Abstract
While the gram-negative bacterium Stenotrophomonas maltophilia is used in biotechnology (e.g., for biological control of plant pathogens and for bioremediation), the number of S. maltophilia diseases in humans has dramatically increased in recent years. A total of 40 S. maltophilia isolates from clinical and environmental sources (plant associated and water) was investigated to determine the intraspecies diversity of the group and to determine whether or not the strains could be grouped based on the source of isolation. The isolates were investigated by phenotypic profiling (enzymatic and metabolic activity and antibiotic resistance patterns) and by molecular methods such as temperature-gradient gel electrophoresis of the 16S rRNA gene fragment, PCR fingerprinting with BOX primers, and pulsed-field gel electrophoresis (PFGE) after digestion with DraI. Results of the various methods revealed high intraspecies diversity. PFGE was the most discriminatory method for typing S. maltophilia when compared to the other molecular methods. The environmental strains of S. maltophilia were highly resistant to antibiotics, and the resistance profile pattern of the strains was not dependent on their source of isolation. Computer-assisted cluster analysis of the phenotypic and genotypic features did not reveal any clustering patterns for either clinical or environmental isolates.
Stenotrophomonas maltophilia, previously called Pseudomonas maltophilia and Xanthomonas maltophilia (33), is ubiquitous in the environment. It has been recovered from a number of water sources and from a wide range of nosocomial sources (8, 12, 37). S. maltophilia is often associated with plants and has been isolated from the rhizosphere of wheat, oat, cucumber, maize, oilseed rape, and potato (4, 10, 19, 25). Investigations have indicated a potential role for this species in biotechnology. It has been used as biological control agent of fungal plant pathogens in agriculture (5, 13, 23, 26) and in bioremediation (7, 30, 43). S. maltophilia has also become important in the last decade as a nosocomial pathogen associated with significant case/fatality ratios in certain patient populations, particularly among individuals who are severely debilitated or immunosuppressed (15, 29, 47). Long-term hospitalization, fungal infections, antimicrobial pressure, and catheterization are also contributory factors to the rise in the S. maltophilia infection rate (11). The emergence of new opportunistic pathogenic microorganisms is somehow linked to a multiresistance phenotype that makes them refractory to the antibiotics commonly used in clinical practice (11). The majority of clinical strains of S. maltophilia are characterized by their multiresistance to common antibiotics (2, 41). With the exception of trimethoprim-sulfamethoxazole, many post-therapy isolates of S. maltophilia quickly become resistant to antimicrobial agents (1, 15). Molecular typing methods for this bacterial species, e.g., restriction fragment length polymorphism by pulsed-field gel electrophoresis (PFGE) (6, 47, 48), random amplified polymorphic DNA (RAPD) analysis by arbitrarily primed PCR (48), enterobacterial repetitive intergenic consensus PCR (9, 12), arbitrarily primed PCR (40), ribotyping (16), and repetitive extragenic palindromic PCR (34), have been developed. All of these fingerprinting methods have been used to detect relationships between clinical strains in epidemiological studies. Despite the acknowledged importance of S. maltophilia as a nosocomial pathogen, little is known regarding its epidemiology. Presently, it is unclear how S. maltophilia finds its way to clinical environments (11, 16). Since certain strains of S. maltophilia may have considerable biotechnological potential, it would be desirable to be able to distinguish those strains from ones obtained from clinical sources. Differences in levels of antibiotic resistance and in the ability to macerate onion tissue between clinical and environmental isolates of Burkholderia cepacia, which is also used as biocontrol or bioremediation agent but can be an opportunist pathogen, were reported (44).
In this study, 40 isolates of S. maltophilia (including some strains of biotechnological interest) obtained from different origins (clinical and environmental sources) were investigated by various phenotypic (enzymatic and metabolic activity and antibiotic resistance pattern) and genotypic (temperature gradient gel electrophoresis [TGGE], BOX-PCR, and PFGE) fingerprinting methods in order to find a system that characterizes the variability among this species and that can distinguish between clinical and environmental isolates.
MATERIALS AND METHODS
Bacterial strains.
A total of 40 isolates were investigated in this study. The isolates c1 through c20 were obtained from Britta Bruun and P. Gerner-Smid (Statens Seruminstitut, Copenhagen, Denmark). The strains were isolated in the Rigshospitalet Copenhagen from various sites (tracheal aspirates, sputa, blood, throat, wounds, skin, ulcers, drainage fluids and aspirates, catheters, urine, etc. [16]). Marine isolates e1 and e2 were obtained from Arite Minkwitz (University of Rostock, Rostock, Germany). Strain e3 was used as biocontrol agent against phytopathogenic fungi (4), and strain e8 showed antifungal properties (5). S. maltophilia DSM 50170 (ATCC 13637, type strain t20, isolated from a patient with oral carcinoma [20]) was used as a reference strain for comparison. Environmental strains were isolated on X. maltophilia selective medium (22). The medium contained the following: maltose (10 g liter−1; Sigma, Deisenhofen, Federal Republic of Germany), tryptone (Gibco, Paisley, Scotland), bromthymol blue (4 ml of 2% aqueous solution liter−1; Sigma), and Bacto-Agar (15 g liter−1; Difco, Detroit, Mich.). The medium was adjusted to a pH of 7.1 with 1 N NaOH, and the following antibiotics (all from Sigma) were added: cycloheximide (100 μg liter−1), nystatin (50 μg liter−1), cephalexin (50 μg liter−1), bacitracin (25 μg liter−1), penicillin G (25 μg liter−1), novobiocin (10 μg liter−1), neomycin sulfate (30 μg liter−1), and tobramycin (1 μg liter−1). The environmental strains are available from the culture collection of the Department of Microbiology at the University of Rostock. The control strain used was Pseudomonas aeruginosa ATCC 15441.
