SUMMARY
Pathogenic mycobacteria cause chronic and acute diseases ranging from human tuberculosis (TB) to nontubercular infections. Mycobacterium tuberculosis causes both acute and chronic human tuberculosis. Environmentally acquired nontubercular mycobacteria (NTM) cause chronic disease in humans and animals. Not surprisingly, NTM and M. tuberculosis often use shared molecular mechanisms to survive within the host. The ESX-1 system is a specialized secretion system that is essential for virulence and is functionally conserved between M. tuberculosis and Mycobacterium marinum. M. marinum is an NTM found in both salt water and freshwater that is often used to study mycobacterial virulence. Since the discovery of the secretion system in 2003, the use of both M. tuberculosis and M. marinum has defined the conserved molecular mechanisms underlying protein secretion and the lytic and regulatory activities of the ESX-1 system. Here, we review the trajectory of the field, including key discoveries regarding the ESX-1 system. We highlight the contributions of M. marinum studies and the conserved and unique aspects of the ESX-1 secretion system.
KEYWORDS: Mycobacterium marinum, ESX-1, mycobacterial pathogenesis, protein secretion, type VII secretion
INTRODUCTION
Chronic and acute mycobacterial infections constitute a significant global health burden. Mycobacterium tuberculosis causes human tuberculosis (TB), one of the world’s leading causes of death by infectious disease (1). In addition to M. tuberculosis, many species of nontuberculous mycobacteria (NTM) cause chronic environmentally acquired disease (2, 3). NTM are an emerging public health threat, increasing in incidence as the number of aging and immunocompromised individuals increases (4). The prevalence of NTM infections and their impact on human health are likely underappreciated (5).
Despite differences in host range and transmission, M. tuberculosis and NTMs share most molecular pathways that play critical roles in physiology and pathogenesis, including the ESX-1 (ESAT-6 [6-kDa early secreted antigenic target]) secretion system (see Appendix 1 for nomenclature) (6). The ESX-1 system is essential for the virulence of both M. tuberculosis and Mycobacterium marinum (7–12). M. marinum is an NTM pathogen with a broad host range that includes fish and other ectotherms as well as humans (13, 14). M. marinum has long been used to study mycobacterial pathogenesis and has provided significant insight into the host immune response to mycobacterial infections (15, 16). Overall, investigating the pathogenicity of M. marinum has informed our understanding of M. tuberculosis virulence mechanisms and will continue to serve as an informative model for how mycobacteria use ESX-1 secretion for virulence. Within 1 year of the initial publications demonstrating that ESX-1 was a specialized secretion pathway in M. tuberculosis, publications followed using M. marinum, further expanding our understanding of the ESX-1 system and its role in virulence (see Fig. 1 for a timeline of contributions from M. marinum studies). Indeed, it is difficult to discuss the trajectory of the ESX-1 field without considering studies in M. marinum. In this review, we highlight how the use of M. marinum has informed the molecular mechanisms of substrate secretion, the role of secretion in virulence, and the mechanisms regulating gene expression by the ESX-1 secretion system (type VII secretion).
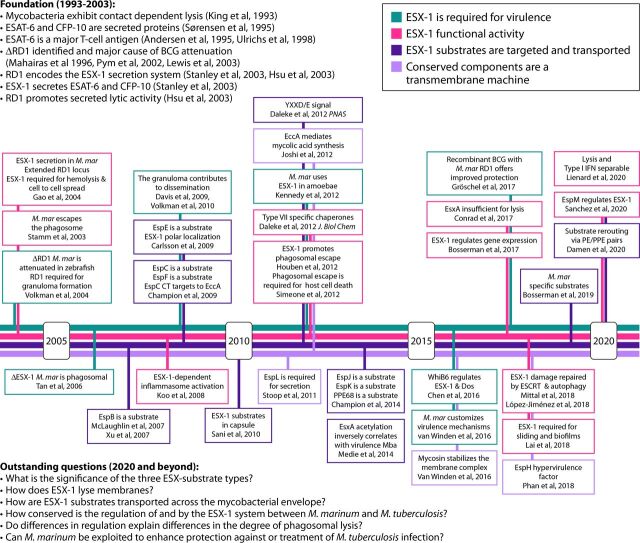
FIG 1.
Timeline of ESX-1 studies performed in M. marinum (7–12, 26, 27, 31, 44, 47, 48, 63, 73–78, 85, 87, 92, 103, 104, 113, 117, 118, 127, 128, 134, 141, 148, 151, 154, 166, 168, 200–206). The timeline shows major contributions from M. marinum (M. mar) studies from ∼2004 to 2020. The four lines indicate major aspects of the ESX-1 field. The list of foundational studies performed on M. tuberculosis and other slow-growing mycobacterial species as well as the highlighted M. marinum studies are not comprehensive. IFN, interferon; CT, C terminus.
TYPE VII SECRETION: A CONSERVED, SPECIALIZED SECRETION PATHWAY DISCOVERED IN MYCOBACTERIA
All bacteria transport proteins from the cytoplasm to the cell surface and into the extracellular environment. In addition to the general protein transport pathways (Sec and Tat [recently reviewed by van Winden et al. {17}]), there are several specialized protein transport systems in bacteria, classified as types I through IX (18, 19). ESX secretion was first discovered in mycobacteria and classified as type VII, reflecting its unique nature from the previous six (20, 21). Since then, type VII systems have been further classified as type VIIa to refer to those in Actinobacteria and type VIIb to refer to those in Firmicutes (22). Type VIIb systems have been widely found in pathogenic and nonpathogenic Gram-positive bacteria (23, 24). Like type VIIa systems, type VIIb systems secrete Esx proteins and include FtsK-SpoIIIE ATPases (22). There has been significant progress in understanding several molecular aspects of protein transport across the Gram-positive cell envelope and the functions of ESX-type secretion in Gram-positive physiology and pathogenesis.
It was well established that mycobacteria secreted ESX-1 proteins into their extracellular environment before the system responsible for transport was identified. In 1999, two of the major ESX-1 substrates, EsxA (ESAT-6) and EsxB (CFP-10 [culture filtrate protein, 10 kDa]), were identified as proteins highly secreted from M. tuberculosis in vitro (25) and facilitated the discovery of ESX systems in mycobacteria. There was interest in EsxA because in 1995 and 1996, it was established as a strong T-cell antigen (26, 27), stimulating interferon gamma production in memory effector cells of previously infected mice (27). As such, EsxA was being considered as a potential diagnostic for TB as early as 1997 (28). In 1998, EsxB was discovered as a novel secreted protein that was encoded in the same operon as the esxA gene (see Fig. 2 for a schematic of the esx-1 locus) (29). Neither protein included obvious secretory signals required for secretion through the general Sec or Tat protein transport pathway, indicating that there were additional protein transport pathways in mycobacteria (25, 30).
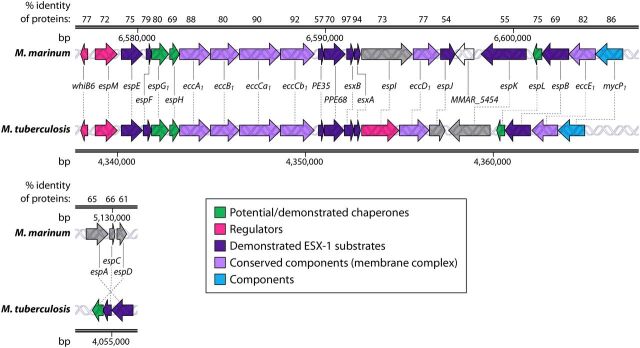
FIG 2.
Syntenic esx-1 loci from M. marinum and M. tuberculosis. The genes and their locations in each genome as well as the percent amino acid identities of the resulting proteins from the conserved esx-1 and espACD loci in M. marinum and M. tuberculosis are shown. MMAR_5454 is a gene found only in M. marinum. Gray shading indicates that the gene function has not been defined. Differences in colors between organisms, for example, for PPE68, indicate that PPE68 is a substrate in M. marinum but is required for secretion and not yet classified as a substrate in M. tuberculosis. The RD1 region is shown (7), as follows. ESX-1 gene products from M. tuberculosis (GenBank accession number NC_000962.3) were searched against those from Mycobacterium marinum (NCBI taxonomy accession number txid1781) using NCBI BLASTP (169). The percent identities of the top orthologous proteins in M. marinum are reported at the top.
A major breakthrough occurred in 1995, when ESAT-6 was found in culture filtrates (secreted protein fractions) from M. tuberculosis, M. marinum, and other pathogenic mycobacterial species but not in culture filtrates from the attenuated Mycobacterium bovis BCG vaccine strain (26). In 1996, a comparison of the M. tuberculosis and M. bovis BCG genomes revealed a large region of difference, named region of difference 1 (RD1), which included the esxBA operon (Fig. 2) (31, 32). Subsequently, in 1999, it was demonstrated that all BCG strains lacked the RD1 region, which includes the esxA gene (33). These studies, coupled with bioinformatic analyses, led to the proposal that the genes at the RD1 locus encoded a specialized secretory apparatus that secreted ESAT-6 and CFP-10 from the mycobacterial cell (23, 25, 34). Sequencing of the M. tuberculosis genome in 1998 revealed five paralogous secretion systems (30), which were named ESX-1 through ESX-5 (35). ESX systems can also be encoded on plasmids (36–38). In this review, we focus on ESX-1. However, significant work on the other ESX systems in several mycobacterial species, including M. marinum, have greatly advanced our understanding of the conserved features of type VII secretion (17, 39, 40).
Comparisons between the genomes of Mycobacterium and several Gram-positive bacteria, including Bacillus subtilis, Staphylococcus aureus, Listeria monocytogenes, and Streptococcus, demonstrated that the ESX-type systems, now termed type VIIa and type VIIb (22), were distinct from previously characterized secretion systems, and they were designated type VII secretion systems (T7SSs), accordingly (21). Further genomic comparisons revealed that T7SSs are widely found in pathogenic and nonpathogenic Gram-positive bacteria (23, 24).
ESX-1 PROTEIN SECRETION IS REQUIRED FOR MYCOBACTERIAL VIRULENCE
Given that the RD1 region was missing from the attenuated M. bovis BCG vaccine strain, it was suspected that the secretory apparatus encoded by the genes within the M. tuberculosis RD1 region was essential for pathogenesis. The links between the RD1 genomic region, protein secretion, and virulence all occurred rapidly between 2002 and 2004, as detailed below.
The genetic deletion of the RD1 locus from the M. tuberculosis genome led to attenuated virulence in mouse and macrophage models of infection (12). Importantly, complementation of the RD1 region from M. tuberculosis increased the virulence of the M. bovis BCG vaccine strain (11, 41) and the interferon gamma response (42), indicating that the deletion of the RD1 region is the major cause of attenuation of the vaccine strain (7, 11, 12).
By 2004, it was apparent that M. marinum required the RD1 region and the ESX-1 system for virulence, similar to M. tuberculosis (10). The genes that make up the RD1 locus, and those surrounding it (the extended RD1 locus), are highly conserved between M. tuberculosis and M. marinum (6, 9). In 2004, Volkman et al. observed that the M. tuberculosis and M. marinum RD1 loci are syntenic and that the EsxB and EsxA gene products share 97 and 91% identities at the amino acid level, respectively (Fig. 2) (10). Transposon (Tn) mutagenesis screens for both M. tuberculosis and M. marinum have consistently identified the RD1 locus as being required for virulence (8, 9, 43–45). The genetic deletion of the orthologous RD1 region in M. marinum resulted in attenuated virulence in both adult and larval zebrafish infection models (10).
Because of its optical transparency, the larval zebrafish model for M. marinum infection gives unique insight into the early stages of disease progression and the role of ESX-1 in this process. Experiments using the M. marinum zebrafish model defined a role for the ESX-1 system in granuloma formation. The granuloma is a characteristic structure of mycobacterial infection, which not only may contain and eradicate the bacterium but also can create optimal conditions for its spread through necrosis and rupture (for a review, see reference 46). Using this model, it was observed that M. marinum cells lacking the ESX-1 secretion system were defective for granuloma formation (10) due, at least in part, to reduced matrix metalloprotease 9 (MMP9)-dependent extracellular matrix remodeling at sites of infection (47, 48).
MECHANISM OF ESX-1 SECRETION
ESX-1 substrates are secreted from the mycobacterial cell, transiting from the cytoplasm through the cytoplasmic membrane and the cell envelope to the cell surface and into the extracellular environment (see Fig. 3 for a model of ESX-1 secretion). The step attributed to the ESX-1 system thus far includes crossing the cytoplasmic membrane. It is not known how any protein crosses the mycobacterial envelope beyond the cytoplasmic membrane. Together, studies in M. tuberculosis and M. marinum have provided complementary insight into the specific mechanisms of how the ESX-1 system recognizes and transports proteins.
FIG 3.
