Abstract
Purpose of Review
Acute allograft rejection is a common cause of morbidity and mortality in heart and lung transplantation. Unfortunately, the current monitoring gold standard—biopsy plus histopathology—has several limitations. Plasma donor-derived cell-free DNA (dd-cfDNA) has emerged as a potentially valuable biomarker for rejection that addresses some of the limitations of biopsy. This review covers the current state of the evidence and future directions for the use of dd-cfDNA in the monitoring of acute rejection.
Recent Findings
The results of several observational cohort studies demonstrate that levels of dd-cfDNA increase in the setting of acute cellular rejection and antibody-mediated rejection in both heart and lung transplant recipients. dd-cfDNA demonstrates acceptable performance characteristics, but low specificity for the detection of underlying injury from rejection or infection. In particular, the high negative predictive value of the test in both heart and lung transplant patients provides the potential for its use as a screening tool for the monitoring of allograft health rather than tissue biopsy alone.
Summary
Existing evidence shows that dd-cfDNA is a safe, convenient, and reliable method of acute rejection monitoring in heart and lung transplant recipients. Further studies are required to validate threshold values for clinical use and determine its role in the diagnosis of alternative forms of allograft injury.
Keywords: Lung transplant, Acute rejection, Rejection monitoring, Heart transplant
Introduction
Acute allograft rejection, including acute cellular rejection (ACR) and antibody-mediated rejection (AMR), remains a substantial cause of morbidity and morbidity in heart and lung transplantation. It is estimated that up to 50% of lung transplant and 12% of heart transplant patients are treated for ACR during the first year post-transplant [1]. The true incidence of AMR remains uncertain, owing to variations in definitions and diagnostic criteria, with estimates ranging from 4 to > 50% in lung transplant recipients and 3 to 85% in heart transplant recipients [1–3]. Acute allograft rejection serves as a risk factor for chronic rejection, defined as chronic lung allograft dysfunction (CLAD) in lung transplant and chronic allograft vasculopathy (CAV) for heart transplant [4–7].
Given the relatively high incidence and serious consequences of acute allograft rejection, close monitoring of allograft health is an essential component of post-transplant care. The standard method of monitoring allograft health is through biopsy of the allograft, either in response to a change in clinical status (clinically indicated biopsy) and/or the performance of routine surveillance biopsy in asymptomatic individuals. The vast majority of heart and lung transplant centers perform routine surveillance biopsy in the form of endomyocardial biopsy (EMB) or transbronchial biopsy (TBBx), respectively, to screen for acute allograft rejection [8, 9]. However, as a screening tool, these procedures may be costly and inconvenient and place the patient at undue risk for procedural complications [10]. Biopsy samples are analyzed for histopathology, which has high interobserver variability and potentially low sensitivity. With these drawbacks in mind, the search for a safer, non-invasive method to assess underlying allograft injury has been the subject of serious investigation.
Plasma donor-derived cell-free DNA (dd-cfDNA) has recently emerged as a potentially valuable biomarker for the non-invasive assessment of allograft injury in heart and lung transplant recipients. Studies demonstrating the ability of dd-cfDNA to detect episodes of acute allograft rejection provide great promise for the development of an accurate, safe, and convenient approach to the diagnosis and monitoring of allograft rejection. This review will cover the current state of the evidence and future directions for the use of dd-cfDNA in the monitoring of acute rejection in heart and lung transplant recipients.
Characteristics of Cell-Free DNA and Detection
When a cell undergoes apoptosis or necrosis, it releases fragments of double-stranded DNA into the bloodstream, and other bodily fluids, known as cell-free DNA (cfDNA). The majority of these fragments are mononucleosomal with lengths of 150–180 base pairs. Lower proportions of di-, tri-, and other multiples of nucleosomal fragments are also detected. This length distribution suggests an apoptotic origin, as this size is roughly consistent with nucleosome length plus variable fragments of linker DNA produced by apoptotic endonucleases [11, 12]. Larger fragments of cfDNA, up to 21 kilobase pairs, have also been identified in the bloodstream presumably due to cellular necrosis or active secretion of cfDNA [13–15]. Upon entering the bloodstream, the half-life of cfDNA has been estimated to range from 16 min to 2.5 h [16, 17]. The majority is excreted in the urine or undergoes degradation by local macrophages in the liver or spleen [18].
Quantitation of dd-cfDNA
Levels of plasma cfDNA may increase in the setting of tissue injury. Transplantation creates an unusual clinical scenario with genomic admixture of donor and recipient genomes. Therefore, it is possible to genetically differentiate cfDNA originating from the donor allograft versus that which originates from the recipient based on genetic differences. Multiple approaches are now available to identify and quantify the amount of donor-derived cfDNA (dd-cfDNA) as a measure of allograft injury.
