Supplemental Digital Content is available in the text.
Keywords: atherosclerosis; carotid artery diseases; lipoprotein(a); myocytes, smooth muscle; RNA, long noncoding; stroke
Background:
Long noncoding RNAs (lncRNAs) are important regulators of biological processes involved in vascular tissue homeostasis and disease development. The present study assessed the functional contribution of the lncRNA myocardial infarction-associated transcript (MIAT) to atherosclerosis and carotid artery disease.
Methods:
We profiled differences in RNA transcript expression in patients with advanced carotid artery atherosclerotic lesions from the Biobank of Karolinska Endarterectomies. The lncRNA MIAT was identified as the most upregulated noncoding RNA transcript in carotid plaques compared with nonatherosclerotic control arteries, which was confirmed by quantitative real-time polymerase chain reaction and in situ hybridization.
Results:
Experimental knockdown of MIAT, using site-specific antisense oligonucleotides (LNA-GapmeRs) not only markedly decreased proliferation and migration rates of cultured human carotid artery smooth muscle cells (SMCs) but also increased their apoptosis. MIAT mechanistically regulated SMC proliferation through the EGR1 (Early Growth Response 1)-ELK1 (ETS Transcription Factor ELK1)-ERK (Extracellular Signal-Regulated Kinase) pathway. MIAT is further involved in SMC phenotypic transition to proinflammatory macrophage-like cells through binding to the promoter region of KLF4 and enhancing its transcription. Studies using Miat–/– and Miat–/–ApoE–/– mice, and Yucatan LDLR–/– mini-pigs, as well, confirmed the regulatory role of this lncRNA in SMC de- and transdifferentiation and advanced atherosclerotic lesion formation.
Conclusions:
The lncRNA MIAT is a novel regulator of cellular processes in advanced atherosclerosis that controls proliferation, apoptosis, and phenotypic transition of SMCs, and the proinflammatory properties of macrophages, as well.
Clinical Perspective.
What Is New?
The contribution of long noncoding RNAs to the development and progression of vascular disease remains largely unknown.
The long noncoding RNA Myocardial Infarction Associated Transcript (MIAT) had previously been discovered in genome-wide association and transcriptomic profiling studies using diseased cardiovascular specimens and experimental models.
We describe a novel role for MIAT in regulating proliferation and transdifferentiation of arterial smooth muscle cells and inflammatory activity in macrophages, as well, during atherosclerotic plaque development and progression.
What Are the Clinical Implications?
Long noncoding RNAs possess key regulatory functions, directly interacting and mediating expression and functionality of proteins, other RNAs, and DNA, as well.
The long noncoding RNA MIAT plays a key role during atherosclerotic plaque development and lesion destabilization. Its expression becomes highly increased in high-risk patients with vulnerable plaques.
Therapeutic targeting of MIAT, using antisense oligonucleotides, offers novel treatment options for patients with advanced atherosclerosis in carotid arteries at risk of stroke.
Atherosclerosis is a chronic disease of intra-arterial atheromatous plaque progression that can remain asymptomatic for decades.1 Pathologically, advanced lesions can be separated into “stable” and “unstable” (also referred to as “vulnerable”) lesions.2 Subclinical data show that plaques are most common in the iliofemoral arteries (44%), followed by the carotid arteries (31%), aorta (25%), and coronary arteries (18%).3 The rupture of lesions in the carotid and coronary vascular beds is the main cause of stroke and myocardial infarction, respectively. Stroke remains a massive public health problem with an increasing need for better strategies to prevent and treat the disease in our aging society.4 Despite surgical removal of the plaque with carotid endarterectomy or stenting for symptomatic patients, obtaining a better understanding of the cellular mechanisms and regulatory networks driving carotid plaque development and progression is essential to identifying novel therapeutic targets.
The pathogenesis of carotid atherosclerotic lesions is orchestrated by different vascular cell subtypes.5 Vascular smooth muscle cell (SMC) accumulation is considered a hallmark of atherosclerosis and vascular injury.6 It is well documented that the aberrant proliferation of SMCs can promote plaque formation.6,7 Recent genetic lineage tracing studies show that SMC phenotypic switching into macrophage-like cells can promote vascular inflammation and the progression of atherosclerosis.8,9 In this advanced disease process, lipid loading and phagocytosis play a crucial role.10,11 Accumulating data suggest that constant uptake of oxidized low-density lipoprotein (oxLDL) in macrophages and SMCs can trigger foam cell formation while activating an inflammatory cascade that enhances plaque vulnerability.12,13
Long noncoding RNAs (lncRNAs), defined as nonprotein coding transcripts longer than 200 nucleotides, have emerged as important regulators in various biological processes and during disease development.14 However, the functional relevance during disease progression remains elusive for most of these transcripts.15 The lncRNA Myocardial Infarction Associated Transcript (MIAT, also known as Gomafu) was initially identified in a genome-wide association study in Japanese patients with myocardial infarction.16 Recent evidence suggests that MIAT regulates endothelial cell inflammation in diabetic retinopathies by competing as an endogenous RNA in a feedback loop with the vascular endothelial growth factor and miR-150-5p.17 Whether and how MIAT participates in atherosclerosis and mechanisms that destabilize advanced plaques has yet to be experimentally addressed.
We performed RNA profiling in 2 independent patient cohorts with advanced carotid plaques undergoing carotid endarterectomy. We further analyzed carotid plaques from a Yucatan LDLR–/– mini-pig model of advanced atherosclerosis and performed mechanistic studies using vascular injury (carotid ligation) and inducible plaque rupture (ligation and cuff) in mice to establish functional relevance for MIAT during lesion progression and destabilization. We show that MIAT not only promotes proliferation of carotid SMCs through the ERK (extracellular signal-regulated kinase)-ELK1 (ETS transcription factor ELK1)-EGR1 (early growth response 1) pathway, but also regulates macrophage activation through Nuclear Factor κ-light-chain enhancer of activated B cells (NF-κB) signaling. In addition, it regulates the phenotypic transition of SMCs into macrophage-like cells through direct interaction with the promoter region of Krüppel-like factor 4 (KLF4) to accelerate its transcriptional activation.
Methods
The authors declare that all data that support the findings of this study are available within the article and its Supplemental Material. Detailed descriptions of materials and methods used in this study, including the preparation and transcriptomic analysis of human plaques, RNA scope in murine and human tissue, laser capture microdissection of human carotid lesions, the inducible plaque rupture model in mice, the murine carotid artery ligation model, cholesterol and triglyceride measurements in mouse plasma, in vitro cell culture and transfection, proliferation, migration, apoptosis, and oxLDL uptake assays using the kinetic live-cell imaging in murine and human cell lines, the porcine atherosclerosis model, in situ hybridization, immunohistochemical and immunofluorescent analysis, RNA immunoprecipitation, luciferase reporter assays, MIAT modulation in vitro and in vivo, protein extraction, and immunoblotting can be found in the Supplemental Material.
Human Advanced Atherosclerotic Carotid Artery Plaques
Advanced carotid artery atherosclerotic lesions (carotid plaques) and nonatherosclerotic iliac artery controls were obtained from the Biobank of Karolinska Endarterectomies as previously published.18 RNA profiling and gene expression analysis were performed as described.18 Microarray data related to this study are available from Gene Expression Omnibus data sets (accession number GSE21545). Paraffin-embedded sections were used for in situ hybridization and immunostaining as described in the Supplemental Material. Another set of independent carotid atherosclerotic plaques originate from the Munich Vascular Biobank.18 From this biobank, plaques with either a ruptured or stable plaque phenotype were analyzed and used for laser-capture microdissection experiments as described in the Supplemental Material. All studies were approved by the Ethical Committee of Stockholm and Klinikum Rechts der Isar, Technical University Munich; patient informed consent was obtained according to the Declaration of Helsinki.
Statistics
Data are presented as mean±SEM, unless stated differently. Groups were compared by using the Student t test (2-tailed) for parametric data. Normality was tested to ensure that parametric testing was appropriate. Gene expression data were analyzed using log2 expression values and then transformed into fold-change for presentation purposes. When comparing multiple groups, data were analyzed using ANOVA with post hoc multiple comparisons using the Tukey method. For the IncuCyte experiments, normalized data were compared by 2-way repeated-measures ANOVA (for confluence data) or area under the curve (for count data). For analysis of multiple gene expression panels, t tests corrected for multiple comparisons using the Holm-Sidak method were performed. Categorical data were analyzed by using the χ2 test or Fisher exact test when 1 category was <3. A value of P<0.05 was considered statistically significant. All analyses were performed using GraphPad Prism version 9, except that microarray data set analyses were performed with GraphPad Prism versions 6 and 7. A linear regression model adjusted for age and sex and a 2-sided Student t test assuming nonequal deviation with correction for multiple comparisons according to Bonferroni were used for analyses of the noncoding RNAs from microarrays.
