRA treatment in vivo (Figure 5F-G).
Figure 5.
Gata2 acts as a tumor suppressor in mouse APL. (A) Leukemia-free survival of recipient Ly5.1 mice transplanted with nonleukemic lineage-depleted PML-RARA × Cas9 bone marrow in which Rosa26 intron 1 or Gata2 exon 2 was targeted with CRISPR guide RNAs prior to transplantation (n = 20 mice in Rosa26 group from 2 cohorts of 10 mice per cohort; n = 25 mice in Gata2-targeted PML-RARA × Cas9 bone marrow group from 3 cohorts of 5 to 10 mice per cohort; n = 6 mice in Gata2-targeted WT Cas9 bone marrow group from 1 cohort of mice). P < .0001, log-rank (Mantel-Cox) test. (B) Protein expression, detected using a Jess by ProteinSimple apparatus, of GATA2 and β-ACTIN from the bone marrow of Gata2-sufficient (Gata2+/+) or Gata2-targeted (Gata2−/−) Ctsg-PML-RARA APL mice. Blots for GATA2 and β-ACTIN were performed on the same samples using 2 antibodies in 2 spectral channels: chemiluminescence (GATA2) and near infrared (β-ACTIN). Data are representative of 3 or 4 independent biological replicates per genotype. (C) Quantification of the relative GATA2/β-ACTIN protein expression from the bone marrow of Gata2+/+ or Gata2−/− Ctsg-PML-RARA APL mice. Data are from 3 or 4 independent biological replicates per genotype. (D) CD34 and Gr-1 expression by flow cytometry on bone marrow from 8- to 12-week-old WT mice, Gata2+/+ Ctsg-PML-RARA APL spleen cells, and Gata2−/− Ctsg-PML-RARA APL spleen cells. Data are representative of 3 or 4 independent biological replicates. (E) Wright-Giemsa staining of blood, bone marrow, and spleen from Gata2+/+ (Rosa26-targeted) or Gata2−/− (Gata2-targeted) Ctsg-PML-RARA APL mice. Images are representative of 5 to 8 independent biological replicates for each genotype. Images were captured using an Olympus BX-53 microscope and an Olympus DP-72 camera (original magnification ×100). (F) Leukemia-free survival of recipient Ly5.1 mice transplanted with fully transformed Gata2−/− Ctsg-PML-RARA APL spleen cells and then treated with surgically implanted ATRA pellets (black lines) or control pellets (Mock; red lines). N = 12 control-treated mice; N = 15 ATRA-treated mice. Gata2−/− APLs were derived from 3 biologically independent mice. P = .0003, log-rank (Mantel-Cox) test. (G) Leukemia-free survival of recipient Ly5.1 mice transplanted with Gata2+/+ Ctsg-PML-RARA APL spleen cells and then treated with surgically implanted ATRA pellets (black lines) or control pellets (Mock; blue lines). N = 3 control-treated mice, N = 3 ATRA-treated mice. Gata2+/+ APLs were derived from 1 mouse. P = .0246, log-rank (Mantel-Cox) test.
Tumor-suppressor activity of Gata2WT in APL cells
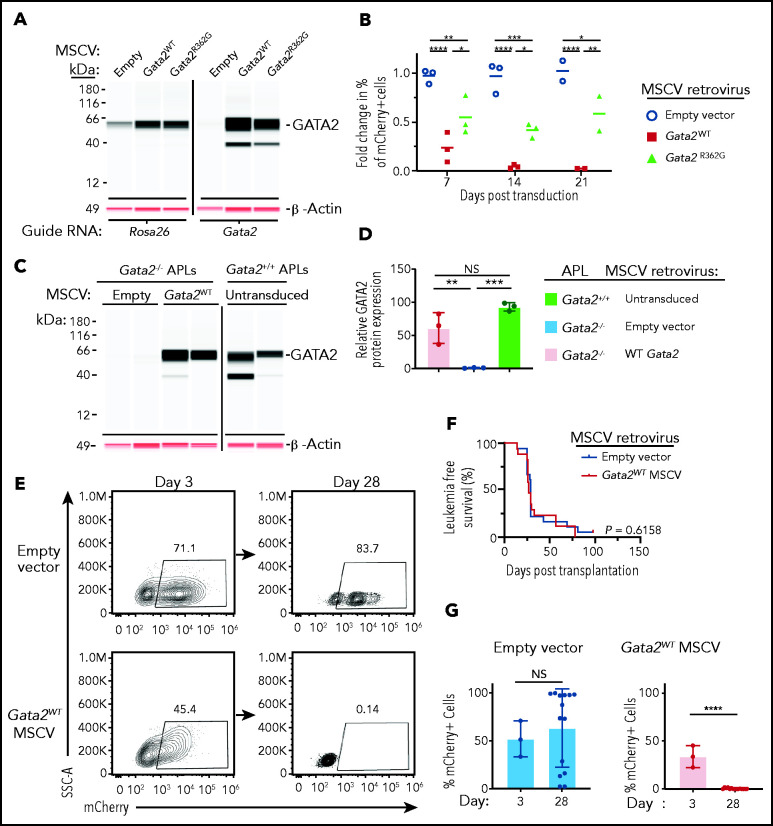
We next wished to determine whether Gata2 overexpression could suppress the replating phenotype in PML-RARA–expressing cells and whether the R362G mutation (found in APL and non-APL AML patients) had gain- or loss-of-function properties. We used MSCV-based retroviruses tagged with IRES-mCherry to express Gata2WT or Gata2R362G (or an empty vector control) in Gata2-deficient PML-RARA × Cas9 cells that had been replated in methylcellulose for 8 weeks (when Gata2-targeted clones with biallelic loss-of-function mutations predominate; Figure 4B-C; supplemental Figure 4A). We then serially replated