Identification and metabolic fingerprinting.
All isolates were identified by API (BioMérieux, Marcyl' Etoile, France) and BIOLOG (Biolog Inc., Hayward, Calif.). In addition, some strains were identified by fatty acid analysis and 16S rRNA gene fragment sequencing (data not shown). Bacterial cells cultivated at 30°C on nutrient agar (Sifin, Berlin, Germany) for 24 h were transferred into the API 20 NE gallery (system for nonenteric rods) and incubated for 24 h at 30°C. Results were read visually and compared to the statistical databank (BioMérieux). To obtain the metabolic fingerprints by BIOLOG, strains were cultivated on tryptic soy agar (Gibco, Eggenstein, Germany) for 24 h at 30°C. Bacterial cells were harvested and suspended in a 0.85% NaCl solution. A 125-μl volume of the suspension with an optical density of 0.2 was transferred with a multichannel pipette into BIOLOG GN microplates (system for gram-negative bacteria). The results were read visually after 24 h of incubation at 30°C and compared to the statistical databank (MicroLog system). All strains were tested in duplicate.
Antibiotic resistance pattern.
The assay (breakpoint determination) to determine the susceptibility of bacteria to relevant antibiotics with a semisolid medium, which was similar to the reference method (agar dilution method), was carried out with ATB Antibiogram PSE 1 (BioMérieux) according to the manufacturer's recommendations. Two concentrations of each antibiotic were used, as follows (in mg liter−1): azlocillin, 16 and 64; piperacillin, 4 and 32; piperacillin-tazobactam, 4 and 32; ticarcillin-clavulanic acid, 8 and 32; cefsulodin, 2 and 16; ceftazidime, 4 and 16; gentamicin, 1 and 4; tobramycin, 1 and 4; amikacin, 4 and 16; doxycycline, 1 and 4; ofloxacin, 1 and 2; ciprofloxacin, 1 and 2; imipenem, 1 and 4; aztreonam, 2 and 16; and cotrimoxazol, 16 and 128. Strains were classified as either susceptible (no growth or resultant turbidity at either concentration), intermediate (growth and turbidity only at the lower concentration), or resistant (growth or turbidity at both concentrations). The disk diffusion test was performed with antibiotic sensitivity disks (BioMérieux) on Mueller-Hinton media (Difco). The final antibiotic concentrations were as follows (in micrograms disk−1): chloramphenicol, 30; kanamycin, 30; tetracycline, 30; and erythromycin, 15 (all from Sigma).
Molecular typing by TGGE.
Total DNA was extracted from the bacterial pellet as previously described (46). PCR amplification of the bacterial 16S rRNA fragment was done with primers spanning V6 to V8 (F968 and R1401 [Escherichia coli numbering system]). Separation of the PCR products was done by TGGE analysis as previously described (18). Approximately equal amounts of PCR products (1 to 2 μl), as determined from an ethidium bromide-stained agarose gel, were applied to TGGE gels. A temperature gradient from 38 to 52°C was used for TGGE (Qiagen, Hilden, Germany) which was performed in TAE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]) at a constant voltage of 180 V for 4 h. DNA was visualized in TGGE gels by acid silver staining (36).
Molecular typing by BOX-PCR.
Genomic DNA from each strain was extracted by the method of Wilson (46). BOX element oligonucleotide primers with the sequence of 5′-CTACGGCAAGGCGACGCTGACG-3′ were synthesized by MWG Biotech (Ebersberg, Germany). The PCR were performed as previously described by Rademaker and De Bruijn in duplicate for each isolate (35).
Molecular typing by PFGE.
Strains were grown overnight in 10-ml volumes of Luria broth (Difco). After centrifugation at 13,600 × g for 1 min, each cell pellet was washed (twice) and resuspended in 1 ml of SE buffer (25 mM EDTA [pH 7.4], 75 mM NaCl). Agarose plugs were made from a 1:1 mixture of 1.6% low-melting-point agarose (Biometra, Göttingen, Germany) and the cell suspension. Each plug was placed in 5 ml of lysis buffer (10 mM Tris-HCl [pH 7.6], 100 mM EDTA [pH 8.0], 50 mM NaCl, 0.2% deoxycholic acid, 1% N-lauroyl sarcosine, 2 mg of lysozyme) for 3 h at 35°C. Samples were then treated for 16 h at 42°C with the same volume of proteinase K solution containing 50 μg of proteinase K per ml, 100 mM EDTA (pH 8.0), 0.2% deoxycholic acid, and 1% N-lauroyl sarcosine. After three 1-h washes with TE buffer (10 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]), the agarose plugs were stored in TE buffer at 4°C for subsequent PFGE. Agarose plugs were digested with restriction enzyme DraI (New England Biolabs, Schalbach, Germany) for 20 h at 35°C according to the manufacturer's recommendations. DNA fragments were separated on a 1% PFGE agarose (peqLab, Erlangen, Germany) gel in a contour-clamped homogeneous electrical field by using the Rotaphor V system of Biometra with 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA [pH 8.0]) at 12°C. The voltage was set at 200 V/cm, and pulse times ranged from 5 to 45 s over 20 h, with linear ramping. The procedure was repeated at least twice for each isolate to determine the reproducibility of the results. The gel was stained with ethidium bromide.
Statistical analysis.
Differences between the antibiotic resistance patterns of clinical and environmental isolates were determined by a two-sided test of binomial proportion (P < 0.05). Data were converted to a binary code, and interisolate relationships were measured by the Euclidian metric unweighted pair-group average method by using the STATISTICA program (StatSoft, Hamburg, Germany). Molecular fingerprint patterns generated for each strain were compared and grouped by using the Gelcompare program (Kortrijk, Belgium). Data describing susceptibility against antibiotics were analyzed by BioMath GmbH company (Rostock, Germany).
