Abstract
Sparse Identification of Nonlinear Dynamics (SINDy) is a method of system discovery that has been shown to successfully recover governing dynamical systems from data [6, 39]. Recently, several groups have independently discovered that the weak formulation provides orders of magnitude better robustness to noise. Here we extend our Weak SINDy (WSINDy) framework introduced in [28] to the setting of partial differential equations (PDEs). The elimination of pointwise derivative approximations via the weak form enables effective machine-precision recovery of model coefficients from noise-free data (i.e. below the tolerance of the simulation scheme) as well as robust identification of PDEs in the large noise regime (with signal-to-noise ratio approaching one in many well-known cases). This is accomplished by discretizing a convolutional weak form of the PDE and exploiting separability of test functions for efficient model identification using the Fast Fourier Transform. The resulting WSINDy algorithm for PDEs has a worst-case computational complexity of for datasets with N points in each of D + 1 dimensions. Furthermore, our Fourier-based implementation reveals a connection between robustness to noise and the spectra of test functions, which we utilize in an a priori selection algorithm for test functions. Finally, we introduce a learning algorithm for the threshold in sequential-thresholding least-squares (STLS) that enables model identification from large libraries, and we utilize scale invariance at the continuum level to identify PDEs from poorly-scaled datasets. We demonstrate WSINDy’s robustness, speed and accuracy on several challenging PDEs. Code is publicly available on GitHub at https://github.com/MathBioCU/WSINDy_PDE.
Keywords: data-driven model selection, partial differential equations, weak solutions, sparse recovery, Galerkin method, convolution
1. Introduction
Stemming from Akaike’s seminal work in the 1970’s [1, 2], research into the automatic creation of accurate mathematical models from data has progressed dramatically. In the last 20 years, substantial developments have been made at the interface of applied mathematics and statistics to design data-driven model selection algorithms that are both statistically rigorous and computationally efficient (see [5, 22, 23, 48, 55, 56] for both theory and applications). An important achievement in this field was the formulation and subsequent discretization of the system discovery problem in terms of a candidate basis of nonlinear functions evaluated at the given dataset, together with a sparsification measure to avoid overfitting [9]. In [50] the authors extended this framework to the context of catastrophe prediction and used compressed sensing techniques to enforce sparsity. More recently, this approach has been generalized as the SINDy algorithm (Sparse Identification of Nonlinear Dynamics) [6] and successfully used to identify a variety of discrete and continuous dynamical systems.
The wide applicability, computational efficiency, and interpretability of the SINDy algorithm has spurred an explosion of interest in the problem of identifying nonlinear dynamical systems from data [8, 34, 10, 11, 15, 49, 27]. In addition to the sparse regression approach adopted in SINDy, some of the primary techniques include Gaussian process regression [31, 36], deep neural networks [26, 25, 24, 40, 51, 22], Bayesian inference [61, 62, 54] and classical methods from numerical analysis [16, 19, 57]. The variety of approaches for model discovery from data qualitatively differ in the interpretability of the resulting data-driven dynamical system, the computational efficiency of the algorithm, and the robustness to noise, scale separation, etc. For instance, a neural-network based data-driven dynamical system does not easily lend itself to physical interpretation1. The SINDy algorithm allows for direct interpretations of the dynamics from identified differential equations and uses sequential-thresholding least-squares (STLS) to enforce a sparse solution to a linear system Ax = b. STLS has been proven to converge to a local minimizer of the non-convex functional in at-most n iterations [60].
The aim of the present article is to extend the Weak SINDy method (WSINDy) for recovering ordinary differential equations (ODEs) from data to the context of partial differential equations (PDEs) [28]. WSINDy is a Galerkin-based data-driven model selection algorithm that utilizes the weak form of the dynamics in a sparse regression framework. By integrating in time against compactly-supported test functions, WSINDy avoids approximation of pointwise derivatives which are known to result in low robustness to noise [39]. In [28] we showed that by integrating against a suitable choice of test functions, correct ODE model terms can be identified together with machine-precision recovery of coefficients (i.e. below the tolerance of the data simulation scheme) from noise-free synthetic data, and for datasets with large noise, WSINDy successfully recovers the correct model terms without explicit data denoising. The use of integral equations for system identification was proposed as early as the 1980’s [9] and was carried out in a sparse regression framework in [42] in the context of ODEs, however neither works utilized the full generality of the weak form.
Sparse regression approaches for learning PDEs from data have seen a tremendous spike in activity in the years since 2016. Pioneering works include [41], [39], and [43], where the Douglas-Rachford algorithm, sequential-thresholding least squares (STLS), and basis pursuit with denoising, respectively, are used to regularize the NP-hard problem of finding an optimal sparse solution. Many other predominant approaches for learning dynamical systems (Gaussian processes, deep learning, Bayesian inference, etc.) have since been extended to the discovery of PDEs [7, 30, 21, 22, 52, 53, 59, 58, 45, 55]. A significant disadvantage for the vast majority of PDE discovery methods is the requirement of pointwise derivative approximations. Steps to alleviate this are taken by the authors of [35] and [58], where neural network-based recovery schemes are combined with integral and abstract evolution equations to recover PDEs, and in [53], where the finite element-based method Variational System Identification (VSI) is introduced to identify reaction-diffusion systems and uses backward Euler to approximate the time derivative.
WSINDy is a method for discovering PDEs without the use of any pointwise derivative approximations, black-box routines or conventional noise filtering. Through integration by parts in both space and time against smooth compactly-supported test functions, WSINDy is able to recover PDEs from datasets with much higher noise levels, and from truly weak solutions (see Figure 3 in Section 5). This works suprisingly well even as the signal-to-noise ratio approaches one. Furthermore, as in the ODE setting, WSINDy achieves high-accuracy recovery in the low-noise regime. These overwhelming improvements resulting from a fully weak2 identification method have also been discovered independently by other groups [37, 13]. WSINDy offers several advantages over these alternative frameworks. Firstly, we use a convolutional weak form which enables efficient model identification using the Fast Fourier Transform (FFT). For measurement data with N points in each of the D + 1 space-time dimensions (ND + 1 total data points), the resulting algorithmic complexity of WSINDy in the PDE setting is at worst , in other words floating point operations per data-point. Subsampling further reduces the cost. Furthermore, our FFT-based approach reveals a key mechanism behind the observed robustness to noise, namely that spectral decay properties of test functions can be tuned to damp noise-dominated modes in the data, and we develop a learning algorithm for test function hyperparameters based on this mechanism. WSINDy also utilizes scale invariance of the PDE and a modified STLS algorithm with automatic threshold selection to recover models from (i) poorly-scaled data and (ii) large candidate model libraries.
Figure 3.
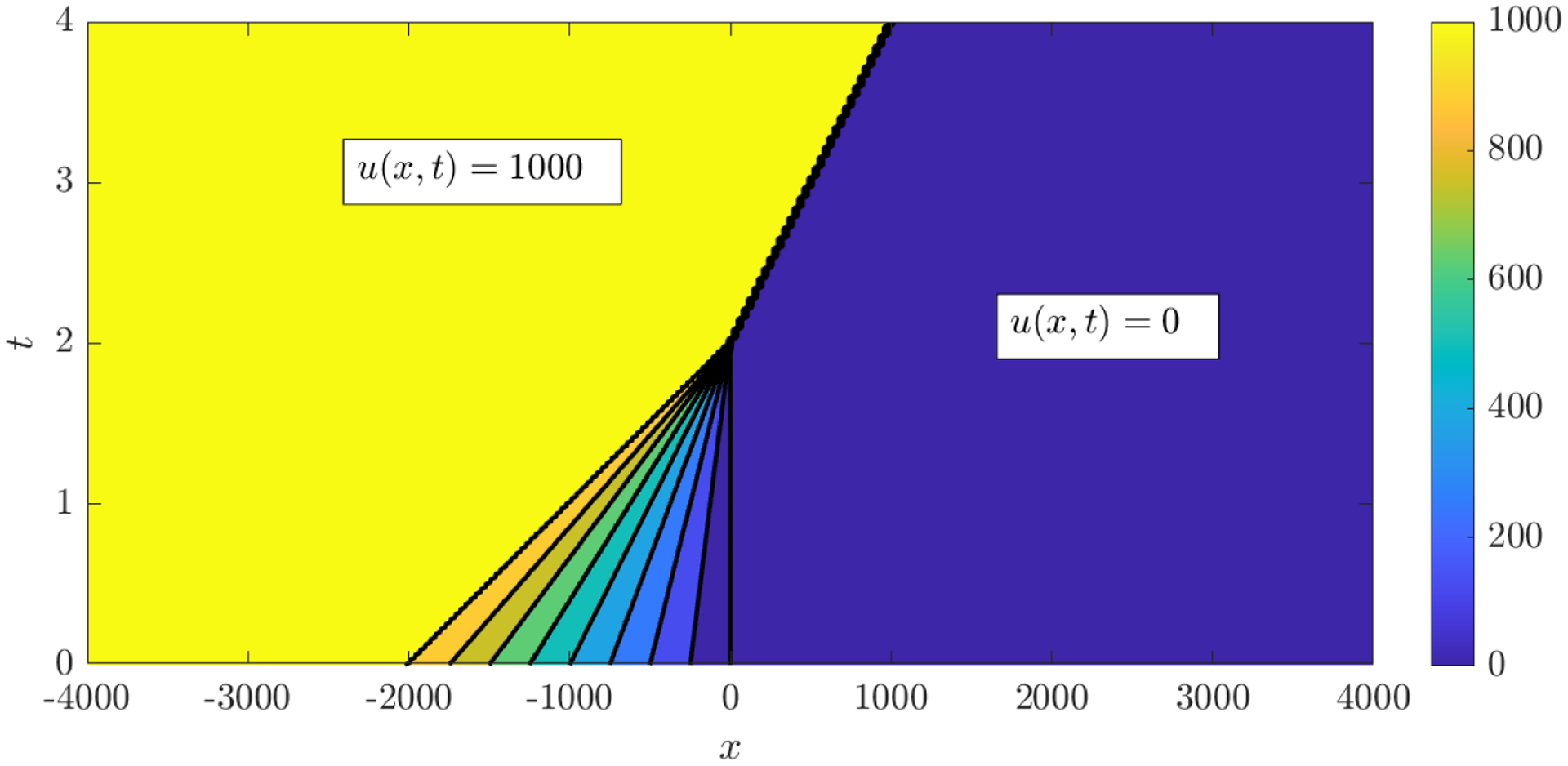
Characteristics of the shock-forming solution (B.2) used to identify the inviscid Burgers equation. A shock forms at time t = 2 and travels along the line x = 500(t − 2).
The outline of the article is as follows. In Section 2 we define the system discovery problem that we aim to solve and the notation to be used throughout. We then introduce the convolutional weak formulation along with our FFT-based discretization in Section 3. Key ingredients of the WSINDy algorithm for PDEs (Algorithm 4.2) are covered in Section 4, including a discussion of spectral properties of test functions and robustness to noise (4.1), our modified sequential thresholding scheme (4.2), and regularization using scale invariance of the underlying PDE (4.3). Section 5 contains numerical model discovery results for a range of nonlinear PDEs, including several vast improvements on existing results in the literature. We conclude the main text in Section 6 which summarizes the exposition and includes natural next directions for this line of research. Lastly, additional numerical details are included in the Appendix.
2. Problem Statement and Notation
Let U be a spatiotemporal dataset given on the spatial grid over timepoints t ⊂ [0, T] where Ω is an open, bounded subset in , D ≥ 1. In the cases we consider here, Ω is rectangular and the spatial grid is given by a tensor product of one-dimensional grids X = X1 ⊗⋯⊗XD, where each for 1 ≤ d ≤ D has equal spacing Δx, and the time grid has equal spacing Δt. The dataset U is then a (D + 1)-dimensional array with dimensions N1 × ⋯ × ND + 1. We write h(X, t) to denote the (D + 1)-dimensional array obtained by evaluating the function at each of the points in the computational grid (X, t). Individual points in (X, t) will often be denoted by (xk, tk) ∈ (X, t) where
In a mild abuse of notation, for a collection of points {(xk, tk)}k∈[K] ⊂ (X, t), the index k plays a double role as a single index in the range [K] := {1, …, K} referencing the point (xk, tk) ∈ {(xk, tk)}k∈[K] and as a multi-index on , where kd references the dth coordinate. This is particularly useful for defining a matrix of the form
(as in equation (3.6) below) where (hj)j∈[J] is a collection of J functions evaluated at the set of K points {(xk, tk)}k∈[K] ⊂ (X, t).
We assume that the data satisfies U = u(X, t) + ϵ for i.i.d. noise3 ϵ and weak solution u of the PDE
| (2.1) |
The problem we aim to solve is the identification of functions (gs)s∈[S] and corresponding differential operators that govern the evolution4 of u according to given the dataset U and computational grid (X, t). Here and throughout we use the multi-index notation to denote partial differentiation5 with respect to x = (x1, … , xD) and t, so that
We emphasize that a wide variety of PDEs can be written in the form (2.1). In particular, in this paper we demonstrate our method of system identification on inviscid Burgers, Korteweg-de Vries, Kuramoto-Sivashinsky, nonlinear Schrödinger’s, Sine-Gordon, a reaction-diffusion system and Navier-Stokes. The list of admissable PDEs that can be transformed into a weak form without any derivatives on the state variables includes many other well-known PDEs (Allen-Cahn, Cahn-Hilliard, Boussinesq,…).
3. Weak Formulation
To arrive at a computatonally tractable model recovery problem, we assume that the set of multi-indices (αs)s∈[S] together with α0 enumerates the set of possible true differential operators that govern the evolution of u and that (gs)s∈[S] ⊂ span(fj)j∈[J] where the family of functions (fj)j∈[J] (referred to as the trial functions) is known beforehand. This enables us to rewrite (2.1) as
| (3.1) |
so that discovery of the correct PDE is reduced to a finite-dimensional problem of recovering the true vector of coefficients , which is assumed to be sparse.
To convert the PDE into its weak form, we multiply equation (3.1) by a smooth test function ψ(x, t), compactly-supported in Ω × (0, T), and integrate over the spacetime domain,
where the L2-inner product is defined and ψ* denotes the complex conjugate of ψ, although in what follows we integrate against only real-valued test functions and will omit the complex conjugation. Using the compact support of ψ and Fubini’s theorem, we then integrate by parts as many times as necessary to arrive at the following weak form of the dynamics:
| (3.2) |
where is the order of the multi-index6. Using an ensemble of test functions (ψk)k∈[K], we then discretize the integrals in (3.2) with fj(u) replaced by fj(U) (i.e. evaluated at the observed data U) to arrive at the linear system
defined by
| (3.3) |
where , and are referred to throughout as the left-hand side, Gram matrix and model coefficients, respectively. In a mild abuse of notation, we use the inner product both in the sense of a continuous and exact integral in (3.2) and a numerical approximation in (3.3) which depends on a chosen quadrature rule. Building off of its success in the ODE setting, we use the trapezoidal rule throughout, as it has been shown to yield nearly negligible quadrature error with the test functions employed below (see Section 4.1 and [28]). In this way, solving b = Gw★ for the model coefficients w★ allows for recovery of the PDE (3.1) without pointwise derivative approximations. The Gram matrix and left-hand side defined in (3.3) conveniently take the same form regardless of the spatial dimension D, as their dimensions only depend on the number of test functions K and the size SJ of the model library, composed of J trial functions (fj)j∈[J] and S candidate differential operators enumerated by the multi-index set α := (αs)1≤s≤S.