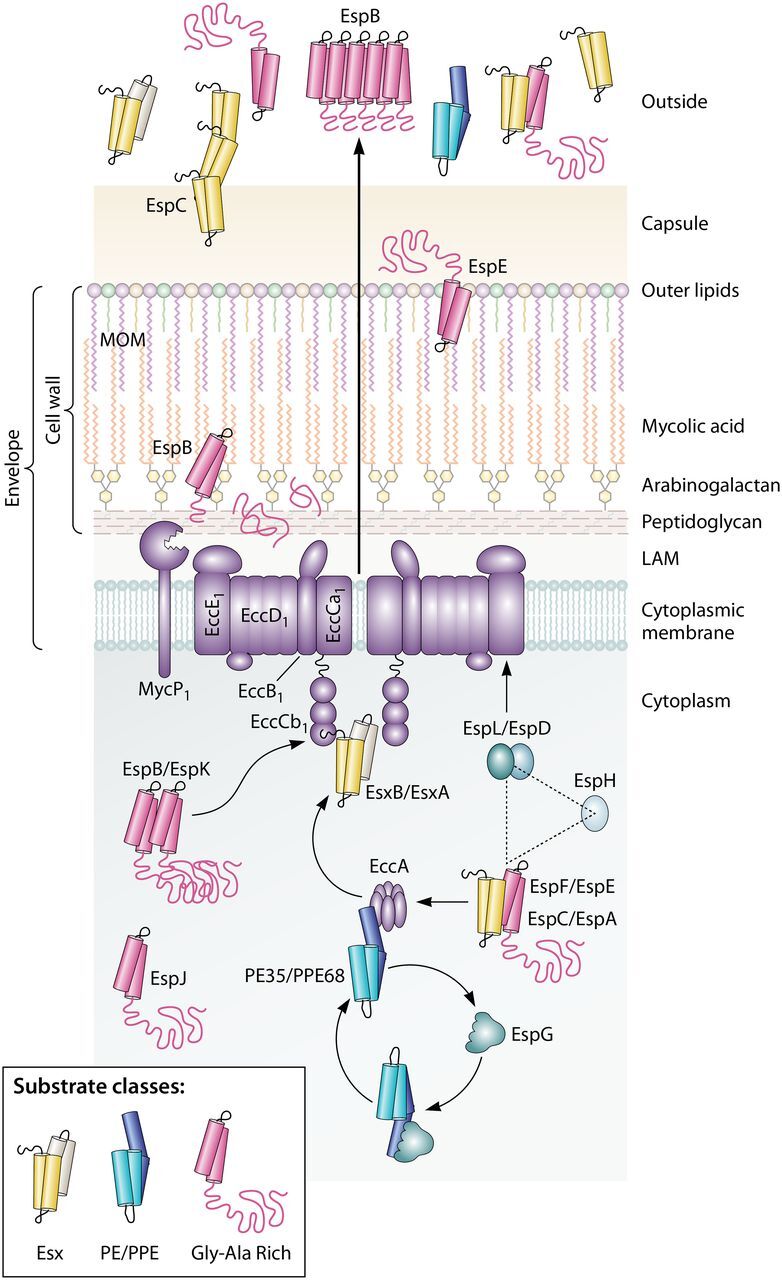
Conserved aspects of ESX-1 protein transport in M. tuberculosis and M. marinum. The schematic shows that the ESX-1 conserved components EccA to EccE interact with substrates and provide a channel across the cytoplasmic membrane. The three classes of conserved ESX-1 substrates are shown. The EccCb1 ATPase directly interacts with the EsxB and EspK substrates; the EccA ATPase interacts with the PPE68 and EspF substrates. Substrates are often targeted in pairs. The EspG chaperone interacts with PE/PPE substrate pairs and likely brings these substrates to the EccA component before being recycled. It is not known how substrates are transferred from the EccA component to the membrane complex. EspL and EccE interact with EspD. EspL and EspD are required for the stability of EspE, EspF, and EspH. Following transport through the membrane complex, ESX-1 substrates are transported across the envelope via an unknown mechanism. EspB is cleaved by the MycP1 protease. Substrates can be targeted to the MOM (mycolate outer membrane), to the capsule, and out of the cell. Some substrates, including EspC and EspB, form higher-order structures following secretion, which may indicate that they are part of the secretory machinery. LAM, lipoarabinomannan.
Model for Protein Transport
The ESX-1 system is made up of component and substrate proteins. Components are part of the machinery, which include membrane-associated proteins known as ESX conserved components (Eccs), chaperones, and other proteins of unknown function that are not secreted by the ESX-1 system (Appendix 1). ESX-1 substrates are primarily recognized by virtue of their extracellular secretion in an ESX-1-dependent manner in vitro. Substrates and other nonconserved components are known as ESX-associated proteins (Esps) (20).
Studies reported in 2003 and 2004 in M. tuberculosis and using BCG and Mycobacterium microti complementation models demonstrated that the genes within and surrounding the RD1 region encoded a specialized secretion system required for the secretion of EsxA and EsxB and that this system was required for virulence in macrophage and animal models of infection (7, 8, 42, 49). Recombinant M. bovis BCG and M. microti strains expressing the M. tuberculosis genes in the extended RD1 region demonstrated that these genes encoded a system that secretes EsxA and EsxB. Moreover, the reintroduction of EsxA and EsxB secretion by these strains led to increased protection against M. tuberculosis infection in animal models (42). At the same time, genetic approaches aimed at understanding M. tuberculosis virulence generated attenuated transposon insertion strains (50), which, when characterized, revealed three components (EccCa1, EccCb1, and EccD1 [the subscript “1” indicates that they are part of the ESX-1 system]) and two substrates (EsxA and EsxB) of the ESX-1 system encoded within the RD1 region (Fig. 2) (8). Direct interactions were found between components (EccCa1 and EccCb1), between substrates (EsxA and EsxB), and, importantly, between components and substrates (EccCb1 and EsxB). These data supported a previous study demonstrating a direct interaction between EsxA and EsxB (51). Interestingly, although under the majority of biological conditions, EsxA and EsxB are likely secreted as a tight 1:1 complex, under certain conditions, EsxB was secreted without EsxA from M. tuberculosis (52, 53). Likewise, a complementary study demonstrated that the insertion of a transposon into the esxBA operon resulted in a loss of secreted lytic activity required for host cell death and mycobacterial tissue invasion (7). Together, these studies linked the ESX-1 system to virulence and resulted in the first models showing that ESX-1 components form a specialized secretion system that interacts with and transports substrates (Fig. 3) (7, 8, 42). Further work demonstrated that individual genes in the RD1 region were required for the secretion of EsxA and EsxB proteins from M. tuberculosis (49), strengthening and expanding the initial model.
Following the initial studies in M. tuberculosis, the genes required for ESX-1 secretion were rapidly expanded using other mycobacterial species, demonstrating the conserved nature of ESX-1 secretion. In 2004, a genetic screen for nonhemolytic M. marinum revealed that several additional genes in the extended RD1 region were required for EsxA secretion and cell-to-cell spread (9). Mycobacterium smegmatis is a nonpathogenic fast-growing mycobacterial species that is widely used as a model to study mycobacterial physiology (54–56). In 2005, the genetic deletion of genes within a region orthologous to RD1 demonstrated that the ESX-1 system was functional in M. smegmatis and that many of the genes required for secretion were conserved (57). M. microti has a natural genetic deletion that partially overlaps the RD1 region (58). In 2006, complementation of the M. microti deletion with genes from M. tuberculosis revealed several additional genes important for EsxA secretion and further linked the secretion system to virulence and immunogenicity (59). Based on the genes collectively identified by these studies, in 2009, a yeast two-hybrid analysis of the gene products encoded from within the extended RD1 locus from M. tuberculosis added further interactions between components and substrates (60). Together, studies in M. tuberculosis, M. marinum, M. microti, and M. smegmatis led to the proposal that several conserved membrane components formed a channel across the cytoplasmic membrane, allowing the secretion of EsxA and EsxB through the cytoplasmic membrane and into the extracellular environment (Fig. 3).
The ESX-1 Membrane Complex
Building on the initial characterization of the extended RD1 locus as a novel class of secretory systems, there remains a great focus on understanding the mechanisms of protein transport across the mycobacterial cell envelope (61). The studies described above revealed that there were five membrane-associated ESX-1 conserved components (EccB1, EccCa1, EccCb1, EccD1, and EccE1) and one cytoplasmic Ecc (EccA1) that together make up the machinery that forms a channel across the cytoplasmic membrane and provides energy for transport and substrate selection through several conserved AAA ATPase domains (Fig. 3). In addition to these components, a conserved membrane-bound mycosin protease, MycP1, processes specific ESX-1 substrates (62) and stabilizes the core membrane complex (63).
All of the known conserved ESX-1 components were identified in Tn insertion studies in both M. tuberculosis and M. marinum in 2003 and 2004 (8, 9, 49). In 2012, a study performed using both M. marinum and M. bovis BCG contributed the first biochemical isolation of the conserved core ESX-5 membrane complex, showing direct interactions between membrane components (64). Because components of the membrane complex are conserved between ESX systems and across species, these findings are considered generalizable to additional ESX systems, including ESX-1 (64).
Studies using proteins from several mycobacterial species and related actinobacteria provided insight into specific domains of individual ESX-1 core components. From 2013 to 2020, structures were reported for the soluble domains of EccB1 and EccD1 from M. tuberculosis (65, 66) and M. smegmatis (65). The study of the individual domains suggested that EccB1 anchors the ESX-1 membrane complex into the periplasm and that EccD1 forms a dimer with a ubiquitin-like fold (65, 66). The N-terminal domain of EccA1 from M. tuberculosis included six tetratricopeptide repeats that likely mediate direct interactions with ESX-1 substrates (67). The structures of EccC (the ortholog of EccCa1 and EccCb1) from Thermomonospora curvata (52) and the C terminus of EccCb1 from M. tuberculosis (68) revealed a hexameric structure with three interlocking AAA ATPase domains, the third of which directly binds ESX-1 substrates. The structures of the mycosins from M. smegmatis (69) and M. tuberculosis (70) uncovered mechanistic insight into how the membrane-anchored protease interacts with and cleaves the EspB substrate (62). Between 2017 and 2019, substantial work yielded structural insight into the intact membrane components of ESX-type transporters. Based on the structures of the ESX-3 systems from M. smegmatis (71, 72) and ESX-5 systems from Mycobacterium xenopi, ESX systems have a conserved multimeric architecture that spans the cytoplasmic membrane and protrudes into the periplasm and cytoplasm that is not shared by other types of secretion systems. These findings should be applicable to the M. tuberculosis ESX-1 system because the components are conserved.
The ESX-1-Secreted Substrates
Identification of conserved ESX-1 substrates.
While the components of the ESX-1 system were being elucidated, beginning in 2005, studies in both M. tuberculosis and M. marinum contributed to our current understanding of which substrates were secreted by the ESX-1 system. Importantly, M. marinum provided complementary and unique insight into the molecular mechanisms of protein secretion to those discovered in M. tuberculosis. In particular, M. marinum has been and continues to be used to identify and study new ESX-1 substrates (73–77), some of which have been confirmed in M. tuberculosis. Moreover, the expression of M. tuberculosis genes in M. marinum has provided a means to broaden our understanding of functional conservation between ESX-1 proteins.
There are several general types of ESX-1 substrates (Fig. 3), including small 100-amino-acid (aa) proteins (similar to EsxA and EsxB [73]), PE/PPE proteins (PE is proline-glutamic acid and PPE is proline-proline-glutamic acid) (45, 74, 75, 78), and alanine-rich proteins (74, 76, 77). It is unclear how the three general groups of substrates contribute to ESX-1 secretion and virulence. However, all three substrate classes are secreted by both M. tuberculosis and M. marinum.
Although most of the initial studies in the early 2000s focused on the RD1 region and the secretion of EsxA and EsxB, it was clear by as early as 2005 that there were additional ESX-1 substrates, some of which were also encoded at unlinked genomic loci. In some cases, substrate genes were identified as being important for ESX-1 secretion and virulence before the resulting proteins were classified as secreted substrates. In 2005, the first substrates outside the RD1 locus were identified at the espACD locus (Fig. 2), which is required for M. tuberculosis virulence and ESX-1 secretion (79, 80). The espACD locus encodes two ESX-1 substrates in M. tuberculosis, EspA and EspC (73, 79, 80). EspA is an alanine- and glycine-rich 392-aa protein (81). EspC is a 103-aa protein (81). EspD, also encoded at the espACD locus, is required for the stability of EspA and EspC but is not itself strictly secreted as an ESX-1 substrate (82). Work by Fortune et al. revealed that EspA exhibited mutually dependent secretion with EsxA and EsxB in M. tuberculosis. The deletion of the espA gene resulted in the loss of EsxA and EsxB secretion, and the deletion of the esxA gene resulted in the loss of EspA secretion (80). It has not yet been determined if EspA is secreted from M. marinum or if it is required for M. marinum virulence.