unsorted mixtures of mCherry+ and mCherry− cells in MethoCult and quantified mCherry-expressing cells. This strategy led to a robust expression of GATA2WT and GATA2R362G protein within 3 days (Figure 6A). Similarly, the mCherry reporter, which identified the transduced cells, was expressed in ≥25% of the cells at day 3 posttransduction (Figure 6B). mCherry+ cells with Gata2WT were strongly selected against within 2 weeks of replating (40.7-fold reduction in the percentage of mCherry+ cells, compared with day 3 post transduction; P < .0001 by ANOVA; Figure 6B). In contrast, cells that were transduced with an empty vector showed no change in the fraction of mCherry+ cells during the same time period (1.03-fold reduction compared with day 3 posttransduction; P = .934, ANOVA). Cells transduced with the Gata2R362G vector developed a small decrease in the percentage of mCherry+ cells (1.7-fold reduction; P < .001, ANOVA), suggesting that this mutation confers a loss-of-function phenotype. We found similar results in Rosa26-targeted PML-RARA × Cas9 cells (Figure 6A; supplemental Figure 4F), suggesting that Gata2WT suppresses serial replating regardless of whether the cells are Gata2 sufficient or deficient. Consistent with these data, Gata2R362G showed a decreased capacity to suppress serial replating in Gata2-sufficient (Rosa26-targeted) PML-RARA × Cas9 cells.
Figure 6.
Restoration of Gata2 expression inhibits PML-RARA-induced aberrant self-renewal and leukemogenesis. (A) Retroviral “addback” of Gata2 in preleukemic PML-RARA–expressing cells. The initial protein expression detected using a Jess apparatus by ProteinSimple of GATA2 and β-ACTIN from preleukemic lineage-depleted PML-RARA × Cas9 bone marrow cells that were electroporated with CRISPR guide RNAs targeting Rosa26 intron 1 or Gata2 exon 2, serially replated in MethoCult M3534 for 8 weeks, and then transduced with empty vector, Gata2WT, or Gata2R362G MSCV-IRES-mCherry retroviruses. Samples are from cells that were grown in suspension culture with SCF, FLT3L, interleukin-3 (IL-3), and TPO (to minimize clonal selection pressures) for 3 days posttransduction, and then flow purified on mCherry+ cells. The Rosa26-targeted samples were run on a separate blot from the Gata2-targeted samples. Blots for GATA2 and β-ACTIN were performed using the same samples with 2 antibodies in 2 spectral channels: chemiluminescence (GATA2) and near infrared (β-ACTIN). (B) Fold change in the percentage of mCherry+ cells by flow cytometry in Gata2 exon 2–targeted PML-RARA × Cas9 bone marrow cells from panel A that were serially replated in MethoCult for 8 weeks, then retrovirally transduced with Gata2WT, Gata2R362G, or empty MSCV-IRES-mCherry vectors, and replated in MethoCult for an additional 2 to 3 weeks. Fold change is relative to the mCherry expression at day 3 posttransduction (n = 3 at days 7 and 14; n = 2 at day 21; independent biological replicates). *P < .05, **P < .01, ***P < .001,****P < .0001; 2-way ANOVA . (C) Retroviral “addback” of Gata2 in Gata2−/− APLs. Protein expression was detected using a Jess apparatus by ProteinSimple of GATA2 and β-ACTIN at day 3 following the transduction of Gata2−/− APL cells with empty vector, Gata2WT MSCV-IRES mCherry retroviruses, or untransduced Gata2+/+ APL cells. Cells were grown in suspension culture ex vivo with SCF, FLT3L, IL-3, and TPO (to minimize clonal selection pressures) for 3 days posttransduction. At day 3 posttransduction, APL cells were transplanted into sublethally irradiated Ly5.1 mice. Antibodies against GATA2 and β-ACTIN were used in the same samples in 2 spectral channels: chemiluminescence (GATA2) and near infrared (β-ACTIN). (D) Quantification of the relative GATA2/β-ACTIN protein expression in untransduced Gata2+/+ APLs (green bar), empty vector–transduced Gata2−/− APLs (blue bar), or Gata2WT MSCV–transduced Gata2−/− APLs (red bar) from the blots in panel C. **P < .01, ***P < .001; 2-way ANOVA. (E) mCherry expression by flow cytometry in Gata2−/− APL samples transduced with empty vector (upper panels) or Gata2 MSCV-IRES-mCherry (lower panels) retroviruses. Samples are from APLs that were grown ex vivo for 3 days posttransduction with cytokines (left panels) or those that were subsequently transplanted into Ly5.1 mice and harvested on day 28 (right panels). Day-28 samples are gated on CD45.2+ peripheral blood APL cells. (F) Leukemia-free survival of recipient Ly5.1 mice that were transplanted with unsorted mCherry− and mCherry+ Gata2−/− Ctsg-PML-RARA APL cells following transduction with empty vector or Gata2 MSCV-IRES-mCherry retroviruses. P = .6158; log-rank (Mantel-Cox) test. (G) Quantification of mCherry expression at day 3 and 28 in empty vector–transduced (left panel) and Gata2WT MSCV–transduced (right panel) APLs from panel D. In panels E-G, each of 3 independently derived Gata2−/− APLs was transduced with empty vector or Gata2 MSCV retroviruses and then transplanted into 5-6 Ly5.1 recipient mice (18 recipient mice received empty vector–transduced Gata2−/− APLs; 17 recipient mice received Gata2 MSCV-mCherry–transduced Gata2−/− APLs). ****P < .0001; unpaired, 2-tailed Student t test. NS, not significant.
We next asked whether a similar strategy could suppress the growth of fully transformed APL cells deficient for Gata2 in vivo. We transduced Gata2-deficient APL samples with Gata2WT or empty vector MSCV-IRES-mCherry–based retroviruses, cultured these cells for 3 days ex vivo, and then transplanted unsorted mixtures of mCherry+ and mCherry− APL cells into sublethally irradiated mice. Western blotting showed that GATA2 protein was expressed at high levels 3 days posttransduction (the day of transplantation) in samples that were transduced with Gata2WT (Figure 6C-D). mCherry was similarly expressed in a large percentage of cells transfected with the empty vector or the Gata2WT vector on the day of transplantation (Figure 6E). As expected, mice transplanted with either vector developed APL with similar penetrance and latency (because both cohorts received a mixture of untransduced and transduced cells; P = .6158 by log-rank [Mantel-Cox] test; Figure 6F). However, the APL samples transduced with the Gata2 vector contained no mCherry+ cells at day 28, suggesting that these cells were eliminated because they expressed Gata2 (Figure 6C-E,G). Conversely, APLs transduced with the empty vector continued to contain mCherry+ cells. Together, these data show that addback of Gata2 expression to Gata2-deficient APLs can suppress the growth of fully transformed APL cells.
