Abstract
Shigella spp. are known primarily as a cause of bacillary dysentery. However, in an initial phase, numerous patients exhibit watery diarrhea that may or may not be followed by dysentery. New virulence factors associated with the species of Shigella have recently been described. These are enterotoxins 1 and 2 of Shigella (ShET-1 and ShET-2, respectively). The aim of the present study was to determine the prevalence of ShET-1 and ShET-2 in species of Shigella isolated from patients with traveler's diarrhea. During the period from 1993 to 1998, stool samples from 500 travelers with diarrhea were cultured for the isolation of Shigella spp. and other enteropathogens. The detection of ShET-1 and ShET-2 was performed by a PCR technique with specific primers. Among a total of 51 strains of Shigella isolated during this period (22 S. flexneri, 26 S. sonnei, and 3 S. dysenteriae strains), at least one enterotoxin was detected in 31 (60.78%) strains; 2 (9.09%; both of which were S. flexneri strains) produced only ShET-1, while 21 (41.17%; 3 S. flexneri, 15 S. sonnei, and 3 S. dysenteriae strains) produced ShET-2. Furthermore, 8 (15.69%) of 22 S. flexneri strains presented both enterotoxins. Our results show that the prevalence of ShET-2 was high in all the Shigella species studied and confirm that ShET-1 is detected only in S. flexneri.
Shigella species are an important cause of diarrheal disease in developing countries (2) and in travelers to tropical countries (16, 18). About 50% of Spanish travelers who visit these countries develop diarrhea, and Shigella species are among the main etiological agents (6–8). Among the Shigella species, Shigella dysenteriae is especially known as a cause of bacillary dysentery; this has also been shown with S. flexneri (4). However, in a considerable number of patients, watery diarrhea is shown in the first phase of the infection, which may or may not be followed by dysentery, similar to what is seen for the species S. flexneri, S. sonnei, and S. boydii, although the last two species generally produce a self-limited, watery diarrhea (4, 9).
Several virulence factors have been associated with Shigella spp., the most common being the ability to colonize and invade the intestinal cells. This phenomenon is mediated, in part, by the invasion-associated locus (ial), which is carried on a plasmid of 120 to 140 MDa (5), and the invasion plasmid antigen H (ipaH) gene, which is present in multiple copies in both the plasmids and the chromosomes of these organisms (17).
Another virulence factor related to S. dysenteriae is its capacity to produce an exotoxin called Shiga toxin (Stx), which is not excreted by the bacteria but is released only during cell lysis (1). Despite its clear toxigenicity, the role of Stx in shigellosis is not clear, since it is known that Stx is not essential for invasion or cellular lysis.
Two new enterotoxins have recently been described in S. flexneri 2a. The first toxin is called Shigella enterotoxin 1 (ShET-1), which is encoded in the set1 chromosomal gene. It has been suggested that the active toxin of ShET-1 has a configuration of one A subunit and several B subunits (A1-Bn) (13).
The second enterotoxin, called Shigella enterotoxin 2 (ShET-2), is encoded in the sen gene. This gene is located on a plasmid of 140 MDa which is associated with invasion in these pathogens (12).
The main aim of this study was to determine the prevalence of ShET-1 and ShET-2 in Shigella species isolated from patients with traveler's diarrhea.
MATERIALS AND METHODS
Patients.
During the period from 1993 to 1998, stool specimens from 500 patients with traveler's diarrhea (TD) were analyzed. TD was defined as the occurrence of three or more episodes of watery diarrhea within a 24-h period, with or without other symptoms, between 12 h after arrival in and 5 days after departure from the country visited or as the occurrence of unformed stools accompanied by one of the following: cramps, tenesmus, vomiting, nausea, fever, chills, and prostration.
Microbiological tests.
To isolate Shigella species, stool samples were inoculated onto MacConkey agar and Salmonella-Shigella agar (Becton Dickinson, Heidelberg, Germany), and the resulting colonies which exhibited characteristics of Shigella spp. were identified by conventional biochemical methods (11). Subsequently, the species were identified with specific polyvalent antisera against S. flexneri, S. sonnei, and S. dysenteriae (Diagnostics Pasteurs, Marnes-la-Coquette, France) and S. boydii (Difco Laboratories, Detroit, Mich.).
