ABSTRACT
Mycobacterium smegmatis has two isocitrate lyase (ICL) isozymes (MSMEG_0911 and MSMEG_3706). We demonstrated that ICL1 (MSMEG_0911) is the predominantly expressed ICL in M. smegmatis and plays a major role in growth on acetate or fatty acid as the sole carbon and energy source. Expression of the icl1 gene in M. smegmatis was demonstrated to be strongly upregulated during growth on acetate relative to that in M. smegmatis grown on glucose. Expression of icl1 was shown to be positively regulated by the RamB activator, and three RamB-binding sites (RamBS1, RamBS2, and RamBS3) were identified in the upstream region of icl1 using DNase I footprinting analysis. Succinyl coenzyme A (succinyl-CoA) was shown to increase the affinity of binding of RamB to its binding sites and enable RamB to bind to RamBS2, which is the most important site for RamB-mediated induction of icl1 expression. These results suggest that succinyl-CoA serves as a coinducer molecule for RamB. Our study also showed that cAMP receptor protein (Crp1; MSMEG_6189) represses icl1 expression in M. smegmatis grown in the presence of glucose. Therefore, the strong induction of icl1 expression during growth on acetate as the sole carbon source relative to the weak expression of icl1 during growth on glucose is likely to result from combined effects of RamB-mediated induction of icl1 in the presence of acetate and Crp-mediated repression of icl1 in the presence of glucose.
IMPORTANCE Carbon flux through the glyoxylate shunt has been suggested to affect virulence, persistence, and antibiotic resistance of Mycobacterium tuberculosis. Therefore, it is important to understand the precise mechanism underlying the regulation of the icl gene encoding the key enzyme of the glyoxylate shunt. Using Mycobacterium smegmatis, this study revealed the regulation mechanism underlying induction of icl1 expression in M. smegmatis when the glyoxylate shunt is required. The conservation of the cis- and trans-acting regulatory elements related to icl1 regulation in both M. smegmatis and M. tuberculosis implies that a similar regulatory mechanism operates for the regulation of icl1 expression in M. tuberculosis.
KEYWORDS: acetate, cAMP, Crp, isocitrate lyase, glyoxylate shunt, Mycobacterium, regulation of gene expression, RamB, succinyl-CoA, tuberculosis
INTRODUCTION
Isocitrate lyase (ICL) catalyzes the cleavage of isocitrate to succinate and glyoxylate. The ICL-catalyzing reaction forms the glyoxylate shunt together with the reaction of malate synthase catalyzing the condensation of glyoxylate with acetyl coenzyme A (acetyl-CoA) to malate. The glyoxylate shunt forms a reaction cycle with reactions of the tricarboxylic acid (TCA) cycle, in which two molecules of acetyl-CoA yield one molecule of succinate as the net result. Therefore, the glyoxylate shunt is required for anaplerosis replenishing the reaction intermediates of the TCA cycle from acetyl-CoA when molecules such as acetate, fatty acids, or ketogenic amino acids are used as the sole carbon source (1). The glyoxylate shunt has been shown to play a crucial role in survival and persistence of Mycobacterium tuberculosis in both activated macrophages and mice (2, 3), implying that a metabolic shift in the principal carbon source from carbohydrates to fat might occur in M. tuberculosis residing within the host. Indeed, several lines of evidence have been reported which suggest that M. tuberculosis preferentially relies on fatty acids rather than carbohydrates when persisting inside the host (3–6). Based on the length and domain organization, ICLs are divided into three groups (3). The small prokaryotic ICLs belonging to group I consist of two domains (domains I and III), while the ICLs of group II, found in eukaryotes, have an additional domain (domain II) between the domains I and III. The mycobacterial ICLs of group III, which are similar to the group II ICLs and found in mycobacteria, are classified into an independent group due to the presence of the additional domain IV at their C termini. M. tuberculosis has two ICL isoforms. ICL1 is a prokaryotic type, while ICL2 is a mycobacterial ICL of group III (2, 3). The ICLs found in mycobacteria also possess the activity of methylcitrate lyase and thus function as key enzymes for the methylcitrate cycle that is involved in propionyl-CoA assimilation (5, 7–9). Since the isocitrate lyase and methylcitrate lyase activities of ICL2 were demonstrated to be substantially activated upon acetyl-CoA and propionyl-CoA binding, it was suggested that ICL2 might act as a gatekeeping enzyme to regulate the carbon flux between the TCA cycle and the glyoxylate shunt (10).
The genes (icl1 or aceA) encoding ICL in M. tuberculosis, Mycobacterium avium, and Mycobacterium smegmatis have been shown to be upregulated when acetate or fatty acid is supplied as the sole carbon source (11–13). Interestingly, expression of the gene in M. tuberculosis has been shown to be also induced under hypoxic and low-pH conditions, as well as during infection of macrophages and mice (9, 14–17). Mycobacteria belong to the order Corynebacteriales, as does Corynebacterium glutamicum, which is a fast-growing environmental saprophyte (18). It was first demonstrated in C. glutamicum that RamB (regulator of acetate metabolism B) is involved in the negative regulation of the aceA gene encoding ICL (19, 20). Likewise, the RamB homolog of M. tuberculosis was also shown to act as a transcriptional repressor in the regulation of icl1 (13). At the posttranscriptional level, the activity of ICL1 has been suggested to be controlled by acetylation and succinylation in M. tuberculosis (21–23).
Since carbon flux through the glyoxylate shunt has been suggested to affect virulence, persistence, and antibiotic resistance of M. tuberculosis (2, 3, 5, 24), it is important to understand the precise mechanism underlying the regulation of the icl gene encoding the key enzyme of the glyoxylate shunt. Although RamB has been demonstrated to be the master regulator functioning as a repressor in the regulation of icl1 (aceA) expression in M. tuberculosis and C. glutamicum, there has been no report clearly revealing the nature of the effector molecule that modulates the functionality of RamB. Here, we demonstrate that succinyl-CoA serves as a coinducer for RamB and that RamB functions as a transcriptional activator for icl1 expression in M. smegmatis, in contrast to the cases in M. tuberculosis and C. glutamicum. We also provide evidence that Crp1 (cAMP receptor protein; MSMEG_6189) is also implicated in the negative regulation of icl1 expression in M. smegmatis during growth on glucose.
RESULTS
M. smegmatis has two ICLs, and ICL1 plays a major role in utilization of acetate and fatty acids.
M. smegmatis has two genes that encode ICL isozymes (MSMEG_0911 and MSMEG_3706). The MSMEG_0911 (icl1) gene encodes a protein composed of 428 amino acids, which is closely related to the prokaryotic ICLs of group I, while the predicted MSMEG_3706 (icl2) gene product, consisting of 769 amino acids, is closely homologous to the mycobacterial ICLs belonging to group III. ICL1 and ICL2 of M. smegmatis show 92% and 77% sequence identity to ICL1 (MT0483) and ICL2 (MT1966) of M. tuberculosis CDC1551, respectively.
