ABSTRACT
Francisella tularensis is the causative agent of tularemia. Because of its extreme infectivity and high mortality rate, this pathogen was classified as a biothreat agent. Francisella spp. are strict aerobes, and ubiquinone (UQ) has been previously identified in these bacteria. While the UQ biosynthetic pathways were extensively studied in Escherichia coli, allowing the identification of 15 Ubi proteins to date, little is known about Francisella spp. In this study, and using Francisella novicida as a surrogate organism, we first identified ubiquinone 8 (UQ8) as the major quinone found in the membranes of this bacterium. Next, we characterized the UQ biosynthetic pathway in F. novicida using a combination of bioinformatics, genetics, and biochemical approaches. Our analysis disclosed the presence in Francisella of 10 putative Ubi proteins, and we confirmed 8 of them by heterologous complementation in E. coli. The UQ biosynthetic pathways from F. novicida and E. coli share similar patterns. However, differences were highlighted: the decarboxylase remains unidentified in Francisella spp., and homologs of the Ubi proteins involved in the O2-independent UQ pathway are not present. This is in agreement with the strictly aerobic niche of this bacterium. Next, via two approaches, i.e., the use of an inhibitor (3-amino-4-hydroxybenzoic acid) and a transposon mutant, both of which strongly impair the synthesis of UQ, we demonstrated that UQ is essential for the growth of F. novicida in respiratory medium and contributes to its pathogenicity in Galleria mellonella used as an alternative animal model.
IMPORTANCE Francisella tularensis is the causative bacterium of tularemia and is classified as a biothreat agent. Using multidisciplinary approaches, we investigated the ubiquinone (UQ) biosynthetic pathway that operates in F. novicida used as a surrogate. We show that UQ8 is the major quinone identified in the membranes of Francisella novicida. We identified a new competitive inhibitor that strongly decreased the biosynthesis of UQ. Our demonstration of the crucial roles of UQ for the respiratory metabolism of F. novicida and for the involvement in its pathogenicity in the Galleria mellonella model should stimulate the search for selective inhibitors of bacterial UQ biosynthesis.
KEYWORDS: ubiquinone biosynthesis, coenzyme Q, quinone, aerobic respiration, Francisella tularensis, Francisella novicida
INTRODUCTION
Francisella tularensis is a Gram-negative, strictly aerobic, facultative intracellular pathogen responsible for tularemia. Infection can occur by inhalation, ingestion, transmission from arthropod vectors, or exposure to infected animals (1). After its entry into macrophages, the bacteria are sequestered into phagosomes and prevent further endosomal maturation. Francisella cells then disrupt the phagosome and are released into the cytosol, in which they rapidly proliferate (2). Eventually, the infected cells undergo apoptosis or pyroptosis, and the progeny bacteria are released to initiate new rounds of infection (2). Currently, there is no suitable vaccine against tularemia, and due to its extreme infectivity and high virulence, the species F. tularensis has been classified as a biothreat agent (3). The genus Francisella includes three species: F. tularensis, F. novicida, and F. philomiragia (4). Moreover, F. tularensis is further divided into F. tularensis subsp. tularensis (type A strains) and F. tularensis subsp. holarctica (type B strains), which are the most virulent strains responsible for human disease, whereas F. philomiragia and F. novicida are avirulent in healthy humans (4). F. novicida type strain U112 is commonly used as a surrogate for Francisella tularensis in virulence studies using animal models (5).
The development of genome-scale genetic methods allowed the identification of hundreds of genes participating to various extents in Francisella virulence (6). However, the specific contribution of only a limited number of these genes was demonstrated at the molecular level. Although an important proportion of the identified genes are related to metabolic functions, the relationship between metabolism and the life cycle of Francisella is still poorly understood. However, global analysis of genes essential for the growth in culture of F. novicida U112 (7) and, more recently, that of F. tularensis subsp. tularensis Schu S4 (8) highlighted the involvement of several ubiquitous pathways found in proteobacteria. Among the most significant are the folate pathway, the heme synthesis pathway, the methylerythritol phosphate pathway involved in isoprenoid synthesis, the chorismate pathway, and the ubiquinone (UQ) synthesis pathway, on which this work is focused.
Isoprenoid quinones are conserved in most respiratory and photosynthetic organisms and function primarily as electron and proton carriers in the electron transfer chains. Quinones are composed of a polar redox-active head group linked to a lipid side chain, which varies in both length and the degree of saturation (9). Proteobacteria contain two main types of quinone, i.e., benzoquinones and naphthoquinones, represented by UQ (or coenzyme Q) and menaquinone (MK)/demethylmenaquinone (DMK), respectively (9). UQ is the major electron carrier used for the reduction of dioxygen by various cytochrome oxidases, whereas MK and DMK function predominantly in anaerobic respiratory chains (9). However, as demonstrated recently in Pseudomonas aeruginosa, UQ can also be produced and used as a main respiratory quinone under anaerobic conditions (10). Besides its role in bioenergetics, UQ was also reported to be involved in gene regulation, oxidative stress, virulence, and resistance to antibiotics (11, 12). More recently, new functions for UQ in bacteria were discovered, such as its requirement for Escherichia coli to grow on medium containing long-chain fatty acids as a carbon source (13). UQ biosynthesis under aerobic conditions has been widely studied in E. coli (14). The classical UQ biosynthetic pathway requires 12 proteins (UbiA to UbiK and UbiX). UbiC catalyzes the first committed step in the biosynthesis of UQ, the conversion of chorismate to the 4-hydroxybenzoate (4HB) precursor. Next, UbiA, UbiD to UbiI, and UbiX catalyze the prenylation, decarboxylation, hydroxylations, and methylations of the phenyl ring of 4HB to synthesize UQ. In addition, UbiB and UbiK are accessory proteins, while UbiJ is involved in the assembly and/or the stability of the aerobic Ubi complex, which was recently characterized in E. coli (15). The latter is also able to synthesize UQ under anoxic conditions, and we identified three proteins, UbiU, UbiV, and UbiT, that are required for UQ biosynthesis only under anoxic conditions (16).
Here, we show that ubiquinone 8 (UQ8) is the major quinone of F. novicida U112. We identified candidate Ubi proteins in F. novicida U112 and validated their functions by heterologous complementation in E. coli mutant strains. Our results show that UQ biosynthesis in Francisella spp. is mostly similar to that of E. coli, with the notable absence of UbiX and UbiD for the decarboxylation step. Genetic and chemical inactivation of UQ biosynthesis thanks to a transposon (Tn) mutant and a new inhibitor (3-amino-4-hydroxybenzoic acid), respectively, demonstrated that UQ8 is crucial for the growth of F. novicida in respiratory medium and that UQ deficiency impairs the pathogenicity of F. novicida against Galleria mellonella. Altogether, our results shed light on the role of UQ in the life cycle of Francisella and show that UQ contributes to its pathogenicity.
RESULTS
UQ8 is the major quinone of F. novicida.