Results
MIAT Was Increased in Advanced Human Carotid Plaques and Important for SMC Survival
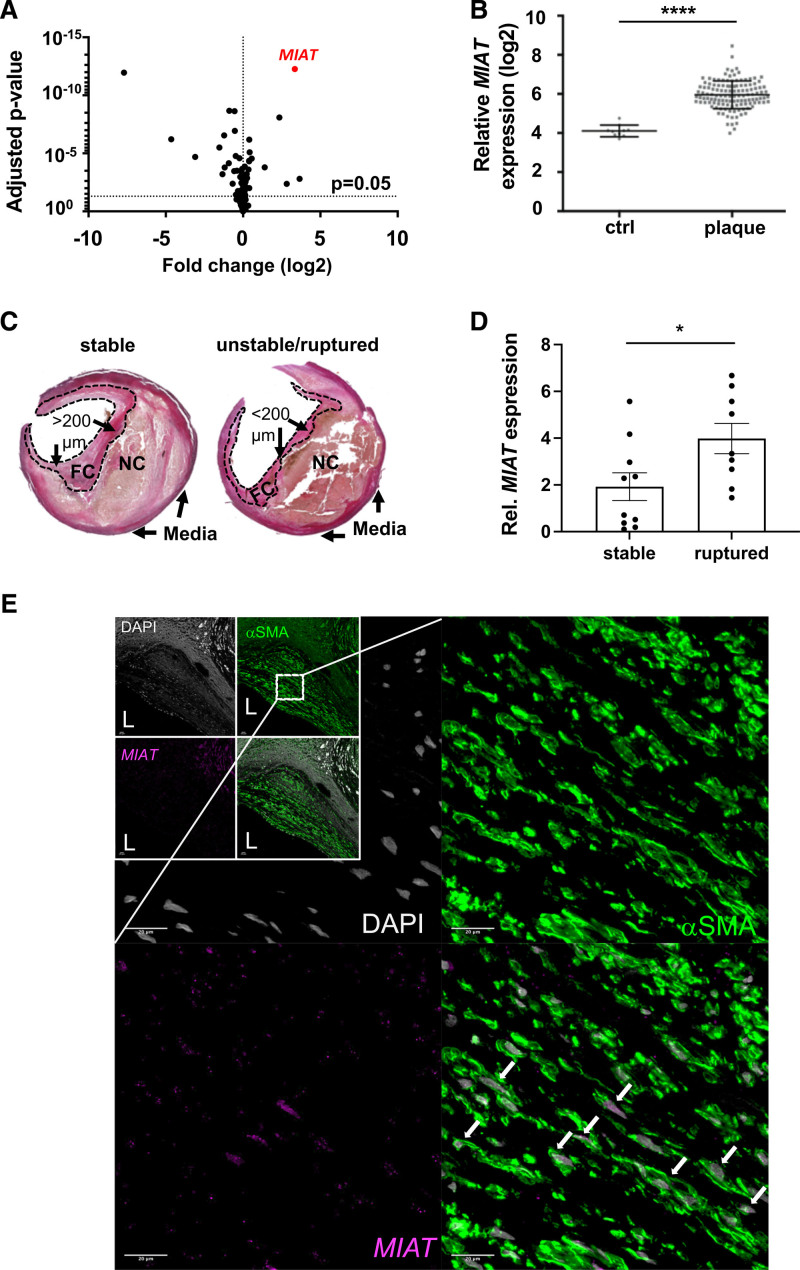
The lncRNA MIAT was identified as the most upregulated noncoding RNA transcript in microarray profiling comparing human carotid plaques (n=127) to nonatherosclerotic controls (n=10) from the Biobank of Karolinska Endarterectomies (Figure 1A and 1B). This result was validated by quantitative real-time polymerase chain reaction using plaques from a nonoverlapping set of patients in Biobank of Karolinska Endarterectomies (Figure S1A in the Supplemental Material, 77 human carotid plaques versus 13 nonatherosclerotic controls; patient demographics are in Table S1 in the Supplemental Material).
Figure 1.
MIAT expression is increased in advanced stages of human carotid artery disease. A, Scatter plot of deregulated noncoding transcripts in human carotid plaques (n=127) compared with nonatherosclerotic controls (n=10) from the Biobank of Karolinska Endarterectomies. B, Expression plot for MIAT based on the results presented in Figure 1A. C, Hematoxylin and eosin–stained stable and unstable/ruptured atherosclerotic lesions from the Munich Vascular Biobank. Dotted line indicates the laser-catapulted fibrous cap (FC). NC represents necrotic core. FC >200 µm was considered a stable lesion; FC <200 µm was unstable/ruptured. D, MIAT expression in laser captured microdissected stable versus ruptured fibrous caps (mean±SEM; unpaired nonparametric Student t test). E, Colocalization analysis of MIAT and αSMA protein was performed by RNAscope protocol. MIAT and αSMA protein were detected through Opal 690 and Opal 520, visualized in purple and green color, respectively. Nuclei are stained with DAPI (gray). Upper Left, the boxes represent overview images of the carotid plaques, and the zoomed-in images are magnifications acquired within the plaque shoulder region. White arrows highlight the nuclear detection of MIAT in cells expressing the smooth muscle cell marker (αSMA). Bar=100 µm. *P<0.05; ****P<0.0005. Data are presented as mean±SD (SEM in D) with Student t test and corrected for multiple comparison (A) using Bonferroni correction. Ctrl indicates control; DAPI, 4′,6-diamidino-2-phenylindole; Rel., relative; and αSMA, smooth muscle cell α-actin.
Next, we assessed MIAT expression in laser-captured microdissected fibrous caps from ruptured/unstable (cap thickness ≤200 µm, n=10) compared with stable (cap thickness ≥200 µm; n=10) advanced atherosclerotic lesions from the Munich Vascular Biobank19 (Figure 1C). In these SMC-enriched fibrous caps, MIAT was expressed 2-fold higher (P=0.031) in ruptured than in stable lesions from laser-dissected tissue (Figure 1D).
RNAscope revealed coexpression of MIAT with SMC α-actin (αSMA)–positive cells, and to a lesser extent with CD68 (Figure 1E, Figure S1B in the Supplemental Material, positive and negative controls in Figure S2A and S2D in the Supplemental Material). Conventional in situ hybridization confirmed MIAT being predominantly present in the medial layer of nonatherosclerotic control arteries (Figure S3A in the Supplemental Material: ctrl_MIAT). In stable plaques, MIAT expression was substantially elevated (Figure S3A in the Supplemental Material, plaque_MIAT) and detected predominantly in αSMA-positive cells, and to a lesser extent coexpressed with CD68 (Figure 1E, Figures S1B and S4A in the Supplemental Material).
MIAT Was Significantly Increased in Advanced Murine and Porcine Carotid Plaques and Regulated Proliferation in Vascular SMCs
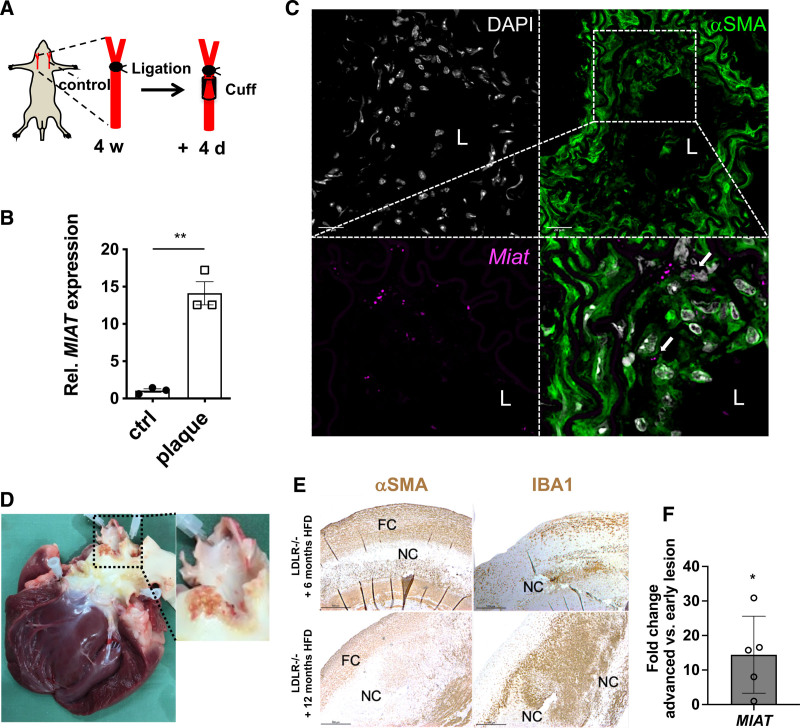
To determine the mechanistic role of MIAT in carotid plaque development and progression, we used a murine inducible plaque rupture model using incomplete ligation and cuff placement in ApoE–/– mice as previously described20,21 (Figure 2A). Consistent with the human data discussed earlier, a significant induction of Miat was observed in advanced murine carotid plaques compared with contralateral uninjured controls (Figure 2B). During early plaque formation using incomplete carotid ligation for 4 weeks (without cuff placement), Miat expression was substantially increased (Figure S4B in the Supplemental Material).
Figure 2.
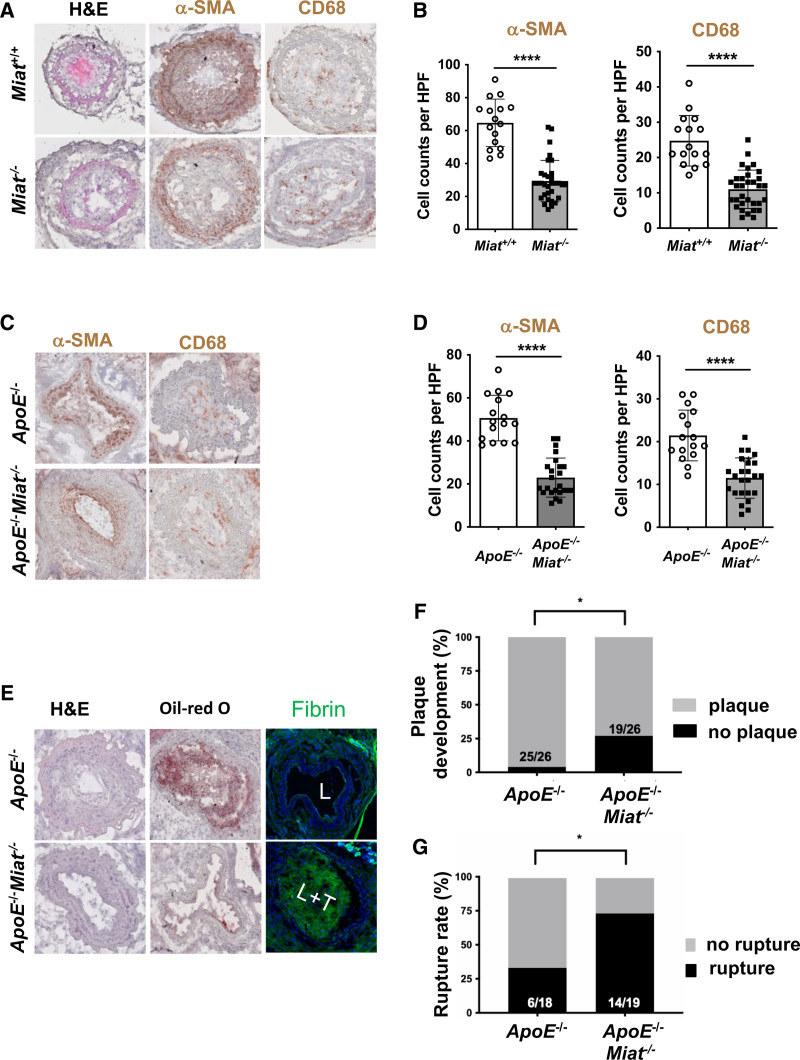
MIAT is significantly increased in murine carotid plaques and regulates proliferation in vascular smooth muscle cells. A, Illustration of the inducible plaque rupture model using incomplete ligation and cuff placement in ApoE–/– mice. B, Expression of Miat determined by quantitative real-time polymerase chain reaction from advanced plaques and contralateral controls from the inducible plaque rupture model presented in Figure 2B. C, Colocalization analysis of Miat and αSMA protein was performed by RNAscope in murine carotid plaques. Miat and αSMA protein were detected through Opal 690 (purple) and Opal 520 (green). Nuclei are stained with DAPI (gray). D, Ruptured plaque in the coronary ostia from an advanced lesion from LDLR–/– Yucatan mini-pigs being fed 12 months of high-fat diet (HFD). E, Immunostaining of smooth muscle cell α-actin (αSMA) and macrophage marker-ionized calcium-binding adapter molecule 1 (IBA1) in plaques from early (6 months HFD) and advanced (12 months HFD) carotid artery lesions (FC indicates fibrous cap; NC, necrotic core). F, Relative MIAT expression in advanced (12 months HFD) versus early (6 months HFD) carotid plaques. Data were analyzed by Student t test. *P<0.05; ***P<0.001. Ctrl indicates control; DAPI, 4′,6-diamidino-2-phenylindole; L, lumen; and Rel., relative.