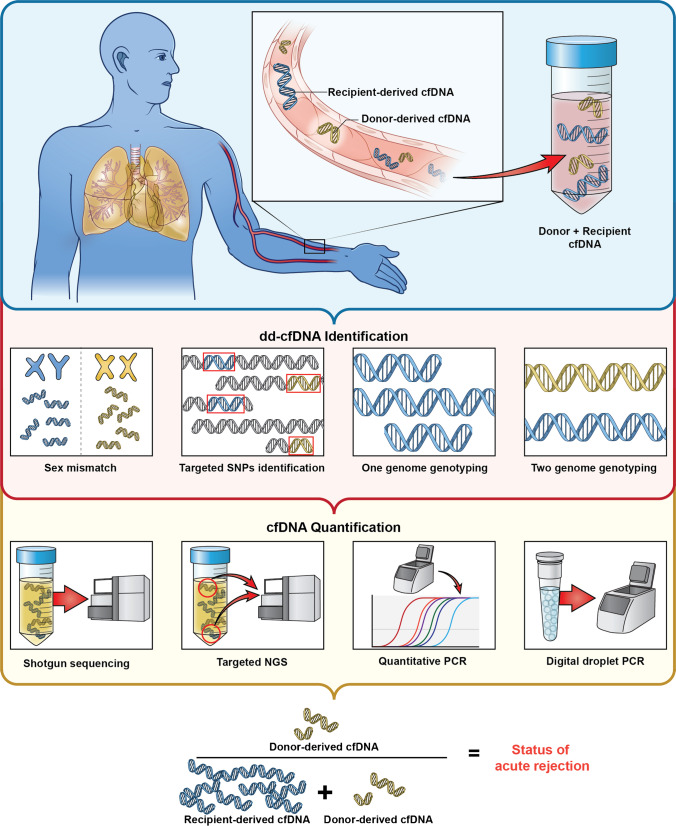
These assays generally have two components: an approach to identify donor versus recipient genetic material and an approach to quantitate or semi-quantitate donor-derived cell-free DNA (Fig. 1). Initial methods leveraged sex-specific chromosomal differences utilizing chromosome Y specific to cfDNA and quantitative PCR to quantitate donor-derived cfDNA [19]. This method was obviously limited by the requirement for a gender mismatch between the donor and recipient. Another early approach identified donor cfDNA based on human leukocyte antigen (HLA) mismatch between the donor and recipients [20]. This method was more broadly applicable than the prior method, but required HLA mismatches between transplant donors and recipients. Recent approaches leverage specific single nucleotide polymorphisms (SNPs) between donor and recipient. The first iteration of the SNP-based approaches relied on whole-genome genotyping using pre-transplant donor and recipient DNA. The genotyped data are compared for each donor-recipient pair to identify informative SNPs. However, genotyping is expensive and increases assay turnaround time and it is often not practical to reliably obtain donor blood for genotyping in clinical practice. Therefore, a second iteration known as the one-genome approach requires only recipient but not donor genotype. This approach uses the recipient genotype and a computational algorithm to estimate informative donor and recipient SNP [21]. A third iteration leverages publicly available population genomic data such as the 1000 Genomes Project to define a set of common and informative SNPs in the population, and therefore, potentially present in a transplant donor-recipient pair. The latter approach completely obviates the need for genotyping or any pre-transplant material. These approaches are generally broadly applicable and now commercially available [22, 23].
Fig. 1.
An illustration of the various approaches to identify donor versus recipient genetic material and an approach to quantitate or semi-quantitate levels of donor-derived cell-free DNA. cfDNA, cell-free DNA; dd-cfDNA, donor-derived cell-free DNA; NGS, next-generation sequencing; PCR, polymerase chain reaction; SNP, single nucleotide polymorphism
After the identification of donor vs recipient genetic material, amplification and quantitation are the next important steps for the various cfDNA testing approaches. These methods vary and include whole-genome sequencing, targeted genome sequencing, quantitative PCR (qPCR), and digital droplet PCR (ddPCR) [22, 24, 25]. These methods estimate the absolute copies or amount of donor versus recipient cfDNA in the sample. However, given that allograft size varies between patients, it is assumed that the absolute amount of dd-cfDNA also varies between patients significantly. Therefore, dd-cfDNA is traditionally reported as a percentage of donor to total (donor plus recipient) SNPs. The latter reporting scheme relies on a stable recipient-derived cfDNA to obtain reliable data, a potential limitation in interpretation of the test. The clinical utility of these two reporting approaches of dd-cfDNA as absolute amount per mL of plasma or as a percentage of total cfDNA is under investigation.