RESULTS
Phenotypic characterization and identification.
A total of 40 S. maltophilia isolates from clinical and environmental sources were compared by their enzymatic and metabolic activity by using the API and the BIOLOG systems. Both methods identified all 40 strains correctly to the species level. With the API system, the positive identification rate ranged from 97.7 to 99.9% (Table 1). Seven different API profiles were detected (Table 1), and the isolates were grouped according to these profiles. The largest profile groups (I, II, and IV) comprised 34 isolates and contained isolates from clinical and environmental sources. The oxidative utilization of 95 different carbon sources was tested for each isolate with BIOLOG GN plates. The isolates exhibited heterogeneity in their carbon utilization profiles. Most isolates varied from the typical Stenotrophomonas pattern in the utilization of 1 to 10 carbon sources, as determined by their individual carbon utilization profiles compared to the average pattern of the investigated isolates (data not shown). Of the carbon sources tested, 34 were not utilized by any of the S. maltophilia strains analyzed. All the other carbon sources (61) were utilized by the majority (>80%) of isolates. However, the BIOLOG system identified all strains as S. maltophilia (Table 1). Relationships between isolates were analyzed statistically by cluster analysis. On the basis of similarity it was possible to arrange all the isolates into six groups (Table 1). Three of these groups were homogenous, and each contained only a single isolate of clinical origin; the other three were heterogenous groups with isolates of both origins. Altogether, the grouping of isolates was independent of origin.
TABLE 1.
Taxonomic, phenotypic, and genotypic classification of S. maltophilia isolates
| Isolate | % Identity | Group by:
|
|||||
|---|---|---|---|---|---|---|---|
| Phenotype
|
Antibiogramsc | Genotype
|
|||||
| API | BIOLOG | APIa | BIOLOGb | TGGEa | BOX-PCRd | ||
| c1 | 99.9 | 0.881 | I | I | VI | I | V |
| c2 | 99.8 | 0.948 | II | III | VI | I | III |
| c3 | 99.9 | 0.715 | I | III | VI | I | V |
| c4 | 99.9 | 0.945 | I | III | V | I | III |
| c5 | 99.2 | 0.900 | IV | IV | VI | I | IV |
| c6 | 99.9 | 0.918 | I | III | VI | I | I |
| c7 | 99.9 | 0.798 | I | III | V | I | IV |
| c8 | 99.9 | 0.827 | I | III | VI | I | IV |
| c9 | 99.9 | 0.880 | I | III | V | II | I |
| c10 | 99.9 | 0.871 | I | III | VI | III | IV |
| c11 | 98.6 | 0.804 | VI | III | V | V | V |
| c12 | 98.6 | 0.906 | VI | I | V | I | IV |
| c13 | 99.9 | 1.000 | I | I | V | I | V |
| c14 | 99.9 | 0.911 | I | III | VI | I | III |
| c15 | 99.9 | 0.978 | I | III | V | I | II |
| c16 | 99.9 | 0.731 | I | III | VI | I | III |
| c17 | 99.2 | 0.807 | IV | III | V | I | III |
| c18 | 99.9 | 0.875 | I | III | V | I | III |
| c19 | 99.9 | 0.884 | I | III | IV | I | II |
| c20 | 99.9 | 0.901 | I | I | IV | I | III |
| e1 | 99.9 | 0.762 | I | IV | VI | I | III |
| e2 | 99.9 | 0.821 | I | V | III | I | III |
| e3 | 99.9 | 0.765 | I | III | III | I | I |
| e4 | 99.9 | 0.869 | I | I | VI | I | II |
| e5 | 99.9 | 0.915 | I | I | V | I | II |
| e6 | 99.2 | 0.836 | IV | I | II | II | I |
| e7 | 99.9 | 0.798 | I | III | I | II | II |
| e8 | 99.2 | 0.823 | IV | III | VI | I | III |
| e9 | 99.2 | 0.807 | IV | I | I | II | IV |
| e10 | 99.4 | 0.752 | III | III | III | I | III |
| e11 | 97.7 | 0.946 | VII | III | V | III | II |
| e12 | 99.9 | 0.853 | I | I | V | I | I |
| e13 | 99.0 | 0.836 | V | II | III | I | III |
| e14 | 99.4 | 0.928 | III | III | VI | IV | I |
| e15 | 99.9 | 0.835 | I | III | VI | IV | V |
| e16 | 99.9 | 0.836 | I | VI | I | I | III |
| e17 | 99.9 | 0.836 | I | III | I | II | V |
| e18 | 99.8 | 0.904 | II | III | VI | I | I |
| e19 | 99.9 | 0.895 | I | I | VI | V | II |
| t20 | 99.9 | 0.720 | I | I | III | I | I |
Grouping on the basis of 100% similarity according to cluster analysis.
Grouping on the basis of 85% similarity according to cluster analysis.
Grouping on the basis of 42% similarity according to cluster analysis.
Grouping on the basis of 90% similarity according to cluster analysis.
Antibiotic resistance pattern.