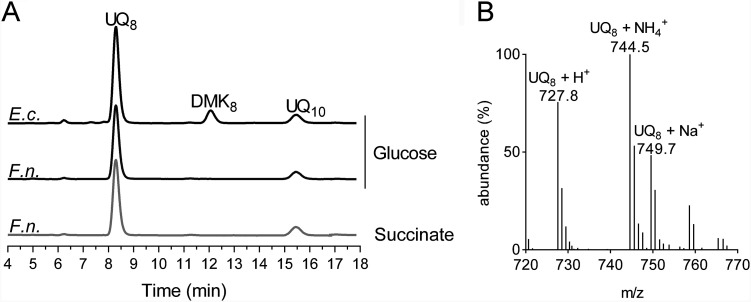
The quinone content of F. novicida grown under ambient air at 37°C in Chamberlain medium supplemented with either glucose (fermentative medium) or succinate (respiratory medium) as the only carbon source was determined and compared with that of E. coli MG1655 grown in the same fermentative medium. In the electrochromatograms of lipid extracts from F. novicida, a single peak was observed at around 8.3 min, the same retention time as that of UQ8 in E. coli extracts (Fig. 1A). Note that in these analyses, UQ10 was used as an internal standard, which was added to the samples. Mass spectrometry (MS) analysis of the major peak in F. novicida extracts showed a predominant ammonium adduct (M+ NH4+) at m/z 744.5, together with minor adducts, such as Na+ (m/z 749.7) and H+ (m/z 727.8) (Fig. 1B). These masses identify UQ8 (monoisotopic mass, 726.5) as the major quinone produced by F. novicida. Interestingly, the carbon source in the culture medium did not greatly affect the UQ8 content (Fig. 1A). The F. novicida extracts did not contain any naphthoquinones, unlike E. coli, which showed predominantly demethylmenaquinone (DMK8) eluting at around 12 min. The absence of detectable levels of naphthoquinones in F. novicida lipid extracts (Fig. 1A) is in agreement with the absence of menaquinone biosynthesis (Men or futalosine)-encoding genes in its genome. Together, our results establish that E. coli and F. novicida share UQ8 as a main quinone under aerobic conditions.
FIG 1.
UQ8 is the major quinone used by F. novicida. (A) HPLC-ECD analysis of lipid extracts from 1 mg of E. coli MG1655 (E.c.) and F. novicida (F.n.) cells grown aerobically in Chamberlain medium with 0.4% (wt/vol) glucose or succinate as the sole carbon source. The chromatograms are representative of results from three independent experiments. The peaks corresponding to UQ8, DMK8, and the UQ10 standard are indicated. (B) Mass spectrum of the quinone eluting at 8.30 min from extracts of F. novicida grown in Chamberlain medium. H+, NH4+, and Na+ adducts of UQ8 are indicated.
Identification of Ubi proteins in Francisella spp.
To identify candidate Ubi proteins in F. novicida, UbiX and UbiA to UbiK from E. coli MG1655 were screened for homologs in the protein sequence data set, available at MicroScope (www.genoscope.cns.fr/agc/microscope), using BLASTP software. As listed in Table S1 in the supplemental material, this analysis identified eight homologous proteins in F. novicida, i.e., UbiA to UbiC, UbiE, UbiG to UbiI, and UbiK, called UbiAFn to UbiCFn, UbiEFn, UbiGFn to UbiIFn, and UbiKFn, respectively, here. Genes ubiAFn and ubiCFn on the one hand and genes ubiIFn and ubiHFn on the other hand present organizations similar to those of the ubiC-ubiA and ubiH-ubiI operons from E. coli, respectively (12). As reported previously for Pseudomonas aeruginosa (17) and Xanthomonas campestris (18), F. novicida possesses a Coq7 hydroxylase, which is a functional homolog of the UbiF protein found in E. coli and other species (19). The detection of a homolog for E. coli UbiJ required less restrictive BLAST parameters. We noticed that the gene coding for the putative UbiJ candidate FTN_0460, called ubiJFn here, lies between ubiEFn and ubiBFn, an organization similar to that of the ubiE-ubiJ-ubiB operon from E. coli (20). UbiJFn has 21% amino acid identity with UbiJ from E. coli, and both proteins contain a sterol carrier protein 2 domain in their N-terminal regions (http://pfam.xfam.org/) (21). The same Ubi proteins were identified in the highly virulent strain F. tularensis subsp. tularensis Schu S4 (Table S1).
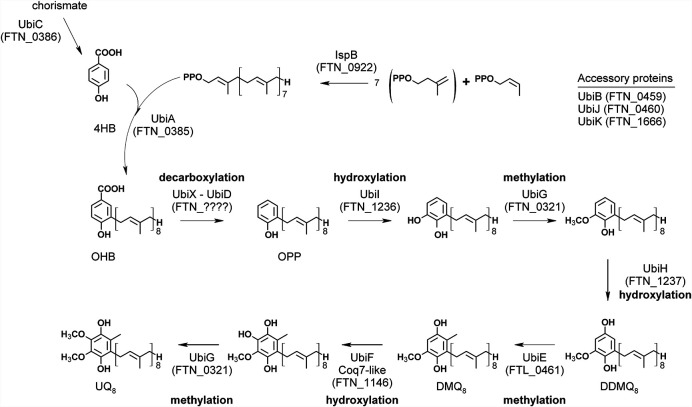
Homologs of UbiD and UbiX were not yet identified, and the counterparts of these two proteins in Francisella spp. remain to be determined. Work is in progress in our laboratory. Under anaerobic conditions, E. coli still synthesizes UQ, and we recently identified three genes, which we called ubiT, ubiU, and ubiV, as being essential for this process (16). Homologs of ubiT, ubiU, and ubiV, which participate in the O2-independent UQ biosynthetic pathway, were not identified in the screened Francisella genomes (Table S1), in agreement with the strictly aerobic metabolism of Francisella spp. Overall, these data show that the O2-dependent UQ biosynthetic pathways in F. novicida, F. tularensis, and E. coli are related, with the major difference being the absence of UbiX-UbiD for the decarboxylation step (Fig. 2).
FIG 2.
Proposed UQ8 biosynthetic pathway in F. novicida deduced from the one characterized in E. coli. The corresponding protein identifiers in F. novicida are indicated in parentheses. There are no identified counterparts of UbiD and UbiX in the F. novicida proteome. UbiF is identified only in E. coli, and its functional homolog in F. novicida is a Coq7 hydroxylase. Abbreviations: 4HB, 4-hydroxybenzoic acid; OHB, 3-octaprenyl-4-hydroxybenzoic acid; OPP, octaprenylphenol; DMQ8, C6-demethoxy-ubiquinone 8; DDMQ8, C1-demethyl-C6-demethoxy-ubiquinone 8; UQ8, ubiquinone 8.
Functional characterization of UbiFn proteins in E. coli.