The same genetic screen that identified specific genes in the ESX-1 locus as being important for M. tuberculosis virulence in mice (50) also identified a Tn insertion in the rv3615c (espC) gene, which was reported in 2005 (79). Based on the similarity between the espE (rv3864), espF (rv3865), and espH (rv3867) genes within the extended RD1 locus, it was hypothesized that the espACD genes were also required for ESX-1 secretion (79). A disruption of the espC gene resulted in an inability of M. tuberculosis to suppress the proinflammatory response in macrophages, similar to a disruption in the eccD1 gene, which encodes a conserved membrane component of the ESX-1 system. Finally, using yeast two-hybrid analysis, this study demonstrated an interaction between EspD (Rv3614c) and the EccE1 membrane component (79), further linking the espACD locus to the ESX-1 machinery. In 2009, the first demonstration that EspC from M. tuberculosis was secreted came from the expression of EspC from M. tuberculosis (EspCMT) in M. marinum strains in the presence and absence of a functional ESX-1 system (73). Using this model, in addition to the ESX-1-dependent secretion of EspCMT by M. marinum, a direct interaction between the C terminus of EspCMT and the EccA component was defined (73). Recent studies in M. tuberculosis in 2017 suggested that EspC forms a filament, which may mean that EspC functions as a “needle” for the ESX-1 system (Fig. 3) (83). However, the role of EspC or its requirement for secretion has not yet been investigated in M. marinum. It is also important to note that despite the conservation between the EspA, EspC, and EspD proteins, the espACD loci are found at different locations and in opposite orientations in the M. tuberculosis and M. marinum genomes (84).
Within the extended RD1 locus, there are 8 ESX-1 substrate genes, which, in addition to EsxA and EsxB, encode EspB, EspE, EspF, PE35, PPE68, EspJ, and EspK (Fig. 3). The EspB protein is an ∼460-aa alanine- and glycine-rich protein (81) that was identified as a secreted substrate in both M. marinum and M. tuberculosis in 2007 (76, 85). EspB is secreted independently of EspA and EspC in M. tuberculosis (86). The structure of the EspB protein from M. tuberculosis was solved and revealed an N-terminal PE/PPE with an unstructured C terminus (87, 88). The PE/PPE domain oligomerizes (87, 88) and forms rings (89). The EspF protein was first identified as a substrate in M. marinum in 2009 (73) and is also secreted from M. tuberculosis (90). EspF is ∼103 aa long, similar to the EspC protein (81). It was previously demonstrated that the espF gene was important for virulence in the M. microti-M. tuberculosis complementation model (59), which was later confirmed in M. tuberculosis (91). EspE was identified as a cell-associated substrate of the ESX-1 system in M. marinum in 2009 (77) and is also secreted from M. tuberculosis (90). The EspE protein is an ∼402-aa protein, similar to the EspA protein (81). The requirement of the espE gene in virulence is still unresolved, as the espE gene has been considered essential for virulence in some studies (43, 77, 92) and dispensable in others (59). Both EspJ and EspK were identified as ESX-1 substrates in M. marinum through proteomic profiling in 2014 (74). EspJ and EspK are both alanine- and glycine-rich proteins of 280 aa and 729 aa, respectively (81). EspK was previously linked to the ESX-1 system in M. marinum as a potential targeting partner for EspB; it was demonstrated biochemically that EspK interacts directly with the EccCb1 component and the EspB substrate (76). Neither EspK nor EspJ has been studied as the substrate in M. tuberculosis. PPE68 and PE35 constitute a PE/PPE pair at the ESX-1 locus. The PE/PPE proteins have a wide variety of roles, including but not limited to immunomodulation and host cell death (93), maintaining mycobacterial cell wall and capsule integrity (94, 95), antivirulence (96), and nutrient transport across the mycobacterial outer membrane (97, 98). PPE68 was first shown to be an immunogenic protein encoded by the RD1 locus in M. tuberculosis (99). In 2014 and 2019, PPE68 and PE35 were identified as ESX-1 substrates in M. marinum (74, 100) but have not yet been confirmed as substrates in M. tuberculosis.
ESX-1 substrates unique to M. marinum.
Although the earliest identified ESX-1 substrates at the esx-1 and espACD loci are highly conserved between M. tuberculosis and M. marinum (see above), several additional substrates have recently been identified in M. marinum, which are absent from M. tuberculosis. In part, diverse substrates between M. marinum and M. tuberculosis could indicate that M. marinum customizes its virulence to survive an expanded host range relative to that of M. tuberculosis (Appendix 2) (45). Accordingly, consistent with its more restrictive niche, it is interesting to note that there have not been ESX-1 substrates reported thus far that are specific to M. tuberculosis and absent from M. marinum.
The M. marinum genome includes 281 pe/ppe genes (6). Many PE/PPE proteins are known to be secreted by the paralogous ESX-5 system in M. marinum (94, 95, 101). However, there is evidence that there are PE/PPE proteins secreted by the ESX-1 system whose genes are located at unlinked chromosomal loci in the M. marinum genome. In 2018 and 2019, MMAR_2894 was identified as an ESX-1 substrate through proteomic screens (78) and verified experimentally (75). MMAR_2894 is required for the optimal secretion of the additional conserved ESX-1 substrates (75). Importantly, the secretion of ESX-1 substrates in the absence of MMAR_2894 was reduced relative to the wild-type (WT) strain (75). However, MMAR_2894, although required for the hemolytic activity of M. marinum, was dispensable for the lysis of host cells in a macrophage model of infection (75). The study of this unique M. marinum substrate has potentially revealed basic mechanistic differences between hemolysis and host cell lysis. Alternatively, the levels of ESX-1 secretion from strains lacking MMAR_2894 were sufficient to promote phagosomal escape and host cell death but were insufficient for hemolysis.
Additional PE/PPE proteins that are unique to M. marinum are secreted by ESX-1. For example, M. marinum has a partial ESX locus, known as ESX-6. In the ESX-6 locus, there are genetic duplications of several genes at the ESX-1 locus, including pe35_1 and ppe68_1 as well as esxB and esxA. EsxB_1, EsxA_3, PE35_1, and PPE68_1 are all secreted by the ESX-1 system in M. marinum (102–105). The ESX-6 locus, although containing several genes that appear to be duplications of ESX-1 genes, is dispensable for M. marinum virulence (105). Likewise, the EsxB_1 and EsxA_3 proteins encoded at this locus can be secreted by the ESX-1 system (104, 105) but must also have alternative routes out of the cell, as they are detected at high levels in secreted protein fractions from M. marinum strains lacking functional ESX-1 secretion systems (73, 74, 105, 106).
Substrate recognition by the ESX-1 system.
ESX-1 substrates are recognized through direct interactions with membrane components and chaperones of the ESX-1 system. Work in both M. tuberculosis and M. marinum resulted in our current understanding of substrate targeting. Importantly, as indicated by early studies of EsxA and EsxB (8, 24, 29, 51, 107, 108), some ESX-1 substrates are likely targeted and secreted in pairs. Moreover, there are substrate-specific and general secretory signals required for targeting proteins for secretion. Overall, the data suggest that different types of ESX-1 substrates have different targeting rules.
Using M. tuberculosis, it was demonstrated in 2006 that the C terminus of the EsxB substrate mediates a direct interaction with the C-terminal half of the EccCb1 protein (8, 107). The terminal 7 amino acids of EsxB (LSSQMGF) were shown to be sufficient for the interaction with EccCb1 and for the targeting of EsxB and EsxA substrates for secretion to the ESX-1 system. The sufficiency of the direct interaction between the terminal 7 aa of EsxB and EccCb1 has since been confirmed by structural studies in both T. curvata in 2015 (52) and M. tuberculosis in 2020 (68). Importantly, it was demonstrated that the third AAA ATPase domain of EccCb1 directly interacts with the EsxB C-terminal 7 amino acids (52, 68). This interaction promotes the oligomerization of EccCb1, which may link the energy required for transport to substrate recognition (52).
M. marinum has been widely used to further define how specific ESX substrates are selected by and targeted to ESX systems. Similar to EsxB, it was shown in 2009 that the C terminus of the EspC substrate from M. tuberculosis directly interacts with the EccA component from M. marinum (73). EccA is a cytosolic ATPase that interacts with several ESX-1 substrates, including EspF (73) and PPE68 (60), which may function to remove substrates from the EspG1 chaperone (see below) prior to secretion (67, 109, 110).
Using M. marinum, additional regions of ESX-1 substrates have since been recognized to be required for substrate targeting, including the conserved YXXXD/E motif, which was reported in 2012 (87). Accordingly, mutation of this motif prevents substrate secretion. However, the YXXXD/E motif is insufficient to confer secretion through an ESX system (87) and does not confer specificity for ESX secretion (102).
The PE/PPE substrate pairs from both M. tuberculosis and M. marinum directly interact with EspG proteins, which are chaperones specific to each ESX system (103, 109–111). Chaperones escort substrates to the secretory apparatus but are not secreted from the cell. A comprehensive review of ESX chaperones was recently published (110). The EspG-substrate interaction has been defined structurally and genetically (103, 109–112). Interestingly, it was demonstrated in 2017 that the interaction between PE/PPE substrate pairs and the EspG chaperone dictates which ESX system secretes these proteins (Fig. 3) (102). Moreover, in 2020, it was demonstrated that the targeting of the PE/PPE heterodimers can also impact the targeting of Esx-type substrates (104). Therefore, the targeting of individual substrate pairs can be influenced by other substrates. Together, these studies in both M. tuberculosis and M. marinum suggest that substrate selection is complex, relying on the recognition of multiple signals present on substrates by both chaperones and secretory components through direct interactions.
ESX-1 LYSES HOST MEMBRANES
One long-standing and well-accepted hypothesis is that ESX-1 is essential for virulence because it lyses host membranes (7). In 1993, King and others observed that the virulent H37Rv strain of M. tuberculosis lysed red blood cells in a contact-dependent manner, while the avirulent strain H37Ra and the M. bovis BCG vaccine strain were defective for contact-dependent lysis (113). Although this study identified specific genetic loci required for lysis, it did not identify the ESX-1 system, in part because this study was reported prior to the observation that both M. tuberculosis H37Ra (114) and M. bovis BCG lack ESX-1 secretion and prior to the sequencing of the M. tuberculosis genome (30). Contact-dependent hemolysis of M. tuberculosis has been observed in numerous clinical isolates in addition to laboratory strain H37Rv (115). The King laboratory also discovered that M. tuberculosis exhibits contact-dependent lysis of pneumocytes in vitro (116). Using this lysis assay, it was discovered in 2003 that the insertion of a transposon into the esxBA operon resulted in a loss of cytolysis of pneumocytes and macrophages and reduced tissue invasiveness by M. tuberculosis (7). All activities were restored upon genetic complementation, linking the secretion of EsxA and EsxB to host cell lysis for the first time.
ESX-1-mediated lysis of host membranes is hypothesized to promote the rupture of the phagosomal membrane and to be required for intracellular growth (117). The first mycobacterial species shown to lyse the phagosome was M. marinum (118, 119). In 2003, M. marinum was shown to robustly enter the cytosol and polymerize host actin for motility (118), reminiscent of other intracellular pathogens, including Listeria monocytogenes (120). Consistent with early studies in M. marinum, M. tuberculosis was first observed to disrupt phagosomal membranes in macrophages by electron microscopy in 2007 (121). Phagosomal permeabilization was subsequently quantified using fluorescence resonance energy transfer (FRET). ESX-1-competent bacteria (M. tuberculosis, ESX-1-complemented BCG, and M. marinum) accessed the cytosol, whereas ESX-1-defective mycobacteria (BCG and M. tuberculosis ΔRD1) did not. Quantification of phagosome permeabilization by electron microscopy confirmed these data (117). It is important to note that although both M. tuberculosis and M. marinum access the cytoplasm, M. marinum is distinct in its use of actin-based motility downstream of phagosomal lysis (118, 122).
Mechanisms of Host Membrane Lysis
The mechanism of ESX-1-mediated host membrane lysis remains elusive and controversial. Since 2003, it was hypothesized that EsxA was necessary and sufficient for membrane lysis (7, 9, 123–126), stemming from the initial demonstration in M. tuberculosis that the ESX-1 system promoted host cell lysis (7). The requirement of EsxA for membrane lysis stemmed from genetic studies, while the sufficiency of EsxA for membrane lysis stemmed from biochemical approaches. The deletion or disruption of the esxA gene resulted in a loss of lytic activity in M. tuberculosis (7, 8), hemolysis in M. marinum (9), and phagosomal lysis and the translocation of mycobacteria to the cytosol (117, 121, 127). Together, these studies demonstrated that that EsxA was required for the lytic activity of the ESX-1 system. However, the interpretation of the results of these genetic experiments is complicated by the fact that EsxA is required for the secretion of the additional ESX-1 substrates (74, 76, 78, 80, 90) and may be a secreted component of the secretory apparatus (74, 78, 128–130). Therefore, the role of EsxA in the lytic activity of the ESX-1 system may be indirect.
Biochemical studies demonstrated that purified EsxA exhibited lytic activity in vitro (7, 124, 125, 131–133), suggesting sufficiency. However, subsequent work in 2015 demonstrated that EsxA exhibited lytic activity only when treated with detergent (133). Moreover, in 2017, it was demonstrated that contaminating detergent was the source of lytic activity from widely used protocols and preparations of the EsxA protein (134). Therefore, either EsxA demonstrates lytic activity under conditions other than those tested in the two previous studies or additional ESX-1 substrates or other virulence factors promote phagosomal lysis. Over the past several years, it has become clear that mycobacterial virulence lipids, including phthiocerol dimycocerosates (PDIMs), work in concert with the ESX-1 system to promote optimal membrane lysis (135–137).
