ABSTRACT
Francisella tularensis is a Gram-negative bacterium that causes a fatal human disease known as tularemia. The Centers for Disease Control and Prevention have classified F. tularensis as a category A tier 1 select agent. The virulence mechanisms of Francisella are not entirely understood. Francisella possesses very few transcription regulators, and most of these regulate the expression of genes involved in intracellular survival and virulence. The F. tularensis genome sequence analysis reveals an AraC (FTL_0689) transcriptional regulator homologous to the AraC/XylS family of transcriptional regulators. In Gram-negative bacteria, AraC activates genes required for l-arabinose utilization and catabolism. The role of the FTL_0689 regulator in F. tularensis is not known. In this study, we characterized the role of FTL_0689 in the gene regulation of F. tularensis and investigated its contribution to intracellular survival and virulence. The results demonstrate that FTL_0689 in Francisella is not required for l-arabinose utilization. Instead, FTL_0689 specifically regulates the expression of the oxidative and global stress response, virulence, metabolism, and other key pathways genes required by Francisella when exposed to oxidative stress. The FTL_0689 mutant is attenuated for intramacrophage growth and virulence in mice. Based on the deletion mutant phenotype, FTL_0689 was termed osrR (oxidative stress response regulator). Altogether, this study elucidates the role of the osrR transcriptional regulator in tularemia pathogenesis.
IMPORTANCE The virulence mechanisms of category A select agent Francisella tularensis, the causative agent of a fatal human disease known as tularemia, remain largely undefined. The present study investigated the role of a transcriptional regulator and its overall contribution to the oxidative stress resistance of F. tularensis. The results provide an insight into a novel gene regulatory mechanism, especially when Francisella is exposed to oxidative stress conditions. Understanding such Francisella- specific regulatory mechanisms will help identify potential targets for developing effective therapies and vaccines to prevent tularemia.
KEYWORDS: AraC/XylS, Francisella tularensis, oxidative stress, pathogenesis, transcriptional regulation, virulence
INTRODUCTION
Francisella tularensis is a Gram-negative bacterium that causes a fatal human disease known as tularemia. Based on its extreme virulence and potential to be used as a bioweapon or a bioterror agent, the Centers for Disease Control and Prevention have classified F. tularensis as category A tier 1 select agent (1). The genus Francisella is divided into two species, F. tularensis and F. philomiragia (2, 3). F. tularensis has four major subspecies; F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B) are virulent for humans, while the other two subsp., F. tularensis subsp. novicida and F. tularensis subsp. mediasiatica, do not cause disease in immunocompetent individuals (3–5). Type A F. tularensis strains are found in North America and Central Europe (6–8), while type B strains are less common in North America but are prevalent throughout Europe and Asia. Both type A and type B strains of F. tularensis can be transmitted through insect bites, direct contact with infected animals, ingestion of contaminated food or water, or inhaling bioaerosols. The live vaccine strain (LVS) is derived from F. tularensis subsp. holarctica.
F. tularensis is an intracellular bacterium and has a unique intracellular life cycle. Francisella can infect macrophages, neutrophils, dendritic cells, and several other cell types (9–12); however, macrophages are targeted primarily to initiate the infection. Upon phagocytosis, Francisella remains in phagosomes for a short duration, prevents the maturation of phagosomes, disintegrates the phagosomal wall, and escapes into the cytosol, where the replication occurs (12). The genes required to escape from the phagosome are encoded on a Francisella pathogenicity island (FPI) (13). The regulation of virulence mechanisms of Francisella is not entirely understood. Francisella possesses very few transcription regulators. The three well-characterized transcriptional regulators, the macrophage growth locus protein A (MglA), the stringent starvation protein A (SspA), and the pathogenicity island gene regulator (PigR), regulate the expression of genes encoded on the FPI (14–16). The other transcriptional regulators, PmrA and QseC, function as response regulators of the two-component system and regulate the expression of virulence-associated genes in F. tularensis (17, 18). Additionally, F. tularensis encodes specialized transcriptional regulators such as Fur, which regulate the expression of genes involved in iron uptake (19), and OxyR, which plays a central role in regulating the essential genes required for oxidative stress resistance and virulence (20).
The F. tularensis genome sequence analysis revealed a transcriptional regulator homologous to the AraC/XylS family of regulators encoded by the FTL_0689 gene. The AraC/XylS family of transcriptional regulators in several Gram-negative bacteria exhibit high amino acid sequence homology. They contain two unique and highly conserved helix-turn-helix DNA binding motifs. The first well-characterized AraC/XylS transcriptional regulator is the one that controls the l-arabinose operon in Escherichia coli (21, 22). It functions as an activator of the araBAD and other operons, the araFGH and araE (21), that are required for l-arabinose utilization and catabolism in E. coli. Like E. coli, the AraC regulator also functions in arabinose utilization in Erwinia chrysanthemum, Citrobacter freundii, and Salmonella (23, 24).
AraC/XylS regulators also regulate the expression of genes involved in metabolic processes, secretion of siderophores, urease, and virulence factors in several Gram-negative bacteria. Members of the AraC/XylS transcriptional regulators also regulate the bacterial responses to alkylating agents and oxidative and other stresses, including antibiotics (25). The role of the AraC/XylS regulator encoded by the FTL_0689 gene of F. tularensis is not known. In this study, we elucidated the role of FTL_0689 in the regulation of oxidative stress response and its contribution to intracellular survival and virulence of F. tularensis. We named FTL_0689 an oxidative stress response regulator (osrR) based on its deletion mutant phenotype. We report a novel role of osrR in the oxidative stress response of F. tularensis.
RESULTS
Sequence analysis, confirmation of gene deletion, and transcomplementation of the osrR gene of F. tularensis LVS.
The scanning of the PROSITE motif database (expasy.org) with the OsrR (FTL_0689) motif confirmed that osrR belongs to the AraC/XylS family of transcriptional regulators. The AraC/XylS family comprises over 260 members with a characteristic C-terminal helix-turn-helix DNA binding domain. Further bioinformatic analysis using the UniProtKB database did not identify additional hits or matches in the F. tularensis genome, indicating that Francisella possesses only one AraC-like transcriptional regulator. Multiple sequence alignment using ClustalW demonstrated that OsrR of F. tularensis has a conserved C-terminal domain with a consensus motif GXXXXXXFXXXXXXXXXXXP (GX6FX11P). This consensus sequence starts at amino acid 244 and ends at amino acid 263 of OsrR (see Fig. S1A in the supplemental material). The sequence comparison of osrR DNA and amino acid sequences within Francisella subspecies revealed high similarity, ranging from 98% to 99.65%. However, OsrR of F. tularensis LVS showed less homology when aligned with the transcriptional regulator present in other pathogenic Gram-negative bacteria. OsrR of F. tularensis exhibits 33%, 26%, 28%, and 20% amino acid sequence homology with the transcriptional regulators of E. coli, Salmonella enterica serovar Typhi, Yersinia enterocolitica, and Pseudomonas aeruginosa, respectively (Fig. S1B). Multiple Em for Motif Elicitation (MEME) software identified the C-terminal and N-terminal conserved sequences of OsrR protein of F. tularensis LVS similar to those observed in AraC of E. coli and VirF of Yersinia pestis (Fig. S1C). Collectively, bioinformatic analysis of OsrR of F. tularensis demonstrates conserved features of the AraC/XylS transcriptional regulators from other bacterial pathogens. The osrR gene deletion and transcomplementation were confirmed by PCR (Fig. S1D).
OsrR of F. tularensis is not required for arabinose utilization.
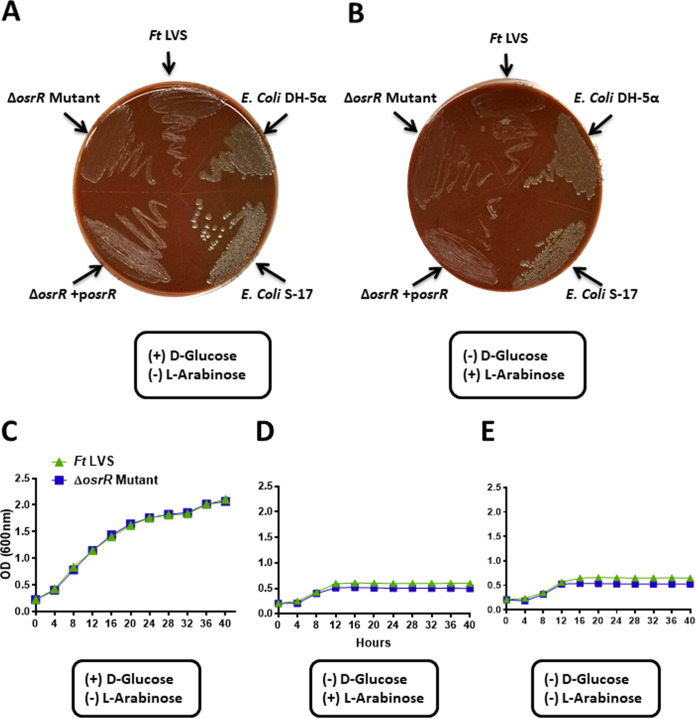
AraC activates araBAD and other operons required for l-arabinose utilization and catabolism in E. coli (21). To determine if OsrR of F. tularensis is required for l-arabinose utilization, we tested the ability of the ΔosrR mutant to grow in the presence of l-arabinose-containing media. We prepared Mueller-Hinton (MH)–chocolate agar plates containing 10 g (1%) of either the d-glucose or l-arabinose. The plates were then streaked with F. tularensis LVS, the ΔosrR mutant, or the transcomplemented strain. Two strains of E. coli, DH-5α and S17.1, were used as positive controls, as both these strains can grow in the presence of arabinose. Our results demonstrate that the ΔosrR mutant, the wild-type F. tularensis LVS, and the transcomplemented strain, as well as E. coli DH-5α and S17.1 strains, all grew sufficiently in the presence of d-glucose (Fig. 1A). Scant growth for the wild-type F. tularensis, the ΔosrR mutant, and the transcomplemented strain was observed when grown on MH-chocolate agar plates containing l-arabinose as the sole sugar source (Fig. 1B). These results indicated that either the Francisella strains poorly utilize l-arabinose or components of the MH-agar were supporting this scant growth. To address this notion, we generated bacterial growth curves using Chamberlain’s chemically defined medium (CDM) with three different compositions containing d-glucose only, l-arabinose only, and neither d-glucose nor l-arabinose. Both the ΔosrR mutant and the wild-type F. tularensis LVS grew very well in glucose-containing CDM (Fig. 1C). Both strains grew at a similar rate and reached an approximate optical density at 600 nm (OD600) of 2.4 after 40 h of growth. However, when the CDM was supplemented with l-arabinose only (Fig. 1D) or with no sugar (Fig. 1E), both the wild type and the mutant strains reached an OD600 of 0.5 after a short period of growth and then entered a stationary phase, indicating their growth was largely affected by the lack of glucose in the medium. Collectively, these results demonstrate that F. tularensis cannot utilize l-arabinose that and d-glucose is essential for its growth. Furthermore, these results also show that OsrR of F. tularensis LVS is not involved in l-arabinose metabolism.
FIG 1.
OsrR of F. tularensis is not required for arabinose utilization. (A and B) The wild-type F. tularensis (Ft) LVS, the ΔosrR mutant, and the transcomplemented strain (ΔosrR +posrR) were grown in MH-chocolate agar containing d-glucose (A) or l-arabinose (B). E. coli DH-5α and E. coli S17.1 were used as positive controls. (C to E) Growth curves of the wild-type F. tularensis LVS and the ΔosrR mutant in Chamberlain’s defined medium supplemented with d-glucose (C) or l-arabinose (D) or in the absence of both sugars (E). The results shown are representative of three independent experiments conducted with similar results.
