Fibronectin binding to collagen type II is crucial for articular chondrocytes to stay in a healthy steady state.
Abstract
Osteoarthritis is the most prevalent joint disease worldwide, yet progress in development of effective disease-modifying treatments is slow because of lack of insight into the underlying disease pathways. Therefore, we aimed to identify the causal pathogenic mutation in an early-onset osteoarthritis family, followed by functional studies in human induced pluripotent stem cells (hiPSCs) in an in vitro organoid cartilage model. We demonstrated that the identified causal missense mutation in the gelatin-binding domain of the extracellular matrix protein fibronectin resulted in significant decreased binding capacity to collagen type II. Further analyses of formed hiPSC-derived neo-cartilage tissue highlighted that mutated fibronectin affected chondrogenic capacity and propensity to a procatabolic osteoarthritic state. Together, we demonstrate that binding of fibronectin to collagen type II is crucial for fibronectin downstream gene expression of chondrocytes. We advocate that effective treatment development should focus on restoring or maintaining proper binding between fibronectin and collagen type II.
INTRODUCTION
Osteoarthritis (OA) is an age-related, degenerative, heterogeneous disease of the articular joints, characterized by pathological changes of the articular cartilage, synovium, and subchondral bone (1). Currently, OA is the most prevalent joints disease worldwide and affects more than 40 million people in Europe alone (2, 3). Patients with OA suffer pain and stiffness in the articular joints, resulting in significant disability in everyday life (4). Despite these detrimental consequences, to date, no effective treatment is available except pain relief medication and, in the final stage, joint replacement surgery. Lacking insight into underlying OA pathophysiology has considerably contributed to failures in disease-modifying OA drug development.
For that matter, next-generation exome sequencing has been highly successful in identifying likely causal mutations in patients with familial, more severe phenotypes (5) or Mendelian disorders (6). Because of their strong effect, these mutations can provide direct clues to genotype-phenotype relations and are more actionable to express a particular disease state in experimental human in vitro tissue models. Since the pathways in which these genes operate are likely extrapolated to confer risk to common OA, these mutations are destined to elucidate OA-specific aspects of the disease. Using cells carrying high-impact mutations in tailored human in vitro tissue models may thus elucidate previously unknown, likely causal underlying pathways of OA (7, 8). Moreover, human induced pluripotent stem cell (hiPSC) technology allows reprogramming of primary cells into a sustainable pluripotent cell source (9), which is subsequently used for the directed differentiation into specialized cells of interest (10, 11). Even more, because of novel advancements in genomic engineering with CRISPR-Cas9 technology, cells are now easily edited or modulated (12). As a result, the creation of isogenic lines can give insight in the functional characterization of genetic variation. Since OA-relevant cells, such as chondrocytes, are not readily available, the combination of hiPSCs and CRISPR-Cas9 holds immense potential for translational human OA disease modeling to uncover the role of high-impact mutations (8).
By applying a genome-wide linkage scan in seven extended early-onset (EO)–OA families without dysplasia, we previously identified significant linkage on chromosome 2q33.3, with one family (family 2) that substantially contributed to this linkage area (13). However, Sanger sequencing of 20 positional candidate genes located in the confined linkage region of these seven families failed to definitively identify the causal mutation in these families. More recently, an exome sequencing dataset was established in these OA families, resulting in the successful identification of a high-impact mutation in one of the families (14). By applying a pathogenic prioritization scheme to the exome sequencing dataset of family 2, we identified a previously unidentified mutation and used hiPSCs and CRISPR-Cas9 technology to develop an in vitro organoid cartilage model to functionally characterize the effects of the causal mutation in this family.
RESULTS
Identification of heterozygous missense mutation in FN1
Whole-exome sequencing was applied to an affected individual of an EO generalized OA family (Fig. 1A, subject 10) with an average age of onset at 37 years and without dysplasia (table S1). This resulted in the identification of 73,407 variants after quality control, after which a prioritization scheme was applied to identify pathogenic variants (table S2). Hence, intergenic variants, intron variants, synonymous variants, and tolerated missense variants as determined by PolyPhen or SIFT (Sorting Intolerant From Tolerant) were excluded, as well as variants present in dbSNP build 124 (including 1000 Genomes pilot project data). In addition, variants were excluded that were previously detected by whole-genome sequencing projects, in-house (N = 221), and by means of the BBMRI-Genome of The Netherlands project (N = 473) (15). As a consequence, the number of variants was reduced to 122 missense variants (table S3) that were predicted to have a functional impact on the gene.
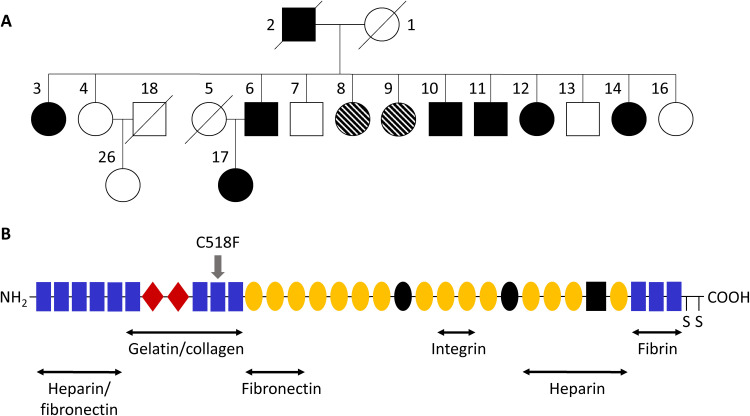
Fig. 1. High-impact mutation identified in FN1 in EO-OA family.
(A) Pedigree of investigated EO-OA family. Individual 10 was used for exome sequencing. Circles represent females, and squares represent males. Closed and open shapes represent affected healthy individuals, and striped shapes represent uncertain phenotypes, i.e., some complaints, but not confirmed by radiography. The diagonal line indicates deceased status. (B) Schematic diagram of a fibronectin subunit. Fibronectin consists of three types of repeats: type I (rectangle), type II (diamond), and type III (sphere). The C-terminal cysteines are involved in dimerization. The alternatively spliced sites are black shapes. Main binding sites for heparin, collagen, fibronectin, and integrin are shown. Position of the identified C518F mutation is indicated by the gray arrow.
Previously, our group showed strong linkage on 2q33.3 with seven extended EO-OA families, including the family described in this study (13). This family showed substantial contribution to this linkage area [LOD (logarithm of odds) score, 2.93]; hence, we investigated the chromosomal location of the previously mentioned 122 missense variants. Of these variants, three were located around the linkage area on chromosome 2, namely, ALS2, FN1, and ABCB6. To explore functionality of these genes in OA-relevant tissue, gene expression levels in our previously published RNA sequencing data of lesioned and preserved cartilage and bone samples of patients undergoing joint replacement surgery due to end-stage OA, from the Research osteoArthritis and Articular Cartilage study (RAAK study), were examined (16, 17). FN1 showed the highest gene expression in both cartilage and bone samples and showed a high, significant up-regulation (fold change, 2.12; false discovery rate, 1.17 × 10−3) in lesioned compared with preserved OA cartilage (Table 1). Furthermore, ALS2 was expressed at low levels in both cartilage and bone samples, while ABCB6 expression was even lower in cartilage and not detected in bone. Therefore, de novo genotyping was performed for the ALS2 and FN1 variant, showing that the ALS2 variant was not detected, yet confirming that the FN1 variant was carried in seven affected individuals, but not in six unaffected family members. These data indicate complete linkage to robustly affected and unaffected subjects in the family (table S4). The FN1 variant was neither present in the additional 1467 OA cases [Genetics Osteoarthritis and Progression (GARP), RAAK, and Patients Prospectively Recruited in Knee and hip Arthroplasty (PAPRIKA) study] and 744 random controls [Leiden Longevity Study (LLS)], nor in the Genome Aggregation Database (v2.1.1) and is therefore likely specific for this EO-OA family. Together, these data indicate that the G-to-T nucleotide change (c1,819) resulting in an amino acid change from cysteine to phenylalanine (C518F; Fig. 1B) in the FN1 gene is likely causal to the EO phenotype in this family.