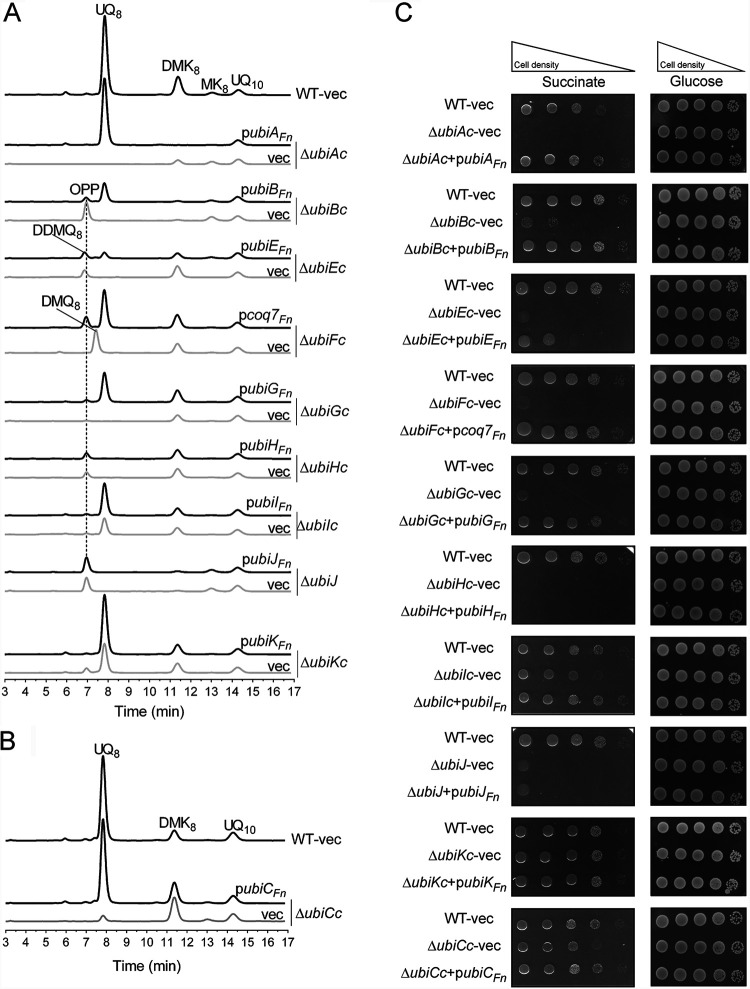
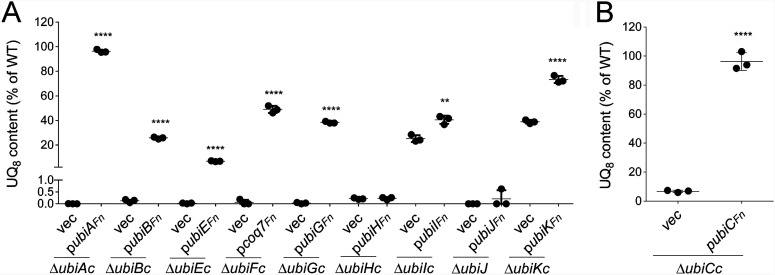
To test whether the candidate Ubi proteins identified in F. novicida were indeed involved in UQ biosynthesis, we assessed their capacity to functionally complement E. coli strains in which the UQ protein-encoding genes were inactivated (ΔubiAc, ΔubiBc, ΔubiCc, ΔubiEc, ΔubiFc, ΔubiGc, ΔubiHc, ΔubiIc, ΔubiJ, and ΔubiKc) (Table S2). We assessed the quinone content and the capacity to grow on solid minimal medium containing fermentable (glucose) or respiratory (succinate) carbon sources. E. coli ΔubiAc, ΔubiBc, ΔubiGc, ΔubiHc, and ΔubiJ transformed with an empty vector are unable to synthesize UQ8 (Fig. 3A) and are thus unable to grow on respiratory medium (Fig. 3C). In contrast, their growth on fermentative medium is not affected (Fig. 3C). Except for the ΔubiAc mutant strain, in which the prenylation reaction of 4HB is impaired, most mutants accumulate an early intermediate corresponding to octaprenylphenol (OPP) (Fig. 2 and Fig. 3A). E. coli ΔubiEc and ΔubiFc cells accumulate C2-demethyl-C6-demethoxy-UQ8 (DDMQ8) and C6-demethoxy-UQ8 (DMQ8), which are the substrates of UbiE and UbiF, respectively (Fig. 2 and Fig. 3A). We found that UbiAFn, UbiBFn, UbiEFn, Coq7Fn, and UbiGFn restored the growth of E. coli ΔubiAc, ΔubiBc, ΔubiEc, ΔubiFc, and ΔubiGc cells on respiratory medium (Fig. 3C) and allowed UQ8 biosynthesis in lysogeny broth (LB) medium to 96, 26, 7, 49, and 38% of the level of UQ8 present in wild-type (WT) cells, respectively (Fig. 3A and Fig. 4A). Concomitantly, the OPP content decreased, and Coq7Fn abolished the accumulation of DMQ8 in ΔubiFc cells (Fig. 3A). As we previously reported, E. coli ΔubiIc and ΔubiKc cells displayed a strong decrease in UQ8 (22, 23), but the residual UQ8 content was sufficient to support growth on succinate (Fig. 3A and C). Similar results were obtained with ΔubiCc cells grown in minimal M9 medium (Fig. 3B and C), which had to be used instead of LB since the latter contains 4HB that restores normal UQ8 content in ΔubiCc cells (data not shown). In all three strains, the expression of the corresponding Ubi proteins, UbiCFn, UbiIFn, and UbiKFn, increased the UQ8 content significantly (Fig. 4A and B). Since the increase obtained in ΔubiIc cells was moderate (from 25 to 40%), we further confirmed the ability of UbiIFn to catalyze C5-hydroxylation by using an E. coli ΔubiIc ΔubiF strain. This deletion mutant lacks C5- and C6-hydroxylation activities and consequently accumulates 3-octaprenyl-4-hydroxyphenol (4HP8) (22). We found that UbiIFn was able to restore DMQ8 biosynthesis in E. coli ΔubiIc ΔubiF cells (Fig. S1), i.e., to catalyze C5-hydroxylation, concomitantly with a strong decrease in 4HP8. Taken together, all these results confirm unambiguously that UbiAFn, UbiBFn, UbiCFn, Coq7Fn, UbiEFn, UbiGFn, UbiIFn, and UbiKFn are the functional counterparts of the E. coli Ubi proteins, and we propose that they compose the biosynthetic pathway of UQ8 in F. novicida. Only two proteins, UbiJFn and UbiHFn, did not complement E. coli ΔubiJ and ΔubiHc (Fig. 3A and C and Fig. 4A). The low percentage of identity between UbiJ and UbiH from E. coli and their homologs in F. novicida (21 and 27%, respectively) could explain these results (Table S1).
FIG 3.
Complementation analysis of E. coli UQ8 biosynthesis mutants with the putative Ubi proteins from F. novicida. (A and B) The Δubi E. coli mutant strains transformed with pTrc99a (vector [vec]) or pTrc99a encompassing the ubiFn genes were grown overnight at 37°C in LB medium (A) or M9 minimal medium (B) with 0.4% (wt/vol) glucose as the sole carbon source. The expression of the UbiFn proteins was induced by the addition of IPTG to a final concentration of 100 μM. E. coli wild-type (WT) strain MG1655 transformed with the pTrc99a empty vector was used as a control. HPLC-ECD analysis of lipid extracts from 1 mg of cells was performed. The chromatograms are representative of results from three independent experiments. The peaks corresponding to OPP, DDMQ8, DMQ8, UQ8, MK8, DMK8, and the UQ10 standard are indicated. (C) Serial dilutions were spotted onto plates containing M9 minimal medium with 0.4% (wt/vol) glucose or succinate as the sole carbon source and IPTG (100 μM final concentration). The plates were incubated overnight at 37°C.
FIG 4.
Quantification of cellular UQ8 contents of Δubi E. coli mutant strains expressing the UbiFn proteins. The Δubi E. coli mutant strains transformed with pTrc99a (vec) or pTrc99a encompassing the ubiFn genes were grown overnight at 37°C in LB medium (A) or M9 minimal medium (B) with 0.4% (wt/vol) glucose as the sole carbon source. Expression of the UbiFn proteins is described in the legend of Fig. 3. Quantifications are expressed as percentages of the control value, which corresponds to the UQ8 content of the wild-type strain (n = 3). ****, P < 0.0001; **, P < 0.005 (by unpaired Student’s t test).
UQ8 biosynthesis is essential for the growth of F. novicida in respiratory medium.