RNAscope, in situ hybridization, and immunostaining showed that Miat expression was induced predominantly in αSMA+ cells in the SMC-rich fibrous cap, and in the medial layer structures of progressed lesions, as well (Figure 2C, Figure S4C in the Supplemental Material). This enhanced expression pattern was associated with increased cell proliferation (Ki-67+ cells) but not apoptosis (Caspase 3+ cells; Figure S5A in the Supplemental Material, negative controls for RNAscope in Figure S5B in the Supplemental Material).
Next, we evaluated MIAT in advanced lesions from LDLR–/– Yucatan mini-pigs after 6 months (early soft-plaque stage of atherosclerosis in this model) and 12 months (late and advanced atherosclerotic state) of being fed a high-fat diet (HFD; Figure 2D). Similar to the inducible plaque rupture model in mice and the expression changes in human plaques, advanced lesions (after 12 months of HFD) displayed more macrophages (indicated by the microglia/macrophage-specific ionized calcium-binding adapter molecule 1/IBA-1–positive cells) and fewer αSMA-positive cells than pigs that were on the shorter-term HFD (Figure 2E). MIAT expression was elevated in the more destabilized lesion phenotype (Figure 2F).
Modulation of MIAT Affected Proliferation and Apoptosis in Human Carotid Artery SMCs
To evaluate the cellular effect of MIAT modulation in human carotid smooth muscle cells (hCASMCs), we used a live-cell imaging system in combination with conventional immunohistochemical analysis. First, 3 different customized anti-MIAT GapmeRs were tested for their knockdown efficiency, and the most potent one was chosen for functional studies (Figure S5C and S5D in the Supplemental Material). Cells were dynamically monitored for rates of proliferation and apoptosis over time. Knockdown of MIAT (KD) limited proliferation rates and substantially enhanced apoptosis (Casp3+, green) of SMCs in a time- and dose-dependent manner (Figure 3A and 3B; Figure S5E in the Supplemental Material). Immunostaining with the proliferation marker Ki-67 and apoptosis marker Caspase 3 after MIAT knockdown confirmed these results (Figures 3c,d).
Figure 3.
MIAT regulates human carotid artery smooth muscle cell proliferation through the ERK/ELK1/EGR1 pathway. A and B, Proliferation (A) or apoptosis (B) of human carotid smooth muscle cells (hCASMCs) on knockdown of MIAT (KD) monitored through live-cell imaging over time (0–72 hours). Data were analyzed by 2-way ANOVA. C and D, Proliferation (C) or apoptosis (D) of hCASMCs on KD of MIAT determined by Ki-67 or Caspase 3 immunofluorescent staining (white arrows indicate Ki-67/Caspase 3–positive cells). Bar=50 µm. E, In silico analysis of transcription factors associated with MIAT (predicted binding free energy of <–30 Kcal/mol) using the regulatory RNA elements/RegRNA tool. F, MIAT fragment containing 2 predicted ELK1 binding sites (MIAT-ELK1_BS1_BS2) was transiently transfected and coimmunoprecipitated with endogenous ELK1 in hCASMCs. IgGs were used as control of immunoprecipitation (IP) specificity. MIAT or GAPDH (as unrelated target) enrichment in ELK1 IP fraction was quantified with quantitative real-time polymerase chain reaction and expressed as (2ΔCt)×100 ELK1 IP÷(2ΔCt)×100 IgG. ΔCt was calculated based on input. RNA content in IP or IgG was normalized on RPLPO mRNA. ELK1 IP efficiency was monitored by Western blot using an anti-ELK1 antibody. G, Transfected cells were lysed after 24 hours, and total protein was extracted. P-ERK normalized to total ERK was monitored using Western Blot. Quantification of the Western Blot was done with Fiji Image J software. H, MIAT-ELK1_BS1_BS2 was transiently transfected, and proliferation was monitored using the IncuCyte Live Cell Imaging System. Bar=50 µm. Data were analyzed by 2-way repeated-measures ANOVA (A), area under the curve (B), and Student t test (C, D, F, G). *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Casp indicates caspase; Ctrl, control; DAPI, 4′,6-diamidino-2-phenylindole; EV, empty vector (control); and IgG, immunoglobulin G.
MIAT regulated hCASMC Proliferation Through the ERK/ELK1/EGR1 Pathway
We next assessed the specific molecular mechanism through which MIAT mediates carotid plaque advancement and lesion destabilization. Previous data suggested that MIAT may act as a sponge for miR-150, which forms a negative feedback loop in the endothelium,17 cardiomyocytes,22 epithelial cells,23 and lung cancer cells.24 However, we did not detect significant changes in miR-150 in human carotid plaques or cultured hCASMCs after MIAT modulation (Figure S6A and S6B in the Supplemental Material), suggesting that the influence of MIAT on SMCs in advanced atherosclerosis is not mediated through its miR-150 interaction.
Transcription factors are master regulators controlling gene expression, chromatin stability, and cell homeostasis.25–27 To explore how MIAT regulates SMC proliferation, we hypothesized that it might affect molecular processes through alteration of transcription factors. On the basis of an in silico analysis using the regulatory RNA elements/RegRNA tool,28 we identified several transcription factors that potentially interact with MIAT but still require experimental validation: transcription factor CP2 (TFCP2), early growth response 1 (EGR1), vitamin D receptor (VDR), E1A binding protein 300 (P300/EP300), myeloid zinc finger 1 (MZF1), nuclear receptor subfamily 3 group C member 1 (GR/NR3C1), ETS transcription factor (ELK1), ETS pro- to-oncogene 1 (C-ETS/ETS1), and poly (AD-Ribose) polymerase (PARP). According to RegRNA, these transcription factors have a predicted binding free energy of < –30 kcal/mol (Figure 3E). To determine whether these transcription factors are regulated by MIAT, a custom-designed mRNA array investigating changes in expression for these transcription factors in SMCs after knockdown of MIAT was performed as previously described.29
Results revealed that EGR1, ELK1, and GR/NR3C1 were downregulated after knockdown of MIAT (Figure S6C in the Supplemental Material). EGR1 deficiency is associated with reduced plaque development in hyperlipidemic mice highlighting its importance in human atherosclerosis.30 Increased ERK signaling and p-ELK1/ELK1 binding to the proximal EGR1 promoter region influence EGR1 transcription.31,32 Because both components of this pathway, EGR1 and ELK1, were downregulated by MIAT inhibition, we investigated a potential direct interaction. Inhibition of MIAT limited phosphorylation of ERK (p-ERK), augmented accumulation of p-ELK1 in the nucleus, and subsequently reduced EGR1 expression, which overall accounted for a loss in SMC proliferation (Figure S6D and S6E in the Supplemental Material). Similar results could be observed in PDGF-BB (platelet-derived growth factor BB) prestimulated SMCs (Figure S7A in the Supplemental Material). Isolation of the nuclear and cytoplasmic fraction of hCASMCs followed by quantitative real-time polymerase chain reaction revealed a predominant nuclear expression of MIAT (Figure S7B in the Supplemental Material), leading to the hypothesis of a possible direct interaction of the lncRNA with transcriptionally active complexes. To investigate the contribution of MIAT to ERK signaling, we assessed its binding with ELK1 through RNA immunoprecipitation. Enrichment of MIAT in the ELK1-immunoprecipitated fraction (Figure 3F) supports the hypothesis of a MIAT-mediated transcriptional regulation at the EGR1 locus. In line with this, MIAT overexpression increased ERK phosphorylation (Figure 3G) and augmented hCASMCs proliferation (Figure 3H).
Moreover, increased MIAT levels correlated positively with EGR1 expression in proliferative SMCs (αSMA and Ki-67+) in human carotid plaques (Figure S7C in the Supplemental Material). The upregulation of EGR1 and ELK1 were also confirmed using quantitative real-time polymerase chain reaction in human and mouse carotid plaques (Figure S7D and S7E in the Supplemental Material).