Discussion
In this study we demonstrate that Gata2 acts as a tumor suppressor in the context of PML-RARA–driven aberrant self-renewal and leukemogenesis. We initially showed that Gata2 is aberrantly expressed in human and mouse models of APL, suggesting that PML-RARA may upregulate Gata2 expression, perhaps promoting self-renewal and APL initiation. However, we found that biallelic loss-of-function Gata2 mutations increased the efficiency of aberrant serial replating caused by PML-RARA, RUNX1-RUNX1T1, or Cebpa inactivation. Gata2 deficiency also increased the penetrance and decreased the latency of mouse APL. In addition, retroviral addback of Gata2WT expression was able to suppress PML-RARA–driven aberrant self-renewal and APL development, whereas addback of the Gata2R362G mutant did not, suggesting that this recurrent AML-associated mutation has a loss-of-function phenotype.
Why does PML-RARA upregulate a tumor suppressor in preleukemic cells? Proliferative stress caused by PML-RARA36 or other AML-initiating mutations may induce Gata2 expression indirectly, as a physiologic “brake” to limit an aberrant proliferative response. Gata2 has indeed been shown to inhibit the proliferation of hematopoietic progenitors in several contexts.16,20,21 Subsequent inactivating mutations of GATA2 may release this brake and allow for rapid AML progression. The high levels of GATA2 expression detected in nearly all AML samples6 strongly suggest that this may be a widespread phenomenon in this disease.
Alternatively, GATA2 may facilitate early steps in transformation by acting as an oncogene. In this model, after transformation has occurred, high GATA2 levels would be toxic, so that inactivation of GATA2 would be required for the leukemic cells to survive. Consistent with this model, Saida et al53 showed that Gata2 haploinsufficiency increases the latency of CBFB-MYH11–initiated AML in mice; however, the leukemias arising in these mice are more aggressive in transplantation experiments,53 suggesting that the residual Gata2WT allele may have been inactivated during tumor progression (although this was not reported in the article). In our study, only cells with biallelic Gata2 inactivation were selected for during in vitro and in vivo expansion, suggesting that the main function of GATA2 during AML progression is that of a tumor suppressor. This model fits with previous reports that described AML patients with heterozygous missense mutations in GATA2,3,8,15,22-27 where expression of the residual GATA2WT allele is nearly always lost by epigenetic silencing.8,22,24,30 Similarly, in AMLs with inv(3)/t(3;3) or atypical 3q26/MECOM translocations (which disrupt the −110 kb GATA2 enhancer), 1 GATA2 allele is silenced as a result of the translocation, and the remaining nonrearranged allele frequently acquires an inactivating mutation or its expression is lost by epigenetic silencing or deletion.8,16,23,31 Previous data also suggest that AML progression occurs when monoallelic expression of GATA2WT is lost in patients with germline GATA2 mutations.22
The molecular mechanisms responsible for high GATA2 expression levels in PML-RARA–expressing cells are unknown. It is most likely a transcriptional mechanism, but it is not yet clear whether GATA2 is a direct or an indirect target of PML-RARA. PML-RARA does bind near the GATA2 gene in APL cells and cell lines,54 but this observation does not establish cause and effect. In the MRP8-PML-RARA model of APL, Gata2 expression was found to be lower in “early” promyelocytes55; it is not clear whether this is due to a difference in the timing of PML-RARA expression during myeloid development or is related to the stage of development of the tested cells. Regardless, in the different models presented in this study, all were associated with increased levels of Gata2 expression, which corroborated the finding of persistently high GATA2 expression in most APL patients tested to date.
Several studies have now implied that the epigenetic repression of the residual WT allele in AMLs with heterozygous missense mutations in GATA2 is associated with methylation of the GATA2 locus,8,22,24 suggesting that hypomethylating agents may have a role in treating GATA2-mutant AML patients.24 One caveat, however, is that GATA2 missense mutations are nearly always progression events that occur in AML subclones.9,11,56 In this article, we have shown that somatically acquired Gata2 deficiency is not sufficient to directly cause AML in mice, similar to findings by other investigators.32,35 Therefore, GATA2 mutations are probably not initiating mutations in AML, and restoring GATA2 expression may only diminish the growth of GATA2-deficient subclones. However, we also noted that Gata2 overexpression reduced the growth of Gata2-sufficient hematopoietic cells, suggesting that it may have broad nonspecific effects on cell growth that could potentially affect nonmutant AML cells as well. Gata2 overexpression also promotes myeloid differentiation in WT hematopoietic progenitor cells36; therefore, enhancing its expression in AML cells may reduce growth and promote differentiation.