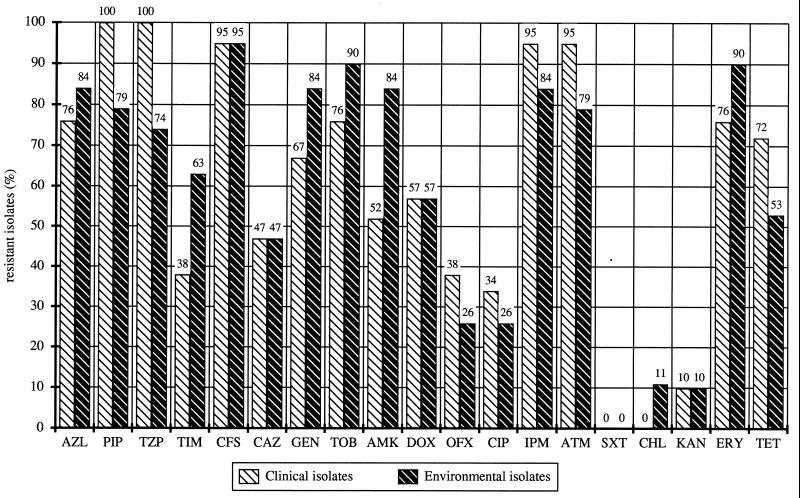
All 40 of the selected strains were resistant to several antibiotics (Fig. 1). The e13 strain isolated from the rhizosphere of oilseed rape was most susceptible to antibiotics. Two clinical isolates (c2 and c4) and two environmental isolates (e5 and e19) were found to have resistance to 16 antibiotics. Three of the multiresistant isolates (c4, e5, and e19) had identical antibiotic resistance profiles. On average, strains were susceptible to 11 of the antibiotics tested. No significant differences (P = 0.624071) in the number of resistances between the two groups of isolates, clinical isolates (mean, 11.45; standard deviation, ± 2.85) and environmental isolates (mean, 11.30; standard deviation, ± 4.0), were found. The higher standard deviation obtained for the environmental isolates indicated the heterogeneity of resistance profiles in this group. Altogether, 34 different antibiotic resistance patterns were observed. Thirty profiles were unique, but four equal profiles were determined for two or more strains. Identical resistance patterns were observed for the clinical strain c3 and the environmental strains e1 and e2 with resistance against 14 antibiotics. None of the strains had resistance to both trimethoprim and sulfamethoxazole (co-trimoxazole). The percentages of resistant strains varied among the antibiotics (Fig. 1).
FIG. 1.
Percentages of clinical and environmental isolates resistant to antibiotics. AZL, azlocillin; PIP, piperacillin; TZP, piperacillin-tazobactam; TIM, ticarcillin-clavulanic acid; CFS, cefsoludin; CAZ, ceftazidime; GEN, gentamicin; TOB, tobramycin; AMK, amikacin; DOX, doxycycline; OFX, ofloxacin; CIP, ciprofloxacin; IPM, imipenem; ATM, aztreonam; SXT, trimethoprim-sulfamethoxazole (co-trimoxazole); CHL, chloramphenicol; KAN, kanamycin; ERY, erythromycin; TET, tetracycline.
The profiles were compared by numerical methods, and the resultant dendrogram based on percent similarity between isolates demonstrated a high degree of diversity. Six different clusters were found on the basis of 42% similarity (Table 1). The largest groups, VI (16 isolates) and V (11 isolates), comprised clinical and environmental isolates. The other groups were homogenous and contained either environmental (groups I, II, and III) or clinical (group IV) isolates.
Genotypic characterization.
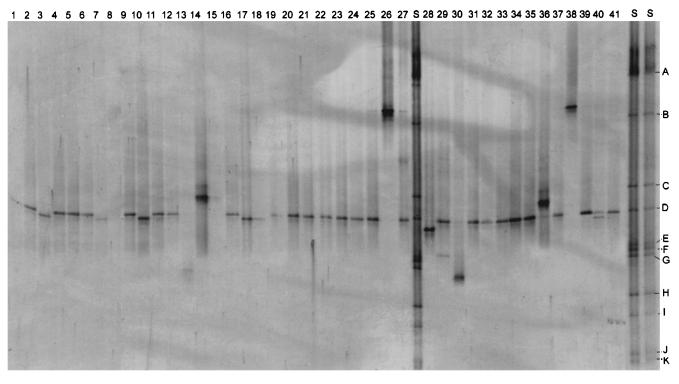
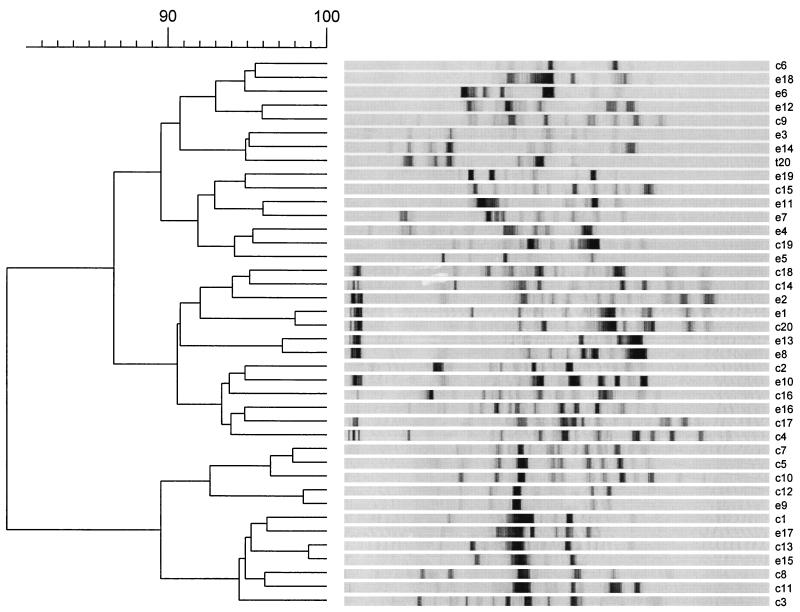
Three different DNA-based fingerprinting methods were used to compare the isolates at the molecular level. With TGGE it was possible to separate 16S rRNA gene fragments of the same length but of different sequences according to their melting properties. A linearly increasing temperature gradient run in the presence of a constantly high concentration of urea and formamide was used for separation of PCR products in TGGE. Each isolate investigated had one band (Fig. 2). The isolates were arranged into five groups according to the position of the band on the gel (Table 1). Group I, the largest group, comprised 85% of the isolates. In most cases groups I, II, III, and V contained isolates from clinical as well as environmental sources. The BOX-PCR method was also used to differentiate the isolates. The PCR products obtained with BOX primers yielded DNA profiles with sufficient numbers of DNA bands to differentiate the 40 isolates (Fig. 3). The method was more discriminating than TGGE of 16S rRNA gene fragments, and most of the isolates showed unique PCR fingerprints. Very similar BOX-PCR banding patterns (relative position and intensity of bands) were observed for the environmental strain e9 and the clinical strain c12. The different BOX-PCR profiles were compared by numerical methods and the resultant dendrogram (Fig. 3), based on percent similarity between the isolates, showed a high degree of genetic diversity. At a 90% similarity level five major groups were defined. All groups were heterogenous and contained environmental and clinical isolates.