Table 1. Reproduced gene expression analysis of detected previously unidentified variants located around linkage area 2q33.3 in osteoarthritic cartilage and bone.
AA sub.*, amino acid substitution; Base, base change; FC, fold change; FDR, false discovery rate of differential expression; ND, not detected.
| Cartilage† | Bone† | |||||||||
| Position | Gene | Codon | AA sub. | Base | Quartile | FC | FDR | Quartile | FC | FDR |
| 202590176 | ALS2 | TAT-gAT | Y1084D | A/C | Q3 | 1.08 | 4.95 × 10−1 | Q3 | 0.98 | 8.14 × 10−1 |
| 216285518 | FN1 | TGC-TtC | C518F | C/A | Q4 | 2.12 | 1.17 × 10−3 | Q4 | 1.19 | 6.04 × 10−1 |
| 220075737 | ABCB6 | CGT-gGT | R688G | G/C | Q1 | 1.05 | 8.72 × 10−1 | ND | ND | ND |
*Variants are novel, missense damaging, nonsense, readthrough, frame error, or splice sites not present in dbSNPv124, as well as not detected in in-house genome sequencing projects (LLS; N = 221) and analyzed human genomes by means of the Genome of The Netherlands (N = 473) (15).
†Expression levels in quartiles, with Q1 being the lowest and Q4 being the highest, fold changes, and false discovery rate of differential expression levels of identified variants reproduced from RNA sequencing dataset of articular cartilage (16) and subchondral bone (17).
FN1 encodes fibronectin, which is a dimeric glycoprotein, highly abundant in the extracellular matrix (ECM) of articular cartilage and more specifically localized in the pericellular matrix surrounding the chondrocyte. Fibronectin can bind to matrix proteins by multiple binding domains, such as the gelatin-binding domain, which binds to collagen type II, as well as to cell surface integrins, thereby functioning as an adhesion molecule facilitating signals from the ECM to chondrocytes (18, 19). Next, we aimed to investigate the molecular implications of the identified FN1 mutation for the pathophysiology of OA.
Targeted gene editing of FN1 mutation using CRISPR-Cas9 in hiPSCs
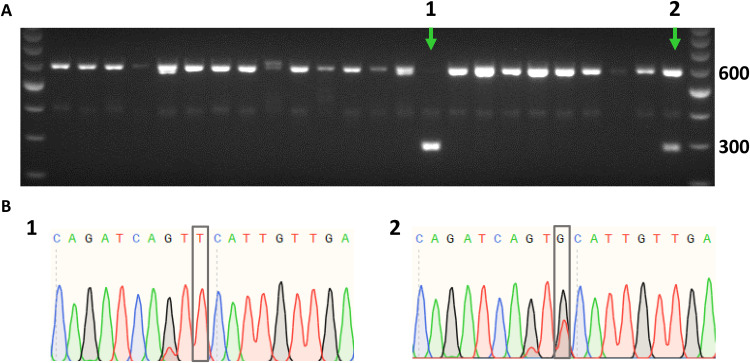
To elucidate the potential pathogenic mechanism of the identified FN1 mutation, we sought to introduce it into wild-type hiPSCs, resulting in an isogenic pair of mutated and nontargeted hiPSCs. Gene editing was achieved by using CRISPR-Cas9, a guide RNA (gRNA) binding in close proximity to the target site, as well as a 141-mer single-stranded oligodeoxynucleotide (ssODN) containing the specific FN1 mutation (table S5). The creation of a Hinc II target site by introducing the mutation was used for screening of successfully targeted clones (Fig. 2A). The presence of the introduced mutation was confirmed by Sanger sequencing (Fig. 2B). A total of 274 hiPSC clones were screened, of which 14 (5.1%) were heterozygous and 3 (1.1%) were homozygous for the FN1 mutation. Last, one heterozygous and one homozygous FN1 mutant clone were selected for further experiments. Both clones had a normal karyotype, and the predicted off-target site allowing two mismatches was not altered (fig. S1).
Fig. 2. Introducing identified FN1 mutation in hiPSCs.
Clonal screen for FN1 mutation in hiPSCs by genomic polymerase chain reaction (PCR) followed by Hinc II digestion (A), confirmed by Sanger sequencing (B). The PCR product is 600 base pairs (bp) in size. Upon integration of the provided ssODN, Hinc II digestion results in 303- and 297-bp products. (A) Representative gel image, where FN1 mutant clones are indicated by a green arrow, showing a homozygous clone (lane 1) and a heterozygous clone (lane 2). (B) Sanger sequencing results of homozygous (1) and heterozygous (2) clones, respectively. Gray boxes represent FN1 mutation.
Effect of missense mutation FN1 on chondrogenesis
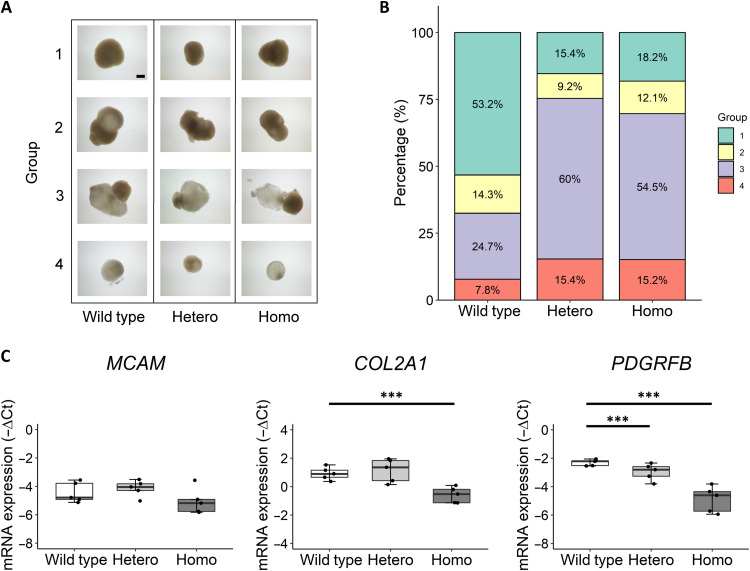
The wild-type, heterozygous, and homozygous FN1 hiPSC clones were differentiated stepwise to the mesodermal lineage and further toward human induced chondroprogenitor cells (hiCPCs). Subsequently, hiCPC aggregates were manually picked, and chondrogenesis was initiated in our organoid cartilage model. After 5 weeks of chondrogenesis, the chondrogenic pellets were collected, where we observed different morphologies of the pellets. We categorized the pellets in groups based on shape (round or protrusive) and opaqueness of the matrix (Fig. 3A). The abundance of round, dense pellets (Fig. 3A, group 1) was 53% of the pellets in the wild-type group, while this decreased substantially to 15% in the heterozygous group and to 18% in the homozygous group (Fig. 3B). Furthermore, the abundance of protrusive pellets with more transparent matrix (Fig. 3A, group 3) in the heterozygous (60%) and homozygous (54%) groups was increased compared with the wild-type group (25%). When considering group 1 as “normal” pellets and groups 2 to 4 as “not-normal” pellets, we found a significant association between the occurrence of not-normal pellets and the presence of the FN1 mutation for both the heterozygous group [Beta = 1.84; 95% confidence interval (CI), 2.8 to 14.1; P = 9 × 10−6] and the homozygous group (Beta = 1.63; 95% CI, 2.4 to 11.1; P = 3.1 × 10−5). These data show that there is a dominant effect of the mutation on the formation of dense, round pellets, thus indicating an inhibited chondrogenic potential of the mutant hiCPCs.