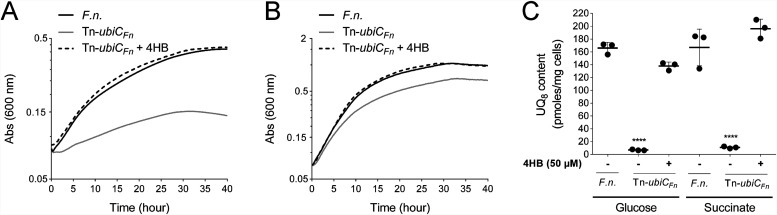
To evaluate the physiological importance of UQ for F. novicida, we screened ubi genes in the F. novicida transposon (Tn) mutant library available at the Manoil Laboratory (7). Only the Tn mutant of ubiCFn (called Tn-ubiCFn here) was available in the library, and we compared this mutant strain to its isogenic control strain U112 (Table S2). Recall that UbiC catalyzes the first committed step in the biosynthesis of UQ, i.e., the conversion of chorismate to 4HB (Fig. 2). First, we showed that the growth of Tn-ubiCFn cells under ambient air in respiratory Chamberlain medium was severely impaired compared to the WT (Fig. 5A). In contrast, the growth of F. novicida in fermentative medium was less affected (Fig. 5B). In parallel, the UQ8 content was strongly lowered in Tn-ubiCFn cells from 166 to 7 pmol/mg cells in fermentative medium and from 134 to 11 pmol/mg cells in respiratory medium (Fig. 5C). As expected, the addition of 4HB to the culture rescued the growth of Tn-ubiCFn cells in respiratory medium and increased the UQ8 content to WT levels (Fig. 5A and C). Taken together, these results show the overall requirement of UQ8 for the growth of F. novicida, especially in respiratory medium.
FIG 5.
UQ8 is essential for the growth of F. novicida in respiratory medium. (A and B) F. novicida (F.n.) and a transposon mutant of ubiCFn (Tn-ubiCFn) were grown aerobically in Chamberlain medium with 0.4% (wt/vol) succinate (A) or glucose (B) as the sole carbon source. Growth (average from sextuplicate growth curves) was monitored as the change in the absorbance at 600 nm in a Tecan plate reader. (C) Cellular UQ8 contents were quantified for F. novicida and the Tn-ubiCFn mutant according to methods described in Materials and Methods. 4HB was added to rescue the growth and UQ8 biosynthesis of the Tn-ubiCFn mutant. Quantifications are expressed as picomoles per milligram of cells (n = 3). ****, P < 0.0001 (by unpaired Student’s t test).
3A4HB inhibits UQ8 biosynthesis and impairs the growth of F. novicida in respiratory medium.
Besides genetic inactivation of the UQ pathway, we were interested in the possibility of decreasing UQ levels by chemical inhibition. Since we had found UQ to be particularly important for the growth of F. novicida in respiratory medium (Fig. 5A), we screened for compounds that could inhibit growth in such medium. We tested several compounds: 3-amino-4-hydroxybenzoic acid (3A4HB), 4-amino-benzoic acid (pABA), 4-amino-2-methoxy-benzoic acid (pA2MBA), and 4-amino-3-methoxy-benzoic acid (pA3MBA). All these molecules are analogs of 4HB, the native precursor of UQ (Fig. S2A). We observed that bacterial growth was slightly affected in respiratory medium in the presence of pABA and pA2MBA, while pA3MBA inhibited growth in both fermentative and respiratory media (Fig. S2B and C). Interestingly, 3A4HB strongly impaired bacterial growth in respiratory medium, while inhibition was milder in fermentative medium (Fig. S2B and C). Based on these results, we followed up on this compound.
We then examined how and to what extent 3A4HB could affect UQ8 biosynthesis in F. novicida. Bacteria were cultured under ambient air in fermentative Chamberlain medium supplemented with 3A4HB (from 10 μM to 1 mM, final concentration). The endogenous UQ8 content was measured in bacterial cells and compared to control conditions in which only dimethyl sulfoxide (DMSO) was added. Figure 6A shows that the UQ8 content decreased with increasing concentrations of 3A4HB in the medium, with 0.5 mM yielding an ∼90% decrease in the UQ8 content. Concomitantly, we confirmed that the growth of F. novicida in the presence of 1 mM 3A4HB was strongly impaired in respiratory medium (Fig. 6B) but less so in fermentative medium (Fig. 6C). Control experiments showed that the addition of 4HB to the growth medium counteracted the negative effect of 3A4HB, in terms of both UQ8 biosynthesis and bacterial growth (Fig. 6A to C).
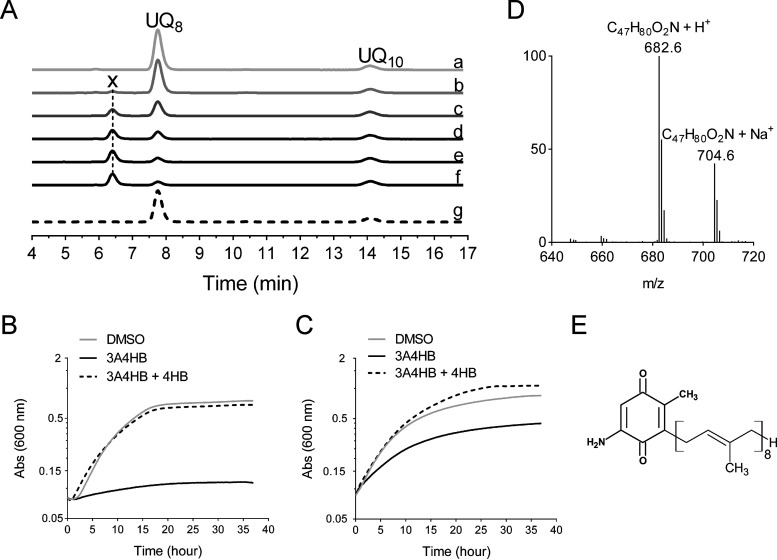
FIG 6.
Effect of 3A4HB on UQ8 biosynthesis and growth of F. novicida. (A) HPLC-ECD analysis of lipid extracts from 1 mg of F. novicida cells grown aerobically in Chamberlain medium with 0.4% (wt/vol) glucose as the sole carbon source and in the presence of different concentrations of 3A4HB solubilized in DMSO (a, DMSO; b, 0.01 mM; c, 0.1 mM; d, 0.25 mM; e, 0.5 mM; f, 1 mM; g, 1 mM 3A4HB plus 1 mM 4HB). The chromatograms are representative of results from three independent experiments. The peaks corresponding to UQ8 and the UQ10 standard are indicated. Compound X eluting at 6.5 min is marked. (B and C) Growth curves for F. novicida cultured under aerobic conditions in Chamberlain medium with 0.4% (wt/vol) succinate (B) or glucose (C) as the sole carbon source and in the presence of either DMSO (control), 1 mM 3A4HB, or 1 mM 3A4HB plus 100 μM 4HB. The growth under each condition (average from sextuplicate growth curves) was monitored as the change in the absorbance at 600 nm in a Tecan plate reader. (D) Mass spectrum of compound X eluting from extracts of F. novicida grown in the Chamberlain medium with 1 mM 3A4HB. H+ and Na+ adducts corresponding to this molecule are indicated. (E) Proposed structure of compound X in its oxidized form.
Treatment with 3A4HB caused the accumulation of a redox compound that eluted at 6.5 min (compound X in Fig. 6A). MS analysis of this peak showed a predominant proton adduct (M+ H+) at m/z 682.6, together with a minor sodium adduct (M+ Na+) at m/z 704.6 (Fig. 6D). Both species are compatible with a monoisotopic mass of 681.7 g · mol−1, which could correspond to that of 2-octaprenyl-3-methyl-6-amino-1,4-benzoquinone (Fig. 6E). According to the sequence of reactions proposed in Fig. 2, the formation of compound X would result from the prenylation of 3A4HB, decarboxylation and hydroxylation at C-1, and then methylation at C-3. Thus, 3-octaprenyl-2-methyl-5-amino-1,4-benzoquinone seems to be the “dead-end” product of the UQ8 pathway in F. novicida cells treated with 3A4HB. Collectively, these results demonstrate unequivocally that 3A4HB acts as a competitive inhibitor of UQ8 biosynthesis and affects particularly the respiratory metabolism of F. novicida.