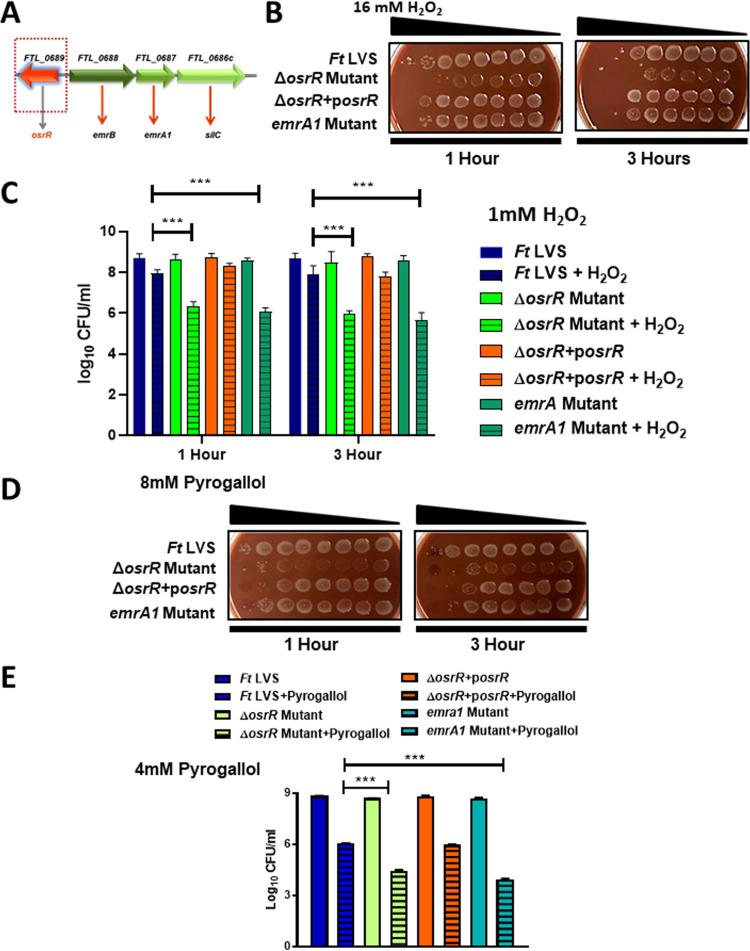
The ΔosrR mutant of F. tularensis is susceptible to oxidative stress.
The genomic organization revealed that the osrR of F. tularensis is divergently transcribed from the genes encoding major facilitator superfamily (MFS) type Emr multidrug efflux pump (MEP) genes—emrB, emrA1, and silC (Fig. 2A). Such a unique organization indicated that osrR might be involved in the regulation of these downstream genes. Both the emrA1 and silC mutants characterized previously exhibit extreme sensitivity to oxidants (26, 27). We hypothesized that if OsrR regulates the expression of MEP genes, the ΔosrR mutant should mirror the oxidant-sensitive phenotype of the emrA1 mutant of F. tularensis LVS. We investigated the sensitivity of the ΔosrR mutant to peroxides and superoxide (O2−.)-generating compounds. The emrA1 mutant was used for comparison. We performed a spot assay by exposing wild-type F. tularensis LVS, the ΔosrR mutant, the ΔosrR+posrR transcomplemented strain, and the emrA1 mutant to various concentrations of H2O2, incubating them for 1 and 3 h, and plating them on MH-chocolate agar to determine the bacterial viability. The results showed enhanced killing of the ΔosrR mutant when exposed to increasing concentrations of H2O2 for 1 and 3 h compared to the wild-type F. tularensis LVS. The emrA1 mutant showed enhanced killing compared to the wild-type F. tularensis LVS after 1 h of exposure to H2O2. However, the ΔosrR mutant exhibited a higher sensitivity to H2O2 than the emrA1 mutant. The transcomplementation restored the wild-type phenotype (Fig. 2B). Similar results were observed in a bacterial killing assay. Wild-type F. tularensis LVS, the ΔosrR mutant, the transcomplemented strain, and the emrA1 mutant (adjusted to an OD600 of 0.2) were exposed to 1 mM H2O2. The results showed a significant reduction in the numbers of ΔosrR and emrA1 mutant bacteria compared to the wild-type F. tularensis LVS after 1 and 3 h of exposure to H2O2. Transcomplementation restored the wild-type phenotype (Fig. 2C). As reported earlier for both the emrA1 and ΔsilC mutants of F. tularensis (26, 27), the ΔosrR mutant also showed enhanced sensitivity to the organic peroxides cumene hydroperoxide (CHP) and tert-butyl hydroquinone (TBH) compared to wild-type F. tularensis LVS (Fig. S2A and B).
FIG 2.
The ΔosrR mutant of F. tularensis is sensitive to hydrogen peroxide and pyrogallol. (A) Genomic organization of the osrR gene of F. tularensis LVS. (B) Wild-type F. tularensis (Ft) LVS, the ΔosrR mutant, the ΔosrR +posrR strain, and the emrA1 mutant were exposed to H2O2 diluted 2-fold from a starting concentration of 16 mM for 1 and 3 h in a spot assay. The results shown are representative of two independent experiments. (C) Bacterial killing assay; equal amounts of the indicated bacterial strains were either left untreated or treated with 1 mM H2O2 for 1 and 3 h. The cultures were diluted 10-fold and plated on MH-chocolate agar plates, and the colonies were counted. (D) Wild-type F. tularensis LVS, the ΔosrR mutant, the ΔosrR +posrR strain, and the emrA1 mutant were exposed to pyrogallol diluted 2-fold with a starting concentration of 8 mM for 1 h and 3 h in a spot assay. The results shown are representative of two independent experiments. (E) A bacterial killing assay was performed using 4 mM pyrogallol as described in panel C. The data for the bacterial killing assay experiments in panels C and E are represented as the mean ± SEM from three independent experiments, each conducted with three technical replicates. The data were analyzed using one-way ANOVA. **, P < 0.01; ***, P < 0.001.
We next tested the sensitivity of the ΔosrR mutant to the superoxide-generating compound pyrogallol and compared it with wild-type F. tularensis LVS and the emrA1 mutant. Enhanced killing of the ΔosrR mutant was observed upon exposure to increasing concentrations of pyrogallol, at 1 and 3 h postexposure in the spot assay, and bacterial killing assay compared to the wild-type F. tularensis LVS. The emrA1 mutant, as reported earlier, also showed enhanced sensitivity compared to the wild-type F. tularensis LVS after one and 3 h of exposure to pyrogallol in the spot and bacterial killing assays (Fig. 2D and E). The sensitivity of the ΔosrR mutant to the superoxide-generating compound paraquat, which generates superoxide radicals intracellularly, was also tested. However, the ΔosrR mutant did not exhibit any enhanced susceptibility compared to that observed for wild-type F. tularensis LVS (Fig. S2C). Similar results were obtained when the sensitivity of the ΔosrR mutant was tested against another superoxide-generating compound, menadione (Fig. S2D). Collectively, these results demonstrate that loss of osrR is associated with enhanced sensitivity of the ΔosrR mutant to oxidants such as H2O2, organic peroxides, and pyrogallol compared to wild-type F. tularensis LVS. These results also demonstrate that the ΔosrR mutant is more sensitive to H2O2 and pyrogallol than the emrA1 mutant.
OsrR regulates the expression of MEP genes under conditions of oxidative stress.
Our preceding results demonstrated that the oxidant-sensitive phenotype of the ΔosrR mutant mirrored the MEP gene mutant phenotype, specifically the emrA1 mutant. We hypothesized that the OsrR of Francisella might regulate the expression of MEP genes under oxidative stress conditions. Based on our observation that the ΔosrR mutant did not exhibit any enhanced sensitivity toward the superoxide-generating compound menadione, we chose menadione to induce the oxidative stress, as both the wild-type and the mutant bacteria exhibit similar menadione sensitivity. Wild-type F. tularensis LVS and the ΔosrR mutant bacteria were left untreated or treated with 1.25 mM menadione and incubated for 1 h at 37°C. The expression of emrB, emrA1, and silC genes in the ΔosrR mutant and the wild-type F. tularensis LVS was determined by quantitative real-time PCR (qRT-PCR). No changes in the expression of these genes were observed in the untreated ΔosrR mutant, and the levels remained similar to those observed for the wild-type F. tularensis LVS. However, upon treatment with menadione, the expression of all the MEP genes was significantly downregulated in the ΔosrR mutant compared to the wild-type F. tularensis LVS. The expression of these genes was restored to a level similar to those observed for the wild-type F. tularensis LVS in the transcomplemented strain (Fig. 3). Collectively, these results demonstrate that OsrR regulates the expression of the Emr MEP genes only under conditions of oxidative stress.
FIG 3.
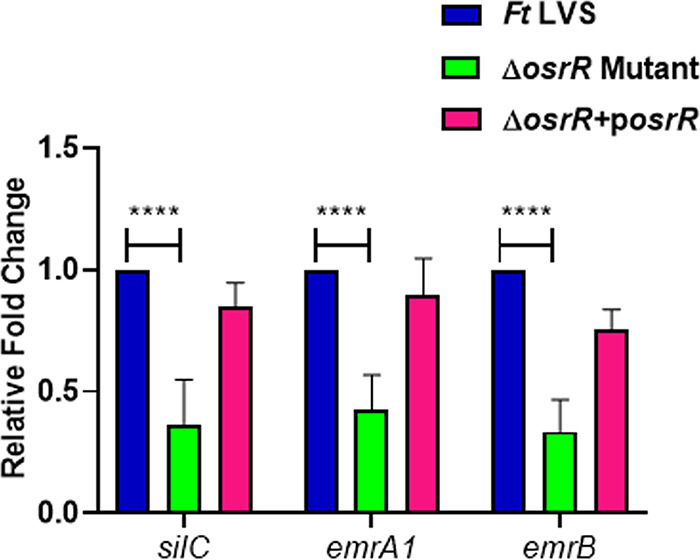
OsrR regulates the expression of Emr multidrug efflux pump (MEP) genes under conditions of oxidative stress. Wild-type F. tularensis (Ft) LVS, ΔosrR mutant, and ΔosrR+posrR transcomplemented strains were exposed to 1.25 mM menadione for 1 h to induce oxidative stress. The RNA was isolated and quantitated for the expression of MEP genes silC, emrA1, and emrB by qRT-PCR. The data are represented as the relative fold change compared to F. tularensis LVS. The data are represented as the mean ± SEM from three independent experiments, each conducted with three technical replicates and analyzed using one-way ANOVA. ****, P < 0.0001.
Except for osrR, expression of other genes is not significantly altered in the ΔosrR mutant in the absence of oxidative stress.