Fig. 3. Decreased chondrogenic potential of hiCPCs as a result of the FN1 mutation.
(A) Representative images of the observed morphology of wild-type, FN1 C518F heterozygous (Hetero), and C518F homozygous (Homo) hiCPC-derived chondrogenic pellets after 5 weeks of chondrogenesis in five independent differentiations. Pellets were categorized in four groups based on shape and opaqueness of the matrix: (1) round and dense, (2) protrusive and dense, (3) protrusive and transparent, and (4) round and transparent. Scale bar, 400 μm. (B) Distribution of pellets in groups 1 to 4 of wild-type (N = 77), FN1 heterozygous (N = 65), and FN1 homozygous pellets (N = 66) (N = 5 to 20 pellets per differentiation). (C) Gene expression levels depicted by −ΔCt values of hiCPC markers MCAM, COL2A1, and PDGFRβ (10) at day 0 of chondrogenesis of pooled wild-type and FN1 mutant hiCPCs (N = 5). The box plots represent 25th, 50th, and 75th percentiles, and whiskers extend up to 1.5 times the interquartile range. Individual samples are depicted by black dots in each graph. P values were determined by generalized estimation equations, with gene expression levels (−ΔCt) as dependent variable and genotype as factor. ***P < 0.001.
To investigate whether the chondrogenic potential of the mutant hiCPCs was also inhibited before the chondrogenesis, we measured expression of the previously identified chondroprogenitor and chondrogenic markers MCAM, PDGFRβ, and COL2A1 of hiCPCs at day 0 of chondrogenesis (Fig. 3C) (10, 20–22). MCAM was not differentially expressed between the groups, yet PDGFRβ and COL2A1 expression was significantly down-regulated in the homozygous group, compared with wild type. Together, these data show that the chondrogenic potential is decreased as a result of the FN1 mutation, with the effect being most pronounced in the homozygous hiCPCs.
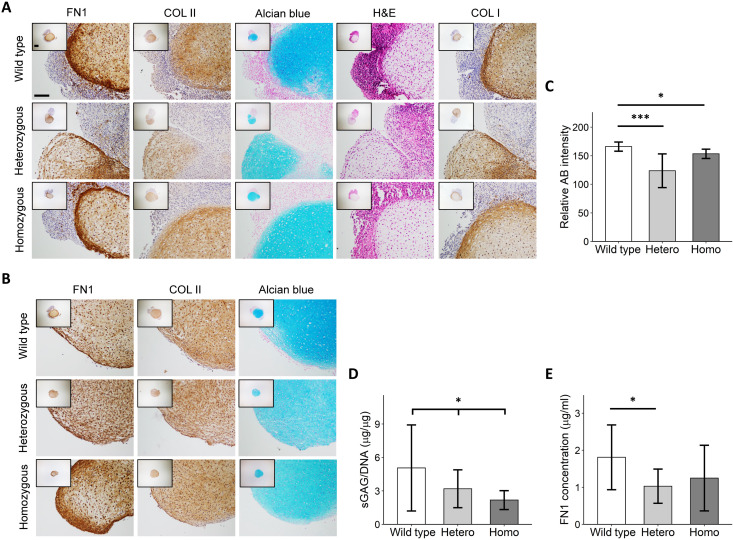
To characterize the differences between the normal and the not-normal chondrogenic pellets, we performed histological and immunohistochemical analyses (Fig. 4A). Hematoxylin and eosin staining of chondrogenic pellets from group 3 showed that there was a clear difference in structure and composition between the dense and transparent matrix. Moreover, Alcian blue, fibronectin, and collagen type I and II staining revealed that the transparent matrix was noncartilaginous tissue. When only considering normal pellets (group 1), visually there was no clear difference in matrix intensity staining of collagen type II and fibronectin (Fig. 4B). However, quantification of the Alcian blue staining in the chondrogenic parts of pellets from groups 1 to 3 revealed a significant decrease in intensity in the FN1 mutant chondrogenic pellets (Fig. 4C). Quantification of the neo-cartilage sulfated glycosaminoglycan (sGAG) deposition normalized to DNA content showed a significant linear reduction (Beta = −1.44; 95% CI, 0.07 to 0.86; P = 2.8 × 10−2) with the presence of the FN1 mutation (Fig. 4D). Moreover, fibronectin concentration was decreased in the medium of the FN1 mutant pellets, albeit not significantly in the homozygous group (Fig. 4E). Together, our data show that the FN1 mutation results in a negative effect on chondrogenic potential and neo-cartilage deposition.
Fig. 4. Decreased neo-cartilage deposition in C518F FN1 mutant chondrogenic pellets.
(A) Representative images of wild-type and FN1 mutant hiCPC-derived chondrogenic pellets categorized in groups 2 to 4 analyzed by hematoxylin and eosin (H&E), Alcian blue, fibronectin (FN1), collagen type I (COL I), and collagen type II (COL II). Scale bar, 200 μm. (B) Representative images of wild-type and FN1 mutant hiCPC-derived chondrogenic pellets categorized in group 1 analyzed by Alcian blue, fibronectin (FN1), and collagen type II (COL II). Scale bar, 200 μm. (C) Quantification of Alcian blue (AB) staining of Alcian blue–positive areas in wild-type and FN1 mutant hiCPC-derived chondrogenic pellets. (N = 3). (D) Dose-response reduction (Beta = −1.44, P = 2.8 × 10−2) of sGAG normalized to DNA in wild-type, FN1 heterozygous (Hetero), and homozygous (Homo) chondrogenic pellets analyzed by dimethylmethylene blue (N = 8). (E) Fibronectin concentration in conditioned medium from FN1 mutant pellets is decreased compared with wild-type, as determined by enzyme-linked immunosorbent assay (N = 3) . Data are means ± SD. P values were determined by generalized estimation equations, with experimental readout (relative Alcian blue intensity, sGAG/DNA, and fibronectin concentration) as dependent variable and genotype as covariate (sGAG/DNA) or factor (relative Alcian blue intensity and fibronectin concentration). *P < 0.05 and ***P < 0.005.