MIAT Was Increased in Patients With Elevated Circulating Lipoprotein(a) Levels
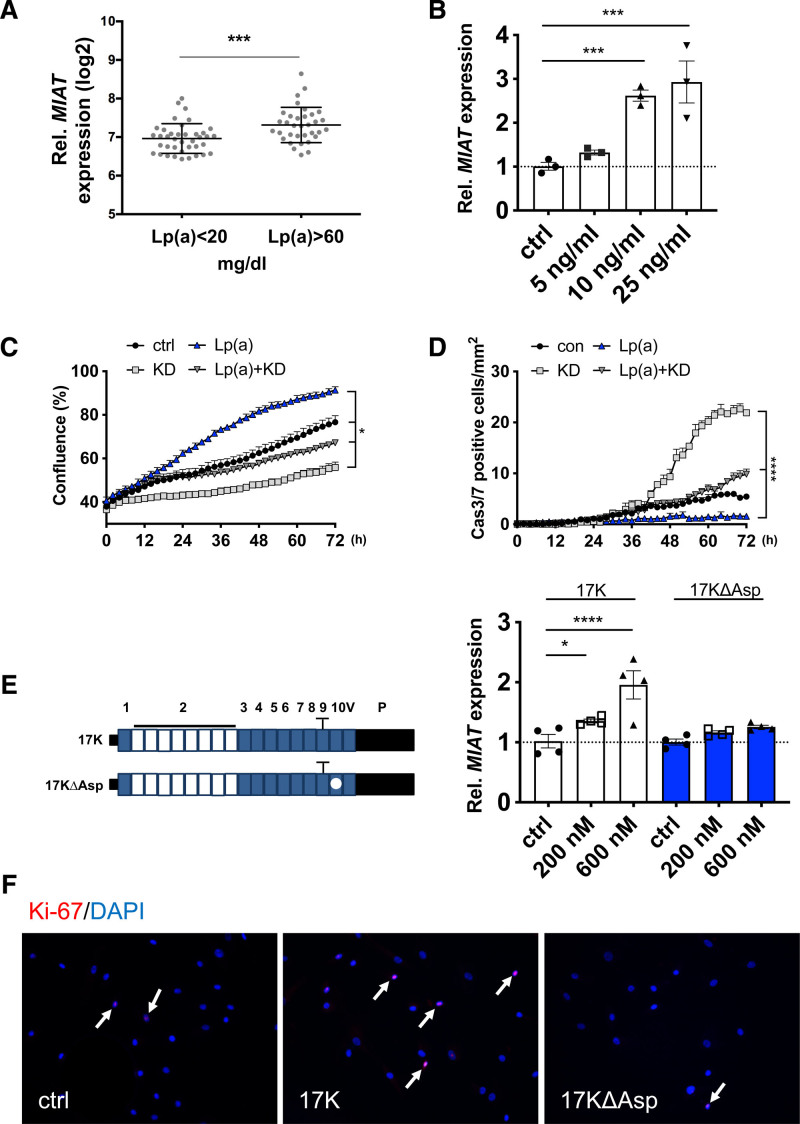
To further dissect the role of MIAT in atherosclerosis, we examined its expression in advanced human carotid plaques in relation to various clinical parameters of patients with carotid artery disease (plasma lipid profile, C-reactive protein, body mass index, hemoglobin A1c, creatinine, medications, etc; see Table S2 in the Supplemental Material). We discovered that MIAT expression in plaques was substantially elevated and correlated with higher plasma levels of lipoprotein(a) (Lp[a]), an important risk factor and potential novel therapeutic target in cardiovascular disease33–35 (Figure 4A). No significant correlation was observed for MIAT and other determinants of plasma lipid levels (triglycerides, low-density lipoproteins, and high-density lipoproteins; Table S2 in the Supplemental Material). We further assessed this finding in vitro, where treatment with lipoprotein(a) (Lp[a]) significantly increased MIAT in a dose-dependent manner (Figure 4B) and induced proliferation and limited apoptosis of SMCs. The knockdown of MIAT attenuated these effects in SMCs (Figure 4C and 4D), indicating that MIAT crucially regulates SMC dynamics and thus fibrous cap stability when elevated Lp(a) levels are present in atherosclerotic plaques.
Figure 4.
MIAT is increased in patients with elevated circulating lipoprotein(a) levels. A, Expression of MIAT in patients in Biobank of Karolinska Endarterectomies with low (<20 mg/dL) and high (>60 mg/dL) lipoprotein(a) (Lp[a]) serum levels. B. Expression of MIAT in hCASMCs on Lp(a) stimulation. C and D, Proliferation (C) or apoptosis (D) of hCASMCs on MIAT knockdown (KD), Lp(a) or combined treatment monitored live-cell imaging over time (data were analyzed by 2-way ANOVA). E, Left, Schematic of APO(a) peptide constructs with (17K) or without (17KDelta) an attached oxidized phospholipid. E, Right, Expression of MIAT in hCASMCs on 17K or 17KDelta stimulation. F, Immunostaining of Ki-67 in hCASMCs with 17K or 17KDelta stimulation (white arrows indicate Ki-67–positive cells). Data were analyzed by Student t test (A), 2-way repeated-measures ANOVA (C), area under the curve (D), or 1-way ANOVA (B, E). *P<0.05; ***P<0.001; ****P<0.0001. Cas indicates caspase; ctrl, control; DAPI, 4′,6-diamidino-2-phenylindole; and Rel., relative.
Last, we used 2 apo(a) peptide constructs with and without an attached oxidized phospholipid34 (17K and 17KDelta, respectively; Figure 4E) to determine their specific importance for Lp(a)-dependent stimulation of MIAT and subsequent regulation of SMCs. The wildtype 17K containing oxidized phospholipid (17K) significantly stimulated MIAT production (Figure 4E) and enhanced SMC proliferation. This effect was not observed using the 17KDelta variant (Figure 4F) lacking oxidized phospholipids. Previous findings have determined that downstream Lp(a) effects appear largely executed by the oxidation of phospholipids.36 Here, we investigated the effect of blocking oxidized phospholipids on Lp(a) by treating SMCs with a novel humanized mouse antibody targeting the oxidized phosphocholine head of phospholipids ATH3G10.37 ATH3G10 blocked MIAT expression in Lp(a)-prestimulated SMCs (Figure S8A in the Supplemental Material), thereby confirming the key role oxidized phospholipids play in Lp(a)-mediated vascular disease exacerbation.
MIAT Triggered Inflammation and Macrophage Activity in Advanced Atherosclerotic Lesions
Inflammation, matrix degradation, and macrophage activity play a decisive role in atherosclerotic lesion formation.10,11,35 A significant positive correlation between MIAT and inflammatory and matrix degradation indicators, such as transforming growth factor β1 (TGFβ1) and matrix metalloproteinase-2 (MMP2) was detected in advanced lesions (Figure S8B and S8C in the Supplemental Material).
The plaque microarray data consistently revealed that MIAT was not only significantly correlated with SMC marker genes such as actin alpha 2 (ACTA2), smoothelin (SMTN), and myosin heavy chain 11 (MYH11), but also several indicators of macrophage activity (such as CD80, CD36, CD163, CD40), cell proliferation (PCNA and NF-κB1), and inflammatory cytokines and growth factors including interleukins 6, 10 and 1β (IL6, IL10, and IL1β), TGFβ1, platelet-derived growth factor β (PDGFβ), and tumor necrosis factor α, as well (TNFα; Table S3 in the Supplemental Material).
To confirm that MIAT contributes to macrophage activity and is of relevance to inflammation in advanced lesions, we first evaluated their ability to internalize oxLDL and transform into foam cells. The knockdown of MIAT caused a reduction in oxLDL uptake in human monocyte–derived macrophages (Figure 5A and 5B). In addition, MIAT was markedly elevated in THP1-derived macrophages, which exerted a proinflammatory phenotype following lipopolysaccharide stimulation (Figure 5C, Figure S8D in the Supplemental Material). In contrast, lower expression rates of MIAT were seen in conjunction with IL4 (interleukin 4) pretreatment (Figure 5D, Figure S8E in the Supplemental Material), linking MIAT to a more proinflammatory macrophage phenotype in atherosclerosis. Lipid loading using oxLDL also significantly increased MIAT expression levels (Figure 5E).
Figure 5.
MIAT triggers inflammation and macrophage activity in advanced atherosclerotic lesions. A, Expression of MIAT in human monocyte-derived macrophages on MIAT knockdown (KD). B, Uptake of florescence-labeled oxLDL in human monocyte-derived macrophages on MIAT KD monitored through live-cell imaging and analyzed by 1-way ANOVA. C, Expression of MIAT in human monocyte-derived macrophages on stimulation with oxLDL. D and E, Expression of MIAT in THP1-derived macrophages stimulated with LPS (D) or IL4 (E). F, Immunostaining of NF-κB in human monocyte-derived macrophages on oxLDL stimulation with or without MIAT KD. Quantification is shown on the right side. G, Binding of endogenous MIAT to NFKB2/p52/p100 in THP1 cells. MIAT expression in THP1 cells was triggered by oxLDL stimulation and immunoprecipitation with NFKB2/p52/p100 or control IgG performed. MIAT or GAPDH (as unrelated target) enrichment in NFKB2 IP fraction were quantified with real-time quantitative polymerase chain reaction. Data were analyzed by Student t test (A, C–E, G), AUC (B), or Mann-Whitney U test (F). Bar=50 µm. **P<0.01; ***P<0.001; ****P<0.0001. ctrl indicates control; DAPI, 4′,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; IgG, immunoglobulin G; IL4, interleukin 4; IP, immunoprecipitation; LPS, lipopolysaccharide; NF-κB, Nuclear Factor κ-light-chain enhancer of activated B cells; oxLDL, oxidized low-density lipoprotein; and Rel., relative.
Next, we explored how MIAT affects molecular processes and inflammatory signaling in macrophages. We discovered that under nonstimulated (control) conditions, NF-κB activity remained at relatively low levels (Figure 5F). When challenged with oxLDL, NF-κB became activated, as indicated by its translocation into the nuclei of macrophages. This nuclear translocation was completely attenuated by MIAT knockdown (Figure 5F). OxLDL uptake was consistently substantially impaired after treatment with an NF-κB translocation inhibitor (Figure S8F in the Supplemental Material). MIAT knockdown further limited expression of transporters involved in cellular oxLDL uptake (CD36; steroid receptor RNA activator, SRA), whereas mediators of cholesterol efflux (ATP Binding Cassette Subfamily Members: ABCA1 and ABCG1) remained unchanged (Figure S8G and S8H in the Supplemental Material). These data suggest that MIAT facilitates NF-κB activation and translocation during plaque progression and destabilization. Similar changes in NF-κB activation and translocation were present in peritoneal mouse macrophages (Figure S9A and S9B in the Supplemental Material). A direct MIAT-NF-κB interaction (with NFκB2/p52/p100) was confirmed by RNA immunoprecipitation in THP1 cells (Figure 5G), where MIAT expression was induced through oxLDL stimulation. The NF-κB interaction with NF-κB–interacting lncRNA (NKILA)38 served as a positive control for the RNA immunoprecipitation experiment (Figure S8G in the Supplemental Material).
MIAT Participated in SMC Transdifferentiation Into Inflammatory Macrophage-Like Cells Through KLF4
SMC phenotypic switching plays a key role during the development and progression of atherosclerotic lesions.8,39 OxLDL stimulation induced the SMC phenotypic transition to inflammatory macrophage-like cells, evidenced by downregulation of SMC markers (ACTA2, Transgelin, Myosin heavy chain 11) and elevation of phagocytosis and macrophage markers (CD68, Galectin 3; Figure 6A). This shift in cellular content and marker gene expression was also evident in stable versus ruptured human carotid arteries (Figure S9C and S9D in the Supplemental Material). MIAT inhibition in oxLDL-primed SMCs did alter expression of the cholesterol sensing gene low density lipoprotein receptor (LDLR), but not of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR; Figure S9E in the Supplemental Material).