Our data complement other studies that have suggested that GATA2 normally acts as a tumor suppressor in AML cells and that the loss of this activity, by genetic or epigenetic means, accelerates AML progression. In AML cells in which GATA2 has been completely inactivated by the mutation of 1 allele and the residual WT allele has been repressed by DNA methylation, hypomethylating agents represent a rational therapeutic strategy to increase its activity. Alternative strategies will require a better understanding of the tumor-suppressive activities of this important hematopoietic transcription factor.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Eric Duncavage for providing images of Wright-Giemsa–stained bone marrow cells, Ling Tian for providing samples for scRNA-seq, and David A. Russler-Germain for the construction of the original MSCV-based PML-RARA WT and C88A vectors. The Siteman Cancer Center Flow Cytometry Core (National Institutes of Health [NIH] National Cancer Institute grant P30CA91842) provided expert support for all flow sorting studies.
This work was supported by NIH National Cancer Institute projects 5P01CA101937 and 5R35CA197561, and the Foundation for Barnes-Jewish Hospital (all to T.J.L.), NIH National Heart, Lung, and Blood Institute Institutional National Research Service Award T32HL007088 (to support C.D.S.K.), and NIH Specialized Program of Research Excellence (SPORE) in Leukemia grant P50CA171063 (D.C. Link, PI, with a developmental research project to C.D.S.K.).
Footnotes
All sequencing data for the mouse studies were deposited to the Sequence Read Archive at the National Center for Biotechnology Information database (accession PRJNA699863).
Data sharing requests should be sent to Timothy J. Ley (timley@wustl.edu).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: C.D.S.K. and T.J.L. designed research ; C.D.S.K, O.R.S.R., R.B.D., M.A.C., T.P.R., N.M.H., M.H., and L.D.W. performed research; C.D.S.K, S.M.R., C.A.M., S.N.S., O.R.S.R., M.A.C., T.P.R., and T.J.L. analyzed data; and C.D.S.K. and T.J.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy J. Ley, Washington University School of Medicine, 660 S Euclid Ave, Box 8007, St. Louis, MO 63110; e-mail: timley@wustl.edu.
REFERENCES
- 1.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostergaard P, Simpson MA, Connell FC, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome). Nat Genet. 2011;43(10):929-931. [DOI] [PubMed] [Google Scholar]
- 3.Hahn CN, Chong CE, Carmichael CL, et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43(10):1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, Döhner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med. 2016;375(9):900-901. [DOI] [PubMed] [Google Scholar]
- 5.Metzeler KH, Herold T, Rothenberg-Thurley M, et al. ; AMLCG Study Group . Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128(5):686-698. [DOI] [PubMed] [Google Scholar]
- 6.Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiba N, Funato M, Ohki K, et al. Mutations of the GATA2 and CEBPA genes in paediatric acute myeloid leukaemia. Br J Haematol. 2014;164(1):142-145. [DOI] [PubMed] [Google Scholar]
- 8.Mulet-Lazaro R, van Herk S, Erpelinck CAJ, et al. Allele-specific expression of GATA2 due to epigenetic dysregulation in CEBPA double mutant AML [published online ahead of print 8 Apr 2021]. Blood. blood.2020009244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tien FM, Hou HA, Tsai CH, et al. GATA2 zinc finger 1 mutations are associated with distinct clinico-biological features and outcomes different from GATA2 zinc finger 2 mutations in adult acute myeloid leukemia. Blood Cancer J. 2018;8(9):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greif PA, Dufour A, Konstandin NP, et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood. 2012;120(2):395-403. [DOI] [PubMed] [Google Scholar]
- 12.Fasan A, Eder C, Haferlach C, et al. GATA2 mutations are frequent in intermediate-risk karyotype AML with biallelic CEBPA mutations and are associated with favorable prognosis. Leukemia. 2013;27(2):482-485. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann L, Dutta S, Opatz S, et al. ZBTB7A mutations in acute myeloid leukaemia with t(8;21) translocation. Nat Commun. 2016;7(1):11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen L, Xu Y, Yao L, et al. Clinical and molecular features of acute promyelocytic leukemia with variant retinoid acid receptor fusions. Haematologica. 2019;104(5):e195-e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Genua C, Valletta S, Buono M, et al. C/EBPα and GATA-2 mutations induce bilineage acute erythroid leukemia through transformation of a neomorphic neutrophil-erythroid progenitor. Cancer Cell. 2020;37(5):690-704.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama S, Suzuki M, Yamaoka A, et al. GATA2 haploinsufficiency accelerates EVI1-driven leukemogenesis. Blood. 2017;130(7):908-919. [DOI] [PubMed] [Google Scholar]
