Graphical abstract
Abbreviations: AI, Artificial Intelligence; SARS-CoV-2, Severe Acute Respiratory Syndrome-Associated Coronavirus 2; FDA, Food and Drug Administration; GAN, Generative Adversarial Network; DTI, Drug-Target Interaction; DDI, Drug-Drug Interaction; DUD-E, Database of Useful (Docking) Decoys Enhanced; AVP, Antiviral Peptide
Keywords: Artificial intelligence, Infectious diseases, Antimicrobial agents, COVID-19
Abstract
The search for effective drugs to treat new and existing diseases is a laborious one requiring a large investment of capital, resources, and time. The coronavirus 2019 (COVID-19) pandemic has been a painful reminder of the lack of development of new antimicrobial agents to treat emerging infectious diseases. Artificial intelligence (AI) and other in silico techniques can drive a more efficient, cost-friendly approach to drug discovery by helping move potential candidates with better clinical tolerance forward in the pipeline. Several research teams have developed successful AI platforms for hit identification, lead generation, and lead optimization. In this review, we investigate the technologies at the forefront of spearheading an AI revolution in drug discovery and pharmaceutical sciences.
Artificial intelligence has been transformative in several areas of human endeavor
Exponential progress in AI and its applications has occurred during the past 15 years.1 AI-based conversational assistants are now powering consumer devices, such as Amazon’s Alexa; self-driving cars have been registering hundreds of thousands of miles on American roads2; AI has beaten world champions in GO, chess, and other games3, 4; AI-based systems are assisting doctors in medical diagnosis and treatment5, 6, 7, 8; AI is helping map the canopy cover across the continental United States9, 10, 11; and a combination of AI and immersive virtual reality is assisting construction engineers to design energy-efficient buildings12, 13. In summary, AI is influencing every aspect of human life from transportation14 to stock trading.15 However, the influence of AI on drug discovery and development has been minimal thus far.
AI currently lacks impact on drug discovery
Although it is undeniable that the application of AI in pharmaceutical sciences holds tremendous promise, the current limited impact of AI on drug discovery can be attributed to multiple factors. A lack of standardized labeled benchmark data sets has been one of the major hurdles of AI-driven drug discovery. The recent AI revolution has been fueled by the availability of cheap computing power and large volumes of data that can be easily shared through the internet. For example, progress in computer vision has been dramatically accelerated by the creation of the benchmark ImageNet data set.16 Despite several attempts, including DrugBank,17 BindingDB,18 KEGG,19 Supertarget,20 and DUD-E,21 an all-encompassing benchmark labeled data set, such as ImageNet, has not yet been created in the pharmaceutical sciences. This lack of a standardized data set means that it is difficult to follow existing transfer learning strategies, in which one fine-tunes a model pretrained on a standard data set for a new task. Hence, it is difficult to transition models trained for discovering drugs for one disease to do the same for another. For AI to be impactful in drug discovery, one needs to develop general techniques and patterns that apply to a range of tasks involving different diseases.
Although deep learning22 has had a central role in the ongoing AI revolution, models developed based on this technique are notorious for their opacity. Deep neural networks essentially behave like black boxes23 and do not provide any insight into their underlying decision-making process. This also makes their application in drug discovery onerous. When a drug is flagged by a neural network as being efficacious for a disease, one needs to understand its mechanism of action, the interaction of the drug with the host–protein network, whether the interaction is inhibitory, the pharmacokinetics, the dose–response curve, any potential cytotoxicity, as well as the epistemic and the aleatoric uncertainty associated with the decision of the network. An off-target decision can entail unnecessary costs incurred not only in failed tests in vitro and in vivo, but also in consequent clinical trials where the loss of reputation is likely.
The current pandemic is driving use of AI in drug discovery
Although the above discussion paints a bleak picture of the suitability of AI in drug discovery, there appears to be hope on the horizon. The current COVID-19 pandemic has become the main driving force behind the use of AI to accelerate preclinical drug discovery. At present, a few drugs, such as remdesivir, have been approved by the US Food and Drug Administration (FDA) for off-label use in treating severe acute respiratory syndrome-associated coronavirus 2 (SARS-CoV-2) infections. Most of these proposed treatments have been discovered through trial-and-error experiences by physicians and researchers around the world. It is well documented that the average pharmaceutical company’s in-house preclinical discovery cost for a new drug compound is US$209 522 157 (adjusted for inflation) over 3 years (only ∼ 12% of all drugs developed eventually get approved by FDA, whereas failed attempts significantly increase the average cost and time requirement of preclinical drug discovery).24, 25 These expenses do not include the costs of basic research at the university level focused on the identification of molecular targets as well as the development of research methods and technologies. The efficiency of drug development, as defined by the successful approval of new pharmaceuticals within the rate of acceptable financial investment, has significantly declined.25, 26 The existing process of creating drugs is slow, inefficient, and costly. Hit identification, lead generation, and lead optimization are key steps at the outset of any drug discovery process. Compounds showing promising activity identified by high-throughput screening as initial hits are filtered and modified to generate lead compounds that satisfy basic drug-likeness properties.27 These lead compounds are further optimized to enhance their potency toward the target protein or mechanism as well as to reduce nonselectivity and toxicity. Conventional hit identification is expensive and requires time-consuming screening experiments. Under the circumstances of the current pandemic, the world cannot afford such an inefficient pipeline. What is needed is a principled approach to drug discovery and repurposing that can rapidly address large data sets. This capability will thereby create an improved method for identifying drugs and/or drug combinations that are likely to succeed.