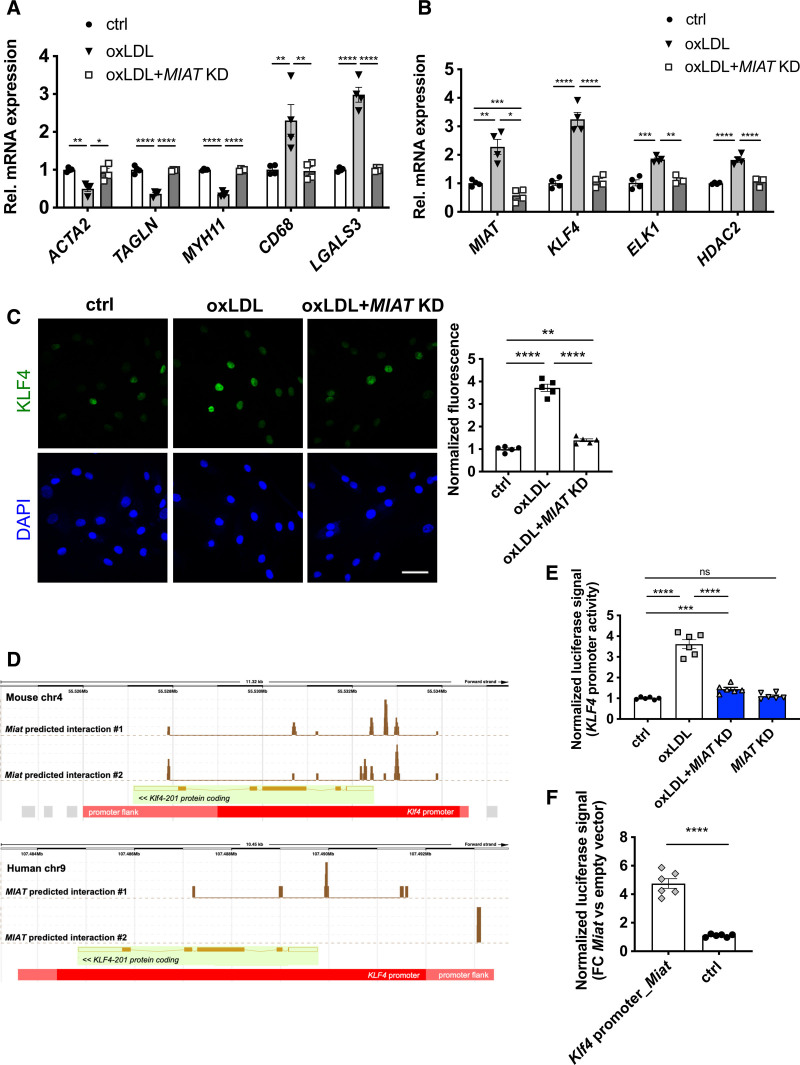
Figure 6.
MIAT participates in smooth muscle cell transdifferentiation into inflammatory macrophage-like cells through KLF4 activation. A, Expression of SMCs, phagocytosis and/or macrophage markers in hCASMCs stimulated with oxLDL and with or without MIAT knockdown (KD). B, Expression of MIAT, transcription factor KLF4, coeffectors ELK1 and HDAC2 in hCASMCs stimulated with oxLDL and with or without MIAT KD. C, Left, Immunostaining of KLF4 in hCASMCs stimulated with oxLDL and with or without MIAT KD; Right, fluorescence quantification. D, Interaction of Miat/MIAT with Klf4/KLF4 promoter as predicted by LongTarget v2.1. Peaks indicate predicted binding sites of Miat/MIAT within Klf4/KLF4 promoter region. E, Luciferase reporter assay with human KLF4 promoter on oxLDL stimulation/MIAT KD in hCASMCs. F, Luciferase reporter assay with murine Klf4 promoter (containing Miat predicted binding sites; Klf4 promoter_Miat) or promoter flanking regions (harboring no predicted Miat binding sites; Ctrl) in mouse aortic SMCs on Miat overexpression (pCAG-Miat, murine). Bar=50 µm. Data were analyzed by 1-way ANOVA.*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; P value >0.05 indicates no significance. ctrl indicates control; DAPI, 4′,6-diamidino-2-phenylindole; FC, fibrous cap; hCASMC, human carotid smooth muscle cell; KLF4, Krüppel-like factor 4; oxLDL, oxidized low-density lipoprotein; Rel., relative; and SMC, smooth muscle cell.
The proinflammatory transdifferentiation process could be blocked in vitro by inhibiting MIAT expression (Figure 6A). Recent studies identified Krüppel-like factor 4 (KLF4) as a key transcription factor mediating the SMC-macrophage transition.8,9 We observed a remarkable induction of KLF4 after oxLDL stimulation in hCASMCs (Figure 6B and 6D), and in advanced human (Figure S9F in the Supplemental Material) and mouse carotid plaques (Figure S9G in the Supplemental Material). The coeffectors ELK1 and HDAC2 were also increased (Figure 6B). The knockdown of MIAT successfully reversed these changes and decreased KLF4, ELK1, and HDAC2 expression (Figure 6B). MIAT knockdown was further able to limit KLF4 on Lp(a) stimulation (Figure S10A in the Supplemental Material). Thus, MIAT participates in oxLDL/Lp(a)-induced SMC-macrophage transition through inducing KLF4 expression.
To thoroughly dissect the molecular mechanism of how MIAT interacts with KLF4, we used the lncRNA:DNA interaction prediction tool LongTarget,40 which predicted a putative MIAT-KLF4 promoter-binding pattern for human and mouse (Figure 6D). To assess the transcriptional activity, we conducted a luciferase reporter assay using the KLF4/Klf4 promoter region. Human CASMCs were transfected with the pLS-KLF4 promoter luciferase vector, with or without oxLDL stimulation. OxLDL stimulation led to an induction of luciferase activity, representing the transcriptional activity of KLF4, here serving as a positive control (Figure 6E). Knockdown of MIAT (MIAT KD) was able to block the oxLDL-induced transcription of KLF4 completely. In contrast, overexpression of Miat enhanced the transcriptional activity of the Klf4 promoter (Klf4 promoter_Miat) in mouse aortic SMCs (Figure 6F) compared with control (promoter-flanking region) without predicted Miat binding sites. In summary, these experiments suggest that MIAT acts as an enhancer of KLF4 transcription through direct interaction with its promoter.
Miat Deletion Affected SMC Proliferation and Plaque Vulnerability in Vivo
For in vivo investigations into Miat-mediated SMC proliferation, Miat–/– mice and their respective wildtype controls were exposed to carotid ligation, enabling us to investigate their responsiveness to stenotic flow obstruction. Miat–/– mice presented with substantially less proliferation and increased apoptosis in their SMC-enriched intima-medial layers. αSMA-positive cells were significantly decreased in Miat–/– mice after vascular injury (Figure 7A and 7B). SMCs derived from Miat–/– mice also displayed a decreased migratory capacity and a higher rate of apoptosis compared with Miat+/+ controls when assessed with live-cell imaging over time (Figure S10B and S10C in the Supplemental Material). Overall, Miat-depleted murine SMCs displayed changes similar to human carotid SMCs after MIAT knockdown.
Figure 7.
MIAT deletion affects smooth muscle cell proliferation and plaque vulnerability in vivo. A, Morphology (H&E), immunostaining for CD68 and smooth muscle cell α-actin (αSMA) in Miat–/– mice versus littermate Miat wildtype controls on carotid ligation injury. B, Analysis of cell counts (8 times/animal) per high-power field (HPF) for αSMA and CD68 from A. C, Morphology (H&E), immunostaining for CD68 and αSMA in ApoE–/–Miat–/– and ApoE–/– (Miat wildtype) controls on exposure to the inducible plaque rupture model (incomplete ligation and cuff placement). D, Cell counts (8 times/animal) per HPF for αSMA and CD68 from C. E, H&E, Oil-Red O (indicating lipid deposition), and cross-linked immunofluorescent fibrin staining (indicating an atherothrombotic event) in ApoE–/–Miat–/– versus Apo–/– Miat+/+ littermate controls (L indicates lumen; L+T, lumen with thrombus). F, Plaque development (in %) when using the inducible plaque rupture model in ApoE–/–-Miat–/– versus ApoE–/–Miat+/+ mice. G, Rupture ratio (in %) in the inducible plaque rupture model comparing ApoE–/–Miat–/– versus ApoE–/– (Miat wildtype) controls. Data were analyzed by Student t test (B, D). Fisher exact test was used to determine plaque development and rupture ratio. *P<0.05; **P<0.01. H&E indicates hematoxylin and eosin.
To further assess the role of MIAT in plaque destabilization, we cross-bred Miat–/– with ApoE–/– mice and exposed them to the inducible plaque rupture model. Here, Miat–/–ApoE–/– mice presented with thinner fibrous caps and a more unstable plaque phenotype when assessing plaque rupture rates (Figure 7C and 7G). The direct Miat targets Egr1 and Elk1 were substantially repressed in advanced lesions of Miat–/–ApoE–/– mice compared with littermate Miat+/+ApoE–/– control mice (Figure S10D in the Supplemental Material), which explains the less proliferative SMC phenotype in response to ligation and cuff placement. Atherogenesis and overall plaque development were however significantly augmented in Miat–/–ApoE–/– mice compared with their Miat+/+ApoE–/– littermate controls (Figure 7F), which we attribute to reduced expression levels of Klf4 in double-knockout mice (Figure S10D in the Supplemental Material). Lack of Klf4 in SMCs has previously been shown to augment the development of atherosclerosis.9 Lipid levels and lipid profile (total cholesterol, low density lipoprotein, very low-density lipoprotein, high density lipoprotein, triglycerides) were not altered between Miat–/–ApoE–/– and ApoE–/– mice (Figure S10E in the Supplemental Material). This strongly suggests that limited plaque development in the double-knockout mice is a distinct process that initiates within the vascular wall.
Similar to the murine data described earlier, we were able to detect higher expression levels of MIAT, KLF4, EGR1, and ELK1 (Figure S10F in the Supplemental Material) in Yucatan LDLR-deficient mini-pigs with more severe lesions (12 versus 6 months HFD).
Discussion
Increasing evidence for lncRNAs suggests that they are involved in numerous important biological events, such as serving as scaffolding proteins, shaping nuclear architecture, imprinting genomic loci, and regulating transcriptional activity.41 The aberrant expression or mutation of lncRNA genes has been implicated in various human diseases.14 In particular, in vascular biology and disease, several lncRNAs have been reported as important functional regulators of endothelial cells and SMCs, such as lincRNA-p21,42 ANRIL,43,44 SENCR,45,46 MALAT1,47 SNHG12,48 Tie-1AS,49,50 and CARMN.51 In our present study, we identified a novel molecular mechanism through which the lncRNA MIAT controls proliferation, apoptosis, and phenotypic transition of SMCs, and drives proinflammatory macrophage activity during foam cell and necrotic core formation, as well.