Investigation of M. marinum ESX-1-mediated membrane lysis has revealed that this lysis is pH independent. Moreover, as observed by electron microscopy, ESX-1-competent mycobacteria cause gross membrane disruptions at sites of direct contact with host membranes (134). These gross disruptions were observed in an extracellular hemolysis assay conducted at neutral pH. Furthermore, ESX-1-competent M. marinum disrupts macrophage phagosomal membranes independent of pH. Inhibiting phagosomal acidification using bafilomycin did not affect ESX-1-dependent cytosolic access as observed by a cytosolic CCF4 assay, in which bacterial contact with the host cytoplasm results in CCF4 cleavage and the loss of FRET (134).
Although the precise mechanism of membrane lysis remains elusive, it is clear that the downstream consequences of phagosomal rupture promote the host response to infection and the survival of mycobacterial pathogens. The consequences of phagosomal lysis include the release of nucleic acid (138–140) and secreted proteins (141, 142) into the cytosol, the activation of the host immune response (8, 143), inflammasome activation, autophagy, host cell death (117, 127, 144–146), and cell-to-cell spread (147). This list is far from comprehensive, and the downstream consequences of phagosomal lysis continue to be the subject of great interest to the field. (For an extensive review of the downstream impact of phagosomal lysis, see reference 142.)
EspH is required for the secretion of the known virulence factors EspE and EspF in M. marinum (78). EspH was found to be essential for infection of RAW macrophages, but espH mutants were hypervirulent in M. marinum zebrafish embryo infections (78). Because M. marinum encounters a greater diversity of environments than the obligate human pathogen, its transcriptional and posttranscriptional regulatory pathways may vary slightly to adapt to this organism’s changing needs.
ESX-1 REGULATES GENE EXPRESSION
In addition to its well-established role in protein transport, it was demonstrated that the ESX-1 system regulates gene expression in M. marinum in 2017 (148) and in M. tuberculosis in 2019 (100). RNA sequencing of M. marinum and M. tuberculosis strains in the presence or absence of the ESX-1 system revealed widespread changes in gene expression, indicating a second, recently appreciated role for this protein transport system (100, 148), as discussed below. M. marinum studies have contributed the majority of our understanding of this new topic of research.
Feedback Control by the ESX-1 System
It was long established for several mycobacterial species that in the absence of a functional ESX-1 secretion system, ESX-1 substrates failed to accumulate within the mycobacterial cell (8, 9, 57, 59, 74, 77). This observation implied that the expression of ESX-1 substrates was regulated. In 2017, studies in M. marinum demonstrated that the ESX-1 system is subject to feedback regulation in which the levels of ESX-1 substrate production are linked to the assembly of the ESX-1 apparatus (148). In the absence of the ESX-1 membrane complex, the expression of the whiB6 gene was reduced 50-fold (148). WhiB6 is an iron-sulfur cluster transcription factor (149) that positively regulates the expression of many genes encoding ESX-1 substrates (148, 150, 151). Reduced whiB6 gene expression caused a corresponding decrease in the expression of the ESX-1 substrate genes at the esx-1 locus (148). ESX-1-dependent changes in whiB6 gene expression were also reported in M. tuberculosis albeit with a lower degree of repression (∼5-fold) in the absence of ESX-1 secretion (90, 100).
The precise signals underlying feedback control of the ESX-1 system remain an open question. In M. marinum, at least one signal is the assembly of the ESX-1 membrane complex (148). It is impossible to separate the loss of the membrane complex from the loss of secretion. The deletion of individual genes encoding ESX-1 components in both M. tuberculosis and M. marinum destabilizes the entire complex (63, 64, 148). However, the expression of EccCb1 alleles that result in a stable nonfunctional ESX-1 membrane complex (52, 152) failed to repress whiB6 gene expression in M. marinum (148). These data are consistent with the possibility that the loss of the membrane complex was the signal resulting in the loss of whiB6 expression in M. marinum. It is also possible that ESX-1-associated mycosin also contributes to the signal for feedback control of ESX-1 because mycosins stabilize ESX membrane complexes in M. marinum (63), although this remains to be tested. The assembly of the secretory apparatus as a signal is reminiscent of the feedback control mechanism of flagella in Campylobacter jejuni (153). It remains possible that additional signals may also contribute to feedback control of the ESX-1 system.
In 2020, further studies in M. marinum demonstrated that the repression of whiB6 gene expression in the absence of the ESX-1 system was at least partly dependent on a conserved transcription factor named EspM (154). The EspM protein directly binds the whiB6-espM intergenic region, repressing whiB6 gene expression. An M. marinum strain lacking the espM gene revealed elevated levels of WhiB6, which were restored or repressed by the expression of the espM gene from M. marinum, M. tuberculosis, and M. smegmatis. Cross-species complementation of the espM gene confirms the functional conservation of the EspM protein across all three species (154). Interestingly, the espM gene is divergently transcribed from the whiB6 gene and is immediately upstream of the start of the esx-1 locus, expanding the extended ESX-1 locus (Fig. 2). However, the espM gene may be essential in M. tuberculosis, revealing a potential difference in regulation between M. marinum and M. tuberculosis (154).
In addition to this pathway, two-component systems, including PhoPR, control ESX-1 secretion by regulating whiB6 and espR gene expression in M. tuberculosis (114, 150, 155, 156). Regulation of the whiB6 gene by PhoPR in M. marinum has not been reported. The EspR transcription factor positively regulates the expression of ESX-1 substrates at the espACD locus in M. tuberculosis (157). The deletion of the espR gene in M. tuberculosis resulted in the loss of ESX-1 secretion. The loss of secretion was caused by the reduced expression of the espACD operon, which is required for secretion in M. tuberculosis (157). All of the studies characterizing the role and mechanisms of EspR in regulating gene expression have been performed in M. tuberculosis (157–162). However, a screen aimed at understanding M. marinum pathogenesis in cell lines derived from different hosts and amoebae indicated that EspR was disadvantageous for M. marinum infection of fish-derived cells but not human cells (45). This study demonstrates that regulators, including EspR, may be differentially required for M. marinum virulence, depending on the host (45). Together, the studies focused on feedback control of the ESX-1 system demonstrate that fine-tuning ESX-1 is an important part of virulence.
Regulation of Gene Expression by the ESX-1 System
It was demonstrated in 2017 and 2018 that the deletion of the eccCb1 gene resulted in widespread changes in gene expression in both M. marinum and M. tuberculosis (100, 148). Based on studies in M. marinum, the WhiB6 and EspM transcription factors regulate a subset of genes affected by the loss of the eccCb1 gene, indicating that there are likely additional transcription factors contributing to the control of gene expression by the ESX-1 system (148, 154). It is unclear why the ESX-1 system controls genes that are not obviously linked to the secretion system in both M. marinum and M. tuberculosis. However, functional analyses from M. marinum studies reveal that ESX-1-regulated genes include those involved in cellular metabolism, lipid synthesis, and stress response pathways, suggesting a potential link to adaptation to cytoplasmic exposure following the lysis of the phagosome by the ESX-1 system (154). In this way, work in M. marinum not only revealed novel roles for the ESX-1 system but also shed light on potential pathways required to respond to phagosomal lysis.
Posttranscriptional Regulation of the ESX-1 System
In addition to these transcriptional mechanisms, it has been widely established that specific ESX-1-associated proteins regulate protein secretion posttranscriptionally on several levels. Based on studies in both M. tuberculosis and M. marinum, ESX-1 substrates are regulated at the level of protein stability through direct interactions. For example, the interaction between EsxA and EsxB is required for the stability of both proteins (8, 51, 108, 163). Several ESX-1-associated proteins stabilize substrate proteins and may act as substrate-specific chaperones. For example, studies in M. tuberculosis demonstrated that the EspD protein is required for the stability of the EspC and EspA substrates (82). Likewise, the EspL protein (which interacts with EspD) is required for the stability of the EspE and EspF substrates and the EspH component in M. tuberculosis (90). Studies in M. marinum demonstrated that EspH is required for the secretion of EspE and EspF (78). It is unclear whether EspL or EspH is the chaperone or if there is a difference in how these proteins function in M. marinum and M. tuberculosis.
In 2015, it was reported that EspI negatively regulates ESX-1 secretion in response to ATP levels in M. tuberculosis (164). The depletion of cellular ATP levels blocked ESX-1 secretion in an EspI-dependent manner. EspI is an ATP-binding protein that is highly conserved between M. tuberculosis and M. marinum. However, EspI has not been studied in M. marinum. EspI is predicted to have an FlhG domain, which is involved in the negative regulation of flagellar biosynthesis in Vibrio cholerae (165). The precise mechanism used by EspI to regulate the ESX-1 system is not known.
In M. tuberculosis, it was shown in 2010 that MycP1 regulates secretion by posttranslationally cleaving EspB. MycP1 is required for infection in mice, and its deletion results in increased secretion and hyperactivation of innate immune pathways (62). Based on the conservation of the membrane complex between M. tuberculosis and M. marinum, it is likely that the role of the mycosin proteases is conserved.
CONCLUDING REMARKS
As we highlight above, it is impossible to fully appreciate the state of the ESX-1 field by considering work in M. tuberculosis or M. marinum alone. Studies using M. marinum complement the work in M. tuberculosis. M. marinum continues to provide key insight into the conserved genes required for secretion and the functional relationships between ESX-1 components and substrates. Studies in M. marinum continue to reveal ESX-1 substrates. One of the major contributions of the M. marinum system in recent years has been defining new conserved roles of the ESX-1 system in pathogenesis and physiology. In addition to what is covered here regarding cell lysis and regulation, there have interesting publications on M. marinum linking the ESX-1 system to additional functions, including motility and biofilm formation (166) and downregulating microRNAs (miRNAs) (167). Moreover, several aspects of M. marinum physiology and pathogenesis, from its robust hemolytic activity to its unique interaction with zebrafish, make this species a powerful tool to study ESX-1-mediated secretion and virulence. Importantly, understanding the conserved aspects between the ESX-1 systems in M. tuberculosis and M. marinum is likely as important as understanding how these systems differ (Fig. 2, Appendix 2, and Table 1). It is clear that M. marinum customizes both the arsenal of ESX-1 substrates as well as regulation by the ESX-1 system to promote virulence in a variety of natural hosts.
TABLE 1.
Comparison between M. tuberculosis and M. marinum
| Property | Description (reference[s])a
|
|
|---|---|---|
| M. tuberculosis | M. marinum | |
| Disease burden | 1.5 million people worldwide in 2018 (1) | Pulmonary NTM now surpass M. tuberculosis infections in the US, Canada, and western Europe (3); for M. marinum, the incidence rate in Denmark from 2010–2016 was 0.05–0.13 per 100,000 person-years (170) |
| Top 10 cause of mortality and leading cause from a single infectious agent (1) | ||
| Host range | Humans are the only reservoir for TB (171), with some sublineage specificity (172) | Amphibians, fish, and humans (173) |
| Suspected free-living amoebae (174) | ||
| Aerobe? | Obligate aerobe (175) | Suspected obligate aerobe (176) |
| Cell wall properties | Acid fast (177) | Acid fast (15) |
| Contains mycolic acids, PDIM, TDM, PGL, and SL1 (178) | Similar lipid composition; lacks the nonvirulent SL1 (178); unique LOS (179) | |
| Generation time (h) | 17–18 (180) | 4–6 (181) |
| Optimal temp (°C) | 37 (182) | 30 (181) |
| Biosafety level | 3 (183) | 2 (183) |
| Role of phagolysosome? | Resisted (184) | Similar to M. tuberculosis (185) |
| Role of TNF-α? | Increased susceptibility by anti-TNF treatment (186) | Higher incidence than TB during anti-TNF therapy (189) |
| Increased susceptibility in TNF KO (187) | Increased susceptibility in TNF receptor knockdown (190) | |
| Excessive TNF-α increases susceptibility (188) | Excessive TNF-α increases susceptibility (191) | |
| Susceptibility to adaptive immunity? | Controlled by polyfunctional CD4+ and CD8+ T cells (192) | Rag1 KO zebrafish are hypersusceptible (193) |
| Unknown what T-cell responses protect against infection (192) | IL-10 mutant zebrafish resist M. marinum infection via interferon gamma (194) | |
| Granuloma formation | Forms necrotic granulomas (46) | Forms necrotic granulomas (46) |
| Presence of ESX-1 secretion system? | Yes (7, 8, 31, 33) | Yes (6, 9) |
| Susceptibility to first-line antibiotics, by MIC (μg/ml) | Isoniazid, 0.1 | Isoniazid, 16 |
| Rifampicin, 0.5 | Rifampicin, 1.0 | |
| Ethambutol, 4 (195) | Ethambutol, 1.0 (197, 198) | |
| Pyrazinamide, 25 (196) | Pyrazinamide, intrinsically resistant (4) | |
LOS, lipooligosaccharide; TNF-α, tumor necrosis factor alpha; KO, knockout; IL-10, interleukin-10; TDM, trehalose-6-6-dimycolate; PGL, phenolic glycolipids.