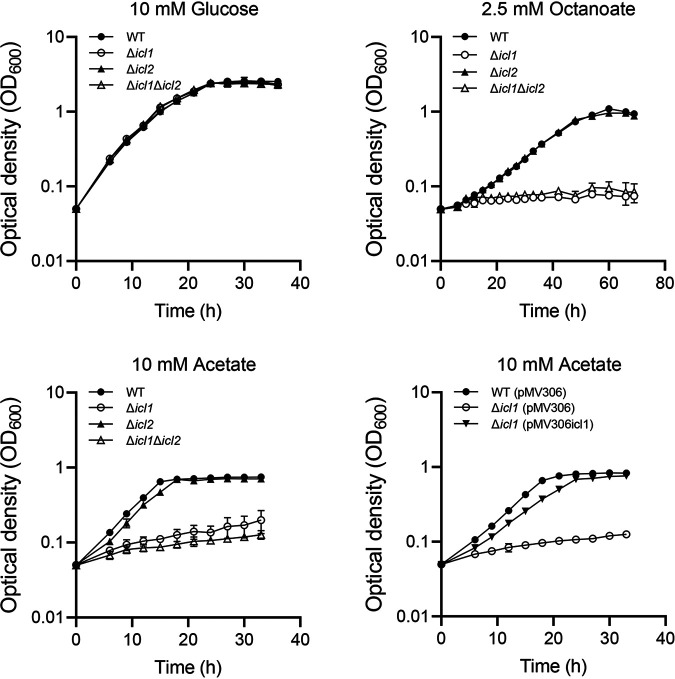
To evaluate the functional roles of the icl1 and icl2 genes in utilization of acetate and fatty acids in M. smegmatis, null mutants of M. smegmatis carrying deletions within icl1, icl2, or both icl1 and icl2 were constructed, and the wild-type (WT) and mutant strains were examined for their growth in medium supplemented with glucose, acetate, or octanoate as the sole carbon source (Fig. 1). No difference between the WT and mutant strains was observed with regard to growth rate when glucose was supplied as a carbon source. When acetate was supplied as the sole carbon source, the Δicl1 mutant strain showed significantly impaired growth, with a doubling time of 21.4 h, compared to the WT strain, showing a doubling time of 4.2 h. In contrast, growth of M. smegmatis on acetate was marginally affected by icl2 mutation. Cumulative effects of the icl1 and icl2 mutations on growth on acetate were observed for the Δicl1 Δicl2 mutant strain. Introduction of the pMV306icl1 plasmid carrying the intact icl1 gene into the Δicl1 mutant restored growth of the mutant on acetate, indicating that the severe defect in acetate utilization observed for the Δicl1 mutant was the result of icl1 inactivation. When octanoate was supplied as the sole carbon source, the WT and Δicl2 mutant strains showed the same growth rate. In contrast, the Δicl1 and Δicl1 Δicl2 mutant strains did not grow under the same condition, indicating that ICL1 is indispensable for growth of M. smegmatis on octanoate. Taken together, the results presented in Fig. 1 suggest that ICL1 serves as the major ICL in M. smegmatis during growth on acetate and fatty acids. The relative expression levels of icl1 and icl2 could be extrapolated from the reads per kilobase pair per million (RPKM) of icl1 and icl2 obtained from RNA sequencing analysis on M. smegmatis grown aerobically on glucose (25). The RPKM value of icl1 was found to be 43-fold greater than that of icl2, indicating that ICL1 is the predominantly expressed ICL in M. smegmatis (the RPKM values of icl1 and icl2 are 895.5 and 20.7, respectively).
FIG 1.
Growth of the WT, Δicl1, Δicl2, and Δicl1 Δicl2 strains of M. smegmatis on various carbon sources. The Δicl1, Δicl2, and Δicl1 Δicl2 mutant strains, as well as the WT strain as a control, were grown aerobically at 37°C in 7H9 medium supplemented with 10 mM glucose, 10 mM acetate, or 2.5 mM octanoate as the sole carbon source. For complementation of the Δicl1 mutant, pMV306icl1 (a pMV306-derived plasmid carrying the intact icl1 gene and its own promoter) was introduced into the Δicl1mutant. The Δicl1 strain harboring pMV306icl1, as well as the WT and Δicl1 strains with the empty vector pMV306, was grown aerobically in 7H9-acetate at 37°C. All values provided were determined from three biological replicates. The error bars indicate the standard deviations.
Expression of icl1 is induced in the presence of acetate and fatty acids.
Acetate and fatty acids are metabolized to acetyl-CoA through acetate activation and β-oxidation, respectively (11). When M. smegmatis utilizes acetate or fatty acid as the sole carbon source, the glyoxylate shunt is required to replenish TCA cycle intermediates from acetyl-CoA. We assessed whether expression of icl1 is induced in M. smegmatis grown on acetate or fatty acids. The expression level of the icl1 gene in M. smegmatis grown on glucose, acetate, or octanoate was examined using the WT strain carrying the icl1::lacZ translational fusion plasmid pNCIIicl1. As shown in Fig. 2, expression of icl1 was induced 7-fold in M. smegmatis grown on acetate relative to that in M. smegmatis grown on glucose, while only a 1.6-fold increase in icl1 expression was observed in M. smegmatis grown on octanoate relative to M. smegmatis grown on glucose. We also determined icl2 expression using the WT strain carrying the icl2::lacZ translational fusion plasmid pNCIIicl2. The icl2 gene was expressed only marginally irrespective of whether glucose or acetate was supplied as the sole carbon source (data not shown).
FIG 2.
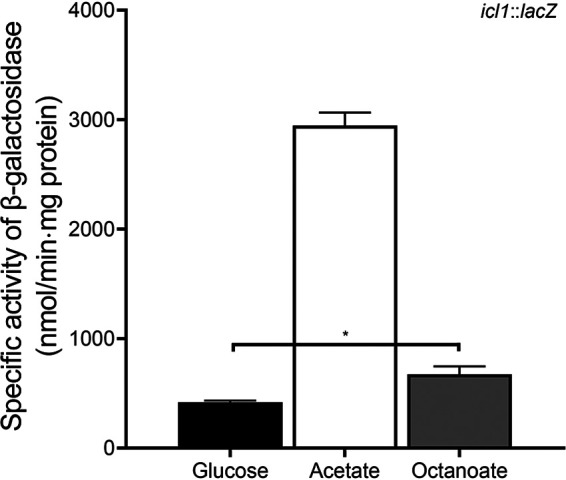
Expression levels of the icl1 gene in M. smegmatis grown on glucose, acetate, or octanoate. The WT strain of M. smegmatis strain containing the icl1::lacZ translational fusion plasmid pNCIIicl1 was grown aerobically to an OD600 of 0.45 to 0.5 in 7H9 medium supplemented with 10 mM glucose, 10 mM acetate, or 2.5 mM octanoate as the sole carbon source. Cell-free crude extracts were used to measure β-galactosidase activity. All values provided were determined from three biological replicates. The error bars indicate the standard deviations. *, P < 0.01.
RamB serves as a transcriptional activator for icl1 expression.
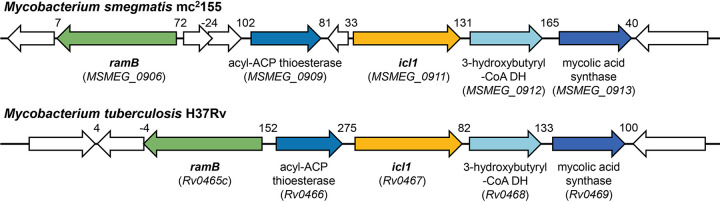
To understand the regulation mechanism underlying induction of icl1 expression by acetate in M. smegmatis, we first searched for the gene homologous to the ramB gene, whose product had been found to regulate expression of several genes related to acetate metabolism in C. glutamicum and M. tuberculosis (13, 19). A BLAST analysis revealed that the chromosome of M. smegmatis contains a gene (MSMEG_0906) the product of which shows 86% and 55% sequence identity to RamB (Rv0465c and Cg0444) of M. tuberculosis and C. glutamicum, respectively. The ramB gene is located in the vicinity of the icl1 gene on the chromosomes of both M. smegmatis and M. tuberculosis (Fig. 3).
FIG 3.
Genetic organizations of the loci associated with the icl1 and ramB genes in M. smegmatis and M. tuberculosis. The open reading frames are shown as arrows, whose size and orientation indicate their relative length and direction, respectively. The lengths of the overlapping and intergenic regions are given as negative and positive nucleotide numbers above the schematic diagrams, respectively.
To gain insight into the functional role of the ramB gene in utilization of acetate in M. smegmatis, a ΔramB mutant of M. smegmatis was constructed, and the mutant was compared with the WT strain for growth in medium supplemented with glucose or acetate (see Fig. S1 in the supplemental material). No difference between the WT and ΔramB mutant strains was observed with respect to growth rate when glucose was supplied as the sole carbon source (the doubling times of the WT and mutant strains were 4.8 h). When acetate was the sole carbon source, growth of the ΔramB mutant strain was retarded compared to that of the WT strain (the doubling times of the WT and mutant strains were 4.8 h and 6.8 h, respectively), indicating that RamB is necessary for the optimal growth of M. smegmatis on acetate. To determine the functional role of RamB in the regulation of icl1 expression in M. smegmatis, expression of the icl1 gene was examined in the WT and ΔramB mutant strains of M. smegmatis carrying pNCIIicl1 (Fig. 4). Consistent with the result in Fig. 2, the expression level of icl1 was shown to be increased 6.5-fold in the WT strain grown on acetate relative to that in the WT strain grown on glucose. The icl1 gene was significantly less expressed in the ΔramB mutant grown on acetate than in the WT strain grown under the same condition, and the lower expression level of icl1 in the mutant relative to the WT strain was also observed when glucose was supplied as the sole carbon source. These results indicate that RamB serves as a transcriptional activator for the icl1 gene in M. smegmatis. Introduction of the intact ramB gene into the ΔramB mutant led to restoration of the expression level of icl1 to that observed in the WT strain when the strains were grown on acetate. We also found that RamB of M. smegmatis negatively regulated its own gene irrespective of whether it was grown on glucose or acetate (Fig. S2).
FIG 4.
