Abstract
Three Saccharomyces cerevisiae proteins (Yng1/YOR064c, Yng2/YHR090c, and Pho23) and two Schizosaccharomyces pombe proteins (Png1/CAA15917 and Png2/CAA21250) share significant sequence identity with the human candidate tumor suppressor p33ING1 in their C-terminal regions. The homologous regions contain PHD finger domains which have been implicated in chromatin-mediated transcriptional regulation. We show that GFP-Yng2, like human Ing1, is localized in the nucleus. Deletion of YNG2 results in several phenotypes, including an abnormal multibudded morphology, an inability to utilize nonfermentable carbon sources, heat shock sensitivity, slow growth, temperature sensitivity, and sensitivity to caffeine. These phenotypes are suppressed by expression of either human Ing1 or S. pombe Png1, suggesting that the yeast and human proteins are functionally conserved. Yng1- and Pho23-deficient cells also share some of these phenotypes. We demonstrated by yeast two-hybrid and coimmunoprecipitation tests that Yng2 interacts with Tra1, a component of histone acetyltransferase (HAT) complexes. We further demonstrated by coimmunoprecipitation that HA-Yng1, HA-Yng2, HA-Pho23, and HA-Ing1 are associated with HAT activities in yeast. Genetic and biochemical evidence indicate that the Yng2-associated HAT is Esa1, suggesting that Yng2 is a component of the NuA4 HAT complex. These studies suggest that the yeast Ing1-related proteins are involved in chromatin remodeling. They further suggest that these functions may be conserved in mammals and provide a possible mechanism for the human Ing1 candidate tumor suppressor.
Several observations suggest that mammalian p33ING1 is involved in the regulation of cell proliferation and apoptosis (18, 21, 28). NIH 3T3 cells transformed by infection with a retrovirus containing a region of the Ing1 cDNA in the antisense orientation exhibit anchorage-independent growth in soft agar, and they form tumors in nude mice. Furthermore, microinjection of constructs that express Ing1 in the sense orientation results in inhibition of DNA synthesis and cell cycle progression in human diploid fibroblasts. Ing1 levels are also increased upon the induction of apoptosis in P19 cells by serum deprivation, and overexpression of Ing1 in P19 and rodent fibroblasts enhances Myc-dependent apoptosis (28). Evidence indicates that expression of Ing1 is repressed in a majority of breast and lymphoid cancer cell lines and glioblastomas and is mutated in some neuroblastoma cell lines, breast cancers, and brain tumors (21, 52, 74). Together, these observations suggest that Ing1 acts as a tumor suppressor and that it is involved in regulating apoptosis. This is further supported by reports that Ing1 and the p53 tumor suppressor form a complex and functionally cooperate to control cell growth (20, 83).
The carboxyl-terminal 70 amino acid residues of Ing1 contain the Cys4-His-Cys3 sequence of a PHD finger domain. This evolutionarily conserved domain is predicted to chelate two Zn2+ ions and is similar to, but distinct from, other zinc binding motifs such as the RING finger (Cys3-His-Cys4) and LIM domain (Cys2-His-Cys5). PHD finger domains have been found in many different proteins, including transcription factors and other proteins implicated in chromatin-mediated transcriptional regulation (1). In particular, PHD fingers are found in the Drosophilia melanogaster polycomb (Pc-G) and trithorax (trx-G) group proteins, which are thought to reside in large multiprotein complexes. Pc-G and trx-G are required for the expression of homeotic genes, and evidence suggests that they exert their effects through chromatin modification or interaction. Thus, it has been proposed that PHD finger domains may be involved in complex formation or recognition of nuclear targets related to chromatin structure and chromatin regulation (1).
In eukaryotes, DNA metabolism is strongly influenced by the packaging of DNA into higher-order chromatin. In general, chromatin structure is repressive to transcription (53, 54), and gene activation or silencing often requires remodeling of nucleosomes in promoter regions (38, 56). Covalent modifications, including acetylation, of core histones have been known for some time to be correlated with the activity of genetic loci (9, 75). Lysines in the amino-terminal extensions of histones are the targets of histone acetyltransferases (HATs) and histone deacetylases (HDACs). It has been hypothesized that neutralization of the positively charged histone N-terminal tails by acetylation lower their affinity for DNA, alter chromatin structure, and/or increase the interaction of histones with transcription factors (8, 31, 41, 44, 79).
Several previously identified transcriptional coactivators or corepressors have been shown to possess the ability to acetylate or deacetylate histones (38, 56, 72). In Saccharomyces cerevisiae, proteins that possess HAT activity include Hat1 (36), Gcn5 (11), and Esa1 (68). Hat1 is localized in both the cytosol and nucleus and acetylates primarily newly synthesized histone H4 prior to its assembly into nucleosomes (36, 55, 60, 78). Gcn5 is a nuclear HAT that preferentially acetylates H3, and to a lesser extent it acetylates H2B and H4 (25). Gcn5 is not essential, but it is required for transcriptional regulation of some genes (22), and mutations that impair Gcn5 HAT activity correlate with decreased transcriptional activity (39, 81). Esa1 is an essential gene that was recently shown to possess HAT activity with a preference for H2A and H4 (13). Several mammalian transcription regulators have also been shown to possess HAT activity, including Gcn5 and Esa1 homologs (8), p300 and CREB-binding protein (5, 51), pCAF (82), ACTR (12), Src-1 (69), and TAFII250 (48).
HATs function as components of large, evolutionarily conserved macromolecular assemblies, five of which have been identified in S. cerevisiae (16, 24, 25, 63). These include the 1.8-MDa SAGA (Spt-Ada-Gcn5-acetyltransferase), 0.8-MDa ADA, NuA3, 1.3-MDa NuA4 (nucleosomal H2A.H4), and the novel SLIK (SAGA-like) complexes. Esa1 was recently shown to be the HAT subunit of NuA4 (3), whereas Gcn5 is the catalytic HAT in the SAGA and ADA complexes (25). The HAT of the NuA3 complex has not been characterized aside from its substrate preference for histone H3 (25).
Purified SAGA promotes acetyl coenzyme A (acetyl-CoA)-dependent transcription from nucleosomal promoter templates, but not free DNA, in vitro (70). This observation is consistent with the requirement for Gcn5 HAT activity for both promoter-directed histone acetylation and Gcn5-mediated transcriptional activation in vivo (39, 81). Furthermore, acidic activators such as Gcn4 and the VP16 activation domain can physically interact with purified native SAGA complex, and GAL4-VP16 targets acetylation and transcriptional stimulation by SAGA (76). Like SAGA, NuA4 is recruited to promoters by acidic activator proteins to promote histone acetylation and transcriptional stimulation (3, 13, 76). Unlike SAGA, which preferentially acetylates the N termini of histones H3 and H2B, NuA4 targets mainly the N termini of histone H4 and to a lesser extent H2A (3, 13).
The SAGA complex contains at least four protein modules, including the Ada and Spt subgroups of transcription regulators, the histone-fold subgroup of TATA-binding protein-associated factors, and the essential 433-kDa Tra1 protein (24, 25). Tra1 has also been shown to be a component of the SLIK and NuA4 HAT complexes, and it also coelutes in a high-molecular-weight region distinct from the nucleosomal HAT complexes, indicating that it is present in uncharacterized protein complexes (24). Tra1 has been highly conserved among eukaryotes (47), and the mammalian homolog, TRRAP, is associated with the PCAF HAT complex (77). TRRAP was identified as a cofactor that interacts with c-Myc and E2F-1 and is required for transformation by c-Myc and E1A (47). The identification of TRRAP as an essential cofactor for these oncogenic transcription factors suggests that it regulates gene expression.
Tra1 and TRRAP belong to the phosphatidyl inositol-3 (PI3) kinase family of serine/threonine protein kinases that includes mammalian DNA-PK, ATM, FRAP, Schizosaccharomyces pombe Rad3, and S. cerevisiae Vps34, Pik1, Stt4, Tor1, Tor2, Tel1, and Mec1 (63; reviewed in reference 8). These proteins appear to be involved in processes including cell cycle control, DNA repair, and transcription (35, 42). Although Tra1 and TRRAP are closely related to the PI3 kinases, they do not contain the DXXXXN and DFG motifs conserved in the catalytic site of PI3 kinases (47). The association of Tra1 and TRRAP with HAT complexes suggests that they regulate transcriptional activation through the recruitment of HAT activity to activator-bound promoters (24, 76). Although a scaffolding role of Tra1 has been suggested (8, 24), the molecular function of Tra1 and TRRAP are not known.
