Abstract
This study evaluated correlation and agreement between version 3 of the Quantiplex human immunodeficiency virus type 1 (HIV-1) RNA assay (v3 branched DNA [bDNA]) and a sensitized Amplicor HIV-1 Monitor assay (reverse transcription [RT]-PCR) for the measurement of HIV RNA. Three hundred eighteen samples from 59 randomly selected, HIV-1-seropositive persons on various drug protocols from the National Institute of Allergy and Infectious Diseases HIV outpatient clinic were studied. The results indicate that v3 bDNA and RT-PCR are highly correlated (r = 0.98) and are in good agreement (mean difference in log10 copies/ml ± 2 standard deviations = 0.072 ± 0.371). The relationship between values obtained by both assays is given by the following equation: log10v3 bDNA = −0.0915 + 1.0052 · log10RT-PCR. This represents a 1.026-fold difference between log10RT-PCR values and log10v3 bDNA values.
The use of highly active antiretroviral therapy in patients with human immunodeficiency virus type 1 (HIV-1) infection has resulted in an increase in the number of CD4+ T lymphocytes as well as in a reduction in plasma virus load (2, 4–6). In order to assess the magnitude of viral load reduction and the occurrence of rebound virus due to drug failure or noncompliance, it is necessary to use as sensitive a viral quantitation assay as is available. There are two widely used techniques to achieve these ends: the Amplicor HIV-1 Monitor assay (reverse transcription [RT]-PCR) and the Quantiplex HIV-1 RNA assay (branched DNA [bDNA]). The Amplicor HIV-1 Monitor assay is an in vitro nucleic acid target amplification (PCR) test for the quantitation of HIV-1 RNA in human plasma, while the Quantiplex HIV-1 RNA assay is a signal amplification nucleic acid probe assay for HIV-1 quantitation. The most current versions of these assays have a quantitative detection limit of 50 copies (3, 7, 8, 11). Because patients may be tested with both of these assays at some point in their treatment, it is useful to determine how the resulting values are related to each other and, ideally, to develop some formula for their comparison. We hypothesize that tests designed to measure the same thing, albeit in different ways, will show a quantifiable relationship to each other; not only will they correlate, but they will show agreement. Previous reports comparing Amplicor with Quantiplex version 2.0 have indicated that although the results show a high correlation, they show poor agreement (9, 10). In this study, we compared two of the commercially available assays for the measurement of HIV RNA, the Quantiplex version 3 assay (3) and a sensitized version of the HIV-1 Amplicor Monitor assay (7, 8, 11), to assess their quantitative relationship.
MATERIALS AND METHODS
Patient population.
The study group consisted of 59 randomly selected HIV-1-seropositive persons on various drug protocols from the outpatient population of the National Institute of Allergy and Infectious Diseases HIV clinic. A total of 318 samples from these patients were analyzed.
Informed consent.
Informed consent was obtained from all patients. Human experimentation guidelines of the U.S. Department of Health and Human Services were followed in the conduct of the clinical research.
Sample collection and processing. (i) v3 bDNA.
Whole, EDTA-preserved blood was separated within 2 h of collection, and plasma was frozen immediately at −70°C in 1-ml aliquots. The samples were then thawed, spun at 23,000 × g for 60 min at 4°C, and aspirated without disturbing the virus pellet. Pellets were then frozen at −70°C for no more than 2 days until they could be tested with the bDNA assay (Bayer Corporation, Diagnostics Division, Norwood, Mass.).
(ii) RT-PCR.
Whole, EDTA-preserved blood was separated within 2 h of collection, and plasma was frozen immediately at −70°C in 1-ml aliquots. Aliquots were thawed and then spun at 23,000 × g for 75 min at 4°C, and 800 μl was drawn off without disturbing the pellet. The resulting pellets in 200 μl of plasma were then frozen at −70°C for no more than 2 days until they could be tested with the RT-PCR assay (Roche Diagnostics Corporation, Indianapolis, Ind.).
(iii) usRT-PCR.
Five-hundred-microliter aliquots of EDTA-preserved plasma were spun at 23,000 × g for 75 min at 4°C and aspirated without disturbing the virus pellet. Pellets were then frozen at −70°C for no more than 2 days until they could be tested with the ultrasensitive (usRT-PCR) Amplicor HIV-1 Monitor assay (Roche Diagnostics Corporation), as described in the manufacturer's insert.
Quantitation of HIV-RNA. (i) v3 bDNA.