Mechanism of reduced chondrogenesis in mutant chondrogenic pellets
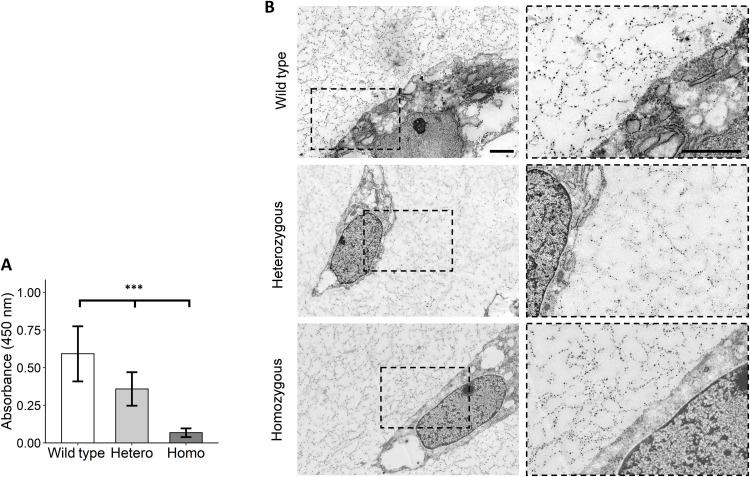
The identified C518F mutation is located in the gelatin-binding domain of fibronectin, which is particularly involved in binding to collagen type II (19). The cysteine is involved in intramolecular disulfide bond formation and was shown to be highly conserved among multiple species, highlighting the importance of this amino acid (fig. S2A). The change from a polar cysteine to a nonpolar phenylalanine resulted in a predicted conformational change of the protein as determined by RaptorX (fig. S2B) (23). When replaced by the phenylalanine, the disulfide bond in this part of the domain cannot be formed. Therefore, we hypothesized that the mutation resulted in a conformational change in the gelatin-binding domain, with concurrent decreased binding of fibronectin to collagen type II. To explore this, we performed a collagen type II binding assay with wild-type, heterozygous, and homozygous mutant fibronectin. As a result, we observed a linear reduction (Beta = −0.26; 95% CI, 0.71 to 0.84; P = 2.3 × 10−9) in binding of mutant fibronectin to collagen type II relative to wild-type fibronectin, thereby confirming our hypothesis (Fig. 5A). To further investigate whether we could identify structural differences in the neo-cartilage as a result of this decreased binding between fibronectin and collagen type II, we visualized cartilage by transmission electron microscopy. We observed electron dense particles that were present both attached to the plasma membrane and to a branching fibrillar network obviously devoid of striations emanating from the plasma membrane. The fibrillar network suggests a network of hyaluronic acid aggregates (24), and the electron dense particles are proteoglycans containing granules (Fig. 5B) (25). However, we could not identify any clear differences in the abundance and structure of observed structures between the wild-type and FN1 mutant.
Fig. 5. FN1 mutant fibronectin showed a dose-response decrease in binding capacity to collagen type II.
(A) Solid-phase binding assay with collagen type II–coated wells and wild-type and C518F mutant fibronectin isolated from chondrogenic pellet cultures. Heterozygous (Hetero) and homozygous (Homo) mutant fibronectin showed linear reduction (Beta = −0.26, P = 2.26 × 10−9) of binding to collagen type II compared to wild-type fibronectin. Data are means ± SD (N = 3). P value was determined by generalized estimation equations, with absorbance as dependent variable and genotype as covariate. ***P < 0.005. (B) Representative transmission electron microscopy images showing a cell surrounded by cartilaginous matrix of wild-type and FN1 mutant hiCPC-derived chondrogenic pellets. The fibrillar network surrounding the cells is suggestive of hyaluronic acid aggregates, and the dense particles are proteoglycans containing granules. Scale bars, 2 μm.
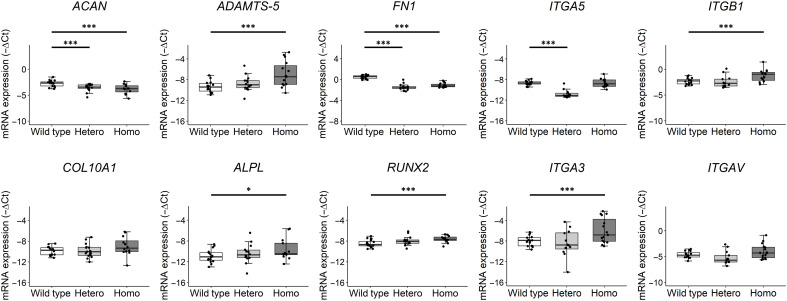
To study the effect of the mutation on markers reflecting chondrocytes in health or disease state, we next explored chondrocyte gene expression in the pellets as a result of the mutation by performing reverse transcription quantitative polymerase chain reaction (RT-qPCR) for 10 relevant genes. Relative gene expression levels were first calculated by normalizing to expression levels of GAPDH and SDHA (fig. S3). The observed difference in formation of neo-cartilage between the genotypes (Fig. 4A) was potentially a confounding factor in the gene expression analysis. Therefore, we aimed to correct for the differences in efficiency of the neo-cartilage formation between the genotypes. Hence, we selected cartilage-specific markers that were highly expressed in our lesioned and preserved osteoarthritic cartilage samples (RAAK study), but without significant differential expression (16), and that we showed to be specific markers for the neo-cartilage (Fig. 4A). As a result, COL1A1 and COL2A1 were selected as reference genes. The anabolic cartilage markers ACAN and FN1 were significantly down-regulated in the FN1 mutant chondrocytes as compared with wild type, while the catabolic and hypertrophic markers ADAMTS-5, ALPL, and RUNX2 were significantly up-regulated in the FN1 homozygous group (Table 2 and Fig. 6). These data indicate that the mutant chondrocytes were in a more OA disease state, where the effect was stronger in the homozygous mutant group. To investigate the downstream effects of the mutation on the fibronectin-binding chondrocyte cell surface integrins, we investigated the gene expression levels of ITGA5, ITGB1, ITGA3, and ITGAV. Integrin α5β1 is the main fibronectin-binding integrin, while α3β1 and αVβ1 can also bind fibronectin (26). ITGA3 and ITGB1 were significantly up-regulated in FN1 homozygous chondrocytes (Fig. 6). Notably, ITGA5 was down-regulated in the heterozygous group, while its expression was not changed in the homozygous group.