Three proteins, Yng1, Yng2, and Pho23, in the budding yeast S. cerevisiae share significant sequence identity in their PHD finger domains with mammalian Ing1. We show that Yng2 is associated with Tra1, and we further demonstrate that Yng1, Yng2, and Pho23 are associated with HAT activities. We also provide strong evidence that the Yng2-associated HAT is Esa1, suggesting that Yng2 is a component of the NuA4 complex. Our results suggest that Yng1, Yng2, and Pho23 are involved in chromatin remodeling and possibly transcriptional regulation. We also report genetic and biochemical evidence suggesting that human and yeast Ing1 homologs have been functionally conserved.
MATERIALS AND METHODS
Yeast strains and genetic analysis.
The S. cerevisiae strain L40 (MATa his3 trp1 leu2 ade2 LYS2::lexA-HIS3 URA3::lexA-lacZ) has been previously described (30, 80). The genotypes of other yeast strains used in this study are listed in Table 1. S. cerevisiae culture, transformation, mating, tetrad analysis, and other genetic manipulations were performed as previously described (2, 17).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| JC1 | MATα ade8 his3 leu2 lys2 trp1 ura3 can1 | J. Colicelli |
| JC2 | MATa ade8 his3 leu2 lys2 trp1 ura3 can1 | J. Colicelli |
| RL 3-17 | MATα ade8 his3 leu2 lys2 trp1 ura3 can1 pho23::TRP1 | This study |
| RL 3-29 | MATa ade8 his3 leu2 lys2 trp1 ura3 can1 pho23::TRP1 | This study |
| RL 3-32 | MATα ade8 his3 leu2 lys2 trp1 ura3 can1 yng1::HIS3 | This study |
| RL 3-49 | MATa ade8 his3 leu2 lys2 trp1 ura3 can1 yng1::HIS3 | This study |
| RL 3-53 | MATα ade8 his3 leu2 lys2 trp1 ura3 can1 yng2::URA3 | This study |
| RL 3-56 | MATa ade8 his3 leu2 lys2 trp1 ura3 can1 yng2::URA3 | This study |
| RL 4-70 | MATα ade8 his3 leu2 lys2 trp1 ura3 can1 yng1::HIS3 yng2::URA3 | This study |
| RL 4-80 | MATα ade8 his3 leu2 lys2 trp1 ura3 can1 yng2::URA3 pho23::TRP1 | This study |
| RL 5-34 | MAT?a ade8 his3 leu2 lys2 trp1 ura3 can1 yng1::HIS3 pho23::TRP1 | This study |
| RL 5-36 | MAT? ade8 his3 leu2 lys2 trp1 ura3 can1 yng1::HIS3 yng2::URA3 pho23::TRP1 | This study |
| 1788 | MATa/MATα leu2/leu2 ura3/ura3 trp1/trp1 his4/his4 can1r/can1r | I. Herskowitz |
| RL 1-37 | MATa leu2 ura3 trp1 his4 can1r pho23::HIS3 | This study |
| RL 2-28 | MATα leu2 ura3 trp1 his4 can1r yng2::URA3 | This study |
| RL 2-80 | MAT? leu2 ura3 trp1 his4 can1r yng1::LEU2 | This study |
| LPY3498 | MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 | L. Pillus |
| LPY3500 | MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1::HIS3 esa1-L254P::URA3 | L. Pillus |
| LPY3291 | MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1::HIS3 pLP863(esa1-414/TRP1) | L. Pillus |
| FY105 | MATα leu2 ura3 trp1 | G. Thireos |
| gcn5Δ | MATα leu2 ura3 trp1 gcn5Δ | G. Thireos |
?, mating type not determined.
DNA manipulation and analysis.
Procedures used for DNA manipulation and analysis (purification, cloning, electrophoresis, transformation, etc.) have been previously described (64). PCR was performed as described (46).
Plasmids.
The yeast two-hybrid vector pBTM116 contains the DNA-binding LexA coding sequence under the control of the ADH1 promoter, the 2μm origin of replication, and the TRP1 gene (6). pLexA-Lamin has been described previously (6). pLexA-Yng2 was generated by inserting the coding sequence of YNG2 into the polylinker of pBTM116 located 3′ to the LexA coding sequence. pLexA-ΔPHD and pLEXA-PHD were generated by inserting codons 1 to 222 and 222 to 282, respectively, of YNG2 into pBTM116. pADHA-Yng2, pADHA-Png1, and pADHA-Ing1 were generated by cloning the PCR-derived open reading frame (ORF) of YNG2, S. pombe PNG1, or human p33ING1 respectively, into pAD4.H, which contains the 2μm origin of replication and ADH1 promoter (32). YEpPDE2 contains S. cerevisiae PDE2 in YEp13, as previously described (65). pADGFPHA was derived by cloning the sequence of the enhanced green fluorescent protein (eGFP; Clonetech) into the HindIII site of pAD4.H (32) using the primers 5′GTCAGCAAGCTTATGGTGAGCAAGGGCGAG and 3′GATCTCAAGCTTCTTGTACAGCTCGTCCAT. pADGFPHA-Yng2, -Gcn5, and -Esa1 were made by cloning the PCR-derived ORF of YNG2, GCN5, or ESA1, respectively, into pADGFPHA. Codons 1 to 222 of the YNG2 ORF were PCR amplified and cloned into pADGFPHA to make pADGFPHA-Yng2ΔPHD. Codons 222 to 282 of the YNG2 ORF were PCR amplified and cloned into pADGFPHA to make pADGFPHA-Yng2PHD. pADGFPHA-Yng2/1 was constructed as follows. A primer, 5′TTCTCGAGAGAATTCAAAAACAGTAGAAATGGTAAAGGCCAAAACGGTTCCCCTGAAAACGAGGAAGAGGACAAAACGGAGGTTTATTGTTTCTGTAGGAAT, was used to amplify the C-terminal 64 codons of the YNG1 ORF. This PCR product was cloned into pADGFPHA-Yng2 as an EcoRI/SacI fragment to replace the C-terminal PHD finger of Yng2 with that of Yng1. pADmyc-Yng2 was generated by cloning the PCR-derived ORF of YNG2 in pUAD6 (32). pADmyc-Tra1 was derived by shuttling the TRA1 ORF as a NotI(filled-in)/SacI fragment from p1259 (a kind gift from C. Brandl) into the SmaI/SacI sites of a modified pUAD6 (SalI digested, filled in, and religated). pPho23 was generated by inserting a PCR-amplified PHO23 sequence (−480 to +490 with respect to the ORF) into the XhoI/BamHI sites of pBluescript II SK. pPho23target was generated by replacing the 0.75-kb EcoRI/EcoRV fragment of pPho23 with the 0.85-kb TRP1 gene. pYng1 was generated by inserting a PCR-amplified YNG1 sequence (−340 to +350 with respect to the ORF) into the EcoRV site of pBluescript II SK. pYng1Ltarget was generated by replacing the 0.5-kb EcoRV/StuI fragment with the 2.2-kb LEU2 gene. pYng1Htarget was generated by replacing the 0.5-kb EcoRV/StuI fragment with the 1.7-kb HIS3 gene. pYng2 was generated by inserting a PCR-amplified YNG2 sequence (SalI to BglII) into the SalI and BamHI sites of pBluescript II SK. pYng2target was generated by replacing the 0.6-kb BamHI/EcoRI fragment of pYng2 with the 1.2-kb URA3 gene.
Yeast two-hybrid screen.