As we as a field continue to study the molecular mechanisms and consequences of ESX-1 secretion, there are several outstanding questions that will likely be facilitated by the continued use of M. marinum in concert with M. tuberculosis and other mycobacterial species. How are ESX-1 substrates and other secreted proteins transported across the mycobacterial envelope beyond the cytoplasmic membrane? How conserved is the regulation of and by the ESX-1 system between M. marinum and M. tuberculosis? Do differences in the regulation of and by the ESX-1 system between M. marinum and M. tuberculosis explain differences in the degree of phagosomal lysis? How does the ESX-1 system promote membrane lysis, and are these mechanisms conserved between M. tuberculosis and M. marinum? Finally, can M. marinum be exploited to enhance protection against or treatment of M. tuberculosis infection? A recent study exploited the M. marinum RD1 region to increase the efficacy of the BCG vaccine (168), indicating that studies in M. marinum may have more translational applications. In the end, it is clear that M. marinum will remain an important organism for revealing the mechanism, regulation, and role of ESX-1 secretion.
ACKNOWLEDGMENTS
The Champion laboratory is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers AI149235 and AI142127 to P.A.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. A.B. and W.C. are supported by Lake Forest College.
We thank Micah Ferrell and Lalita Ramakrishnan for the critical reading of the manuscript.
Biographies

Alexandra E. Chirakos graduated from The Ohio State University in 2015 with a bachelor’s degree in Microbiology and a minor in French. Her passion for research on globally relevant human pathogens led her to join Dr. Champion’s laboratory at the University of Notre Dame, pursuing a Ph.D. in Biological Sciences. Here, she has used molecular genetic approaches to study the regulation of a transcription factor that controls ESX-1 substrate production in M. marinum and M. tuberculosis. She is now a fifth-year Ph.D. Candidate preparing to graduate and has recently accepted a position as a 2021 Ohio Legislative Services Commission Fellow, where she hopes to work on health policy.

Ariane Balaram graduated from Lake Forest College, IL, with a B.A. in biology in May 2020. She minored in neuroscience. She grew up in Ahmedabad, India, as a result of which she developed an interest in studying tuberculosis. As the founding member of William Conrad’s Lake Forest College laboratory, she compared Mycobacterium tuberculosis with M. marinum, facilitating the consideration of this as a safer model of the disease. She also genetically engineered mammalian cells with the goal of improving resistance to mycobacterial infection. She is presently pursuing opportunities in clinical microbiology and molecular biology research.

William Conrad is an assistant professor of chemistry at Lake Forest College in Lake Forest, IL. He received his B.A. in biology at Macalester College, a Ph.D. in pharmacology with Randall Moon at the University of Washington, and postdoctoral fellowships in microbiology and medicine at the University of Washington and Cambridge University with Prof. Lalita Ramakrishnan. He began work on the ESX-1 secretion system in the laboratory of Lalita Ramakrishnan, where he discovered that ESAT-6 is not sufficient for host membrane lysis and demonstrated that ESX-1 causes gross membrane disruptions in a pH-independent manner. He continues to use the M. marinum zebrafish model of tuberculosis to identify and characterize conserved mycobacterial virulence mutants, including the ESX-1 secretion system.

Patricia A. Champion is an Associate Professor in the Department of Biological Sciences at the University of Notre Dame in Notre Dame, IN. She earned her B.S. in Biological Sciences at Carnegie Mellon University. She earned her Ph.D. in Molecular Biology with Dr. Thomas J. Silhavy, where she used genetics and molecular biology to study two-component signaling and stress responses in Escherichia coli. Her postdoctoral training was performed at UCSF with Dr. Jeffery S. Cox, where she began studying how proteins were targeted for secretion by the ESX-1 system in 2003. Since opening her own laboratory in 2009, she has had a continued interest in using genetics, molecular biology, and proteomic approaches to define the fundamental mechanisms of ESX-1 secretion and regulation, primarily using M. marinum.
APPENDIX 1
The glossary below serves as a reference to tie these diverse names and nomenclature referring to type VII/ESX secretion together.
- CFP-10
Ten-kilodalton culture filtrate antigen, also known as EsxB (26).
- Ecc proteins
The secretory machinery is made up of ESX-1 conserved components, or Ecc proteins, which span the cytoplasmic membrane and reside in the cytoplasm (Fig. 2 and 3). Subscripts in the Ecc or Esp names refer to the ESX system with which the protein is associated. For example, EccD1 is the EccD protein associated with the ESX-1 system. (For further nomenclature information, see reference 20.)
- ESAT-6
Six-kilodalton early secreted antigenic target, also known as EsxA (26).
- Esp proteins
Esx secretion-associated proteins that can be secreted substrates or components of the secretion system, such as EspB (76) or EspL (90). These proteins are not conserved across ESX systems.
- ESX-1
The name reflects that these specialized systems secrete proteins similar to EsxA, which was originally named ESAT-6, or 6-kDa early secreted antigenic target.
- Esx proteins
Small ∼100-aa substrates of ESX systems, which include a WXG domain (24), including EsxA and EsxB.
- gene identifiers
Genes identified from sequencing the laboratory strain M. tuberculosis H37Rv are labeled in descending order of genome locus, with a “c” if the gene is present on the complementary strand. For instance, the espJ gene has the identifier rv3878 and is found on the forward strand. In comparison, the neighboring gene espK has the gene identifier rv3879c and is found on the complementary strand.
- PE or PPE proteins
Proline-glutamic acid or proline-proline-glutamic acid proteins secreted by ESX systems, such as PPE68 (99, 199).
- RD1 locus
The region of difference 1 (RD1) locus is a genomic region lost in the vaccine strain M. bovis BCG (31, 33).
- Snm proteins
Esx-1 proteins were initially referred to as Snm proteins, which stands for “secretion in mycobacteria.” One example is Snm4, which is synonymous with EccD1 (8).
- type VII secretion
The ESX secretion systems were discovered to be distinct from the six other classes of secretion systems and were categorized as type VII secretion systems (T7SSs) (21).
APPENDIX 2
What is different about M. marinum? Despite the conservation between the ESX-1 systems, there are important differences between M. marinum and M. tuberculosis. M. marinum has an expanded host range relative to M. tuberculosis, which likely impacts how it uses conserved virulence pathways, including ESX-1 (45). Accordingly, M. marinum has a significantly larger genome than M. tuberculosis (6). M. marinum has plasmids that are absent in M. tuberculosis (6), and some strains of M. marinum carry an additional ESX system on a conjugative plasmid that is absent from the M strain (36). M. marinum has an expanded ESX-1 substrate repertoire that is not conserved in M. tuberculosis. M. marinum has an ESX-6 locus that includes a duplication of several genes from the ESX-1 locus, including the genes encoding demonstrated substrates, PE35_1, PPE68_1, EsxB_1, and potentially EsxA_3 (78, 103–105). Likewise, additional substrates, similar to MMAR_2894 (75, 78), are likely to be identified. Importantly, following phagosomal lysis, M. marinum uses actin-based motility (118), which has not been observed for M. tuberculosis. For additional differences, see Table 1.
REFERENCES
- 1.World Health Organization. 2019. Global tuberculosis report. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Dartois V, Sizemore C, Dick T. 2019. Editorial: NTM—the new uber-bugs. Front Microbiol 10:1299. doi: 10.3389/fmicb.2019.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratnatunga CN, Lutzky VP, Kupz A, Doolan DL, Reid DW, Field M, Bell SC, Thomson RM, Miles JJ. 2020. The rise of non-tuberculosis mycobacterial lung disease. Front Immunol 11:303. doi: 10.3389/fimmu.2020.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Diseases Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 5.Biondi G, Sotgiu G, Dore S, Molicotti P, Ruggeri M, Aliberti S, Satta R. 2017. Beyond pulmonary nontuberculous mycobacteria disease: do extra-pulmonary forms represent an emerging clinical and public health threat? ERJ Open Res 3:00091-2017. doi: 10.1183/23120541.00091-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley SA, Raghavan S, Hwang WW, Cox JS. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc Natl Acad Sci U S A 100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol 53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- 10.Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. 2004. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol 2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol 46:709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 12.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis 187:117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aubry A, Mougari F, Reibel F, Cambau E. 2017. Mycobacterium marinum. Microbiol Spectr 5:TNMI7-0038-2016. doi: 10.1128/microbiolspec.TNMI7-0038-2016. [DOI] [PubMed] [Google Scholar]
- 14.Pozos TC, Ramakrishnan L. 2004. New models for the study of Mycobacterium-host interactions. Curr Opin Immunol 16:499–505. doi: 10.1016/j.coi.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Linell F, Norden A. 1954. Mycobacterium balnei, a new acid-fast bacillus occurring in swimming pools and capable of producing skin lesions in humans. Acta Tuberc Scand Suppl 33:1–84. [PubMed] [Google Scholar]
- 16.Ramakrishnan L. 2020. Mycobacterium tuberculosis pathogenicity viewed through the lens of molecular Koch’s postulates. Curr Opin Microbiol 54:103–110. doi: 10.1016/j.mib.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 17.van Winden VJC, Houben ENG, Braunstein M. 2019. Protein export into and across the atypical diderm cell envelope of mycobacteria. Microbiol Spectr 7:GPP3-0043-2018. doi: 10.1128/microbiolspec.GPP3-0043-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green ER, Mecsas J. 2016. Bacterial secretion systems: an overview. Microbiol Spectr 4:VMBF-0012-2015. doi: 10.1128/microbiolspec.VMBF-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chagnot C, Zorgani MA, Astruc T, Desvaux M. 2013. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol 4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitter W, Houben EN, Bottai D, Brodin P, Brown EJ, Cox JS, Derbyshire K, Fortune SM, Gao LY, Liu J, Gey van Pittius NC, Pym AS, Rubin EJ, Sherman DR, Cole ST, Brosch R. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog 5:e1000507. doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion—mycobacteria show the way. Nat Rev Microbiol 5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 22.Groschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. 2016. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol 14:677–691. doi: 10.1038/nrmicro.2016.131. [DOI] [PubMed] [Google Scholar]
- 23.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol 2:RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallen MJ. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol 10:209–212. doi: 10.1016/S0966-842X(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 25.Tekaia F, Gordon SV, Garnier T, Brosch R, Barrell BG, Cole ST. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis 79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 26.Sørensen AL, Nagai S, Houen G, Andersen P, Andersen AB. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun 63:1710–1717. doi: 10.1128/IAI.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen P, Andersen AB, Sorensen AL, Nagai S. 1995. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J Immunol 154:3359–3372. [PubMed] [Google Scholar]
- 28.Pollock JM, Andersen P. 1997. The potential of the ESAT-6 antigen secreted by virulent mycobacteria for specific diagnosis of tuberculosis. J Infect Dis 175:1251–1254. doi: 10.1086/593686. [DOI] [PubMed] [Google Scholar]
- 29.Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 144(Part 11):3195–3203. doi: 10.1099/00221287-144-11-3195. [DOI] [PubMed] [Google Scholar]
- 30.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 31.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol 178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philipp WJ, Nair S, Guglielmi G, Lagranderie M, Gicquel B, Cole ST. 1996. Physical mapping of Mycobacterium bovis BCG Pasteur reveals differences from the genome map of Mycobacterium tuberculosis H37Rv and from M. bovis. Microbiology 142(Part 11):3135–3145. doi: 10.1099/13500872-142-11-3135. [DOI] [PubMed] [Google Scholar]