3.1. Convolutional Weak Form and Discretization.
We now restrict to the case of each test function ψk being a translation of a reference test function ψ, i.e. ψk(x, t) = ψ(xk − x, tk − t) for some collection of points {(xk, tk)}k∈[K] ⊂ (X, t) (referred to as the query points). The weak form of the dynamics (3.2) over the test function basis (ψk)k∈[K] then takes the form of a convolution:
| (3.4) |
The sign factor appearing in (3.2) after integrating by parts is eliminated in (3.4) due to the sign convention in the integrand of the space-time convolution, which is defined by
Construction of the linear system b = Gw★ as a discretization of the convolutional weak form (3.4) over the query points (xk, tk)k∈[K] can then be carried out efficiently using the FFT as we describe below.
To relate the continuous and discrete convolutions, we assume that the support of ψ is contained within some rectangular domain
where bd = mdΔx for d ∈ [D] and bD + 1 = mD + 1Δt. We then define a reference computational grid for ψ centered at the origin and having the same sampling rates (Δx, Δt) as the data U, where Y = Y1 ⊗·⋯·⊗YD for and . In this way Y contains 2md + 1 points along each dimension d ∈ [D], with equal spacing Δx, and t contains 2mD + 1 + 1 points with equal spacing Δt. As with (X, t), points in (yk, tk) ∈ (Y, t) take the form
where each index kd for d ∈ [D + 1] takes values in the range {−md, … , 0, … , md}, and for valid indices k − j, the two grids (X, t) and (Y, t) are related by
| (3.5) |
We stress that (Y, t) is completely defined by the integers m = (md)d∈[D + 1], specified by the user, and that the values of m have a significant impact on the algorithm. For this reason we develop an automatic selection algorithm for m using spectral properties of the data U (see Appendix A).
The linear system (3.3) can now be rewritten
| (3.6) |
where and the factor ΔxDΔt characterizes the trapezoidal rule. We define the discrete (D + 1)-dimensional convolution between Ψs and fj(U) at a point by
which, substituting the definition of Ψs,
| (3.7) |
truncating indices appropriately and using (3.5),
| (3.8) |
| (3.9) |
| (3.10) |
3.2. FFT-based Implementation and Computational Complexity for Separable ψ.
Convolutions in the linear system (3.6) may be computed rapidly if the reference test function ψ is separable over the given coordinates, i.e.
for univariate functions (ϕd)d∈[D + 1]. In this case,
so that only the vectors
need to be computed for each 0 ≤ s ≤ S and the multi-dimensional arrays (Ψs)s=0, …, S are never directly constructed. Convolutions can be carried out sequentially in each coordinate7, so that the overall cost of computing each column Ψs * fj (U) of G is
| (3.11) |
if the computational grid (X, t) and reference grid (Y, t) have N and n ≤ N points along each of the D + 1 dimensions, respectively. Here CN log(N) is the cost of computing the 1D convolution between vectors vectors and using the FFT,
| (3.12) |
where , ⊙ denotes element-wise multiplication and projects onto the first N − n + 1 components. The discrete Fourier transform is defined
with inverse
The projection ensures that the convolution only includes points that correspond to integrating over test functions ψ that are compactly supported in (X, t), which is necessary for integration by parts to hold in the weak form. The spectra of the test functions can be precomputed and in principle each convolution Ψs * fj(U) can be carried out in parallel8, making the total cost of the WSINDy Algorithm (4.2) in the PDE setting equal to (3.11) (ignoring the cost of the least-squares solves which are negligible in comparison to computing (G, b)). In addition, subsampling reduces the term (N − n + 1) in (3.11) to (N − n + 1)/s where s ≥ 1 is the subsampling rate such that (N − n + 1)/s points are kept along each dimension.
For most practical combinations of n and N, (say n > N/10 and N > 150) using the FFT and separability provides a considerable reduction in computational cost. See Figure 1 for a comparison between TI and the naive cost TII of an (N + 1)-dimensional convolution:
| (3.13) |
For example, with n = N/4 (a typical value) we have and , hence exploiting separability reduces the computational complexity by a factor of ND + 1/log(N).
Figure 1.
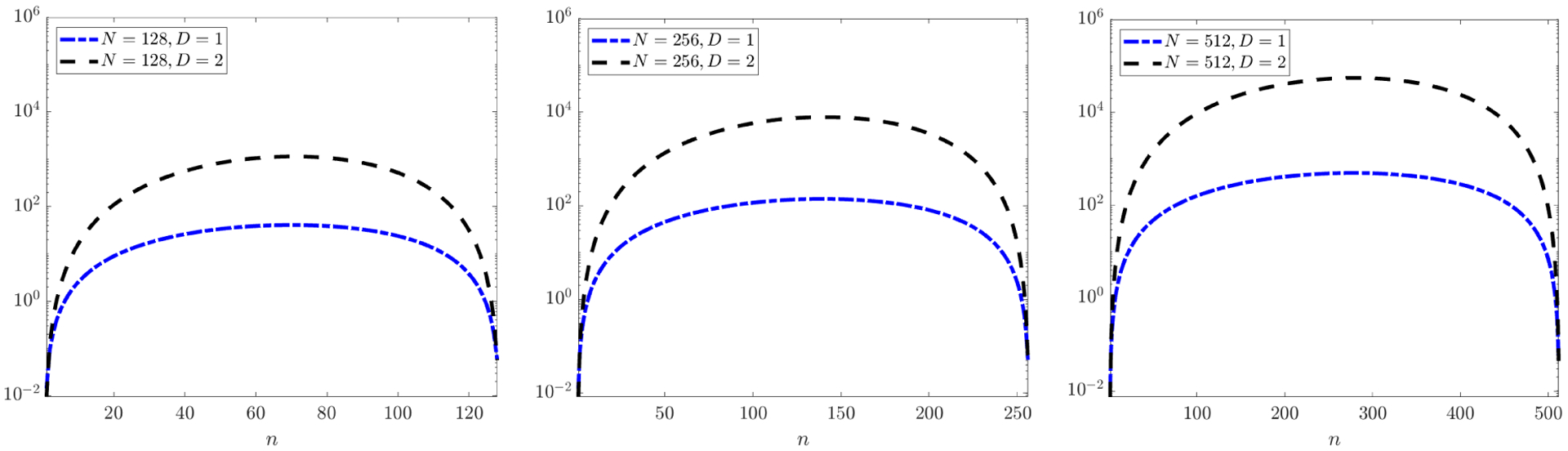
Reduction in computational cost of multi-dimensional convolution Ψs * fj (U) when Ψs and fj(U) have n and N points in each of D + 1 dimensions, respectively. Each plot shows the ratio TII/TI (equations (3.13) and (3.11)), i.e. the factor by which the separable FFT-based convolution reduces the cost of the naive convolution, for D + 1 = 2 and D + 1 = 3 space-time dimensions and n ∈ [N]. The right-most plot shows that when N = 512 and D + 1 = 3, the separable FFT-based convolution is 104 times faster for 100 ≤ n ≤ 450.
4. WSINDy Algorithm for PDEs and Hyperparameter Selection
WSINDy for PDE discovery is given in Algorithm 4.2, where the user must specify each of the hyperparameters in Table 1. The key pieces of the algorithm are (i) the choice of reference test function ψ, (ii) the method of a sparsification, (iii) the method of regularization, (iv) selection of convolution query points {(xk, tk)}k∈K, and (v) the model library. At first glance, the number of hyperparameters is quite large. We now discuss several simplifications that either reduce the number of hyperparameters or provide methods of choosing them automatically. In Section 4.1 we discuss connections between the convolutional weak form and spectral properties of ψ that determine the scheme’s robustness to noise and inform the selection of test function hyperparameters. In Section 4.2 we introduce a modified sequential-thresholding least-squares algorithm (MSTLS) which includes automatic selection of the threshold λ and allows for PDE discovery from large libraries. In Section 4.3 we describe how scale invariance of the PDE is used to rescale the data and coordinates in order to regularize the model recovery problem in the case of poorly-scaled data. In Sections 4.4 and 4.5 we briefly discuss selection of query points and an appropriate model library, however these components of the algorithm will be investigated more thoroughly in future research.
Table 1.
Hyperparameters for the WSINDy Algorithm 4.2. Note that fj piecewise continuous is sufficient (we just need convergence of the trapezoidal rule), m may be replaced by a spectral-decay tolerance if test functions are automatically selected from the data using the method in Appendix A, and K is determined from m and s using (4.10).
| Hyperparameter | Domain | Description |
|---|---|---|
| (fj)j∈[J] | trial function library | |
| α = (αs)s=0,…,S | partial derivative multi-indices | |
| m = (md)d∈[D + 1] | discrete support lengths of 1D test functions (ϕd)d∈[D + 1] | |
| s = (sd)d∈[D + 1] | subsampling frequencies for query points {(xk, tk)}k∈[K] | |
| λ | [0, ∞) | search space for sparsity threshold |
| τ | (0, 1] | ψ real-space decay tolerance |
4.1. Selecting a Reference Test Function ψ.
4.1.1. Convolutional Weak Form and Fourier Analysis.
Computation of G and b in (3.6) with ψ separable requires the selection of appropriate 1D coordinate test functions (ϕd)d∈[D + 1]. Computing convolutions using the FFT (3.12) suggests a mechanism for choosing appropriate test functions. Define the Fourier coefficients of a function u ∈ L2([0, T]) by
Consider data for a T-periodic function u, , and i.i.d. noise . The discrete Fourier transform of the noise is then distributed . In addition, there exist constants C > 0 and ℓ > 1/2 such that for each . There then exists a noise-dominated region of the spectrum determined by the noise-to-signal ratio
where ‘≈’ corresponds to omitting the aliasing error. For NSRk ≥ 1 the kth Fourier mode is by definition noise-dominated, which corresponds to wavenumbers
| (4.1) |
If the critical wavenumber k* between the noise dominated (NSRk ≥ 1) and signal-dominated (NSRk ≤ 1) modes can be estimated from the dataset U, then it is possible to design test functions ψ such that the noise-dominated region of lies in the tail of . The convolutional weak form (3.6) can then be interpreted as an approximate low-pass filter on the noisy dataset, offering robustness to noise without altering the frequency content of the data9.
In summary, spectral decay properties of the reference test function ψ serve to damp high-frequency noise in the convolutional weak form, which acts together with the natural variance-reducing effect of integration, as described in [13], to allow for quantification and control of the scheme’s robustness to noise. Specifically, coordinate test functions ϕd with wide support in real space (larger md) will reduce more variance, but will have a faster-decaying spectrum , so that signal-dominated modes may not be resolved, leading to model misidentification. On the other hand, if ϕd decays too swiftly in real space (smaller md), then the spectrum will decay more slowly and may put too much weight on noise-dominated frequencies. In addition, smaller md may not sufficiently reduce variance. A balance must be struck between (a) effectively reducing variance, which is ultimately determined by the decay of ψ in physical space, and (b) resolving the underlying dynamics, determined by the decay of in Fourier space.
4.1.2. Piecewise-Polynomial Test Functions.
Many test functions achieve the necessary balance between decay in real space and decay in Fourier space in order to offer both variance reduction and resolution of signal-dominated modes (defined by (4.1)). For simplicity, in this article we use the same test function space used in the ODE setting [28] and leave an investigation of the performance of different test functions to future work. Define to be the space of functions
| (4.2) |
where p, q ≥ 1 and v is a variable in time or space. The normalization
ensures that ‖ϕ‖∞ = 1. Functions are non-negative, unimodal, compactly-supported in [a, b], and have ⌊min{p, q}⌋ weak derivatives10. Larger p and q imply faster decay towards the endpoints (a, b) and for p = q we refer to p as the degree of ϕ. See Figure 2 for a visualization of ψ and partial derivatives constructed from tensor products of functions from . In addition to having nice integration properties combined with the trapezoidal rule (see Lemma 1 of [28]), (a, b, p, q) can be chosen to localize around signal-dominated frequencies in using the fact that for any reference domain length L ≥ |b − a|,
Figure 2.
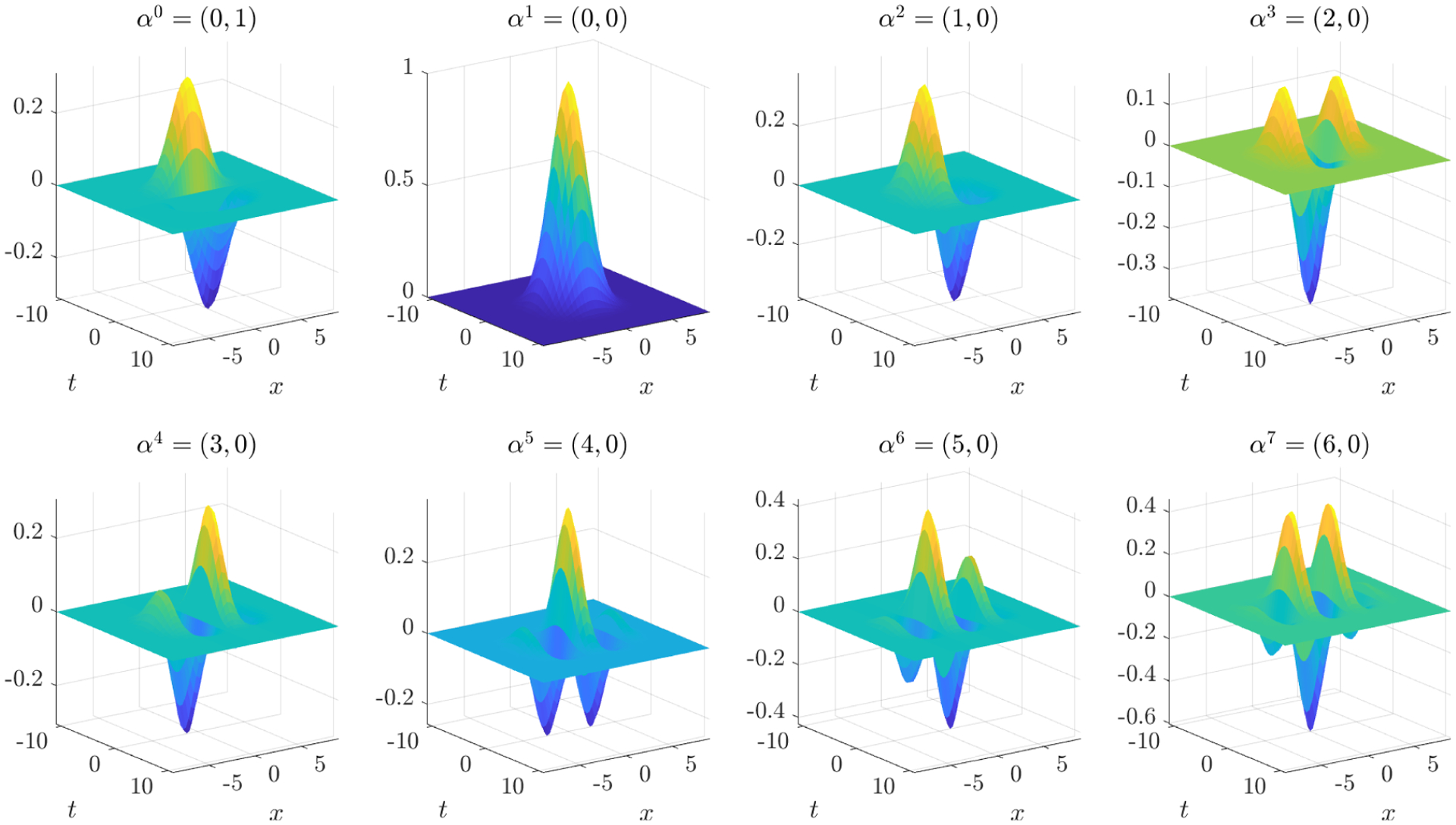
Plots of reference test function ψ and partial derivatives used for identification of the Kuramoto-Sivashinsky equation. The upper left plot shows ∂tψ, the bottom right shows . See Tables 2–4 for more details.