Table 2. Differences in gene expression between C518F FN1 heterozygous and homozygous hiCPC-derived chondrogenic pellets compared with wild type after 5 weeks of chondrogenesis.
| HETERO vs. WT* | HOMO vs. WT* | |||||||
| Gene | FC | Beta | SE | P | FC | Beta | SE | P |
| ACAN | 0.57 | −0.81 | 0.23 | 4.52 × 10−4 | 0.57 | −1.10 | 0.24 | 2.62 × 10−4 |
| ADAMTS-5 | 1.55 | 0.63 | 0.48 | 1.93 × 10−1 | 5.14 | 2.36 | 0.68 | 5.45 × 10−4 |
| ALPL | 1.41 | 0.50 | 0.56 | 3.79 × 10−1 | 2.45 | 1.30 | 0.63 | 4.03 × 10−2 |
| COL10A1 | 1.03 | 0.05 | 0.41 | 9.11 × 10−1 | 1.86 | 0.90 | 0.56 | 1.09 × 10−1 |
| FN1 | 0.25 | −1.98 | 0.16 | 1.00 × 10−4 | 0.33 | −1.59 | 0.13 | 1.00 × 10−4 |
| ITGA3 | 1.18 | −0.41 | 0.77 | 5.93 × 10−1 | 3.91 | 1.97 | 0.65 | 2.51 × 10−3 |
| ITGA5 | 0.23 | −2.13 | 0.23 | 1.00 × 10−4 | 1.01 | 0.02 | 0.24 | 9.34 × 10−1 |
| ITGAV | 0.66 | −0.60 | 0.37 | 1.03 × 10−1 | 1.53 | 0.61 | 0.37 | 1.03 × 10−1 |
| ITB1 | 0.92 | −0.12 | 0.33 | 7.20 × 10−1 | 2.09 | 1.07 | 0.33 | 1.11 × 10−3 |
| RUNX2 | 1.42 | 0.50 | 0.28 | 6.75 × 10−2 | 1.79 | 0.84 | 0.26 | 9.48 × 10−4 |
*Fold change (FC), Beta value, SE, and P value for expression levels (−ΔCt) of 10 genes as determined by generalized estimation equations, with gene expression levels (−ΔCt) as dependent variable and genotype as factor, when comparing FN1 heterozygous (HETERO) and FN1 homozygous (HOMO) pellets to wild-type (WT) pellets.
Fig. 6. FN1 mutation results in aberrant chondrocyte gene expression.
Box plots of −ΔCt values of ACAN, ADAMTS-5, FN1, ITGA5, ITGB1, COL10A1, ALPL, RUNX2, ITGA3, and ITGAV in wild-type and C518F FN1 heterozygous (Hetero) and homozygous (Homo) chondrogenic pellets. −ΔCt values shown were corrected for COL1A1 and COL2A1 expression levels to correct for the difference in efficiency of chondrogenic differentiation between genotypes. The box plots represent 25th, 50th, and 75th percentiles, and whiskers extend to 1.5 times the interquartile range. Individual samples are depicted by black dots in each graph. P values were determined by generalized estimation equations, with gene expression levels (−ΔCt) as dependent variable and genotype as factor. *P < 0.05 and ***P < 0.005
DISCUSSION
In the current study, we identified a heterozygous missense mutation (c1,819 G> T; C518F) in FN1 that is likely causal to the EO phenotype in an extended EO-OA family (Fig. 1). By introducing this high-impact mutation in hiPSCs using CRISPR-Cas9 gene editing and using them in an established in vitro organoid cartilage model, we showed that the chondrogenic potential (Fig. 3) and deposition of neo-cartilage (Fig. 4) of the FN1 mutant chondrocytes were decreased. Moreover, we demonstrated that the underlying pathogenic mechanism of the mutation was caused by a decreased binding of fibronectin to the surrounding ECM protein collagen type II (Fig. 5). As a downstream effect, the formed neo-cartilage was prone to a hypertrophic and procatabolic (OA) state, as reflected by the changed chondrocyte gene expression pattern, particularly of ADAMTS-5 and RUNX2 (Fig. 6). This is further confirmed by the up-regulation of ITGA3 and ITGB1, which is comparable to an OA disease state of chondrocytes (27). Since fibronectin is an important adhesion protein between ECM and chondrocytes via binding to the cell surface integrins, we advocate that the identified C518F mutation in fibronectin hampers essential interaction between chondrocytes and the ECM; hence, plasticity of chondrocytes to adequately respond to cues from the ECM is decreased. In osteoarthritic joints, it has been shown that fibronectin is degraded proteolytically into fragments (FN-fs), which amplify catabolic processes in articular cartilage (28–31). These FN-fs have distinct activities due to different binding capacities and thereby lead to changes in communication between the ECM and the chondrocyte. Therefore, it is tempting to speculate that the phenomenon of changed binding of fibronectin with ECM and chondrocytes could be directly contributing to the onset of age-related OA in the population.
One of the findings in this study is that the FN1 mutant hiCPCs compared to wild type had both reduced chondrogenic potential (Fig. 3). These results are in line with previous findings showing that the presence of fibronectin matrix is essential for mesenchymal stromal cell condensation and chondrogenic differentiation (32, 33). The results of our study therefore suggest that decreased binding between fibronectin and collagen type II may negatively affect these processes. To elucidate the exact mechanism during differentiation, longitudinal analysis of the differentiation would have to be performed in future studies.
The less efficient formation of dense, round pellets as a result of the mutation (Fig. 3), hence variable quantity of neo-cartilage formed in the FN1 mutant pellets, was a confounding factor in the gene expression data analyses. We observed a lower amount of cartilage-producing cells relative to the total amount of cells in FN1 mutated pellets compared with wild type, as well as a decrease in the deposition of neo-cartilage (Fig. 4). As a result, an overall stepwise decrease in cartilage-specific gene expression was observed in the FN1 mutant pellets. To adjust for the relatively lower amount of cartilage formed in the FN1 mutant chondrogenic pellets, we used the cartilage-specific markers COL2A1 and COL1A1 as reference genes. We chose COL2A1 and COL1A1 for their typically high expression in cartilage, as well as their stability in ongoing OA pathophysiology, as reflected by a previously published large RNA sequencing dataset of autologous cartilage (16), and that we showed to be specific markers for the neo-cartilage (Fig 4A). Although we are confident that the observed significant increase in ADAMTS-5, ALPL, and RUNX2 and the significant decrease in ACAN and FN1 expression reflected true biological effects (Fig. 6), the accuracy of the fold changes provided may be affected. There are different possibilities to overcome this problem in the future. Recently, Adkar et al. (10) engineered a knock-in COL2A1-GFP reporter system in hiPSCs to purify chondroprogenitors, consequently improving chondrogenic capacity during differentiation. To be able to further investigate the effect of the mutation on chondrogenesis without the decrease in chondrogenic potential, the COL2A1 reporter system could be used for our FN1 OA disease model to circumvent this effect. On the other hand, single-cell RNA sequencing could identify the chondroprogenitor subpopulation in the heterogeneous pellets, so that gene expression analysis of only that subpopulation would be possible (21).
Notably, even after correcting for COL2A1 and COL1A1 as cartilage-specific markers, FN1 and ACAN expression was significantly decreased in FN1 mutant pellets, likely affecting quality of deposited cartilage matrix. Moreover, ITGA3 and ITGB1 were significantly up-regulated in the FN1 homozygous pellets. Since α3β1 integrin was shown to be up-regulated in OA chondrocytes (27), the here observed switch from α5β1 to α3β1 integrins in FN1 homozygous chondrocytes may reflect an unbeneficial disease state of the FN1 homozygous chondrocytes (27). Conversely, ITGA5 was down-regulated only in the FN1 heterozygous pellets, which could have been a direct result of the formation of wild type–mutant fibronectin dimers, thereby also potentially being responsible for the decreased FN1 and ACAN expression. Recently, it has been shown that the fibronectin-α5β1 adhesion is essential for cartilage regeneration in mice (34). These data indicate that the mutation results in aberrant chondrocyte gene expression, potentially via a change in integrin subunit expression levels toward a more hypertrophic, catabolic state, possibly due to decreased binding between fibronectin and collagen type II. However, this should be confirmed by more investigative studies on protein level, e.g., by Western blot to quantify protein levels of the integrin subunits.