PCR assay.
The detection of ShET-1 (A and B subunits) was performed by amplifying both set1A and set1B genes by PCR. A PCR technique was also used to detect the sen, ipaH, ial, and stx genes. The isolated Shigella species were grown on MacConkey agar overnight. One colony of each isolate was suspended in 25 μl of a reaction mixture containing 20 mM Tris-HCl (pH 8.8), 100 mM KCl, 3 mM MgCl2, 0.1% gelatin, 400 μM (each) deoxynucleoside triphosphate, and 1 μM primer together with 2.5 U of Taq polymerase (GIBCO-BRL). The reaction mixture was overlaid with mineral oil and was subjected to the following program: 30 cycles at 95°C for 50 s, 55°C for 1.5 min, and 72°C for 2 min, with a final extension at 72°C for 7 min. Seven sets of primers, obtained from Boehringer Mannheim (Mannheim, Germany), were used to amplify the set1A, set1B, sen, ipaH, ial, and stx genes, as indicated in Table 1.
TABLE 1.
Primers used for identification of virulence factors of Shigella spp.
| Gene encoding virulence factor | Primer | Oligonucleotide sequence (5′ to 3′) | Location within gene | Size of amplified product (bp) | Reference(s) |
|---|---|---|---|---|---|
| set1A | ShET-1A upper | TCACGCTACCATCAAAGA | 460–477 | ||
| ShET-1A lower | TATCCCCCTTTGGTGGTA | 751–768 | 309 | 3 | |
| set1B | ShET-1B upper | GTGAACCTGCTGCCGATATC | 70–89 | ||
| ShET-1B lower | ATTTGTGGATAAAAATGACG | 197–216 | 147 | 3 | |
| sen | ShET-2 upper | ATGTGCCTGCTATTATTTAT | 380–399 | ||
| ShET-2 lower | CATAATAATAAGCGGTCAGC | 1158–1178 | 799 | 12 | |
| ipaH | Shig-1 | TGGAAAAACTCAGTGCCTCT | 1063–1083 | ||
| Shig-2 | CCAGTCCGTAAATTCATTCT | 1466–1485 | 423 | 10 | |
| ial | ial upper | CTGGATGGTATGGTGAGG | 5340–5357 | ||
| ial lower | GGAGGCCAACAATTATTTCC | 5640–5659 | 320 | 5 | |
| Stx | stx upper | CAGTTAATGTGGTTGCGAAG | 50–69 | ||
| stx lower | CTGCTAATAGTTCTGCGCATC | 924–944 | 895 | 14, 15 |
PCR assays were performed in a DNA thermal cycler (model 480; Perkin-Elmer Cetus, Emeryville, Calif.). A reagent blank, which contained all components of the reaction mixture with the exception of the bacteria, was included in every PCR procedure.
Amplification products were subjected to gel electrophoresis in 2% agarose and were detected by staining with ethidium bromide.
RESULTS
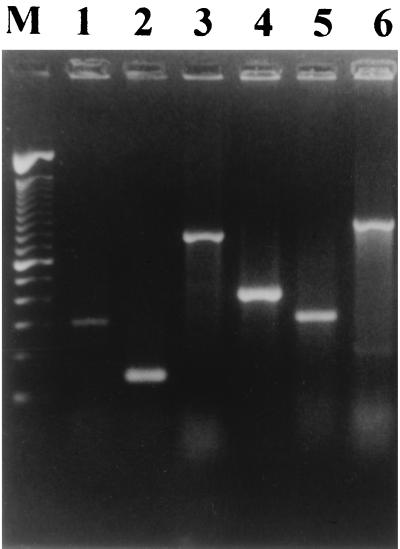
The products from amplification DNA by the different PCRs are shown in Fig. 1. In the amplification reaction with the set1A and set1B genes, which encode the ShET-1 A and B subunits, respectively, bands of 309 bp (A subunit) and 147 bp (B subunit), respectively, were observed. The product amplified from the sen gene, which encodes ShET-2, was 799 bp. The PCR product of the ipaH gene was 423 bp, and that of the ial gene was 320 bp. Amplification of the stx gene produced a PCR product of 895 bp. All strains that were ShET-1A positive were also found to be ShET-1B positive. The DNA sequences of the PCR products were determined to confirm amplification of the correct gene. The geographic distributions of the Shigella spp. studied are provided in Table 2.