The Chiron 3.0 bDNA assay was performed according to the manufacturer's instructions provided with the assay kit.
(ii) RT-PCR.
The pellets in 200 μl of plasma were resuspended in lysis buffer and tested according to the test procedure of the Amplicor HIV-1 Monitor test. No further modification of the procedure was performed.
(iii) usRT-PCR.
The viral pellets were processed according to the “UltraSensitive specimen preparation” procedure provided with the assay kit. With the exception of the alternate specimen preparation procedures, the test procedure remains the same as that of the Amplicor HIV-1 Monitor test.
Statistical analysis.
Because viral load tends to change in a logarithmic fashion and because data are evaluated on the basis of half-log changes, values were transformed into common (log10) logarithms and expressed as log10v3 bDNA or log10RT-PCR. Linear regression and correlation were used to determine the relationship between the v3 bDNA and RT-PCR values. The methods of Bland and Altman (1) were employed to assess agreement between the values.
RESULTS
Correlation and regression.
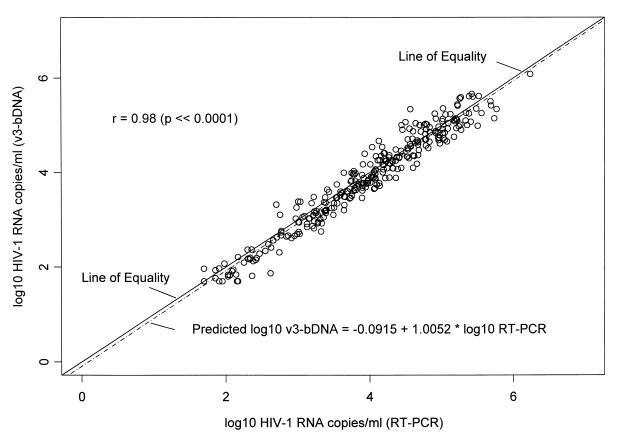
Assay values were transformed to common logs. In this analysis, log10RT-PCR is taken as the predictor variable and log10v3 bDNA is taken as the response variable. The results are highly correlated. Figure 1 shows a plot of log10v3 bDNA versus log10RT-PCR with the fitted regression line described by the equation y = −0.0915 + 1.0052 · x, with r = 0.98. The estimate of the intercept is −0.0915, with a standard error of 0.04. This value differs from the ideal value of 0.0 at the 0.05 level of confidence. The value of the slope is 1.0052, with a standard error of 0.01. This value is statistically equivalent to an ideal value of 1.0. A 95% confidence interval for the estimate of the slope is 0.98 to 1.03, a range that includes 1.0, indicating that the assays are in good agreement (discussed below).
FIG. 1.
Scatter plot of log10v3 bDNA versus log10RT-PCR with the line of equality (solid) and the fitted regression line (hatched). The equation for the fitted regression line is given in the lower-right-hand corner, and the correlation coefficient (r) is given in the upper-left-hand corner. There is good agreement between the two assays.
Agreement.
Bland and Altman have pointed out that when a comparison of two clinical measurements of the same criterion is made, the use of the correlation may be misleading (1). The correlation coefficient measures the strength of the relationship between the variables, but does not necessarily measure the agreement between them. Figure 2 shows the difference in log10 assay results (log10RT-PCR − log10v3 bDNA) versus the average of assay results [(log10RT-PCR + log10v3 bDNA)/2]. The plot shows that the differences in values are fairly homogeneously distributed between 1.96 standard deviations above and below the mean (0.072) and are in good agreement (mean difference in log10 copies/ml ± 2 standard deviations = 0.072 ± 0.371). A one-sample t test demonstrates that the mean is significantly different from zero (P < 0.0001). This implies that the log10RT-PCR values, on average, are higher than the log10v3 bDNA values. If the difference values were normally distributed, one would expect to observe approximately 95% of them between 1.96 standard deviations of the mean. In fact, 95.28% (303 of 318) of the values fell in this range.
FIG. 2.
Difference in log10 assay results versus average of log10 assay results. The mean difference (solid line) ± 1.96 standard deviations (dashed lines) is shown. The plot shows that the difference values are fairly homogeneously distributed between 2 standard deviations (SD) above and below the mean (0.072). A total of 95.28% (303 of 318) of the values fall between the hatched lines, indicating normal distribution.