Current state of antimicrobial drug discovery
The enormous time and cost incurred in discovering a new compound as well as developing it through the approval process have been so overwhelming in recent times that the pharmaceutical industry has repeatedly shown reduced interest in bringing new drug products to market. The inactivity is most notable in less profitable market segments, such as infectious diseases.28 Over the past 20 years, the pharmaceutical industry has put infectious disease and antimicrobial drug discovery and development on the backburner. The COVID-19 pandemic has been a distressing reminder of the lack of infrastructure to develop treatments for emerging infectious diseases. The pandemic has been a global reckoning, highlighting the importance of antiviral and antimicrobial drug research for potential future outbreaks. In recent history, there have been meagre enthusiasm and scarcity of growth in the field of infectious diseases. Case in point, for bacterial infections, every new antibiotic brought to market over the past few decades has been only a slight variation on existing drugs discovered before 1984.29 Only one of the top 50 pharmaceutical companies has antibiotics in clinical development and nearly 75% of the companies currently developing antimicrobials can be regarded as prerevenue, with no approved products in the market.30, 31 Market analysis has shown that drug-resistant forms of these diseases will grow significantly by 2025, with very few new drug strategies in the near future.32
The rise of new AI techniques and their application to drug discovery
Recent advances in AI with the development of fundamentally new techniques, such as graph neural networks,33, 34 graph embeddings,35 geometric deep learning,36 attention networks,37 self-supervised38 and unsupervised39, 40 learning, Monte-Carlo graph search,41 neural networks for protein folding,42 explainable AI,43 and generative adversarial networks (GANs),44 have spurred renewed interest in applyications to accelerate drug discovery. These techniques promise to mitigate the above-mentioned drawbacks of previous-generation AI. They allow for the development of an efficient drug discovery pipeline by leveraging mathematical representations of all interactions between proteins in the host cell.
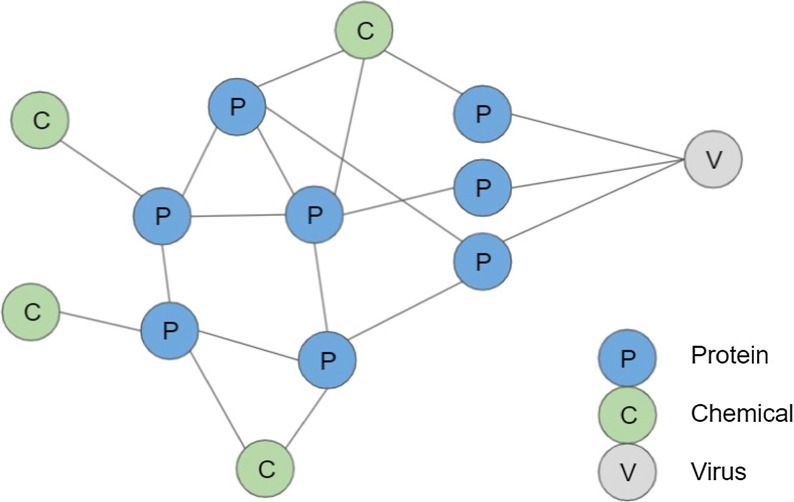
Using such a model, we can accurately predict whether a particular microbial mechanism will be inhibited by a certain drug. For example, in discovering antivirals, understanding the effects of a drug on viral mechanisms, such as viral entry, RNA transcription, and viral exit, can be crucial for predicting the effectiveness of a therapy involving the drug. Databases, such as HU.MAP,45 HPIDB,46 and STRING,47 provide both human–human and human–virus protein interactions that can be exploited by the above-mentioned techniques. These interactions can be used to provide explanations for why a particular drug compound is efficacious against a disease both in terms of the proteins targeted by the compound and subsequent protein–protein interaction cascades. For instance, a graph neural network33, 34 can take a graph structure and a feature description for every node as input, to comprehensively model the interactions of a drug within the human interactome, that is, the protein–protein interactions of the human cell. The network learns and operates on the graph structure of the input and ground truth data. Each protein is represented as a node in the graph and the neighborhood of each node is assigned from the set of neighboring nodes in the structure of the protein. Chemical nodes can correspond to existing drugs (including 131 nutraceuticals) in Drugbank, which contains data on 13 580 approved and experimental drugs, or SuperTarget, a large data set of 332 828 drug–target interactions (DTIs). The edges of the graph represent protein interactions. Each protein node could also have features computed from its amino sequence and structure, whereas edges have weights describing interactions experimentally derived between residues. Such a network would be a predominantly encompassing mathematical representation of all physical contacts between proteins within a cell (Fig. 1 ). ProtVec,48 a vector representation of protein sequences, would constitute the input features of each protein node. ProtVec is an unsupervised data-driven distributed representation of the protein k-mer sequences as an n-dimensional vector in a context-aware manner, useful for neural network predictions or analyses. Target mechanisms would be represented with edges to all proteins associated with them.
Figure 1.
Visualization of protein–protein and protein–chemical graphs. The blue dots represent protein nodes, the green dots represent chemical nodes, the gray dot represents a virus protein, and the lines represent edges in the graph (protein–protein or chemical–protein interactions).
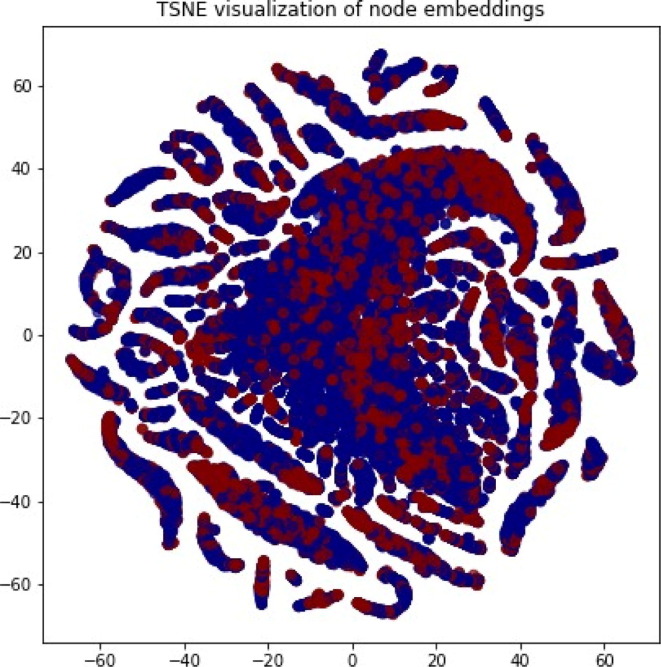
The output of such a graph neural network would be node embeddings for each node in the graph. A node embedding characterizes the context of a node with respect to its interaction with other nodes in the graph. Fig. 2 visualizes the embeddings of such a graph in 2D using t-SNE.49 The red clusters in Fig. 2 show how drugs are clustered, whereas blue clusters show the clustering of the proteins. Overlap of the blue clusters with the red clusters indicates drug–protein interactions.
Figure 2.
Visualization of node embeddings in two dimensions using t-SNE. The red clusters show how the drugs are clustered, whereas the blue clusters show the clustering of the proteins. Overlap of the blue clusters with the red clusters represent drug–protein interactions.