dd-cfDNA in Lung Transplant Recipients
Increases in levels of dd-cfDNA have been detected in response to allograft injury in lung transplant patients [20, 26–30]. De Vlaminck et al. conducted a single-center, prospective cohort study in which 51 patients were enrolled. Serial plasma samples were collected at specified time points after transplantation, including at the time of bronchoscopy [27]. Their approach used genotyped donor and recipient pre-transplant DNA to identify informative SNPs and used whole-genome sequencing to amplify cfDNA. They reported %ddcfDNA and showed that levels were high after transplant surgery and decayed logarithmically with a two-phase decay to reach stable baseline levels. %ddcfDNA significantly increased at the time of histopathological grade moderate or severe ACR (ACR grade ≥ 3) vs stable controls. A threshold dd-cfDNA level of > 1% demonstrated 100% sensitivity and 73% specificity for the diagnosis of moderate or severe ACR with an area under the receiver-operating characteristic curve (AUC) of 0.9.
Khush et al. also performed a single-center, prospective cohort study analyzing 107 plasma dd-cfDNA samples in 38 patients with corresponding histopathology by TBBx and clinical data to adjudicate for ISHLT grade “probable” AMR [31]. The commercially available dd-cfDNA approach used targeted sequencing around a set of pre-established informative SNPs that were established from population genomic data. They reported %ddcfDNA and showed that median dd-cfDNA levels were significantly elevated in 29 ACR events vs. 28 stable controls (0.91% vs. 0.38%; p = 0.021). While median levels of dd-cfDNA were higher in samples with “probable” AMR (n = 9), this did not meet statistical significance, potentially owing to a small sample size (1.34% vs 0.38%; p = 0.07). Notably, dd-cfDNA levels were also elevated in the setting of obstructive CLAD vs stable controls (2.06% vs 0.38%; p = 0.02). In this study, dd-cfDNA was not elevated in patients with positive BAL microbiology (reflecting both infection and colonization) in comparison to stable controls.
Jang et al. recently estimated the performance characteristics of dd-cfDNA for the diagnosis of acute rejection (both ACR and AMR) [29]. This multicenter prospective cohort study enrolled 148 lung transplants and serially collected plasma samples simultaneously with all bronchoscopy (both clinical and surveillance) and pulmonary function testing (PFT). Their approach used whole-genome genotyping and sequencing to quantitate and report %ddcfDNA. The presented post-transplant decay kinetics of %ddcfDNA that followed a logarithmic decay. Their data indicate that %ddcfDNA levels are elevated before 45 days of transplantation as they undergo natural decay, making them potentially less reliable for interpretation during this timeframe. The median dd-cfDNA for episodes of acute rejection (n = 87) was 1.95%. A dd-cfDNA threshold of > 1% yielded a sensitivity of 77%, specificity of 84%, positive predictive value (PPV) of 60%, and negative predictive value (NPV) of 90% with an AUC of 0.89. Furthermore, levels of dd-cfDNA were often observed to be elevated months before the initial diagnosis, particularly in the case of AMR at the onset of identification of donor-specific antibodies (DSA). This finding suggests that dd-cfDNA may indicate the onset of allograft injury prior to the clinical or histopathologic manifestations of the disease. This study also demonstrated no significant elevation in dd-cfDNA in patients with positive BAL microbiology alone; however, dd-cfDNA was significantly elevated in patients with positive microbiology and concurrent decline in PFT and abnormal histopathology. This may indicate that dd-cfDNA is elevated in the setting of clinically relevant infection rather than solely colonization.
Taking the available evidence into account, dd-cfDNA, reported as %ddcfDNA appears to demonstrate good diagnostic accuracy for the diagnosis of acute rejection in lung transplant patients (Table 1). In particular, its high negative predictive value for acute rejection suggests the potential to non-invasively rule outpatients with underlying allograft injury. This may obviate the need for routine surveillance bronchoscopy, and instead, provide a non-invasive method of screening patients who would most benefit from bronchoscopy with transbronchial biopsy.
Table 1.