To assemble the reference test function ψ from one-dimensional test functions along each coordinate, we must determine the parameters (ad, bd, pd, qd) in the formula (4.2) for each ϕd. For convenience we center (Y, t) at the origin so that each ϕd is supported on a centered interval [ad, bd] = [−bd, bd], where bd = mdΔx for d ∈ [D] and bD + 1 = mD + 1Δt, and set pd = qd so that ψ is symmetric11. In this way only m ≔ (md)d∈[D + 1] and degrees p ≔ (pd)d∈[D + 1] need to be specified, hence the vectors can be computed from an analogous function with support [−1, 1],
using
where is an associated scaled grid and Δ ∈ {Δx, Δt}.
The discrete support lengths m and degrees p determine the smoothness of ψ, as well as its decay in real and in Fourier space, hence are critical to the method’s performance. The degrees p can be chosen from m to ensure necessary smoothness and decay in real space using
| (4.3) |
where is the maximum derivative along the dth coordinate and τ is a chosen (real-space) decay tolerance. By enforcing that ϕd decays to τ at the first interior gridpoint of its support, (4.3) controls the integration error (specifically, ensures integration error for noise-free data), while ensures that , which is necessary to integrate by parts as many times as required by the multi-index set α. The steps for arriving at the test function values on the reference grid are contained in Algorithm 4.1.
In the examples below, we set τ = 10−10 throughout12 and we use the method introduced in Appendix A to choose m, which involves estimating the critical wavenumber k* (defined in (4.1)) between noise-dominated and signal-dominated modes of . We also simplify things by choosing a single test function for all spatial coordinates, ϕx = ϕ1 = ϕ2 = ⋯ = ϕD where ϕx has degree px and support mx, and a (possibly different) test function ϕt = ϕD + 1 for the time axis with degree pt and support mt (recall that subscripts x and t are indices, not partial derivatives). This convention is used in the following sections.
4.2. Sparsification.
To enforce a sparse solution we present a modified sequential-thresholding least-squares algorithm MSTLS(G, b; λ), defined in (4.6), which accounts for terms that are outside of the dominant balance physics of the data, as determined by the left-hand side b, as well as terms with small coefficients. We then utilize the loss function
| (4.4) |
to select an optimal threshold , where wλ is the output of MSTLS(G, b; λ) defined in equation (4.6), #{·} denotes cardinality, is the index set of non-zero coefficients of wλ, wLS ≔ (GTG)−1GTb is the least squares solution, and SJ is the total number of terms in the library (S differential operators and J functions of the data). The first term in penalize the distance between GwLS (the projection of b onto the range of G) and Gwλ (the projection of b onto the columns of G restricted to ), while the second term penalizes the number of nonzero terms in the resulting model. The normalization simply enforces .
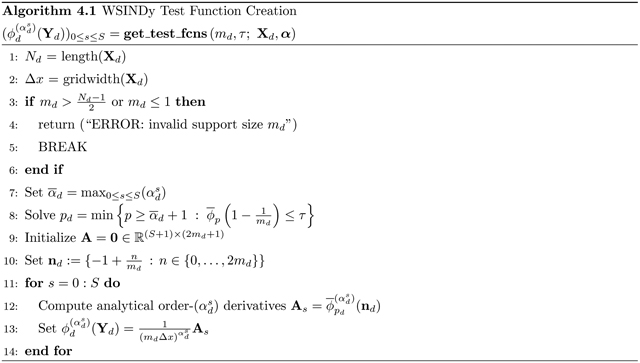
The MSTLS(G, b; λ) iteration is as follows. For a given λ ≥ 0, define the set of lower bounds Lλ and upper bound Uλ by
| (4.5) |
Then with w0 = wLS, define the iterates
| (4.6) |
The constraint is clearly more restrictive than standard sequential thresholding, but it enforces two desired qualities of the model: (i) that the coefficients wλ do not differ too much from 1, since 1 is the coefficient of the “evolution” term (assumed known), and (ii) that the ratio ‖wiGi‖2/‖b‖2 lies in [λ, λ−1], enforcing an empirical dominant balance rule (e.g. λ = 0.01 allows terms in the model to be at most two orders of magnitude from . Using previous results on the convergence of STLS [60], for MSTLS(G, b; λ) we employ the stopping criteria , which must occur for some ℓ ≤ SJ. The overall sparsification algorithm MSTLS(G, b; , λ) is
| (4.7) |
where λ is a finite set of candidate thresholds13. The learned threshold is the smallest minimizer of over the range λ and hence marks the boundary between identification and misidentification of the minimum-cost model, such that results in overfitting. A similar learning method for combining STLS and Tikhonov regularization (or ridge regression) was developed in [39]. We have found that our approach of combining MSTLS(G, b; , λ) with rescaling, as introduced in the next section, regularizes the sparse regression problem in the case of large model libraries without adding hyperparameters14 and definitely deserves further study.
4.3. Regularization through Rescaling.
Construction of the linear system b = Gw involves taking nonlinear transformations of the data fj(U) and then integrating against , which oscillates for large |αs|. This can lead to a large condition number κ(G) and prevent accurate inference of the true model coefficients w★, especially when the underlying data is poorly scaled15. In particular, identification of polynomial terms such as ∂x(u2) from a large library of polynomial terms is ill-conditioned for large (or small) amplitude data. Naively rescaling the data can easily lead to unreliable inference of model coefficients, since characteristic scales often effect the dynamics in nontrivial ways. For example, solution amplitude determines the wavespeed in the inviscid Burgers and Korteweg-de Vries equations, hence the solution and space-time coordinates must be rescaled in a principled manner in order to preserve the dynamics. To overcome this problem we propose to rescale the data using scale invariance of the PDE and choose scales that achieve a lower condition number, as described below. This approach allows for reliable identification of the Burgers and KdV equations from highly-corrupted large-amplitude data , see Section 5.4).
First, we note that the true model is scale invariant in the following way. If u solves (3.1), then for any scales γx, γt, γu > 0, the rescaled function
solves
where denotes differentiation with respect to . For homogeneous functions fj with power βj, we have , otherwise (in which case we set βj = 0). The linear system in the rescaled coordinates is constructed by discretizing the convolutional weak form as before but with a reference test function on the rescaled grid . We recover the coefficients16 at the original scales by setting , where M = diag(μ) is the diagonal matrix with entries
| (4.8) |
There is flexibility in choosing the scales γu, γx, γt, and a natural choice is to enforce that the columns of are similar in norm. Motivated by this, we find that for polynomial and trigonometric libraries, the scales17
| (4.9) |
are sufficient to regularize ill-conditioning due to poor scaling. Here and are the maximum spatial and temporal derivatives appearing in the library and is the highest monomial power of the functions (fj)j∈[J]. From (4.9) we get that
and
hence, using Young’s inequality for convolutions,
This shows that with scales γu, γx, γt set according to (4.9), the columns of are close in norm to the original dataset U. Similar scales γx, γt, γu can be chosen for different model libraries and reference test functions, and a more refined analysis will lead to scales that achieve closer agreement in norm. In the examples below we rescale the data and coordinates according to (4.9), which results in a low condition number (see Table 4). Throughout what follows, quantities defined over scaled coordinates will be denoted by tildes.
Table 4.
Additional specifications resulting from the choices in Table 3. The last column shows the start-to-finish walltime of Algorithm 4.2 with all computations in serial measured on a laptop with an 8-core Intel i7-2670QM CPU with 2.2 GHz and 8 GB of RAM.
| PDE | (Px, Pt) | (γu, γx, γt) | Walltime (sec) | ||
|---|---|---|---|---|---|
| IB | 784 × 43 | 1.4 × 106 | (7, 7) | (4.5 × 10−4, 0.0029, 1.1) | 0.12 |
| KdV | 1443 × 43 | 3.2 × 106 | (8, 7) | (5.7 × 10−4, 8.3, 1250) | 0.39 |
| KS | 1806 × 43 | 3.7 × 103 | (10, 10) | (0.26, 0.74, 0.091) | 0.24 |
| NLS | 1804 × 190 | 1.2 × 105 | (11, 10) | (0.33, 3.1, 9.4) | 2.5 |
| PM | 4608 × 65 | 2.4 × 104 | (8, 10) | (1.6, 2.7, 3.2) | 16 |
| SG | 13000 × 73 | 1.3 × 104 | (8, 10) | (0.23, 8.1, 8.1) | 29 |
| RD | 11638 × 181 | 4.5 × 103 | (13, 12) | (0.86, 6.5, 1.4) | 75 |
| NS | 3872 × 50 | 8.2 × 102 | (9, 12) | (0.53, 0.72, 2.4) | 12 |
4.4. Query Points and Subsampling.
Placement of {(xk, tk)}k∈[K] determines which regions of the observed data will most influence the recovered model18. In WSINDy for ODEs ([28]), an adaptive algorithm was designed for placement of test functions near steep gradients along the trajectory. Improvements in this direction in the PDE setting are a topic of active research, however, for simplicity in this article we uniformly subsample {(xk, tk)}k∈[K] from (X, t) using subsampling frequencies s = (s1, … , sD + 1) along each coordinate, specified by the user. That is, along each one-dimensional grid points are selected with uniform spacing sdΔx for d ∈ [D] and sD + 1Δt for d = D + 1. This results in a (D + 1)-dimensional coarse grid with dimensions , which determines the number of query points
| (4.10) |
4.5. Model Library.
The model library is determined by the nonlinear functions (fj)j∈[J] and the partial derivative indices α and is crucial to the well-posedness of the recovery problem. In the examples below we choose (fj)j∈[J] to be polynomials and trigonometric functions as these sets are dense in many relevant function spaces. When the true PDE does not contain cross derivatives (e.g. ), we remove them from the derivative library α and note that including these terms does not have a significant impact on the results.
5. Examples
We now demonstrate the effectiveness of WSINDy by recovering the PDEs listed in Table 2 over a range of noise levels. These examples show that WSINDy provides orders of magnitude improvements over derivative-based methods [39], with reliable and accurate recovery of four out of the eight PDEs under noise levels as high as 100% (defined in (5.1) and (5.2)) and for all examples under 20% noise. In contrast to the weak recovery methods in [37, 13], WSINDy uses (i) the convolutional weak form (3.6) and FFT-based implementation (3.12), (ii) improved thresholding and automatic selection of the sparsity threshold via (4.6) and (4.7), and (iii) rescaling using (4.9). The effects of these improvements are discussed in Sections 5.4 and 5.5.
Table 2.
PDEs used in numerical experiments, written in the form identified by WSINDy. Domain specification and boundary conditions are given in Appendix B.
| Inviscid Burgers (IB) | |
| Korteweg-de Vries (KdV) | |
| Kuramoto-Sivashinsky (KS) | |
| Nonlinear Schrödinger (NLS) | |
| Anisotropic Porous Medium (PM) | ∂tu = (0.3) ∂xx (u2) − (0.8) ∂xy (u2) + ∂yy (u2) |
| Sine-Gordon (SG) | ∂ttu = ∂xxu + ∂yyu − sin (u) |
| Reaction-Diffusion (RD) | |
| 2D Navier-Stokes (NS) |
To test robustness to noise, a noise ratio σNR is specified and a synthetic “observed” dataset
is obtained from a simulation U★ of the true PDE19 by adding i.i.d. Gaussian noise with variance σ2 to each data point, where
| (5.1) |
We examine noise ratios σNR in the range [0, 1] and often refer to the noise level as σNR or equivalently that the data contains 100σNR% noise. We note that the resulting true noise ratio
| (5.2) |
matches the specified σNR to at least four significant digits in all cases and so we only list σNR. When the state variable is vector-valued, as with the nonlinear Schrödinger, reaction-diffusion, and Navier-Stokes equations (see Table 2), a separate noise variance σ2 is computed for each vector component so that the noise ratio σNR of each component satisfies (5.2).
5.1. Performance Measures.
To measure the ability of the algorithm to correctly identify the terms having nonzero coefficients, we use the true positivity ratio (introduced in [22]) defined by
| (5.3) |
where TP is the number of correctly identified nonzero coefficients, FN is the number of coefficients falsely identified as zero, and FP is the number of coefficients falsely identified as nonzero. Identification of the true model results in a TPR of 1, while identification of half of the correct nonzero terms and no falsely identified nonzero terms results in TPR of 0.5 (e.g. the 2D Euler equations ∂tω = −∂x(ωu) − ∂y(ωv) result in a TPR of 0.5 if the underlying true model is the 2D Navier-Stokes vorticity equation). We will see that in several cases that the average TPR remains above 0.95 even as the noise level approaches 1. The loss function (defined in (4.4)) and the resulting learned sparsity threshold (defined in (4.7)) provide additional information on the algorithm’s ability to identify the correct model terms with respect to the noise level. In particular, sensitivity to the sparsity threshold suggests that automatic selection of is essential to successful recovery in the relatively large noise regime.