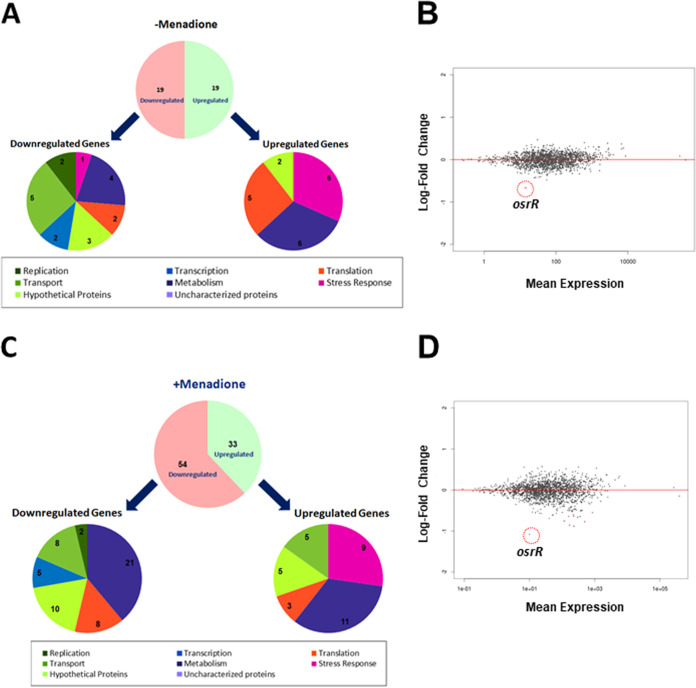
We performed RNA sequencing using the RNA isolated from untreated and menadione-treated F. tularensis LVS and the ΔosrR mutant to determine the differential expression of genes. A total of 38 genes were differentially expressed in the ΔosrR mutant in the absence of oxidative stress (Fig. 4A; Fig. S3A and B). The expression of the osrR gene (FTL_0689) was most significantly downregulated (–0.67 log2 fold change) (Fig. 4B and Fig. S3A) in the untreated ΔosrR mutant compared to the wild-type F. tularensis LVS. A total of 87 genes were differentially expressed in the ΔosrR mutant compared to F. tularensis LVS in the presence of oxidative stress induced by menadione. The expression of 33 genes was upregulated, whereas 54 genes were downregulated in the ΔosrR mutant (Fig. 4C; Fig. S4 and S5). The expression of the osrR gene was downregulated further (–1.08 log2 fold change) when exposed to oxidative stress caused by menadione compared to the level observed for the untreated ΔosrR mutant (Fig. 4D and Fig. S4). These results collectively, together with those observed for the MEP genes emrB, emrA1, and silC, demonstrate that OsrR may not be required for gene regulation of F. tularensis under normal growth conditions. However, OsrR does play a regulatory role under oxidative stress conditions. This notion was investigated further.
FIG 4.
Differential expression of genes in the ΔosrR mutant of F. tularensis LVS in the absence and presence of oxidative stress induced by menadione. (A and C) Profile of differentially expressed genes involved in various cellular functions in ΔosrR mutant compared to F. tularensis (Ft) LVS as determined by RNA sequence analysis in the absence (A) and presence (C) of oxidative stress induced by exposure to menadione. (B and D) Dot plots of gene expression profiles of differentially expressed genes in the ΔosrR mutant compared to those in F. tularensis LVS as determined by RNA sequence analysis in the absence (B) and presence (D) of oxidative stress induced by exposure to menadione. The data shown are cumulative of three independent RNA sequencing experiments.
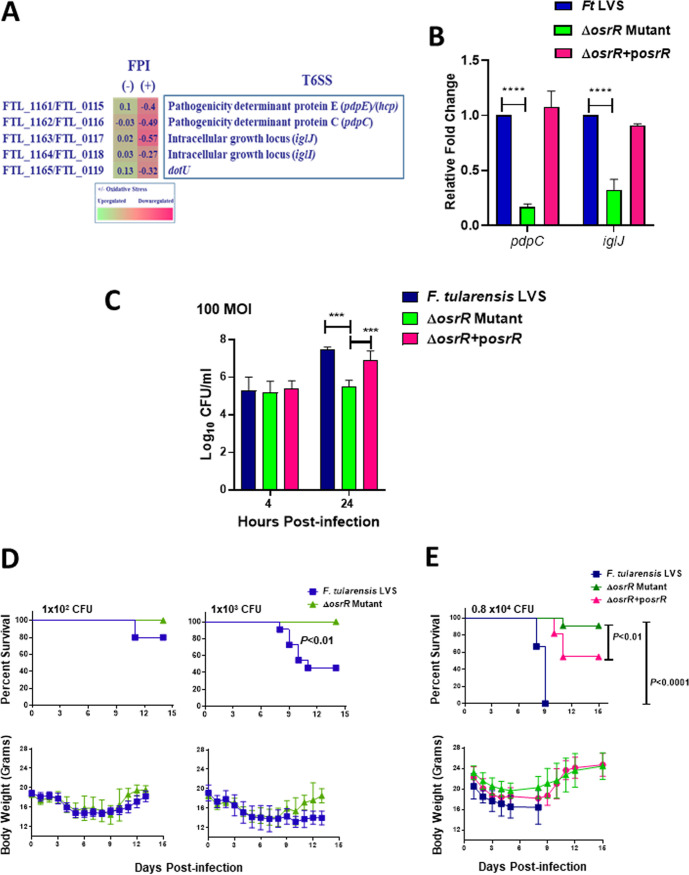
Exposure of the ΔosrR mutant to oxidative stress results in downregulation of genes encoded on the FPI.
We next determined the differentially expressed genes in the ΔosrR mutant compared to the wild-type F. tularensis LVS when the bacteria were exposed to oxidative stress for 1 h. The RNA sequencing results identified five downregulated genes, pdpE, pdpC, iglJ, iglI, and dotU, encoded within the FPI. The expression of these genes remained unaltered when the ΔosrR mutant was not exposed to oxidative stress (Fig. 5A). These FPI genes encode components of the type VI secretion system (T6SS) of F. tularensis (28–30). The qRT-PCR analysis confirmed the downregulated expression of two of the most downregulated genes, pdpC and iglJ, in the ΔosrR mutant in the presence of oxidative stress. Transcomplementation of the osrR gene in the ΔosrR mutant restored the expression levels of both pdpC and iglJ genes to levels similar to those observed for the wild-type F. tularensis LVS in the presence of oxidative stress (Fig. 5B). Overall, these results indicate that OsrR regulates the expression of FPI genes under oxidative stress conditions.
FIG 5.
Exposure of the ΔosrR mutant to oxidative stress results in downregulation of genes encoded on the Francisella pathogenicity island (FPI), attenuated intramacrophage survival, and virulence in mice. (A) Expression profile of FPI genes in the ΔosrR mutant in the absence of (–) or upon exposure to (+) menadione compared to the wild-type F. tularensis (Ft) LVS. The data shown are cumulative of three independent RNA sequencing experiments and expressed as the log2 fold change. (B) Quantitation of RNA transcripts of pdpC and iglJ genes in the indicated Francisella strains by qRT-PCR. The data are represented as the relative fold change compared to F. tularensis LVS. The data are represented as the mean ± SEM from three independent experiments, each conducted with three technical replicates and analyzed using one-way ANOVA. ****, P < 0.0001. (C) Attenuation of intramacrophage growth of the ΔosrR mutant compared to F. tularensis LVS and the transcomplemented ΔosrR+posrR strain at the indicated multiplicity of infection at 4 and 24 h postinfection. The data are represented as the mean ± SEM from two independent experiments, each conducted with four technical replicates and analyzed using one-way ANOVA. ***, P < 0.001. (D) C57BL/6 mice inoculated intranasally with the indicated CFU of F. tularensis LVS or the ΔosrR mutant. The morbidity and mortality of infected mice (n = 5 mice/group) were recorded for 15 days. (E) C57BL/6 mice inoculated intranasally with the 0.8 × 104 CFU of F. tularensis LVS (n = 6), the ΔosrR mutant (n = 8), or the transcomplemented ΔosrR+posrR strain (n = 11). The morbidity and mortality of infected mice were recorded for 15 days. The data shown are cumulative of two independent experiments. The survival curves shown in the top of panels D and E are plotted as Kaplan-Meier survival curves, and the data were analyzed using the log rank test. Body weights represented as the mean ± SD for the indicated doses of infection are shown in the bottom of panels D and E.
The ΔosrR mutant exhibits attenuated intramacrophage growth and virulence in mice.
During its intracellular residence, F. tularensis is exposed to oxidative stress. We next verified if the downregulated expression of FPI genes in the ΔosrR mutant, when exposed to oxidative stress, is associated with its impaired survival in macrophages. We infected murine primary bone marrow-derived macrophages (BMDMs) with the wild-type F. tularensis LVS, the ΔosrR mutant, and the transcomplemented strain at a multiplicity of infection (MOI) of 100. The BMDMs were lysed at 4 and 24 h postinfection to determine the numbers of bacteria that invaded BMDMs and replicated intracellularly, respectively. The results revealed that all three Francisella strains invaded the macrophages with equal efficacy, and identical bacterial numbers were recovered from all the three strains tested at 4 h postinfection. At 24 h, the differences were more prominent. Significantly fewer ΔosrR mutant bacteria (5.49 ± 0.3 log10 CFU/ml) were recovered from the infected macrophages than the wild-type F. tularensis LVS (7.46 ± 0.1 log10 CFU/ml). Transcomplementation partially restored the wild-type phenotype with significantly higher numbers (6.89 ± 0.5 log10 CFU/ml) of transcomplemented than ΔosrR mutant bacteria recovered from BMDMs 24 h postinfection (Fig. 5C). These results demonstrate that the ΔosrR mutant is attenuated for intramacrophage growth and that OsrR contributes to intramacrophage survival of F. tularensis.
To further establish that OsrR is required not only for intramacrophage growth and survival but also for virulence in mice, we performed in vivo experiments in a mouse model of tularemia. We infected C57BL/6 mice intranasally with incremental doses (1 × 102 and 1 × 103 CFU) of F. tularensis LVS or the ΔosrR mutant and monitored their survival and body weights daily for 15 days. It was observed that the ΔosrR mutant was attenuated for virulence compared to the wild-type F. tularensis LVS. All the mice infected with 1 × 102 and 1 × 103 CFU of the ΔosrR mutant survived the infection, whereas 20% of mice infected with 1 × 102 CFU and 40% of those infected with 1 × 103 CFU of the wild-type F. tularensis LVS succumbed to infection. Furthermore, all infected mice lost body weight during the first 3 to 7 days and then gradually regained their original body weight, indicating that all mice received the infection (Fig. 5D). In another experiment, C57BL/6 mice were infected intranasally with 0.8 × 104 CFU of F. tularensis LVS, the ΔosrR mutant, or the transcomplemented ΔosrR+posrR strain. They were monitored for their survival and body weights daily for 15 days. All mice infected with 0.8 × 104 CFU of F. tularensis LVS succumbed to the infection by day 9 postinfection. However, 90% (1/8) of mice infected with an equal dose of the ΔosrR mutant survived the infection. Transcomplementation partially restored the virulence of the ΔosrR mutant, and 45% (5/11) of mice infected with the transcomplemented strain succumbed to infection. All mice that survived the infection with either the ΔosrR mutant or the transcomplemented strain after an initial loss of their body weight regained their preinfection body weight by day 12 postinfection (Fig. 5E). These results demonstrate that mice infected with the ΔosrR mutant survive better than those infected with wild-type F. tularensis LVS and that OsrR contributes to the virulence of F. tularensis.
Exposure of the ΔosrR mutant to oxidative stress results in differential expression of stress response genes.
RNA sequencing analysis also revealed downregulated expression of three stress response genes encoded on an operon in the ΔosrR mutant treated with menadione. These genes were also marginally downregulated when the ΔosrR mutant was not exposed to oxidative stress. These three downregulated genes included aroA, a gene encoding a protein of the shikimate pathway required for the synthesis of aromatic amino acids from glucose, a gene encoding a hypothetical protein, and the rnhA gene, which encodes RNase H. These findings indicate that OsrR modulates the expression of genes of F. tularensis involved in metabolism as well as RNase H abundance under oxidative stress conditions (Fig. 6A).
FIG 6.