- 33.Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 34.Gey van Pittius NC, Warren RM, van Helden PD. 2002. ESAT-6 and CFP-10: what is the diagnosis? Infect Immun 70:6509–6510. doi: 10.1128/iai.70.11.6509-6511.2002. (Reply, 70:6511). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brodin P, Rosenkrands I, Andersen P, Cole ST, Brosch R. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol 12:500–508. doi: 10.1016/j.tim.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 36.Ummels R, Abdallah AM, Kuiper V, Aajoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5:e01744-14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newton-Foot M, Warren RM, Sampson SL, van Helden PD, Gey van Pittius NC. 2016. The plasmid-mediated evolution of the mycobacterial ESX (type VII) secretion systems. BMC Evol Biol 16:62. doi: 10.1186/s12862-016-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dumas E, Boritsch EC, Vandenbogaert M, Rodriguez de la Vega RC, Thiberge JM, Caro V, Gaillard JL, Heym B, Girard-Misguich F, Brosch R, Sapriel G. 2016. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol Evol 8:387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaziri F, Brosch R. 2019. ESX/type VII secretion systems—an important way out for mycobacterial proteins. Microbiol Spectr 7:PSIB-0029-2019. doi: 10.1128/microbiolspec.PSIB-0029-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottai D, Groschel MI, Brosch R. 2017. Type VII secretion systems in Gram-positive bacteria. Curr Top Microbiol Immunol 404:235–265. doi: 10.1007/82_2015_5015. [DOI] [PubMed] [Google Scholar]
- 41.Sherman DR, Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Smith S. 2004. Mycobacterium tuberculosis H37Rv:ΔRD1 is more virulent than M. bovis bacille Calmette-Guerin in long-term murine infection. J Infect Dis 190:123–126. doi: 10.1086/421472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, Griffiths KE, Marchal G, Leclerc C, Cole ST. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med 9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- 43.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoop EJ, Schipper T, Huber SK, Nezhinsky AE, Verbeek FJ, Gurcha SS, Besra GS, Vandenbroucke-Grauls CM, Bitter W, van der Sar AM. 2011. Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis Model Mech 4:526–536. doi: 10.1242/dmm.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerdenburg EM, Abdallah AM, Rangkuti F, Abd El Ghany M, Otto TD, Adroub SA, Molenaar D, Ummels R, Ter Veen K, van Stempvoort G, van der Sar AM, Ali S, Langridge GC, Thomson NR, Pain A, Bitter W. 2015. Genome-wide transposon mutagenesis indicates that Mycobacterium marinum customizes its virulence mechanisms for survival and replication in different hosts. Infect Immun 83:1778–1788. doi: 10.1128/IAI.03050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagan AJ, Ramakrishnan L. 2018. The formation and function of granulomas. Annu Rev Immunol 36:639–665. doi: 10.1146/annurev-immunol-032712-100022. [DOI] [PubMed] [Google Scholar]
- 47.Davis JM, Ramakrishnan L. 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, Ramakrishnan L. 2010. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science 327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox JS, Chen B, McNeil M, Jacobs WR, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 51.Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Hewinson RG, Williamson RA, Carr MD. 2002. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J Biol Chem 277:21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg OS, Dovala D, Li X, Connolly L, Bendebury A, Finer-Moore J, Holton J, Cheng Y, Stroud RM, Cox JS. 2015. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell 161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguilo N, Gonzalo-Asensio J, Alvarez-Arguedas S, Marinova D, Gomez AB, Uranga S, Spallek R, Singh M, Audran R, Spertini F, Martin C. 2017. Reactogenicity to major tuberculosis antigens absent in BCG is linked to improved protection against Mycobacterium tuberculosis. Nat Commun 8:16085. doi: 10.1038/ncomms16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pitulle C, Dorsch M, Kazda J, Wolters J, Stackebrandt E. 1992. Phylogeny of rapidly growing members of the genus Mycobacterium. Int J Syst Bacteriol 42:337–343. doi: 10.1099/00207713-42-3-337. [DOI] [PubMed] [Google Scholar]
- 55.Stahl DA, Urbance JW. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol 172:116–124. doi: 10.1128/jb.172.1.116-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobs WR, Jr, Tuckman M, Bloom BR. 1987. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature 327:532–535. doi: 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 57.Converse SE, Cox JS. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol 187:1238–1245. doi: 10.1128/JB.187.4.1238-1245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brodin P, Eiglmeier K, Marmiesse M, Billault A, Garnier T, Niemann S, Cole ST, Brosch R. 2002. Bacterial artificial chromosome-based comparative genomic analysis identifies Mycobacterium microti as a natural ESAT-6 deletion mutant. Infect Immun 70:5568–5578. doi: 10.1128/iai.70.10.5568-5578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, Cole ST, Brosch R. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun 74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teutschbein J, Schumann G, Mollmann U, Grabley S, Cole ST, Munder T. 2009. A protein linkage map of the ESAT-6 secretion system 1 (ESX-1) of Mycobacterium tuberculosis. Microbiol Res 164:253–259. doi: 10.1016/j.micres.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 61.Bosserman RE, Champion PA. 2017. ESX systems and the mycobacterial cell envelope: what’s the connection? J Bacteriol 199:e00131-17. doi: 10.1128/JB.00131-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. 2010. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Winden VJC, Ummels R, Piersma SR, Jimenez CR, Korotkov KV, Bitter W, Houben ENG. 2016. Mycosins are required for the stabilization of the ESX-1 and ESX-5 type VII secretion membrane complexes. mBio 7:e01471-16. doi: 10.1128/mBio.01471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol Microbiol 86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 65.Wagner JM, Chan S, Evans TJ, Kahng S, Kim J, Arbing MA, Eisenberg D, Korotkov KV. 2016. Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system. BMC Struct Biol 16:5. doi: 10.1186/s12900-016-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xie XQ, Zhang XL, Qi C, Li DF, Fleming J, Wang DC, Bi LJ. 2016. Crystallographic observation of the movement of the membrane-distal domain of the T7SS core component EccB1 from Mycobacterium tuberculosis. Acta Crystallogr F Struct Biol Commun 72:139–144. doi: 10.1107/S2053230X16000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wagner JM, Evans TJ, Korotkov KV. 2014. Crystal structure of the N-terminal domain of EccA ATPase from the ESX-1 secretion system of Mycobacterium tuberculosis. Proteins 82:159–163. doi: 10.1002/prot.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S, Zhou K, Yang X, Zhang B, Zhao Y, Xiao Y, Yang X, Yang H, Guddat LW, Li J, Rao Z. 2020. Structural insights into substrate recognition by the type VII secretion system. Protein Cell 11:124–137. doi: 10.1007/s13238-019-00671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Solomonson M, Huesgen PF, Wasney GA, Watanabe N, Gruninger RJ, Prehna G, Overall CM, Strynadka NC. 2013. Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J Biol Chem 288:17782–17790. doi: 10.1074/jbc.M113.462036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner JM, Evans TJ, Chen J, Zhu H, Houben EN, Bitter W, Korotkov KV. 2013. Understanding specificity of the mycosin proteases in ESX/type VII secretion by structural and functional analysis. J Struct Biol 184:115–128. doi: 10.1016/j.jsb.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poweleit N, Czudnochowski N, Nakagawa R, Trinidad DD, Murphy KC, Sassetti CM, Rosenberg OS. 2019. The structure of the endogenous ESX-3 secretion system. Elife 8:e52983. doi: 10.7554/eLife.52983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Famelis N, Rivera-Calzada A, Degliesposti G, Wingender M, Mietrach N, Skehel JM, Fernandez-Leiro R, Böttcher B, Schlosser A, Llorca O, Geibel S. 2019. Architecture of the mycobacterial type VII secretion system. Nature 576:321–325. doi: 10.1038/s41586-019-1633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Champion PA, Champion MM, Manzanillo P, Cox JS. 2009. ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol Microbiol 73:950–962. doi: 10.1111/j.1365-2958.2009.06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Champion MM, Williams EA, Pinapati RS, Champion PA. 2014. Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial Esx-1 substrates. J Proteome Res 13:5151–5164. doi: 10.1021/pr500484w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bosserman RE, Nicholson KR, Champion MM, Champion PA. 2019. A new ESX-1 substrate in Mycobacterium marinum that is required for hemolysis but not host cell lysis. J Bacteriol 201:e00760-18. doi: 10.1128/JB.00760-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McLaughlin B, Chon JS, MacGurn JA, Carlsson F, Cheng TL, Cox JS, Brown EJ. 2007. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog 3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carlsson F, Joshi SA, Rangell L, Brown EJ. 2009. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog 5:e1000285. doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Phan TH, van Leeuwen LM, Kuijl C, Ummels R, van Stempvoort G, Rubio-Canalejas A, Piersma SR, Jiménez CR, van der Sar AM, Houben ENG, Bitter W. 2018. EspH is a hypervirulence factor for Mycobacterium marinum and essential for the secretion of the ESX-1 substrates EspE and EspF. PLoS Pathog 14:e1007247. doi: 10.1371/journal.ppat.1007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacGurn JA, Raghavan S, Stanley SA, Cox JS. 2005. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol 57:1653–1663. doi: 10.1111/j.1365-2958.2005.04800.x. [DOI] [PubMed] [Google Scholar]
- 80.Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. 2005. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci U S A 102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kapopoulou A, Lew JM, Cole ST. 2011. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 82.Chen JM, Boy-Rottger S, Dhar N, Sweeney N, Buxton RS, Pojer F, Rosenkrands I, Cole ST. 2012. EspD is critical for the virulence-mediating ESX-1 secretion system in Mycobacterium tuberculosis. J Bacteriol 194:884–893. doi: 10.1128/JB.06417-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lou Y, Rybniker J, Sala C, Cole ST. 2017. EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol Microbiol 103:26–38. doi: 10.1111/mmi.13575. [DOI] [PubMed] [Google Scholar]
- 84.Ates LS, Brosch R. 2017. Discovery of the type VII ESX-1 secretion needle? Mol Microbiol 103:7–12. doi: 10.1111/mmi.13579. [DOI] [PubMed] [Google Scholar]
- 85.Xu J, Laine O, Masciocchi M, Manoranjan J, Smith J, Du SJ, Edwards N, Zhu X, Fenselau C, Gao LY. 2007. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol Microbiol 66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]
- 86.Chen JM, Zhang M, Rybniker J, Boy-Rottger S, Dhar N, Pojer F, Cole ST. 2013. Mycobacterium tuberculosis EspB binds phospholipids and mediates EsxA-independent virulence. Mol Microbiol 89:1154–1166. doi: 10.1111/mmi.12336. [DOI] [PubMed] [Google Scholar]
- 87.Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, Bitter W. 2012. General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci U S A 109:11342–11347. doi: 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Solomonson M, Setiaputra D, Makepeace KAT, Lameignere E, Petrotchenko EV, Conrady DG, Bergeron JR, Vuckovic M, DiMaio F, Borchers CH, Yip CK, Strynadka NCJ. 2015. Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism. Structure 23:571–583. doi: 10.1016/j.str.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 89.Piton J, Pojer F, Wakatsuki S, Gati C, Cole ST. 2 July 2020. High resolution cryoEM structure of the ring-shaped virulence factor EspB from Mycobacterium tuberculosis. J Struct Biol doi: 10.1016/j.yjsbx.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sala C, Odermatt NT, Soler-Arnedo P, Gulen MF, von Schultz S, Benjak A, Cole ST. 2018. EspL is essential for virulence and stabilizes EspE, EspF and EspH levels in Mycobacterium tuberculosis. PLoS Pathog 14:e1007491. doi: 10.1371/journal.ppat.1007491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bottai D, Majlessi L, Simeone R, Frigui W, Laurent C, Lenormand P, Chen J, Rosenkrands I, Huerre M, Leclerc C, Cole ST, Brosch R. 2011. ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J Infect Dis 203:1155–1164. doi: 10.1093/infdis/jiq089. [DOI] [PubMed] [Google Scholar]
- 92.Lienard J, Nobs E, Lovins V, Movert E, Valfridsson C, Carlsson F. 2020. The Mycobacterium marinum ESX-1 system mediates phagosomal permeabilization and type I interferon production via separable mechanisms. Proc Natl Acad Sci U S A 117:1160–1166. doi: 10.1073/pnas.1911646117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brennan MJ. 2017. The enigmatic PE/PPE multigene family of mycobacteria and tuberculosis vaccination. Infect Immun 85:e00969-16. doi: 10.1128/IAI.00969-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, Bitter W, Brennan MJ, Leclerc C, Batoni G, Campa M, Brosch R, Esin S. 2012. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol 83:1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- 95.Ates LS, van der Woude AD, Bestebroer J, van Stempvoort G, Musters RJP, Garcia-Vallejo JJ, Picavet DI, van de Weerd R, Maletta M, Kuijl CP, van der Wel NN, Bitter W. 2016. The ESX-5 system of pathogenic mycobacteria is involved in capsule integrity and virulence through its substrate PPE10. PLoS Pathog 12:e1005696. doi: 10.1371/journal.ppat.1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ates LS, Dippenaar A, Ummels R, Piersma SR, van der Woude AD, van der Kuij K, Le Chevalier F, Mata-Espinosa D, Barrios-Payán J, Marquina-Castillo B, Guapillo C, Jiménez CR, Pain A, Houben ENG, Warren RM, Brosch R, Hernández-Pando R, Bitter W. 2018. Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat Microbiol 3:181–188. doi: 10.1038/s41564-017-0090-6. [DOI] [PubMed] [Google Scholar]