The DeepDrug team (see below) developed such node embeddings to be inputted into a Siamese network.50 Siamese networks project embeddings into multidimensional space and calculate distance between them within that dimensionality. The closer the prediction is to zero, the higher the interaction between a pair of embeddings. A Siamese network will take embeddings of a pair of drug–protein nodes as input. The network would output a distance metric indicating the effect of the drug on target proteins and viral mechanisms involving them. For example, for the nutraceutical biotin, the Siamese network predicts Abelson kinase 1 (ABL1) as a target protein. It is known from the literature51 that Abelson kinase inhibitors can have effectiveness against SARS and Middle East respiratory syndrome-associated (MERS) coronavirus infections. Similarly, the nutraceutical levomenol, a chamomile extract, is predicted to target signal transducer and activator of transcription 3 (STAT3). The literature52 shows that inhibition of STAT3 can help reduce cytokine storms (i.e., severe cytokine release) and acute respiratory distress syndrome (ARDS) during COVID-19 infection. One could use a Bayesian Siamese network53 with weights sampled from a Gaussian distribution to further provide uncertainty estimates for its predictions. Geometric deep-learning techniques can also generalize such graph neural networks and can efficiently extract representations of chemical features.54
The resulting weights with their uncertainty estimates can be used to prioritize drugs and filter the top drug candidates by taking their respective toxicities and synthetic accessibilities24 into consideration using a multicriterion optimization algorithm. This multicriteria optimization algorithm can: (i) rank all FDA-approved drugs according to the weight/uncertainty estimates as obtained from the Siamese network; and (ii) solve an optimization problem that will shortlist drugs with the highest weight/certainty, lowest toxicity score, and highest synthetic accessibility score.
Another important advancement in AI that can significantly impact drug discovery is explainable AI (XAI). Confidence-aware networks55, 56, 57 have helped lift the veil on the opaque decision-making process of deep neural networks. It is now possible to understand the epistemic and aleatoric uncertainty associated with the decision of a deep neural network. Indeed, when a confidence-aware neural network predicts that a drug is efficacious against a particular disease, it will also provide a measure of its own confidence in its prediction. High confidence predictions can proceed for in vitro validation, whereas low confidence predictions can be filtered out. Recent advances in transfer learning58, 59, 60 also bode well for drug discovery. Domain adaptation61 now allows models trained to predict drugs targeting one disease to be repurposed for other indications. Transfer learning together with low-shot techniques62, 63 alleviate the need for large, labeled data sets in model training. Currently, tasks such as predicting toxicity or drug–drug interactions (DDIs) require large volumes of labeled training data. Acquiring such data in the pharmaceutical field is difficult because labeling requires domain knowledge. This markedly hinders the development of essential tools for drug discovery. Modern unsupervised39, 40 and self-supervised learning techniques38 can ease the problem by exploiting vast amounts of available unlabeled data.
Renewed efforts in applying AI to drug discovery
Stunning advances in AI, as described above, have spurred renewed interest in using AI to accelerate preclinical drug discovery. Several teams have been working with AI platforms to repurpose existing drugs and re-engineer new drugs in the pursuit of finding life-saving medicines. Here, we highlight platforms with state-of-the-art machine learning and AI technology that are spearheading new methods for drug discovery. Recently, Bender and Cortés-Ciriano published a paper discussing whether AI was having an impact on drug discovery and limitations of this approach to date.64, 65 Here, we address the concerns raised by these authors and provide a brief introduction to the implementation, strategy, and successes of each team. Each of these methods can lead to both theoretical and practical applications in drug discovery.
BenevolentAI
The BenevolentAI team is working on a drug discovery approach that involves the use of biological knowledge graphs to identify new treatments.66 Using an AI technique called natural language processing (NLP),67, 68 knowledge graphs are extracted from the scientific literature to identify previously unknown correlations.69 The resulting graph represents an interlinked network of concepts that places scientific data in context by linking semantic metadata. This framework allows the BenevolentAI team to integrate previously unconnected research to identify links that could be targets for drug development. This network was used to identify baricitinib,70 a drug approved for the treatment of rheumatoid arthritis, as a repurposed treatment for COVID-19 in mitigating the cytokine storm through inhibition of adaptor-associated protein kinase 1 (AAK1). By making use of this knowledge base, the team was able to complete this analysis by February 2020, only weeks after the first COVID-19 case was reported in the USA. By November of the same year, BenevolentAI and Eli Lilly had completed clinical trials and received an Emergency Use Authorization from the FDA as a treatment for COVID-19.
BenevolentAI also has a secondary project71 to analyze and compare 3D binding sites in which both positive and negative binding pairs of protein-pockets and ligands are used to train a network for protein-pocket matching. By encoding the 3D shapes of the binding sites, BenevolentAI’s network is able to learn which features of a protein-pocket representation predict binding affinity and can screen many pockets to identify novel drug targets. This machine learning approach is called ‘field of distance metric learning’,72 and enables the BenevolentAI team to predict results of previously unknown DTIs.
Atomwise
Another emerging platform is Atomwise, which uses an AI technique called convolutional neural networks (CNNs) to analyze the biological activity to predict the binding affinity of small molecules.73 CNNs are a class of neural networks mainly used to understand imagery. Molecular shape analysis of small molecules using CNNs can predict binding affinity measurements of different molecules to protein structures. This allows Atomwise to predict the biological activity and pharmacology of small molecules for drug discovery. The Atomwise networks apply feature locality and hierarchical composition to model pharmacological activity and chemical interactions. Their networks showed promising results for the Database of Useful (Docking) Decoys Enhanced (DUD-E), achieving an area under the curve (AUC) greater than 0.9 on 57.8% of the docking targets in DUD-E.21, 73 Atomwise used this technology to screen millions of molecules against known SARS-CoV-2 proteins to explore broad-spectrum therapies for the treatment of COVID-19 and other coronavirus infections.
Insilico Medicine
The Insilico Medicine team proposed a unique generative adversarial network (GAN)-based approach for synthesizing new drugs for individual diseases.74 GANs function by discovering patterns in input data from which the model can generate new samples that could have plausibly been drawn from the original data set. Insilico Medicine’s GAN network synthesizes new compounds by iteratively generating molecules; while analyzing certain molecular parameters, such as biological activity and synthetic feasibility. The system then optimizes across its set parameters and generates new molecules until it reaches a local maximum. Such a network can generate molecules with certain properties or activities against a pharmacological target, making the network useful for initial discovery. However, only a few examples of generative drug design have achieved validation in in vitro or in vivo experiments.