FIG. 1.
Agarose gel electrophoresis showing PCR amplification products. Lane M, DNA molecular size markers (100-bp DNA ladder from Gibco-BRL). The products for detection of the set1A gene (lane 1), set1B gene (lane 2), sen gene (lane 3), ipaH gene (lane 4), ial gene (lane 5), and stx gene (lane 6) are shown.
TABLE 2.
Geographic distribution of Shigella isolates from patients with TD
| Geographic area | Total no. of isolates testeda | No. of isolates that produced the indicated toxin:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. flexneri(n = 22)
|
S. sonnei(n = 26)
|
S. dysenteriae(n = 3)
|
|||||||||||
| ShET-1 | ShET-2 | ShET-1 + ShET-2 | None | ShET-1 | ShET-2 | ShET-1 + ShET-2 | None | ShET-1 | ShET-2 | ShET-1 + ShET-2 | None | ||
| America (17)b | |||||||||||||
| Central America | 15 | 1 | 1 | 1 | 2 | 5 | 5 | ||||||
| South America | 2 | 1 | 1 | ||||||||||
| Africa (16) | |||||||||||||
| West Africa | 9 | 1 | 4 | 2 | 1 | 1 | |||||||
| East Africa | 4 | 1 | 3 | ||||||||||
| North Africa | 3 | 3 | |||||||||||
| Asia (18) | |||||||||||||
| India | 14 | 1 | 1 | 4 | 5 | 3 | |||||||
| Middle East | 2 | 1 | 1 | ||||||||||
| Southeast Asia | 2 | 1 | 1 | ||||||||||
| Total | 51 | 2 | 3 | 8 | 9 | 15 | 11 | 3 | |||||
Total number of strains in each geographic area.
Values in parentheses are total number of strains from each continent.
Among the 51 strains of Shigella isolated, 31 (60.78%) were found to produce ShET-1 and/or ShET-2 (Table 3). Among the toxin producers, 2 S. flexneri strains (3.92%) produced only ShET-1 (A and B subunit positive), 21 (41.17%; 3 S. flexneri, 15 S. sonnei, and 3 S. dysenteriae strains) produced only ShET-2, and 8 (15.69%) S. flexneri strains produced both enterotoxins. Furthermore, all strains of Shigella were also ipaH positive, whereas only 29 (57%) were ial positive. Only the three S. dysenteriae isolates were positive for the stx gene. If the detection of ShET-2 is considered in correlation with the detection of ial, 3 of 25 Shigella strains (2 S. flexneri strains and 1 S. sonnei strain) isolated during the period from 1993 to 1995 were positive for both the sen and the ial genes, whereas 26 of 26 Shigella strains (9 S. flexneri, 14 S. sonnei, and 3 S. dysenteria strains) isolated during the period from 1996 to 1998 were positive for both genes.
TABLE 3.