- 97.Wang Q, Boshoff HIM, Harrison JR, Ray PC, Green SR, Wyatt PG, Barry CE, III. 2020. PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367:1147–1151. doi: 10.1126/science.aav5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ates LS, Ummels R, Commandeur S, van de Weerd R, Sparrius M, Weerdenburg E, Alber M, Kalscheuer R, Piersma SR, Abdallah AM, Abd El Ghany M, Abdel-Haleem AM, Pain A, Jiménez CR, Bitter W, Houben ENG. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet 11:e1005190. doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Demangel C, Brodin P, Cockle PJ, Brosch R, Majlessi L, Leclerc C, Cole ST. 2004. Cell envelope protein PPE68 contributes to Mycobacterium tuberculosis RD1 immunogenicity independently of a 10-kilodalton culture filtrate protein and ESAT-6. Infect Immun 72:2170–2176. doi: 10.1128/iai.72.4.2170-2176.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abdallah AM, Weerdenburg EM, Guan Q, Ummels R, Borggreve S, Adroub SA, Malas TB, Naeem R, Zhang H, Otto TD, Bitter W, Pain A. 2019. Integrated transcriptomic and proteomic analysis of pathogenic mycobacteria and their esx-1 mutants reveal secretion-dependent regulation of ESX-1 substrates and WhiB6 as a transcriptional regulator. PLoS One 14:e0211003. doi: 10.1371/journal.pone.0211003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jimenez C, Parra M, Cadieux N, Brennan MJ, Appelmelk BJ, Bitter W. 2009. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol 73:329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 102.Phan TH, Ummels R, Bitter W, Houben EN. 2017. Identification of a substrate domain that determines system specificity in mycobacterial type VII secretion systems. Sci Rep 7:42704. doi: 10.1038/srep42704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, Piersma SR, Jimenez CR, Luirink J, Bitter W, Houben EN. 2012. Specific chaperones for the type VII protein secretion pathway. J Biol Chem 287:31939–31947. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Damen MPM, Phan TH, Ummels R, Rubio-Canalejas A, Bitter W, Houben ENG. 2020. Modification of a PE/PPE substrate pair reroutes an Esx substrate pair from the mycobacterial ESX-1 type VII secretion system to the ESX-5 system. J Biol Chem 295:5960–5969. doi: 10.1074/jbc.RA119.011682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bosserman RE, Thompson CR, Nicholson KR, Champion PA. 2018. Esx paralogs are functionally equivalent to ESX-1 proteins but are dispensable for virulence in Mycobacterium marinum. J Bacteriol 200:e00726-17. doi: 10.1128/JB.00726-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Champion MM, Williams EA, Kennedy GM, Champion PA. 2012. Direct detection of bacterial protein secretion using whole colony proteomics. Mol Cell Proteomics 11:596–604. doi: 10.1074/mcp.M112.017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Champion PA, Stanley SA, Champion MM, Brown EJ, Cox JS. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- 108.Renshaw PS, Lightbody KL, Veverka V, Muskett FW, Kelly G, Frenkiel TA, Gordon SV, Hewinson RG, Burke B, Norman J, Williamson RA, Carr MD. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J 24:2491–2498. doi: 10.1038/sj.emboj.7600732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ekiert DC, Cox JS. 2014. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A 111:14758–14763. doi: 10.1073/pnas.1409345111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Phan TH, Houben ENG. 2018. Bacterial secretion chaperones: the mycobacterial type VII case. FEMS Microbiol Lett 365:fny197. doi: 10.1093/femsle/fny197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Korotkova N, Freire D, Phan TH, Ummels R, Creekmore CC, Evans TJ, Wilmanns M, Bitter W, Parret AH, Houben EN, Korotkov KV. 2014. Structure of the Mycobacterium tuberculosis type VII secretion system chaperone EspG5 in complex with PE25-PPE41 dimer. Mol Microbiol 94:367–382. doi: 10.1111/mmi.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tuukkanen AT, Freire D, Chan S, Arbing MA, Reed RW, Evans TJ, Zenkeviciutė G, Kim J, Kahng S, Sawaya MR, Chaton CT, Wilmanns M, Eisenberg D, Parret AHA, Korotkov KV. 2019. Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3, and ESX-5 type VII secretion systems. J Mol Biol 431:289–307. doi: 10.1016/j.jmb.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.King CH, Mundayoor S, Crawford JT, Shinnick TM. 1993. Expression of contact-dependent cytolytic activity by Mycobacterium tuberculosis and isolation of the genomic locus that encodes the activity. Infect Immun 61:2708–2712. doi: 10.1128/IAI.61.6.2708-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frigui W, Bottai D, Majlessi L, Monot M, Josselin E, Brodin P, Garnier T, Gicquel B, Martin C, Leclerc C, Cole ST, Brosch R. 2008. Control of M. tuberculosis ESAT-6 secretion and specific T cell recognition by PhoP. PLoS Pathog 4:e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brzychcy M, Andrzejczyk Z, Zalewska N, Zwolska Z, Rudnicka W. 1997. Haemolytic activity of Mycobacterium spp. Acta Microbiol Pol 46:377–385. [PubMed] [Google Scholar]
- 116.Dobos KM, Spotts EA, Quinn FD, King CH. 2000. Necrosis of lung epithelial cells during infection with Mycobacterium tuberculosis is preceded by cell permeation. Infect Immun 68:6300–6310. doi: 10.1128/iai.68.11.6300-6310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Houben D, Demangel C, van Ingen J, Perez J, Baldeon L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 118.Stamm LM, Morisaki JH, Gao LY, Jeng RL, McDonald KL, Roth R, Takeshita S, Heuser J, Welch MD, Brown EJ. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J Exp Med 198:1361–1368. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stamm LM, Pak MA, Morisaki JH, Snapper SB, Rottner K, Lommel S, Brown EJ. 2005. Role of the WASP family proteins for Mycobacterium marinum actin tail formation. Proc Natl Acad Sci U S A 102:14837–14842. doi: 10.1073/pnas.0504663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tilney LG, Portnoy DA. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. 2007. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 122.Singh VK, Berry L, Bernut A, Singh S, Carrere-Kremer S, Viljoen A, Alibaud L, Majlessi L, Brosch R, Chaturvedi V, Geurtsen J, Drancourt M, Kremer L. 2016. A unique PE_PGRS protein inhibiting host cell cytosolic defenses and sustaining full virulence of Mycobacterium marinum in multiple hosts. Cell Microbiol 18:1489–1507. doi: 10.1111/cmi.12606. [DOI] [PubMed] [Google Scholar]
- 123.Williams EA, Mba Medie F, Bosserman RE, Johnson BK, Reyna C, Ferrell MJ, Champion MM, Abramovitch RB, Champion PA. 2017. A nonsense mutation in Mycobacterium marinum that is suppressible by a novel mechanism. Infect Immun 85:e00653-16. doi: 10.1128/IAI.00653-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Derrick SC, Morris SL. 2007. The ESAT6 protein of Mycobacterium tuberculosis induces apoptosis of macrophages by activating caspase expression. Cell Microbiol 9:1547–1555. doi: 10.1111/j.1462-5822.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 125.Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, McDonald KL, Szyk A, LaRonde-LeBlanc N, Gao LY. 2008. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun 76:5478–5487. doi: 10.1128/IAI.00614-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, Honore N, Marchal G, Jiskoot W, England P, Cole ST, Brosch R. 2007. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol 189:6028–6034. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mba Medie F, Champion MM, Williams EA, Champion PAD. 2014. Homeostasis of N-α-terminal acetylation of EsxA correlates with virulence in Mycobacterium marinum. Infect Immun 82:4572–4586. doi: 10.1128/IAI.02153-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aly KA, Anderson M, Ohr RJ, Missiakas D. 2017. Isolation of a membrane protein complex for type VII secretion in Staphylococcus aureus. J Bacteriol 199:e00482-17. doi: 10.1128/JB.00482-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Casabona MG, Buchanan G, Zoltner M, Harkins CP, Holden MTG, Palmer T. 2017. Functional analysis of the EsaB component of the Staphylococcus aureus type VII secretion system. Microbiology 163:1851–1863. doi: 10.1099/mic.0.000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Peng X, Jiang G, Liu W, Zhang Q, Qian W, Sun J. 2016. Characterization of differential pore-forming activities of ESAT-6 proteins from Mycobacterium tuberculosis and Mycobacterium smegmatis. FEBS Lett 590:509–519. doi: 10.1002/1873-3468.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.De Leon J, Jiang G, Ma Y, Rubin E, Fortune S, Sun J. 2012. Mycobacterium tuberculosis ESAT-6 exhibits a unique membrane-interacting activity that is not found in its ortholog from non-pathogenic Mycobacterium smegmatis. J Biol Chem 287:44184–44191. doi: 10.1074/jbc.M112.420869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Refai A, Haoues M, Othman H, Barbouche MR, Moua P, Bondon A, Mouret L, Srairi-Abid N, Essafi M. 2015. Two distinct conformational states of Mycobacterium tuberculosis virulent factor early secreted antigenic target 6 kDa are behind the discrepancy around its biological functions. FEBS J 282:4114–4129. doi: 10.1111/febs.13408. [DOI] [PubMed] [Google Scholar]
- 134.Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, Hopkinson-Woolley D, Brosch R, Ramakrishnan L. 2017. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci U S A 114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Augenstreich J, Arbues A, Simeone R, Haanappel E, Wegener A, Sayes F, Le Chevalier F, Chalut C, Malaga W, Guilhot C, Brosch R, Astarie-Dequeker C. 2017. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol 19:e12726. doi: 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- 136.Quigley J, Hughitt VK, Velikovsky CA, Mariuzza RA, El-Sayed NM, Briken V. 2017. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. mBio 8:e00148-17. doi: 10.1128/mBio.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barczak AK, Avraham R, Singh S, Luo SS, Zhang WR, Bray MA, Hinman AE, Thompson M, Nietupski RM, Golas A, Montgomery P, Fitzgerald M, Smith RS, White DW, Tischler AD, Carpenter AE, Hung DT. 2017. Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog 13:e1006363. doi: 10.1371/journal.ppat.1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Watson RO, Manzanillo PS, Cox JS. 2012. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cheng Y, Schorey JS. 2018. Mycobacterium tuberculosis-induced IFN-beta production requires cytosolic DNA and RNA sensing pathways. J Exp Med 215:2919–2935. doi: 10.1084/jem.20180508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mittal E, Skowyra ML, Uwase G, Tinaztepe E, Mehra A, Koster S, Hanson PI, Philips JA. 2018. Mycobacterium tuberculosis type VII secretion system effectors differentially impact the ESCRT endomembrane damage response. mBio 9:e01765-18. doi: 10.1128/mBio.01765-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Upadhyay S, Mittal E, Philips JA. 2018. Tuberculosis and the art of macrophage manipulation. Pathog Dis 76:fty037. doi: 10.1093/femspd/fty037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. 2007. The type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol 178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 144.Mishra BB, Moura-Alves P, Sonawane A, Hacohen N, Griffiths G, Moita LF, Anes E. 2010. Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell Microbiol 12:1046–1063. doi: 10.1111/j.1462-5822.2010.01450.x. [DOI] [PubMed] [Google Scholar]
- 145.Wong KW, Jacobs WR, Jr. 2011. Critical role for NLRP3 in necrotic death triggered by Mycobacterium tuberculosis. Cell Microbiol 13:1371–1384. doi: 10.1111/j.1462-5822.2011.01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, Sherman DR, Gerner MY, Urdahl KB. 2018. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe 24:439–446.e4. doi: 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Aguilo JI, Alonso H, Uranga S, Marinova D, Arbues A, de Martino A, Anel A, Monzon M, Badiola J, Pardo J, Brosch R, Martin C. 2013. ESX-1-induced apoptosis is involved in cell-to-cell spread of Mycobacterium tuberculosis. Cell Microbiol 15:1994–2005. doi: 10.1111/cmi.12169. [DOI] [PubMed] [Google Scholar]
- 148.Bosserman RE, Nguyen TT, Sanchez KG, Chirakos AE, Ferrell MJ, Thompson CR, Champion MM, Abramovitch RB, Champion PA. 2017. WhiB6 regulation of ESX-1 gene expression is controlled by a negative feedback loop in Mycobacterium marinum. Proc Natl Acad Sci U S A 114:E10772–E10781. doi: 10.1073/pnas.1710167114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alam MS, Garg SK, Agrawal P. 2009. Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. FEBS J 276:76–93. doi: 10.1111/j.1742-4658.2008.06755.x. [DOI] [PubMed] [Google Scholar]