Previous studies regarding familial, high-impact FN1 mutations have been associated with glomerulopathy with fibronectin deposits (GDND), which is a hereditary kidney disease with proteinuria, microscopic hematuria, and hypertension, as well as spondylometaphyseal dysplasia with “corner fractures” (35, 36). These mutations were located in multiple heparin-binding domains, which play an essential role in regulating fibronectin assembly into organized fibrils in ECM. When comparing the phenotypes associated to the FN1 mutations, it is notable that the phenotype in our family is less severe since glomerulopathy results in renal failure and the spine and growth plates are affected in spondylometaphyseal dysplasia, resulting in, among others, shortened trunk and scoliotic posture. Apparently, a mutation in the gelatin-binding domain results in a more specific pathology involving only the joints. The previously identified mutations also often involved cysteine residues, confirming that proper folding of fibronectin, e.g., via disulfide bonds, is crucial for the binding of fibronectin to other matrix components. Together, these data highlight the importance of proper binding of fibronectin to collagen in articular cartilage.
Together, by combining exome sequencing and linkage analysis, we demonstrated that the identified FN1 mutation is causal to human primary OA at an age of onset around 20 to 40 years. Moreover, the mutation does not cause severe developmental aberrations; yet, it has sufficient impact on cartilage tissues to allow measurable functional in vitro effects. Furthermore, by precise genetic engineering of a registered hiPSC line without off-target effects (fig. S1), we established both a hetero- and homozygous isogenic clone, with the OA causing FN1 mutation (Fig. 2). By subsequently applying an established differentiation protocol able to produce biomimetic human neo-cartilage, we generated multiple reliable biological replicates on how aberrant function of fibronectin in cartilage is causal to OA onset. Hence, we are confident that our approach was able to create reliable data highly translating to the human in vivo situation while contributing to the societal need to reduce animal studies.
In conclusion, our approach highlights the immense potential of combining exome sequencing, hiPSCs, CRISPR-Cas9, and organoid disease modeling to uncover previously unknown underlying disease mechanisms that are readily extrapolated to novel therapeutic avenues in common, complex human genetic diseases. Particularly, we here show that the underlying pathogenic mechanism of the mutation was caused by a decreased binding of fibronectin to the surrounding ECM protein collagen type II. Moreover, as reflected by the changes in chondrocyte gene expression, particularly of ADAMTS-5 and RUNX2, such impaired binding results in the cartilage becoming prone to enter an OA disease state. In addition, we observed a switch in expression levels of the main fibronectin-binding receptor integrin α5β1 to integrin α3β1, reflecting a more OA disease state of the chondrocytes. Together, our work merits further exploration of fibronectin as potential target for therapeutic interventions. We advocate that restoring or maintaining proper binding between fibronectin and collagen type II should be the focus of such a quest.
MATERIALS AND METHODS
Experimental design
The objective of the current study was to identify a likely causal pathogenic mutation in an EO-OA family and underlying disease pathways by functional analyses of isogenic hiPSCs. Exome sequencing was applied to identify a pathogenic mutation, whereas linkage analysis was applied to confirm cosegregation of the mutation and the OA phenotype. To study underlying mechanisms, the mutation was introduced in hiPSCs using CRISPR-Cas9 genome engineering, after which they were used in an in vitro organoid cartilage model, followed by functional analyses. A solid-phase binding assay was performed to quantify binding capacity of mutant and wild-type fibronectin to collagen type II.
Study populations
EO-OA family members
Previously, we reported on an EO family with primary generalized OA without dysplasia (fig. S4) (13, 37). The proband was collected through a rheumatology outpatient clinic; family members were recruited via the proband. The age of onset of OA was between 23 and 45, and the phenotype is characterized by progressive OA with symptoms at multiple joints simultaneously. Two family members reported OA-related complaints; however, no radiographic evidence was available to confirm the diagnosis. An overview of the phenotypes is shown in table S1. The study was approved by the medical ethical committee, and written and informed consent was obtained from all participants.
Controls
Occurrence of the FN1 missense mutation was tested in the general population by de novo genotyping in healthy participants and OA cases. Middle-aged partners (N = 744) of the offspring of nonagenarian siblings from the LLS (38) were considered as controls for OA cases and were named “random controls.” OA cases included were selected from the GARP study (N = 177) (39), the PAPRIKA study (N = 1137) (40), and the ongoing RAAK study (N = 153) (41). For more details on ascertainment of RAAK study biospecimen, see Ramos et al. (14). Ethical permission for the described studies was obtained from the appropriate medical ethical committee under protocol numbers P08.239 and P19.013. Written informed consent was obtained from all participants.
Exome sequencing
Exome sequencing of an affected EO-OA family member was performed by Illumina HiSeq 2000 technology (Beijing Genome Institute). The sequences were generated as 100–base pair paired-end reads, after enrichment of 44-Mb exonic sequences by NimbleGen EZ (Roche NimbleGen). Raw imaging files were processed by Illumina base calling software v1.7 with default parameters. SOAPaligner/SOAP2.21 was used to align reads to the GRCh37 reference genome at the UCSC Genome Browser website (http://genome.ucsc.edu/). Approximately 70% of bases originated from the targeted exome, resulting in a mean coverage of 55.7-fold for the proband. More than 83% of the targeted exons were covered more than 10 times. Single-nucleotide variants were subsequently called by the SOAPsnps. The variant-filtering scheme, which resulted in 122 pathogenic mutations, is detailed in table S2.
Genotyping
Single-nucleotide polymorphisms were genotyped by mass spectrometry (MassARRAY System; Sequenom, San Diego, CA) using standard conditions as described by Meulenbelt et al. (42). Variants were genotyped across available EO-OA family members (see table S4), OA case studies (GARP, PAPRIKA, and RAAK study), and random controls (LLS).
hiPSC line and cell culture
The hiPSC line LUMC0004iCTRL10 (“LUMC0004” here) was generated from skin fibroblasts of a male donor without known genetic diseases (“wild type”), as described previously (43), and registered at the Human Pluripotent Stem Cell Registry. The generation of the hiPSC line was approved by the Leiden University Medical Center ethical committee under P13.080. The hiPSCs were maintained under standard conditions (37°C, 5% CO2) on Vitronectin-XF (STEMCELL Technologies)–coated plates and refreshed daily with TeSR-E8 medium (STEMCELL Technologies). Cells were passaged in aggregates using Gentle Cell Dissociation Reagent (STEMCELL Technologies) upon reaching approximately 80% confluency.
Genome editing of hiPSCs and clonal isolation
hiPSCs were dissociated with Gentle Cell Dissociation Reagent (STEMCELL Technologies), and 1 × 105 cells were transfected with the Alt-R Cas9 RNP complex and the ssODN (both IDT) using the Neon Transfection System (Invitrogen) at 1200 V/30 ms/1 pulse (for gRNA and ssODN sequence, see table S5). Cells were plated in a Synthemax II-SC (Corning)–coated plate using TeSR-E8 supplemented with CloneR (STEMCELL Technologies). After recovery, 1000 cells were plated in a Synthemax II-SC–coated 10-cm dish in TeSR-E8 supplemented with CloneR and refreshed daily. After ~14 days, single cell–derived hiPSC colonies were manually picked, and one-half was used for DNA isolation with QuickExtract Solution (Lucigen). Positive clones were identified by PCR screening of the region of interest, followed by successful digestion by restriction enzyme Hinc II (New England Biolabs), according to the manufacturer’s instructions. Primer sequences are shown in table S6. The introduced mutation was confirmed by Sanger sequencing, performed by the Leiden Genome Technology Center.