- 17.Katsumura KR, Ong IM, DeVilbiss AW, Sanalkumar R, Bresnick EH. GATA factor-dependent positive-feedback circuit in acute myeloid leukemia cells. Cell Rep. 2016;16(9):2428-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menendez-Gonzalez JB, Sinnadurai S, Gibbs A, et al. Inhibition of GATA2 restrains cell proliferation and enhances apoptosis and chemotherapy mediated apoptosis in human GATA2 overexpressing AML cells. Sci Rep. 2019;9(1):12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vicente C, Vazquez I, Conchillo A, et al. Overexpression of GATA2 predicts an adverse prognosis for patients with acute myeloid leukemia and it is associated with distinct molecular abnormalities. Leukemia. 2012;26(3):550-554. [DOI] [PubMed] [Google Scholar]
- 20.Tipping AJ, Pina C, Castor A, et al. High GATA-2 expression inhibits human hematopoietic stem and progenitor cell function by effects on cell cycle. Blood. 2009;113(12):2661-2672. [DOI] [PubMed] [Google Scholar]
- 21.Heyworth C, Gale K, Dexter M, May G, Enver T. A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev. 1999;13(14):1847-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al Seraihi AF, Rio-Machin A, Tawana K, et al. GATA2 monoallelic expression underlies reduced penetrance in inherited GATA2-mutated MDS/AML. Leukemia. 2018;32(11):2502-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gröschel S, Sanders MA, Hoogenboezem R, et al. Mutational spectrum of myeloid malignancies with inv(3)/t(3;3) reveals a predominant involvement of RAS/RTK signaling pathways. Blood. 2015;125(1):133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Celton M, Forest A, Gosse G, et al. Epigenetic regulation of GATA2 and its impact on normal karyotype acute myeloid leukemia. Leukemia. 2014;28(8):1617-1626. [DOI] [PubMed] [Google Scholar]
- 25.Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369-381. [DOI] [PubMed] [Google Scholar]
- 26.Chong CE, Venugopal P, Stokes PH, et al. Differential effects on gene transcription and hematopoietic differentiation correlate with GATA2 mutant disease phenotypes. Leukemia. 2018;32(1):194-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortés-Lavaud X, Landecho MF, Maicas M, et al. GATA2 germline mutations impair GATA2 transcription, causing haploinsufficiency: functional analysis of the p.Arg396Gln mutation. J Immunol. 2015;194(5):2190-2198. [DOI] [PubMed] [Google Scholar]
- 28.Katsumura KR, Yang C, Boyer ME, Li L, Bresnick EH. Molecular basis of crosstalk between oncogenic Ras and the master regulator of hematopoiesis GATA-2. EMBO Rep. 2014;15(9):938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ping N, Sun A, Song Y, et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia. 2017;31(1):195-202. [DOI] [PubMed] [Google Scholar]
- 30.Hsu AP, Johnson KD, Falcone EL, et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121(19):3830-3837, S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ottema S, Mulet-Lazaro R, Beverloo HB, et al. Atypical 3q26/MECOM rearrangements genocopy inv(3)/t(3;3) in acute myeloid leukemia. Blood. 2020;136(2):224-234. [DOI] [PubMed] [Google Scholar]
- 32.Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371(6494):221-226. [DOI] [PubMed] [Google Scholar]
- 33.Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7(4):532-544. [DOI] [PubMed] [Google Scholar]
- 34.Johnson KD, Hsu AP, Ryu MJ, et al. Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J Clin Invest. 2012;122(10):3692-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Pater E, Kaimakis P, Vink CS, et al. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210(13):2843-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nandakumar SK, Johnson K, Throm SL, Pestina TI, Neale G, Persons DA. Low-level GATA2 overexpression promotes myeloid progenitor self-renewal and blocks lymphoid differentiation in mice. Exp Hematol. 2015;43(7):565-577.e1-10. [DOI] [PubMed] [Google Scholar]
- 37.Westervelt P, Lane AA, Pollock JL, et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARalpha expression. Blood. 2003;102(5):1857-1865. [DOI] [PubMed] [Google Scholar]