Prevalence of ShET-1 and ShET-2 in Shigella spp. isolated from patients with TD
| Strain | Total no. of strains tested | No. (%) of strains that produced the following:
|
||
|---|---|---|---|---|
| ShET-1 | ShET-2 | ShET-1 and ShET-2 | ||
| S. flexneri | 22 | 2 (9.09) | 3 (13.64) | 8 (36.36) |
| S. sonnei | 26 | 0 (0) | 15 (57.69) | 0 (0) |
| S. dysenteriae | 3 | 0 (0) | 3 (100) | 0 (0) |
| Total | 51 | 2 (3.92) | 21 (41.17) | 8 (15.69) |
DISCUSSION
Acute dysentery is a common disease in many developing countries, whereas in developed countries diarrhea is often acquired during travel abroad and hence is TD. In our study, different virulence factors associated with the pathogenicity of Shigella spp. were investigated. In the colon, the bacteria invade the mucosal cells. Some of the genes involved in the invasion have been identified and have been designated ipa, for invasion plasmid antigen. However, one of these genes, the ipaH gene, which encodes a 60-kDa antigen, is found in multiple copies on both the invasion-related plasmid and the chromosome. All Shigella species that we studied were positive for this gene. Another virulence factor gene associated with the invasion of the cell by Shigella species is the ial gene, which is plasmid encoded. In our study, 57% of the Shigella strains were positive for ial. This fact is discussed below. The geographic distribution (by continent) of the different species of Shigella, as well as those of the ShET-1 and/or ShET-2-producing strains, was homogeneous.
S. dysenteriae, as is already known, was the only species of the genus Shigella that produces Stx.
In a considerable number of patients, watery diarrhea is observed prior to the onset of dysentery. This is likely explained by the synthesis of two recently described enterotoxins, ShET-1 and ShET-2. However, in our study a significant difference was not observed when clinical symptoms and the isolation of ShET-1- and/or ShET-2-producing Shigella strains were compared (data not shown). Noriega et al. (13) found ShET-1 almost exclusively in S. flexneri 2a. In our study, although no serotyping was performed, ShET-1 was found in 45% of S. flexneri strains but was not found in either S. sonnei or S. dysenteriae, confirming the previous results. In 36% of S. flexneri strains, ShET-1 was found together with ShET-2. ShET-2 was also found alone in 14% of S. flexneri strains, 57% of S. sonnei strains, and 100% of S. dysenteriae strains. The ShET-1 toxin shows an A-B structure. The PCRs with primers designed to amplify either the set1A gene (which encodes the A subunit) or the set1B (which encodes the B subunit) showed complete correlation, although the intensity of the PCR product was stronger for the set1B gene than for the set1A gene. Although the strains carry the genes for the toxins, they may not express the toxin.
Nataro et al. (12) found the sen gene (which encodes ShET-2), using a DNA probe, in 73% of the S. flexneri strains that they studied, whereas we found this gene in 50% of the S. flexneri strains that we studied. However, our results showed bias because some of these strains have been stored since 1993, and as this gene is plasmid encoded, these strains could have lost the plasmid, which is known to occur frequently upon repeated subculturing and prolonged storage (10, 17). This is demonstrated by the fact that only 3 (12%) of 25 Shigella strains isolated from 1993 to 1995 were positive for both the sen and the ial plasmid-encoded genes, whereas 100% of Shigella strains isolated from 1996 to 1998 were positive for both genes.
In conclusion, this study shows the high prevalence of ShET-1 and ShET-2 in S. flexneri strains and the high prevalence of ShET-2 in S. sonnei and S. dysenteriae strains isolated from patients with TD.
ACKNOWLEDGMENT
This work was supported in part by grant FIS 94/0980 from Fondo de Investigaciones Sanitarias de la Seguridad Social of Spain.