An S. cerevisiae two-hybrid genomic library (kindly provided by I. Sadowski) was transformed into the S. cerevisiae strain L40 containing pLexA-Yng2 using a high-efficiency transformation method (29, 66, 80). These transformants were grown in synthetic medium (YC-Trp-Ura-Leu-Lys) for 16 h at 30°C to obtain efficient expression of the HIS3 reporter gene. The transformants were then plated on synthetic plates (YC-Trp-His-Ura-Leu-Lys) to select for cells that express His3. Approximately 2 × 103 His+ transformants were obtained from 2 × 106 primary transformants. A subset of these His+ transformants were further analyzed, yielding 15 library clones that transactivated His3 and LacZ expression in pLex-Yng2 harboring strains but not strains containing pLexA-lamin.
Gene disruptions.
PHO23 (YNL097c), YNG1 (YOR064c), and YNG2 (YHR090c) were disrupted in both 1788 diploid and JC1 and JC2 haploid strains by the gene replacement method (59, 61). PHO23 was disrupted in JC1, JC2, and 1788 by transformation of a 1.7-kb BspDI/MluI fragment of pPho23target in which the PHO23 coding sequence had been replaced with TRP1. YNG1 was disrupted in JC1 and JC2 by transformation of a 2.5-kb fragment derived by PCR from pYng1Htarget in which the YNG1 coding sequence had been replaced with HIS3. YNG1 was disrupted in 1788 by transformation of a 3.0-kb fragment derived by PCR from pYng1Ltarget in which the YNG1 coding sequence had been replaced with LEU2. YNG2 was disrupted by transformation of a 2.2-kb AatII/XbaI fragment from pYng2target in which the YNG2 coding sequence had been replaced with URA3. Gene disruptions were confirmed by Southern blot analysis. In almost all cases, asci derived from sporulated diploid heterozygotes segregated the auxotrophic marker 2:2. Haploid strains of opposite mating types were crossed, and the resulting diploids were sporulated using standard methods (2) to derive double (yng1Δyng2Δ and pho23Δyng2Δ) and triple (yng1Δyng2Δpho23Δ) deletion mutants.
Protein biochemistry.
Proteins were isolated from yeast cultures grown in synthetic medium to an optical density at 600 nm of approximately 1.0. For temperature shift experiments, cells were grown to an optical density at 600 nm of approximately 0.3; half of the culture was allowed to continue to grow at 30°C for 4 h, while the other half was collected by centrifugation and resuspended in prewarmed medium and grown for 4 h at 37°C. Briefly, cells from 250 ml of culture were collected by centrifugation, washed once with lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.2% Triton X-100) and resuspended in 4 ml of lysis buffer with protease inhibitors (1 μg of pepstatin A per ml, 200 μM phenylmethylsulfonyl fluoride, 500 μM benzamidine HCl, 10 μg of aprotinin per ml, 1 μg of leupeptin per ml). Cell suspensions were aliquoted into four 2-ml tubes with 1.5 g of glass beads (diameter, 425 to 600 nm; Sigma) and shaken for 5 min in a Mini-BeadBeater (Biospec Products) at 4°C. Cell debris was removed by a 2-min centrifugation at 450 × g, and lysates were further clarified by centrifugation for 5 min at 10,600 × g. Protein concentrations (typically 10 to 15 mg/ml) were determined at this point using a Bio-Rad Protein assay. Ten milligrams of protein was used in 1- to 2-ml immunoprecipitation reactions (IPs) in lysis buffer with protease inhibitors at 4°C with gentle rotation. IPs were precleared with 40 μl of protein A-Sepharose beads for 20 min. After removal of the beads, 5 μl of 12CA5 (antihemagglutinin [anti-HA]) ascites fluid was added and the reaction mixtures were incubated overnight. Immune complexes were collected by addition of 50 μl of protein A-Sepharose followed by a 2-h incubation. Beads were collected by a 1-min centrifugation at 2,200 rpm and washed five or six times with lysis buffer. Half of the beads were removed at this point for HAT assays. For coimmunoprecipitation analyses, 20 μl of protein sample buffer was added and the beads were boiled and aliquoted prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (half for the 12CA5 blot, half for the 9E10 [anti-myc] blot). Sixty micrograms of lysate was separated by SDS-PAGE (8% acrylamide–0.075% bisacrylamide) to observe the expression of myc-Tra1. Thirty micrograms of lysate was analyzed to observe expression of myc-Yng2. After SDS-PAGE, proteins were electroblotted onto nitrocellulose membranes (1.5 h; 100 V), blocked for 1 h or overnight in 5% milk in Tris-buffered saline, and incubated 1 h with primary antibody (1:2,000 12CA5; 1:30 9E10) in Tris-buffered saline–0.05% Tween 20. After washing, tagged proteins were detected with horseradish peroxidase-conjugated anti-mouse secondary antibodies and enhanced chemiluminescence reagents (Amersham).
HAT assays.
HAT assays were performed as previously described (49). Immunoprecipitates were incubated at 30°C for 30 min with 2 μg of calf thymus total histones (type IIA; Sigma) and 0.3 μCi of [3H]-acetyl-CoA (ICN) in buffer containing 50 mM Tris-HCl (pH 8.0), 30 mM KCl, 10% glycerol, 10 mM sodium butyrate, 1 mM dithiothreitol, and 1.0 μM acetyl-CoA in a total volume of 30 μl. Reactions were started by the addition of acetyl-CoA and stopped by the addition of 10 μl of 4× SDS sample buffer. Aliquots (20 μl) were fractionated on 18% acrylamide–0.48% bisacrylamide SDS gels, stained with Coomassie blue, and destained by standard methodology. Destained gels were then subjected to fluorography by treatment with En3Hance (Dupont-NEN) according to the manufacturer's instructions. Gels were dried, placed in cassettes with X-ray film (Kodak XAR), and exposed at −80°C.
DAPI staining.
Nuclear and mitochondrial DNA was visualized by DAPI (4′,6′-diamidino-2-phenylindole; Sigma) staining following established protocols (2) with slight modifications. Briefly, ∼107 cells were pelleted and resuspended in 70% ethanol, incubated for 30 s, and washed twice with water. After the final wash, cells were resuspended in mounting medium (p-phenylenediamine [1 mg/ml] in PBS [pH 9], 90% glycerol) containing DAPI (0.025 mg/ml) and viewed with UV optics. GFP images were visualized with fluorescein isothiocyanate optics.
RESULTS
Comparison of yeast proteins with human Ing1.
We performed a search of the public sequence databases for proteins related to the human candidate tumor suppressor p33ING1. This comparison revealed that three budding yeast proteins (Yng1, Yng2, and Pho23) and two fission yeast proteins (Png1 and Png2) share significant sequence identity with Ing1 (Fig. 1). The budding and fission yeast genes encode small proteins that are similar in length (219 to 330 residues) to human Ing1 (279 residues). The N-terminal regions of these proteins have not been well conserved, but the C-terminal regions are very similar (50 to 60% identity) (Fig. 1). These homologous regions contain PHD finger domains, which have been found in proteins implicated in chromatin-mediated transcriptional regulation (1). The strong similarity among the PHD finger domains of these yeast proteins and human Ing1 suggests that they were derived from the same common ancestral gene and that their functions may have been conserved during evolution.
FIG. 1.
Alignment of PHD finger domains. The C-terminal ends of human Ing1 (AAC00501), S. pombe Png1 (CAA15917) and Png2 (CAA21250), and S. cerevisiae Yng1 (P50947), Yng2 (P38806), and Pho23 (S66947) were aligned to fit the PHD finger consensus sequence (1). The symbols $ and # indicate semiconserved and highly conserved hydrophobic residues, respectively. Regions of protein sequence identity with Ing1 are shaded. Numbers at the right of each sequence indicate the length of each protein.
Deletion of YNG1, YNG2, and PHO23 results in pleotrophic phenotypes.