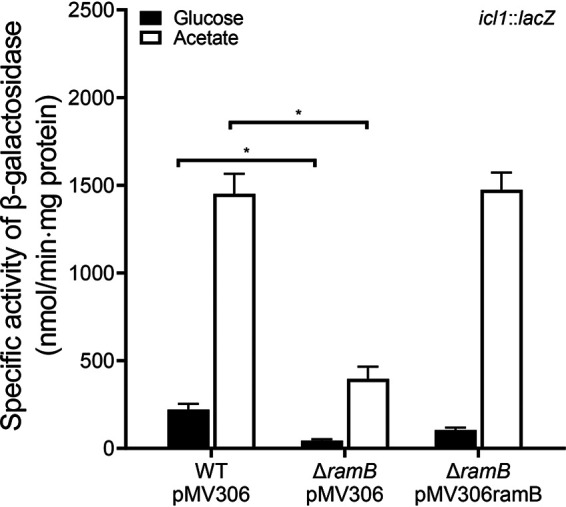
Expression levels of the icl1 gene in the WT and ΔramB mutant strains of M. smegmatis. The WT and ΔramB mutant strains carrying both the empty vector pMV306 and the icl1::lacZ translational fusion plasmid pNCIIicl1 were used in the experiment. For complementation of the ΔramB mutant, pMV306ramB (a pMV306-derived plasmid carrying the intact ramB gene and its own promoter) was introduced into the mutant in place of pMV306. The strains were grown aerobically to an OD600 of 0.45 to 0.5 in 7H9 medium supplemented with 10 mM glucose or 10 mM acetate as the carbon source. Cell-free crude extracts were used to measure β-galactosidase activity. All values provided were determined from three biological replicates. The error bars indicate the standard deviations. *, P < 0.01.
It was previously demonstrated that PrpR (propionate regulator), a homolog of RamB, positively regulated the icl1 gene in M. tuberculosis and M. smegmatis when they were grown on propionate (26). PrpR (MSMEG_6643) of M. smegmatis shows 48% sequence identity and 65% similarity to RamB (MSMEG_0906), and the amino acid sequences of their DNA binding helix-turn-helix domains are similar to each other. Since the PrpR-binding sequence was demonstrated to be similar to the RamB-binding sequence (26), it is possible that PrpR is involved in induction of icl1 expression in M. smegmatis during growth on acetate. To verify this assumption, null mutants of M. smegmatis carrying deletions within prpR or both ramB and prpR were constructed, and the expression level of the icl1 gene was determined using the WT, ΔramB, ΔprpR, and ΔramB ΔprpR strains of M. smegmatis carrying pNCIIicl1. As shown in Fig. 5, inactivation of the prpR gene in the background of the WT and ΔramB mutant strains resulted in only a marginal decrease in icl1 expression when the strains were grown on acetate. In conjunction with the previous report (26), this result indicates that PrpR plays a specific role in icl1 induction by propionate but not by acetate.
FIG 5.
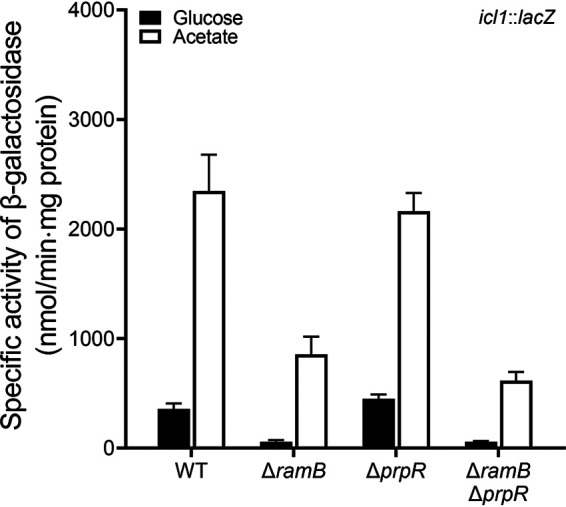
Expression levels of the icl1 gene in the WT, ΔramB, ΔprpR, and ΔramB ΔprpR mutant strains of M. smegmatis. The WT, ΔramB, ΔprpR, and ΔramB ΔprpR mutant strains containing the icl1::lacZ translational fusion plasmid pNCIIicl1 were grown aerobically to an OD600 of 0.45 to 0.5 in 7H9 medium supplemented with 10 mM glucose or 10 mM acetate as the sole carbon source. Cell-free crude extracts were used to measure β-galactosidase activity. All values provided were determined from three biological replicates. The error bars indicate the standard deviations.
Identification of the RamB-binding sites in the upstream region of icl1.
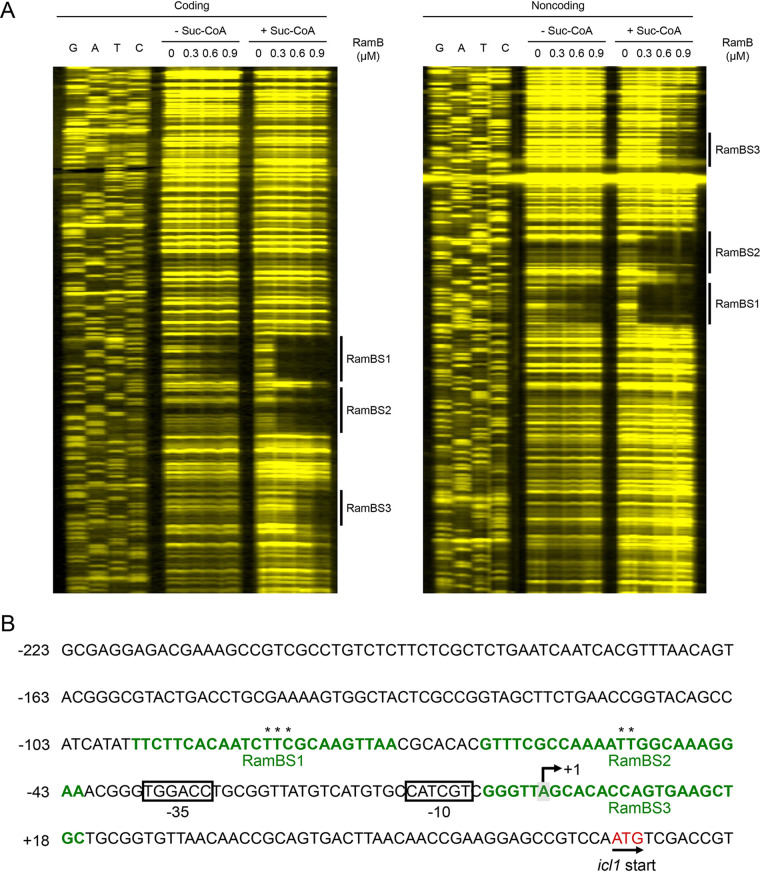
Despite the importance of RamB in the regulation of aceA (icl1), the nature of its effector molecule has remained elusive. Based on structural and mutational studies on PrpR, Tang et al. have recently predicted that succinyl-CoA might be an effector molecule that can bind to RamB (27). To define precisely the number and location of RamB-binding sites in the icl1 upstream region as well as the effect of succinyl-CoA on binding of RamB to the DNA region, DNase I footprinting analysis was conducted with purified RamB and 6-carboxytetramethylrhodamine (TAMRA)-labeled DNA fragments containing the icl1 upstream region in the presence or absence of succinyl-CoA (Fig. 6). In the absence of succinyl-CoA in the reaction mixtures, binding of RamB protected the 24-bp DNA region from DNase I cleavage at positions −96 to −73 (RamBS1) with respect to the transcriptional start point (TSP). The addition of 200 μM succinyl-CoA to the reaction mixtures resulted in a more clearly protected window for RamBS1 and protection of two additional DNA regions at positions −65 to −42 (RamBS2) and −5 to +19 (RamBS3). The binding affinity of RamB for the binding sites appeared to be in descending order of RamBS1, RamBS2, and RamBS3. The sequence of RamBS1 (TTCTTCACAATCTTCGCAAGTTAA; the conserved nucleotides in both RamBS1 and RamBS2 are underlined) is similar to that of RamBS2 (GTTTCGCCAAAATTGGCAAAGGAA), while the sequence of RamBS3 (GGGTTAGCACACCAGTGAAGCTGC) deviates from those of RamBS1 and RamBS2, which explains the low binding affinity of RamB for RamBS3. We also examined whether succinate, acetate, and acetyl phosphate (an intermediate of acetate activation) alter the binding affinity of RamB for the icl1 regulatory region using electrophoretic mobility shift assay (EMSA). We did not perform EMSA with acetyl-CoA, because it had been reported that acetyl-CoA did not change the binding affinity of RamB for its binding site (19). As shown in Fig. S3, the presence of succinate, acetate, or acetyl phosphate in the EMSA reaction mixtures did not lead to noticeable changes in binding of RamB to the icl1 regulatory region, in contrast to the presence of succinyl-CoA. Taken together, the DNase I footprinting and EMSA results clearly suggest that succinyl-CoA leads to changes in both binding affinity and binding patterns of RamB for the icl1 regulatory region.