To assess the accuracy of the recovered coefficients we use two metrics. We measure the maximum error in the true non-zero coefficients using
| (5.4) |
where |·| denotes absolute value, and the ℓ2 distance in parameter space using
| (5.5) |
E∞ determines the number of significant digits in the recovered true coefficients while E2 provides information about the magnitudes of coefficients that are falsely identified as nonzero. Often when a term is falsely identified and the resulting nonzero coefficient is small, a larger sparsity factor will result in idenfitication of the true model.
Finally, when , we report the prediction accuracy between the true data U★ and a numerical solution Udd to the data-driven PDE using the same initial conditions. We compute the relative L2 error at time t = 0.5T (i.e. at the half-way point in time) defined by
| (5.6) |
Where , denote the numerical solutions over the spatial domain at time t. Since solutions to the data-driven dynamics and the true dynamics will eventually drift apart, we also measure
| (5.7) |
or, the first time t (relative to the final time T) that the numerical solution reaches a relative L2 distance of tol from the truth. The minimum in (5.7) is computed over t ∈ t and we set tol = 0.1. We provide results for and averaged over the weights satisfying .
For each system in Table 2 and each noise level σNR ∈ {0.025q : q ∈ {0, … , 40}} we run WSINDy on 200 instantiations of noise20 and average the results of error statistics (5.3)–(5.7). Computations were carried out on a University of Colorado Boulder Blanca Condo cluster21.
5.2. Implementation Details.
The hyperparameters used in WSINDy applied to each of the PDEs in Table 2 are given in Table 3. To select test function discrete support lengths we used a combination of the changepoint method22 described in Appendix A and manual tuning. Across all examples the real-space decay tolerance for test functions is fixed at τ = 10−10.
Table 3.
WSINDy hyperparameters used to identify each example PDE.
| PDE | U | f j | α | (mx, mt) | (sx, st) |
|---|---|---|---|---|---|
| IB | 256 × 256 | (uj−1)j∈[7] | ((ℓ, 0))0≤ℓ≤6 | (60, 60) | (5, 5) |
| KdV | 400 × 601 | (uj−1)j∈[7] | ((ℓ, 0))0≤ℓ≤6 | (45, 80) | (8, 12) |
| KS | 256 × 301 | (uj−1)j∈[7] | ((ℓ, 0))0≤ℓ≤6 | (23, 22) | (5, 6) |
| NLS | 2 × 256 × 251 | (uivj)0≤i + j≤6 | ((ℓ, 0))0≤ℓ≤6 | (19, 25) | (5, 5) |
| PM | 200 × 200 × 128 | (ui−1)i∈[5] | (37, 20) | (8, 5) | |
| SG | 129 × 403 × 205 | (uj−1)i∈[5], (sin(ju), cos(ju))j=1, 2 | ((ℓ, 0, 0), (0, ℓ, 0))0≤ℓ≤4 | (40, 25) | (5, 8) |
| RD | 2 × 256 × 256 × 201 | (uivj)0≤i + j≤4 | ((ℓ, 0, 0), (0, ℓ, 0))0≤ℓ≤5 | (13, 14) | (13, 12) |
| NS | 3 × 324 × 149 × 201 | ((ℓ, 0, 0), (0, ℓ, 0))0≤ℓ≤2 | (31, 14) | (12, 8) |
In computing a sparse solution (see equation (4.7)), the search space λ for the learned threshold is fixed for all examples at:
in other words λ contains 50 points with log10(λ) equally spaced from −4 to 0. This implies a stopping criteria of 50SJ thresholding iterations23.
We fix the subsampling frequencies (sx, st) to for PDEs in one spatial dimension and to for two spatial dimensions, where the dimensions (N1, N2, N3) depend on the dataset. Additional information about the convolutional weak discretization is included in Table 4, such as the dimensions and condition number of the rescaled Gram matrix (computed from a typical dataset with 20% noise), test function polynomial degrees (px, pt), scale factors (γu, γx, γt), and start-to-finish walltime of Algorithm 4.2 with all computations performed serially on a laptop with an 8-core Intel i7-2670QM CPU with 2.2 GHz and 8 GB of RAM.
5.3. Comments on Chosen Examples.
The primary reason for choosing the examples in Table 2 is to demonstrate that WSINDy can successfully recover models over a wide range of physical phenomena such as spatiotemporal chaos, nonlinear waves, nonlinear diffusion, shock-forming solutions, complex limit cycles, and pattern formation in reaction diffusion equations.
Recovery of the inviscid Burgers and anisotropic porous medium equations demonstrates (i) that WSINDy can discover PDEs from solutions that can only be understood in a weak sense and (ii) that discovery in this case is just as accurate and robust to noise and scaling as with smooth data (i.e. no special modifications of the algorithm are required to discover models from non-smooth data, as conjectured in [13]). We use analytical weak solutions, with inviscid Burgers data forming a shock which propagates at constant speed (see Figure 3 for plots of the characteristic curves) and porous medium data having a jump in the gradient ∇u. In addition, we discover the porous medium equation using an anisotropic diffusivity tensor to demonstate that WSINDy can identify the cross-diffusion term ∂xy(u2) to high accuracy from a large candidate model library.
The inviscid Burgers and Korteweg-de Vries equations demonstrate that WSINDy successfully recovers the correct models for nonlinear transport data with large amplitude. Both datasets have mean amplitudes on the order of 103 (in addition KdV is given over a short time window of t ∈ [0, 10−3]), and hence are not identifiable from large polynomial libraries using naive approaches. The sparsification and rescaling measures in Sections 4.2 and 4.3 are essential to removing this barrier.
The Sine-Gordon equation24 is used to show both that trigonometric library terms can easily be identified alongside polynomials and that hyperbolic problems do not seem to present further challenges. Discovery of the Sine-Gordon equation also appears to be particularly robust to noise, which suggests that the added complexity of having multiple spatial dimensions is not in general a barrier to identification.
For the nonlinear Schrödinger and reaction-diffusion systems, we test the ability of WSINDy to select the correct monomial nonlinearities from an excessively large model library. Using a library of 190 terms for nonlinear Schrödinger’s and 181 terms for reaction-diffusion (see the dimensions of in Table 4), we demonstrate successful identification of the correct nonzero terms. Moreover, for the reaction-diffusion system, misidentified terms directly reflect the existence of a limit cycle25. Finally, the vortex-shedding limit cycle for the 2D Navier-Stokes equations is used primarily to compare to previous results in [39], and we find that at large-noise WSINDy conveniently selects the Euler equations.
5.4. Results: Model Identification.
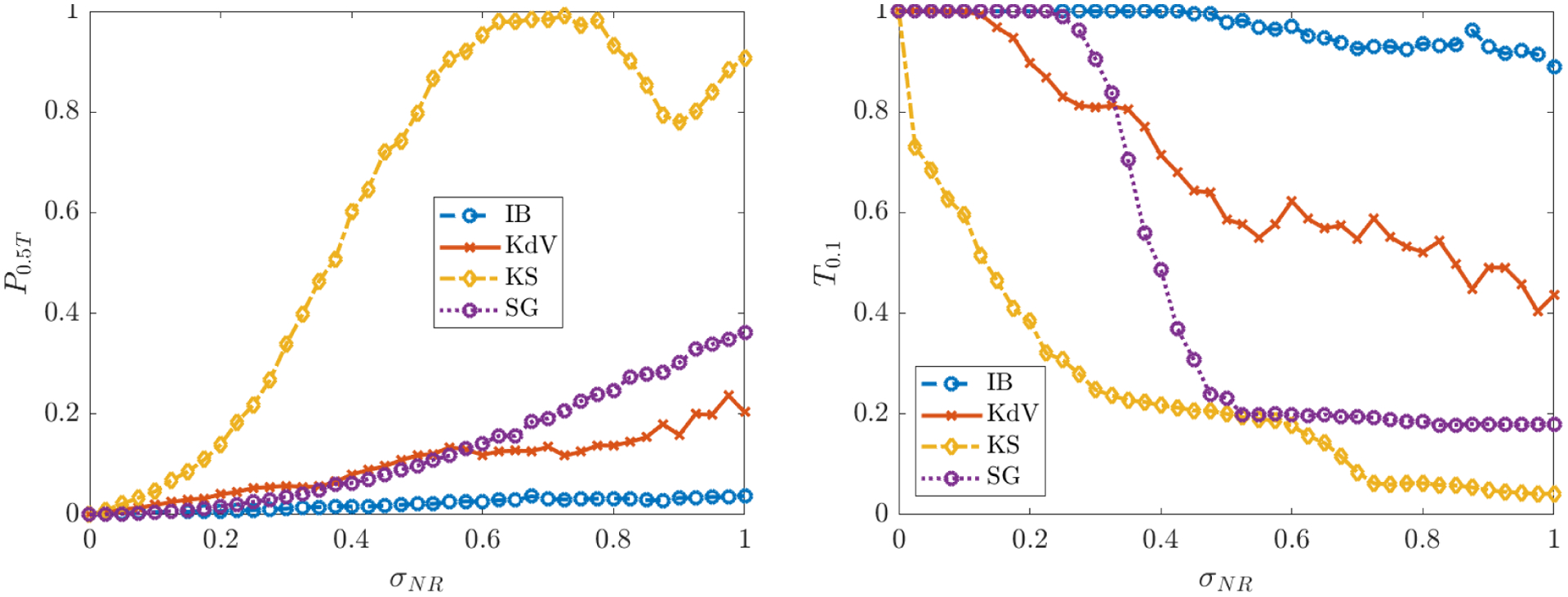
Performance regarding the identification of correct nonzero terms in each model is reported in Figures 4 and 5, which include plots of the average TPR, the learned threshold , and the loss function (defined in (5.3), (4.7), and (4.4), respectively). As we will discuss, significant decreases in average TPR are often accompanied by transitions in the identified .
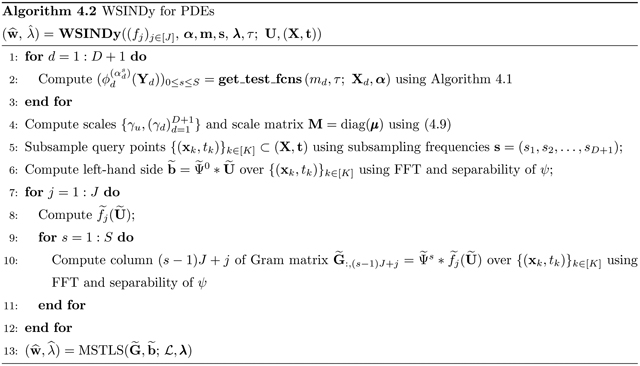
Figure 4.
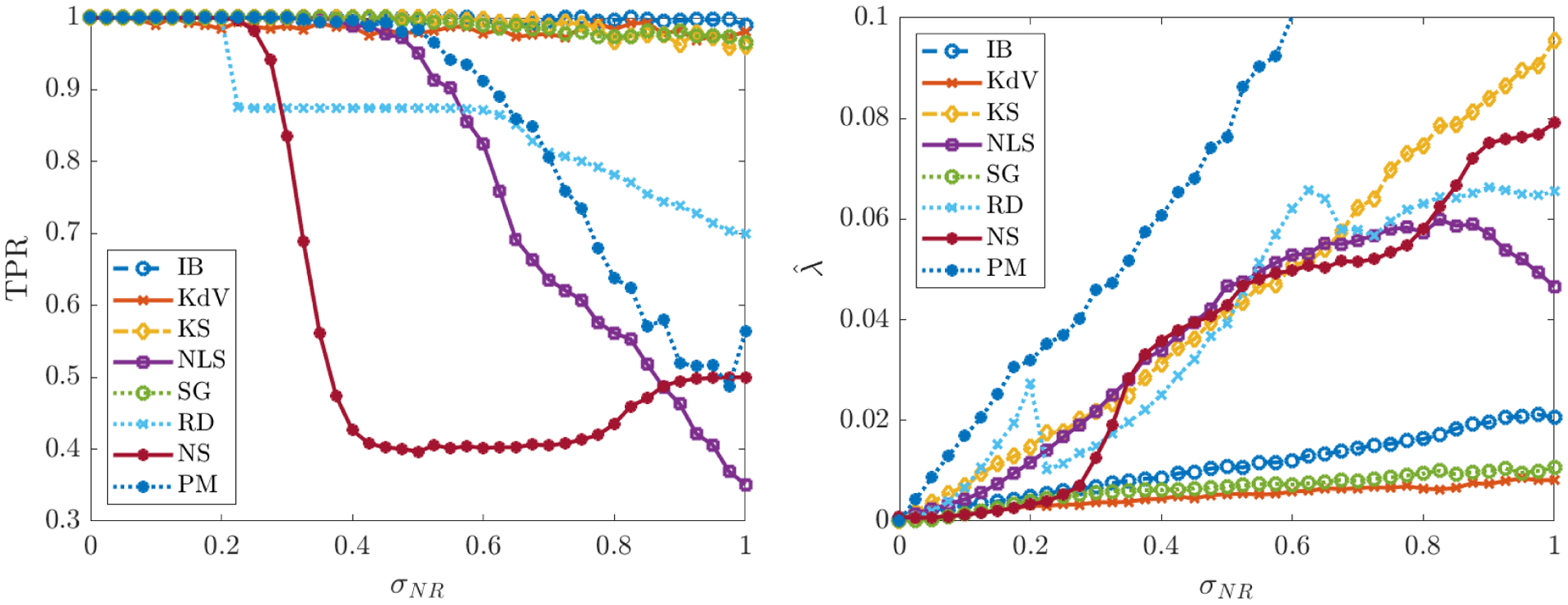
Left: average TPR (true positivity ratio, defined in (5.3)) for each of the PDEs in Table 2 computed from 200 instantiations of noise for each noise level σNR. Right: average learned threshold (defined in (4.7)). For the porous medium equation (PM), increases to 0:2 as σNR approaches 1 (we omit this from the plot in order to make visible the trends for the other systems).
Figure 5.