FIG. 2.
TGGE profiles of S. maltophilia strains. Lane 1, c5; lane 2, c2; lane 3, e10; lane 4, e16; lane 5, c13; lane 6, c10; lane 7, c15; lane 8, c17; lane 9, c12; lane 10, c20; lane 11, e19; lane 12, e3; lane 13, e8; lane 14, c9; lane 15, c2; lane 16, e15; lane 17, c11; lane 18, c8; lane 19, c7; lane 20, e5; lane 21, c4; lane 22, e12; lane 23, c14; lane 24, e7; lane 25, e17; lane 26, c6; lane 27, standard; lane 28, c10; lane 29, e13; lane 30, e18; lane 31, e1; lane 32, e9; lane 33, e4; lane 34, c16; lane 35, e6; lane 36, c1; lane 37, c19; lane 38, t20; lane 39, e11; lane 40, c3; lane 41, c18; lanes S, standard. A, Clostridium pasteurianum; B, Erwinia carotovora; C, Agrobacterium tumefaciens; D, Pseudomonas fluorescens; E, Pantoea agglomerans; F, Nocardia asteroides; G, Rhizobium leguminosarum; H, Actinomadura malachitica; I, Kineosporia aurantiaca; J, Nocardiopsis atra; K, Actinoplanes philippinensis.
FIG. 3.
BOX-PCR profiles of S. maltophilia strains including statistical analysis and dendrogram showing the genetic relationship between strains.
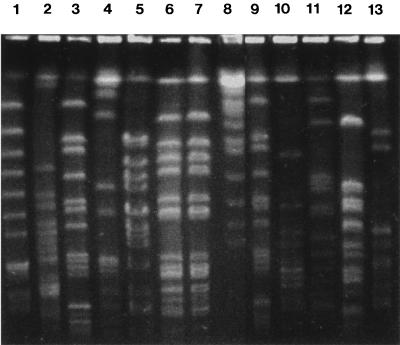
The restriction endonuclease DraI was used to determine restriction fragment length polymorphism (RFLP) patterns for all the isolates. The rare cutting restriction endonucleases, such as DraI, produce DNA profiles with numbers of large DNA fragments suitable for differentiating isolates by PFGE. The RFLP patterns of 12 representative isolates are shown in Fig. 4. With the exception of e1 and e2, RFLP analysis of total DNA obtained after digestion with DraI revealed unique patterns for each isolate.
FIG. 4.
Examples of genomic DNA macrorestriction profiles of S. maltophilia produced by PFGE after DraI digestion. Lane 1, c12; lane 2, c1; lane 3, c7; lane 4, c17; lane 5, c5; lane 6, e1; lane 7, e2; lane 8, lambda ladder marker (size range, 225 to 1,900 kb); lane 9, e9; lane 10, e10; lane 11, t20; lane 12, e18; lane 13, e5.
DISCUSSION
Each of the DNA-based fingerprinting methods was suitable for distinguishing and grouping the isolates, although the sensitivity of the methods varied. Of the two physiological methods tested, BIOLOG was more discriminatory than the API system. However, the systems did prove useful for the accurate identification of S. maltophilia strains. This is in accordance with findings of previous studies (28, 31).
Molecular fingerprinting methods yielded rapid, reproducible, and discriminatory fingerprints for clinical and environmental isolates of S. maltophilia. However, the methods displayed different discriminatory effects. Macrorestriction analysis of digested DNA by PFGE and BOX-PCR typing was found to be more discriminatory than TGGE of the 16S rRNA gene fragments. The majority of strains were shown to possess unique genotypes by PFGE and BOX-PCR (38). Different discriminatory effects of molecular typing methods have also been described by others (35, 48). TGGE analysis revealed some sequence variation in the 16S rRNA gene fragment containing the variable V6 to V8 region. Not surprisingly, TGGE analysis of 16S rRNA was less discriminatory than the PFGE and BOX-PCR methods. However, TGGE has great potential for differentiation of species within natural microbial communities with PCR-amplified 16S rRNA gene fragments obtained from total bacterial community DNA (17). A strategy for linking 16S rRNA from bacterial community fingerprints to pure culture isolates from the same habitat has been recently developed (17).
In recent epidemiological studies of S. maltophilia, PFGE yielded reproducible and easily identifiable patterns (24, 48). Thus, PFGE may be the more superior method for epidemiological typing of S. maltophilia. Although Yao et al. (48) have compared S. maltophilia DNA by PFGE and RAPD analysis methods, this study has demonstrated, for the first time, the intraspecies diversity of S. maltophilia by PCR-dependent fingerprinting with BOX primers and TGGE analysis. In conclusion, all three molecular methods proved useful for typing S. maltophilia. However, PCR analysis with BOX primers was the fastest method used.