Characterization of gene-edited hiPSCs
In silico analysis with the Cas-OFFinder tool (44) was used to identify most likely sites for off-target mutations. When allowing two mismatches, one potential off-target was identified, which was sequenced as described above (for primer sequences, see table S6). G-banding analysis was conducted at the Laboratory of Clinical Genetics Leiden according to standard procedures. A total of 18 metaphases were analyzed for one heterozygous clone, and 20 metaphases were analyzed for one homozygous clone.
hiPSC differentiation to induced chondroprogenitor cells
Generation of induced chondroprogenitor cells (hiCPCs) was based on a protocol previously described (10), which was shown to produce similar neo-cartilage to that produced by human primary articular chondrocytes (11). When hiPSCs reached 60% confluence, the culture medium was switched to mesodermal differentiation (MD) medium, composed of IMDM GlutaMAX (IMDM; Thermo Fisher Scientific) and Ham’s F12 Nutrient Mix (F12; Sigma-Aldrich) with 1% chemically defined lipid concentrate (Gibco), 1% insulin/human transferrin/selenous (ITS+; Corning), 0.5% penicillin-streptomycin (P/S; Gibco), and 450 μM 1-thioglycerol (Sigma-Aldrich). Before induction of anterior primitive streak (day 0), hiPSCs were washed with wash medium (IMDM/F12 and 0.5% P/S) and then fed with MD medium supplemented with activin A (30 ng/ml; Stemgent), 4 μM CHIR99021 (CHIR; Stemgent), and human fibroblast growth factor (20 ng/ml; FGF-2; R&D Systems) for 24 hours. Subsequently, the cells were washed again with wash medium, and paraxial mesoderm was induced on day 1, by MD medium supplemented with 2 μM SB-505124 (Tocris), 3 μM CHIR, FGF-2 (20 ng/ml), and 4 μM dorsomorphin (Tocris) for 24 hours. Before induction of early somite (day 2), cells were washed with wash medium, and then cells were fed with MD medium supplemented with 2 μM SB-505124, 4 μM dorsomorphin, 1 μM C59 (Cellagen Technology), and 500 nM PD173074 (Tocris) for 24 hours. Subsequently, cells were washed with wash medium, and for induction of sclerotome, cells (days 3 to 5) were fed daily with MD medium supplemented with 2 μM purmorphamine (Stemgent) and 1 μM C59. To induce chondroprogenitor cells (days 6 to 14), cells were washed briefly with wash medium and fed daily with MD medium supplemented with human bone morphogenetic protein 4 (BMP-4; 20 ng/ml; Miltenyi Biotec). Five independent differentiations were done per clone.
Chondrogenic differentiation
Monolayer cultured hiCPC aggregates present at day 14 of the differentiation were washed with MD medium, manually picked, and centrifuged for 4 min at 1200 rpm. Cell aggregates were subsequently maintained in Dulbecco’s modified Eagle’s medium/F12 (Gibco), supplemented with 1% ITS+, 55 μM 2-mercaptoethanol (Gibco), 1% non-essential amino acids (Gibco), 0.5% P/S, l-ascorbate-2-phosphate (50 μg/ml; Sigma-Aldrich), l-proline (40 μg/ml; Sigma-Aldrich), and transforming growth factor–β1 (10 ng/ml; PeproTech) for 35 days while refreshing medium every 3 to 4 days. The remaining hiCPC aggregates in the well were pooled and lysed with 500 μl of TRIzol reagent (Thermo Fisher Scientific) and stored at −80°C until further processing.
sGAG measurement
sGAG concentration in the chondrogenic pellets was measured with the 1,9-dimethylmethylene blue (DMMB) assay (45). Pellets were digested with papain from papaya (Sigma-Aldrich) at 60°C overnight. Shark chondroitin sulfate (Sigma-Aldrich) was used as a reference standard. The absorbance was measured at 525 and 595 nm in a microplate reader (SpectraMax iD3; Molecular Devices).
Histology and immunohistochemistry
Chondrogenic pellets were fixed in 4% formaldehyde overnight and stored in 70% ethanol at 4°C. They were then embedded in paraffin and sectioned at 5 μm. After sectioning, slides were deparaffinized and rehydrated. Overall cellular and tissue structure was visualized with hematoxylin and eosin staining. Sections were stained with 1% Alcian blue 8GX (Sigma-Aldrich) and Nuclear Fast Red (Sigma-Aldrich) to visualize glycosaminoglycan deposition. To detect fibronectin (1:400; ab2413, Abcam), collagen type I (1:1000; ab34710, Abcam), and collagen type II (1:100; ab34712, Abcam), immunohistochemistry was performed with 3-diaminobenzidine (DAB) solution (Sigma-Aldrich) and hematoxylin (Klinipath) as described before (46). Alcian blue staining was quantified by loading the images in Fiji and splitting the color channels. Subsequently, gray values were measured of three to five separate squares per pellet and corrected for the gray value of the background.
RNA isolation and RT-qPCR
For RNA isolation, two pellets were pooled and lysed in 200 μl of TRIzol reagent (Thermo Fisher Scientific) and homogenized using micro pestles. RNA was extracted with chloroform and purified from the supernatant using the RNeasy Mini Kit (QIAGEN). Synthesis of cDNA was performed with 150 ng of total RNA using a First Strand cDNA Synthesis kit according to the manufacturer’s protocol (Roche Applied Science). cDNA was diluted five times, and preamplification with TaqMan PreAmp master mix (Thermo Fisher Scientific) was performed for eight genes of interest. Primer sequences are shown in table S6. Gene expression was measured with the Fluidigm Biomark HD machine using a 96.96 IFC (Integrated Fluidic Circuit) chip or with QuantStudio 6 Real-Time PCR system (Applied Biosystems) using FastStart SYBR Green Master reaction mix (Roche Applied Science). Quality control of the data was performed, and nondetected values were removed. Relative gene expression levels were calculated with the 2−∆∆Ct method as follows: The average of triplicate raw cycle threshold (Ct) values of the genes were averaged. Ct values of GAPDH and SDHA were averaged to function as reference genes (RG). Next, ΔCt values of the gene of interest (GOI) were calculated as CtGOI − CtRG. Subsequently, we averaged the ΔCt values of COL1A1 and COL2A1 as chondrogenic controls (CC) to calculate final ΔCt values of the GOI as follows: ΔCtGOI new = ΔCtGOI − ΔCtCC. Fold changes were calculated with 2−∆∆Ct, with the wild type as reference.
Solid-phase binding assay
Conditioned medium of wild-type and FN1 mutant pellets was collected and concentrated in preparation for the binding assay. To this end, 450 μl of medium was collected in 100 K molecular weight cutoff Pierce Protein Concentrators (Thermo Scientific) and centrifuged for 10 min at 12,000g. Subsequently, fibronectin concentration was determined using the Human Fibronectin ELISA Kit (Invitrogen) according to the manufacturer’s protocol.