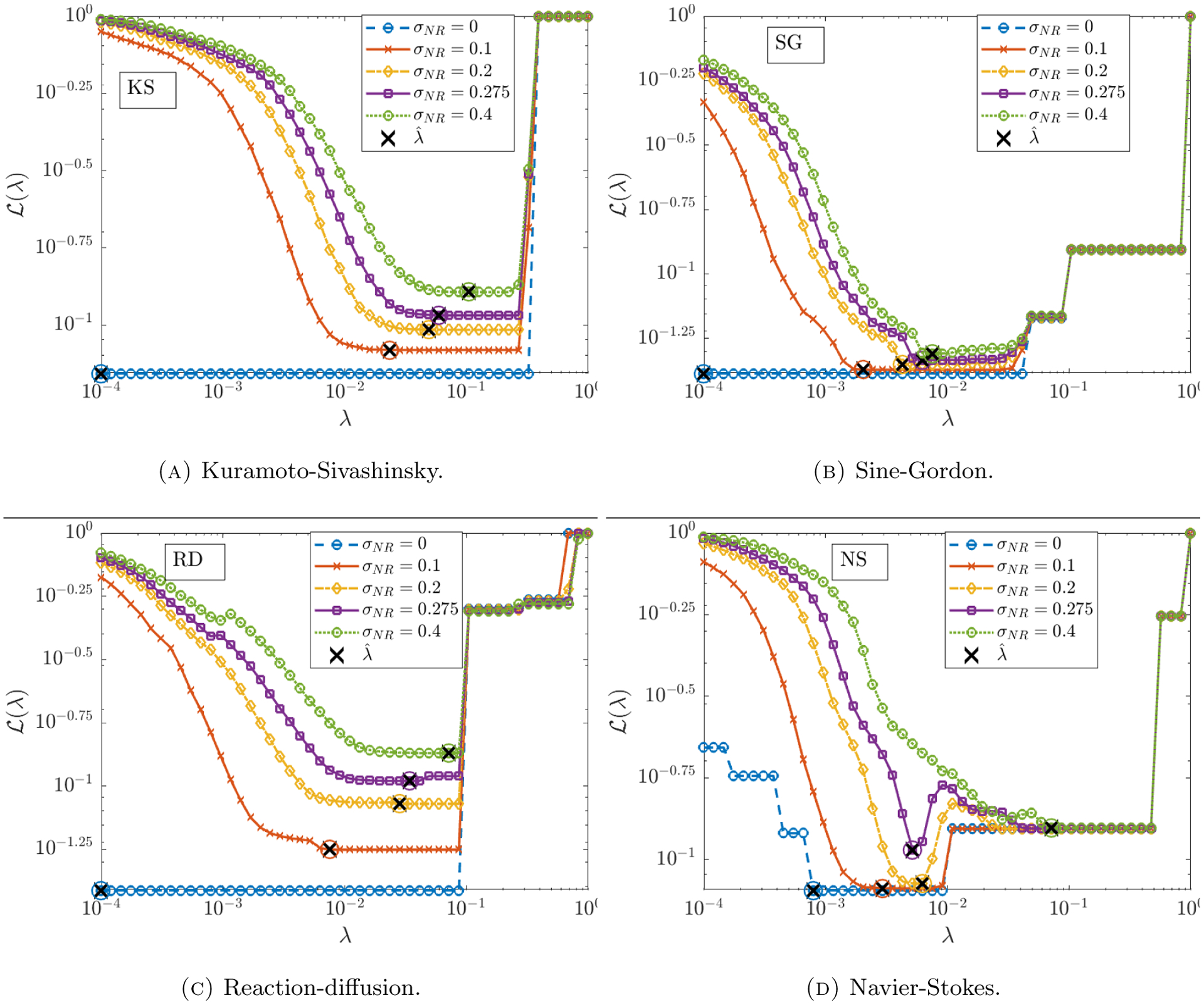
Plots of the average loss function and resulting optimal threshold for the Kuramoto Sivashinsky, Sine-Gordon, Reaction diffusion and Navier-Stokes equations.
Figure 4 (left) shows that for inviscid Burgers, Korteweg-de Vries, Kuramoto-Sivashinsky and Sine-Gordon, the average TPR stays above 0.95 even for noise levels as high as 100% (i.e. WSINDy reliably identifies these models in the presence of noise that has the same L2-norm as the underlying clean data). The average TPR for the nonlinear Schrödinger and porous medium equations stays above 0.95 until 50% noise, after which identification of the correct monomial nonlinearity is not as reliable. For NLS, this is a drastic improvement over previous studies [39], especially considering the large library of 190 terms used.
We observe in Figure 4 (right) that the learned threshold increases with σNR, suggesting that automatic selection of in the learning algorithm (4.7) is crucial to the algorithm’s robustness to noise. For example, the Kuramoto-Sivashinsky equation has a minimum nonzero coefficient of 0.5 (multiplying ∂x(u2)), and we find that approaches 0.1 as σNR approaches 1, which implies that at higher noise levels the range of values that is necessary26 for correct model identification is approximately (~ 0.1, ~ 0.5). Since it is highly unlikely that this range of admissible values would be known a priori, the chances of manually selecting a feasible for Kuramoto-Sivashinsky are prohibitively low in the large noise regime (see Figure 5a for visualizations of the loss applied to KS data). This effect is even greater for the porous medium equation. Automatic selection of thus removes this sensitivity. In contrast, is largely unaffected by increases in σNR for Burgers, Korteweg-de Vries and Sine-Gordon. In particular, Figure 5b shows little qualitative changes in the loss landscape for Sine-Gordon in the range 0.1 ≤ σNR ≤ 0.4.
Intriguingly, for reaction-diffusion, the average TPR falls below 0.95 at 22% noise, after which WSINDy falsely identifies linear terms in u and v. If the true model is given by the compact form for u = (u, v)T, then the misidentified model in all trials for noise levels in the range 0.25 ≤ σNR ≤ 0.55 is given by
| (5.8) |
for some α > 0 and β ≈ 1 dependent on σNR. This is explainable by the fact that the underlying solution settles into a limit cycle, which means that at every point in space the solution oscillates. Indeed, the falsely identified nonzero terms in (5.8) exactly convey that at each point in space the solution is oscillating at a uniform frequency (albeit with variable amplitude and phase determined by the initial conditions27). Hence, in the presence of certain lower-dimensional structures (in this case a limit cycle), higher noise levels result in a mixture of the true model with a spatially-averaged reduced model. This shift between detection of the correct model and the oscillatory version (5.8) is also detectable in the learned threshold , which decreases at σNR = 0.22 (see RD data in Figure 4 (right)), and in the loss function (Figure 5c). At σNR = 0.275 we see that in Figure 5c is minimized for λ in the approximate range (~ 0.02, ~ 0.05) but also has a near-minimum for λ ∈ (~ 0.05, ~ 0.1). These two regions correspond to discovery of the oscillatory model (5.8) and the true model, respectively, but since the true model has a slightly higher loss at σNR = 0.275, model (5.8) is selected. For σNR ≥ 0.4 there is no longer (on average) a region of λ that results in discovery of the true model, and WINSDy returns (5.8) to compensate for noise.
For Navier-Stokes we see an averaging effect at higher noise, similar to the reaction-diffusion system. TPR drops below 0.95 for noise levels above 27% with the resulting misidentified model being simply Euler’s equations in vorticity form:
This is due primarily to the small viscosity ν = 0.01 which prevents identification of the viscous forces at higher noise levels. Examining the loss function , Figure 5d shows that above σNR ≈ 0.275, minimizers of are above 0.01, hence the viscous terms will be thresholded out. Another possible explanation is the low-accuracy simulation used for the clean dataset: in the noise-free setting, Table 5 shows that WSINDy recovers the model coefficients of Navier-Stokes to less than 3 significant digits in the absence of noise, which is the same level of accuracy exhibited on each of the other systems under 5% noise (see Figure 6). Nevertheless, with reliable recovery up to 27% noise, WSINDy makes notable improvements on previous results ([39]). Moreover, recovery of the Euler equations at high noise is desirable as this can be seen as the correct leader-order model.
Table 5.
Accuracy of WSINDy applied to noise-free data (σNR = 0).
| IB | KdV | KS | NLS | PM | SG | RD | NS | |
|---|---|---|---|---|---|---|---|---|
| E ∞ | 4.3 × 10−5 | 3.1 × 10−7 | 8.1 × 10−7 | 9.4 × 10−8 | 2.2 × 10−6 | 4.3 × 10−5 | 3.9 × 10−10 | 1.1 × 10−3 |
Figure 6.
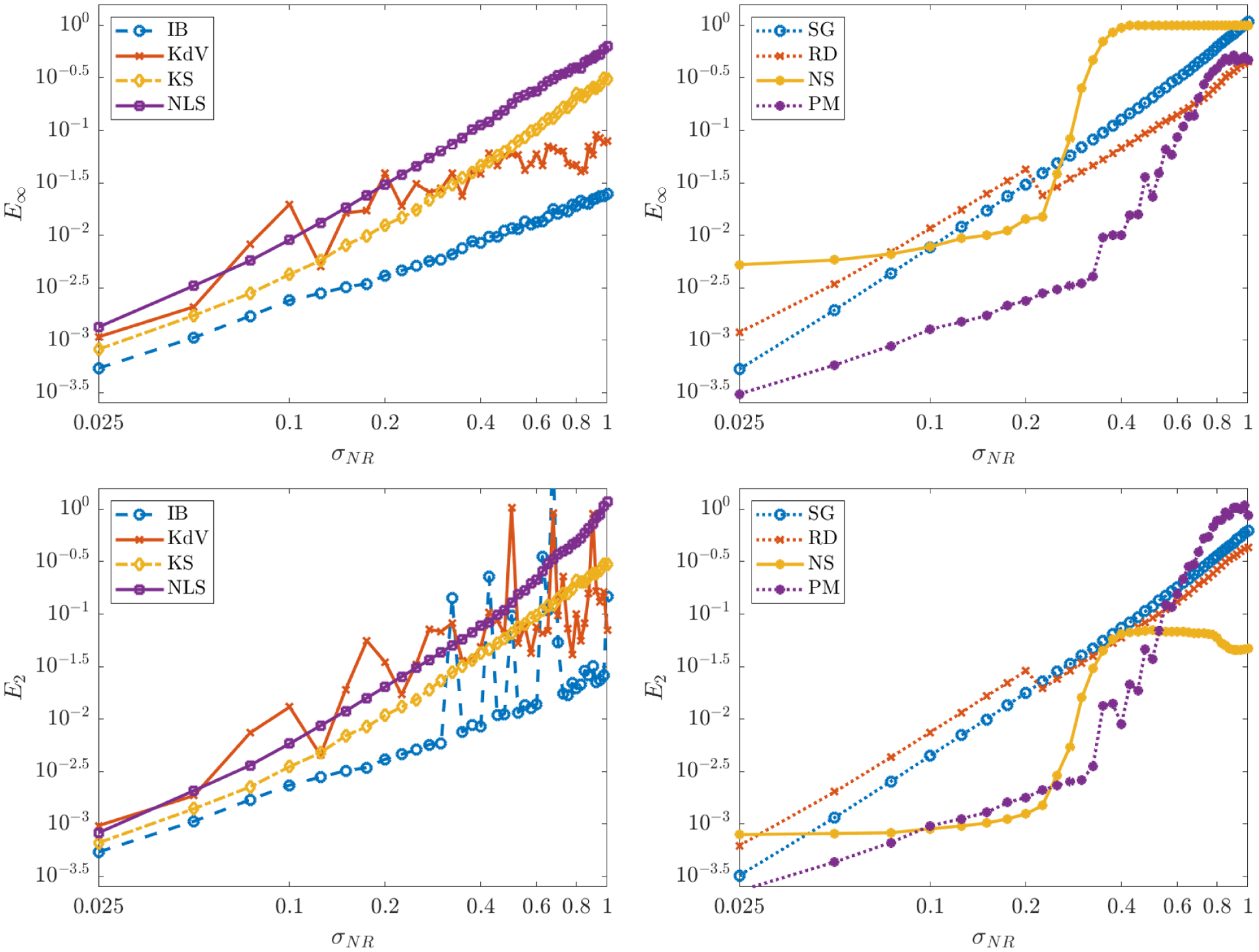
Coefficient errors E∞ and E2 (equations (5.4) and (5.5)) for each of the seven models Table 2. Models in one and two spatial dimensions are shown on the left and right, respectively.
5.5. Results: Coefficient Accuracy.
Accuracy in the recovered coefficients is measured by E∞ and E2 (defined in (5.4) and (5.5), respectively) and shown in Table 5 for σNR = 0 and in Figure 6 for σNR > 0. As in the ODE case, the coefficient error E∞ for smooth, noise-free data is determined by the order of accuracy of the numerical simulation method28, since the error resulting from the trapezoidal rule is of lower order for the values (px, pt) used in Table 4 (see [28], Lemma 1). Table 5 also shows that the algorithm returns reasonable accuracy for non-smooth data, with E∞ = 4.3 × 10−5 and E∞ = 2.2 × 10−6 for the inviscid Burgers and porous medium equations, respectively. For reference, Table 6 shows that WSINDy improves over PDE-FIND by about two digits29.
Table 6.
Accuracy comparison between WSINDy and PDE-FIND with σNR = 0.01 (results for PDE-FIND reproduced from [39]).
| KdV | KS | NLS | RD | NS | |
|---|---|---|---|---|---|
| WSINDy | 6.7 × 10−4 | 1.8 × 10−4 | 2.9 × 10−4 | 6.0 × 10−4 | 1.2 × 10−3 |
| PDE-FIND | 7.0 × 10−2 | 0.52 | 3.0 × 10−2 | 3.8 × 10−2 | 7.0 × 10−2 |
For σNR > 0, in Figure 6 it is apparent that E∞ scales approximately as a power law for some r approximately in the range (~ 1, ~ 2) in all systems except Navier-Stokes. It was observed in [13] that E∞ will approximately scale linearly with σNR for Kuramoto-Sivashinsky, however our results show that in general, for larger σNR, the rate will be superlinear and dependent on the reference test function and the nonlinearities present. A simple explanation for this in the case of normally-distributed noise is the following: linear terms Ψs * U will be normally-distributed with mean Ψs * U★ and approximate variance , hence are unbiased30 and lead to perturbations that scale linearly with σNR. On the other hand, general monomial nonlinearities31 Ψs *Uj with j > 1 are biased and have approximate variance for p2j a polynomial of degree 2j. Hence, nonlinear terms Ψs * fj(U) lead to biased columns of the Gram matrix G with variance scaling with σ2r for some r > 1 and proportional to . Thus, for larger noise and higher-degree monomial nonlinearities, we expect superlinear growth of the error, as observed in particular with nonlinear Schrödinger’s, Sine-Gordon, and reaction-diffusion. Nevertheless, Figure 6 suggests that a conservative estimate on the coefficient error is , indicating 1 − log10(σNR) significant digits (e.g. for σNR = 0.1 we have E∞ ≤ 10−2 for each system except KdV, indicating two significant digits), which is consistent with the ODE case [28].
For Burgers and Korteweg-De Vries, the average error E2 at higher noise levels is affected by outliers containing a falsely-identified advection term ∂xu. This is due to the large amplitude datasets used, which lead to the closest pure-advection model for each system being given by32
Hence, a falsely identified ∂xu term will have a large coefficient compared to the true model coefficients which have magnitudes 0.5 or 1. In all other cases, the values of E2 and E∞ are comparable, which implies that misidentified terms do not have large coefficients and might be removed with a larger threshold. Lastly, the sigmoidal shape of E∞ and E2 for Navier-Stokes is due again to the unidentified diffusive terms at larger noise. It is interesting to note that for σNR ≤ 0.27 the coefficient error for Navier-Stokes is relatively constant, in contrast to the other systems, and does not exhibit a power-law. However, at present, we do not have a concrete explanation for this behavior.