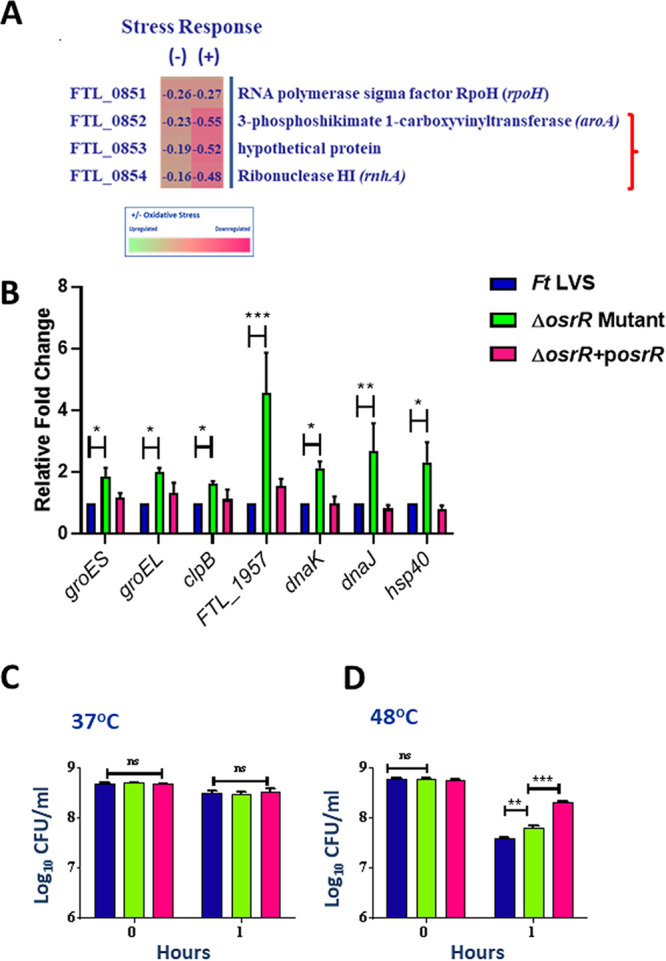
Exposure of the ΔosrR mutant to oxidative stress results in differential expression of stress response genes. (A) Expression profile of stress response genes in the ΔosrR mutant in the absence of (–) or upon exposure to (+) menadione compared to the wild-type F. tularensis (Ft) LVS. The data shown are cumulative of three independent RNA sequencing experiments and expressed as the log2 fold change. (B) Wild type F. tularensis LVS, ΔosrR mutant, and ΔosrR+posrR transcomplemented strains were exposed to 1.25 mM menadione for 1 h to induce oxidative stress. The RNA was isolated and quantitated for the expression of the indicated heat-stress response genes by qRT-PCR. The data are represented as the relative fold change compared to F. tularensis LVS. The data are represented as the mean ± SEM from three independent experiments, each conducted with three technical replicates and analyzed using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C and D) The viability of the indicated strains was determined after exposure to 37 and 48°C for 1 h. The data are represented as the mean ± SEM from three independent experiments, each conducted with three technical replicates. The data were analyzed using one-way ANOVA. **, P < 0.01; ***, P < 0.001; ns, not significant.
Interestingly, another gene, rpoH, which encodes a sigma factor that shares homology with the σ32 heat shock family protein, was also downregulated in the ΔosrR mutant irrespective of its exposure to oxidative stress (31). RpoH negatively regulates several genes, including stress response genes groES, gorEL, clpB, FTL_1957, dnaK, dnaJ, and hsp40 (32). The RNA sequence analysis revealed that all these genes were moderately upregulated in the ΔosrR mutant not exposed to oxidative stress (Fig. S3B). However, the expression of these genes was further upregulated upon exposure to oxidative stress in the ΔosrR mutant compared to the wild-type F. tularensis LVS (Fig. S5). These findings were further confirmed by determining the expression of groES, groEL, clpB, FTL_1957 dnaK, dnaJ, and hsp40 genes by qRT-PCR. The expression of all these genes that are primarily involved in heat shock response were significantly upregulated in the ΔosrR mutant upon exposure to oxidative stress. Transcomplementation restored the expression levels of these genes to levels similar to those observed for the wild-type F. tularensis (Fig. 6B).
Further, to verify if OsrR regulates rpoH-mediated heat shock response, we conducted a thermal resistance experiment in which bacteria were exposed to a temperature (48°C) higher than the physiological temperature (37°C). The transcomplemented strain was used as a positive control, as the osrR gene in the transcomplemented vector is under the control of the GroEL promoter, which is activated at high temperatures. It was observed that the viabilities of all these three Francisella strains were not affected when exposed to a temperature of 37°C for 1 h, indicating that at physiological temperature, OsrR does not affect the growth of F. tularensis (Fig. 6C).
Next, we induced high-temperature stress by growing the bacteria at 48°C (Fig. 6D). After 1 h of incubation, all three strains exhibited a significant decrease in the viable bacterial count. However, the numbers of the ΔosrR mutant bacteria (7.8 ± 0.0 log10 CFU/ml) were significantly higher than those of the wild-type F. tularensis LVS (7.5 ± 0.0 log10 CFU/ml). The transcomplemented strain survived better than the former two Francisella strains tested, with significantly higher numbers of bacteria (8.3 ± 0.0 log10 CFU/ml) remaining viable at the higher temperature. Overall, these results indicate that OsrR of F. tularensis LVS regulates the expression of heat shock proteins negatively and facilitates survival under extreme stress conditions.
OsrR of F. tularensis positively regulates key components of the TCA cycle during oxidative stress.
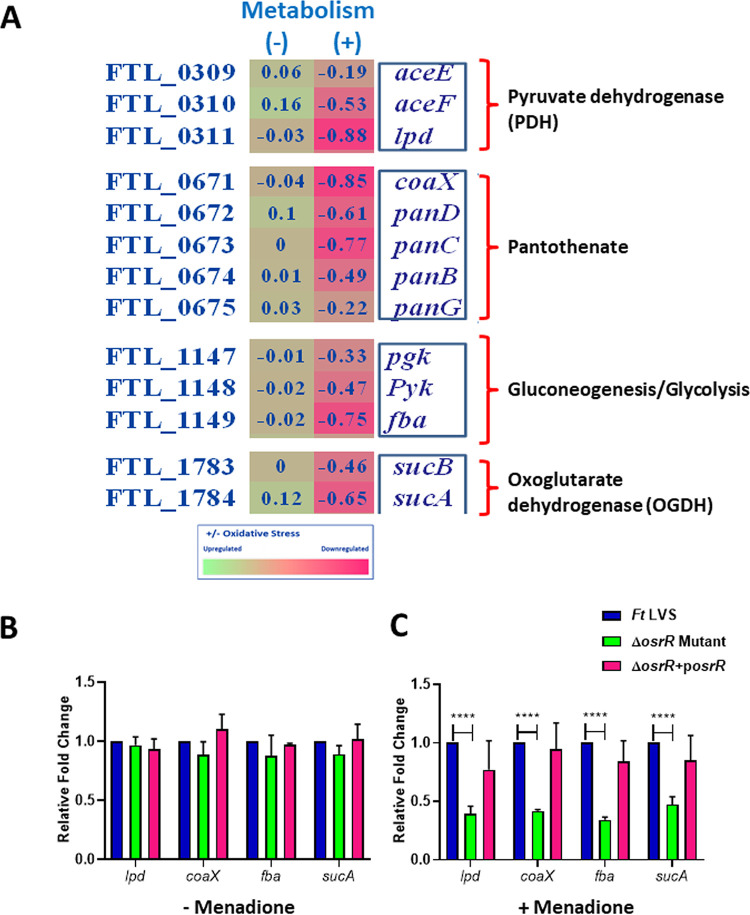
Exposure of the ΔosrR mutant to menadione downregulated expression of genes involved in metabolism encoded on four different operons. One of these operons encodes the enzyme pyruvate dehydrogenase subunits, E1 (aceE), E2 (aceF), and E3 (lpd), required for the biosynthesis of acetyl coenzyme A (acetyl-CoA), an enzyme that initiates the tricarboxylic acid (TCA) cycle (33). Among the three subunits, E3 or dihydrolipoyl dehydrogenase encoded by lpd was downregulated more in the ΔosrR mutant exposed to oxidative stress compared to the wild-type F. tularensis LVS. The other two subunits, E1 pyruvate dehydrogenase (encoded by aceE) and E2 dihydrolipoyl transacetylase (encoded by aceF), were also downregulated in the ΔosrR mutant. These results indicated that OsrR potentially contributes to the activation of the TCA cycle in F. tularensis under oxidative stress conditions (Fig. 7A).
FIG 7.
OsrR of F. tularensis positively regulates key components of the TCA cycle during oxidative stress. (A) Expression profile of genes of the indicated metabolic pathways encoded as operons in the ΔosrR mutant in the absence of (–) or upon exposure to (+) menadione compared to the wild-type F. tularensis (Ft) LVS. The data shown are cumulative of three independent RNA sequencing experiments and expressed as the log2 fold change. (B and C) Quantitation of transcripts of lpd, coaX, fba, and sucA genes in the indicated Francisella strains by qRT-PCR in the absence (B) or the presence (C) of oxidative stress induced by menadione. The data are represented as relative the fold change compared to F. tularensis LVS. The data are represented as the mean ± SEM from three independent experiments, each conducted with four technical replicates and analyzed using one-way ANOVA. ****, P < 0.0001.
The expression of genes encoded on another operon was downregulated in the ΔosrR mutant. This operon comprises the genes required for the biosynthesis of pantothenate (vitamin B5), which is the central component of CoA. The RNA sequencing data analysis revealed that the coaX gene encoding type III pantothenate kinase was downregulated in the ΔosrR mutant compared to the wild-type F. tularensis LVS when subjected to oxidative stress. The analysis also exhibited downregulation of the aspartate-1-decarboxylase (panD) gene, required for the formation of β-alanine from l-aspartate, and the panC gene encoding the pantothenate synthase that employs β-alanine and l-pantoate to produce pantothenate. The expression levels of the panB (ketopantoate hydroxymethyltransferase) and panG (ketopantoate reductase) genes were also downregulated in the menadione-treated ΔosrR mutant compared to wild-type F. tularensis LVS. Enzymes, PanB and PanG, catalyze the conversion of 2-oxoisovalerate to 2-dehydropantoate and pantoate, respectively. Altogether, these results indicate that, under oxidative stress, OsrR of F. tularensis positively regulates the pantothenate biosynthesis pathway required for CoA formation to initiate the TCA cycle (Fig. 7A).
Furthermore, exposing the ΔosrR mutant to oxidative stress altered the expression of genes encoded on an additional metabolic operon. This operon harbors three genes—pgk, pyk, and fba. The pgk gene encodes phosphoglycerate kinase, an enzyme that catalyzes the formation of ATP from ADP. The pyk gene encodes pyruvate kinase involved in the production of ATP and pyruvate, which is crucial for several metabolic pathways, including the TCA cycle (34). Moreover, the fba gene was downregulated to a greater extent in the ΔosrR mutant post-exposure to menadione. This gene encodes fructose-bisphosphate aldolase class II (FBA), which is required for the formation of fructose 1,6-bisphosphate (FBP) through gluconeogenesis and glycolysis pathways in Francisella (35). FBA is required for bacterial virulence and is essential for intracellular survival, indicating the significance of these metabolic pathways in bacterial pathogenesis (35) (Fig. 7A).
In addition to the genes described above, RNA expression levels of a set of metabolic genes, sucA and sucB, were significantly altered in the ΔosrR mutant when treated with menadione. These genes encode oxoglutarate dehydrogenase (OGDH) subunits E1 and E2, respectively, which are the key enzymes of the TCA cycle (Fig. 7A). Collectively, the RNA sequencing data analysis indicated that inducing oxidative stress in the ΔosrR mutant resulted in differential expression of genes encoding key elements of the metabolic pathways, TCA cycle, and energy production.