The great diversity of S. maltophilia isolates observed in this study is consistent with findings of other previous typings. Palleroni and Bradbury (33) mentioned the diversity of the species in the type description of S. maltophilia. A wide range of heterogeneity in physiological parameters was also shown by Swings et al. (37). This heterogeneity was confirmed by genotypic studies. In further epidemiological studies, the majority of patients had unique types, and only occasionally have small clusters of indistinguishable strains been identified (11, 24, 40). In a recent study, ribotyping was used to characterize the 20 clinical strains used in this study which were isolated from a Danish hospital environment. Considerable diversity among the ribotypes of hospital strains was found, and no single-strain outbreak was detected (16). Potential reservoirs for these strains were not determined. Results by Chatelut et al. (9) obtained with ERIC-PCR and RAPD-PCR showed 29 different profiles among 38 isolates. A consistent observation of all the genotypic studies has been that a wide diversity of strains has been isolated from patients (11). Presently, little is known regarding the source of harmful S. maltophilia strains occurring in hospital environments (12). Investigations by other authors have reported no evidence of patient-to-patient transmission, and they suggest that multiple independent acquisitions from environmental sources could be an important mode of transmission of S. maltophilia (12). The most common sites of contamination were blood sampling tubes, dialysis machines, ice-making machines, nebulizers, shower heads, sink traps, water faucets, and other items frequently in contact with water (12).
Our results indicate that the antibiotic resistance profile of S. maltophilia isolates was not associated with their origin (e.g., clinical and environmental). Such findings suggest that strains of S. maltophilia did not acquire their antibiotic resistance during antibiotic therapy in the clinical or hospital environment. Wüst et al. (47) demonstrated that a single strain of S. maltophilia (typed by PFGE and ribotyping) became increasingly resistant to antimicrobial agents during 15 months after rigorous antimicrobial combination therapy. Their results suggested that isolates became resistant through antibiotic therapy with imipenem. Interestingly, our environmental isolates exhibited a high level of resistance to the antibiotic imipenem.
S. maltophilia is often a dominant member of the rhizosphere microbial community of plants (22) and is known to be a plant root-associated bacterium. S. maltophilia can also produce high amounts of the plant growth hormone indole-3-acetic acid (3, 5). Many of the strains investigated in this study were isolated from the rhizosphere. This particular microenvironment is rich in nutrients due to the exudation of organic compounds from plants (27). Thus, the competition between microorganisms for these ecological sites is very high. Many other rhizobacteria, like the fluorescent pseudomonads and Streptomyces species, produce an extended list of antibiotics (32). Antibiotics produced by rhizobacteria include pyrrolnitrin, pyoluteorin, and herbicolin A, which have also been detected in the rhizosphere (39). When antibiotics have been detected in nature, it has been in material obtained from these microhabitats, which are localized areas of intense microbial interaction (39). Furthermore, S. maltophilia produces two macrocyclic lactam antibiotics, alteramid A and maltophilin (21). Rhizobacteria including S. maltophilia are protected from their own antibiotics and those produced by other rhizobacteria by resistance mechanisms most likely to enhance competition in such natural microenvironments. Perhaps not surprisingly, strains of S. maltophilia often exhibit multiple antibiotic resistances (41). As has been previously reported, we have found that the combination of trimethoprim and sulfamethoxazole has an exceptionally high activity against S. maltophilia (2, 11, 14). Thus, it is the drug(s) of choice for the treatment of severe S. maltophilia infections. Presently, the mechanisms of drug resistance in S. maltophilia are not well characterized. β-Lactam drugs, including imipenem, are not effective against this bacterium because S. maltophilia produces several β-lactamases (metallo-β-lactamase and L2-cephalosporinase) (42). Tetracycline resistance in S. maltophilia is the consequence of an active efflux of the antibiotics, and it is associated with resistance to quinolone and chloramphenicol but not to aminoglycosides or β-lactam antibiotics (1). Temperature-regulated gentamicin resistance has been correlated with the expression of outer membrane proteins (45).
In conclusion, isolates from diseased humans (c1 to c20 [16]), biological control agents (e3 and e4), marine strains (e1 and e2), the type strain (t20), and other plant-associated bacteria (e4 to e19) have exhibited a high intraspecific diversity and did not cluster by origin, as determined by the different DNA-based fingerprinting methods used.
ACKNOWLEDGMENTS
We thank Hella Goschke for valuable technical assistance; Britta Bruun (Copenhagen), Arite Minkwitz, Petra Marten, and Jana Lottmann (Rostock) for providing Stenotrophomonas strains; and J. Hacker (Würzburg) for helpful discussion. We thank J. Jungkurth (Braunschweig) for reading the manuscript.