REFERENCES
- 1.Cantey J R. Shiga toxin—an expanding role in the pathogenesis of infectious diseases. J Infect Dis. 1985;151:766–771. doi: 10.1093/infdis/151.5.766. [DOI] [PubMed] [Google Scholar]
- 2.Division of Health Promotion and Disease Prevention and Division of International Health, Institute of Medicine. New vaccine development: establishing priorities. II. Diseases of importance in developing countries. Washington, D.C: National Academy Press; 1986. The burden of disease resulting from various diarrheal pathogens, appendix C. [Google Scholar]
- 3.Fasano A, Noriega F, Manevel D R, Chanasongcram S, Rossell R, Guandalini S, Levine M M. Shigella enterotoxin 1: an enterotoxin of Shigella flexneri 2a active in rabbit small intestine in vivo and in vitro. J Clin Investig. 1995;95:2853–2861. doi: 10.1172/JCI117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Formal S B, Levine M M. Shigellosis. In: Germanier R, editor. Bacterial vaccines. New York, N.Y: Academic Press, Inc.; 1984. pp. 167–186. [Google Scholar]
- 5.Frankel G, Giron J A, Valmassoi J, Schoolnik G K. Multi-gene amplification: simultaneous detection of three virulence genes in diarrhoeal stool. Mol Microbiol. 1989;3:1729–1734. doi: 10.1111/j.1365-2958.1989.tb00158.x. [DOI] [PubMed] [Google Scholar]
- 6.Gascón J, Ruiz L, Canela J, Mallart M, Corachan M. Epidemiología de la diarrea del viajero en turistas españoles a países tropicales desarrollados. Med Clin. 1993;100:365–367. [PubMed] [Google Scholar]
- 7.Gascón J, Vila J, Valls M E, Ruiz L, Vidal J, Corachán M, Prats G, Jiménez de Anta M T. Etiology of traveller's diarrhea in Spanish travellers to developing countries. Eur J Epidemiol. 1993;9:217–223. doi: 10.1007/BF00158796. [DOI] [PubMed] [Google Scholar]
- 8.Gascón J, Vargas M, Quintó L, Corachán M, Jimenez de Anta M T, Vila J. Enteroaggregative Escherichia coli strains as a cause of traveler's diarrhea: a case-control study. J Infect Dis. 1998;177:1409–1412. doi: 10.1086/517826. [DOI] [PubMed] [Google Scholar]
- 9.Keush G T. Shigella. In: Gorbach S L, editor. Infectious diarrhea. Boston, Mass: Blackwell Scientific Publications; 1986. pp. 31–50. [Google Scholar]
- 10.Lüscher D, Athwegg M. Detection of shigellae, enteroinvasive and enterotoxigenic Escherichia coli using the polymerase chain reaction (PCR) in patients returning from tropical countries. Mol Cell Probes. 1994;8:285–290. doi: 10.1006/mcpr.1994.1040. [DOI] [PubMed] [Google Scholar]
- 11.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 12.Nataro J P, Seriwatana J, Fasano A, Maneval D R, Guers L D, Noriega F, Dubovsky F, Levine M M, Morris J G., Jr Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect Immun. 1995;63:4721–4728. doi: 10.1128/iai.63.12.4721-4728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noriega F R, Liao F M, Formal S B, Fassano A, Levine M M. Prevalence of Shigella enterotoxin 1 among Shigella clinical isolates of diverse serotypes. J Infect Dis. 1995;172:1408–1410. doi: 10.1093/infdis/172.5.1408. [DOI] [PubMed] [Google Scholar]
- 14.Olvik O, Strockbine N A. In: PCR detection of heat-stable, heat-labile, and Shiga-like toxin genes in Escherichia coli. Persing S H, Smith T F, Tenover F C, White T J, editors. Rochester, Minn: Diagnostic molecular microbiology. Principles and applications. Mayo Foundation; 1993. pp. 271–276. [Google Scholar]
- 15.Strockbine N A, Jackson M P, Sung L M, Holmes R K, O'Brien A D. Cloning and sequencing of the genes for Shiga toxin from Shigella dysenteriae type 1. J Bacteriol. 1988;170:1116–1122. doi: 10.1128/jb.170.3.1116-1122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tauxer R, Puhr N D, Well J G. Antimicrobial resistance of Shigella isolates in the USA: the importance of international travelers. J Infect Dis. 1990;162:1107–1111. doi: 10.1093/infdis/162.5.1107. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesan M M, Buysee J M, Kopecko D J. Use of Shigella flexneri ipaC and ipaH gene sequences for the general identification of Shigella spp. and enteroinvasive Escherichia coli. J Clin Microbiol. 1989;27:2687–2691. doi: 10.1128/jcm.27.12.2687-2691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vila J, Gascón J, Abdalla S, Gomez J, Marco F, Moreno A, Corachán M. Antimicrobial resistance of Shigella isolates causing traveller's diarrhea. Antimicrob Agents Chemother. 1994;38:2668–2670. doi: 10.1128/aac.38.11.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]