5.6. Results: Prediction Accuracy.
Lastly, Figure 7 shows the prediction accuracy on a subset of the systems in Table 2 as measured by and (defined in (5.6) and (5.7), respectively). We report that data-driven solutions attain greater than 90% accuracy in the L2 sense up to time 0.8T (80% of the trajectory) for noise levels as high as 40%. (This excludes the KS equation, which exhibits spatiotemporal chaos and cannot be expected to remain close to the noise-free data.) Data-driven solutions to the KS equation, while eventually divergent, also attain 90% accuracy up to time 0.5T for noise levels below 15%. Lastly, we note that for lower noise levels (up to 10%), the accuracy of data-driven solutions to the inviscid Burgers, Korteweg-de Vries and Sine-Gordon equations is on average above 96% along the entire trajectory (not shown in the figures).
Figure 7.
Prediction accuracy measured by and (defined in (5.6) and (5.7), respectively).
6. Conclusion
We have extended the WSINDy algorithm to the setting of PDEs for the purpose of discovering models for spatiotemporal dynamics without relying on pointwise derivative approximations, black-box closure models (e.g. deep neural networks), dimensionality reduction, or other noise filtering. We have provided methods for learning many of the algorithm’s hyperparameters directly from the given dataset, and in the case of the threshold , demonstrated the necessity of avoiding manual hyperparameter tuning. The underlying convolutional weak form (3.4) allows for efficient implementation using the FFT. This naturally leads to a selection criterion for admissable test functions based on spectral decay, which is implemented in the examples above. In addition, we have shown that by utilizing scale invariance of the PDE together with a modified sparsification measure, models may be recovered from large candidate model libraries and from data that is poorly scaled. When unsuccessful, WSINDy appears to discover a nearby sparse model that captures the dominant spatiotemporal behavior (see the discussions surrounding misidentification of the reaction-diffusion and Navier-Stokes equations in Section 5.4).
We close with a summary of possible future directions. In Section 4.1 we discussed the significance of decay properties of test functions in real and in Fourier space, as well as general test function regularity. We do not make any claim that the class defined by (4.2) is optimal, but it does appear to work very well, as demonstrated above (as well as in the ODE setting [28]) and also observed in [37, 13]. A valuable tool for future development of weak identification schemes would be the identification of optimal test functions. A preliminary step in this direction is our use of the changepoint method described in Appendix A.
In the ODE setting, adaptive placement of test functions provided increased robustness to noise. Convolution query points can similary be strategically placed near regions of the dynamics with high information content, which may be crucial for model selection in higher dimensions. Defining regions of high information content and adaptively placing query points accordingly would allow for identification from smaller datasets.
Ordinary least squares makes the assumption of i.i.d. residuals and should be replaced with generalized least squares to accurately reflect the true error structure. The current framework could be vastly improved by incorporating more precise statistical information about the linear system (G, b). The first step in this direction is the derivation of an approximate covariance matrix as in WSINDy for ODEs [28]. Previous results on generalized sensitivity analysis for PDEs may provide improvements in this direction [18, 46].
Accuracy in the recovered coefficients is still not entirely understood and is needed to derive recovery guarantees. It is claimed in [13] that at higher noise levels the scaling will approximately be linear in σNR, while we have demonstrated that this is not the case in general: the scaling depends on the nonlinearities present in the true model, the decay properties of the test functions, and accuracy of the underlying clean data. Analysis of coefficient error dependence (on noise, amplitudes, number of datapoints, etc.) could occur in tandem with development of a generalized least-squares framework.
The examples above show that WSINDy is very robust to noise for problems involving nonlinear waves (Burgers, Korteweg de-Vries, nonlinear Schrödinger, Sine-Gordon), spatiotemporal chaos (Kuramoto-Sivashinsky), and even nonlinear diffusion (porous medium), but is less robust for data with limit cycles (reaction-diffusion, Navier-Stokes). Further, identification of Burgers, Korteweg de-Vries, and Sine-Gordon appears robust to changes in the sparsity threshold (see Figure 4 (right)). A structural identifiability criteria for measuring uncertainty in the recovery process based on identified structures (transport processes, mixing, spreading, limit cycles, etc.) would also be invaluable for general model selection.
Highlights.
We present the WSINDy algorithm for data-driven discovery of partial differential equations in weak form. Pointwise derivative approximations are replaced by convolution against test functions for robust, high-accuracy model selection in the presence of large measurement noise.
Reformulating the weak dynamics in terms of convolutions allows for fast FFT-based implementation and reveals that test function spectra plays a crucial role in guaranteeing robustness to noise.
Scale invariance is used together with an improved thresholding algorithm with automatic threshold selection to enable PDE identification from poorly-scaled data and large candidate libraries.
Successful discovery is demonstrated on the inviscid Burgers, Korteweg-de Vries, Kuramoto-Sivashinksy, nonlinear Schrödinger, anisotropic porous medium, Sine-Gordon, reaction-diffusion, and Navier-Stokes equations from highly corrupted datasets, and in the case of inviscid Burgers and porous medium, from non-classical (weak) solutions.
7. Acknowledgements
This research was supported in part by the NSF/NIH Joint DMS/NIGMS Mathematical Biology Initiative grant R01GM126559 and in part by the NSF Computing and Communications Foundations Division grant CCF-1815983. This work also utilized resources from the University of Colorado Boulder Research Computing Group, which is supported by the National Science Foundation (awards ACI-1532235 and ACI-1532236), the University of Colorado Boulder, and Colorado State University. Code used in this manuscript is publicly available on GitHub at https://github.com/MathBioCU/WSINDy_PDE. The authors would like to thank Prof. Vanja Dukic (University of Colorado at Boulder, Department of Applied Mathematics), Kadierdan Kaheman (University of Washington, Department of Applied Mathematics), Samuel Rudy (Massachusetts, Department of Mechanical Engineering), and Zofia Stanley (University of Colorado at Boulder, Department of Applied Mathematics), for helpful discussions.
Appendix A. Learning Test Functions From Data
We present the following algorithm for automatic selection of test functions which utilizes the implicit smoothing of high-frequency noise afforded by the convolution. This approach is useful in practice but we leave rigorous justification of it to future work. We proceed in two steps: (1) estimation of critical wavenumbers separating noise- and signal-dominated modes in each coordinate and (2) enforcing decay in real and in Fourier space.
We will describe the process for detecting from data given over the one-dimensional spatial grid at timepoints . Figures 8–9 illustrate this approach using Kuramoto-Sivashinsky data with 50% noise. Below, and denote the discete Fourier transform (DFT) along the x and t coordinates, respectively, while denotes the full two-dimensional DFT.
A.1. Detection of Critical Wavenumbers.
Assume the data has additive white noise U = U★ + ϵ with and that decays. The power spectrum of the noise is then i.i.d, hence as discussed in Section 4.1, there will be a critical wavenumber in the power spectrum of the data after which the modes become noise-dominated. To detect , we collapse into a one-dimensional array by averaging in time and then take the cumulative sum in x:
| (A.1) |
where is the time-average of the jth mode of the discrete Fourier transform along the x-coordinate. Since is i.i.d., Hx will be approximately linear over the noise-dominated modes, which is an optimal setting for locating as a changepoint, or in other words the corner point of the best piecewise-linear approximation33 to Hx using two pieces (see Figure 8). An algorithm for this is given in [20] and implemented in MATLAB using the function findchangepts.
A.2. Enforcing Decay.
Having detected the changepoints and , we compute hyperparameters for the coordinate test functions ϕx and ϕt using user-specified hyperparameters τ and . As in Section 4.1, τ specifies the rate of decay of ϕx and ϕt in real space through equation (4.3). The hyperparameter is introduced to specify the rate of decay of ϕx and ϕt in Fourier space. Specifically, for a chosen we enforce that the changepoints and fall approximately standard deviations into the tail of the spectra and . This is done by utilizing that ϕx and ϕt are functions of the form
(i.e. centered, symmetric functions in the class defined in (4.2)) which are well-approximated by Gaussians for large enough p and appropriate scaling C. Indeed, letting C be such that ‖ϕa,p‖1 = 1 and setting we have that ϕa,p matches the first three moments of the Gaussian
which provides a bound on the error in the Fourier transforms and and for small frequencies ξ in terms of their 4th moments34:
This implies that for small ξ and a and large p, it suffices to use to estimate . Hence, we enforce decay of (and similarly for ) by choosing mx and px such that
| (A.2) |
so that is standard deviations into the tail of , where . To solve (4.3) and (A.2) simultaneously, we compute mx as a root of
F(m) has a unique root mx ≥ 2 in the nonempty interval
on which F monotonically decreases and changes sign, provided N1 > 4, τ ∈ (0, 1) and , constraints which are easily satisfied. After finding mx we can solve for px using either (4.3) or (A.2).
Figure 9 illustrates the implicit filtering of this process using the Burgers-type nonlinearity ∂x(U2) and the same KS dataset as in Figure 9 with 50% noise. The top panel compares a one-dimensional slice in x taken at fixed time t = 99 of the clean data (U★)2 and noisy data (U)2. The middle panel shows the Fourier transforms of (U★)2 and (U)2 along the given slice, showing that modes after become noise-dominated. Finally, the bottom panel shows that after convolution with , where mx and kx are chosen with τ = 10−10 and , the clean and noisy spectra agree well, indicating successful filtering of noise-dominated modes (note that (U)2 is highly-corrupted, nonlinearly-transformed, and biased from the noise-free term (U★)2, making this agreement in spectrum nontrivial).
Appendix B. Simulation Methods
We now review the numerical methods used to simulate noise-free datasets for each of the PDEs in Table 2 (note that dimensions of the datasets are given in Table 3). Resolutions in space and time were chosen to limit computational overhead while exemplifying the dominant features of the solution. With the exception of the Navier-Stokes equations, which was simulated using the immersed boundary projection method in C + + [44], all computations were performed in MATLAB 2019b. An interesting extension for future work would be to examine the dependence of WSINDy on the resolution, similar to the work in [30].
B.1. Inviscid Burgers.
| (B.1) |
We take for exact data the shock-forming solution
| (B.2) |
which becomes discontinuous at t = α−1 with a shock travelling along (see Figure 3). We choose α = 0.5 and an extreme value of A = 1000 to demonstrate that WSINDy still has excellent performance for large amplitude data. The noise-free data consists of (B.2) evaluated at the points (xi, tj) = (−4000 + iΔx, jΔt) with Δx = 31.25 and Δt = 0.0157 for 1 ≤ i, j ≤ 256.
Figure 8.
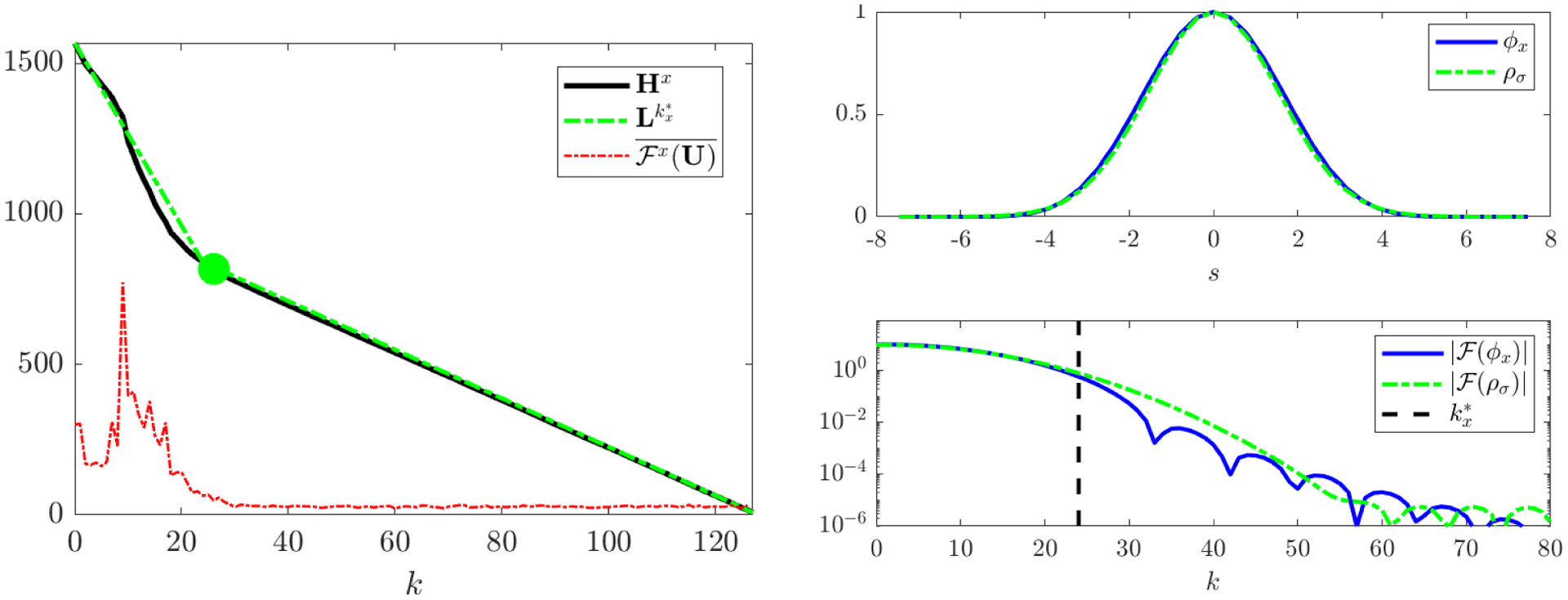
Visualization of the changepoint algorithm for KS data with 50% noise. Left: Hx (defined in (A.1)) and best two-piece approximation along with resulting changepoint . The noise-dominated region of Hx (k > 24) is approximately linear as expected from the i.i.d. noise. (The time-averaged power spectrum is overlaid and magnified for scale). Right: resulting test function ϕx and power spectrum along with reference Gaussian ρσ with . The power spectra and are in agreement over the signal-dominated modes (k ≤ 24). (Note that the power spectrum is symmetric about zero.)
B.2. Korteweg-de Vries.
| (B.3) |
A solution is obtained for (x, t) ∈ [−π, π] × [0, 0.006] with periodic boundary conditions using ETDRK4 timestepping and Fourier-spectral differentiation [17] with N1 = 400 points in space and N2 = 2400 points in time. We subsample 25% of the timepoints for system identification and keep all of the spatial points for a final resolution of Δx = 0.0157, Δt = 10−5. For initial conditions we use the two-soliton solution
B.3. Kuramoto-Sivashinsky.
| (B.4) |
A solution is obtained for (x, t) ∈ [0, 32π] × [0, 150] with periodic boundary conditions using ETDRK4 timestepping and Fourier-spectral differentiation [17] with N1 = 256 points in space and N2 = 1500 points in time. For system identification we subsample 20% of the time points for a final resolution of Δx = 0.393 and Δt = 0.5. For initial conditions we use
Figure 9.