This study was partially supported by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Alonso A, Martínez J L. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1140–1142. doi: 10.1128/aac.41.5.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpi M, Victor M A, Mortensen I, Gottschau A, Bruun B. In vitro susceptibility of 124 Xanthomonas maltophilia (Stenotrophomonas maltophilia) isolates: comparison of the agar dilution method with the E-test and two agar diffusion methods. APMIS. 1996;104:108–114. [PubMed] [Google Scholar]
- 3.Berg G, Ballin G. Bacterial antagonists of Verticillium dahliae KLEB. J Phytopathol. 1994;141:99–110. [Google Scholar]
- 4.Berg G, Knaape C, Ballin G, Seidel D. Biological control of Verticillium dahliae KLEB by naturally occurring rhizosphere bacteria. Arch Phytopathol Dis Prot. 1994;29:249–262. [Google Scholar]
- 5.Berg G, Marten P, Ballin G. Stenotrophomonas maltophilia in the rhizosphere of oilseed rape—occurrence, characterization and interaction with phytopathogenic fungi. Microbiol Res. 1996;151:19–27. [Google Scholar]
- 6.Bingen E H, Denamur E, Lambert-Zechovsky N Y, Bourdois A, Mariani-Kurkdjian P, Cezard J P, Elion J. DNA restriction fragment length polymorphism differentiates crossed from independent infections in nosocomial Xanthomonas maltophilia bacteremia. J Clin Microbiol. 1991;29:1348–1350. doi: 10.1128/jcm.29.7.1348-1350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binks P R, Nicklin S, Bruce N C. Degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Stenotrophomonas maltophilia PB1. Appl Environ Microbiol. 1995;61:1318–1322. doi: 10.1128/aem.61.4.1318-1322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottone E J, Madayag R M, Qureshi M N. Acanthamoeba keratitis: synergy between amebic and bacterial cocontaminants in contact lens care systems as a prelude to infection. J Clin Microbiol. 1992;30:2447–2450. doi: 10.1128/jcm.30.9.2447-2450.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatelut M, Dournes J L, Chabanon G, Marty N. Epidemiological typing of Stenotrophomonas (Xanthomonas) maltophilia by PCR. J Clin Microbiol. 1995;33:912–914. doi: 10.1128/jcm.33.4.912-914.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debette J, Blondeau R. Présence de Pseudomonas maltophilia dans la rhizosphère de quelque plantes cultivée. Can J Microbiol. 1980;26:460–463. [PubMed] [Google Scholar]
- 11.Denton M, Kerr K G. Microbiological and clinical aspects of infections associated with Stenotrophomonas maltophilia. Clin Microbiol Rev. 1998;11:7–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denton M, Todd N J, Kerr K G, Hawkey P M, Littlewood J M. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J Clin Microbiol. 1998;36:1953–1958. doi: 10.1128/jcm.36.7.1953-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elad Y, Chet I, Baker R. Increased growth response of plants induced by rhizobacteria antagonistic to soilborne pathogenic fungi. Plant Soil. 1987;98:325–339. [Google Scholar]
- 14.Fang F C, Madinger N E. Resistant nosocomial gram-negative bacillary pathogens: Acinetobacter baumanii, Xanthomonas maltophilia, and Pseudomonas cepacia. Curr Top Infect Dis Clin Microbiol. 1996;116:52–83. [PubMed] [Google Scholar]
- 15.Garrison M W, Anderson D E, Campbell D M, Carroll K C, Malone C L, Anderson J D, Hollis R J, Pfaller M A. Stenotrophomonas maltophilia: emergence of multidrug resistant strains during therapy and in an in vitro pharmacodynamic chamber model. Antimicrob Agents Chemother. 1996;40:2859–2864. doi: 10.1128/aac.40.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerner-Smidt P, Bruun B, Arpi M, Schmidt J. Diversity of nosocomial Xanthomonas maltophilia (Stenotrophomonas maltophilia) as determined by ribotyping. Eur J Clin Microbiol Infect Dis. 1995;14:137–140. doi: 10.1007/BF02111874. [DOI] [PubMed] [Google Scholar]
- 17.Heuer H, Hartung K, Wieland G, Kramer I, Smalla K. Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. Appl Environ Microbiol. 1999;65:1045–1049. doi: 10.1128/aem.65.3.1045-1049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuer H, Smalla K. Bacterial phyllosphere communities of Solanum tuberosum L and T4-lysozyme producing genetic variants. FEMS Microbiol Ecol. 1999;28:357–371. [Google Scholar]
- 20.Hugh R, Ryschenko E. Pseudomonas maltophilia, an Alcaligenes like species. J Gen Microbiol. 1961;26:123–132. doi: 10.1099/00221287-26-1-123. [DOI] [PubMed] [Google Scholar]
- 21.Jacobi M, Kaiser D, Berg G, Jung G, Winkelmann G, Bahl H. Maltophilin—a new antifungal compound produced by Stenotrophomonas maltophilia R3089. J Antibiot. 1996;49:1101–1104. doi: 10.7164/antibiotics.49.1101. [DOI] [PubMed] [Google Scholar]
- 22.Juhnke M E, Des Jardin E. Selective medium for isolation of Xanthomonas maltophilia from soil and rhizosphere environments. Appl Environ Microbiol. 1989;55:747–750. doi: 10.1128/aem.55.3.747-750.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwok O C H, Fahy P C, Hoitink H A J, Kuter G A. Interactions between bacteria and Trichoderma hamatum in suppression of Rhizoctonia damping-off in bark compost media. Phytopathology. 1987;77:1206–1212. [Google Scholar]
- 24.Laing F P Y, Ramotar K, Read R R, Alfieri N, Kureishi A, Henderson E A, Louie T J. Molecular epidemiology of Xanthomonas maltophilia colonization and infection in the hospital environment. J Clin Microbiol. 1995;33:513–518. doi: 10.1128/jcm.33.3.513-518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert B, Joos H. Fundamental aspects of rhizobacterial plant growth promotion research. Trends Biotechnol. 1989;7:215–219. [Google Scholar]