FIG 6.
Binding of RamB to the icl1 regulatory region. (A) DNase I footprinting analysis of the icl1 regulatory region bound by purified RamB. The DNA fragments containing the coding and noncoding strands labeled with TAMRA at their 5′ ends were incubated with increasing concentrations of purified RamB (0.3, 0.6, and 0.9 μM) in the absence (- Suc-CoA) or presence (+ Suc-CoA) of 200 μM succinyl-CoA and then subjected to DNase I footprinting reactions. The amounts of RamB protein used are given above the lanes. The regions protected by RamB, which indicate the RamB-binding sites, are marked by thick black lines (RamBS1, RamBS2, and RamBS3). Lanes G, A, T, and C represent the sequence ladders. (B) The upstream sequence of the icl1 gene, including its putative promoter region and the identified cis-acting regulatory elements involved in the regulation of the icl1 gene. The RamB-binding sites (RamBS1, RamBS2, and RamBS3) are indicated in green font. The nucleotides in RamBS1 and RamBS2 which were mutated for the promoter assay in Fig. 7 are indicated with asterisks. The TSP of the icl1 gene is marked by +1. The putative promoter region (−35 and −10) of the icl1 gene is boxed. The start codon of icl1 is indicated both in red font and by the arrow indicating the transcriptional direction. The numbers on the left side of the sequences show the positions of the leftmost nucleotides relative to the TPS of the icl1 gene.
In order to ascertain the roles of the major RamB-binding sites (RamBS1 and RamBS2) in the regulation of icl1 expression in M. smegmatis, we determined the effect of RamBS1 and RamBS2 mutations on icl1 expression in the WT strain of M. smegmatis grown on glucose or acetate by using pNCIIicl1 derivatives that contain mutations within either RamBS1 or RamBS2 (Fig. 7). Transversion mutations were introduced into RamBS1 (CAATCTTCGCAAG to CAATCGAGGCAAG) and RamBS2 (CAAAATTGGCAAA to CAAAAGAGGCAAA) on pNCIIicl1 to construct pNCIIicl1BM1 and pNCIIicl1BM2, respectively (the mutated nucleotides are underlined). As expected, expression of icl1 was strongly induced by acetate in the control WT strain with pNCIIicl1. Expression of icl1 from pNCIIicl1BM2 was significantly reduced compared to that from pNCIIicl1 when the M. smegmatis strains were grown on acetate. Mutations in RamBS1 affected icl1 expression to a lesser extent than those in RamBS2, indicating that RamBS2 is more important in activation of icl1 transcription than RamBS1.
FIG 7.
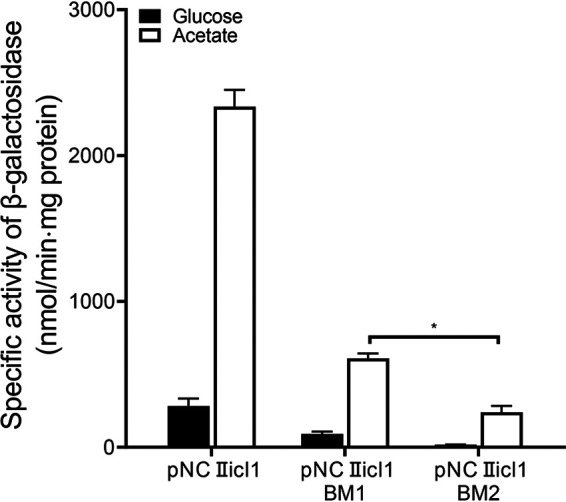
Effects of mutations in the RamB-binding sites (RamBS1 and RamBS2) on icl1 expression. The icl1 promoter activities were determined by using the pNCIIicl1-derived translational fusion plasmids (pNCIIicl1BM1 and pNCIIicl1BM2) with mutations in RamBS1 and RamBS2, respectively. As a control, pNCIIicl1 was included in the experiment. The WT strain of M. smegmatis harboring the translational fusion plasmids was grown aerobically to an OD600 of 0.45 to 0.5 in 7H9 medium supplemented with 10 mM glucose or 10 mM acetate as the sole carbon source. Cell-free crude extracts were used to measure β-galactosidase activity. All values provided were determined from three biological replicates. The error bars indicate the standard deviations. *, P < 0.01.
By means of 1H nuclear magnetic resonance (NMR) analysis, we tried to determine the level of succinyl-CoA in the WT strain of M. smegmatis grown on glucose or acetate to see whether the level of succinyl-CoA correlates with the expression level of icl1. We failed to detect succinyl-CoA using this analytical method. However, a 2-fold increase in succinate levels was observed in M. smegmatis grown on acetate relative to M. smegmatis grown on glucose (Fig. S4). This finding might indirectly imply an increased cellular level of succinyl-CoA in M. smegmatis grown on acetate, given that succinyl-CoA is the preceding intermediate of succinate in the TCA cycle.
Crp is involved in repression of icl1 in the presence of glucose.
The ΔramB ΔprpR mutant of M. smegmatis still showed the residual acetate-inducible regulation of icl1 (Fig. 5), which implies that a regulatory system(s) other than RamB and PrpR still operates to increase icl1 expression in M. smegmatis during growth on acetate (Fig. 5). The genome of M. smegmatis has two genes (crp1 [MSMEG_6189] and crp2 [MSMEG_0539]) encoding the cAMP receptor protein (Crp) paralogs that show 78% sequence identity to each other at the amino acid level (28, 29). Crp1 was demonstrated to be the major Crp that is predominantly expressed in M. smegmatis (30). Interestingly, it was observed in our previous RNA sequencing analysis that the Δcrp1 mutant of M. smegmatis exhibited higher expression of icl1 than the isogenic WT strain when both strains were grown on glucose (30). Consistent with the RNA sequencing result, expression of icl1 was shown to be derepressed in the Δcrp1 mutant grown on glucose relative to that in the WT strain grown on glucose, and this derepression was not observed during growth on acetate (Fig. 8). This result indicates that icl1 is under the negative regulation of Crp1 in M. smegmatis during growth on glucose. Since Crp is a regulatory protein that controls gene expression in response to changes in cAMP levels, we determined whether the intracellular level of cAMP in M. smegmatis is changed depending on the carbon source in growth medium. As shown in Fig. 9, the cellular levels of cAMP in the WT strain of M. smegmatis grown on glucose and octanoate were found to be 1.7- and 1.8-fold higher than that in the same strain grown on acetate, respectively, which suggests that increased levels of cAMP might lead to Crp-mediated repression of icl1 expression in M. smegmatis during growth on glucose or octanoate.
FIG 8.
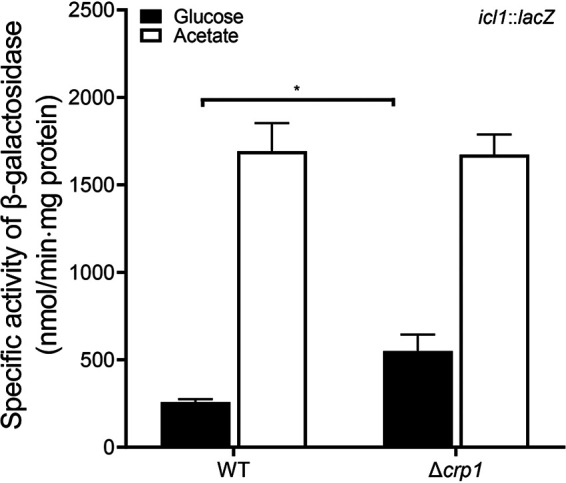
Expression levels of the icl1 gene in the WT and Δcrp1 mutant strains of M. smegmatis. The WT and Δcrp1 mutant strains containing the icl1::lacZ translational fusion plasmid pEMIIicl1 were grown aerobically to an OD600 of 0.45 to 0.5 in 7H9 medium supplemented with 10 mM glucose or 10 mM acetate as the sole carbon source. Cell-free crude extracts were used to measure β-galactosidase activity. All values provided were determined from three biological replicates. The error bars indicate the standard deviations. *, P < 0.01.
FIG 9.