To validate the RNA sequencing results, the expressions of lpd, coaX, fba, and sucA genes were determined by qRT-PCR analysis in the absence or the presence of oxidative stress induced by menadione. The expression profile of all these genes remained unaltered in the absence of oxidative stress in wild-type F. tularensis LVS, the ΔosrR mutant, and the transcomplemented strain (Fig. 7B). However, in the presence of oxidative stress, the expression of all these genes was significantly downregulated in the ΔosrR mutant compared to the wild-type F. tularensis LVS. Transcomplementation restored the expression of these genes to levels similar to those observed for the wild-type strain (Fig. 7C). Together, these results demonstrate that OsrR of F. tularensis positively regulates components of the TCA cycle, pantothenate, and glucose metabolic pathways during oxidative stress to sustain intracellular survival of F. tularensis.
Expression of regulatory proteins and several transcription units is downregulated in the ΔosrR mutant exposed to oxidative stress.
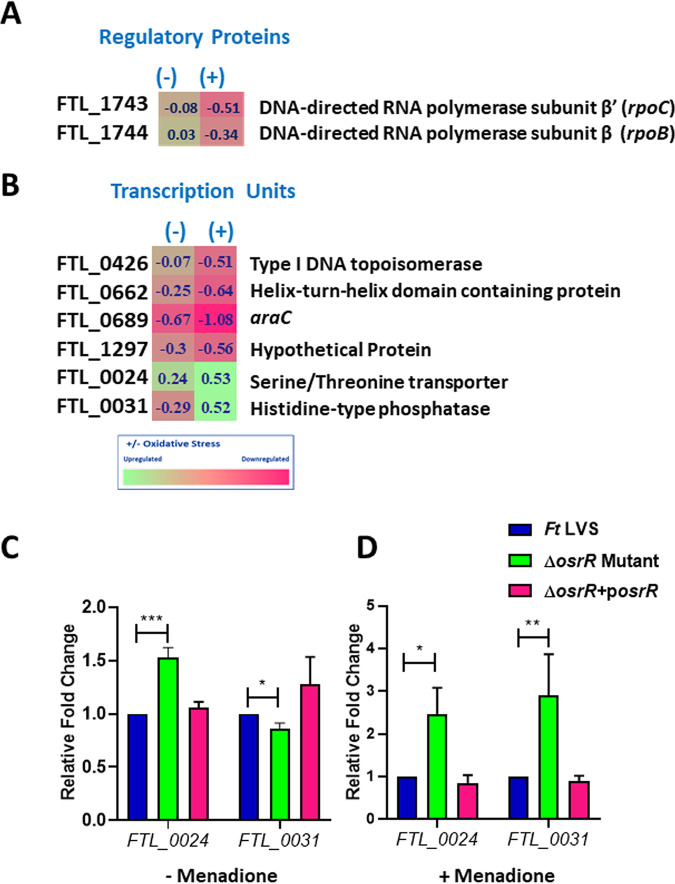
We identified an additional set of genes that were differentially expressed in the ΔosrR mutant compared to the wild-type F. tularensis LVS after induction of oxidative stress. The expression level of two genes, rpoB and rpoC, which encoded an operon, was significantly downregulated in the menadione-treated ΔosrR mutant. These genes encode two of the RNA polymerase core proteins, the β (RpoB) and β′ (RpoC) subunits (31) (Fig. 8A), indicating that OsrR is also involved in regulating the F. tularensis transcriptional process during oxidative stress.
FIG 8.
Differential expression of regulatory proteins and several transcription units in the ΔosrR mutant exposed to oxidative stress. (A and B) Expression profile of regulatory genes (A) and transcription units (B) in the ΔosrR mutant in the absence of (–) or upon exposure to (+) menadione compared to the wild-type F. tularensis (Ft) LVS. The data shown are cumulative of three independent RNA sequencing experiments and expressed as the log2 fold change. (C and D) Quantitation of FTL_0024 and FTL_0031 gene transcripts in the indicated Francisella strains by qRT-PCR in the absence (C) or the presence (D) of oxidative stress induced by menadione. The data are represented as the relative fold change compared to F. tularensis LVS. The data are represented as the mean ± SEM from three independent experiments, each conducted with three technical replicates and analyzed using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Besides regulating the RNA expression levels of genes encoded on operons, OsrR of F. tularensis also exhibited a significant regulatory effect on a few transcriptional units during oxidative stress. The osrR gene, which is transcribed as a single transcription unit, remained downregulated to the greatest extent irrespective of the treatment with menadione in the ΔosrR mutant. The pyrH gene that encodes the uridylate kinase enzyme to catalyze pyrimidine synthesis was also downregulated, suggesting that OsrR is also involved in nucleotide damage repair response during oxidative stress. Another gene that was largely affected in the ΔosrR mutant by menadione treatment was FTL_0426 coding for type I DNA topoisomerase. This enzyme is required for DNA supercoil repair, indicating that OsrR is also involved in regulating DNA repair mechanisms in F. tularensis during oxidative stress.
Furthermore, two genes, FTL_0024 and FTL_0031, were significantly upregulated in the ΔosrR mutant compared to the wild-type strain. These genes encode serine/threonine transporter and acid phosphatase, respectively, indicating that OsrR negatively regulates genes required to transport amino acids (serine and threonine) during the conditions of oxidative stress (Fig. 8B). qRT-PCR analysis validated the transcription profile of both FTL_0024 and FTL_0031 obtained by RNA sequencing in the absence or presence of oxidative stress induced by menadione (Fig. 8C and D). These results indicate that, since acid phosphatase is essential for glucose and G6P phosphorylation and dephosphorylation mechanisms in bacteria, OsrR of F. tularensis is possibly involved in the negative regulation of G6P to suppress the glucose metabolism.
DISCUSSION
The AraC/XylS family is one of the largest families of bacterial transcriptional regulators (25). Members of the AraC/XylS family can alter gene expression, mostly positively (36). In rare cases, gene expression can be either negatively altered by family members such as the CelD of Y. pestis (37) or both positively and negatively by the AraC of E. coli and YbtA of Y. pestis (38). AraC/XylS transcriptional regulators are additionally classified into two categories based on the regulation of their own transcription (25). The first category has signal receptors within the same protein that respond to the availability of substrates to activate or repress gene expression. Examples of this type of transcriptional regulators are AraC, XylS, RhaR, and UreR (25, 39, 40). The second category requires the activity of another regulator (activator or repressor) to control gene expression in a two-component regulatory system manner, such as MarA, SoxS, and TcpN (41, 42). The AraC/XylS family proteins regulate three unique processes—regulation of virulence factors, stress response, and metabolism of carbons (25). The present study characterized the role of OsrR, a member of the AraC/Xyls family, in oxidative stress response, intramacrophage survival, and virulence of F. tularensis.
Sugar utilization experiments revealed that OsrR transcriptional regulator, unlike its reported role in other bacteria, is not involved in l-arabinose utilization in F. tularensis LVS. The genomic organization of the Emr type MEP of F. tularensis is unique, comprising three adjacent genes, emrB, emrA1, and silC, that constitute an operon (26, 27). The location of the osrR gene in proximity to and its transcription divergent from emrB, emrA1, and silC genes suggested a possible involvement of OsrR in the regulation of these MEP genes. A similar genomic organization of the MEP genes is also present in Pseudomonas aeruginosa, with the MexR transcriptional regulator transcribed divergently from the mexAB-oprM multidrug efflux system (43). In response to oxidative stress, the MexR transcriptional regulator detaches from the promoter and functions as an activator to initiate the MEP gene expression. Moreover, Campylobacter jejuni possesses a cmeGH multidrug efflux pump that is controlled by a Fur repressor transcribed divergently upstream of the operon (44). Downregulated expression of the MEP genes observed in this study suggests that the OsrR controls the functioning of the Emr MEP only under conditions of oxidative stress. These findings were further confirmed by the enhanced sensitivity of the ΔosrR mutant to peroxides and superoxide-generating compounds compared to those observed for the emrA mutant.
The role of the OsrR in oxidative stress response is further cemented by RNA sequencing results. In the absence of oxidative stress, the transcription profiles of both the ΔosrR mutant and the wild type F. tularensis LVS, except for the osrR gene expression, were more or less identical, indicating that OsrR does not exert its regulatory role when Francisella is not exposed to oxidative stress. A differential expression of several genes in the ΔosrR mutant indicated that OsrR functions when the bacteria are exposed to oxidative stress conditions. It has been reported that SoxS, an AraC/XylS family transcriptional regulator of E. coli, is induced by oxidants such as paraquat and menadione to activate the transcription of the SoxRS regulon (25, 41). However, SoxRS homologs are absent in F. tularensis. The transcriptome analysis of the ΔosrR mutant treated with menadione does confirm the notion that OsrR is activated in the presence of an oxidant to exert its regulatory effect. Thus, it is probable that OsrR of F. tularensis functions similarly to the SoxRS regulon present in E. coli and other bacterial pathogens.
Our results demonstrate that oxidative stress induced by menadione resulted in the differential expression of a multitude of genes in the ΔosrR mutant of F. tularensis transcribed as operons as well as transcriptional units. The results from this study demonstrate the involvement of OsrR in the positive regulation of the FPI genes pdpE, pdpC, iglJ, iglI, and dotC in response to oxidative stress. These genes constitute the components of the type VI secretion system of F. tularensis. Thus far, several studies have published the fundamental role of these FPI genes in the intramacrophage virulence of F. tularensis (12, 13, 45, 46). Specifically, the pdpC gene encodes a pathogenicity determinant protein that does not contribute to intracellular survival but is required for virulence in mice (47). On the other hand, pdpE is required for neither virulence nor intracellular survival. The iglJ gene product is required for intracellular growth, cytopathogenicity, and virulence in mice, whereas iglI plays an important role in the phagosomal escape, cytopathogenicity, and virulence in mice (48). The dotU gene product is an essential structural component of the T6SS of F. tularensis and is required for the functioning of T6SS. The ΔosrR mutant of F. tularensis also exhibits defective intramacrophage survival and attenuated virulence in mice.
The transcriptome analysis of the ΔosrR mutant revealed the involvement of OsrR of F. tularensis LVS in the regulation of two genes, aroA, and rnhA, that are a part of the heat stress-response-related operon. The AroA is involved in the shikimate pathway that catalyzes the synthesis of aromatic amino acids from glucose (49). OsrR of F. tularensis positively regulated the rnhA gene that encodes RNase H. In Yersinia, Salmonella, Helicobacter pylori, Shigella flexneri, and Mycobacterium smegmatis ribonucleases are known to function as virulence factors (50) and protect bacteria against oxidative stress (51). We observed that the expression level of the rpoH gene was not altered in the ΔosrR mutant in the absence or the presence of oxidative stress and remained marginally downregulated. RpoH negatively regulates the expression of several genes, including heat shock response genes (32). The RNA sequence analysis revealed that expression of groEL, groES, clpB, FTL_1957, dnaK, dnaJ, and hsp40 were moderately upregulated in the ΔosrR mutant not exposed to oxidative stress and were further upregulated upon exposure to oxidative stress. These findings were also substantiated by qRT-PCR analysis. Collectively, these results indicate a broader role for OsrR in the regulation of oxidative as well as overall stress response of F. tularensis.