Insilico Medicine originally focused their efforts on generating chemotypes targeting the SARS-CoV-2 main protease. By 4 February, 2020, Insilico Medicine released their first potential de novo protease inhibitor. The Insilico Medicine team recently published ten representative structures of protease inhibitors for potential development against COVID-19.75 Even so, the greatest complication of using a GAN lies in the nature of the network itself. Any output from such a GAN is derived within a ‘black box’ system, giving researchers little to no explanation or understanding of the underlying analyses. Given that the patterns and regularities identified in the data are known only by the AI system, extensive laboratory testing is required to confirm any findings from this technique.
ComboNet
The ComboNet team at the Broad Institute (Cambridge, MA, USA) leveraged DTIs to identify synergistic combinations against SARS-CoV-2.76 The ComboNet system predicts DTIs from the molecular structures of the compounds analyzed. The ComboNet architecture comprises two major components: a graph convolutional network (GCN), which is trained to represent the molecular structure of the compound, and a model for target–disease association. The advantage of using this methodology is the ability to predict from compounds with incomplete DTI information. The second model learns how biological targets and molecular structure features interact to present antiviral activity and synergy. The team used training data from the National Center for Advancing Translational Sciences (NCATS) at the National Institutes of Health (NIH) cytopathic effect assay against SARS-CoV-2 as well as SARS-CoV-2 drug combination assays with synergy scored using the BLISS model.77
DeepDrug
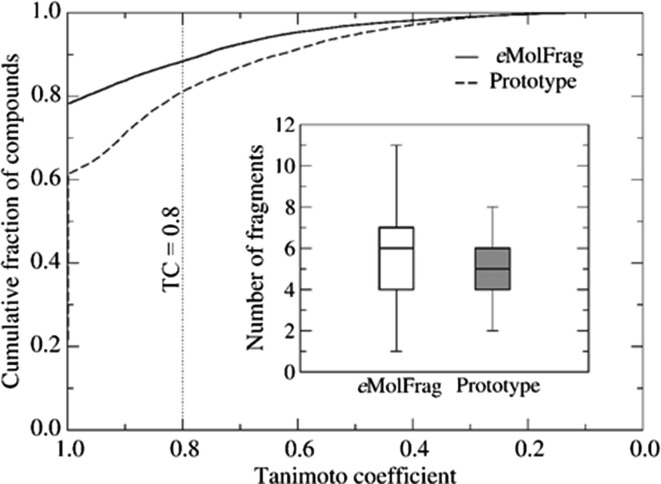
The DeepDrug team, a semifinalist in the IBM Watson Artificial Intelligence XPRIZE competition, created an efficient AI-based platform to design new compounds and repurpose existing drugs for emerging infectious diseases.24, 26, 78 The DeepDrug pipeline is capable of automatically synthesizing targeted drug molecules using beam search techniques,79 as well as filtering candidates based on chemical criteria (e.g., Lipinski’s Rule of Five)27 and potential adverse effects. This allows the team to predict the candidates that are most likely to succeed in the patient population. The pipeline is modular in nature and currently comprises eMolFrag,26 eSynth,78 eToxPred,24 eDrugRes, eVir, eComb and several other AI-based filters. Given a collection of molecules, eMolFrag generates a set of unique fragments and pharmacophores that act as ‘building blocks’. Fig. 3 shows the ability of eMolFrag to identify bioactive building blocks from known drugs. eSynth78 uses beam search techniques79 to combine these molecular fragments into novel molecules de novo. It assembles millions of molecules in minutes, while logging the associated chemical reactions used to construct each molecule. This trace of chemical reactions can be used to synthesize any of these molecules in a wet lab setting. These molecules can be then further filtered for toxicity, specificity, and ease of manufacturing.
Figure 3.
Bioactive compounds from the Database of Useful (Docking) Decoys Enhanced (DUD-E) database fragmented with eMolFrag. eMolfrag was able to generate an average of six fragments per molecule. eSynth uses beam search techniques to create new drug molecules by combining the building blocks generated by eMolFrag in a chemically comprehensive way. By using fragments generated by eMolFrag, eSynth reconstructed 78.3% of active compounds with a Tanimoto coefficient (TC) of 1.0 and 88.4% with a TC ≥ 0.8. Adapted from Liu et al.26
Figure 4.
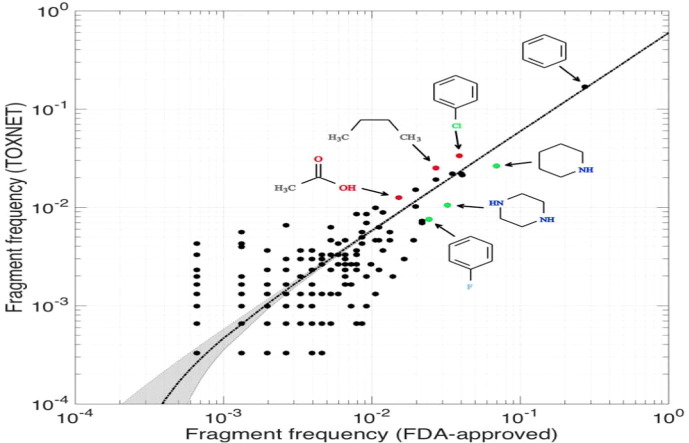
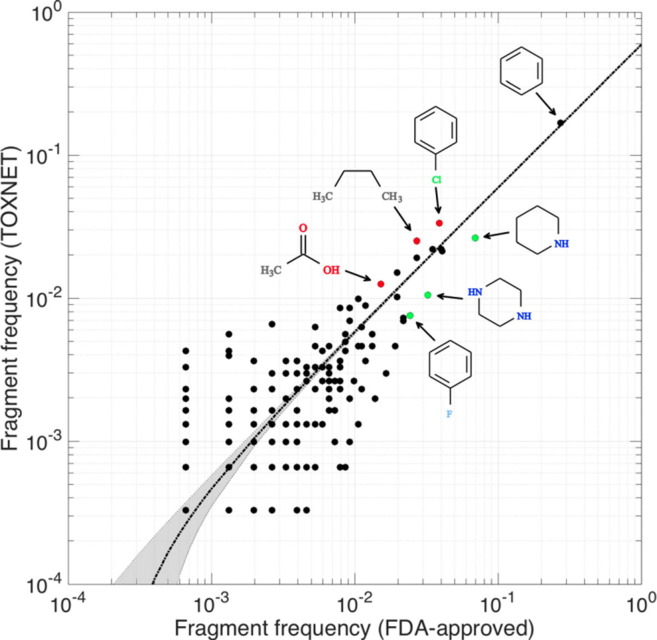
Composition of nontoxic and toxic compounds. The scatter plot shows the frequencies of eMolFrag-extracted chemical fragments from US FDA-approved (nontoxic) and TOXNET (toxic) molecules. The dotted black line is the line of regression, and the gray area represents the corresponding confidence intervals. Examples of three commonly found FDA-approved fragments (piperidine, piperazine, and fluorophenyl) are in green, whereas fragments of more commonly toxic fragments from the TOXNET data set (chlorophenyl, n-butyl, and acetic acid) are in red. Adapted from Figure 8 in Pu et al.24
Using two of these modules, the DeepDrug team synthesized an adenosine receptor from components acquired by decomposing four adenosine receptor antagonists.26 Adenosine receptor antagonists have important roles in inflammation, pain, and immune responses, making them attractive targets for pharmacotherapy.