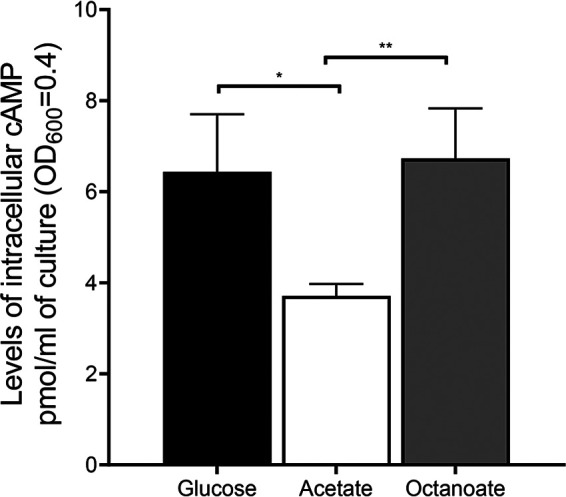
Effects of the carbon source on intracellular levels of cAMP in M. smegmatis. The WT strain of M. smegmatis was grown aerobically to an OD600 of 0.45 to 0.5 in 7H9 medium supplemented with 10 mM glucose, 10 mM acetate, or 2.5 mM octanoate as the sole carbon source. cAMP quantification was performed as described in Materials and Methods. All values provided are the average of the results from three biological replicates. The error bars indicate the standard deviations. *, P < 0.05; **, P < 0.01.
DISCUSSION
In this study, we demonstrated that ICL1 functions as the major ICL in the glyoxylate shunt that is required for growth of M. smegmatis on acetate or fatty acid as the sole carbon source, which is probably due to the very marginal expression of icl2 relative to icl1. In contrast to the constitutive expression of icl2, expression of icl1 was shown to be strongly (∼7-fold) induced in M. smegmatis when acetate was used as the sole carbon source. This result is consistent with the previous report that 8-fold-higher ICL activity was detected in M. smegmatis grown on acetate than in M. smegmatis grown on glucose (31). In C. glutamicum and M. tuberculosis, the aceA and icl1 genes are also upregulated during growth on acetate, which is achieved by RamB-mediated negative regulation in the absence of acetate (13, 19). In contrast to the RamB repressor in M. tuberculosis, the RamB homolog of M. smegmatis serves as an activator for icl1 expression in the presence and absence of acetate despite a high (86%) sequence identity between the RamB proteins of M. smegmatis and M. tuberculosis. Using DNase I footprinting analysis, we identified three RamB-binding sites (RamBS1, RamBS2, and RamB3), two (RamBS1 and RamBS2) of which contain the sequences similar to the known RamB-binding motif (AA/GAACTTTGCAAA) (19). In the absence of succinyl-CoA, RamB was shown to preferentially bind to RamBS1, indicating that RamBS1 is the high-affinity binding site for RamB. Importantly, the presence of succinyl-CoA was shown to cause the binding of RamB to two additional sites, RamBS2 and RamBS3, as well as an increase in the binding affinity of RamB for RamBS1. The binding affinity of RamB for RamBS3 is noticeably weaker than that for RamBS1 and RamBS2 in the presence of succinyl-CoA, and RamBS3 overlaps the TSP of icl1. Therefore, it is unlikely that RamBS3 is involved in the positive regulation of icl1. We found that mutations in RamBS2 resulted in a more severe defect in icl1 expression than those in RamBS1. The RamBS2 site was found to be located at positions −65 to −42 relative to the TSP of icl1, which is in line with the widely accepted principle that transcriptional activators generally bind upstream of their target promoters between positions −30 and −80 to enhance the binding of RNA polymerase to the promoters or the formation of an open complex (32, 33). Given both the location of RamBS2 and the result of site-directed mutagenesis on RamBS1 and RamBS2 (Fig. 7), it can be assumed that binding of RamB to RamBS2 might be directly involved in transcriptional activation of icl1 and that RamB bound at the high-affinity binding site RamBS1 might facilitate binding of RamB to RamBS2. Given that the centers of RamBS1 and RamBS2 are separated by three helical turns (31 bp), two RamB proteins are expected to bind to RamBS1 and RamBS2 on the same phase of DNA, which probably enables the cooperative binding of RamB to RamBS1 and RamBS2. Taken together, the results of DNase I footprinting and mutagenesis analyses indicate that succinyl-CoA serves as a coinducer for the RamB activator in the regulation of icl1 expression. It is especially noteworthy that the positions of three RamB-binding sites relative to the TSP as well as the icl1 promoter sequences are also well conserved upstream of icl1 in M. tuberculosis (see Fig. S5 in the supplemental material). We cannot explain why expression of icl1 is not positively but negatively regulated by RamB in M. tuberculosis despite the conservation of the cis-acting regulatory elements for icl1 in M. smegmatis and M. tuberculosis. To explain this disparity, a reassessment of the regulation of icl1 expression in M. tuberculosis appears to be necessary.
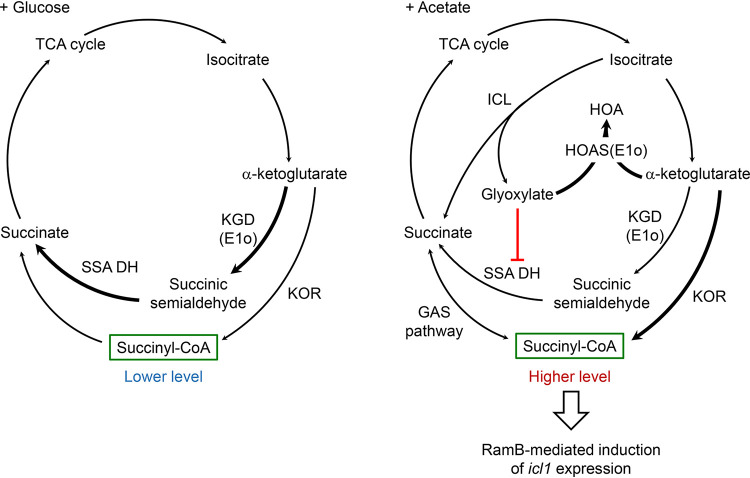
The cellular level of succinyl-CoA is determined by a balance between its synthesis and consumption for biosynthesis of cellular constituents. Succinyl-CoA can be produced from α-ketoglutarate (αKG) and succinate in the TCA cycle, as well as from methylmalonyl-CoA by methylmalonyl-CoA mutase in propionyl-CoA metabolism (34, 35). The oxidation of αKG to succinate in the TCA cycle of M. smegmatis and M. tuberculosis has been suggested to occur in two separate metabolic routes. αKG is oxidized to succinate via succinyl-CoA through two sequential reactions catalyzed by αKG:ferredoxin (Fd) oxidoreductase and succinyl-CoA synthetase (35). The conventional αKG dehydrogenase complex composed of E1o, E2 (DlaT or SucB), and E3 (Lpd) has been suggested to play, at most, a minor role in the oxidation of αKG in the TCA cycle of M. smegmatis and M. tuberculosis (36, 37). As an alternative route for the oxidation of αKG to succinate, E1o alone without association with E2 and E3 catalyzes decarboxylation of αKG to succinic semialdehyde (SSA), which is subsequently oxidized to succinate by SSA dehydrogenase (37). In the presence of glyoxylate, the E1o protein was demonstrated to function mainly as a carboligase that catalyzes the ligation of αKG and glyoxylate with the concomitant release of CO2 to yield 2-hydroxy-3-oxoadipate (HOA) (38), indicating that E1o serves as an HOA synthase or αKG decarboxylase, depending on the presence or absence of glyoxylate. Furthermore, it was suggested that αKG decarboxylase is predominantly utilized in oxidation of αKG to succinate in the TCA cycle when M. smegmatis is grown on glucose or glycerol, while αKG:Fd oxidoreductase was suggested to be important for mycobacterial growth under conditions that requires the glyoxylate shunt at ambient CO2 levels (35, 39). Additionally, the GAS (glyoxylate shunt, anaplerotic fixation of CO2, and succinyl-CoA synthetase for the generation of succinyl-CoA) pathway was suggested to operate in mycobacteria, which connects the glyoxylate shunt with the generation of succinyl-CoA (40). Carbon flux through the glyoxylate shunt was shown to be negligible in M. smegmatis grown on glucose (31), which is in line with our observation that the WT and Δicl1 Δicl2 mutant strains of M. smegmatis showed the same growth rate when they were grown on glucose (Fig. 1). The cellular levels of glyoxylate and succinate were demonstrated to rise in M. smegmatis when icl1 expression was increased (Fig. S4) (41). Taken together, the above-described findings allow us to present a model for RamB-dependent induction of icl1 expression in M. smegmatis grown on acetate (Fig. 10). When M. smegmatis is grown on acetate, the glyoxylate shunt is required for anaplerosis of the TCA cycle intermediates. As a result of increased carbon flux through the glyoxylate shunt, the cellular level of glyoxylate rises, which is expected to make E1o preferentially function as an HOA synthase rather than as an αKG decarboxylase. Due to the diminished αKG decarboxylase activity and inhibition of SSA dehydrogenase by glyoxylate (42), the oxidation of αKG is likely to proceed mainly by the succinyl-CoA-generating αKG:Fd oxidoreductase in the TCA cycle. On the other hand, the glyoxylate shunt is not necessary for growth of M. smegmatis on glucose, and thus, carbon flux through the glyoxylate shunt is minimal. The marginal cellular level of glyoxylate allows E1o to serve as an αKG decarboxylase that converts αKG to SSA. As suggested previously (35), αKG decarboxylase, which does not produce succinyl-CoA, is predominantly utilized in oxidation of αKG to succinate in the TCA cycle under these conditions. Therefore, the cellular level of succinyl-CoA is assumed to be lower in M. smegmatis during growth on glucose than in M. smegmatis grown on acetate wherein αKG:Fd oxidoreductase and the GAS pathway produce succinyl-CoA. The increased cellular level of succinyl-CoA in M. smegmatis during growth on acetate enables RamB-mediated induction of icl1 expression.