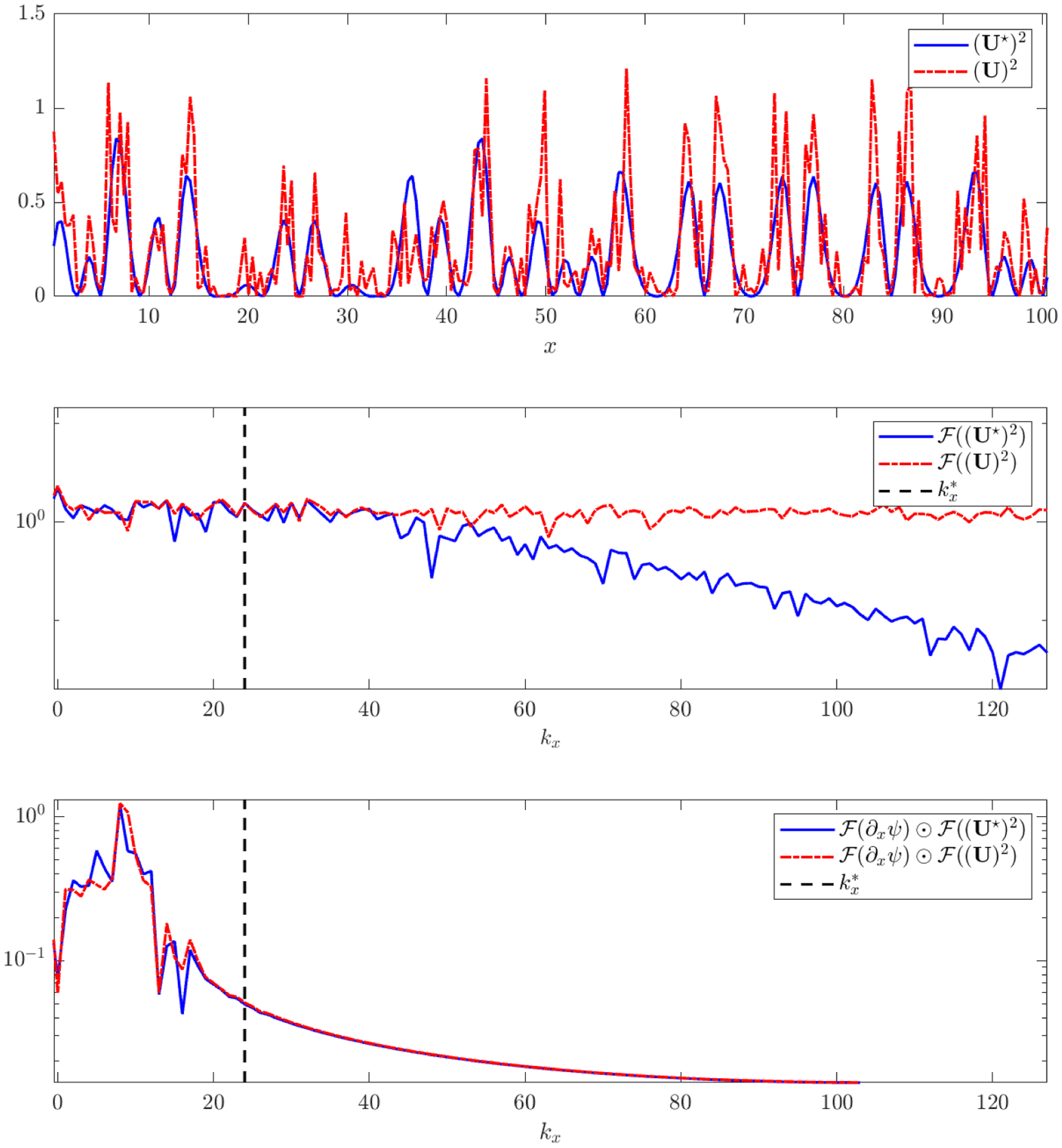
Illustration of the test function learning algorithm using computation of ∂xψ*(U2) along a slice in x at fixed time t = 99 for the same dataset used in Figure 8. From top to bottom: (i) clean U★ and noisy U variables, (ii) power spectra of the clean vs. noisy data along with the learned corner point , (iii) power spectra of the element-wise products and (recall that these computations are embedded in the FFT-based convolution (3.12)).
B.4. Nonlinear Schrödinger.
| (B.5) |
Figure 10.
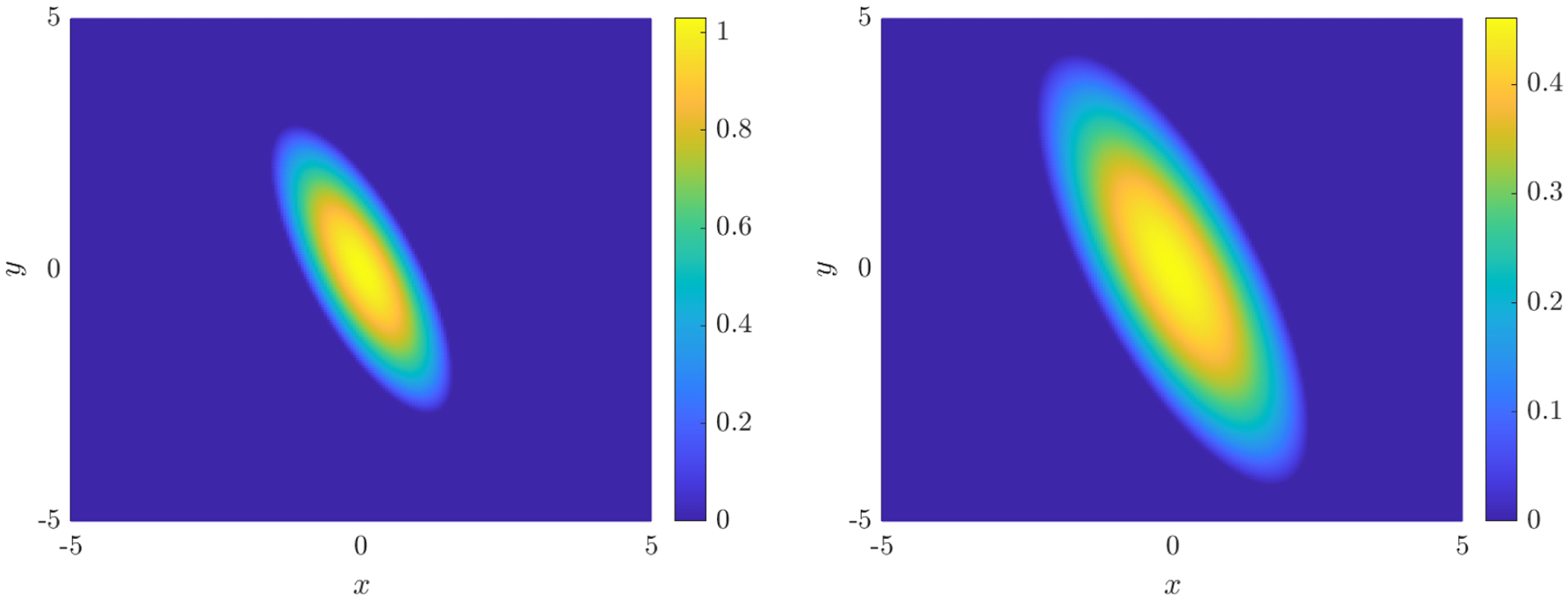
Noise-free data used for the anisotropic porous medium equation (B.7) at the initial time t = 0.5 (left) and final time t = 2.5 (right).
For the nonlinear Schrödinger equation (NLS) we reuse the same dataset from [39], containing N1 = 512 points in space and N2 = 502 timepoints, although we subsample 50% of the spatial points and 50% of the time points for a final resolution of Δx = 0.039, Δt = 0.0125. For system identification, we break the data into real and imaginary parts (w = u + iv) and recover the system
| (B.6) |
B.5. Anisotropic Porous Medium.
| (B.7) |
The equation can be rewritten
for diffusivity tensor
For noise-free data we use the analytical weak solution
where x = (x, y)T and is chosen to enforce that for all time. The solution has a finite jump in the gradient ∇u. For reference, this is the anisotropic version of the classical Barenblatt-Pattle solution to the (isotropic) porous medium equation [3, 32]. For the computation grid we use 200 points equally spaced from −5 to 5 in both x and y and 128 timepoints equally spaced from 0.5 to 2.5. The resolution is then Δx = 0.05 and Δt = 0.0157.
B.6. Sine-Gordon.
| (B.8) |
A numerical solution is obtained using a pseudospectral method on the spatial domain [−π, π] × [−1, 1] with 64 equally-spaced points in x and 64 Legendre nodes in y. Periodic boundary conditions are enforced in x and homogeneous Dirichlet boundaries in y. Geometrically, waves can be thought of as propagating on a right cylindrical sheet with clamped ends. Leapfrog time-stepping is used to generate the solution until T = 5 with Δt = 6e−5. We then subsample 0.25% of the timepoints and interpolate onto a uniform grid in space with N1 = 403 points in x and N2 = 129 points in y. The final resolution is Δx = 0.0156, Δt = 0.025. We arbitrarily use Gaussian data for the initial wave disturbance:
It is worth noting that when STLS is used instead of MSTLS (see Section 4.2) for sparsity enforcement, WSINDy returns a combination of sin(u) and terms from Taylor expansion of sin(u),
| (B.9) |
MSTLS removes this problem. Furthermore, the test function selection method in Appendix A is essential for allowing robust recovery of the Sine-Gordon equation as σ → 1 (see Figure 4).
B.7. Reaction-Diffusion.
| (B.10) |
The system (B.10) is simulated over a doubly-periodic domain (x, y) ∈ [−10, 10]×[−10, 10] with t ∈ [0, 10] using Fourier-spectral differentiation in space and method-of-lines time integration via MATLAB’s ode45 with default tolerance. The computational domain has dimensions N1 = N2 = 256 and N3 = 201, for a final resolution of Δx = 0.078, Δt = 0.0498. For initial conditions we use the spiral data
where θ(z) is the principle angle of . Note that this is an unstable spiral which breaks apart over time but still settles into a limit cycle.
Using the traditional (stable) spiral wave data [39] (differing only from the dataset used here in that the term in the initial conditions above is replaced by ) we noticed an interesting behavior in that for high noise the resulting model is purely oscillatory. In other words, the stable spiral limit cycle happens to be well-approximated by the pure-oscillatory model
| (B.11) |
with α ≈ 0.91496. A comparison between this purely oscillatory reduced model and the full model simulated from the same initial conditions is shown in Figure 11. For σNR ≤ 0.1 WSINDy applied to the stable spiral dataset returns the full model, while for σNR > 0.1 the oscillatory reduced model is more frequently detected. This suggests that although the stable spiral wave is a hallmark of the λ-ω reaction-diffusion system, from the perspective of data-driven model selection it is not an ideal candidate for identification of the full model.
Figure 11.
Comparison between the full reaction-diffusion model (B.10) (left) and the pure-oscillatory reduced model (B.11) (right) at the final time T = 10 with both models simulated from the same initial conditions leading to a spiral wave (only the v component is shown, results for u are similar). The reduced model provides a good approximation away from the boundaries.
B.8. Navier-Stokes.
| (B.12) |
A solution is obtained on a spatial grid (x, y) ⊂ [−1, 8] × [−2, 2] with a “cylinder” of diameter 1 located at (0, 0). The immersed boundary projection method [44] with 3rd-order Runge-Kutta timestepping is used to simulate the flow at spatial and temporal resolutions Δx = Δt = 0.02 for 2000 timesteps following the onset of the vortex shedding limit cycle. The dataset (U, V, W) contains the velocity components as well as the vorticity for points away from the cylinder and boundaries in the rectangle (x, y) ∈ [1, 7.5] × [−1.5, 1.5]. We subsample 10% of the data in time for a final resolution of Δx = 0.02 and Δt = 0.2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The underlying true solution need only have bounded variation and the only derivatives approximated are weak derivatives.
Here ϵ is used to denote a multi-dimensional array of i.i.d. random variables and has the same dimensions as U.
Commonly is a time derivative ∂t or ∂tt, although this is not required.
We will avoid using subscript notation such as ux to denote partial derivatives, instead using or ∂xu. For functions f(x) of one variable, f(n)(x) denotes the nth derivative of f.
For example, with , integration by parts occurs twice with respect to the x-coordinate and once with respect to y, so that and .
The technique of exploiting separability in high-dimensional integration is not new (see [33] for an early introduction) and is frequently utilized in scientific computing (see [4, 14] for examples in computational chemistry).
For the examples in Section 5 the walltimes are reported for serial computation of (G, b).
This is in contrast to explicit data-denoising, where a filter is applied to the dataset prior to system identification and may fundamentally alter the underlying clean data. The implicit filtering of the convolutional weak form is made explicit by the FFT-based implementation (3.12).
can also be seen as a scaled subset of the Bernstein polynomials, which, among other considerations, are used in the construction of B-Splines [12].
Test function asymmetry may provide an advantage in some cases, for instance along the time axis, however we do not investigate this here.
WSINDy appears not to be particularly sensitive to τ, similar results were obtained for τ = 10−6, 10−10, 10−16.
Other methods of minimizing can be used, however minimizers are not unique (there exists a set of minimizers - see Figure 5). Our approach is efficient and returns the minimizer which has the useful characterization of defining the thresholds λ that result in overfitting.
Tikhonov regularization involves solving
A common remedy for this is to scale G to have columns of unit 2-norm, however this has no connection with the underlying physics.
Note that thresholding in equation (4.6) occurs on and the terms in the bounds (4.5) become .
Here ‖U‖2′ is the 2-norm of U stretched into a column vector (and similarly for ‖·‖1′).
Note that the projection operation in (3.12) restricts the admissable set of query points to those for which ψ(xκ – x, tκ − t) is compactly supported within Ω × [0, T], which is necessary for integration by parts to be valid.
Details on the numerical methods and boundary conditions used to simulate each PDE can be found in Appendix B.
We find that 200 runs sufficiently reduces variance in the results.
2X Intel Xeon 5218 at 2.3 GHz with 22 MB cache, 16 cores per cpu, and 384 GB ram.
For Burgers, KdV, and KS we set (defined in Appendix A.2) while for NLS, PM, SG, RD, and NS we used . For KS and NLS we chose (mx, mt) values nearby that had better performance.
In the examples shown here we observed an average of 5 thresholding iterations and a maximum of 14 in any given inner MSTLS(G, b; λ) loop (i.e. for each λ ∈ λ as in equation (4.6)), hence in practice the full MSTLS(G, b; , λ) algorithm requires far fewer iterations than the theoretical maximum of #{λ}SJ.