MIAT, also known as RNCR2 (retinal noncoding RNA 2) and Gomafu,52,53 shows extensive evolutionary and functional conservation among mammals. It was first identified as a novel transcript that confers a genetic risk of myocardial infarction in a Japanese cohort.16 Later, it was shown to associate with cardiac hypertrophy,22 diabetic cardiomyopathy,54 and cardiac fibrosis,55 mainly by acting as a sponge for different miRNAs. In addition, MIAT regulates diabetes-induced microvascular dysfunction by competing with endogenous vascular endothelial growth factor and miR-150 in retinal endothelial cells. Qin et al56 also confirmed that Miat promotes neurovascular remodeling in the eye and brain. Miat knockdown leads to cerebral microvascular degeneration, progressive neuronal loss and neurodegeneration, and behavioral deficits in Alzheimer disease, as well.56 Our current data suggest that MIAT may not only play a crucial role in microvascular diseases, but also macrovascular diseases such as carotid stenosis and ischemic forms of stroke (Figure 8D).
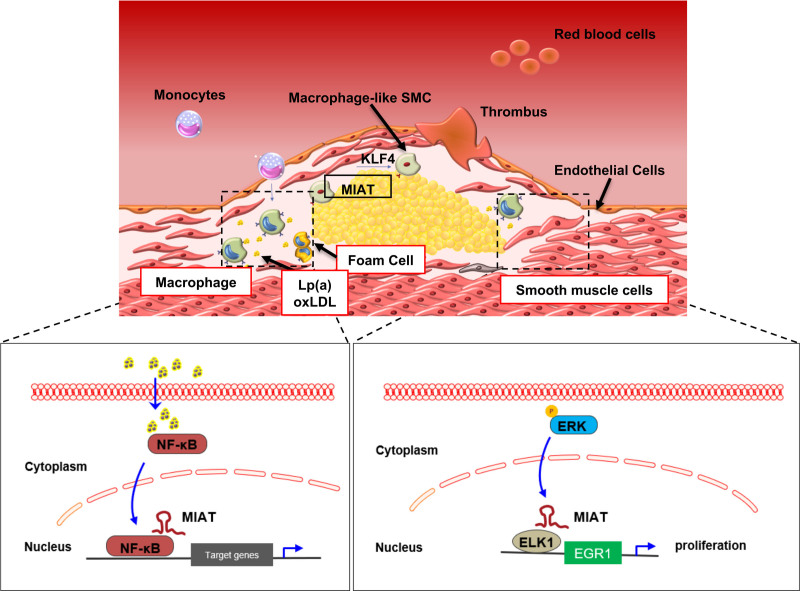
Figure 8.
Proposed mechanism of action for MIAT in advanced atherosclerosis and plaque destabilization. KLF4 indicates Krüppel-like factor 4; Lp(a), lipoprotein(a); NF-κB, Nuclear Factor κ-light-chain enhancer of activated B cells; oxLDL, oxidized low-density lipoprotein; and SMC, smooth muscle cell.
We discovered an upregulation of MIAT in human and murine advanced carotid plaques, where it was coexpressed and relevant for both, lesional SMCs and macrophages. The aberrant proliferation of SMCs is considered a hallmark of carotid plaque formation and progression.6,7 SMCs with high MIAT expression displayed a more proliferative status in our murine vulnerable plaque rupture model, and in human advanced lesions, as well, than those with low expression. Furthermore, increased MIAT expression levels or Lp(a) treatment significantly promoted proliferation of in vitro cultured hCASMCs, whereas the loss of MIAT manifested the opposite phenotype. Increased Lp(a) levels are closely linked to atherosclerosis development and progression.32,57 Indeed, our data from advanced human carotid lesions indicate a positive correlation for high Lp(a) levels and MIAT expression. Inhibition of MIAT strongly mediated the consequences of Lp(a) stimulation in SMCs.
EGR1 is an inducible protein that belongs to the early growth response gene family and has been implicated in cellular proliferation, differentiation, and engagement with cell death pathways in vascular cells.58 A previous study shows that EGR1 mediates platelet-derived growth factor–induced sphingosine kinase 1 (SphK1) expression to promote pulmonary artery SMC proliferation.59 Recent publications further demonstrate that serine/arginine-rich splicing factor 1 promotes vascular SMC proliferation through an EGR1/KLF5-mediated signaling pathway.60 In addition, EGR1 activation is dependent on MEK-ERK signaling.31,32 Constitutively bound ELK1 at the EGR1 proximal promoter occurs, which leads to increased pELK1 binding and de novo recruitment of RNA polymerase II to this region.32 In our study, we first focused on a panel of transcription factors predicted to interact with MIAT based on an in silico analysis. This analysis revealed ELK1 and EGR1 as relevant targets. Activation of ERK signaling increased cell proliferation, whereas blocking ERK signaling abolished this phenotype. In line with this concept and previous studies,30 we confirmed that EGR1 expression was highly increased in advanced carotid plaques and positively correlated with MIAT expression and SMC proliferation rates.
Macrophage infiltration and activation around lipid pools is crucial for necrotic core formation and progressing plaque vulnerability.61 In atherosclerotic lesions, macrophages scavenge oxLDL, which has strong proinflammatory and immunogenic properties.62,63 NF-κB signaling is thought to be involved in this process64 and inhibition of NF-κB prevents foam cell formation and plaque destabilization.65 Our current discovery that knockdown of MIAT impaired oxLDL uptake in human monocyte-differentiated and murine peritoneal macrophages suggests that MIAT regulates inflammation and lesion progression in carotid plaques. Furthermore, oxLDL-induced activation and translocation of NF-κB were attenuated after inhibiting MIAT and blocking its proinflammatory contribution to plaque progression through direct targeting of the NF-κB subunit NFκB2/p52/p100.
Conventionally, phenotypic switching of SMCs from quiescent and contractile to proliferative and synthetic states occurs during vascular development and in response to injury.66 Recent studies suggest that SMCs that undergo phenotypic switching can acquire macrophage-like properties, which is of fundamental importance to atherosclerosis.67 This transdifferentiation process, which plays a key role in plaque pathogenesis, seems to be heavily dependent on KLF4 activity.9 Cooperative binding of KLF4, pELK-1, and HDAC2 to a G-C repressor element in the SM22α promoter mediates this process.8 SMC-specific conditional knockout of KLF4 results in reduced numbers of SMC-derived mesenchymal stem cells and macrophage-like cells, and marked reductions in lesion size, as well, and increases in multiple indices of plaque stability.9 Consistent with our results, KLF4, ELK1, and HDAC2 are induced by increased MIAT expression or oxLDL stimulation. Elevated MIAT levels and oxLDL induction, as well, resulted in reduced expression of SMC markers (ACTA2) and increased macrophage determinants (CD68). The knockdown of MIAT reversed these changes, suggesting that MIAT is crucially involved in oxLDL-induced SMC-macrophage transdifferentiation through control of KLF4. Our data suggest direct binding of MIAT to the KLF4 promoter, which is sufficient to induce its transcription. MIAT is not the only lncRNA with an enhancer-like function. Similar roles have been described previously for H19 in abdominal aortic aneurysm29 and, more recently, for REG1CP in promoting tumorigenesis through REG3A.68
The effect of experimental MIAT modulation appears to be quite complex. Miat–/–ApoE–/– mice seem largely protected from developing advanced lesions compared with littermate Miat++-ApoE–/– controls. During atherogenesis, genetic depletion of Miat limited Klf4-mediated SMC-macrophage transition and inflammation. However, once lesions were established in Miat–/–ApoE–/– animals, they appeared unstable and more likely to rupture than Miat wildtype (but ApoE–/–) littermate controls in our experimental plaque rupture model. The downregulation of the direct Miat targets Erk, Elk1, and Egr1 was responsible for the limited SMC proliferation and the resulting fibrous cap thinning.
In conclusion, we identified a key functional role for the lncRNA MIAT in atherosclerotic plaque development and destabilization. In high-risk individuals with advanced atherosclerotic plaques, increased MIAT expression has 3 main effects after exposure to elevated oxLDL and Lp(a) levels: (1) it can promote SMC proliferation through the ERK-ELK1-EGR1 pathway; (2) it activates proinflammatory macrophages through NF-κB signaling, and (3) it mediates SMC phenotypic transdifferentiation to inflammatory macrophage-like cells through increasing the transcriptional activity of KLF4. Knockdown of MIAT in vitro and in vivo mitigated these disease-relevant consequences. Ultimately targeting MIAT can serve as a novel molecular treatment strategy to limit vascular inflammation and atherosclerosis progression under certain conditions.
Article Information
Acknowledgments
The authors greatly appreciate the pCAG_Miat vector that Dr Blackshow (The Johns Hopkins University, Baltimore, MD) and Prof Qin (University of Alabama at Birmingham) kindly provided. Miat-deficient mice (originally denoted Gomafu CDB1347K) were a kind gift from Prof Nakagawa at the RIKEN Center for Life Science Technologies, Kobe, Japan. The authors further thank Prof Engelhardt, Dr Dueck, and Dr Ramanujaran (Institute of Pharmacology and Toxicology, Technical University Munich) for critical discussions and sharing of expertise and equipment, and Prof Misgeld, as well, for providing access to confocal microscopy. All have provided permission to be named in the article.
Sources of Funding
This study is supported by the Swedish Heart-Lung-Foundation (20180680), the Swedish Research Council (Vetenkapsrådet, 2019-01577), the European Research Council (ERC-StG NORVAS), a DZHK Junior Research Group (JRG_LM_MRI), the SFB1123 and TRR267 of the German Research Council (DFG), the National Institutes of Health (NIH; 1R011HL150359-01), the Bavarian State Ministry of Health and Care through the research project DigiMed Bayern (all to L. Maegdefessel). Dr Winski has received support from The Committee for Doctoral Education at Karolinska Institutet (CSTP/Research Internship programmes). Dr Matic is the recipient of fellowships and awards from the Swedish Research Council (VR, 2019–02027), Swedish Heart-Lung Foundation (HLF, 20200621, 20200520, 20180244, 201602877, 20180247), Swedish Society for Medical Research (SSMF, P13-0171). Dr Matic also acknowledges funding from Sven and Ebba-Christina Hagberg, Tore Nilsson’s, Magnus Bergvall’s and Karolinska Institute research (KI Fonder) and doctoral education (KID) foundations. Dr Koschinsky is supported by the Heart and Stroke Foundation of Canada.
Disclosures
None.
Supplemental Materials
Supplemental Methods
Figures S1–S10
Tables S1–S4
References 69–74
Supplementary Material
Nonstandard Abbreviations and Acronyms
- EGR1
- early growth response 1
- ELK1
- ETS transcription factor ELK1
- ERK
- extracellular signal-regulated kinase
- hCASMC
- human carotid smooth muscle cell
- KLF4
- Krüppel-like factor 4
- lncRNAs
- long noncoding RNAs
- Lp(a)
- lipoprotein(a)
- MIAT
- Myocardial Infarction Associated Transcript
- NF-κB
- nuclear factor κ-light-chain-enhancer of activated B cells
- oxLDL
- oxidized low-density lipoprotein
- SMC
- smooth muscle cell
F. Fasolo, H. Jin, and G. Winski contributed equally.
V. Paloschi and L. Maegdefessel contributed equally.
Supplemental Material is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.120.052023.