FIG 10.
Model for RamB-mediated induction of icl1 expression in M. smegmatis during growth on acetate. The simplified TCA cycle and glyoxylate shunt in M. smegmatis grown on glucose (+ Glucose) or acetate (+ Acetate) are depicted. When two or more alternative pathways are possible, the major pathways are indicated by thick arrows. Succinyl-CoA, which was used as a coinducer of RamB, is highlighted by green boxes. Abbreviations: HOA, 2-hydroxy-3-oxoadipate; HOAS, HOA synthase; ICL, isocitrate lyase; KOR, α-ketoglutarate:ferredoxin oxidoreductase; KGD, α-ketoglutarate decarboxylase; SSA DH, succinic semialdehyde dehydrogenase.
Expression of icl1 in M. tuberculosis has been demonstrated to be induced under respiration-inhibitory conditions such as hypoxic and low-pH conditions (9, 15–17). Inhibition of the respiratory electron transport chain shifts the redox state of the NADH/NAD+ pool toward a more reduced state (43). Under these conditions, it could be possible that the glyoxylate shunt is favored to bypass the oxidative decarboxylation reactions of the TCA cycle between isocitrate and succinate to maintain NADH/NAD+ homeostasis, which might lead to an increase in carbon flux via the glyoxylate shunt, followed by induction of icl1 expression.
Our RNA sequencing analysis (30) and reporter gene assay revealed that expression of icl1 was derepressed in the Δcrp1 mutant relative to that in the WT strain when the strains were grown on glucose (Fig. 8). This derepression did not occur during growth on acetate. Since we failed to obtain a Δcrp1 Δcrp2 double mutant of M. smegmatis, the involvement of Crp in the regulation of icl1 expression was examined in the Δcrp1 mutant, in which the crp1 gene encoding the major Crp is inactivated. Due to the presence of Crp2 in the Δcrp1 mutant, the extent of icl1 repression by Crp in the WT strain of M. smegmatis grown on glucose might be greater than that extrapolated from the result in Fig. 8 using the Δcrp1 mutant. The cellular level of cAMP in the WT strain grown on glucose or octanoate was shown to be approximately 2-fold higher than that in the WT grown on acetate (Fig. 9). Increased cAMP levels in glucose-grown cells are expected to enhance the functionality of Crp, leading to repression of icl1 in glucose-grown M. smegmatis. Both a decrease in the cellular level of cAMP in acetate-grown cells relative to glucose-grown cells and the Crp (GlxR)-mediated negative regulation of the ICL gene have been previously reported for C. glutamicum (44, 45). Taken together, our findings suggest that the strong induction of icl1 expression during growth on acetate as the sole carbon source relative to the weak expression of icl1 in M. smegmatis grown on glucose is likely to result from combined effects of Crp-mediated repression of icl1 in the presence of glucose and RamB-mediated induction of icl1 in the presence of acetate. It is also conceivable that Crp might be to some extent responsible for the weak induction of icl1 expression in M. smegmatis grown on octanoate relative to M. smegmatis grown on acetate.
It has been demonstrated in C. glutamicum that the LuxR-type transcriptional regulator RamA (Cg2831: regulator of acetate metabolism A) serves as an activator in the regulation of genes involved in acetate metabolism such as aceA, pta, and ack (46). M. smegmatis possesses a RamA homolog (MSMEG_5651) that shares 38% sequence identity and 57% similarity with RamA of C. glutamicum. We constructed a ramA deletion mutant of M. smegmatis and comparatively examined icl1 expression in the WT and ΔramA mutant strains. Inactivation of ramA did not result in changes in icl1 expression in M. smegmatis grown on glucose or acetate, indicating that RamA is not involved in the regulation of icl1 expression in M. smegmatis (Fig. S6).
Repression of icl1 expression under particular conditions, such as inorganic phosphate (Pi)- or nitrogen-limiting conditions, has been recently reported for M. smegmatis. The icl1 gene was demonstrated to be under the negative regulation of the Pi-sensing SenX3-RegX3 two-component system (47). We found that the suggested RegX3-binding operator is located immediately upstream of RamBS1, which implies that RegX3 bound at the operator might interfere with binding of RamB to RamBS1, leading to repression of icl1 expression (Fig. S7). Analogously, GlnR, an orphan response regulator, was reported to negatively regulate icl1 expression in M. smegmatis under nitrogen-limiting conditions. The reported GlnR-binding site overlaps RamBS2, indicating that GlnR represses icl1 expression by occluding RamB binding to RamBS2 (41).
In conclusion, expression of the icl1 gene encoding the major isocitrate lyase in M. smegmatis is regulated by RamB, which serves as the main regulator reflecting the cellular necessity of the glyoxylate shunt. Additionally, the cellular levels of cAMP as well as the availability of nitrogen sources and Pi are integrated by Crp1, GlnR, and the SenX3-RegX3 two-component system to exquisitely fine-tune expression of icl1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C. M. smegmatis strains were grown in Middlebrook 7H9 medium (Difco, Sparks, MD) supplemented with 0.2% (wt/vol) or 10 mM glucose, 10 mM acetate, or 2.5 mM octanoate as a carbon source and 0.02% (vol/vol) Tween 80 as an anticlumping agent at 37°C. M. smegmatis strains were grown aerobically in a 500-ml flask filled with 100 ml of growth medium on a gyratory shaker (200 rpm). Ampicillin (100 μg/ml for E. coli), chloramphenicol (34 μg/ml for E. coli), kanamycin (50 μg/ml for E. coli and 30 μg/ml for M. smegmatis), and hygromycin (200 μg/ml for E. coli and 50 or 25 μg/ml for M. smegmatis) were added to the growth medium when required. The construction of the mutants and the plasmids used in this study is described in the supplemental material.
DNA manipulation and electroporation.
Standard protocols and manufacturers’ instructions were followed for recombinant DNA manipulations (48). Transformation of M. smegmatis with plasmids was carried out by electroporation as described elsewhere (49). The primers used for PCR and site-directed mutagenesis are listed in Table S2.
β-Galactosidase assay.
The β-galactosidase activity was measured spectrophotometrically as described previously (50). The protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as a standard protein.
Protein purification.
The E. coli BL21-Codonplus (DE3) strain harboring pET29bramB was grown aerobically at 37°C in LB medium containing 50 μg/ml of kanamycin and 34 μg/ml of chloramphenicol to an optical density at 600 nm (OD600) of 0.6 to 0.7. Expression of the ramB genes was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to the cultures to a final concentration of 0.5 mM, and then cells were further grown for 6 h at 37°C. Cells were harvested from 500-ml cultures and resuspended in 10 ml of buffer A (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, and 1 mM dithiothreitol [DTT]) containing DNase I (10 U/ml) and 10 mM MgCl2. The resuspended cells were disrupted twice using a French pressure cell, and cell-free crude extracts were obtained by centrifugation twice at 20,000 × g for 15 min. The crude extracts were loaded into a column packed with 500 μl of the 80% (vol/vol) slurry of Ni-Sepharose high-performance resin (GE Healthcare, Piscataway, NJ). The resin was washed with 40 bed volumes of buffer A containing 5 mM imidazole and washed further with 40 bed volumes of buffer A containing 75 mM imidazole. His6-tagged RamB was eluted from the resin with 6 bed volumes of buffer A containing 250 mM imidazole and 10% (vol/vol) glycerol. Imidazole and NaCl were removed from purified RamB by means of a PD-10 desalting column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 8.0) containing 1 mM DTT and 10% (vol/vol) glycerol. His6-tagged RamB was purified to near homogeneity (Fig. S8).