eToxPred,24 the third module in the DeepDrug pipeline, is used to estimate toxicity and synthetic accessibility of small molecules. Estimating toxicity is a key component of the overall DeepDrug pipeline, to rapidly and proactively filter out compounds with undesirable or adverse effects. In contrast to other approaches that use manually crafted descriptors,80 eToxPred uses the molecular fingerprints of the chemical compounds to model toxicity directly, making it more effective against highly diverse data sets. Fig. 4 shows eToxPred using machine-learning techniques to filter the candidate drug molecules with respect to their potential toxicity based on structural properties. The output eToxPred value is a score between zero and one, with zero being the least toxic and one indicating a high likelihood for toxicity. FDA-approved drugs have the lowest median Tox-score of 0.34, whereas the toxicity of active compounds from the DUD-E data set is slightly higher, with a median Tox-score of 0.46. Compounds in both natural products and traditional herbal medicine data sets show higher toxicity scores with a median Tox-score of ∼ 0.55. These results are validated by other studies that examine the potentially toxic constituents, which include alkaloids, glycosides, polypeptides, amino acids, phenols, organic acids, terpenes, and lactones.
eDrugRes was created to identify effective chemicals against antibiotic-resistant bacteria by exploring drug effects and mutations within microbial protein–protein interaction networks. This system uses GCNs to predict whether a specific chemical compound would have therapeutic activity against certain strains of bacteria.
Several new modules have recently been added to the DeepDrug pipeline. The first is eVir, which can determine viral specificity of drugs with the goal of repurposing existing drugs. It uses an AI technique to generate a fingerprint for drugs and known antiviral peptides (AVPs)81 that captures their properties and context within a mathematical representation of all cellular protein interactions. By comparing these fingerprints in the context of the data, the system provides separate predictions for three mechanisms of viral infection (e.g., entry, fusion, and replication), which affords a higher degree of specificity in drug selection. This enables eVir to explain its predictions based on specific correlated mechanisms and protein interactions. The DeepDrug team has used eVir to identify multiple drugs and drug therapies with high likelihood of efficacy against SARS-CoV-2. These therapies have demonstrated their effectiveness against SARS-CoV-2 infection, both in in vitro studies (with Vero E682 and Calu-383 cells) as well as in vivo studies using transgenic mice. Finally, the DeepDrug AI platform can predict the DDIs in drug combinations as well as the synergy of specific drug combination therapies with the latest module, eComb. Recently, an oral drug combination therapy for COVID-19, discovered by the DeepDrug AI platform, started clinical trials at the Riverside University Health System, California.84 In addition, the nutraceuticals biotin and levomenol were identified to have potential effects against SARS-CoV-2, based on the AI analysis above. The DeepDrug team combined these two nutraceuticals with other essential vitamins and minerals to create a dietary supplement known as Inhibinol.85
Comparison of technologies
Drug discovery is associated with complex workflows that have multiple spanning aspects. The innovative teams mentioned above (Table 1 ) are each working on specific verticals pertinent to drug discovery. Depending on their particular use cases, the teams use diverse techniques, each of which has their own advantages and disadvantages. For instance, the Insilico Medicine team uses GANs, the underlying analysis of which is difficult to explain. However, when applied in the context of COVID-19, the team identified ten proteasomal inhibitors that are currently being testing in labs by several research groups worldwide. Unlike Insilico Medicine, Atomwise’s system is only capable of repurposing known molecules. However, their approach requires a large volume of experimental and structural data. By contrast, BenevolentAI leveraged a massive data set and previously developed knowledge graphs to become the first team to identify a possible inhibitor for reducing the severity of a cytokine storm: baricitinib. The disadvantage of BenevolentAI’s system is its limited capability in discovering known molecules based only on natural language processing from a corpus of existing literature. BenevolentAI also has protein-binding prediction networks still in early phase testing. ComboNet is designed to predict drug synergy by modeling compound and biological target structural features with a GCN. The advantage of this technique is the ability to predict DTIs for compounds with incomplete experimental data. The disadvantage is that the structural training set is hyperspecific to a few key viral SARS-CoV-2 proteins, whereas the drug combinations are based on old curated data with previously tested drugs, such as remdesivir. Without extensive testing on a disjoint test set, it is unclear whether such a training set would be able to predict accurately for compounds outside the training set. Unfortunately, ComboNet has only tested their predicted combination therapies for SARS-CoV-2 against Vero E6 cells in vitro. Finally, DeepDrug is capable of both synthesizing new molecules de novo and repurposing drugs, while predicting their likelihood of human toxicity, manufacturing difficulty, and target specificity.
Table 1.