We have not included experiments involving multiple-soliton solutions to Sine-Gordon, however the success of WSINDy applied to KdV, nonlinear Schrödinger and Sine-Gordon suggests that the class of integrable systems could be a fruitful avenue for future research.
We note that discovery of the same reaction-diffusion system from a much smaller library of terms is shown in [39, 37], but with different initial conditions that result in a spiral wave limit cycle. Our choice of initial conditions is motivated below in Appendix B.
By definition (4.7), is the minimum value in λ that minimizes the loss (4.7), hence values in λ below are precisely the thresholds that result in misidentification of the correct model by overfitting, while thresholds above necessarily underfit the model.
This is discussed further in Appendix B.7.
For example, Sine-Gordon and Navier-Stokes are both integrated in time using second-order methods, hence have lower accuracy than the other examples (see Appendix B for more details).
Results shown for σNR = 0.01 reproduced from [39] (note that PDE-FIND is unreliable at higher noise levels).
In other words, equal to the noise-free case in expectation (recall that U★ is the underlying noise-free data).
With the exception of j = 2 and odd |αs|, due to the fact that .
This is found by projecting the left-hand side b onto the column ∂x * U★ (i.e. in the noise-free case).
In the weighted least-squares sense with weights .
This also shows that with , if we take then we get pointwise convergence ϕa,p → ρ1 as p → ∞.
References
- [1].Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control, 19(6):716–723, December 1974. [Google Scholar]
- [2].Akaike Hirotugu. On entropy maximization principle. In Krishnaiah PR, editor, Applications of Statistics, pages 27–41. North Holland, Amsterdam, Netherlands, 1977. [Google Scholar]
- [3].Barenblatt GI. On some unsteady fluid and gas motions in a porous medium. Prikl. Mat. Mekh, 16(1):67–78, 1952. [Google Scholar]
- [4].Beylkin Gregory and Mohlenkamp Martin J. Algorithms for numerical analysis in high dimensions. SIAM Journal on Scientific Computing, 26(6):2133–2159, 2005. [Google Scholar]
- [5].Bortz DM and Nelson PW. Model Selection and Mixed-Effects Modeling of HIV Infection Dynamics. Bulletin of Mathematical Biology, 68(8):2005–2025, November 2006. [DOI] [PubMed] [Google Scholar]
- [6].Brunton Steven L, Proctor Joshua L, and Kutz J Nathan. Discovering governing equations from data by sparse identification of nonlinear dynamical systems. Proceedings of the national academy of sciences, 113(15):3932–3937, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Xiaoli, Daun Jinqiao, and Karniadakis George. Learning and meta-learning of stochastic advection–diffusion–reaction systems from sparse measurements. European Journal of Applied Mathematics, pages 1–24. [Google Scholar]
- [8].Cortiella Alexandre, Park Kwang-Chun, and Doostan Alireza. Sparse identification of nonlinear dynamical systems via reweighted ℓ1-regularized least squares. Computer Methods in Applied Mechanics and Engineering, 376:113620, 2021. [Google Scholar]
- [9].Crutchfield James P and McNamara Bruce S. Equations of motion from a data series. Complex systems, 1(417–452):121, 1987. [Google Scholar]
- [10].Dai Min, Gao Ting, Lu Yubin, Zheng Yayun, and Duan Jinqiao. Detecting the maximum likelihood transition path from data of stochastic dynamical systems. Chaos: An Interdisciplinary Journal of Nonlinear Science, 30(11):113124, 2020. [DOI] [PubMed] [Google Scholar]
- [11].de Silva Brian M, Champion Kathleen, Quade Markus, Loiseau Jean-Christophe, Kutz J Nathan, and Brunton Steven L. PySINDy: A python package for the sparse identification of nonlinear dynamics from data. arXiv, pages arXiv–2004, 2020. [Google Scholar]
- [12].Ershov SN. B-splines and bernstein basis polynomials. Physics of Particles and Nuclei Letters, 16(6):593–601, 2019. [Google Scholar]
- [13].Gurevich Daniel R, Reinbold Patrick AK, and Grigoriev Roman O. Robust and optimal sparse regression for nonlinear pde models. Chaos: An Interdisciplinary Journal of Nonlinear Science, 29(10):103113, 2019. [DOI] [PubMed] [Google Scholar]
- [14].Harrison Robert J, Beylkin Gregory, Bischoff Florian A, Calvin Justus A, Fann George I, Fosso-Tande Jacob, Galindo Diego, Hammond Jeff R, Hartman-Baker Rebecca, Hill Judith C, et al. MADNESS: A multiresolution, adaptive numerical environment for scientific simulation. SIAM Journal on Scientific Computing, 38(5):S123–S142, 2016. [Google Scholar]
- [15].Hoffmann Moritz, Christoph Fröhner, and Frank Noé. Reactive sindy: Discovering governing reactions from concentration data. The Journal of Chemical Physics, 150(2):025101, 2019. [DOI] [PubMed] [Google Scholar]
- [16].Sung Ha Kang Wenjing Liao, and Liu Yingjie. Ident: Identifying differential equations with numerical time evolution. arXiv preprint arXiv:1904.03538, 2019. [Google Scholar]
- [17].Kassam Aly-Khan and Trefethen Lloyd N. Fourth-order time-stepping for stiff pdes. SIAM Journal on Scientific Computing, 26(4):1214–1233, 2005. [Google Scholar]
- [18].Keck Dustin D and Bortz David M. Generalized sensitivity functions for size-structured population models. Journal of Inverse and Ill-posed Problems, 24(3):309–321, 2016. [Google Scholar]
- [19].Keller Rachael T and Du Qiang. Discovery of dynamics using linear multistep methods. SIAM Journal on Numerical Analysis, 59(1):429–455, 2021. [Google Scholar]
- [20].Killick Rebecca, Fearnhead Paul, and Eckley Idris A. Optimal detection of changepoints with a linear computational cost. Journal of the American Statistical Association, 107(500):1590–1598, 2012. [Google Scholar]
- [21].Lagergren John H, Nardini John T, Baker Ruth E, Simpson Matthew J, and Flores Kevin B. Biologically-informed neural networks guide mechanistic modeling from sparse experimental data. PLoS computational biology, 16(12):e1008462, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lagergren John H., Nardini John T., Lavigne G. Michael, Rutter Erica M., and Flores Kevin B.. Learning partial differential equations for biological transport models from noisy spatio-temporal data. Proc. R. Soc. A, 476(2234):20190800, February 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lillacci Gabriele and Khammash Mustafa. Parameter Estimation and Model Selection in Computational Biology. PLoS Comput Biol, 6(3):e1000696, March 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Long Zichao, Lu Yiping, and Dong Bin. Pde-net 2.0: Learning pdes from data with a numeric-symbolic hybrid deep network. Journal of Computational Physics, 399:108925, 2019. [Google Scholar]
- [25].Long Zichao, Lu Yiping, Ma Xianzhong, and Dong Bin. Pde-net: Learning pdes from data. In International Conference on Machine Learning, pages 3208–3216. PMLR, 2018. [Google Scholar]
- [26].Lu Yiping, Zhong Aoxiao, Li Quanzheng, and Dong Bin. Beyond finite layer neural networks: Bridging deep architectures and numerical differential equations. In International Conference on Machine Learning, pages 3276–3285. PMLR, 2018. [Google Scholar]
- [27].Niall M Mangan J Kutz Nathan, Brunton Steven L, and Proctor Joshua L. Model selection for dynamical systems via sparse regression and information criteria. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences, 473(2204):20170009, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Messenger Daniel A and Bortz David M. Weak SINDy: Galerkin-based data-driven model selection. arXiv preprint arXiv:2005.04339, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Montavon Grégoire, Samek Wojciech, and Müller Klaus-Robert. Methods for interpreting and understanding deep neural networks. Digital Signal Processing, 73:1–15, 2018. [Google Scholar]
- [30].Nardini John T, Lagergren John H, Hawkins-Daarud Andrea, Curtin Lee, Morris Bethan, Rutter Erica M, Swanson Kristin R, and Flores Kevin B. Learning equations from biological data with limited time samples. Bulletin of Mathematical Biology, 82(9):1–33, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Owhadi Houman. Bayesian numerical homogenization. Multiscale Modeling & Simulation, 13(3):812–828, 2015. [Google Scholar]
- [32].Pattle RE. Diffusion from an instantaneous point source with a concentration-dependent coefficient. The Quarterly Journal of Mechanics and Applied Mathematics, 12(4):407–409, 1959. [Google Scholar]
- [33].Pereyra Victor and Scherer G. Efficient computer manipulation of tensor products with applications to multidimensional approximation. Mathematics of Computation, 27(123):595–605, 1973. [Google Scholar]
- [34].Qin Tong, Chen Zhen, Jakeman John, and Xiu Dongbin. Data-driven learning of non-autonomous systems. arXiv preprint arXiv:2006.02392, 2020. [Google Scholar]
- [35].Qin Tong, Wu Kailiang, and Xiu Dongbin. Data driven governing equations approximation using deep neural networks. Journal of Computational Physics, 395:620–635, 2019. [Google Scholar]
- [36].Raissi Maziar, Perdikaris Paris, and Karniadakis George Em. Machine learning of linear differential equations using Gaussian processes. Journal of Computational Physics, 348:683–693, 2017. [Google Scholar]
- [37].Reinbold Patrick AK, Gurevich Daniel R, and Grigoriev Roman O. Using noisy or incomplete data to discover models of spatiotemporal dynamics. Physical Review E, 101(1):010203, 2020. [DOI] [PubMed] [Google Scholar]
- [38].Rudin Cynthia. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nature Machine Intelligence, 1(5):206–215, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rudy Samuel H, Brunton Steven L, Proctor Joshua L, and Kutz J Nathan. Data-driven discovery of partial differential equations. Science Advances, 3(4):e1602614, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Samuel H Rudy J Kutz Nathan, and Brunton Steven L. Deep learning of dynamics and signal-noise decomposition with time-stepping constraints. Journal of Computational Physics, 396:483–506, 2019. [Google Scholar]
- [41].Schaeffer Hayden. Learning partial differential equations via data discovery and sparse optimization. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences, 473(2197):20160446, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schaeffer Hayden and McCalla Scott G. Sparse model selection via integral terms. Physical Review E, 96(2):023302, 2017. [DOI] [PubMed] [Google Scholar]
- [43].Schaeffer Hayden, Tran Giang, and Ward Rachel. Extracting sparse high-dimensional dynamics from limited data. SIAM Journal on Applied Mathematics, 78(6):3279–3295, 2018. [Google Scholar]
- [44].Taira Kunihiko and T. Colonius. The immersed boundary method: A projection approach. J. Comput. Phys, 225(2):2118–2137, August 2007. [Google Scholar]
- [45].Thaler Stephan, Paehler Ludger, and Nikolaus A Adams. Sparse identification of truncation errors. Journal of Computational Physics, 397:108851, 2019. [Google Scholar]
- [46].Thomaseth Karl and Cobelli Claudio. Generalized sensitivity functions in physiological system identification. Annals of biomedical engineering, 27(5):607–616, 1999. [DOI] [PubMed] [Google Scholar]
- [47].Toms Benjamin A, Barnes Elizabeth A, and Ebert-Uphoff Imme. Physically interpretable neural networks for the geosciences: Applications to earth system variability. Journal of Advances in Modeling Earth Systems, 12(9):e2019MS002002, 2020. [Google Scholar]
- [48].Toni Tina, Welch David, Strelkowa Natalja, Ipsen Andreas, and Stumpf Michael P.H. Approximate Bayesian computation scheme for parameter inference and model selection in dynamical systems. J. R. Soc. Interface, 6(31):187–202, February 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tran Giang and Ward Rachel. Exact recovery of chaotic systems from highly corrupted data. Multiscale Modeling & Simulation, 15(3):1108–1129, 2017. [Google Scholar]
- [50].Wang Wen-Xu, Yang Rui, Lai Ying-Cheng, Kovanis Vassilios, and Grebogi Celso. Predicting catastrophes in nonlinear dynamical systems by compressive sensing. Physical review letters, 106(15):154101, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang Yating, Cheung Siu Wun, Chung Eric T, Efendiev Yalchin, and Wang Min. Deep multiscale model learning. Journal of Computational Physics, 406:109071, 2020. [Google Scholar]
- [52].Wang Z, Huan X, and Garikipati K. Variational system identification of the partial differential equations governing microstructure evolution in materials: Inference over sparse and spatially unrelated data. Computer Methods in Applied Mechanics and Engineering, 377:113706, 2021. [Google Scholar]
- [53].Wang Zhenlin, Huan Xun, and Garikipati Krishna. Variational system identification of the partial differential equations governing the physics of pattern-formation: inference under varying fidelity and noise. Computer Methods in Applied Mechanics and Engineering, 356:44–74, 2019. [Google Scholar]
- [54].Wang Zhenlin, Wu Bowei, Garikipati Krishna, and Huan Xun. A perspective on regression and bayesian approaches for system identification of pattern formation dynamics. Theoretical and Applied Mechanics Letters, 10(3):188–194, 2020. [Google Scholar]
- [55].Warne David J., Baker Ruth E., and Simpson Matthew J.. Using Experimental Data and Information Criteria to Guide Model Selection for Reaction–Diffusion Problems in Mathematical Biology. Bull Math Biol, 81(6):1760–1804, June 2019. [DOI] [PubMed] [Google Scholar]
- [56].Wu Hulin and Wu Lang. Identification of significant host factors for HIV dynamics modelled by non-linear mixed-effects models. Statist. Med, 21(5):753–771, March 2002. [DOI] [PubMed] [Google Scholar]
- [57].Wu Kailiang and Xiu Dongbin. Numerical aspects for approximating governing equations using data. Journal of Computational Physics, 384:200–221, 2019. [Google Scholar]
- [58].Wu Kailiang and Xiu Dongbin. Data-driven deep learning of partial differential equations in modal space. Journal of Computational Physics, 408:109307, 2020. [Google Scholar]
- [59].Xu Hao, Chang Haibin, and Zhang Dongxiao. Dlga-pde: Discovery of pdes with incomplete candidate library via combination of deep learning and genetic algorithm. Journal of Computational Physics, page 109584, 2020. [Google Scholar]
- [60].Zhang Linan and Schaeffer Hayden. On the convergence of the SINDy algorithm. Multiscale Modeling & Simulation, 17(3):948–972, 2019. [Google Scholar]
- [61].Zhang Sheng and Lin Guang. Robust data-driven discovery of governing physical laws with error bars. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences, 474(2217):20180305, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhang Sheng and Lin Guang. Robust subsampling-based sparse Bayesian inference to tackle four challenges (large noise, outliers, data integration, and extrapolation) in the discovery of physical laws from. arXiv preprint arXiv:1907.07788, 2019. [Google Scholar]