Selected studies evaluating performance characteristics of dd-cfDNA for the diagnosis of acute rejection in adult lung transplant recipients
| Study | Study design | Patients enrolled | Identification technique | %ddcfDNA threshold | Performance characteristics |
|---|---|---|---|---|---|
| De Vlaminck et al. 2015 [27] | Single-center prospective cohort study | 51 | Two-genome genotyping | > 1% | ACR ≥ 2R: Sensitivity: 100% Specificity: 73% AUC: 0.9 |
| Sayah et al. 2020 [26] | Multicenter prospective cohort study | 26 | Targeted SNPs | > 0.85% | ACR ≥ 1R: Sensitivity: 73% Specificity: 53% AUC: 0.72 |
| Khush et al. 2021 [31] | Single-center prospective cohort study | 38 | Targeted SNPs | > 0.85% | ACR ≥ 1 R or AMR or BOS: Sensitivity: 56% Specificity:76% NPV: 84% AUC: 0.67 |
| Jang et al. 2021 [29] | Multicenter prospective cohort study | 148 | Two-genome genotyping | > 1% | ACR ≥ 1R or AMR: Sensitivity:77% Specificity: 84% NPV: 90% AUC: 0.89 |
A previous prospective cohort study by our group also demonstrated that early elevations of %ddcfDNA may be predictive of long-term outcomes [28]. The sample of 108 patients included subjects from two prospective cohorts with a median post-transplant follow-up of 36 months. A plot of %ddcfDNA against time revealed that all subjects showed high %ddcfDNA immediately after surgery. However, the decay kinetics to reach baseline levels varied between patients. Group 1 patients showed fast %ddcfDNA decay and reached low baseline levels within 1 month of transplantations; group 2 reach low baseline levels within 1–3 months, while group 3 patients reached baseline levels after 3 months. Group 3 patients showed ~ 7 times the risk of developing allograft failure, a composite endpoint of CLAD, re-transplantation, or early death. This work provided evidence that high and unresolving %ddcfDNA early after transplantation, indicative of early allograft injury, is a risk factor for chronic allograft failure. Further studies by our lab have also revealed that higher levels of dd-cfDNA at 72 h in patients with primary graft dysfunction are associated with an increased risk of developing CLAD [30].
dd-cfDNA in Heart Transplant Recipients
Elevations in dd-cfDNA are also present in the setting of allograft injury in heart transplant recipients [32–34]. An early retrospective analysis of 7 heart transplant recipients performed by Snyder et al. in 2011 demonstrated increases in dd-cfDNA in the setting of acute rejection with an AUC of 0.84 at a threshold of 1.7% for the detection of ≥ Grade 2R/3A ACR or AMR [35]. This study genotyped both donor and recipient and used unbiased whole-genome sequencing to identify informative SNPs and quantitate %ddcfDNA. A subsequent single-center study from the same group performed a prospective cohort study involving 21 pediatric and 44 adult heart transplant recipients (565 samples) that demonstrated an AUC of 0.83 and 0.95 for the diagnosis of moderate and severe rejection respectively [36]. Notably, this study further evaluated cases of “discordant” results, in which levels of dd-cfDNA were elevated in the absence of histopathological evidence of rejection or elevated out of proportion to cases of mild rejection. Among 5 samples with an elevated dd-cfDNA but biopsy grade 0, 4 of these patients were diagnosed with mild rejection within the subsequent 6 weeks. Likewise, among the 5 most discordant readings in patients diagnosed with mild rejection, 4 of these patients went on to develop moderate-severe ACR or AMR in the subsequent 2 months.
Two recent large, multicenter, prospective cohort studies have further evaluated the performance characteristics for the diagnosis of acute rejection in heart transplant recipients. Using targeted next-generation sequencing, Khush et al. longitudinally sampled plasma dd-cfDNA in 740 heart transplant patients (2447 samples) from 26 centers undergoing both surveillance and for-cause EMB [34]. Levels of plasma dd-cfDNA were paired with histopathological results at the time of biopsy in order to determine the correlation of levels of dd-cfDNA with acute rejection and estimate the performance characteristics for the diagnosis of acute rejection. At a threshold of 0.2%, dd-cfDNA had a sensitivity of 44% and specificity of 80% for the diagnosis of acute rejection (AUC of 0.64) with a NPV of 97.1%.
Utilizing whole-genome shotgun sequencing, Agbor-Enoh et al. prospectively followed 171 heart transplant patients from 5 US centers for a median of 17.7 months post-transplant and collected serial plasma dd-cfDNA (1834 samples) in conjunction with both surveillance and for-cause EMB (1392 biopsies) [33]. Levels of dd-cfDNA were higher in patients with histopathological evidence of acute rejection than in controls with mild or no rejection (0.38% vs. 0.03%, p < 0.001). Levels of dd-cfDNA correlated with the severity of rejection and levels were noted to be higher in AMR vs ACR. A threshold dd-cfDNA of 0.25% demonstrated a sensitivity of 81% and specificity of 85% for the diagnosis of AR (AUC 0f 0.92) with a negative predictive value of 99.2%. It was estimated that utilizing a dd-cfDNA monitoring strategy with a threshold of 0.25% to trigger EMB would have avoided 81% of the EMBs performed. Again, rises in dd-cfDNA were often noted months prior to the histopathologic diagnosis of acute rejection, especially in the case of AMR.