Comparison of emerging AI teams and their respective technologies.
| AI team | Technique | In vitro | In vivo | Clinical trials |
|---|---|---|---|---|
| BenevolentAI | Knowledge graphs and protein pocket analysis | ✓ | ✓ | ✓ |
| Atomwise | Molecular docking prediction; GAN de novo synthesis of chemotypes | ✓ | ||
| Insilico Medicine, ComboNet | GCN in silico analysis of drug combinations | ✓ | ||
| DeepDrug | eMolFrag, eSynth, eToxPred, eDrugRes, eVir, eComb | ✓ | ✓ | ✓ (Approved May 2021) |
Overall, AI in drug discovery is an extremely powerful but nascent tool. Companies and teams have designed systems that handle only specific types of analyses proficiently. Since each team’s respective data sets are meticulously aggregated and collated individually, their frame of reference might only be useful in a narrow vertical. Additionally, such data are considered proprietary and are often siloed within the team. For instance, the recommendations provided by the existing AI pipelines do not consider the pre-existing conditions of patients. Such global contextual information could be provided in the form of deidentified patient electronic health records. Access to such data would allow for more context-sensitive recommendations that can be valuable in a clinical setting. Overall, these emerging AI tools can be utilized to move the ball toward an ultimate goal: rapidly identifying treatments for infectious diseases. Although certain types of analyses, such as drug combination synergy, expected dosage, and adverse drug reactions, are also important, predictive algorithms for these aspects have yet to be extensively developed. From toxicology to DDIs, to drug–protein specificity, scientists are trying to perfect these prediction systems in every aspect of drug discovery. In the long run, these technologies are a first step toward a comprehensive pipeline capable of rapidly identifying key drugs to combat any emerging infectious disease at a fraction of the time and cost.
Concluding remarks and outlook
The current drug development process is slow, inefficient, and costly. There is a dire need to develop new platforms and approaches to combat diseases quickly compared with traditional approaches. AI applications in other sectors are massively improving platform efficiencies, refining targeted results, and transforming labor-intensive processes. Such efficiencies are key in disrupting the current stagnation of the pharmaceutical industry. Big pharma’s insufficient response to emerging pathogens burdens healthcare systems across the globe and ultimately costs many lives. Data projection, mining, and analysis at scale will assist scientists and pharmacologists in identifying the most effective compounds by cross-checking millions of chemical combinations. All of the AI platforms described in this paper are applying cutting-edge techniques to their respective complex pharmacological challenges. These novel approaches for drug discovery and development are a transformative first step. We need to embrace these new technologies and strategies amid the turmoil of the current COVID-19 pandemic.
References
- 1.Mukhopadhyay S., Iyengar S., Madni A.M., Di Biano R. The next generation of artificial intelligence: synthesizable AI. Advances in Intelligent Systems and Computing. 2018;880:659–677. [Google Scholar]
- 2.Daily M., Medasani S., Behringer R., Trivedi M. Self-driving cars. Computer. 2017;50:18–23. [Google Scholar]
- 3.Gibney E. Google AI algorithm masters ancient game of Go. Nature News. 2016;529:445. doi: 10.1038/529445a. [DOI] [PubMed] [Google Scholar]
- 4.Risi S., Preuss M. From chess and Atari to Starcraft and beyond: how game AI is driving the world of AI. KI-Künstliche Intelligenz. 2020;34:7–17. [Google Scholar]
- 5.Gruber K. Is the future of medical diagnosis in computer algorithms? Lancet Digital Health. 2019;1:e15–e16. [Google Scholar]
- 6.Shamsi F., Aneja B., Hasan P., et al. Synthesis, anticancer evaluation and DNA-binding spectroscopic insights of quinoline-based 1,3,4-oxadiazole-1,2,3-triazole conjugates. ChemistrySelect. 2019;4(41):12176–12182. [Google Scholar]
- 7.Q. Liu, S. Mukhopadhyay, M.X.B. Rodriguez, et al., A One-Shot Learning Framework for Assessment of Fibrillar Collagen from Second Harmonic Generation Images of an Infarcted Myocardium. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI). 2020. 10.1109/isbi45749.2020.9098444. [DOI]
- 8.Iyengar S.S., Li X., Xu H., Mukhopadhyay S., Balakrishnan N., Sawant A., et al. Toward more precise radiotherapy treatment of lung tumors. Computer. 2012;45(1):59–65. [Google Scholar]
- 9.Basu S., Ganguly S., Mukhopadhyay S., DiBiano R., Karki M., Nemani R. SIGSPATIAL '15: Proceedings of the 23rd SIGSPATIAL International Conference on Advances in Geographic Information Systems. Association for Computing Machinery; New York: 2015. Deepsat: a learning framework for satellite imagery; p. 37. [Google Scholar]
- 10.Liu Q., Basu S., Ganguly S., Mukhopadhyay S., DiBiano R., Karki M., et al. Deepsat v2: feature augmented convolutional neural nets for satellite image classification. Remote Sensing Letters. 2020;11:156–165. [Google Scholar]
- 11.Basu S., Ganguly S., Nemani R.R., Mukhopadhyay S., Zhang G., Milesi C., et al. A semiautomated probabilistic framework for tree-cover delineation from 1-m NAIP imagery using a high-performance computing architecture. IEEE Transactions on Geoscience and Remote Sensing. 2015;53(10):5690–5708. [Google Scholar]
- 12.Chokwitthaya C., Zhu Y., Mukhopadhyay S., Collier E. Augmenting building performance predictions during design using generative adversarial networks and immersive virtual environments. Automation in Construction. 2020;119 [Google Scholar]
- 13.Chokwitthaya C., Zhu Y., Dibiano R., Mukhopadhyay S. Combining context-aware design-specific data and building performance models to improve building performance predictions during design. Automation in Construction. 2019;107 [Google Scholar]
- 14.A. Nabijiang, S. Mukhopadhyay, Y. Zhu, R. Gudishala, S. Saeidi and Q. Liu, Why do you take that route? arXiv 2019; 2019: arXiv:1905.06463.
- 15.Ferreira F.G., Gandomi A.H., Cardoso R.T. Artificial intelligence applied to stock market trading: a review. IEEE Access. 2021;9:30898–30917. [Google Scholar]
- 16.Deng J., Dong W., Socher R., Li L.J., Li K., Fei-Fei L. IEEE Conference on Computer Vision and Pattern Recognition. IEEE; New York: 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 17.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research. 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Research. 2016;44(D1):D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Research. 2021;49(D1):D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Günther S., Kuhn M., Dunkel M., Campillos M., Senger C., Petsalaki E., et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Research. 2007;36(Suppl. 1):D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. Journal of Medicinal Chemistry. 2012;55(14):6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 23.G. Marcus, Deep learning: a critical appraisal. arXiv 2018; 2018: arXiv:1801.00631.