- 26.Lambert B, Frederik L, Van Rooyen L, Gossele F, Papon Y, Swings J. Rhizobacteria of maize and their antifungal activities. Appl Environ Microbiol. 1987;53:1866–1871. doi: 10.1128/aem.53.8.1866-1871.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch J M, Whipps J M. Substrate flow in the rhizosphere. In: Kleister D L, Cregan P B, editors. The rhizosphere and plant growth. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. pp. 15–24. [Google Scholar]
- 28.Miller J M, Rohden D L. Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J Clin Microbiol. 1991;29:1143–1147. doi: 10.1128/jcm.29.6.1143-1147.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muder R R, Harris A P, Muller S, Edmond M, Chow J W, Papadakis M, Wagener M W, Bodey G P, Steckelberg J M. Bacteremia due to Stenotrophomonas (Xanthomonas) maltophilia: a prospective multi-center study of 91 episodes. Clin Infect Dis. 1996;22:508–512. doi: 10.1093/clinids/22.3.508. [DOI] [PubMed] [Google Scholar]
- 30.Nawaz M S, Franklin W, Cerniglia C E. Degradation of acrylamide by immobilised cells of a Pseudomonas sp. and Xanthomonas maltophilia. Can J Microbiol. 1993;39:207–212. doi: 10.1139/m93-029. [DOI] [PubMed] [Google Scholar]
- 31.O'Hara C M, Tenover F C, Miller J M. Parallel comparison of accuracy of API 20E, Vitek GNI, MicroScan Walk/Away Rapid ID, and Becton Dickinson Cobas Micro ID-E/NF for identification of members of the family Enterobacteriaceae and common gram-negative, non-glucose-fermenting bacilli. J Clin Microbiol. 1993;31:3165–3169. doi: 10.1128/jcm.31.12.3165-3169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Sullivan D J, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palleroni N J, Bradbury J F. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int J Syst Bacteriol. 1993;43:606–609. doi: 10.1099/00207713-43-3-606. [DOI] [PubMed] [Google Scholar]
- 34.Rademaker J L, Louws F J, Schultz M H, Rossbach U, Vauterin L, Swings J, De Bruijn F J. Molecular systematics of Xanthomonads by Rep-PCR genomic fingerprinting and computer-assisted analysis. Phytopathology. 1997;87:81. [Google Scholar]
- 35.Rademaker J L W, De Bruijn F J. Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer-assisted pattern analysis. In: Caetano-Anollés G, Gresshoff P M, editors. DNA markers: protocols, applications and overviews. J. New York, N.Y: Wiley & Sons, Inc.; 1992. pp. 151–171. [Google Scholar]
- 36.Riesner D, Steger G, Zimmat R. Temperature-gradient gel electrophoresis of nucleic acids: analysis of conformational transitions, sequence variations, and protein-nucleic acid interactions. Electrophoresis. 1989;10:377–389. doi: 10.1002/elps.1150100516. [DOI] [PubMed] [Google Scholar]
- 37.Swings J, De Vos P, Van Den Mooter M, De Ley J. Transfer of Pseudomonas maltophilia Hugh 1981 to the genus Xanthomonas as Xanthomonas maltophilia (Hugh 1981) comb. nov. Int J Syst Bacteriol. 1983;33:409–413. [Google Scholar]
- 38.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomashow L S, Bonsall R F, Weller D M. Antibiotic production by soil and rhizosphere microbes in situ. In: Hurst C J, Knudson G R, McInerney M J, Setzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C: ASM Press; 1997. pp. 493–499. [Google Scholar]
- 40.Van Couwenberghe C J, Cohen S H, Tang Y J, Gumerlock P H, Silva J. Genomic fingerprinting of epidemic and endemic strains of Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) by arbitrarily primed PCR. J Clin Microbiol. 1995;33:1289–1291. doi: 10.1128/jcm.33.5.1289-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Den Mooter M, Swings J. Numerical analysis of 295 phenotypic features of 266 Xanthomonas strains and related strains and an improved taxonomy of the genus. Int J Syst Bacteriol. 1990;40:348–369. doi: 10.1099/00207713-40-4-348. [DOI] [PubMed] [Google Scholar]
- 42.Walsh T R, Hall L, Assinder S J, Nichols W W, Cartwright S J, MacGowan A P, Benett P M. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994;1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Li B, Herman P L, Weeks D P. A three-component enzyme system catalyzes the O demethylation of the herbizide dicamba in Pseudomonas maltophilia DI-6. Appl Environ Microbiol. 1997;63:1623–1626. doi: 10.1128/aem.63.4.1623-1626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wigley P, Burton N F. Genotypic and phenotypic relationships in Burkholderia cepacia isolated from cystic fibrosis patients and the environment. J Appl Microbiol. 1999;86:460–468. doi: 10.1046/j.1365-2672.1999.00687.x. [DOI] [PubMed] [Google Scholar]
- 45.Willcox M H, Winstanley T G, Spencer R C. Outer membrane protein profiles of Xanthomonas isolates displaying temperature-dependent susceptibility to gentamicin. J Antimicrob Chemother. 1994;33:663–666. doi: 10.1093/jac/33.3.663. [DOI] [PubMed] [Google Scholar]
- 46.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel J A, Bent R, Kingston R E, Moore D D, Smith J A, Seidmann J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley and Sons Inc.; 1987. pp. 2.10–2.12. [Google Scholar]
- 47.Wüst J, Frei R, Gunthard H, Altwegg M. Analysis of restriction fragment length polymorphism and ribotyping of multiresistant Stenotrophomonas maltophilia isolated from persisting lung infection in a cystic fibrosis patient. Scand J Infect Dis. 1995;27:499–502. doi: 10.3109/00365549509047053. [DOI] [PubMed] [Google Scholar]
- 48.Yao J D C, Louie M, Louie J, Goodfellow J, Simor A E. Molecular typing of Stenotrophomonas (Xanthomonas) maltophilia by DNA macrorestriction analysis and random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:2195–2198. doi: 10.1128/jcm.33.8.2195-2198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]