Similar to lung transplant recipients, initial studies imply that one of the most promising clinical applications of dd-cfDNA in heart transplant patients lies with a non-invasive method of surveillance monitoring (Table 2). The high NPV of the test may allow it to be utilized as a screening tool to effectively rule out the presence of underlying allograft dysfunction and identify patients who would benefit from proceeding to surveillance EMB. Clinical trials are being planned to test the utility of dd-cfDNA as a safe surveillance monitoring tool in place of surveillance biopsies.
Table 2.
Selected studies evaluating performance characteristics of dd-cfDNA for the diagnosis of acute rejection in adult heart transplant recipients
| Study | Study design | Patients enrolled | Identification technique | %ddcfDNA threshold | Performance characteristics |
|---|---|---|---|---|---|
| De Vlaminck et al. 2014 [36] | Single-center prospective cohort study | 65a | Two-genome genotyping | 0.25% | Grade 2R/3A ACR + AMR: Sensitivity: 53% Specificity: 93% AUC 0.83 |
| Khush et al. 2019 [34] | Multicenter prospective cohort study | 740 | Targeted SNPs | 0.2% | ACR 2R + AMR: Sensitivity: 44% Specificity: 80% NPV: 97% AUC: 0.64 |
| Richmond et al. 2020 [32] | Multicenter prospective cohort study | 174b | One-genome genotyping | 0.3% | ACR ≥ 1R: Sensitivity: 73% Specificity: 93% NPV: 91% AUC: 0.86 |
| Agbor-Enoh et al. 2021 [33] | Multicenter prospective cohort study | 171 | Two-genome sequencing | 0.25% | ACR Grade 2R or AMR: Sensitivity: 81% Specificity: 85% NPV: 99.2% AUC: 0.92 |
aIncluded 21 pediatric patients
bIncluded 101 pediatric patients
Limitations
Despite the potential of dd-cfDNA to serve as a promising tool for rejection monitoring in heart and lung transplant recipients, it is important to understand its limitations. Plasma dd-cfDNA, as reported by a percentage, represents the fraction of donor to donor plus recipient cell-free DNA and, therefore, is also influenced by the amount of recipient cell-free DNA in the plasma. In transplantation, immunosuppression drug toxicity, sepsis, and other complications are common. All of these may trigger injury to recipient end organs, particularly kidney, hematopoietic cells, and liver, leading to a rise in recipient-derived cfDNA, and an erroneously low plasma %ddcfDNA even in the presence of concurrent allograft dysfunction and injury [37–42]. Furthermore, the use of dd-cfDNA in patients with multiorgan transplants remains undefined. In patients with multiorgan transplants from a single donor, current assays do not have the ability to distinguish the tissue of origin, and baseline levels differ between solid organ transplants according to tissue mass, cellular composition, and level of perfusion [43, 44]. In multiorgan transplant patients with different donors, many commercially available assays do not distinguish the genotype from each allograft and are limited to single organ, single donor transplants [45].
Another limitation to the use of dd-cfDNA in heart and lung transplant is its poor specificity to distinguish between specific causes of allograft injury. Given that several pathologic conditions may induce allograft cellular apoptosis and necrosis, dd-cfDNA may result from conditions other than acute rejection such as infection or inflammatory disorders. In lung transplant recipients, elevations in dd-cfDNA occur in the presence of clinically relevant lower respiratory tract infections or colonization with high-risk pathogens associated with downstream allograft dysfunction [29, 46, 47]. Along these lines, without corresponding histopathology, dd-cfDNA alone cannot reliably differentiate between ACR vs. AMR.
Future Directions
In both heart and lung transplants, cohort studies have established specific dd-cfDNA threshold values for the identification of clinically relevant transplant complications in surveillance monitoring. Additional studies are needed to validate these thresholds and test their clinical utility. Additionally, while levels of dd-cfDNA may be elevated at the time of acute rejection or infection, changes in levels in response to treatment, and their implications for recovery and prognosis, remain an area of active research. The utility of dd-cfDNA to risk stratify patients with various transplant complications, such as the development of donor-specific antibodies (DSA), is also under investigation. Although the majority of research has focused on the diagnostic potential of dd-cfDNA in the setting of acute rejection, further investigation is required to determine the utility of dd-cfDNA in the development of forms of chronic rejection such as CLAD and CAV.