DNase I footprinting analysis.
DNase I Footprinting was carried out using fluorescence (6-carboxytetramethylrhodamine [TAMRA])-labeled DNA fragments and purified RamB. A 369-bp TAMRA-labeled DNA fragment containing the icl1 upstream region was generated by PCR using the F_TAMRA_pUC19 and R_icl1FootF primers. The pUC19icl1FootF and pUC19icl1FootR plasmids were used for PCR as templates to generate DNA fragments with the TAMRA-labeled coding and noncoding strands, respectively. The PCR products were purified after agarose gel electrophoresis, and the DNA concentration was determined using a Multiskan SkyHigh microplate spectrophotometer (Thermo Fisher, Waltham, MA). DNA binding reaction mixtures were composed of 5 pmol of labeled DNA probes, various amounts of purified RamB, 20 mM Tris-HCl (pH 8.0), 0.2 mM MgCl2, 2.1 mM KCl, 0.04 mM DTT, and 11.1% (vol/vol) glycerol in a final volume of 190 μl. When necessary, the RamB protein was treated with 200 μM succinyl-CoA. The mixture was incubated for 10 min at 25°C prior to DNase I treatment. DNase I treatment, DNA purification, and electrophoresis on 6% (wt/vol) denaturing polyacrylamide gels were performed as described previously (30). Reference sequencing was performed by using a Thermo Sequenase dye primer manual cycle sequencing kit (Thermo Fisher) with the primer F_TAMRA_pUC19 and the template plasmid (pUC19icl1FootF or pUC19icl1FootR).
EMSA.
A 184-bp DNA fragment containing the upstream region of icl1 and a 145-bp control DNA fragment without the RamB-binding site were used in EMSA. The 184-bp DNA fragment was amplified by PCR using pUC19icl1FootF as a template and the F_icl1EMSA and R_icl1EMSA primers. The 145-bp control DNA fragment was generated by PCR using pUC19 as a template and the F_EMSAns and R_EMSAns primers. Purified RamB protein was incubated with 90 fmol of the DNA fragments containing the icl1 upstream region and 90 fmol of the control DNA fragments in binding buffer (20 mM Tris-HCl [pH 8.0], 2.5 mM MgCl2, and 1 mM DTT) in a reaction volume of 10 μl for 20 min at 25°C. To examine the effect of succinyl-CoA, succinate, acetate, and acetyl phosphate on the binding of RamB to the DNA fragments, 200 μM succinyl-CoA, succinate, acetate, and acetyl phosphate were included in the DNA-protein mixtures. After the addition of 2 μl of 6× loading buffer (0.25% [wt/vol] bromophenol blue, 0.25% [wt/vol] xylene cyanol, and 40% [wt/vol] sucrose), the mixtures were subjected to nondenaturing PAGE (6% [wt/vol] acrylamide) using 0.5× TBE buffer (41.5 mM Tris-borate [pH 8.3] and 0.5 mM EDTA) at 70 V for 1 h 45 min at 4°C. The gels were stained with SYBR green staining solution (Invitrogen, Waltham, MA) for 1 h.
Determination of the intracellular cAMP concentration.
M. smegmatis cells corresponding to 1 ml of cultures at an OD600 of 0.4 were harvested. Cell pellets were resuspended in 1 ml of sample diluent concentrate buffer (Arbor Assays, Ann Arbor, MI) and then incubated for 10 min. Cells were disrupted once by using a Fastprep 120 bead beater (Thermo Fisher, Waltham, MA) at 5.0 m/s for 45 s. Cell-free supernatants were obtained by centrifugation at 20,000 × g for 10 min. The concentration of cAMP in the prepared supernatants was determined by using a DetectX cyclic AMP direct immunoassay kit (Arbor Assays) and a microplate reader (Thermo Fisher) following the manufacturers’ instructions.
NMR analysis.
M. smegmatis cells were grown aerobically to an OD600 of 0.5 in 7H9 medium supplemented with 10 mM glucose or 10 mM acetate. Fifty milliliters of culture was harvested by centrifugation at 5,000 × g for 5 min at 4°C. Extraction of intracellular metabolites was performed as described previously (51). Briefly, the cell pellets were resuspended in 1 ml of 95% acetonitrile and 25 mM formic acid and disrupted by vigorous vortexing 10 times for 1 min. Samples were chilled on ice between vortexings for 5 min. Cell-free extracts were obtained following centrifugation at 15,000 × g for 10 min at 4°C. The obtained extracts were mixed with 10 ml of deionized water. To retrieve the residual metabolites from the cell pellet, the cell pellets were washed with 8 ml of deionized water once. After centrifugation at 15,000 × g for 10 min at 4°C, the obtained supernatants were mixed with the extracts and subjected to lyophilization. The lyophilized samples were redissolved using 500 μl of resuspension buffer (25 mM ammonium formate [pH 3.0], 2% methanol). For nuclear magnetic resonance (NMR) measurement, 20-μl volumes of redissolved samples were placed in a 4-mm NMR nanotube (Agilent; 4-mm sample tube) with 20 μl of phosphate buffer (pH 7.4 in deuterated water) containing 4 mM sodium 3-trimethylshily-2,2,3,3-d4 -propionate (TSP-d4) as a reference of chemical shift (0.00 ppm) and quantification. 1H NMR spectra were obtained using a 600-MHz high-resolution magic-angle spinning (HR-MAS) NMR spectrometer with a nano-NMR probe (Agilent Technologies, Santa Clara, CA) at a degree of magic angle of 54.74° and a spinning rate of 2,000 Hz. For NMR data of each sample, the Carr-Purcell-Meiboom-Gill (CPMG) pulse was applied, and spectra were acquired using a 3-s relaxation delay time and 3-s acquisition time at a temperature of 298 K. A total of 128 scans were collected, and total acquisition time was 13 min 9 s. All spectra were processed and assigned using Chenomx NMR Suite 8.4 professional with the Chenomx 600-MHz library database (Chenomx Inc., Edmonton, Canada).
ACKNOWLEDGMENT
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2020R1A2C1005305).
Footnotes
Supplemental material is available online only.