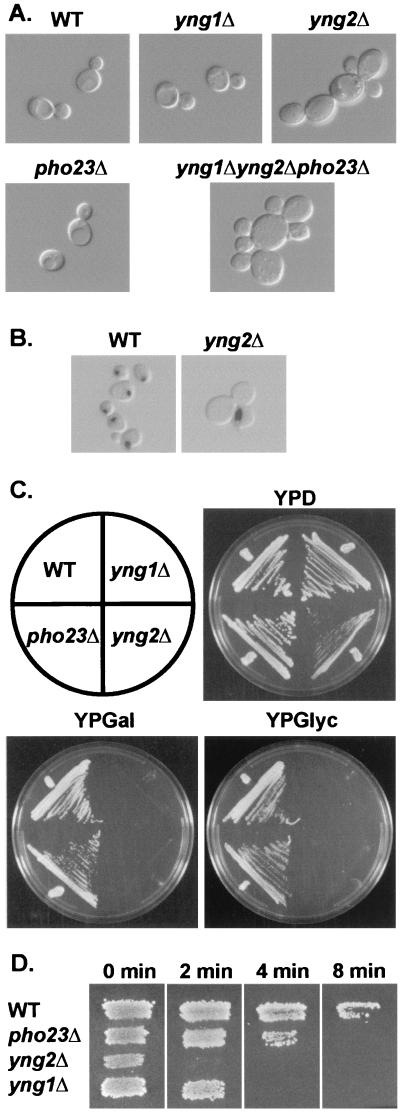
To investigate the function of these proteins we have performed genetic analysis in yeasts. First, we constructed S. cerevisiae strains that contain disruptions of each of the three Ing1 homologous genes, individually and in all possible combinations (described in Materials and Methods) (Table 1). In almost all cases, asci derived from sporulated diploid heterozygotes segregated the auxotrophic marker 2:2, demonstrating that neither Yng1, Yng2, nor Pho23 is required for germination or cell growth. However, yng2Δ cells exhibited slow growth compared with their parental cells (5.2- and 2.8-h doubling times, respectively, in YPD at 30°C). Deletion of YNG1 or PHO23 had little effect on growth or morphology, but yng2Δ cells were swollen and ∼50% of cells were multibudded, containing three to eight large buds (Fig. 2A). Incubation at elevated temperatures increased the severity of the abnormal morphology. DAPI staining demonstrated that ∼80% of multibudded clumps contained large buds that lacked nuclear DNA (Fig. 2B). Disruption of all three genes in combination was not lethal but resulted in more severe growth inhibition (9.7-h doubling time) and a more exaggerated morphological, multibudded phenotype than that of yng2Δ cells (Fig. 2A). Both yng2Δ and yng1Δ cells were unable to utilize nonfermentable carbon sources such as galactose, glycerol (Fig. 2C), and acetate (not shown). yng2Δ, yng1Δ, and pho23Δ cells are all hypersensitive to heat shock, albeit to varying degrees (Fig. 2D). Deletion of YNG2 alone resulted in several other phenotypes, including a slight sensitivity to UV irradiation but not gamma irradiation or treatment with alkylating agents (not shown), temperature sensitivity, and sensitivity for growth on media containing caffeine (Fig. 3B). Thus, there appear to be both functional similarities and differences between these three related yeast proteins.
FIG. 2.
Deletion of YNG1, YNG2, and PHO23 result in various phenotypes. (A) Normal wild-type (WT) (JC1), yng1Δ (RL3-32), yng2Δ (RL3-53), pho23Δ (RL3-17), and triple mutant yng1Δyng2Δpho23Δ (RL5-36) cells were grown in yeast extract-peptone-dextrose (YPD) at 30°C and examined by differential interference contrast (DIC) microscopy. (B) Localization of DNA in normal WT (JC1) and yng2Δ cells (RL3-53) cells grown in YPD was examined by DAPI staining (Materials and Methods). DIC and fluorescent images were overlaid to produce the images shown. (C) The carbon source requirements of normal WT (JC1), yng1Δ (RL3-32), yng2Δ (RL3-53), and pho23Δ (RL3-17) cells were determined by growth on either glucose (YPD), galactose (YPGal), or glycerol (YPGlyc) as the sole carbon source at 30°C for 3 to 5 days. (D) Normal WT (JC1), pho23Δ (RL3-17), yng2Δ (RL3-53), and yng1Δ (RL3-32) were tested for heat shock sensitivity by replica plating onto prewarmed YPD plates and incubated at 55°C for 0, 2, 4, or 8 min. Plates were then allowed to cool to room temperature before incubation at 30°C for 3 days.
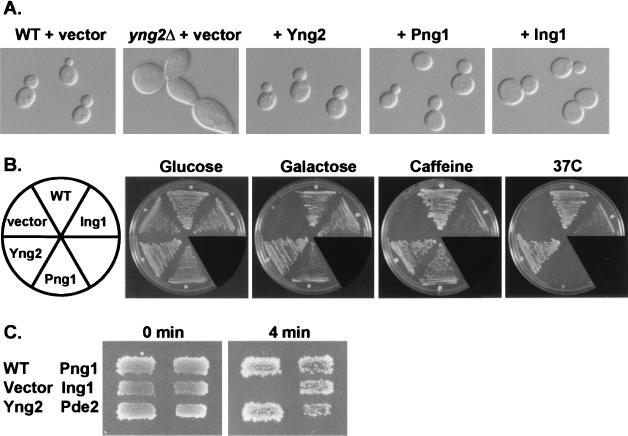
FIG. 3.
Expression of human Ing1 or S. pombe Png1 rescues yng2 null phenotypes. (A) Normal wild-type (WT) (JC1) or yng2Δ (RL3-53) cells harboring a control vector, or yng2Δ cells expressing either Yng2, Png1, or Ing1 were grown at 30°C in SC-L synthetic medium and examined by differential interference contrast microscopy. Plasmids used to express proteins were pAD4.H (+vector), pADHA-Yng2 (+Yng2), pADHA-Png1 (+Png1), and pADHA-Ing1 (+Ing1). (B) These strains were also tested for growth under different conditions. Cells were grown on synthetic complete (SC) medium (Glucose) at 30°C, SC containing galactose (Galactose) instead of glucose as the sole carbon source at 30°C, SC containing 3 mM caffeine (Caffeine) at 30°C, or SC at 37°C (37C). (C) These strains and yng2Δ (RL3-53) overexpressing PDE2 (pYepPDE2) were also tested for hypersensitivity to heat shock at 53°C for 0 or 4 min (see Fig. 2 legend).
Expression of human Ing1 or fission yeast Png1 complements deletion of YNG2.
Since these three yeast proteins share significant sequence identity with human Ing1, we investigated whether the human and yeast genes are also functionally related. We chose to focus our complementation studies on the yng2 null strain since it exhibited the most pleotrophic and severe phenotypes. We found that expression of either human Ing1 or S. pombe Png1 could complement the yng2 mutant phenotypes, including the swollen, multibudded morphology (Fig. 3A), the abnormal DNA distribution (data not shown), carbon source sensitivity, caffeine sensitivity, temperature sensitivity (Fig. 3B), and heat shock sensitivity (Fig. 3C). However, there were some distinct differences in the abilities of these proteins to complement; specifically, caffeine sensitivity was only weakly rescued by Ing1, and temperature sensitivity was very poorly rescued by Png1. Nevertheless, the complementation results suggest that human Ing1, S. pombe Png1, and S. cerevisiae Yng2 share conserved functional properties.
Since some of the phenotypes exhibited by yng2Δ cells are similar to those that result from elevated cyclic AMP (cAMP) levels, we suspected that Yng2 modulates the Ras-cAMP signaling pathway. This was further supported by our observation that overexpression of the high-affinity cAMP phosphodiesterase, Pde2, could suppress the yng2Δ heat shock-sensitive phenotype (Fig. 3C). However, overexpression of Pde2 failed to complement other phenotypes, including the inability to utilize galactose (data not shown), suggesting that these phenotypes are not simply a consequence of elevated cAMP levels.
Yng2 is localized to the nucleus.