Levels of dd-cfDNA may have prognostic significance, with elevations at the time of allograft injury predicting future adverse outcomes. In lung transplant recipients, early levels of dd-cfDNA during the first 3 months post-transplant and at the time of primary graft dysfunction are associated with the development of CLAD [28, 30]. Similar trends in worse long-term outcomes associated with early dd-cfDNA levels have also been suggested in heart transplant recipients and merit further investigation [48–50]. The development of predictive modeling algorithms incorporating dd-cfDNA would allow for the identification of patients at risk of poor long-term outcomes. These patients may require closer monitoring or consideration for inclusion in clinical trials aimed at preventing the onset of chronic rejection.
Recently, epigenetic advances in techniques utilizing DNA methylation patterns as well as fragmentomic and topologic analysis of cell-free DNA have generated the potential to identify tissue-specific cfDNA patterns that may distinguish the tissue of origin [51]. By analyzing tissue-specific DNA methylation signatures with bisulfite sequencing, levels of cfDNA from different tissue sources have been quantitated in a cohort of COVID-19 patients, producing a “map” of tissue injury patterns [52]. These advances may provide further clarity into the role that recipient-derived cfDNA plays in the interpretation of dd-cfDNA values and may also provide further insight into the role that systemic inflammation may play during episodes of acute allograft rejection. Given that different tissue types contribute to AMR, ACR, or infection, different tissue sources of cfDNA likely occur in these conditions. These novel cfDNA approaches may therefore be utilized to increase the specificity of cfDNA for the diagnosis of various types of allograft injury.
Conclusion
As a non-invasive, quantitative marker of allograft injury, dd-cfDNA provides promise as a safe, accurate, and feasible method of acute rejection monitoring in heart and lung transplant recipients. While further studies are required to validate specific threshold values for routine clinical use, dd-cfDNA currently demonstrates the greatest potential as a surveillance monitoring tool, screening patients who would most benefit from preceding to biopsy. The further ongoing investigation will determine its role in the diagnosis of other forms of allograft injury, its potential to serve as a treatment target following episodes of acute rejection of infection, and the ability to serve as a prognostic marker for adverse long-term outcomes. Advances in the use of dd-cfDNA rejection monitoring further realizes our quest for the development of precision medicine techniques in heart and lung transplant recipients.
Compliance with Ethical Standards
Conflict of Interest
Michael B. Keller and Sean Agbor-Enoh declare no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
This article is part of the Topical Collection on Thoracic Transplantation
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lund LH, Edwards LB, Kucheryavaya AY, et al. The registry of the International Society for Heart and Lung Transplantation: thirty-first official adult heart transplant report–2014; focus theme: retransplantation. J Heart Lung Transplant. 2014;33:996–1008. doi: 10.1016/j.healun.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Witt CA, Gaut JP, Yusen RD, et al. Acute antibody-mediated rejection after lung transplantation. J Heart Lung Transplant. 2013;32:1034–1040. doi: 10.1016/j.healun.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colvin MM, Cook JL, Chang P, et al. Antibody-mediated rejection in cardiac transplantation: emerging knowledge in diagnosis and management: a scientific statement from the American Heart Association. Circulation. 2015;131:1608–1639. doi: 10.1161/CIR.0000000000000093. [DOI] [PubMed] [Google Scholar]
- 4.Burton CM, Iversen M, Carlsen J, et al. Acute cellular rejection is a risk factor for bronchiolitis obliterans syndrome independent of post-transplant baseline FEV1. J Heart Lung Transplant. 2009;28:888–893. doi: 10.1016/j.healun.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Bando K, Paradis IL, Similo S, et al. Obliterative bronchiolitis after lung and heart-lung transplantation. an analysis of risk factors and management. J Thorac Cardiovasc Surg. 1995;110:4–13. doi: 10.1016/S0022-5223(05)80003-0. [DOI] [PubMed] [Google Scholar]
- 6.Stoica SC, Cafferty F, Pauriah M, et al. The cumulative effect of acute rejection on development of cardiac allograft vasculopathy. J Heart Lung Transplant. 2006;25:420–425. doi: 10.1016/j.healun.2005.11.449. [DOI] [PubMed] [Google Scholar]