UQ8 is involved in the pathogenesis of F. novicida in the later steps of infection.
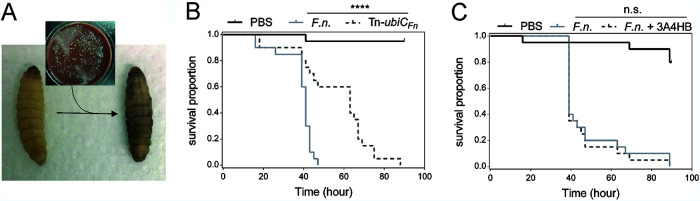
We evaluated the importance of UQ in the pathogenicity of F. novicida by studying the Tn-ubiCFn mutant. To assess the overall virulence of the Tn-ubiCFn strain in a whole organism, we used the wax moth (G. mellonella) infection model, which was previously used in studies of human-pathogenic and closely related opportunistic and nonpathogenic Francisella spp. such as F. novicida (24–27). We monitored the survival of larvae infected with the Tn-ubiCFn strain or with the isogenic strain U112 as a control. When the larvae are turning gray/black and no movement of the larval legs can be observed, they are considered dead (Fig. 7A). The Tn-ubiCFn strain was found to be statistically much less virulent than the wild type but was nevertheless still capable of killing Galleria larvae (Fig. 7B). This result suggests that UQ8 is involved in the virulence potential of F. novicida in G. mellonella. To better understand the role of UQ8 in different stages of infection in G. mellonella, the pathogenicity of the isogenic control strain pretreated with 1 mM 3A4HB was studied in order to mimic acute UQ8 deficiency. Recall that this treatment causes an ∼90% decrease in the UQ8 content (Fig. 6A), but the inhibition should be alleviated over the infection cycle in larvae where 3A4HB is not present. Pretreatment with 3A4HB has no effect on the capacity of F. novicida to kill Galleria larvae (Fig. 7C), suggesting that UQ8 does not contribute to the virulence of Francisella novicida in the early steps of infection but more likely contributes in later ones. This result is in contrast to that obtained with the Tn-ubiCFn strain, which represents a chronic deficiency of UQ8.
FIG 7.
UQ8 contributes to later steps of infection of G. mellonella by F. novicida. (A) The larvae turn gray/black when infected. (B) Survival curve of G. mellonella infected with either F. novicida (F.n.) or the transposon mutant of ubiCFn (Tn-ubiCFn). ****, P < 0.0001 (by a log rank [Mantel-Cox] test). (C) Survival curve of G. mellonella infected with F. novicida pretreated or not with 3A4HB (1 mM final concentration). Each group of G. mellonella larvae (n = 20) was injected with ∼106 CFU/larva, and PBS injection was used as a control. n.s. (not significant), P = 0.6670 (by a log rank [Mantel-Cox] test).
DISCUSSION
The chemical analysis performed in this paper established that UQ8 is the major isoprenoid quinone synthesized by F. novicida. In two representative Francisella genomes, we identified homologs for 9 of the 12 genes that are currently known to contribute to UQ biosynthesis in E. coli under aerobic conditions. We confirmed the function of seven of the nine homologs by heterologous complementation of E. coli Δubi mutants. From these results, we show again that E. coli is a good model to study the function of most exogenous ubi genes (19). We could not confirm the function of UbiHFn and UbiJFn, but the fact that ubiHFn and ubiJFn show the same genetic organization as that in E. coli (a ubiI-ubiH operon and a ubiE-ubiJ-ubiB operon) strongly supports the implication of these genes in the UQ biosynthetic pathway. Interestingly, both proteins are part of the Ubi complex in E. coli (15). We hypothesize that the low identity of UbiHFn and UbiJFn with their E. coli homologs (∼25%) might impair their assembly within the E. coli Ubi complex and thus compromise our in vivo complementation assays. Another possibility relates to the proposed implication in UQ biosynthesis of a noncoding RNA partially overlapping the open reading frame (ORF) of UbiJ from E. coli (28). We note that the expression of UbiJ from X. campestris was also unable to complement an E. coli ΔubiJ strain (18). Francisella spp. share with P. aeruginosa and X. campestris a yeast Coq7 protein homolog, which catalyzes C6-hydroxylation as for UbiF from E. coli (17, 18, 29). As we demonstrated previously, the Coq7 proteins are found in all three subclasses, alpha-, beta-, and gammaproteobacteria. In contrast, homologs of UbiF proteins are limited to the gammaproteobacteria (19). Our analysis also disclosed the presence in Francisella spp. of UbiI- and UbiH-homologous proteins, which catalyze C5- and C1-hydroxylation in E. coli, respectively (22, 30). Consequently, we propose that both E. coli and Francisella spp. share a UQ biosynthetic pathway involving three hydroxylases, i.e., UbiI, UbiH, and UbiF in E. coli and UbiI, UbiH, and Coq7 in Francisella spp. Several studies highlighted that the enzymes involved in multiple steps of the UQ biosynthetic pathway vary between bacterial species (14), like for the hydroxylation steps (19) or for the production of 4HB from chorismate by UbiC or XanB2 proteins (31). The decarboxylation step involves UbiD and UbiX in E. coli (32), but we could not identify homologs in Francisella genomes. A candidate gene, ubiZ, was proposed based on its colocalization with ubiE and ubiB in the genomes of Acinetobacter spp. and Psychrobacter sp. strain PRwf-1, which are also devoid of homologs of UbiD and UbiX (33). However, ubiZ was not confirmed functionally, and this gene is not conserved in Francisella genomes. We demonstrated that UbiI proteins from F. novicida and E. coli shared the same function, i.e., the catalysis of the first hydroxylation of the OPP, which is the product of the decarboxylation step in E. coli (Fig. 2). Consequently, we propose that the decarboxylation step occurring in F. novicida also precedes the first hydroxylation of the OPP. Collectively, these data suggest the existence of another decarboxylation system operating in UQ biosynthesis in Francisella spp. and potentially other bacteria lacking ubiX and ubiD (34).
To assess the essentiality of the UQ biosynthetic pathway in the respiratory metabolism of F. novicida, two different approaches were carried out. First, we showed that a transposon mutation of the ubiCFn gene, which decreases 4HB synthesis, impaired the growth of F. novicida mainly in respiratory medium. Interestingly, among all the ubi genes identified in Francisella genomes, only ubiC was mutated in large-scale studies (7, 8). This supports that the other ubi genes are essential for the viability of Francisella spp. and strengthens the idea that UQ is key for the development of these bacteria. We noted that the mutation of the ubiC gene affects F. novicida more severely than E. coli for growth in respiratory medium despite both mutants producing comparable amounts of UQ (∼7 to 8% compared to the WT) (Fig. 4B and Fig. 5C). As E. coli synthesizes naphthoquinones but F. novicida does not, we propose that the milder phenotype of the E. coli ubiC mutant results from naphthoquinones participating in aerobic respiration, as previously suggested (35). Second, we tested the effect of structural analogs of 4HB, and we showed that 3A4HB impaired the growth of F. novicida mainly in respiratory medium, in agreement with a strong decrease in UQ8 biosynthesis. We demonstrated that 3A4HB competes with endogenous 4HB and progresses through several steps of the UQ biosynthetic pathway to form the redox compound X that we propose to be 3-octaprenyl-2-methyl-5-amino-1,4-benzoquinone. As both the Tn-ubiCFn strain and the control strain U112 treated with 1 mM 3A4HB yielded an ∼90% decrease in the UQ8 content and presented a strong impairment of growth in respiratory medium (Fig. 5 and 6), we propose that compound X would not be used as a quinone in the respiratory chain of F. novicida.