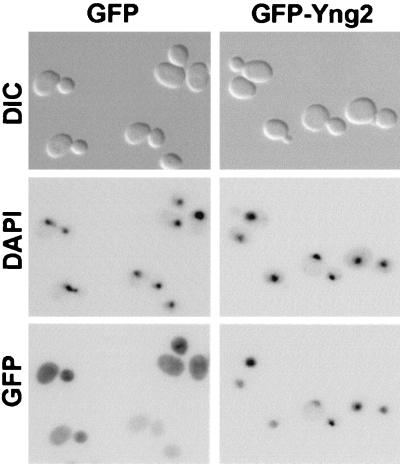
Ing1 has been previously shown to localize in the nucleus (19). To examine the localization of Yng2, we expressed GFP-tagged Yng2 in wild-type yeast cells. Expression of the GFP-tagged Yng2 suppressed the yng2Δ phenotypes, indicating that it was functional (data not shown). Examination of cells expressing GFP-Yng2 by fluorescence microscopy revealed that it colocalized with DAPI staining, suggesting that it was predominately localized in the nucleus, whereas GFP alone was distributed throughout the cell (Fig. 4). This observation is consistent with conserved nuclear functions for Yng2 and Ing1.
FIG. 4.
Yng2 is localized in the nucleus. Normal JC1 cells expressing GFP (left panels) or GFP-Yng2 (right panels) were grown in HC medium, harvested, and briefly fixed before being mounted in medium containing DAPI (Materials and Methods). Cells were visualized by differential interference contrast microscopy (DIC [top row]), DNA was visualized using UV optics (DAPI [middle row]), and GFP localization was visualized using fluorescein isothiocyanate optics (GFP [bottom row]). Images in each column are of the same field of cells. Plasmids used to express proteins were pADGFPHA (GFP) and pADGFPHA-Yng2 (GFP-Yng2).
Yng2 is associated with Tra1 and HAT activity.
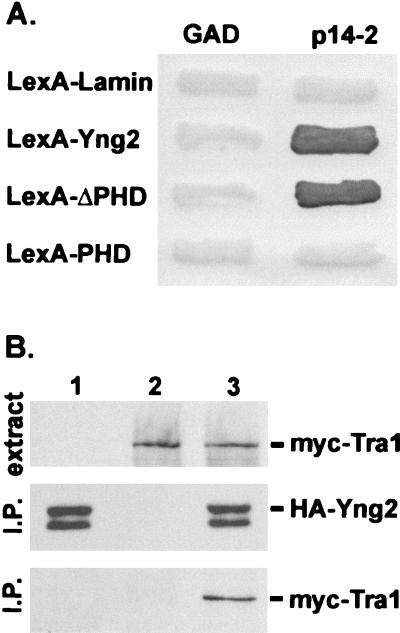
To further investigate the function of Yng2, we performed a yeast two-hybrid screen for proteins that interact with Yng2 (Materials and Methods). One class of proteins that we identified from this screen encoded a short region (amino acid [aa] 1678 to 1740) of Tra1 (Fig. 5A). Tra1 is a 433-kDa protein that is associated with HAT complexes involved in transcriptional regulation (8, 24, 63). To verify the interaction between Yng2 and Tra1, we coexpressed HA-Yng2 and myc-Tra1 in yeast and performed coimmunoprecipitation experiments. Our results confirmed that these proteins interact in vivo (Fig. 5B). This observation suggested that Yng2, and possibly Yng1 and Pho23, are associated with HAT complexes, although neither Yng1, Pho23, nor human Ing1 can interact with Tra1 (aa 1678 to 1740) as shown by two-hybrid analysis (data not shown). To examine this possibility, we performed HAT assays on immunoprecipitated material from yeast expressing HA-Yng2, HA-Yng1, or HA-Pho23. We found that anti-HA immunoprecipitates from such cells had significant levels of HAT activity, while similar immunoprecipitates from control cells expressing GFP-HA did not have detectable HAT activity (Fig. 6A). Interestingly, the patterns of histone acetylation in vitro were different for the samples containing the three yeast proteins, suggesting that they are associated with different HAT complexes. Yng2 is associated with HAT activity that preferentially acetylated H2A and H4, Yng1-associated HAT activity preferentially acetylated H3 and H4, and Pho23 was associated with a relatively weak and nonspecific activity. We also found that HA-Ing1 expressed in yeast is associated with HAT activity. This observation and our complementation data strongly suggest that the functional properties of Yng2 and human Ing1 have been conserved.
FIG. 5.
Yng2 is associated with Tra1. (A) The Gal activation domain (GAD) and a GAD fusion with Tra1 (residues 1678 to 1740) (p14-2) were tested for their ability to interact with LexA fusions with lamin, Yng2, the N-terminal domain (residues 1 to 222) of Yng2 (LexA-ΔPHD), or the C-terminal domain (residues 222 to 282) of Yng2 (LexA-PHD) by the yeast two-hybrid test (Materials and Methods). Each patch represents an independent transformant of the yeast two-hybrid tester strain L40 expressing the indicated proteins. Interaction between fusion proteins was assayed by their ability to induce expression of β-galactosidase by a filter assay (10). (B) Extracts from JC1 cells expressing HA-Yng2 (lane 1), myc-Tra1 (lane 2), or HA-Yng2 and myc-Tra1 (lane 3) were assayed for expression of myc-tagged proteins by Western blot analysis using anti-myc (monoclonal antibody 9E10) antibody (top panel). Ten milligrams of total protein from each extract was immunoprecipitated (I.P.) with anti-HA (monoclonal antibody 12CA5) antibody; half of the immunoprecipitate was probed with anti-HA antibody (middle panel), and half was probed with anti-myc antibody (bottom panel). Plasmids used to express proteins were pADHA-Yng2 and pUAD6 (lane 1), pAD4.H and pADmyc-Tra1 (lane 2), or pADHA-Yng2 and pADmyc-Tra1 (lane 3).
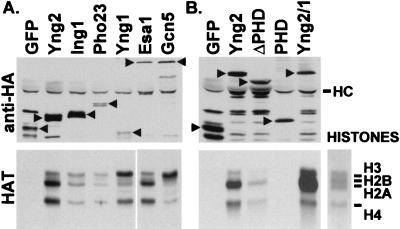
FIG. 6.
Yng1, Yng2, and Pho23 are associated with HAT activities. (A) Extracts from JC1 cells expressing GFP-HA (GFP), HA-Yng2 (Yng2), HA-Ing1 (Ing1), HA-Pho23 (Pho23), HA-Yng1 (Yng1), GFP-HA-Esa1 (Esa1), or GFP-HA-Gcn5 (Gcn5) were immunoprecipitated with anti-HA (12CA5) antibody. Immunoprecipitates were split, and half of each sample was examined by Western blot analysis using anti-HA antibody (top), and half was assayed for HAT activity (bottom panels) (see Materials and Methods). The left lanes (GFP, Yng2, Ing1, Pho23, and Yng1) and right lanes (Gcn5 and Esa1) of the HAT assay were run on the same gel, but they were exposed to film for 15 and 3 days, respectively. (B) Extracts from JC1 cells expressing GFP-HA (GFP), or GFP-HA fusions with either Yng2 (Yng2), the N-terminal domain (residues 1 to 222) of Yng2 (ΔPHD), the C-terminal domain (residues 222 to 282) of Yng2 (PHD), or Yng2/1 (Yng2 residues 1 to 222 fused to Yng1 residues 155 to 219) were immunoprecipitated with anti-HA antibody. Immunoprecipitates were examined by a Western blot using anti-HA antibody (top), or HAT assays (bottom). Plasmids used to express proteins were pADGFPHA (GFP), pADHA-Yng2 (Yng2), pADHA-Ing1 (Ing1), pADHA-Pho23 (Pho23), pADHA-Yng1 (Yng1), pADGFPHA-Esa1 (Esa1), and pADGFPHA-Gcn5 (Gcn5) (in panel A) and pADGFPHA (GFP), pADGFPHA-Yng2 (Yng2), pADGFPHA-Yng2ΔPHD (ΔPHD), pADGFPHA-Yng2PHD (PHD), and pADGFPHA-Yng2/1 (Yng2/1) (in panel B). Arrows denote migration of relevant proteins. HC denotes antibody heavy chain. The panel on the lower right shows a Coomassie-stained lane from the HAT gel and the migration of histones H3, H2B, H2A, and H4.
The Yng2 PHD finger is not required for HAT association.