The results from this study further demonstrated the involvement of OsrR in the regulation of the components of the TCA cycle in F. tularensis during oxidative stress. Several studies have linked the TCA cycle with the virulence of F. tularensis during stress conditions (35, 52, 53). We observed the downregulated expression of key TCA cycle enzymes in the ΔosrR mutant after menadione treatment. The operon encoding pyruvate dehydrogenase enzyme was significantly downregulated. The pyruvate dehydrogenase is an enzyme that catalyzes the synthesis of the first enzyme, acetyl-CoA, of the TCA cycle (33). It has three subunits, E1 (pyruvate dehydrogenase [AceE]), E2 (dihydrolipoamide transacetylase [AceF]), and E3 (dihydrolipoamide dehydrogenase [Lpd]) (52). We also observed the downregulation of another enzyme, oxoglutarate dehydrogenase, encoded on another operon. The oxoglutarate dehydrogenase promotes the conversion of α-ketoglutarate (or 2-oxoglutarate) into succinyl-CoA in the TCA cycle. Oxoglutarate dehydrogenase is also composed of three subunits, E1 (2-oxoglutarate dehydrogenase [SucA]), E2 (dihydrolipoamide succinyltransferase [SucB]), and E3 (lipoamide dehydrogenase [Lpd]). In F. tularensis subsp. novicida, a study has shown the requirement of the E1 (AceE) subunit of the pyruvate dehydrogenase enzyme in intramacrophage growth (54). In addition, another study has demonstrated the contribution of the E3 (Lpd) subunit to F. tularensis LVS virulence (55). Collectively, these results demonstrate that the expression of key TCA cycle enzymes of F. tularensis LVS, especially under oxidative stress conditions, is controlled by OsrR.
Results from this study also demonstrate that OsrR regulates the expression of genes involved in pantothenate metabolism when Francisella is exposed to oxidative stress. Pantothenate, also known as vitamin B5, is an essential central metabolic component of the CoA of the TCA cycle (56). In E. coli and S. enterica serovar Typhimurium, the de novo synthesis of pantothenate molecules relies on their ketopantoate reductase genes (57). The ketopantoate reductase genes in F. tularensis are composed of a group of five genes (coaX, panD, panC, panB, and panG) that promote the enzymatic reaction of pantothenate synthesis. F. novicida requires the coaX gene for the successful dissemination into mouse organs (58). It was observed that all of the five ketopantoate reductase genes were significantly downregulated in the ΔosrR mutant exposed to menadione, indicating a key role of OsrR in the pantothenate pathway, which subsequently is required for the formation of CoA enzyme and the initiation of the TCA cycle.
Our observations further indicate that OsrR is also involved in the positive regulation of glucose metabolism in F. tularensis LVS. Three genes, pgk, pyk, and fba, were significantly downregulated in the ΔosrR mutant after menadione treatment. Phosphoglycerate kinase (PGK) is an essential enzyme for the breakdown of glucose into pyruvate and, subsequently, the release of high energy (59). Pyruvate kinase (PYK) activation is promoted by fructose 1,6-bisphosphate (34), which is then metabolized by the enzyme fructose-bisphosphate aldolase class II (FBA). A study has shown that the FBA plays an important role in Francisella cytosolic replication only if gluconeogenesis substrate is available but is not required for the bacterial escape from the phagosome (35). These results suggest that OsrR plays a regulatory role in gluconeogenesis and glycolysis pathways in F. tularensis.
The results from this study additionally reveal that OsrR positively regulates the expression of other regulatory genes, rpoB, and rpoC. These genes encode two key components, the β (RpoB) and β′ (RpoC), that constitute the catalytic core of RNA polymerase (60). In addition to the role of OsrR in regulating the expression of genes transcribed as operons, the results from whole-transcriptome analysis reveal OsrR-dependent expression of genes transcribed as single transcriptional units. The most prominent ones are type I DNA topoisomerase, required for the repair of damaged DNA and removal of supercoiling when exposed to oxidative stress. Further, in addition to the downregulated expression, several genes were also upregulated in the ΔosrR mutant when exposed to oxidative stress. The most prominently upregulated genes in the ΔosrR mutant were serine/threonine transporter and histidine-type phosphatase, indicating additional amino acid requirements due to the loss of OsrR under oxidative stress conditions.
Collectively, these results provide ample evidence of the regulatory role of the OsrR. However, the mechanism(s) through which OsrR exerts its regulatory role during oxidative stress remained elusive in this study. The notion that the expression of all known transcriptional regulators of Francisella remained unaltered in untreated or menadione-treated ΔosrR mutant compared to wild-type F. tularensis LVS indicates that OsrR may have a direct rather than an indirect role in gene regulation.
To conclude, as described for other bacterial pathogens, OsrR annotated as AraC in the Francisella genome is not required for l-arabinose utilization. Instead, the results obtained from the present study demonstrate that the OsrR transcriptional regulator exerts a global regulatory role on several key pathways of F. tularensis LVS. The OsrR contributes to the oxidative stress response by regulating the expression of Emr MEP genes involved in oxidative stress resistance. OsrR regulates the expression of FPI genes when Francisella is exposed to oxidative stress and contributes to intramacrophage survival and virulence. OsrR also regulates the expression of genes involved in metabolic and other key pathways required by Francisella to overcome oxidative stress. Altogether, this study demonstrates that OsrR belonging to the AraC/XylS family of transcriptional regulators plays an essential role in the oxidative stress response of F. tularensis LVS and assigns a prominent role to a relatively unknown transcriptional regulator in tularemia pathogenesis.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study are listed in Table 1. The wild-type F. tularensis LVS used in this study was obtained from BEI Resources, Manassas, VA. The deletion mutant of the osrR (FTL_0689) gene (ΔosrR) of F. tularensis LVS and its transcomplemented strain (ΔosrR+posrR) were generated in this study. A transposon insertion mutant in the emrA1 gene (FTL_0687) of F. tularensis LVS available in our laboratory and used in previously published works (27, 61) was also used in this study. The E. coli DH-5α and S-17 strains were obtained from Invitrogen.
TABLE 1.
List of bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description | Source or reference |
|---|---|---|
| Strains | ||
| F. tularensis LVS | Wild-type strain | ATCC |
| F. tularensis ΔosrR mutant | Deletion mutant of osrR gene of F. tularensis LVS | This study |
| F. tularensis emrA1 mutant | LVS FTL_0687::Tn5 Kanr | 27 |
| F. tularensis osrR transcomplemented strain (ΔosrR+posrR) | LVS, ΔosrR, pMM09 (pMP822+osrR), Hygr | This study |
| E. coli | F– ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK- mK+) phoA supE44 λ- thi-1 gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| pMP822 | E. coli-Francisella shuttle vector, Hygr | 63 |
| pJC84 | E. coli-Francisella suicide vector, Kanr | 62 |
| pMM03 | pJC84 + fused flanking fragment of osrR gene, Kanr | This study |
| pMM014 | pMP822 + osrR, Hygr | This study |
| Primers for osrR gene deletion | ||
| MP135 | 5′-CAAGGATCCAAATAGATTGTGTTAGCATTGCAC-3′ | |
| MP137 | 5′-TTTATCTGTACTCTTTCACTAGAGCATACTTTGTCCTTTTTTCACCA-3′ | |
| MP139 | 5′-TGGTGAAAAAAGGACAAAGTATGCTCTAGTGAAAGAGTACAGATAAA-3′ | |
| MP140 | 5′-TGATGTCGACGCAATACTCAAGTGGAACAACTGG-3′ | |
| Primers for confirmation of osrR gene deletion | ||
| MP037 | 5′-CCGGATCCATGAAATTTGAATTACCAAAAC-3′ | |
| MP038 | 5′-CGCTGCAGCTAATCAGCGAATTGCTCAGAAAC-3′ | |
| MP192 | 5′-ATGGGTGTTGCCATCAAATAGG-3′ | |
| MP193 | 5′-TGTTGTCGCCAACGTGAAA-3′ | |
| Primers for transcomplementation | ||
| MP372 | 5′-CAAGGTTCCATGATACGGGAAGAGATCACATAT-3′ | |
| MP373 | 5′-TGATCTCGAGTCAGTTCTTATAAATATTTTTTATC-3′ | |
| Primers for qRT-PCR | ||
| silC | MP015 5′-AGCCAAGTTAGTGCTGCATATTT-3′ MP016 5′-AAGCCAAGTTAGTGCTGCATATTT-3′ |
|
| emrA1 | MP017 5′-GTGCATCTTGTAAAGAGCCAGCATC-3′ MP018 5′-CAGCCAAACTAAGCGCACAGTCAT-3′ |
|
| emrB | MP019 5′-TCCTAATCCCTGAATAGCCGTTGT-3′ MP020 5′-AGGTGTTGCCGCTATTATTGGTGC-3′ |
|
| groES | MP280 5′-ACCCATGATATCTTCTTCTCTC-3′ MP281 5′-AGAGTATTAGTTCGTCGTGCAG-3′ |
|
| groEL | MP282 5′-TCTTCAAAGCCTTTGCCTTC-3′ MP283 5′-TGTCACTGCAGGTATGAATCC-3′ |
|
| clpB | MP284 5′-TTTCATAATCCTGACCTTGCA-3′ MP285 5′-GGCGGCTATCTAACTGAACAT-3′ |
|
| FTL_1957 | MP286 5′-AGATATAACAGAAGATGAAGCTGC-3′ MP287 5′-TAGGGATGTTCAAGCTTAGTAC-3′ |
|
| dnaK | MP446 5′-TATGACCTAGGTGGTGGTACATTC-3′ MP447 5′-TTCTCAGCAGCCTCTCTAACTCTT-3′ |
|
| dnaJ | MP448 5′-TGATGGTACAGGCTCTAAATCCAG-3′ MP449 5′-ACAAATCACCGTTCATAGCACC-3′ |
|
| hsp40 | MP450 5′-TTTGGTGGTTTCTCGCAAAGT-3′ MP451 5′-CCCATTGCTGATGGTATTTTAAC-3′ |
|
| pdpC | MP654 5′-GCCTGAGTCATTGCTTGATCTAAA-3′ MP655 5′-AATAACCCCAAGGATCACTCAA-3′ |
|
| FTL_1163 |
MP656 5′-
TGGCTATTGAAATACTTGGATACGG
-3′
MP657 5′- TGTTGTGAATTTGTGCAGTGCTT -3′ |
|
| lpd | MP658 5′-CACGGTTGTTGAGTTTGCTGA-3′ MP659 5′-GCAGGATGATCGCCTTCCATA-3′ |
|
| coaX | MP660 5′-GCAGATCGTGTTGCAAACTCA-3′ MP661 5′-AGCACCGCCGATATAGGTTT-3′ |
|
| fba | MP662 5′-TGAGGGTGAGTTAGGTTGCC-3′ MP663 5′-GCACCATGCGAAGTACCGA-3′ |
|
| sucA | MP664 5′-TGAAATGAAGACGCCACCTG-3′ MP665 5′-TCTCTTCTAATTGCGCACGAG-3′ |
|
| FTL_0024 | MP666 5′-ACTGCAAAAGAGCATTGAGTACAA-3′ MP667 5′-ACGGCTAAAGAAGAGATAATCGGT-3′ |
|
| FTL_0031 | MP668 5′-ACTCACCAGAGTGGCAAAACA-3′ MP669 5′-ACCATGTGCTTGAGCAACAATC-3′ |
The Francisella strains were cultured on Mueller-Hinton (MH)–chocolate agar plates (BD Life Sciences) and incubated for 48 h at 37°C and 5% CO2. Individual colonies were selected and inoculated in MH broth (MHB) enhanced with Isovitalex (BD Biosciences), ferric pyrophosphate, glucose, anhydrous calcium chloride, and hydrous magnesium chloride. The bacterial cultures were incubated at 37°C with constant shaking until the bacterial growth reached the mid-log phase. Bacterial aliquots were snap-frozen in liquid nitrogen and stored at −80°C. The emrA1 mutant was grown on MH chocolate agar plates or MHB supplemented with kanamycin (10 μg/ml). For the transcomplemented strain (ΔosrR+posrR), hygromycin (100 μg/ml) was added to the medium instead of kanamycin. All the work was performed under biosafety containment level 2 (BSL2).