- 7.Raichlin E, Edwards BS, Kremers WK, et al. Acute cellular rejection and the subsequent development of allograft vasculopathy after cardiac transplantation. J Heart Lung Transplant. 2009;28:320–327. doi: 10.1016/j.healun.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Levine SM. A survey of clinical practice of lung transplantation in North America. Chest. 2004;125:1224–1238. doi: 10.1378/chest.125.4.1224. [DOI] [PubMed] [Google Scholar]
- 9.Gradek WQ, D’Amico C, Smith AL, Vega D, Book WM. Routine surveillance endomyocardial biopsy continues to detect significant rejection late after heart transplantation. J Heart Lung Transplant. 2001;20:497–502. doi: 10.1016/S1053-2498(01)00236-4. [DOI] [PubMed] [Google Scholar]
- 10.Lampert BC, Teuteberg JJ, Shullo MA, Holtz J, Smith KJ. Cost-effectiveness of routine surveillance endomyocardial biopsy after 12 months post-heart transplantation. Circ Heart Fail. 2014;7:807–813. doi: 10.1161/CIRCHEARTFAILURE.114.001199. [DOI] [PubMed] [Google Scholar]
- 11.Lichtenstein AV, Melkonyan HS, Tomei LD, Umansky SR. Circulating nucleic acids and apoptosis. Ann N Y Acad Sci. 2001;945:239–249. doi: 10.1111/j.1749-6632.2001.tb03892.x. [DOI] [PubMed] [Google Scholar]
- 12.Heitzer E, Auinger L, Speicher MR. Cell-free DNA and apoptosis: how dead cells inform about the living. Trends Mol Med. 2020;26:519–528. doi: 10.1016/j.molmed.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Gall TMH, Belete S, Khanderia E, Frampton AE, Jiao LR. Circulating tumor cells and cell-free DNA in pancreatic ductal adenocarcinoma. Am J Pathol. 2019;189:71–81. doi: 10.1016/j.ajpath.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142. doi: 10.1016/S0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 15.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 16.Yao W, Mei C, Nan X, Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. Gene. 2016;590:142–148. doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 17.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martuszewski A, Paluszkiewicz P, Król M, Banasik M, Kepinska M. Donor-derived cell-free DNA in kidney transplantation as a potential rejection biomarker: a systematic literature review. Journal of Clinical Medicine. 2021;10(2):193. doi: 10.3390/jcm10020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo YMD, Tein MSC, Pang CCP, Yeung CK, Tong K-L, Hjelm NM. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. The Lancet. 1998;351:1329–1330. doi: 10.1016/S0140-6736(05)79055-3. [DOI] [PubMed] [Google Scholar]
- 20.Zou J, Duffy B, Slade M, et al. Rapid detection of donor cell free DNA in lung transplant recipients with rejections using donor-recipient HLA mismatch. Hum Immunol. 2017;78:342–349. doi: 10.1016/j.humimm.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharon E, Shi H, Kharbanda S, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput Biol. 2017;13:e1005629. doi: 10.1371/journal.pcbi.1005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016;18:890–902. doi: 10.1016/j.jmoldx.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Altuğ Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation. 2019;103:2657–2665. doi: 10.1097/TP.0000000000002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drag MH, Kilpeläinen TO. Cell-free DNA and RNA-measurement and applications in clinical diagnostics with focus on metabolic disorders. Physiol Genomics. 2021;53:33–46. doi: 10.1152/physiolgenomics.00086.2020. [DOI] [PubMed] [Google Scholar]
- 25.Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013;59:1732–1741. doi: 10.1373/clinchem.2013.210328. [DOI] [PubMed] [Google Scholar]
- 26.Sayah D, Weigt SS, Ramsey A, Ardehali A, Golden J, Ross DJ. Plasma donor-derived cell-free DNA levels are increased during acute cellular rejection after lung transplant: pilot data. Transplant Direct. 2020;6:e608. doi: 10.1097/TXD.0000000000001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Vlaminck I, Martin L, Kertesz M, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015;112:13336–13341. doi: 10.1073/pnas.1517494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agbor-Enoh S, Wang Y, Tunc I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. EBioMedicine. 2019;40:541–553. doi: 10.1016/j.ebiom.2018.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang MK, Tunc I, Berry GJ, et al. Donor-derived cell-free DNA accurately detects acute rejection in lung transplant patients, a multicenter cohort study. J Heart Lung Transplant. 2021;40(8):82283. doi: 10.1016/j.healun.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller M, Bush E, Diamond JM, et al. Use of donor-derived-cell-free DNA as a marker of early allograft injury in primary graft dysfunction (PGD) to predict the risk of chronic lung allograft dysfunction (CLAD) J Heart Lung Transplant. 