- 24.Pu L., Naderi M., Liu T., Wu H.C., Mukhopadhyay S., Brylinski M. eToxpred: a machine learning-based approach to estimate the toxicity of drug candidates. BMC Pharmacology and Toxicology. 2019;20(1):1–15. doi: 10.1186/s40360-018-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature Reviews Drug Discovery. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 26.Liu T., Naderi M., Alvin C., Mukhopadhyay S., Brylinski M. Break down in order to build up: decomposing small molecules for fragment-based drug design with eMolFrag. Journal of Chemical Information and Modeling. 2017;57(4):627–631. doi: 10.1021/acs.jcim.6b00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews. 1997;23(1–3):3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 28.Trouiller P., Olliaro P., Torreele E., Orbinski J., Laing R., Ford N. Drug development for neglected diseases: a deficient market and a public-health policy failure. Lancet. 2002;359(9324):2188–2194. doi: 10.1016/S0140-6736(02)09096-7. [DOI] [PubMed] [Google Scholar]
- 29.Pew. Tracking the Global Pipeline of Antibiotics in Development, April 2020. www.pewtrusts.org/en/research-and-analysis/issue-briefs/2020/04/tracking-the-global-pipeline-of-antibiotics-in-development [Accessed 28 October 2021].
- 30.Plackett B. Why big pharma has abandoned antibiotics. Nature. 2020;586(7830):S50–S52. [Google Scholar]
- 31.Pew. A Scientific Roadmap for Antibiotic Discovery. www.pewtrusts.org/en/research-and-analysis/reports/2016/05/a-scientific-roadmap-for-antibiotic-discovery [Accessed 28 October 2021].
- 32.[WHO] World Health Organization (2020). Lack of new antibiotics threatens global efforts to contain drug-resistant infections. www.who.int/news/item/17-01-2020-lack-of-new-antibiotics-threatens-global-efforts-to-contain-drug-resistant-infections [Accessed 28 October 2021].
- 33.Wu Z., Pan S., Chen F., Long G., Zhang C., Philip S.Y. A comprehensive survey on graph neural networks. IEEE Transactions on Neural Networks and Learning Systems. 2020;32(1):4–24. doi: 10.1109/TNNLS.2020.2978386. [DOI] [PubMed] [Google Scholar]
- 34.D. Duvenaud, D. Maclaurin, J. Aguilera-Iparraguirre, R. Gómez-Bombarelli, T. Hirzel, A. Aspuru-Guzik, et al., Convolutional networks on graphs for learning molecular fingerprints. arXiv 2015; 2015: arXiv:1509.09292.
- 35.Z. Yang, W. Cohen and R. Salakhudinov, Revisiting semi-supervised learning with graph embeddings. arXiv 2016; 2016: arXiv:1603.08861v2.
- 36.Bronstein M.M., Bruna J., LeCun Y., Szlam A., Vandergheynst P. Geometric deep learning: going beyond euclidean data. IEEE Signal Processing Magazine. 2017;34(4):18–42. [Google Scholar]
- 37.A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A.N. Gomez, et al., Attention is all you need. arXiv 2017; 2017: arXiv:1706.03762v5.
- 38.Jing L., Tian Y. Self-supervised visual feature learning with deep neural networks: a survey. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2020;XX::XXX–YYY. doi: 10.1109/TPAMI.2020.2992393. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q., Mukhopadhyay S. 2018 International Joint Conference on Neural Networks (IJCNN) IEEE; 2018. Unsupervised learning using pretrained CNN and associative memory bank. [DOI] [Google Scholar]
- 40.T. Chen, S. Kornblith, M. Norouzi and G. Hinton, A simple framework for contrastive learning of visual representations. arXiv 2020; 2020: arXiv:2002.05709v3.
- 41.Silver D., Huang A., Maddison C.J., Guez A., Sifre L., van den Driessche G., et al. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529(7587):484–489. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- 42.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunning D., Stefik M., Choi J., Miller T., Stumpf S., Yang G.Z. XAI: explainable artificial intelligence. Science. Robotics. 2019;4(37):XXX. doi: 10.1126/scirobotics.aay7120. [DOI] [PubMed] [Google Scholar]
- 44.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial networks. Communications of the ACM. 2020;63(11):139–144. [Google Scholar]
- 45.Drew K., Lee C., Huizar R.L., Tu F., Borgeson B., McWhite C.D., et al. Integration of over 9,000 mass spectrometry experiments builds a global map of human protein complexes. Molecular Systems Biology. 2017;13(6):932. doi: 10.15252/msb.20167490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ammari M.G., Gresham C.R., McCarthy F.M., Nanduri B. HPIDB 2.0: a curated database for host–pathogen interactions. Database. 2016;2016:baw103. doi: 10.1093/database/baw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asgari E., Mofrad M.R. Continuous distributed representation of biological sequences for deep proteomics and genomics. PLoS ONE. 2015;10(11) doi: 10.1371/journal.pone.0141287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van der Maaten L., Hinton G. Visualizing data using t-SNE. Journal of Machine Learning Research. 2008;9(11):XXX. [Google Scholar]
- 50.L. Bertinetto, J. Valmadre, J.F. Henriques, A. Vedaldi and P.H. Torr, Fully-convolutional Siamese networks for object tracking. arXiv 2016; 2016: arXiv:1606.09549v2.
- 51.Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Frieman M.B. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. Journal of Virology. 2016;90(19):8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jafarzadeh A., Nemati M., Jafarzadeh S. Contribution of STAT3 to the pathogenesis of COVID-19. Microbial Pathogenesis. 2021;154 doi: 10.1016/j.micpath.2021.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Yeung D.Y. Towards Bayesian deep learning: a framework and some existing methods. IEEE Transactions on Knowledge and Data Engineering. 2016;28(12):3395–3408. [Google Scholar]
- 54.Hop P., Allgood B., Yu J. Geometric deep learning autonomously learns chemical features that outperform those engineered by domain experts. Molecular Pharmaceutics. 2018;15(10):4371–4377. doi: 10.1021/acs.molpharmaceut.7b01144. [DOI] [PubMed] [Google Scholar]
- 55.J. Moon, J. Kim, Y. Shin and S. Hwang, Confidence-aware learning for deep neural networks. arXiv 2020; 2020: arXiv:2007.01458v3.