Generation of the ΔosrR mutant and the transcomplemented strains of F. tularensis LVS.
The plasmid and primers sequences used for generation and confirmation of ΔosrR mutant and the transcomplemented strain of F. tularensis LVS are listed in Table 1. The pJC84 suicide vector that allows for SacB-dependent allele replacement was used as reported earlier (62) for the generation of the in-frame gene deletion mutant of the osrR gene (FTL_0689) in F. tularensis LVS. For the deletion of the osrR gene, a 5′ fragment composed of a 1,152-bp sequence upstream of the start codon of the FTL_0689 and the start codon was amplified by PCR using the MP135 and MP137 end primers. In addition, a 3′ fragment of a 2,081-bp downstream sequence of the FTL_0689 and the stop codon was also amplified by PCR using MP139 and MP140 primers. By overlapping extension PCR using primers MP135 and MP140, both fragments were fused to generate a larger fragment (2,233 bp). The fused fragment was cloned into the BamHI and SalI sites of the pJC84 to generate pMM03. After that, PCR confirmation was performed and followed by pMM03 electroporation into the F. tularensis LVS. The colonies were then selected on MH-chocolate agar plates containing 25 μg/ml kanamycin. Following that, sucrose counterselection was performed by plating bacteria on MH-chocolate agar supplemented with 8% sucrose and incubated at 37°C with 5% CO2 for 48 to 72 h. The sucrose-resistant and kanamycin-sensitive clones were screened for the loss of the osrR gene using PCR. For osrR gene deletion confirmation, a duplex colony PCR was performed using the following primer sets osrR gene-specific (MP192/MP193) and internal control-sodB gene (MP037/MP038). The in-frame gene deletion of osrR was confirmed by DNA sequencing of the PCR-positive clones.
For the generation of the transcomplemented strain (ΔosrR +posrR), the F. tularensis osrR gene was amplified using the primers MP372 and MP373. BamHI and XhoI restriction enzymes were used to digest the amplified sequence and then cloned into E. coli-Francisella shuttle vector pMP822 (63) to generate pMM014. This plasmid was then confirmed using PCR and DNA sequencing and was electroporated into the ΔosrR mutant. To confirm the transcomplementation of the osrR gene, the bacteria were selected in MH-chocolate agar plates containing 200 μg/ml hygromycin. The hygromycin-resistant colonies were screened with PCR.
Determination of the role of OsrR in sugar utilization.
Wild-type F. tularensis LVS or the ΔosrR mutant bacteria were streaked on MH chocolate agar plates supplemented with 1% glucose or 1% arabinose and incubated at 37°C with 5% CO2 for 48 h. The bacterial growth pattern of the ΔosrR mutant was compared to that of the wild-type F. tularensis LVS and the transcomplemented strain. E. coli DH-5α and E. coli S-17 were used as positive controls.
Defined Chamberlain’s medium (CDM) was prepared at three different compositions—containing glucose (4%) and no arabinose, containing arabinose (4%) and no glucose, and containing neither glucose nor arabinose. F. tularensis LVS or the ΔosrR mutant bacteria adjusted to a 0.2 OD600, which corresponded to 1 × 109 CFU/ml, was inoculated into these CDM formulations and incubated at 37°C with shaking. The OD600 was recorded every 4 h, and growth curves were generated using GraphPad Prism 7 software.
Determination of the role of OsrR in oxidative stress resistance.
The sensitivity of the ΔosrR mutant to oxidants was determined by spot assays and bacterial killing assays in the presence or the absence of oxidants as described previously (20, 26, 64, 65). Briefly, peroxides (16 mM H2O2, 1 mM tert-butyl hydroquinone [TBH], and 0.7 mM cumene hydroperoxide [CHP]) or superoxide-generating compounds (1.25 mM menadione, 8 mM pyrogallol, 50 mg/ml paraquat) were serially diluted in a 96 well-plate using MHB. Wild-type F. tularensis LVS, the ΔosrR mutant, and the ΔosrR+posrR cultures were adjusted to 0.2 OD600 (equivalent to 1 × 109 CFU/ml), and equal numbers of bacteria were added to each well containing the serially diluted compounds. The 96-well plate was incubated for 1 and 3 h at 37°C and 5% CO2, and then the bacteria were spotted on MH-chocolate agar plates. In another approach, bacterial suspensions were exposed to 1 mM H2O2 for 1 and 3 h and to 4 mM pyrogallol for 1 h with shaking at 180 rpm and 37°C. Bacteria were serially diluted (10-fold) and plated on MH chocolate agar and incubated for 48 h at 37°C and 5% CO2. Bacterial colonies were counted and plotted as log10 CFU/ml.
Macrophage cell culture assay.
Primary murine BMDMs were isolated from 6- to 8-week old C57BL/6 mice purchased from Jackson Laboratory (Bar Harbor, ME) as previously described (66). Bone marrow cells were differentiated into macrophages by culturing in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2% l-glutamine, 1% sodium pyruvate, 1% HEPES, and 20% medium conditioned with L929 cells. The BMDMs were infected with F. tularensis LVS, the ΔosrR mutant, or the transcomplemented strain at a multiplicity of infection (MOI) of 100. Cells were centrifuged for 10 min at 1,000 rpm to synchronize bacterial infection and incubated at 37°C and 5% CO2 for 2 h. Then, 2 h postinfection, the medium from the infected cells was replaced with DMEM containing gentamicin (250 μg/ml) to kill extracellular bacteria and was then replaced with DMEM without any antibiotics after 1 h of incubation. The BMDMs were lysed with 0.1% sodium deoxycholate at 4 and 24 h postinfection, and then a 10-fold serial dilution of the lysed cells was spotted on MH chocolate agar plates. The plates were incubated at 37°C and 5% CO2 for 48 h, bacterial colonies were counted, and the results were expressed as CFU per milliliter.
Mouse survival studies.
The role of OsrR in virulence was investigated by infecting wild-type C57BL/6 mice with F. tularensis LVS, the ΔosrR mutant, or the ΔosrR+posrR transcomplemented strain intranasally after anesthetizing them through intraperitoneal injection of ketamine and xylazine. Mice were inoculated with 1 × 102 and 1 × 103 CFU of F. tularensis LVS and the ΔosrR mutant or with 0.8 × 104 CFU of F. tularensis LVS, the ΔosrR mutant, and the transcomplemented strain in a volume of 20 μl (10 μl/naris). The morbidity and mortality of the infected mice were observed for 15 days. Body weight and survival were recorded daily. Survival was expressed as the Kaplan-Meier survival curve, and the data were analyzed statistically by the log rank test. All the protocols were approved by the IACUC of New York Medical College.
RNA sequencing.
To determine the role of OsrR as a global transcriptional regulator, we profiled the whole transcriptome of the ΔosrR mutant in the absence or the presence of oxidative stress. The superoxide-generating compound, menadione (final concentration of 1.25 mM), was used as an oxidative stress inducer. This concentration of menadione equally affects the viability of wild-type F. tularensis LVS or the ΔosrR mutant. For untreated control, an equal volume of MHB was added, and bacteria were incubated for 1 h at 37°C with 5% CO2. Bacteria were then centrifuged at 10,000 rpm for 10 min at room temperature, and pellets were resuspended in 990 μl lysis buffer (from Purelink RNA minikit/Invitrogen) supplemented with 10 μl 2-mercaptoethanol, and RNA was purified following the manufacturer’s protocol. RNA samples were treated with DNase using Invitrogen’s Turbo DNA-free kit.
Purified RNA samples were sent to the Genomics Core Laboratory at the New York Medical College for sequencing. Differentially expressed genes were evaluated based on log2 (fold change) value. The differential expression of F. tularensis LVS and ΔosrR mutant genes was analyzed based on the log2 fold change in the expression of genes in the ΔosrR mutant compared to those in wild-type F. tularensis LVS in the untreated and the menadione-treated samples. For the downregulated genes, the expression of the osrR gene, which has been deleted in the ΔosrR mutant, was taken as 100%. Any gene expressed lower or higher than 50% of the log2 fold change of the osrR gene was considered down- or upregulated, respectively. To further narrow the number of differentially expressed genes in the ΔosrR mutant, the adjusted P values of less than 0.05 were considered statistically significant. The adjusted P values were calculated using the DESeq2 package in R Bioconductor, and the Benjamini-Hochberg algorithm was used for the P value adjustment and controlling the false-discovery rate.
Quantitative real-time PCR (qRT-PCR).
The qRT-PCR was used to verify the differential expression of F. tularensis LVS and ΔosrR mutant genes in the untreated and the menadione-treated samples. The gene-specific primers used for qRT-PCR are listed in Table 1. The results were expressed as the relative fold change in gene expression of the ΔosrR mutant compared to the F. tularensis LVS.
Statistical analysis.
All results were analyzed using GraphPad Prism 7 software and were represented as the mean ± standard deviation (SD) or standard error of the mean (SEM). Comparisons among experimental groups were made using one-way analysis of variance (ANOVA). Survival data were plotted as the Kaplan-Meier survival curves, and significance was statistically calculated by the log rank test. A P value of <0.05 was considered statistically significant.
Data availability.
The RNA-sequencing data are available at the NCBI GEO repository under the accession number GSE183001.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R21AI51277 (C.S.B.) and R15AI107698 (M.M.). The funders had no role in study design, data collection and analysis, decision to publish, or manuscript preparation.
No financial conflicts of interest exist regarding the contents of the manuscript and its authors.
Footnotes
Supplemental material is available online only.
Contributor Information
Chandra Shekhar Bakshi, Email: Shekhar_Bakshi@nymc.edu.