We noted that homologs of ubiT, ubiU, and ubiV, which belong to the O2-independent UQ biosynthetic pathway characterized in E. coli and P. aeruginosa (10, 16), were not identified in the screened genomes of Francisella spp. This result is in agreement with the strictly aerobic metabolism of these bacteria. Indeed, the tricarboxylic acid (TCA) cycle and the UQ-dependent electron transfer chain, leading to efficient oxidative phosphorylation, take place in Francisella spp. (36). A possible link between stress defense and the TCA cycle was previously suggested for Francisella pathogenesis (37). Unfortunately, the contribution of UQ and the electron transfer chain to virulence has not been well documented to date in Francisella spp. Using G. mellonella as an infection model at the scale of an entire organism, we demonstrated through the study of the Tn-ubiCFn mutant and the isogenic control strain pretreated with 3A4HB that UQ8 contributes to the virulence of F. novicida and more likely in the later steps of infection, during which the bacteria undergo extensive replication (38). Such a notion supports the view that, as for other facultative intracellular bacteria, Francisella spp. are able to use several substrates in order to grow in various environments, such as macrophages. Glycerol via gluconeogenesis and amino acids were identified as the main sources of carbon during the intracellular replication of Francisella spp. in host cells (36, 39). However, glycerol requires UQ to be efficiently metabolized via the ubiquitous enzyme GlpD (40), and amino acid degradation is closely linked to the TCA cycle, which produces reducing equivalents in Francisella spp. (36). Besides its requirement for bioenergetics, UQ might also contribute to the antioxidant capacity of Francisella since it was shown to be a potent lipid-soluble antioxidant in E. coli (41). During its intracellular life, Francisella is exposed to oxidative stress. Indeed, as a defense mechanism for the clearance of phagocytosed microorganisms, both macrophages and neutrophils produce reactive oxygen species, which in turn trigger bacterial killing by causing damage to macromolecules (42, 43). We propose that the reduced content of UQ in the Tn-ubiCFn mutant could therefore affect F. novicida’s oxidative defense. This hypothesis is in good agreement with recent data showing that reduced expression of UbiCFn decreases the resistance of F. novicida to oxidative stress (44). In a similar way, we showed previously that UbiE, UbiJ, and UbiB proteins were needed for Salmonella enterica serovar Typhimurium intracellular proliferation in macrophages (21). Collectively, all these data assign a role for Ubi proteins in bacterial intracellular proliferation and, more generally, highlight the importance of UQ production for bacterial virulence.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains used in this study are listed in Table S2 in the supplemental material. F. novicida U112 was obtained from the Centre National de Référence des Francisella, CHU Grenoble-Alpes, France. The transposon mutant Tn-ubiCFn of the F. novicida U112 strain was obtained from the Manoil Laboratory, Department of Genome Science, University of Washington (7). Both strains were grown on Polyvitex-enriched chocolate agar (PVX-CHA) plates (bioMérieux, Marcy l’Etoile, France) incubated at 37°C for 48 to 72 h. Liquid cultures were carried out at 37°C with rotary shaking at 200 rpm in Chamberlain medium (45) supplemented with either glucose or succinate (0.4% [wt/vol] final concentration) as the only carbon source. For growth studies, cultures grown overnight were used to inoculate a 96-well plate to obtain a starting optical density at 600 nm (OD600) of around 0.1 and further incubated with shaking at 37°C. Changes in the OD600 were monitored every 10 min for 40 h using the Infinite 200 Pro microplate reader (Tecan, Lyon, France). When required, the medium was supplemented with 4HB in DMSO at a 50 to 100 μM final concentration; pABA, pA2MBA, and pA3MBA in DMSO at a 1 mM final concentration; or 3A4HB at a 10 μM to 1 mM final concentration. For CFU counting, bacteria were suspended in phosphate-buffered saline (PBS), and cell suspensions were serially diluted in PBS. For each sample, 100 μl of at least four different dilutions was plated onto PVX-CHA plates and incubated for 72 h at 37°C, and CFU were counted using a Scan 100 instrument (Interscience).
The E. coli ΔubiA and ΔubiJ mutants were constructed as described previously (46). Briefly, the ubiA::cat and ubiJ::cat mutations were generated by one-step inactivation of the ubiA and ubiJ genes. A DNA fragment containing the cat gene flanked with the 5′ and 3′ regions of the ubiA and ubiJ genes was PCR amplified using pKD3 as a template and oligonucleotides 5′-wannerubiA/3′-wannerubiA and 5′-wannerubiJ/3′-wannerubiJ, respectively (Table S3). The ΔubiB mutant was generated as follows. The cat gene was inserted into the ubiB gene between the two sites of NruI at bp 842 and 1004. Next, ubiB::cat was PCR amplified using oligonucleotide pair 5′-xbaIubiB/3′-xbaIubiB (Table S3). Strain BW25113, carrying the pKD46 plasmid, was transformed by electroporation with the amplified fragments, and Catr colonies were selected. The replacement of chromosomal ubi by the cat gene was verified by PCR amplification in the Catr clones. E. coli K-12 strains JW5713 and JW2226 from the Keio Collection (47) were used as donors in transduction experiments to construct the ΔubiC::kan and ΔubiG::kan mutants of E. coli MG1655 strains. The ΔubiA, ΔubiB, ΔubiC, ΔubiE, ΔubiG, and ΔubiK strains were cured with pCP20 to yield the ΔubiAc, ΔubiBc, ΔubiCc, ΔubiEc, ΔubiGc, and ΔubiKc strains, respectively (Table S2). E. coli strains (K-12, MG1655, or Top10) were grown on lysogeny broth (LB) rich medium or in M9 minimal medium (supplemented with glucose or succinate at a 0.4% [wt/vol] final concentration) at 37°C. Ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (35 μg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG) (100 μM) were added when needed.
Cloning, plasmid construction, and complementation assays.
The plasmids and the primers used in this study are listed in Tables S2 and S3 in the supplemental material, respectively. All the plasmids produced in this work were verified by DNA sequencing (GATC Biotech, Constance, Germany). The FTN_0385 (ubiAFn), FTN_0459 (ubiBFn), FTN_0386 (ubiCFn), FTN_0461 (ubiEFn), FTN_1146 (coq7Fn), FTN_0321 (ubiGFn), FTN_1237 (ubiHFn), FTN_1236 (ubiIFn), FTN_0460 (ubiJFn), and FTN_1666 (ubiKFn) inserts were obtained by PCR amplification using the F. novicida U112 genome as the template and the oligonucleotides described in Table S3. Inserts were EcoRI-BamHI or EcoRI-HindIII digested and inserted into EcoRI-BamHI- or EcoRI-HindIII-digested pTrc99a plasmids, respectively, yielding the pubiAFn, pubiBFn, pubiCFn, pubiEFn, pcoq7Fn, pubiGFn, pubiHFn, pubiIFn, pubiJFn, and pubiKFn plasmids (Table S3). The plasmids were transformed into E. coli MG1655 strains with mutation of the ubiA, ubiB, ubiC, ubiE, ubiF, ubiG, ubiH, ubiI, ubiJ, and ubiK genes (single and double mutations) (Table S2), and complementation of the UQ8 biosynthetic defect was assessed by both measuring the quinone content and plating serial dilutions onto solid M9 minimal medium supplemented with glucose or succinate (0.4% [wt/vol] final concentration) as the only carbon source, with growth overnight at 37°C. Expression of the Ubi proteins was induced by the addition of IPTG to a final concentration of 100 μM.