Since the PHD finger domains are the highly conserved regions of these proteins, we examined whether the Yng2 PHD or N-terminal domains were sufficient or necessary for interaction with Tra1 and HAT activity. Interestingly, we found that the N-terminal domain of Yng2 (aa 1 to 222) was capable of interacting with both Tra1 (aa 1678 to 1740) (Fig. 5A) and HAT activity (Fig. 6B), although the HAT activity was reduced compared with that of full-length Yng2. In contrast, the PHD finger domain (aa 222 to 282) was neither necessary nor sufficient for these interactions. We also found that the N-terminal domain of Yng2, either alone or fused to the PHD finger of Yng1, coimmunoprecipitated HAT activity with a histone acetylation pattern similar to Yng2-associated HAT activity (Fig. 6B). Thus, the PHD finger was not required to maintain HAT specificity but appears to be required for efficient association with, or activity of the HAT complex. However, we found that expression of either the N-terminal domain of Yng2, the PHD finger of Yng2, or a fusion of the N-terminal domain of Yng2 with the PHD finger of Yng1 failed to suppress the inability of yng2Δ cells to utilize galactose (data not shown). Thus, each domain is essential but not sufficient for the normal functions of Yng2, and even though the PHD fingers of Yng1 and Yng2 share a high degree of identity, they are not functionally redundant.
Esa1 is the Yng2-associated HAT.
To explore the nature of the HAT associated with Yng2 we investigated two possible candidates: Esa1 and Gcn5. We expressed and immunoprecipitated GFP-HA-tagged Esa1 and Gcn5 from yeast and performed HAT assays. We found that HA-Yng2-associated HAT exhibited a specificity for H2A and H4 that was remarkably similar to that of GFP-HA-Esa1 (Fig. 6A) (3, 68). In contrast, immunoprecipitated GFP-HA-Gcn5 preferentially acetylated H3, consistent with previous observations (8, 25).
Next, we performed Western blot analysis to examine acetylation of H3 and H4 in cells lacking Yng2, Yng1, Pho23, or Gcn5, or in cells containing an esa1 temperature-sensitive (Ts) allele at both the restrictive (37°C) and nonrestrictive (30°C) temperatures. We found that yng2Δ cells exhibited a significant decrease in the level of acetylation of H4 residues K5, K8, and K12 compared to wild-type cells, whereas the levels of acetylated H3 remained unchanged (Fig. 7). However, yng1Δ or pho23Δ cells did not exhibit significant differences in acetylation of either H3 or H4. esa1(Ts) cells exhibited a decrease in H4-K5, H4-K8, H4-K12, and total H4 acetylation at the restrictive temperature similar to that exhibited by yng2Δ cells. In contrast, gcn5Δ cells did not exhibit a noticeable decrease in H4 acetylation, but there was a decrease in total H3 acetylation.
FIG. 7.
yng2Δ and esa1(Ts) cells exhibit similar H4 acetylation deficiency. Whole-cell extracts from wild-type strain FY105 (WT), gcn5Δ, wild-type strain LPY3498 grown at 30°C (WT 30) or 37°C (WT 37), esa1(Ts) strain LPY3291 grown at 30°C (esa1 30) or 37°C (esa1 37), wild-type strain JC1, yng1Δ, yng2Δ, and pho23Δ cells were separated on an SDS–18% polyacrylamide gel and analyzed by Western blotting using antibodies (Upstate Biochemical) that specifically recognize acetylated H3 (H3), acetylated lysine (H4; histone H4 region of gel shown), and H4 acetylated at lysine residues 12 (H4K12), 8 (H4K8), and 5 (H4K5), or stained with Coomassie brilliant blue (total protein).
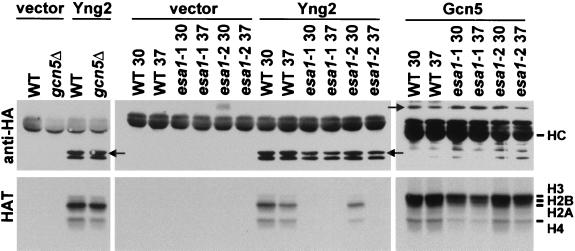
Next, we compared HA-Yng2-associated HAT activity in wild-type, gcn5Δ, and esa1(Ts) cells (Fig. 8). We did not observe a detectable difference in Yng2-associated HAT activity between wild-type and gcn5Δ cells. However, we found that Yng2-associated HAT activity was undetectable in two different esa1(Ts) strains at 37°C, whereas wild-type cells showed similar activity at both 30 and 37°C. The two esa1(Ts) alleles examined were esa1-L254P, which exhibits the most severe phenotypes, and esa1-414, which exhibits more moderate defects at 37°C, as previously described (13). Yng2-associated HAT activity was undetectable in cells containing the more severe esa1-L254P allele at 30°C, suggesting that this mutation disrupts complex formation and/or severely reduces HAT activity. In contrast, immunoprecipitated GFP-HA-Gcn5 exhibited similar levels of activity in either wild-type or esa1(Ts) cells at both temperatures, indicating that HAT activities were not generally impaired in esa1(Ts) cells at elevated temperatures.
FIG. 8.
Yng2-associated HAT activity is deficient in esa1(Ts) cells. Extracts from cells containing a control plasmid pAD4.H (vector), or expressing HA-Yng2 (Yng2) or GFP-HA-Gcn5 (Gcn5) were immunoprecipitated with anti-HA. Immunoprecipitates were split, and half of each sample was examined by Western blot analysis using anti-HA antibody (top), and half was assayed for HAT activity (bottom). Strains used include FY105 (WT), gcn5Δ, LPY3498 grown at 30°C (WT 30) or 37°C (WT 37), LPY3291 grown at 30°C (esa1-1 30) or 37°C (esa1-1 37), and LPY3500 grown at 30°C (esa1-2 30) or 37°C (esa1-2 37).
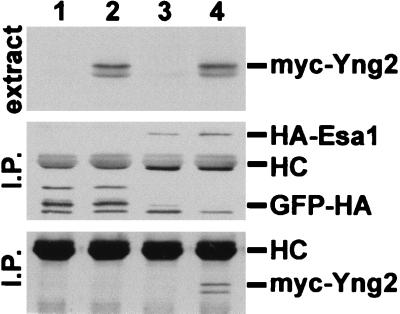
Finally, we examined whether Yng2 associates with Esa1. We coexpressed myc-tagged Yng2 with GFP-HA-Esa1 in yeast and performed coimmunoprecipitation experiments. We found that a detectable level of myc-Yng2 coimmunoprecipitated with GFP-HA-Esa1 but not with GFP-HA (Fig. 9). Together, our observations provide strong evidence that Yng2 is associated with a HAT complex containing Esa1.
FIG. 9.
Yng2 and Esa1 form a complex in vivo. Extracts from JC1 cells expressing GFP-HA (lane 1), GFP-HA and myc-Yng2 (lane 2), GFP-HA-Esa1 (HA-Esa1) (lane 3), or GFP-HA-Esa1 and myc-Yng2 (lane 4) were assayed for expression of myc-tagged proteins by Western blot analysis using anti-myc (mAb 9E10) antibody (top panel). 10 mg of total protein from each extract was immunoprecipitated with anti-HA (monoclonal antibody 12CA5) antibody; half of the immunoprecipitate was probed with anti-HA antibody (middle panel), and half was probed with anti-myc antibody (bottom panel). myc-Yng2 was only precipitated when coexpressed with GFP-HA-Esa1. Plasmids used to express proteins were pADGFPHA and pUAD6 (lane 1), pADGFPHA and pADmyc-Yng2 (lane 2), pADGFPHA-Esa1 and pUAD6 (lane 3), or pADGFPHA-Esa1 and pADmyc-Yng2 (lane 4).
DISCUSSION
Yeast Ing1 homologs are associated with HAT complexes.
We have investigated the functions of three yeast proteins that share strong sequence identity in their C-terminal PHD finger domains with the human candidate tumor suppressor Ing1. Our results provide several clues to the functions of these proteins and suggest that they have a role in chromatin remodeling and transcriptional regulation. Such a role is suggested by our observations that Yng2 is nuclear and associates with Tra1 and that Yng1, Yng2, and Pho23 are complexed with HAT activities. Furthermore, the pleotrophic phenotypes exhibited by yng2 mutant strains are consistent with a role for Yng2 in regulating the expression of a subset of genes. Also, a previous report suggests that Pho23 is involved in the transcriptional regulation of Pho5 (40).