The RT-PCR assay used in these analyses includes a precentrifugation step of 1 ml of plasma before application to the standard Amplicor Monitor assay and was used in this laboratory before the UltraSensitive specimen preparation procedure was included with the Amplicor Monitor kit. In order to address possible concerns that this sensitized RT-PCR assay might yield results discordant with those obtained by the UltraSensitive specimen preparation procedure (11), a subset of 47 samples was reanalyzed by the UltraSensitive specimen preparation procedure (usRT-PCR). The results showed that, in fact, the sensitized and usRT-PCR assays were highly correlated (r = 0.98) and showed good agreement (mean difference in log10 copies/ml ± 2 standard deviations = 0.041 ± 0.176).
The relationship between values obtained by v3 bDNA and usRT-PCR is given by the following equation: log10v3 bDNA = −0.608 + 1.1214 · log10usRT-PCR. This represents a 1.048-fold difference between log10usRT-PCR values and log10v3 bDNA values.
DISCUSSION
In the current environment of HIV-1 treatments, it is essential to be able to quickly and accurately assess viral load. There have been a number of studies comparing the various techniques available for monitoring HIV RNA (9, 10). These studies all failed to find significant agreement between assays, and none revealed a numerical way to compare the results from the different techniques. Because patients may be monitored with more than one assay over the course of their treatment, it is useful to find a quantitative relationship between these assays.
Our comparison of the third-generation bDNA assay and a modification of the Amplicor Monitor assay indicates that these two assays are highly correlated. Although RT-PCR values, on average, are larger than the v3 bDNA values, regression analyses indicate that values are normally distributed, and the two assays show very good agreement.
In conclusion, we were able to determine that, given an RT-PCR value, it is possible to derive the comparable bDNA value by the formula log10v3 bDNA = −0.0915 + 1.0052 · log10RT-PCR.
The knowledge that both assays show good agreement overall and have a reliable mathematical relationship should assist clinicians in their use of both data sets when monitoring patient virus load.
ACKNOWLEDGMENT
This project has been funded completely or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-56000.
REFERENCES
- 1.Bland J M, Altman D G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;i:307–310. [PubMed] [Google Scholar]
- 2.Chun T W, Engel D, Mizell S B, Ehler L A, Fauci A S. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J Exp Med. 1998;188:83–91. doi: 10.1084/jem.188.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins M L, Irvine B, Tyner D, Fine E, Zayti C, Chang C, Horn T, Ahle D, Detmer J, Shen L-P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.d'Arminio Monforte A, Testa L, Adorni F, Chiesa E, Bini T, Moscatelli G C, Abeli C, Rusconi S, Sollima S, Balotta C, Musicco M, Galli M, Moroni M. Clinical outcome and predictive factors of failure of highly active antiretroviral therapy in antiretroviral-experienced patients in advanced stages of HIV-1 infection. AIDS. 1998;12:1631–1637. doi: 10.1097/00002030-199813000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Ferrando S, van Gorp W, McElhiney M, Goggin K, Sewell M, Rabkin J. Highly active antiretroviral treatment in HIV infection: benefits for neuropsychological function. AIDS. 1998;12:F65–F70. doi: 10.1097/00002030-199808000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Li T S, Tubiana R, Katlama C, Calvez V, Ait Mohand H, Autran B. Long-lasting recovery in CD4 T-cell function and viral-load reduction after highly active antiretroviral therapy in advanced HIV-1 disease. Lancet. 1998;351:1682–1686. doi: 10.1016/s0140-6736(97)10291-4. [DOI] [PubMed] [Google Scholar]
- 7.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolte F S, Boysza J, Thurmond C, Clark W S, Lennox J L. Clinical comparison of an enhanced-sensitivity branched-DNA assay and reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:716–720. doi: 10.1128/jcm.36.3.716-720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segondy M, Izopet J, Pellegrin I, Montes B, Dumon B, Pasquier C, Peeters M, Fleury H J A, Puel J, Reynes J. Comparison of the QUANTIPLEX HIV-1 RNA 2.0 assay with the AMPLICOR HIV-1 MONITOR 1.0 assay for quantitation of levels of human immunodeficiency virus type 1 RNA in plasma of patients receiving stavudine-didanosine combination therapy. J Clin Microbiol. 1998;36:3392–3395. doi: 10.1128/jcm.36.11.3392-3395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun R, Ku J, Jayakar H, Kuo J-C, Brambilla D, Herman S, Rosenstraus M, Spadoro J. Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:2964–2969. doi: 10.1128/jcm.36.10.2964-2969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]