Lipid extractions and quinone analysis.
Cultures (5 ml under ambient air) were cooled down on ice 30 min before centrifugation at 3,200 × g at 4°C for 10 min. Cell pellets were washed in 1 ml ice-cold PBS and transferred to preweighed 1.5-ml Eppendorf tubes. After centrifugation at 12,000 × g at 4°C for 1 min, the supernatant was discarded, the cell wet weight was determined (∼5 to 30 mg), and pellets were stored at −20°C. Quinone extraction from cell pellets was performed as previously described (22). Lipid extracts corresponding to 1 mg of cells (wet weight) were analyzed by high-performance liquid chromatography (HPLC)-electrochemical detection (ECD) MS with a BetaBasic-18 column at a flow rate of 1 ml/min with mobile phases composed of 50% methanol, 40% ethanol, and a mix of 90% isopropanol, 10% ammonium acetate (1 M), and 0.1% trifluoroacetic acid (TFA). When necessary, MS detection was performed on an MSQ spectrometer (Thermo Scientific) with electrospray ionization in positive mode (probe temperature, 400°C; cone voltage, 80 V). Single-ion monitoring detected the following compounds: UQ8 (M+ NH4+), m/z 744 to 745, 6 to 10 min, with a scan time of 0.2 s; UQ10 (M+ NH4+), m/z 880 to 881, 10 to 17 min, with a scan time of 0.2 s; DMQ8 (M+ NH4+), m/z 714 to 715, 10 min, with a scan time of 0.4 s; DDMQ8 (M+ NH4+), m/z 700 to 701, 5 to 8 min, with a scan time of 0.4 s; OPP (M+ NH4+), m/z 656.0 to 657, 5 to 9 min, with a scan time of 0.4 s; and compound X (M+ H+), m/z 682 to 683, 5 to 10 min, with a scan time of 0.4 s. MS spectra were recorded between m/z 600 and 900 with a scan time of 0.3 s. ECD and MS peak areas were corrected for sample loss during extraction on the basis of the recovery of the UQ10 internal standard and then normalized to cell wet weight. The peaks of UQ8 obtained by electrochemical detection or MS detection were quantified with a standard curve of UQ10 as previously described (22).
Infections of G. mellonella larvae.
Larvae of the wax moth G. mellonella were purchased from Lombri’carraz SARL, Mery, France. Healthy and uniformly white larvae measuring around 3 cm were selected for infection. The bacteria were grown overnight to an OD600 of ∼3. Culture medium was removed by centrifugation, and bacteria were diluted in PBS to 108 CFU/ml. Insulin cartridges were sterilized before filling with bacterial solutions. Larvae were injected with 10 μl of bacterial suspensions (106 CFU per larva as recommended previously [24]) using an insulin pen or with 10 μl of PBS only. The precise number of bacteria transferred in injections was determined by spotting serial dilutions onto chocolate agar plates and counting CFU after growth at 37°C for 48 h. Infected larvae were placed into petri dishes and maintained at 37°C. The survival of larvae was monitored for 6 days by counting the number of dead larvae each day. A cohort of 20 larvae was used under each condition, and the experiment was performed twice. As a control, an untreated cohort of larvae was also monitored.
ACKNOWLEDGMENTS
This work was supported by the Agence Nationale de la Recherche (ANR), project O2-taboo ANR-19-CE44-0014, project Emergence (TIMC-UGA), the Université Grenoble Alpes (UGA), and the French Centre National de la Recherche Scientifique (CNRS).
We thank Patricia Renesto for constructive discussions and technical assistance and Laurent Aussel for critically reading the paper.
F.B., F.P., and L.P. conceived the project and its design. K.K., M.H.C., and G.H. conducted experiments and performed data analysis. K.K., C.D.B., and Y.C. performed experiments on G. mellonella. L.L. contributed new reagents (strains). All authors edited the manuscript. L.P. wrote the manuscript. L.P. supervised the project.
Footnotes
Supplemental material is available online only.
Contributor Information
Ludovic Pelosi, Email: ludovic.pelosi@univ-grenoble-alpes.fr.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Sjostedt A. 2011. Special topic on Francisella tularensis and tularemia. Front Microbiol 2:86. 10.3389/fmicb.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallet P, Lagrange B, Henry T. 2016. Francisella inflammasomes: integrated responses to a cytosolic stealth bacterium. Curr Top Microbiol Immunol 397:229–256. 10.1007/978-3-319-41171-2_12. [DOI] [PubMed] [Google Scholar]
- 3.Oyston PC, Sjostedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2:967–978. 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 4.Sjostedt A. 2007. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci 1105:1–29. 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 5.Kingry LC, Petersen JM. 2014. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol 4:35. 10.3389/fcimb.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BD, Faron M, Rasmussen JA, Fletcher JR. 2014. Uncovering the components of the Francisella tularensis virulence stealth strategy. Front Cell Infect Microbiol 4:32. 10.3389/fcimb.2014.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci USA 104:1009–1014. 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ireland PM, Bullifent HL, Senior NJ, Southern SJ, Yang ZR, Ireland RE, Nelson M, Atkins HS, Titball RW, Scott AE. 2019. Global analysis of genes essential for Francisella tularensis Schu S4 growth in vitro and for fitness during competitive infection of Fischer 344 rats. J Bacteriol 201:e00630-18. 10.1128/JB.00630-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowicka B, Kruk J. 2010. Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta 1797:1587–1605. 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Vo CD, Michaud J, Elsen S, Faivre B, Bouveret E, Barras F, Fontecave M, Pierrel F, Lombard M, Pelosi L. 2020. The O2-independent pathway of ubiquinone biosynthesis is essential for denitrification in Pseudomonas aeruginosa. J Biol Chem 295:9021–9032. 10.1074/jbc.RA120.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soballe B, Poole RK. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology (Reading) 145(Part 8):1817–1830. 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 12.Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. 2014. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 1837:1004–1011. 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal S, Jaswal K, Shiver AL, Balecha H, Patra T, Chaba R. 2017. A genome-wide screen in Escherichia coli reveals that ubiquinone is a key antioxidant for metabolism of long-chain fatty acids. J Biol Chem 292:20086–20099. 10.1074/jbc.M117.806240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abby SS, Kazemzadeh K, Vragniau C, Pelosi L, Pierrel F. 2020. Advances in bacterial pathways for the biosynthesis of ubiquinone. Biochim Biophys Acta 1861:148259. 10.1016/j.bbabio.2020.148259. [DOI] [PubMed] [Google Scholar]
- 15.Hajj Chehade M, Pelosi L, Fyfe CD, Loiseau L, Rascalou B, Brugiere S, Kazemzadeh K, Vo CD, Ciccone L, Aussel L, Coute Y, Fontecave M, Barras F, Lombard M, Pierrel F. 2019. A soluble metabolon synthesizes the isoprenoid lipid ubiquinone. Cell Chem Biol 26:482–492.e7. 10.1016/j.chembiol.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Pelosi L, Vo CD, Abby SS, Loiseau L, Rascalou B, Hajj Chehade M, Faivre B, Gousse M, Chenal C, Touati N, Binet L, Cornu D, Fyfe CD, Fontecave M, Barras F, Lombard M, Pierrel F. 2019. Ubiquinone biosynthesis over the entire O2 range: characterization of a conserved O2-independent pathway. mBio 10:e01319-19. 10.1128/mBio.01319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenmark P, Grunler J, Mattsson J, Sindelar PJ, Nordlund P, Berthold DA. 2001. A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane-bound hydroxylase involved in ubiquinone biosynthesis. J Biol Chem 276:33297–33300. 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou L, Li M, Wang XY, Liu H, Sun S, Chen H, Poplawsky A, He YW. 2019. Biosynthesis of coenzyme Q in the phytopathogen Xanthomonas campestris via a yeast-like pathway. Mol Plant Microbe Interact 32:217–226. 10.1094/MPMI-07-18-0183-R. [DOI] [PubMed] [Google Scholar]