- 56.T. DeVries and G.W. Taylor, Learning confidence for out-of-distribution detection in neural networks. arXiv 2018; 2018: arXiv:1802.04865.
- 57.B. Lakshminarayanan, D. Tran, J. Liu, S. Padhy, T. Bedrax-Weiss and Z. Lin, Simple and principled uncertainty estimation with deterministic deep learning via distance awareness. arXiv 2020; 2020: arXiv:2006.10108v2.
- 58.J. Yosinski, J. Clune, Y. Bengio and H. Lipson, How transferable are features in deep neural networks?. arXiv 2014; 2014: arXiv:1411.1792.
- 59.Collier E., DiBiano R., Mukhopadhyay S. 2018 International Joint Conference on Neural Networks. IEEE. 2018. Cactusnets: layer applicability as a metric for transfer learning; pp. 1–8. [Google Scholar]
- 60.Collier E., Mukhopadhyay S. 2020 25th International Conference on Pattern Recognition (ICPR). IEEE; 2021. GAP: Quantifying the Generative Adversarial Set and Class Feature Applicability of Deep Neural Networks. pp. 8384–8391. [Google Scholar]
- 61.M. Long, H. Zhu, J. Wang and M.I. Jordan, Unsupervised domain adaptation with residual transfer networks. arXiv 2016; 2016: arXiv:1602.04433 2016.
- 62.Y.X. Wang, R. Girshick, M. Hebert and B. Hariharan, Low-shot learning from imaginary data. arXiv 2018; 2018: arXiv:1801.05401v2.
- 63.Collier E., Mukhopadhyay S., Duffy K., Ganguly S., Madanguit G., Kalia S., et al. Semantic segmentation of high resolution satellite imagery using generative adversarial networks with progressive growing. Remote Sensing Letters. 2021;12(5):439–448. [Google Scholar]
- 64.Bender A., Cortes-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 1: ways to make an impact, and why we are not there yet. Drug Discovery Today. 2021;26(2):511–524. doi: 10.1016/j.drudis.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Bender A., Cortes-Ciriano I. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 2: a discussion of chemical and biological data used for AI in drug discovery. Drug Discovery Today. 2021;26(4):1040–1052. doi: 10.1016/j.drudis.2020.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infectious Diseases. 2020;20(4):400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manning C., Schutze H. MIT Press; Cambridge: 1999. Foundations of Statistical Natural Language Processing. [Google Scholar]
- 68.T. Wolf, J. Chaumond, L. Debut, J. Chaumond, C. Delangue, A. Moi, et al., Transformers: state-of-the-art natural language processing. In: Proceedings of the 2020 EMNLP (Systems Demonstration). Stroudsburg; Association for Computational Linguistics; 2020: 38–45.
- 69.J. Fauqueur, A. Thillaisundaram and T. Togia, Constructing large scale biomedical knowledge bases from scratch with rapid annotation of interpretable patterns. arXiv 2019; 2019: arXiv:1907.01417.
- 70.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223) doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simonovsky M., Meyers J. DeeplyTough: learning structural comparison of protein binding sites. Journal of Chemical Information and Modeling. 2020;60(4):2356–2366. doi: 10.1021/acs.jcim.9b00554. [DOI] [PubMed] [Google Scholar]
- 72.Yang L., Jin R. Thesis Distance metric learning: a comprehensive survey. Michigan State University. 2006;2:4. [Google Scholar]
- 73.I. Wallach, M. Dzamba and A. Heifets A, AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv 2015; 2015: arXiv:1510.02855.
- 74.Kadurin A., Nikolenko S., Khrabrov K., Aliper A., Zhavoronkov A. druGAN: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico. Molecular Pharmaceutics. 2017;14(9):3098–3104. doi: 10.1021/acs.molpharmaceut.7b00346. [DOI] [PubMed] [Google Scholar]
- 75.A. Zhavoronkov, V. Aladinskiy, A. Zhebrak, B. Zagribelnyy, V. Terentiev, D.S. Bezrukov, et al., Potential 2019-nCoV 3C-like protease inhibitors designed using generative deep learning approaches. ChemRxiv. Published online February 19, 2020. 10.26434/chemrxiv.11829102.v2. [DOI]
- 76.O. Akal, Z. Peng and G.H. Valadez, ComboNet: combined 2D & 3D architecture for aorta segmentation. arXiv 2020; 2020: arXiv:2006.05325 2020.
- 77.Bobrowski T., Chen L., Eastman R.T., Itkin Z., Shinn P., Chen C.Z., et al. Synergistic and antagonistic drug combinations against SARS-CoV-2. Molecular Therapy. 2021;29(2):873–885. doi: 10.1016/j.ymthe.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Naderi M., Alvin C., Ding Y., Mukhopadhyay S., Brylinski M. A graph-based approach to construct target- focused libraries for virtual screening. Journal of Cheminformatics. 2016;8(1):1–16. doi: 10.1186/s13321-016-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar A., Vembu S., Menon A.K., Elkan C. Beam search algorithms for multilabel learning. Machine Learning. 2013;92(1):65–89. [Google Scholar]
- 80.Mayr A., Klambauer G., Unterthiner T., Hochreiter S. DeepTox: toxicity prediction using deep learning. Frontiers in Environmental Science. 2016;3:80. [Google Scholar]
- 81.Thakur N., Qureshi A., Kumar M. AVPpred: collection and prediction of highly effective antiviral pep- tides. Nucleic Acids Research. 2012;40(W1):W199–W204. doi: 10.1093/nar/gks450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ng M.L., Tan S.H., See E.E., Ooi E.E., Ling A.E. Proliferative growth of SARS coronavirus in Vero E6 cells. Journal of General Virology. 2003;84(12):3291–3303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- 83.Foster K.A., Avery M.L., Yazdanian M., Audus K.L. Characterization of the Calu-3 cell line as a tool to screen pulmonary drug delivery. International Journal of Pharmaceutics. 2000;208(1–2):1–11. doi: 10.1016/s0378-5173(00)00452-x. [DOI] [PubMed] [Google Scholar]
- 84.PRNewswire. Human studies begin on artificial Intelligence discovered COVID-19 treatment with up to 97 percent effectiveness. https://finance.yahoo.com/news/human-studies-begin-artificial-intelligence-130000945.html [Accessed 28 October 2021].
- 85.https://inhibinol.com/ [Accessed 28 October 2021].