- 150.Solans L, Aguilo N, Samper S, Pawlik A, Frigui W, Martin C, Brosch R, Gonzalo-Asensio J. 2014. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WhiB6 as a novel ESX-1 component. Infect Immun 82:3446–3456. doi: 10.1128/IAI.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen Z, Hu Y, Cumming BM, Lu P, Feng L, Deng J, Steyn AJ, Chen S. 2016. Mycobacterial WhiB6 differentially regulates ESX-1 and the Dos regulon to modulate granuloma formation and virulence in zebrafish. Cell Rep 16:2512–2524. doi: 10.1016/j.celrep.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 152.Ramsdell TL, Huppert LA, Sysoeva TA, Fortune SM, Burton BM. 2015. Linked domain architectures allow for specialization of function in the FtsK/SpoIIIE ATPases of ESX secretion systems. J Mol Biol 427:1119–1132. doi: 10.1016/j.jmb.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Joslin SN, Hendrixson DR. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol 191:2656–2667. doi: 10.1128/JB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sanchez KG, Ferrell MJ, Chirakos AE, Nicholson KR, Abramovitch RB, Champion MM, Champion PA. 2020. EspM is a conserved transcription factor that regulates gene expression in response to the ESX-1 system. mBio 11:e02807-19. doi: 10.1128/mBio.02807-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Solans L, Gonzalo-Asensio J, Sala C, Benjak A, Uplekar S, Rougemont J, Guilhot C, Malaga W, Martín C, Cole ST. 2014. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog 10:e1004183. doi: 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee JS, Krause R, Schreiber J, Mollenkopf HJ, Kowall J, Stein R, Jeon BY, Kwak JY, Song MK, Patron JP, Jorg S, Roh K, Cho SN, Kaufmann SH. 2008. Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe 3:97–103. doi: 10.1016/j.chom.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 157.Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. 2008. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454:717–721. doi: 10.1038/nature07219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Anil Kumar V, Goyal R, Bansal R, Singh N, Sevalkar RR, Kumar A, Sarkar D. 2016. EspR-dependent ESAT-6 protein secretion of Mycobacterium tuberculosis requires the presence of virulence regulator PhoP. J Biol Chem 291:19018–19030. doi: 10.1074/jbc.M116.746289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hunt DM, Sweeney NP, Mori L, Whalan RH, Comas I, Norman L, Cortes T, Arnvig KB, Davis EO, Stapleton MR, Green J, Buxton RS. 2012. Long-range transcriptional control of an operon necessary for virulence-critical ESX-1 secretion in Mycobacterium tuberculosis. J Bacteriol 194:2307–2320. doi: 10.1128/JB.00142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Blasco B, Japaridze A, Stenta M, Wicky BI, Dietler G, Dal Peraro M, Pojer F, Cole ST. 2014. Functional dissection of intersubunit interactions in the EspR virulence regulator of Mycobacterium tuberculosis. J Bacteriol 196:1889–1900. doi: 10.1128/JB.00039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rosenberg OS, Dovey C, Tempesta M, Robbins RA, Finer-Moore JS, Stroud RM, Cox JS. 2011. EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins. Proc Natl Acad Sci U S A 108:13450–13455. doi: 10.1073/pnas.1110242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, Pojer F, Cole ST. 2012. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog 8:e1002621. doi: 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lightbody KL, Renshaw PS, Collins ML, Wright RL, Hunt DM, Gordon SV, Hewinson RG, Buxton RS, Williamson RA, Carr MD. 2004. Characterisation of complex formation between members of the Mycobacterium tuberculosis complex CFP-10/ESAT-6 protein family: towards an understanding of the rules governing complex formation and thereby functional flexibility. FEMS Microbiol Lett 238:255–262. doi: 10.1016/j.femsle.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 164.Zhang M, Chen JM, Sala C, Rybniker J, Dhar N, Cole ST. 2014. EspI regulates the ESX-1 secretion system in response to ATP levels in Mycobacterium tuberculosis. Mol Microbiol 93:1057–1065. doi: 10.1111/mmi.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lai LY, Lin TL, Chen YY, Hsieh PF, Wang JT. 2018. Role of the Mycobacterium marinum ESX-1 secretion system in sliding motility and biofilm formation. Front Microbiol 9:1160. doi: 10.3389/fmicb.2018.01160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zuo X, Wang L, Bao Y, Sun J. 2020. The ESX-1 virulence factors downregulate miR-147-3p in Mycobacterium marinum-infected macrophages. Infect Immun 88:e00088-20. doi: 10.1128/IAI.00088-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gröschel MI, Sayes F, Shin SJ, Frigui W, Pawlik A, Orgeur M, Canetti R, Honore N, Simeone R, van der Werf TS, Bitter W, Cho SN, Majlessi L, Brosch R. 2017. Recombinant BCG expressing ESX-1 of Mycobacterium marinum combines low virulence with cytosolic immune signaling and improved TB protection. Cell Rep 18:2752–2765. doi: 10.1016/j.celrep.2017.02.057. [DOI] [PubMed] [Google Scholar]
- 169.Shiryev SA, Papadopoulos JS, Schaffer AA, Agarwala R. 2007. Improved BLAST searches using longer words for protein seeding. Bioinformatics 23:2949–2951. doi: 10.1093/bioinformatics/btm479. [DOI] [PubMed] [Google Scholar]
- 170.Holden IK, Kehrer M, Andersen AB, Wejse C, Svensson E, Johansen IS. 2018. Mycobacterium marinum infections in Denmark from 2004 to 2017: a retrospective study of incidence, patient characteristics, treatment regimens and outcome. Sci Rep 8:6738. doi: 10.1038/s41598-018-24702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brites D, Gagneux S. 2015. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev 264:6–24. doi: 10.1111/imr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, Fenner L, Rutaihwa L, Borrell S, Luo T, Gao Q, Kato-Maeda M, Ballif M, Egger M, Macedo R, Mardassi H, Moreno M, Tudo Vilanova G, Fyfe J, Globan M, Thomas J, Jamieson F, Guthrie JL, Asante-Poku A, Yeboah-Manu D, Wampande E, Ssengooba W, Joloba M, Boom WH, Basu I, Bower J, Saraiva M, Vaconcellos SEG, Suffys P, Koch A, Wilkinson R, Gail-Bekker L, Malla B, Ley SD, Beck H-P, de Jong BC, Toit K, Sanchez-Padilla E, Bonnet M, Gil-Brusola A, Frank M, Penlap Beng VN, Eisenach K, Alani I, Wangui Ndung’u P, et al. 2016. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet 48:1535–1543. doi: 10.1038/ng.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Broutin V, Banuls AL, Aubry A, Keck N, Choisy M, Bernardet JF, Michel C, Raymond JC, Libert C, Barnaud A, Stragier P, Portaels F, Terru D, Belon C, Dereure O, Gutierrez C, Boschiroli ML, Van De Perre P, Cambau E, Godreuil S. 2012. Genetic diversity and population structure of Mycobacterium marinum: new insights into host and environmental specificities. J Clin Microbiol 50:3627–3634. doi: 10.1128/JCM.01274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.De Jonckheere JF. 1979. Pathogenic free-living amoebae in swimming pools: survey in Belgium. Ann Microbiol (Paris) 130B:205–212. [PubMed] [Google Scholar]
- 175.Cook GM, Berney M, Gebhard S, Heinemann M, Cox RA, Danilchanka O, Niederweis M. 2009. Physiology of mycobacteria. Adv Microb Physiol 55:81–182, 318–319. doi: 10.1016/S0065-2911(09)05502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mariniano SL, Nick JA, Daley CL. 2018. Nontuberculosis mycobacterial disease, p 498–506.e4. In Wilmott RW, Bush A, Deterding RR, Ratjen F, Sly P, Zar H, Li A (ed), Kendig’s disorders of the respiratory tract in children, 9th ed. Elsevier, Philadelphia, PA. [Google Scholar]
- 177.Sakula A. 1983. Robert Koch: centenary of the discovery of the tubercle bacillus, 1882. Can Vet J 24:127–131. [PMC free article] [PubMed] [Google Scholar]
- 178.Tobin DM, Ramakrishnan L. 2008. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol 10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 179.Rombouts Y, Burguiere A, Maes E, Coddeville B, Elass E, Guerardel Y, Kremer L. 2009. Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-alpha secretion in macrophages. J Biol Chem 284:20975–20988. doi: 10.1074/jbc.M109.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wayne LG. 1977. Synchronized replication of Mycobacterium tuberculosis. Infect Immun 17:528–530. doi: 10.1128/IAI.17.3.528-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Clark HF, Shepard CC. 1963. Effect of environmental temperatures on infection with Mycobacterium marinum (Balnei) of mice and a number of poikilothermic species. J Bacteriol 86:1057–1069. doi: 10.1128/JB.86.5.1057-1069.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wayne LG. 1994. Cultivation of Mycobacterium tuberculosis for research purposes, p 73–83. In Bloom BR (ed), Tuberculosis: pathogenesis, protection, and control. ASM Press, Washington, DC. 10.1128/9781555818357.ch6. [DOI] [Google Scholar]
- 183.Wilson DE, Chosewood LC. 2009. Biosafety in microbiological and biomedical laboratories, 5th ed. CDC, Atlanta, GA. https://www.cdc.gov/labs/pdf/CDC-BiosafetyMicrobiologicalBiomedicalLaboratories-2009-P.PDF. [Google Scholar]
- 184.Armstrong JA, Hart PD. 1975. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med 142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Barker LP, George KM, Falkow S, Small PL. 1997. Differential trafficking of live and dead Mycobacterium marinum organisms in macrophages. Infect Immun 65:1497–1504. doi: 10.1128/IAI.65.4.1497-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.British Thoracic Society Standards of Care Committee. 2005. BTS recommendations for assessing risk and for managing Mycobacterium tuberculosis infection and disease in patients due to start anti-TNF-alpha treatment. Thorax 60:800–805. doi: 10.1136/thx.2005.046797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bean AG, Roach DR, Briscoe H, France MP, Korner H, Sedgwick JD, Britton WJ. 1999. Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J Immunol 162:3504–3511. [PubMed] [Google Scholar]
- 188.Tobin DM, Vary JC, Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, Moens CB, Ramakrishnan L. 2010. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell 140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Raychaudhuri SP, Nguyen CT, Raychaudhuri SK, Gershwin ME. 2009. Incidence and nature of infectious disease in patients treated with anti-TNF agents. Autoimmun Rev 9:67–81. doi: 10.1016/j.autrev.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 190.Clay H, Volkman HE, Ramakrishnan L. 2008. Tumor necrosis factor signaling mediates resistance to mycobacteria by inhibiting bacterial growth and macrophage death. Immunity 29:283–294. doi: 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Roca FJ, Ramakrishnan L. 2013. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 153:521–534. doi: 10.1016/j.cell.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jasenosky LD, Scriba TJ, Hanekom WA, Goldfeld AE. 2015. T cells and adaptive immunity to Mycobacterium tuberculosis in humans. Immunol Rev 264:74–87. doi: 10.1111/imr.12274. [DOI] [PubMed] [Google Scholar]
- 193.Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. 2006. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun 74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Harjula SE, Ojanen MJT, Taavitsainen S, Nykter M, Ramet M. 2018. Interleukin 10 mutant zebrafish have an enhanced interferon gamma response and improved survival against a Mycobacterium marinum infection. Sci Rep 8:10360. doi: 10.1038/s41598-018-28511-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Suo J, Chang CE, Lin TP, Heifets LB. 1988. Minimal inhibitory concentrations of isoniazid, rifampin, ethambutol, and streptomycin against Mycobacterium tuberculosis strains isolated before treatment of patients in Taiwan. Am Rev Respir Dis 138:999–1001. doi: 10.1164/ajrccm/138.4.999. [DOI] [PubMed] [Google Scholar]
- 196.Pina RZ, Caleffi-Ferracioli KR, Campanerut-Sa PAZ, Ghiraldi-Lopez LD, Pavan FR, Siqueira VLD, Scodro RBL, Cardoso RF. 2016. Pyrazinamide susceptibility testing in Mycobacterium tuberculosis using the fast resazurin microtiter assay plate. Int J Tuberc Lung Dis 20:1535–1538. doi: 10.5588/ijtld.16.0304. [DOI] [PubMed] [Google Scholar]
- 197.Wallace RJ, Jr, Nash DR, Steele LC, Steingrube V. 1986. Susceptibility testing of slowly growing mycobacteria by a microdilution MIC method with 7H9 broth. J Clin Microbiol 24:976–981. doi: 10.1128/JCM.24.6.976-981.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Aubry A, Jarlier V, Escolano S, Truffot-Pernot C, Cambau E. 2000. Antibiotic susceptibility pattern of Mycobacterium marinum. Antimicrob Agents Chemother 44:3133–3136. doi: 10.1128/aac.44.11.3133-3136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol 6:95. doi: 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.López-Jiménez AT, Cardenal-Muñoz E, Leuba F, Gerstenmaier L, Barisch C, Hagedorn M, King JS, Soldati T. 2018. The ESCRT and autophagy machineries cooperate to repair ESX-1-dependent damage at the Mycobacterium-containing vacuole but have opposite impact on containing the infection. PLoS Pathog 14:e1007501. doi: 10.1371/journal.ppat.1007501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sani M, Houben ENG, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jiménez CR, Daffé M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. 2010. Direct visualization by Cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog 6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ulrichs T, Munk ME, Mollenkopf H, Behr-Perst S, Colangeli R, Gennaro ML, Kaufmann SHE. 1998. Differential T cell responses to Mycobacterium tuberculosis ESAT6 in tuberculosis patients and healthy donors. Eur J Immunol 28:3949–3958. doi:. [DOI] [PubMed] [Google Scholar]
- 203.Koo IC, Wang C, Raghavan S, Morisaki JH, Cox JS, Brown EJ. 2008. ESX-1-dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol 10:1866–1878. doi: 10.1111/j.1462-5822.2008.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Joshi SA, Ball DA, Sun MG, Carlsson F, Watkins BY, Aggarwal N, McCracken JM, Huynh KK, Brown EJ. 2012. EccA1, a component of the Mycobacterium marinum ESX-1 protein virulence factor secretion pathway regulates mycolic acid lipid synthesis. Chem Biol 19:372–380. doi: 10.1016/j.chembiol.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 205.Kennedy GM, Morisaki JH, Champion PAD. 2012. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba Acanthamoeba castellanii. Appl Environ Microbiol 78:2049–2052. doi: 10.1128/AEM.06965-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tan T, Lee WL, Alexander DC, Grinstein S, Liu J. 2006. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol 8:1417–1429. doi: 10.1111/j.1462-5822.2006.00721.x. [DOI] [PubMed] [Google Scholar]