- 19.Pelosi L, Ducluzeau AL, Loiseau L, Barras F, Schneider D, Junier I, Pierrel F. 2016. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems 1:e00091-16. 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF. 2000. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol 182:5139–5146. 10.1128/JB.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aussel L, Loiseau L, Hajj Chehade M, Pocachard B, Fontecave M, Pierrel F, Barras F. 2014. ubiJ, a new gene required for aerobic growth and proliferation in macrophage, is involved in coenzyme Q biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium. J Bacteriol 196:70–79. 10.1128/JB.01065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajj Chehade M, Loiseau L, Lombard M, Pecqueur L, Ismail A, Smadja M, Golinelli-Pimpaneau B, Mellot-Draznieks C, Hamelin O, Aussel L, Kieffer-Jaquinod S, Labessan N, Barras F, Fontecave M, Pierrel F. 2013. ubiI, a new gene in Escherichia coli coenzyme Q biosynthesis, is involved in aerobic C5-hydroxylation. J Biol Chem 288:20085–20092. 10.1074/jbc.M113.480368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loiseau L, Fyfe C, Aussel L, Hajj Chehade M, Hernandez SB, Faivre B, Hamdane D, Mellot-Draznieks C, Rascalou B, Pelosi L, Velours C, Cornu D, Lombard M, Casadesus J, Pierrel F, Fontecave M, Barras F. 2017. The UbiK protein is an accessory factor necessary for bacterial ubiquinone (UQ) biosynthesis and forms a complex with the UQ biogenesis factor UbiJ. J Biol Chem 292:11937–11950. 10.1074/jbc.M117.789164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thelaus J, Lundmark E, Lindgren P, Sjodin A, Forsman M. 2018. Galleria mellonella reveals niche differences between highly pathogenic and closely related strains of Francisella spp. Front Cell Infect Microbiol 8:188. 10.3389/fcimb.2018.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aperis G, Fuchs BB, Anderson CA, Warner JE, Calderwood SB, Mylonakis E. 2007. Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect 9:729–734. 10.1016/j.micinf.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Propst CN, Pylypko SL, Blower RJ, Ahmad S, Mansoor M, van Hoek ML. 2016. Francisella philomiragia infection and lethality in mammalian tissue culture cell models, Galleria mellonella, and BALB/c mice. Front Microbiol 7:696. 10.3389/fmicb.2016.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodmann M, Schnider S, Basler M. 2021. Type VI secretion system and its effectors PdpC, PdpD and OpiA contribute to Francisella virulence in Galleria mellonella larvae. Infect Immun 89:e00579-20. 10.1128/IAI.00579-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Q, Feng M, Xia H, Zhao Y, Hou B, Ye J, Wu H, Zhang H. 2019. Differential quantitative proteomics reveals the functional difference of two yigP locus products, UbiJ and EsrE. J Basic Microbiol 59:1125–1133. 10.1002/jobm.201900350. [DOI] [PubMed] [Google Scholar]
- 29.Jiang HX, Wang J, Zhou L, Jin ZJ, Cao XQ, Liu H, Chen HF, He YW. 2019. Coenzyme Q biosynthesis in the biopesticide shenqinmycin-producing Pseudomonas aeruginosa strain M18. J Ind Microbiol Biotechnol 46:1025–1038. 10.1007/s10295-019-02179-1. [DOI] [PubMed] [Google Scholar]
- 30.Nakahigashi K, Miyamoto K, Nishimura K, Inokuchi H. 1992. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J Bacteriol 174:7352–7359. 10.1128/jb.174.22.7352-7359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L, Wang JY, Wu J, Wang J, Poplawsky A, Lin S, Zhu B, Chang C, Zhou T, Zhang LH, He YW. 2013. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol 87:80–93. 10.1111/mmi.12084. [DOI] [PubMed] [Google Scholar]
- 32.Marshall SA, Payne KAP, Leys D. 2017. The UbiX-UbiD system: the biosynthesis and use of prenylated flavin (prFMN). Arch Biochem Biophys 632:209–221. 10.1016/j.abb.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 33.Ravcheev DA, Thiele I. 2016. Genomic analysis of the human gut microbiome suggests novel enzymes involved in quinone biosynthesis. Front Microbiol 7:128. 10.3389/fmicb.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degli Esposti M. 2017. A journey across genomes uncovers the origin of ubiquinone in cyanobacteria. Genome Biol Evol 9:3039–3053. 10.1093/gbe/evx225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma P, Teixeira de Mattos MJ, Hellingwerf KJ, Bekker M. 2012. On the function of the various quinone species in Escherichia coli. FEBS J 279:3364–3373. 10.1111/j.1742-4658.2012.08608.x. [DOI] [PubMed] [Google Scholar]
- 36.Ziveri J, Barel M, Charbit A. 2017. Importance of metabolic adaptations in Francisella pathogenesis. Front Cell Infect Microbiol 7:96. 10.3389/fcimb.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieppedale J, Gesbert G, Ramond E, Chhuon C, Dubail I, Dupuis M, Guerrera IC, Charbit A. 2013. Possible links between stress defense and the tricarboxylic acid (TCA) cycle in Francisella pathogenesis. Mol Cell Proteomics 12:2278–2292. 10.1074/mcp.M112.024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong A, Celli J. 2010. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol 1:138. 10.3389/fmicb.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brissac T, Ziveri J, Ramond E, Tros F, Kock S, Dupuis M, Brillet M, Barel M, Peyriga L, Cahoreau E, Charbit A. 2015. Gluconeogenesis, an essential metabolic pathway for pathogenic Francisella. Mol Microbiol 98:518–534. 10.1111/mmi.13139. [DOI] [PubMed] [Google Scholar]
- 40.Austin D, Larson TJ. 1991. Nucleotide sequence of the glpD gene encoding aerobic sn-glycerol 3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol 173:101–107. 10.1128/jb.173.1.101-107.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soballe B, Poole RK. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology (Reading) 146(Part 4):787–796. 10.1099/00221287-146-4-787. [DOI] [PubMed] [Google Scholar]
- 42.Kinkead LC, Allen LA. 2016. Multifaceted effects of Francisella tularensis on human neutrophil function and lifespan. Immunol Rev 273:266–281. 10.1111/imr.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steiner DJ, Furuya Y, Jordan MB, Metzger DW. 2017. Protective role for macrophages in respiratory Francisella tularensis infection. Infect Immun 85:e00064-17. 10.1128/IAI.00064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felix J, Siebert C, Ducassou JN, Nigou J, Garcia PS, Fraudeau A, Huard K, Mas C, Brochier-Armanet C, Coute Y, Gutsche I, Renesto P. 2021. Structural and functional analysis of the Francisella lysine decarboxylase as a key actor in oxidative stress resistance. Sci Rep 11:972. 10.1038/s41598-020-79611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chamberlain RE. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl Microbiol 13:232–235. 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2 and Tables S1 to S3. Download JB.00400-21-s0001.pdf, PDF file, 0.4 MB (399.7KB, pdf)