Laurie E. Comstock, Brigham and Women’s Hospital/Harvard Medical School
REFERENCES
- 1.Oyston PCF, Sjöstedt A, Titball RW. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2:967–978. 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 2.Hollis DG, Weaver RE, Steigerwalt AG, Wenger JD, Moss CW, Brenner DJ. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J Clin Microbiol 27:1601–1608. 10.1128/jcm.27.7.1601-1608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsman M, Sandstrom G, Jaurin B. 1990. Identification of Francisella species and discrimination of type A and type B strains of F. tularensis by 16S rRNA analysis. Appl Environ Microbiol 56:949–955. 10.1128/aem.56.4.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svensson K, Larsson P, Johansson D, Byström M, Forsman M, Johansson A. 2005. Evolution of subspecies of Francisella tularensis. J Bacteriol 187:3903–3908. 10.1128/JB.187.11.3903-3908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisen L. 2007. Francisella tularensis: biology, pathogenicity, epidemiology, and biodefense. Emerg Infect Dis 13:1973–1973. 10.3201/eid1312.071169. [DOI] [Google Scholar]
- 6.Keim P, Johansson A, Wagner DM. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci 1105:30–66. 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 7.Mörner T, Mattsson R, Forsman M, Johansson K-E, Sandström G. 1993. Identification and classification of different isolates of Francisella tularensis. Zentralbl Veterinarmed B 40:613–620. 10.1111/j.1439-0450.1993.tb00184.x. [DOI] [PubMed] [Google Scholar]
- 8.Farlow J, Wagner DM, Dukerich M, Stanley M, Chu M, Kubota K, Petersen J, Keim P. 2005. Francisella tularensis in the United States. Emerg Infect Dis 11:1835–1841. 10.3201/eid1112.050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall JD, Woolard MD, Gunn BM, Craven RR, Taft-Benz S, Frelinger JA, Kawula TH. 2008. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect Immun 76:5843–5852. 10.1128/IAI.01176-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven RR, Hall JD, Fuller JR, Taft-Benz S, Kawula TH. 2008. Francisella tularensis invasion of lung epithelial cells. Infect Immun 76:2833–2842. 10.1128/IAI.00043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosio CM, Dow SW. 2005. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J Immunol 175:6792–6801. 10.4049/jimmunol.175.10.6792. [DOI] [PubMed] [Google Scholar]
- 12.Santic M, Molmeret M, Klose KE, Abu Kwaik Y. 2006. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol 14:37–44. 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Nano FE, Schmerk C. 2007. The Francisella pathogenicity island. Ann N Y Acad Sci 1105:122–137. 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 14.Brotcke A, Weiss DS, Kim CC, Chain P, Malfatti S, Garcia E, Monack DM. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun 74:6642–6655. 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charity JC, Blalock LT, Costante-Hamm MM, Kasper DL, Dove SL. 2009. Small molecule control of virulence gene expression in Francisella tularensis. PLoS Pathog 5:e1000641. 10.1371/journal.ppat.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauriano CM, Barker JR, Yoon S-S, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci USA 101:4246–4249. 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sammons-Jackson WL, McClelland K, Manch-Citron JN, Metzger DW, Bakshi CS, Garcia E, Rasley A, Anderson BE. 2008. Generation and characterization of an attenuated mutant in a response regulator gene of Francisella tularensis live vaccine strain (LVS). DNA Cell Biol 27:387–403. 10.1089/dna.2007.0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durham-Colleran MW, Verhoeven AB, van Hoek ML. 2010. Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb Ecol 59:457–465. 10.1007/s00248-009-9586-9. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. 2006. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J Bacteriol 188:3785–3795. 10.1128/JB.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Z, Russo VC, Rabadi SM, Jen Y, Catlett SV, Bakshi CS, Malik M. 2016. Elucidation of a mechanism of oxidative stress regulation in Francisella tularensis live vaccine strain. Mol Microbiol 101:856–878. 10.1111/mmi.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee N, Francklyn C, Hamilton EP. 1987. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc Natl Acad Sci USA 84:8814–8818. 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 23.Pletzer D, Schweizer G, Weingart H. 2014. AraC/XylS family stress response regulators Rob, SoxS, PliA, and OpiA in the fire blight pathogen Erwinia amylovora. J Bacteriol 196:3098–3110. 10.1128/JB.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke KA, Wilcox G. 1987. The araC gene of Citrobacter freundii. Gene 61:243–252. 10.1016/0378-1119(87)90188-0. [DOI] [PubMed] [Google Scholar]
- 25.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev 61:393–410. 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alqahtani M, Ma Z, Ketkar H, Suresh RV, Malik M, Bakshi CS. 2018. Characterization of a unique outer membrane protein required for oxidative stress resistance and virulence of Francisella tularensis. J Bacteriol 200:e00693-17. 10.1128/JB.00693-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Z, Banik S, Rane H, Mora VT, Rabadi SM, Doyle CR, Thanassi DG, Bakshi CS, Malik M. 2014. EmrA1 membrane fusion protein of Francisella tularensis LVS is required for resistance to oxidative stress, intramacrophage survival and virulence in mice. Mol Microbiol 91:976–995. 10.1111/mmi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigard M, Bröms JE, Mosnier A, Hologne M, Martin A, Lindgren L, Punginelli C, Lays C, Walker O, Charbit A, Telouk P, Conlan W, Terradot L, Sjöstedt A, Henry T. 2016. Francisella tularensis IglG belongs to a novel family of PAAR-Like T6SS proteins and harbors a unique N-terminal extension required for virulence. PLoS Pathog 12:e1005821. 10.1371/journal.ppat.1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemens DL, Lee B-Y, Horwitz MA. 2018. The Francisella type VI secretion system. Front Cell Infect Microbiol 8:121. 10.3389/fcimb.2018.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, Celli J, Arulanandam BP, Klose KE. 2009. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol 74:1459–1470. 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grall N, Livny J, Waldor M, Barel M, Charbit A, Meibom KL. 2009. Pivotal role of the Francisella tularensis heat-shock sigma factor RpoH. Microbiology (Reading) 155:2560–2572. 10.1099/mic.0.029058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guisbert E, Herman C, Lu CZ, Gross CA. 2004. A chaperone network controls the heat shock response in E. coli. Genes Dev 18:2812–2821. 10.1101/gad.1219204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Mets F, Van Melderen L, Gottesman S. 2019. Regulation of acetate metabolism and coordination with the TCA cycle via a processed small RNA. Proc Natl Acad Sci USA 116:1043–1052. 10.1073/pnas.1815288116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veith N, Feldman-Salit A, Cojocaru V, Henrich S, Kummer U, Wade RC. 2013. Organism-adapted specificity of the allosteric regulation of pyruvate kinase in lactic acid bacteria. PLoS Comput Biol 9:e1003159. 10.1371/journal.pcbi.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziveri J, Barel M, Charbit A. 2017. Importance of metabolic adaptations in Francisella pathogenesis. Front Cell Infect Microbiol 7:96. 10.3389/fcimb.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker LL, Hall BG. 1990. Characterization and nucleotide sequence of the cryptic cel operon of Escherichia coli K12. Genetics 124:455–471. 10.1093/genetics/124.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fetherston JD, Bearden SW, Perry RD. 1996. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol 22:315–325. 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 39.D’Orazio SE, Thomas V, Collins CM. 1996. Activation of transcription at divergent urea-dependent promoters by the urease gene regulator UreR. Mol Microbiol 21:643–655. 10.1111/j.1365-2958.1996.tb02572.x. [DOI] [PubMed] [Google Scholar]
- 40.Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421–425. 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 41.Amábile-Cuevas CF, Demple B. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res 19:4479–4484. 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins DE, Nazareno E, DiRita VJ. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J Bacteriol 174:6974–6980. 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Yi C, Zhang J, Zhang W, Ge Z, Yang C, He C. 2010. Structural insight into the oxidation‐sensing mechanism of the antibiotic resistance of regulator MexR. EMBO Rep 11:685–690. 10.1038/embor.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon B, Wang Y, Hao H, Barton Y-W, Zhang Q. 2011. Contribution of CmeG to antibiotic and oxidative stress resistance in Campylobacter jejuni. J Antimicrob Chemother 66:79–85. 10.1093/jac/dkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Golovliov I, Sjöstedt A, Mokrievich A, Pavlov V. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol Lett 222:273–280. 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
- 46.de Bruin OM, Ludu JS, Nano FE. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol 7:1. 10.1186/1471-2180-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindgren M, Bröms JE, Meyer L, Golovliov I, Sjöstedt A. 2013. The Francisella tularensis LVS ΔpdpC mutant exhibits a unique phenotype during intracellular infection. BMC Microbiol 13:20. 10.1186/1471-2180-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bröms JE, Sjöstedt A, Lavander M. 2010. The Role of the Francisella Tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front Microbiol 1:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Felgner S, Frahm M, Kocijancic D, Rohde M, Eckweiler D, Bielecka A, Bueno E, Cava F, Abraham W-R, Curtiss R, Häussler S, Erhardt M, Weiss S. 2016. aroA-deficient Salmonella enterica serovar Typhimurium is more than a metabolically attenuated mutant. mBio 7:e01220-16. 10.1128/mBio.01220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawal A, Jejelowo O, Chopra AK, Rosenzweig JA. 2011. Ribonucleases and bacterial virulence. Microb Biotechnol 4:558–571. 10.1111/j.1751-7915.2010.00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta R, Chatterjee D, Glickman MS, Shuman S. 2017. Division of labor among Mycobacterium smegmatis RNase H enzymes: RNase H1 activity of RnhA or RnhC is essential for growth whereas RnhB and RnhA guard against killing by hydrogen peroxide in stationary phase. Nucleic Acids Res 45:1–14. 10.1093/nar/gkw1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dieppedale J, Gesbert G, Ramond E, Chhuon C, Dubail I, Dupuis M, Guerrera IC, Charbit A. 2013. Possible links between stress defense and the tricarboxylic acid (TCA) cycle in Francisella pathogenesis. Mol Cell Proteomics 12:2278–2292. 10.1074/mcp.M112.024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brissac T, Ziveri J, Ramond E, Tros F, Kock S, Dupuis M, Brillet M, Barel M, Peyriga L, Cahoreau E, Charbit A. 2015. Gluconeogenesis, an essential metabolic pathway for pathogenic Francisella. Mol Microbiol 98:518–534. 10.1111/mmi.13139. [DOI] [PubMed] [Google Scholar]
- 54.Asare R, Abu Kwaik Y. 2010. Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ Microbiol 12:2559–2586. 10.1111/j.1462-2920.2010.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, Forsberg Å, Charbit A. 2009. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun 77:1866–1880. 10.1128/IAI.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller CN, LoVullo ED, Kijek TM, Fuller JR, Brunton JC, Steele SP, Taft-Benz SA, Richardson AR, Kawula TH. 2013. PanG, a new ketopantoate reductase involved in pantothenate synthesis. J Bacteriol 195:965–976. 10.1128/JB.01740-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elischewski F, Pühler A, Kalinowski J. 1999. Pantothenate production in Escherichia coli K12 by enhanced expression of the panE gene encoding ketopantoate reductase. J Biotechnol 75:135–146. 10.1016/s0168-1656(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 58.Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, Brittnacher MJ, Manoil C, Skerett SJ, Salama NR. 2009. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun 77:232–244. 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blake CC, Rice DW. 1981. Phosphoglycerate kinase. Philos Trans R Soc London B Biol Sci 293:93–104. [DOI] [PubMed] [Google Scholar]
- 60.Finn RD, Orlova EV, Gowen B, Buck M, van Heel M. 2000. Escherichia coli RNA polymerase core and holoenzyme structures. EMBO J 19:6833–6844. 10.1093/emboj/19.24.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suresh RV, Ma Z, Sunagar R, Bhatty V, Banik S, Catlett SV, Gosselin EJ, Malik M, Bakshi CS. 2015. Preclinical testing of a vaccine candidate against tularemia. PLoS One 10:e0124326. 10.1371/journal.pone.0124326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, Brouwer D, Nair V, Fischer ER, Wicke L, Curda AJ, Kupko JJ, Martens C, Crane DD, Bosio CM, Porcella SF, Celli J. 2009. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol 11:1128–1150. 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LoVullo ED, Sherrill LA, Perez LL, Pavelka MS. 2006. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology (Reading) 152:3425–3435. 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- 64.Alharbi A, Rabadi SM, Alqahtani M, Marghani D, Worden M, Ma Z, Malik M, Bakshi CS. 2019. Role of peroxiredoxin of the AhpC/TSA family in antioxidant defense mechanisms of Francisella tularensis. PLoS One 14:e0213699. 10.1371/journal.pone.0213699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bakshi CS, Malik M, Regan K, Melendez JA, Metzger DW, Pavlov VM, Sellati TJ. 2006. Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J Bacteriol 188:6443–6448. 10.1128/JB.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mahawar M, Atianand MK, Dotson RJ, Mora V, Rabadi SM, Metzger DW, Huntley JF, Harton JA, Malik M, Bakshi CS. 2012. Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J Biol Chem 287:25216–25229. 10.1074/jbc.M112.367672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download jb.00185-21-s0001.pdf, PDF file, 0.9 MB (898.5KB, pdf)
Data Availability Statement
The RNA-sequencing data are available at the NCBI GEO repository under the accession number GSE183001.