Contributor Information
Jeong-Il Oh, Email: joh@pusan.ac.kr.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Dolan SK, Welch M. 2018. The glyoxylate shunt, 60 years on. Annu Rev Microbiol 72:309–330. 10.1146/annurev-micro-090817-062257. [DOI] [PubMed] [Google Scholar]
- 2.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 3.Munoz-Elias EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med 11:638–644. 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz-Elias EJ, McKinney JD. 2006. Carbon metabolism of intracellular bacteria. Cell Microbiol 8:10–22. 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Elias EJ, Upton AM, Cherian J, McKinney JD. 2006. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol Microbiol 60:1109–1122. 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 6.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. 2009. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol 10:943–948. 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upton AM, McKinney JD. 2007. Role of the methylcitrate cycle in propionate metabolism and detoxification in Mycobacterium smegmatis. Microbiology (Reading) 153:3973–3982. 10.1099/mic.0.2007/011726-0. [DOI] [PubMed] [Google Scholar]
- 8.Gould TA, van de Langemheen H, Munoz-Elias EJ, McKinney JD, Sacchettini JC. 2006. Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol Microbiol 61:940–947. 10.1111/j.1365-2958.2006.05297.x. [DOI] [PubMed] [Google Scholar]
- 9.Eoh H, Rhee KY. 2014. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc Natl Acad Sci USA 111:4976–4981. 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhusal RP, Jiao W, Kwai BXC, Reynisson J, Collins AJ, Sperry J, Bashiri G, Leung IKH. 2019. Acetyl-CoA-mediated activation of Mycobacterium tuberculosis isocitrate lyase 2. Nat Commun 10:4639. 10.1038/s41467-019-12614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chopra T, Hamelin R, Armand F, Chiappe D, Moniatte M, McKinney JD. 2014. Quantitative mass spectrometry reveals plasticity of metabolic networks in Mycobacterium smegmatis. Mol Cell Proteomics 13:3014–3028. 10.1074/mcp.M113.034082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honer Zu Bentrup K, Miczak A, Swenson DL, Russell DG. 1999. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J Bacteriol 181:7161–7167. 10.1128/JB.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micklinghoff JC, Breitinger KJ, Schmidt M, Geffers R, Eikmanns BJ, Bange FC. 2009. Role of the transcriptional regulator RamB (Rv0465c) in the control of the glyoxylate cycle in Mycobacterium tuberculosis. J Bacteriol 191:7260–7269. 10.1128/JB.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham JE, Clark-Curtiss JE. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc Natl Acad Sci USA 96:11554–11559. 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb) 84:239–246. 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Saxena A, Srivastava V, Srivastava R, Srivastava BS. 2008. Identification of genes of Mycobacterium tuberculosis upregulated during anaerobic persistence by fluorescence and kanamycin resistance selection. Tuberculosis (Edinb) 88:518–525. 10.1016/j.tube.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Tang YJ, Shui W, Myers S, Feng X, Bertozzi C, Keasling JD. 2009. Central metabolism in Mycobacterium smegmatis during the transition from O2-rich to O2-poor conditions as studied by isotopomer-assisted metabolite analysis. Biotechnol Lett 31:1233–1240. 10.1007/s10529-009-9991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stackebrandt E, Rainey FA, Ward-Rainey NL. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Evol Microbiol 47:479–491. 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- 19.Gerstmeir R, Cramer A, Dangel P, Schaffer S, Eikmanns BJ. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J Bacteriol 186:2798–2809. 10.1128/JB.186.9.2798-2809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auchter M, Cramer A, Huser A, Ruckert C, Emer D, Schwarz P, Arndt A, Lange C, Kalinowski J, Wendisch VF, Eikmanns BJ. 2011. RamA and RamB are global transcriptional regulators in Corynebacterium glutamicum and control genes for enzymes of the central metabolism. J Biotechnol 154:126–139. 10.1016/j.jbiotec.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Xie L, Wang X, Zeng J, Zhou M, Duan X, Li Q, Zhang Z, Luo H, Pang L, Li W, Liao G, Yu X, Li Y, Huang H, Xie J. 2015. Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int J Biochem Cell Biol 59:193–202. 10.1016/j.biocel.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M, Xie L, Yang Z, Zhou J, Xie J. 2017. Lysine succinylation of Mycobacterium tuberculosis isocitrate lyase (ICL) fine-tunes the microbial resistance to antibiotics. J Biomol Struct Dyn 35:1030–1041. 10.1080/07391102.2016.1169219. [DOI] [PubMed] [Google Scholar]
- 23.Bi J, Wang Y, Yu H, Qian X, Wang H, Liu J, Zhang X. 2017. Modulation of central carbon metabolism by acetylation of isocitrate lyase in Mycobacterium tuberculosis. Sci Rep 7:44826. 10.1038/srep44826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 25.Lee HN, Ji CJ, Lee HH, Park J, Seo YS, Lee JW, Oh JI. 2018. Roles of three FurA paralogs in the regulation of genes pertaining to peroxide defense in Mycobacterium smegmatis mc2 155. Mol Microbiol 108:661–682. 10.1111/mmi.13956. [DOI] [PubMed] [Google Scholar]
- 26.Masiewicz P, Brzostek A, Wolański M, Dziadek J, Zakrzewska-Czerwińska J. 2012. A novel role of the PrpR as a transcription factor involved in the regulation of methylcitrate pathway in Mycobacterium tuberculosis. PLoS One 7:e43651. 10.1371/journal.pone.0043651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang S, Hicks ND, Cheng YS, Silva A, Fortune SM, Sacchettini JC. 2019. Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor. Nucleic Acids Res 47:9934–9949. 10.1093/nar/gkz724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma R, Zaveri A, Gopalakrishnapai J, Srinath T, Thiruneelakantan S, Varshney U, Visweswariah SS. 2014. Paralogous cAMP receptor proteins in Mycobacterium smegmatis show biochemical and functional divergence. Biochemistry 53:7765–7776. 10.1021/bi500924v. [DOI] [PubMed] [Google Scholar]
- 29.Aung HL, Dixon LL, Smith LJ, Sweeney NP, Robson JR, Berney M, Buxton RS, Green J, Cook GM. 2015. Novel regulatory roles of cAMP receptor proteins in fast-growing environmental mycobacteria. Microbiology (Reading) 161:648–661. 10.1099/mic.0.000015. [DOI] [PubMed] [Google Scholar]
- 30.Ko EM, Oh JI. 2020. Induction of the cydAB operon encoding the bd quinol oxidase under respiration-inhibitory conditions by the major cAMP receptor protein MSMEG_6189 in Mycobacterium smegmatis. Front Microbiol 11:608624. 10.3389/fmicb.2020.608624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murima P, Zimmermann M, Chopra T, Pojer F, Fonti G, Dal Peraro M, Alonso S, Sauer U, Pethe K, McKinney JD. 2016. A rheostat mechanism governs the bifurcation of carbon flux in mycobacteria. Nat Commun 7:12527. 10.1038/ncomms12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raibaud O, Schwartz M. 1984. Positive control of transcription initiation in bacteria. Annu Rev Genet 18:173–206. 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 33.Collado-Vides J, Magasanik B, Gralla JD. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev 55:371–394. 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner T, Bellinzoni M, Wehenkel A, O’Hare HM, Alzari PM. 2011. Functional plasticity and allosteric regulation of alpha-ketoglutarate decarboxylase in central mycobacterial metabolism. Chem Biol 18:1011–1020. 10.1016/j.chembiol.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Baughn AD, Garforth SJ, Vilcheze C, Jacobs WR, Jr.. 2009. An anaerobic-type alpha-ketoglutarate ferredoxin oxidoreductase completes the oxidative tricarboxylic acid cycle of Mycobacterium tuberculosis. PLoS Pathog 5:e1000662. 10.1371/journal.ppat.1000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian J, Bryk R, Shi S, Erdjument-Bromage H, Tempst P, Nathan C. 2005. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol Microbiol 57:859–868. 10.1111/j.1365-2958.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 37.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci USA 102:10670–10675. 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. 2010. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol 17:1122–1131. 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 39.O’Hare HM, Duran R, Cervenansky C, Bellinzoni M, Wehenkel AM, Pritsch O, Obal G, Baumgartner J, Vialaret J, Johnsson K, Alzari PM. 2008. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol 70:1408–1423. 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 40.Beste DJ, Bonde B, Hawkins N, Ward JL, Beale MH, Noack S, Noh K, Kruger NJ, Ratcliffe RG, McFadden J. 2011. 13C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog 7:e1002091. 10.1371/journal.ppat.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi N, She GL, Du W, Ye BC. 2021. Mycobacterium smegmatis GlnR regulates the glyoxylate cycle and the methylcitrate cycle on fatty acid metabolism by repressing icl transcription. Front Microbiol 12:603835. 10.3389/fmicb.2021.603835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakoby WB, Scott EM. 1959. Aldehyde oxidation. III. Succinic semialdehyde dehydrogenase. J Biol Chem 234:937–940. 10.1016/S0021-9258(18)70207-X. [DOI] [PubMed] [Google Scholar]
- 43.Jeong JA, Park SW, Yoon D, Kim S, Kang HY, Oh JI. 2018. Roles of alanine dehydrogenase and induction of its gene in Mycobacterium smegmatis under respiration-inhibitory conditions. J Bacteriol 200:e00152-18. 10.1128/JB.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HJ, Kim TH, Kim Y, Lee HS. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J Bacteriol 186:3453–3460. 10.1128/JB.186.11.3453-3460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park SY, Moon MW, Subhadra B, Lee JK. 2010. Functional characterization of the glxR deletion mutant of Corynebacterium glutamicum ATCC 13032: involvement of GlxR in acetate metabolism and carbon catabolite repression. FEMS Microbiol Lett 304:107–115. 10.1111/j.1574-6968.2009.01884.x. [DOI] [PubMed] [Google Scholar]
- 46.Cramer A, Gerstmeir R, Schaffer S, Bott M, Eikmanns BJ. 2006. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J Bacteriol 188:2554–2567. 10.1128/JB.188.7.2554-2567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei JF, Qi N, Li YX, Wo J, Ye BC. 2021. RegX3-mediated regulation of methylcitrate cycle in Mycobacterium smegmatis. Front Microbiol 12:619387. 10.3389/fmicb.2021.619387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Green MR. 2012. Molecular cloning: a laboratory manual, 4th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR, Jr.. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 50.Oh JI, Kaplan S. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38:2688–2696. 10.1021/bi9825100. [DOI] [PubMed] [Google Scholar]
- 51.Glaser L, Kuhl M, Jovanovic S, Fritz M, Vogeli B, Erb TJ, Becker J, Wittmann C. 2020. A common approach for absolute quantification of short chain CoA thioesters in prokaryotic and eukaryotic microbes. Microb Cell Fact 19:160. 10.1186/s12934-020-01413-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download JB.00402-21-s0001.pdf, PDF file, 1.4 MB (1.4MB, pdf)