For Sources of Funding and Disclosures, see page 1581.
Contributor Information
Francesca Fasolo, Email: francesca.fasolo@tum.de.
Hong Jin, Email: hong.jin@ki.se.
Greg Winski, Email: greg.winski@ki.se.
Ekaterina Chernogubova, Email: ekaterina.chernogubova@ki.se.
Jessica Pauli, Email: Jessica.pauli@tum.de.
Hanna Winter, Email: hanna.winter@tum.de.
Daniel Y. Li, Email: danielcolumbianyc@gmail.com.
Nadiya Glukha, Email: nadiya.glukha@tum.de.
Sabine Bauer, Email: s.bauer@tum.de.
Susanne Metschl, Email: susanne.metschl@tum.de.
Zhiyuan Wu, Email: zhiyuan.wu@tum.de.
Marlys L. Koschinsky, Email: mlk@robarts.ca.
Muredach Reilly, Email: mpr2144@cumc.columbia.edu.
Jaroslav Pelisek, Email: jaroslav.pelisek@usz.ch.
Wolfgang Kempf, Email: wolfgang.kempf@tum.de.
Hans-Henning Eckstein, Email: Hans-Henning.Eckstein@mri.tum.de.
Oliver Soehnlein, Email: soehnlein@uni-muenster.de.
Ljubica Matic, Email: Ljubica.Matic@ki.se.
Ulf Hedin, Email: ulf.hedin@ki.se.
Alexandra Bäcklund, Email: alexandra.backlund@ki.se.
Claes Bergmark, Email: claesbergmark@hotmail.com.
Valentina Paloschi, Email: Valentina.Paloschi@tum.de.
References
- 1.Sadat U, Jaffer FA, van Zandvoort MA, Nicholls SJ, Ribatti D, Gillard JH. Inflammation and neovascularization intertwined in atherosclerosis: imaging of structural and molecular imaging targets. Circulation. 2014; 130:786–794. doi: 10.1161/CIRCULATIONAHA.114.010369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Lammeren GW, den Ruijter HM, Vrijenhoek JE, van der Laan SW, Velema E, de Vries JP, de Kleijn DP, Vink A, de Borst GJ, Moll FL, et al. Time-dependent changes in atherosclerotic plaque composition in patients undergoing carotid surgery. Circulation. 2014; 129:2269–2276. doi: 10.1161/CIRCULATIONAHA.113.007603 [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Friera L, Peñalvo JL, Fernández-Ortiz A, Ibañez B, López-Melgar B, Laclaustra M, Oliva B, Mocoroa A, Mendiguren J, Martínez de Vega V, et al. Prevalence, vascular distribution, and multiterritorial extent of subclinical atherosclerosis in a middle-aged cohort: the PESA (Progression of Early Subclinical Atherosclerosis) study. Circulation. 2015; 131:2104–2113. doi: 10.1161/CIRCULATIONAHA.114.014310 [DOI] [PubMed] [Google Scholar]
- 4.Donnan GA, Fisher M, Macleod M, Davis SM. Stroke. Lancet. 2008; 371:1612–1623. doi: 10.1016/S0140-6736(08)60694-7 [DOI] [PubMed] [Google Scholar]
- 5.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011; 473:317–325. doi: 10.1038/nature10146 [DOI] [PubMed] [Google Scholar]
- 6.Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jørgensen HF. Extensive proliferation of a subset of differentiated, yet plastic, medial vascular smooth muscle cells contributes to neointimal formation in mouse injury and atherosclerosis models. Circ Res. 2016; 119:1313–1323. doi: 10.1161/CIRCRESAHA.116.309799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Y, Nagel DJ, Zhou Q, Cygnar KD, Zhao H, Li F, Pi X, Knight PA, Yan C. Role of cAMP-phosphodiesterase 1C signaling in regulating growth factor receptor stability, vascular smooth muscle cell growth, migration, and neointimal hyperplasia. Circ Res. 2015; 116:1120–1132. doi: 10.1161/CIRCRESAHA.116.304408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22α promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ Res. 2012; 111:685–696. doi: 10.1161/CIRCRESAHA.112.269811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015; 21:628–637. doi: 10.1038/nm.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finney AC, Funk SD, Green JM, Yurdagul A, Jr, Rana MA, Pistorius R, Henry M, Yurochko A, Pattillo CB, Traylor JG, et al. EphA2 expression regulates inflammation and fibroproliferative remodeling in atherosclerosis. Circulation. 2017; 136:566–582. doi: 10.1161/CIRCULATIONAHA.116.026644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zysset D, Weber B, Rihs S, Brasseit J, Freigang S, Riether C, Banz Y, Cerwenka A, Simillion C, Marques-Vidal P, et al. TREM-1 links dyslipidemia to inflammation and lipid deposition in atherosclerosis. Nat Commun. 2016; 7:13151. doi: 10.1038/ncomms13151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, Duan H, Qian Y, Feng L, Wu Z, Wang F, Feng J, Yang D, Qin Z, Yan X. Macrophagic CD146 promotes foam cell formation and retention during atherosclerosis. Cell Res. 2017; 27:352–372. doi: 10.1038/cr.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarrazy V, Sore S, Viaud M, Rignol G, Westerterp M, Ceppo F, Tanti JF, Guinamard R, Gautier EL, Yvan-Charvet L. Maintenance of macrophage redox status by ChREBP limits inflammation and apoptosis and protects against advanced atherosclerotic lesion formation. Cell Rep. 2015; 13:132–144. doi: 10.1016/j.celrep.2015.08.068 [DOI] [PubMed] [Google Scholar]
- 14.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013; 152:1298–1307. doi: 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014; 157:77–94. doi: 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- 16.Ishii N, Ozaki K, Sato H, Mizuno H, Susumu Saito, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006; 51:1087–1099. doi: 10.1007/s10038-006-0070-9 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Li L, Su Q, Liu T, Ma Z, Yang H. Ultrasound-targeted microbubble destruction enhances gene expression of microRNA-21 in swine heart via intracoronary delivery. Echocardiography. 2015; 32:1407–1416. doi: 10.1111/echo.12876 [DOI] [PubMed] [Google Scholar]
- 18.Perisic L, Aldi S, Sun Y, Folkersen L, Razuvaev A, Roy J, Lengquist M, Åkesson S, Wheelock CE, Maegdefessel L, et al. Gene expression signatures, pathways and networks in carotid atherosclerosis. J Intern Med. 2016; 279:293–308. doi: 10.1111/joim.12448 [DOI] [PubMed] [Google Scholar]
- 19.Pelisek J, Hegenloh R, Bauer S, Metschl S, Pauli J, Glukha N, Busch A, Reutersberg B, Kallmayer M, Trenner M, et al. Biobanking: objectives, requirements, and future challenges-experiences from the Munich Vascular Biobank. J Clin Med. 2019; 8:E251. doi: 10.3390/jcm8020251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eken SM, Jin H, Chernogubova E, Li Y, Simon N, Sun C, Korzunowicz G, Busch A, Bäcklund A, Österholm C, et al. MicroRNA-210 enhances fibrous cap stability in advanced atherosclerotic lesions. Circ Res. 2017; 120:633–644. doi: 10.1161/CIRCRESAHA.116.309318 [DOI] [PubMed] [Google Scholar]
- 21.Jin H, Li DY, Chernogubova E, Sun C, Busch A, Eken SM, Saliba-Gustafsson P, Winter H, Winski G, Raaz U, et al. Local delivery of miR-21 stabilizes fibrous caps in vulnerable atherosclerotic lesions. Mol Ther. 2018; 26:1040–1055. doi: 10.1016/j.ymthe.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu XH, Yuan YX, Rao SL, Wang P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur Rev Med Pharmacol Sci. 2016; 20:3653–3660 [PubMed] [Google Scholar]
- 23.Shen Y, Dong LF, Zhou RM, Yao J, Song YC, Yang H, Jiang Q, Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J Cell Mol Med. 2016; 20:537–548. doi: 10.1111/jcmm.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang HY, Zheng FS, Yang W, Lu JB. The long non-coding RNA MIAT regulates zinc finger E-box binding homeobox 1 expression by sponging miR-150 and promoting cell invasion in non-small-cell lung cancer. Gene. 2017; 633:61–65. doi: 10.1016/j.gene.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 25.Hnisz D, Day DS, Young RA. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016; 167:1188–1200. doi: 10.1016/j.cell.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014; 157:13–25. doi: 10.1016/j.cell.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013; 502:462–471. doi: 10.1038/nature12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013; 14(suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, Sennblad B, Liu S, Lao S, Hofmann P, et al. H19 induces abdominal aortic aneurysm development and progression. Circulation. 2018; 138:1551–1568. doi: 10.1161/CIRCULATIONAHA.117.032184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaffrey TA, Fu C, Du B, Eksinar S, Kent KC, Bush H, Jr, Kreiger K, Rosengart T, Cybulsky MI, Silverman ES, et al. High-level expression of Egr-1 and Egr-1-inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000; 105:653–662. doi: 10.1172/JCI8592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamimura M, Viedt C, Dalpke A, Rosenfeld ME, Mackman N, Cohen DM, Blessing E, Preusch M, Weber CM, Kreuzer J, et al. Interleukin-10 suppresses tissue factor expression in lipopolysaccharide-stimulated macrophages via inhibition of Egr-1 and a serum response element/MEK-ERK1/2 pathway. Circ Res. 2005; 97:305–313. doi: 10.1161/01.RES.0000177893.24574.13 [DOI] [PubMed] [Google Scholar]
- 32.Shan J, Balasubramanian MN, Donelan W, Fu L, Hayner J, Lopez MC, Baker HV, Kilberg MS. A mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK)-dependent transcriptional program controls activation of the early growth response 1 (EGR1) gene during amino acid limitation. J Biol Chem. 2014; 289:24665–24679. doi: 10.1074/jbc.M114.565028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordestgaard BG, Chapman MJ, Ray K, Borén J, Andreotti F, Watts GF, Ginsberg H, Amarenco P, Catapano A, Descamps OS, et al. ; European Atherosclerosis Society Consensus Panel. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur Heart J. 2010; 31:2844–2853. doi: 10.1093/eurheartj/ehq386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho T, Romagnuolo R, Scipione C, Boffa MB, Koschinsky ML. Apolipoprotein(a) stimulates nuclear translocation of β-catenin: a novel pathogenic mechanism for lipoprotein(a). Mol Biol Cell. 2013; 24:210–221. doi: 10.1091/mbc.E12-08-0637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JL, Baker AH, Oka K, Chan L, Newby AC, Jackson CL, George SJ. Suppression of atherosclerotic plaque progression and instability by tissue inhibitor of metalloproteinase-2: involvement of macrophage migration and apoptosis. Circulation. 2006; 113:2435–2444. doi: 10.1161/CIRCULATIONAHA.106.613281 [DOI] [PubMed] [Google Scholar]