2021;40:488–493. doi: 10.1016/j.healun.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khush KK, De Vlaminck I, Luikart H, Ross DJ, Nicolls MR. Donor-derived, cell-free DNA levels by next-generation targeted sequencing are elevated in allograft rejection after lung transplantation. ERJ Open Res. 2021;7(1):00462–2020. doi: 10.1183/23120541.00462-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richmond ME, Zangwill SD, Kindel SJ, et al. Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation. J Heart Lung Transplant. 2020;39:454–463. doi: 10.1016/j.healun.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Agbor-Enoh S, Shah P, Tunc I, et al. Cell-free DNA to detect heart allograft acute rejection. Circulation. 2021;143:1184–1197. doi: 10.1161/CIRCULATIONAHA.120.049098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant. 2019;19:2889–2899. doi: 10.1111/ajt.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A. 2011;108:6229–6234. doi: 10.1073/pnas.1013924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dwivedi DJ, Toltl LJ, Swystun LL, et al. Prognostic utility and characterization of cell-free DNA in patients with severe sepsis. Crit Care. 2012;16:R151. doi: 10.1186/cc11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saukkonen K, Lakkisto P, Pettilä V, et al. Cell-free plasma DNA as a predictor of outcome in severe sepsis and septic shock. Clin Chem. 2008;54:1000–1007. doi: 10.1373/clinchem.2007.101030. [DOI] [PubMed] [Google Scholar]
- 39.Liao W, Zuo X, Lin G, et al. Microbial cell-free DNA in plasma of patients with sepsis: a potential diagnostic methodology. Discov Med. 2020;29:129–137. [PubMed] [Google Scholar]
- 40.Stortz JA, Hawkins RB, Holden DC, et al. Cell-free nuclear, but not mitochondrial, DNA concentrations correlate with the early host inflammatory response after severe trauma. Sci Rep. 2019;9:13648. doi: 10.1038/s41598-019-50044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbany G, Arthur C, Liedén A, et al. Cell-free tumour DNA testing for early detection of cancer - a potential future tool. J Intern Med. 2019;286:118–136. doi: 10.1111/joim.12897. [DOI] [PubMed] [Google Scholar]
- 42.Zemmour H, Planer D, Magenheim J, et al. Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat Commun. 2018;9:1443. doi: 10.1038/s41467-018-03961-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClure T, Goh SK, Cox D, Muralidharan V, Dobrovic A, Testro AG. Donor-specific cell-free DNA as a biomarker in liver transplantation: a review. World J Transplant. 2020;10:307–319. doi: 10.5500/wjt.v10.i11.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bloom RD, Bromberg JS, Poggio ED, et al. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–2232. doi: 10.1681/ASN.2016091034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khush KK. Clinical utility of donor-derived cell-free DNA testing in cardiac transplantation. J Heart Lung Transplant. 2021;40:397–404. doi: 10.1016/j.healun.2021.01.1564. [DOI] [PubMed] [Google Scholar]
- 46.Bazemore K, Rohly M, Permpalung N, et al. 2021 Donor derived cell free DNA% is elevated with pathogens that are risk factors for acute and chronic lung allograft injury. The Journal of Heart and Lung Transplantation [DOI] [PMC free article] [PubMed]
- 47.Keller MB, Mutebi C, Shah P, et al. Performance of donor derived cell-free DNA in routine clinical care of lung transplant recipients, a multi-center study. J Heart Lung Transplant. 2021;40:S148. doi: 10.1016/j.healun.2021.01.451. [DOI] [Google Scholar]
- 48.Zangwill SD, Kindel SJ, Ragalie WS, et al. Early changes in cell-free DNA levels in newly transplanted heart transplant patients. Pediatr Transplant. 2020;24:e13622. doi: 10.1111/petr.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crespo-Leiro M, Hiller D, Woodward R, et al. (161) - Analysis of donor-derived cell-free DNA with 3-year outcomes in heart transplant recipients. J Heart Lung Transplant. 2017;36:S69–S70. doi: 10.1016/j.healun.2017.01.172. [DOI] [Google Scholar]
- 50.Scott JP, Ragalie WS, Stamm KD, et al. Total cell-free DNA predicts death and infection following pediatric and adult heart transplantation. Ann Thorac Surg. 2020;112(4):1282–1289. doi: 10.1016/j.athoracsur.2020.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Lo YMD, Han DSC, Jiang P, Chiu RWK. Epigenetics fragmentomics, and topology of cell-free DNA in liquid biopsies. Science. 2021;372:eaaw3616. doi: 10.1126/science.aaw3616. [DOI] [PubMed] [Google Scholar]
- 52.Andargie TE, Tsuji N, Seifuddin F, et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight. 2021;6(7):e147610. doi: 10.1172/jci.insight.147610. [DOI] [PMC free article] [PubMed] [Google Scholar]