HAT activities are often found in large, modular, multiprotein complexes containing known transcriptional regulators (8, 24, 63). At least five HAT complexes have been identified in S. cerevisiae (25). Our results indicate that Yng1, Yng2, and Pho23 associate with HAT activities with preferences for different histones, suggesting that they are associated with different HAT complexes (58). Based on its association with Tra1, Yng2 may be a component of one or all of the Tra1-associated complexes: SAGA, SLIK, or NuA4. Gcn5 colocalizes with Tra1 to the SAGA complex (25). Deletion of Gcn5 is not lethal but results in various phenotypes, including slow growth, temperature sensitivity, inability to grow on some nonfermentable carbon sources, and sensitivity to amino acid starvation (22, 45). Many of these phenotypes are similar to those exhibited by Yng2 deficient cells. However, in contrast to yng2Δ cells, Gcn5-deficient cells are viable in media with galactose as the sole carbon source. In addition, Yng2 seems to associate with H2A-H4 HAT activity similar to NuA4, whereas Gcn5 predominately acts on H3 and H2B (reviewed in reference 8).
Our studies provide strong evidence that Yng2 is associated with a complex containing Esa1. First, temperature-sensitive esa1 alleles exhibit morphological and cytokinesis defects (swollen, multibudded cells often lacking nuclei) that are very similar to the morphology of yng2Δ cells (13). Second, both Yng2 and Esa1 associate with Tra1. Third, the HA-Yng2-associated HAT activity has an identical substrate preference compared to immunoprecipitated GFP-HA-Esa1. Fourth, in vivo deficiencies in histone H4 acetylation in yng2Δ and esa1(Ts) cells at the nonpermissive temperature are remarkably similar. Fifth, Yng2 fails to coimmunoprecipitate HAT activity in esa1(Ts) strains at the restrictive temperature, or in a severe esa1(Ts) strain at the nonrestrictive temperature. Sixth, myc-Yng2 can be coimmunoprecipitated with GFP-HA-Esa1. Esa1 is the HAT component of NuA4, and all cellular Esa1 is predicted to be associated with the NuA4 complex (3). Thus, it seems likely that Yng2 is a component of the NuA4 complex. Immunopurification of NuA4 demonstrated the tight association of a 32-kDa protein (3), which is identical in size to the predicted molecular mass of Yng2. However, it should be noted that these observations do not exclude the possibility that Yng2 functions in additional HAT complexes.
HA-Yng1 coimmunoprecipitates a HAT activity with a preference for H3 that appears to be very similar to that of immunoprecipitated GFP-HA-Gcn5, raising the possibility that Yng1 is associated with Gcn5 functions. Interestingly, a silver-stained gel of purified SAGA complex indicates the existence of an ∼25-kDa protein similar in size to Yng1. Further studies may reveal if Yng1 is a component of SAGA, ADA, NuA3 (which also acetylates H3), and/or previously unidentified HAT complexes.
HA-Pho23 coimmunoprecipitated a much weaker and nonspecific HAT activity than Yng1 or Yng2. A clue to the macromolecular associations of Pho23 comes from a recent report that Gcn5 regulates the remodeling of chromatin at the Pho5 promoter in vivo (26). Pho23 was originally characterized in a genetic selection to isolate mutants that express Pho5 constitutively (Phoc) (40). Null alleles of PHO23 result in a partial Phoc phenotype; however, the mechanism by which loss of Pho23 leads to a Phoc phenotype is not understood. As both Pho23 and Gcn5 appear to be involved in regulation of Pho5 expression, perhaps they colocalize to the same HAT complex (SAGA or ADA).
There are several possible roles of Yng2, Yng1, or Pho23 in HAT complexes. They may be involved in regulation of acetyltransferase activity, complex formation, or complex stability, or perhaps they serve as specificity factors to direct certain HAT complexes to appropriate targets. Recently, the activities of several HAT complex components other than acetyltransferase have been reported. For example, Ada2 protein has been shown to be required for the assembly of Gcn5 and the potentiation of its HAT activity (73). Other published data also suggests that HATs need to be part of a native complex to work efficiently (3, 25). Interestingly, genetic and biochemical studies suggest that SAGA possesses both HAT-dependent and -independent activities important for transcription (15, 71). Thus, it is possible that yeast Ing1 homologs perform additional functions in HAT complexes in a capacity not related to histone acetylation.
PHD fingers are implicated in transcriptional regulation and cancer.
The strong similarity between Ing1 and the yeast homologs is primarily restricted to the PHD finger domains. This homology was the initial basis for suggesting that these proteins are functionally related. Indeed, our evidence indicates that all three yeast proteins are associated with HAT complexes. Furthermore, Ing1 associates with HAT activity when it is expressed in yeast. These observations support the model that PHD finger domains are involved in multiprotein complexes that modulate chromatin structure, as previously suggested (1). However, we have shown that the PHD finger is not required for the association of Yng2 with either Tra1 or HAT activity. These results suggest that the unique N-terminal domains are the regions that determine the interaction with specific HAT complexes while the PHD fingers provide a more general and conserved function. Exactly how the PHD fingers of these proteins are involved in the formation or function of HAT complexes remains to be determined. PHD fingers have been identified in several genes associated with genetic syndromes, including genes encoding WSTF in Williams syndrome, PHF2 in hereditary sensory neuropathy type 1, AIRE in autoimmune polyglandular syndrome type 1, ATRX in X-linked alpha-thalassemia mental retardation syndrome, and MOZ in chromosomal translocations in acute myeloid leukemias (23, 27, 33, 34, 43, 50, 57). Further characterization of PHD finger function may potentially shed light on the molecular mechanisms of these diseases.
Human, budding yeast, and fission yeast Ing1 homologs are functionally conserved.
The strong similarity among human, budding yeast, and fission yeast Ing1 homologs suggest that these proteins have been structurally conserved throughout eukaryote evolution. Our genetic complementation and biochemical results further suggest that these proteins perform related and functionally conserved roles. Human Ing1 and fission yeast Png1 functionally rescue yng2Δ phenotypes; however, Png1 rescues caffeine lethality more efficiently than Ing1, and Png1 rescues temperature sensitivity very poorly. Thus, Yng2 may perform more than a single discrete biochemical function, and these functions may have been differentially conserved throughout eukaryote evolution.
The structure and function of Ing1 and Yng2 seem to have been particularly well conserved, suggesting that human Ing1 may be associated with HAT activity and TRRAP, the mammalian homolog of Tra1, in mammals. TRRAP has been shown to interact with c-Myc and E2F-1, and inhibition of TRRAP function blocks c-Myc and E1A-mediated oncogenic transformation, suggesting that TRRAP is an essential cofactor for c-Myc and E1A-E2F oncogenic transcription factor pathways (14, 47, 62). Interestingly, Ing1 has been shown to cooperate with c-Myc to induce apoptosis (28). Such cooperation may be mediated by TRRAP and its associated HAT activity.
Approximately one-third of the genes in S. cerevisiae have significant similarity to human cDNAs reported in the expressed sequence tag database (7). However, among those conserved genes are only a few known oncogenes or tumor suppressors. Most notably absent in yeast are homologs of p53, a tumor suppressor associated with over 50% of human cancers. Thus, identification of yeast homologs of the mammalian Ing1 putative tumor suppressor is quite novel and suggests that these proteins function in a conserved primeval mechanism. A model in which Ing1 regulates HAT activity would be consistent with its putative tumor-suppressive role. Several recent reports implicate HATs and HDACs in cell cycle regulation, differentiation, and cancer development (4, 34, 37). Also, the tumor suppressor BRCA2 has been shown to possess intrinsic HAT activity (67).
ACKNOWLEDGMENTS
We thank Chris Brandl, Lorraine Pillus, George Thireos, and John Colicelli for providing strains and plasmids.