- 36.Boffa MB, Koschinsky ML. Oxidized phospholipids as a unifying theory for lipoprotein(a) and cardiovascular disease. Nat Rev Cardiol. 2019; 16:305–318. doi: 10.1038/s41569-018-0153-2 [DOI] [PubMed] [Google Scholar]
- 37.de Vries MR, Ewing MM, de Jong RCM, MacArthur MR, Karper JC, Peters EAB, Nordzell M, Karabina SAP, Sexton D, Dahlbom I, et al. Identification of IgG1 isotype phosphorylcholine antibodies for the treatment of inflammatory cardiovascular diseases. J Intern Med. 2021; 290:141–156. doi: 10.1111/joim.13234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. A cytoplasmic NF- B interacting long noncoding RNA blocks I B phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015; 27:370–381. doi: 10.1016/j.ccell.2015.02.004 [Google Scholar]
- 39.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007; 292:C59–C69. doi: 10.1152/ajpcell.00394.2006 [DOI] [PubMed] [Google Scholar]
- 40.He S, Zhang H, Liu H, Zhu H. LongTarget: a tool to predict lncRNA DNA-binding motifs and binding sites via Hoogsteen base-pairing analysis. Bioinformatics. 2015; 31:178–186. doi: 10.1093/bioinformatics/btu643 [DOI] [PubMed] [Google Scholar]
- 41.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016; 17:756–770. doi: 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 42.Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014; 130:1452–1465. doi: 10.1161/CIRCULATIONAHA.114.011675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho H, Shen GQ, Wang X, Wang F, Archacki S, Li Y, Yu G, Chakrabarti S, Chen Q, Wang QK. Long noncoding RNA ANRIL regulates endothelial cell activities associated with coronary artery disease by up-regulating CLIP1, EZR, and LYVE1 genes. J Biol Chem. 2019; 294:3881–3898. doi: 10.1074/jbc.RA118.005050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan P, Guo YH, Zhan JK, Long LM, Xu ML, Ye L, Ma XY, Cui XJ, Wang HQ. LncRNA-ANRIL inhibits cell senescence of vascular smooth muscle cells by regulating miR-181a/Sirt1. Biochem Cell Biol. 2019; 97:571–580. doi: 10.1139/bcb-2018-0126 [DOI] [PubMed] [Google Scholar]
- 45.Lyu Q, Xu S, Lyu Y, Choi M, Christie CK, Slivano OJ, Rahman A, Jin ZG, Long X, Xu Y, et al. SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc Natl Acad Sci U S A. 2019; 116:546–555. doi: 10.1073/pnas.1810729116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boulberdaa M, Scott E, Ballantyne M, Garcia R, Descamps B, Angelini GD, Brittan M, Hunter A, McBride M, McClure J, et al. A role for the long noncoding RNA SENCR in commitment and function of endothelial cells. Mol Ther. 2016; 24:978–990. doi: 10.1038/mt.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014; 114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265 [DOI] [PubMed] [Google Scholar]
- 48.Haemmig S, Yang D, Sun X, Das D, Ghaffari S, Molinaro R, Chen L, Deng Y, Freeman D, Moullan N, et al. Long noncoding RNA SNHG12 integrates a DNA-PK-mediated DNA damage response and vascular senescence. Sci Transl Med. 2020; 12:eaaw1868. doi: 10.1126/scitranslmed.aaw1868 [DOI] [PubMed] [Google Scholar]
- 49.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015; 116:737–750. doi: 10.1161/CIRCRESAHA.116.302521 [DOI] [PubMed] [Google Scholar]
- 50.Li K, Blum Y, Verma A, Liu Z, Pramanik K, Leigh NR, Chun CZ, Samant GV, Zhao B, Garnaas MK, et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood. 2010; 115:133–139. doi: 10.1182/blood-2009-09-242180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vacante F, Rodor J, Lalwani MK, Mahmoud AD, Bennett M, De Pace AL, Miller E, Van Kuijk K, de Bruijn J, Gijbels M, et al. CARMN loss regulates smooth muscle cells and accelerates atherosclerosis in mice. Circ Res. 2021; 128:1258–1275. doi: 10.1161/CIRCRESAHA.120.318688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007; 120(pt 15):2498–2506. doi: 10.1242/jcs.009357 [DOI] [PubMed] [Google Scholar]
- 53.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010; 10:49. doi: 10.1186/1471-213X-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017; 8:e2929. doi: 10.1038/cddis.2017.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu X, Du Y, Shu Y, Gao M, Sun F, Luo S, Yang T, Zhan L, Yuan Y, Chu W, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. 2017; 7:42657. doi: 10.1038/srep42657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang H, Liu C, Li YJ, Yao J, Li XM, Shen Y, et al. Long non-coding RNA-MIAT promotes neurovascular remodeling in the eye and brain. Oncotarget. 2016; 7:49688–49698. doi: 10.18632/oncotarget.10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hippe DS, Phan BAP, Sun J, Isquith DA, O’Brien KD, Crouse JR, Anderson T, Huston J, Marcovina SM, Hatsukami TS, et al. Lp(a) (lipoprotein(a)) levels predict progression of carotid atherosclerosis in subjects with atherosclerotic cardiovascular disease on intensive lipid therapy: an analysis of the AIM-HIGH (Atherothrombosis Intervention in Metabolic Syndrome With Low HDL/High Triglycerides: Impact on Global Health Outcomes) Carotid Magnetic Resonance Imaging Substudy-brief report. Arterioscler Thromb Vasc Biol. 2018; 38:673–678. doi: 10.1161/ATVBAHA.117.310368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996; 271:1427–1431. doi: 10.1126/science.271.5254.1427 [DOI] [PubMed] [Google Scholar]
- 59.Sysol JR, Natarajan V, Machado RF. PDGF induces SphK1 expression via Egr-1 to promote pulmonary artery smooth muscle cell proliferation. Am J Physiol Cell Physiol. 2016; 310:C983–C992. doi: 10.1152/ajpcell.00059.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie N, Chen M, Dai R, Zhang Y, Zhao H, Song Z, Zhang L, Li Z, Feng Y, Gao H, et al. SRSF1 promotes vascular smooth muscle cell proliferation through a Δ133p53/EGR1/KLF5 pathway. Nat Commun. 2017; 8:16016. doi: 10.1038/ncomms16016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finn AV, Kolodgie FD, Virmani R. Correlation between carotid intimal/medial thickness and atherosclerosis: a point of view from pathology. Arterioscler Thromb Vasc Biol. 2010; 30:177–181. doi: 10.1161/ATVBAHA.108.173609 [DOI] [PubMed] [Google Scholar]
- 62.Kimmel DW, Dole WP, Cliffel DE. Elucidation of the role of lectin-like oxLDL receptor-1 in the metabolic responses of macrophages to human oxLDL. J Lipids. 2017; 2017:8479482. doi: 10.1155/2017/8479482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Z, Zhu H, Dai X, Wang C, Ding Y, Song P, Zou MH. Macrophage liver kinase B1 inhibits foam cell formation and atherosclerosis. Circ Res. 2017. doi: 10.1161/CIRCRESAHA.117.311546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian H, Yao ST, Yang NN, Ren J, Jiao P, Zhang X, Li DX, Zhang GA, Xia ZF, Qin SC. D4F alleviates macrophage-derived foam cell apoptosis by inhibiting the NF- B-dependent Fas/FasL pathway. Sci Rep. 2017; 7:7333. doi: 10.1038/s41598-017-07656-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plotkin JD, Elias MG, Dellinger AL, Kepley CL. NF- B inhibitors that prevent foam cell formation and atherosclerotic plaque accumulation. Nanomedicine. 2017; 13:2037–2048. doi: 10.1016/j.nano.2017.04.013 [DOI] [PubMed] [Google Scholar]
- 66.Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu Rev Physiol. 2012; 74:13–40. doi: 10.1146/annurev-physiol-012110-142315 [DOI] [PubMed] [Google Scholar]
- 67.Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cells in atherosclerosis. Circ Res. 2016; 118:692–702. doi: 10.1161/CIRCRESAHA.115.306361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yari H, Jin L, Teng L, Wang Y, Wu Y, Liu GZ, Gao W, Liang J, Xi Y, Feng YC, et al. LncRNA REG1CP promotes tumorigenesis through an enhancer complex to recruit FANCJ helicase for REG3A transcription. Nat Commun. 2019; 10:5334. doi: 10.1038/s41467-019-13313-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008; 3:1101–1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006; 39:356–362. doi: 10.1016/j.ymeth.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 71.Stary HC, Chandler AB, Dinsmore RE, Fuster V, Glagov S, Insull W, Jr, Rosenfeld ME, Schwartz CJ, Wagner WD, Wissler RW. A definition of advanced types of atherosclerotic lesions and a histological classification of atherosclerosis. A report from the Committee on Vascular Lesions of the Council on Arteriosclerosis, American Heart Association. Circulation. 1995; 92:1355–1374. doi: 10.1161/01.cir.92.5.1355 [DOI] [PubMed] [Google Scholar]
- 72.Redgrave JN, Gallagher P, Lovett JK, Rothwell PM. Critical cap thickness and rupture in symptomatic carotid plaques: the oxford plaque study. Stroke. 2008; 39:1722–1729. doi: 10.1161/STROKEAHA.107.507988 [DOI] [PubMed] [Google Scholar]
- 73.Hermansson A, Ketelhuth DF, Strodthoff D, Wurm M, Hansson EM, Nicoletti A, Paulsson-Berne G, Hansson GK. Inhibition of T cell response to native low-density lipoprotein reduces atherosclerosis. J Exp Med. 2010; 207:1081–1093. doi: 10.1084/jem.20092243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Magné J, Gustafsson P, Jin H, Maegdefessel L, Hultenby K, Wernerson A, Eriksson P, Franco-Cereceda A, Kovanen PT, Gonçalves I, et al. ATG16L1 expression in carotid atherosclerotic plaques is associated with plaque vulnerability. Arterioscler Thromb Vasc Biol. 2015; 35:1226–1235. doi: 10.1161/ATVBAHA.114.304840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.