- 38.Wartman LD, Welch JS, Uy GL, et al. Expression and function of PML-RARA in thehematopoietic progenitor cells of Ctsg-PML-RARA mice. PLoS One. 2012;7(10):e46529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole CB, Verdoni AM, Ketkar S, et al. PML-RARA requires DNA methyltransferase 3A to initiate acute promyelocytic leukemia. J Clin Invest. 2016;126(1):85-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ketkar S, Verdoni AM, Smith AM, et al. Remethylation of Dnmt3a−/− hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing. Proc Natl Acad Sci USA. 2020;117(6):3123-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kogan SC, Brown DE, Shultz DB, et al. BCL-2 cooperates with promyelocytic leukemia retinoic acid receptor alpha chimeric protein (PMLRARalpha) to block neutrophil differentiation and initiate acute leukemia. J Exp Med. 2001;193(4):531-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimonjic DB, Pollock JL, Westervelt P, Popescu NC, Ley TJ. Acquired, nonrandom chromosomal abnormalities associated with the development of acute promyelocytic leukemia in transgenic mice. Proc Natl Acad Sci USA. 2000;97(24):13306-13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter MJ, Park JS, Ries RE, et al. Reduced PU.1 expression causes myeloid progenitor expansion and increased leukemia penetrance in mice expressing PML-RARalpha. Proc Natl Acad Sci USA. 2005;102(35):12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones L, Wei G, Sevcikova S, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. J Exp Med. 2010;207(12):2581-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Beau MM, Davis EM, Patel B, Phan VT, Sohal J, Kogan SC. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice identify cooperating events and genetic pathways to acute promyelocytic leukemia. Blood. 2003;102(3):1072-1074. [DOI] [PubMed] [Google Scholar]
- 46.Wartman LD, Larson DE, Xiang Z, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121(4):1445-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole CB, Russler-Germain DA, Ketkar S, et al. Haploinsufficiency for DNA methyltransferase 3A predisposes hematopoietic cells to myeloid malignancies. J Clin Invest. 2017;127(10):3657-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Yuan H, Peres L, et al. The DNA binding property of PML/RARA but not the integrity of PML nuclear bodies is indispensable for leukemic transformation. PLoS One. 2014;9(8):e104906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross ME, Mahfouz R, Onciu M, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104(12):3679-3687. [DOI] [PubMed] [Google Scholar]
- 50.Chu VT, Weber T, Wefers B, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells [published correction appears in Nat Biotechnol. 2018;36(2):196]. Nat Biotechnol. 2015;33(5):543-548. [DOI] [PubMed] [Google Scholar]
- 51.Mulloy JC, Cammenga J, Berguido FJ, et al. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003;102(13):4369-4376. [DOI] [PubMed] [Google Scholar]
- 52.Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA, Nimer SD. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002;99(1):15-23. [DOI] [PubMed] [Google Scholar]
- 53.Kirstetter P, Schuster MB, Bereshchenko O, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299-310. [DOI] [PubMed] [Google Scholar]
- 54.Jeong EG, Kim MS, Nam HK, et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14(12):3716-3721. [DOI] [PubMed] [Google Scholar]
- 55.Saida S, Zhen T, Kim E, et al. Gata2 deficiency delays leukemogenesis while contributing to aggressive leukemia phenotype in Cbfb-MYH11 knockin mice. Leukemia. 2020;34(3):759-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martens JH, Brinkman AB, Simmer F, et al. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell. 2010;17(2):173-185. [DOI] [PubMed] [Google Scholar]
- 57.Gaillard C, Tokuyasu TA, Rosen G, et al. Transcription and methylation analyses of preleukemic promyelocytes indicate a dual role for PML/RARA in leukemia initiation. Haematologica. 2015;100(8):1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petti AA, Williams SR, Miller CA, et al. A general approach for detecting expressed mutations in AML cells using single cell RNA-sequencing. Nat Commun. 2019;10(1):3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganapathi KA, Townsley DM, Hsu AP, et al. GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood. 2015;125(1):56-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasquet M, Bellanné-Chantelot C, Tavitian S, et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 2013;121(5):822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dickinson RE, Milne P, Jardine L, et al. The evolution of cellular deficiency in GATA2 mutation. Blood. 2014;123(6):863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spinner MA, Sanchez LA, Hsu AP, et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood. 2014;123(6):809-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kazenwadel J, Secker GA, Liu YJ, et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood. 2012;119(5):1283-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vinh DC, Patel SY, Uzel G, et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115(8):1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.