This research was supported by grants from the Alberta Cancer Board and the National Cancer Institute of Canada. R.L. was supported by the National Science and Engineering Research Council of Canada, the Alberta Heritage Foundation for Medical Research, and the University of Calgary Silver Anniversary Scholarship. D.Y. is a Senior Scholar of the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Aasland R, Gibson T J, Stewart A F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics, 1997 ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 3.Allard S, Utley R T, Savard J, Clarke A, Grant P, Brandl C J, Pillus L, Workman J L, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer S Y, Hodin R A. Histone acetylation and cancer. Curr Opin Genet Dev. 1999;9:171–174. doi: 10.1016/s0959-437x(99)80026-4. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Bartel P, Chien C T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 7.Bassett D E, Boguski M S, Spencer F, Reeves R, Kim S, Weaver T, Hieter P. Genome cross-referencing and XREFdb: implications for the identification and analysis of genes mutated in human disease. Nat Genet. 1997;15:339–344. doi: 10.1038/ng0497-339. [DOI] [PubMed] [Google Scholar]
- 8.Berger S L. Gene activation by histone and factor acetyltransferases. Curr Opin Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury E M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992;14:9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- 10.Breeden L, Nasmyth K. Regulation of yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Clarke A S, Lowell J E, Jacobson S J, Pillus L. Esa1 is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole M, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 15.Dudley A M, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberharter A, Sterner D E, Schieltz D, Hassan A, Yates III J R, Berger S L, Workman J L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elble R A. Simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 18.Garkavstev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garkavtsev I, Demetrick D, Riabowol K. Cellular localization and chromosome mapping of a novel candidate tumor suppressor gene (ING1) Cytogenet Cell Genet. 1997;76:176–178. doi: 10.1159/000134539. [DOI] [PubMed] [Google Scholar]
- 20.Garkavtsev I, Grigorian I A, Ossovskaya V S, Chernov M V, Chumakov P M, Gudkov A V. The candidate tumor suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 21.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 22.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbons R J, Bachoo S, Picketts D J, Aftimos S, Asenbauer B, Bergoffen J, Berry S A, Dahl N, Fryer A, Keppler K, Kurosawa K, Levin M L, Masuno M, Neri G, Pierpont M E, Slaney S F, Higgs D R. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 24.Grant P A, Schieltz D, Pray-Grant M G, Yates III J R, Workman J L. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 25.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 26.Gregory P D, Schmid A, Zavari M, Lui L, Berger S L, Hörz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell. 1998;1:495–505. doi: 10.1016/s1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 27.Hasenpusch-Theil K, Chadwick B P, Theil T, Heath S K, Wilkinson D G, Frischauf A M. PHF2, a novel PHD finger gene located on human chromosome 9q22. Mamm Genome. 1999;10:294–298. doi: 10.1007/s003359900989. [DOI] [PubMed] [Google Scholar]
- 28.Helbing C C, Veillette C, Riabowol K, Johnston R N, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- 29.Hill J, Donald K A, Griffiths D E, Donald G. Enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollenberg S M, Sternglanz R, Chenag P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong L, Schroth G P, Matthews H R, Yau P, Bradbury E M. Studies of the DNA binding properties of the histone H4 amino terminus. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 32.Hubberstey A, Yu G, Loewith R J, Lakusta C, Young D. Mammalian CAP interacts with CAP, CAP2 and actin. J Cell Biochem. 1996;61:459–466. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C459::AID-JCB13%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 33.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobson S, Pillus L. Modifying chromatin and concepts of cancer. Curr Opin Gen Dev. 1999;9:175–184. doi: 10.1016/S0959-437X(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 35.Jeggo P A, Carr A M, Lehmann A R. Splitting the ATM: distinct repair and checkpoint defects in ataxia-telangiectasia. Trends Genet. 1998;14:312–316. doi: 10.1016/s0168-9525(98)01511-x. [DOI] [PubMed] [Google Scholar]
- 36.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–25677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 37.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 38.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 39.Kuo M H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Histone acetyltransferase activity of Gcn5p is required for the activation of targeted genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau W W, Schneider K R, O'Shea E K. A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics. 1998;150:1349–1359. doi: 10.1093/genetics/150.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 42.Lees-Miller S P. The DNA-dependent protein kinase, DNA-PK: 10 years and still no ends in sight. Biochem Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 43.Lu X, Meng X, Morris C A, Keating M T. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- 44.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 45.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matviw H, Yu G, Young D. Identification of a human cDNA encoding a protein that is structurally related to the yeast adenylyl cyclase-associated CAP proteins. Mol Cell Biol. 1992;12:5033–5040. doi: 10.1128/mcb.12.11.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMahon S B, Van Buskirk H A, Dugan K A, Copeland T D, Cole M D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 48.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J L, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 49.Mizzen C A, Brownell J E, Cook R G, Allis C D. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 1999;304:675–696. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- 50.Nagamine K, Peterson P, Scott H S, Kudoh J, Minoshima S, Heino M, Krohn K J, Lalioti M D, Mullis P E, Antonarakis S E, Kawasaki K, Asakawa S, Ito F, Shimizu N. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 51.Ogryzko V O, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 52.Ohmori M, Nagai M, Tasaka T, Koeffler H P, Riabowol K, Takahara J. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol. 1999;62:118–119. doi: 10.1002/(sici)1096-8652(199910)62:2<118::aid-ajh11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 53.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:401–441. [PubMed] [Google Scholar]
- 54.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 55.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 56.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 57.Rinderle C, Christensen H M, Schweiger S, Lehrach H, Yaspo M L. AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered subcellular distribution of mutants lacking the PHD zinc fingers. Hum Mol Genet. 1999;8:277–290. doi: 10.1093/hmg/8.2.277. [DOI] [PubMed] [Google Scholar]
- 58.Roth S Y, Allis C D. The subunit-exchange model of histone acetylation. Trends Cell Biol. 1996;6:371–375. doi: 10.1016/0962-8924(96)20032-7. [DOI] [PubMed] [Google Scholar]
- 59.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 60.Ruiz-Garcia A B, Sendra R, Galiana M, Pambianco M, Perez-Ortin J E, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273:12599–12605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 61.Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- 62.Sakamuro D, Prendergast G C. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 63.Saleh A, Schieltz D, Ting N, McMahon S B, Litchfield D W, Yates III J R, Lees-Miller S P, Cole M D, Brandl C J. Tra1 is a component of the yeast Ada-Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 65.Sass P, Field J, Nikawa J, Toda T, Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1986;83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 67.Siddique H, Zou J P, Rao V N, Reddy E S. The BRAC2 is a histone acetyltransferase. Oncogene. 1998;16:2283–2285. doi: 10.1038/sj.onc.1202003. [DOI] [PubMed] [Google Scholar]
- 68.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. Esa1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 70.Steger D J, Eberharter A, John S, Grant P A, Workman J L. Purified histone acetyltransferases stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci USA. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Biol Cell. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 73.Syntichaki P, Thireos G. The Gcn5-Ada complex potentiates the histone acetyltransferase activity of Gcn5. J Biol Chem. 1998;273:24414–24419. doi: 10.1074/jbc.273.38.24414. [DOI] [PubMed] [Google Scholar]
- 74.Toyama T, Hirotaka I, Watson P, Muzik H, Saettler E, Magliocco A, DiFrancesco L, Forsyth P, Garkavtsev I, Kobayashi S, Riabowol K. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- 75.Turner B M, O'Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 76.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 77.Vassilev A, Yamauchi J, Kotani T, Prives C, Avantaggianti M L, Qin J, Nakatani Y. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol Cell. 1998;2:869–875. doi: 10.1016/s1097-2765(00)80301-9. [DOI] [PubMed] [Google Scholar]
- 78.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 79.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 80.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian RAS interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 81.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang X J, Ogryzko V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 83.Zeremski M, Hill J E, Kwek S S S, Grigorian I A, Gurova K V, Garkavtsev I V, Diatchenko L, Koonin E V, Gudkov A V. Structure and regulation of the mouse ing1 gene. J Biol Chem. 1999;274:32172–32181. doi: 10.1074/jbc.274.45.32172. [DOI] [PubMed] [Google Scholar]