Clear multiwell plates (R&D Systems) were coated overnight with 100 μl of purified human collagen type II (10 μg/ml; Merck) in phosphate-buffered saline (PBS) at 4°C, followed by four wash steps with wash buffer (0.05% Tween 20 in PBS). Nonspecific binding was blocked for 1 hour with 3% (w/v) bovine serum albumin (BSA) in PBS. After washing with wash buffer, the plates were incubated with 100 μl of concentrated medium samples at FN1 concentration of 40 ng/ml in assay buffer (0.05% Tween 20 and 0.5% BSA in PBS) for 2 hours. Plates were then washed four times with wash buffer and incubated with rabbit anti-fibronectin biotin-conjugated antibody (Rockland) at 0.2 μg/ml in assay buffer for 1 hour. Plates were washed, after which the plates were incubated with streptavidin–horseradish peroxidase (Thermo Scientific) at 0.1 μg/ml in assay buffer for 1 hour. After washing, color development was performed with 100 μl of tetramethylbenzidine substrate (Thermo Fisher Scientific) for 10 min, reaction was stopped with 100 μl of 1 M HCl, and absorbance was measured at 450 nm. Assays were performed in triplicate. Error bars are SDs.
Transmission electron microscopy
Chondrogenic pellets that where embedded in paraffin were melted from the paraffin for 30 min at 62°C. Subsequently, the samples were deparaffinized and rehydrated with a series of ethanol. The rehydrated samples were fixed again in 1.5% glutaraldehyde/0.1 M cacodylate buffer, and after 3 times rinsing with 0.1 M cacodylate buffer, postfixed in 1% OsO4/0.1 M cacodylate buffer. A dehydration with a series of ethanol and a series of propylene oxide mixed with Epon (LX112, Ladd Research Industries) was performed. After a final step with Epon, the samples were each placed in a BEEM capsule; these were filled with Epon and polymerized at 70°C during 48 hours. Ultrathin sections (90 nm) were made on a Reichert Ultracut S (Leica Microsystems). After the poststaining of the sections with uranyl acetate and lead citrate, the electron microscopy images were taken with a twin transmission electron microscope (Thermo Fisher Scientific, formerly FEI) operating at 120 kV, using a Gatan OneView camera (Gatan, Pleasanton) on binning 2. Overlapping images were collected and stitched together into a big image as previously described (47).
Statistical analysis
Statistical analyses were performed using SPSS version 25 (IBM). Data are shown as means ± SD. With respect to RT-qPCR gene expression data, individual −∆Ct values are shown, as well as box plots per genotype. The box plots represent 25th, 50th, and 75th percentiles, and whiskers represent the lowest and largest data point lying within 1.5 times the interquartile range. Individual samples are depicted by dots in each graph. The reported P values were determined by applying a generalized estimating equation (GEE) to the experimental readout, i.e., −∆Ct values, relative Alcian blue intensity, and FN1 concentration, to effectively adjust for dependencies of the independent differentiations per genotype. In general, we followed a linear GEE model, with the readout data as dependent variable, genotype as factor, and exchangeable working matrix: Read-out ~ Genotype + (1| Differentiation). To determine a dose-response effect, we applied a GEE to the experimental readout, i.e., absorbance and sGAG/DNA, with the readout as dependent variable, genotype as covariate, and exchangeable working matrix. For the analysis of the pellet morphology, we followed a binary logistic GEE model, with normal and not-normal pellets as readout and genotype as factor, and adjusted for independent differentiations. P values <0.05 were considered statistically significant. *P < 0.05, **P < 0.01, and ***P < 0.005.
Acknowledgments
We thank C. Mummery for the initial help with the hiPSC work and M. Gonçalves for the help with the design of the CRISPR-Cas9 components. We thank all the members of the MolEpi Osteoarthritis group for valuable discussion and feedback: E. Houtman, R. Timmermans, N. Bloks, I. Boone, N. Korthagen, and G. Hajmousa. We specifically thank R. Coutinho de Almeida and M. Tuerlings for providing the RNA sequencing data of lesioned and preserved OA cartilage and bone.
Funding: Dutch Scientific Research council NWO/ZonMW VICI scheme grant number 91816631/528 (I.M.), Dutch Arthritis Society grant numbers DAF-16-1-405 and DAF-16-1-406 (I.M.), and U.S. NIH grant number AG15768 (F.G.). Data were generated within the scope of the Medical Delta Programs Regenerative Medicine 4D: Generating complex tissues with stem cells and printing technology and improving mobility with technology.
Author contributions: M.v.H., A.R.R., Y.F.M.R., and I.M. developed the concept of this study; M.v.H., A.R.R., M.B., A.A.M., C.F., H.E.D.S., and F.G. acquired materials and data; M.v.H., A.R.R., R.I.K., C.R.J., A.A.M., Y.F.M.R., and I.M. analyzed the data; and all authors contributed to the writing of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The hiPSC line can be provided by I.M. pending scientific review and a completed material transfer agreement. Requests for the hiPSC line should be submitted to I.M.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 to S6
REFERENCES AND NOTES
- 1.Goldring S. R., Goldring M. B., Changes in the osteochondral unit during osteoarthritis: Structure, function and cartilage-bone crosstalk. Nat. Rev. Rheumatol. 12, 632–644 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Conaghan P. G., Kloppenburg M., Schett G., Bijlsma J. W.; EULAR osteorthritis and hoc committee , Osteoarthritis research priorities: A report from a EULAR ad hoc expert committee. Ann. Rheum. Dis. 73, 1442–1445 (2014). [DOI] [PubMed] [Google Scholar]
- 3.van der Kraan P. M., Berenbaum F., Blanco F. J., Cosimo de B., Lafeber F., Hauge E., Higginbottom A., Ioan-Facsinay A., Loughlin J., Meulenbelt I., Moilanen E., Pitsillidou I., Tsezou A., van Meurs J., Vincent T., Wittoek R., Lories R.; EULAR study group in OA , Translation of clinical problems in osteoarthritis into pathophysiological research goals. RMD Open 2, e000224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litwic A., Edwards M. H., Dennison E. M., Cooper C., Epidemiology and burden of osteoarthritis. Br. Med. Bull. 105, 185–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirulli E. T., Goldstein D. B., Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 11, 415–425 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Rabbani B., Mahdieh N., Hosomichi K., Nakaoka H., Inoue I., Next-generation sequencing: Impact of exome sequencing in characterizing Mendelian disorders. J. Hum. Genet. 57, 621–632 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Goldstein D. B., Allen A., Keebler J., Margulies E. H., Petrou S., Petrovski S., Sunyaev S., Sequencing studies in human genetics: Design and interpretation. Nat. Rev. Genet. 14, 460–470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adkar S. S., Brunger J. M., Willard V. P., Wu C. L., Gersbach C. A., Guilak F., Genome engineering for personalized arthritis therapeutics. Trends Mol. Med. 23, 917–931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S., Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Adkar S. S., Wu C. L., Willard V. P., Dicks A., Ettyreddy A., Steward N., Bhutani N., Gersbach C. A., Guilak F., Step-wise chondrogenesis of human induced pluripotent stem cells and purification via a reporter allele generated by CRISPR-Cas9 genome editing. Stem Cells 37, 65–76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez Ruiz A., Dicks A., Tuerlings M., Schepers K., van Pel M., Nelissen R., Freund C., Mummery C. L., Orlova V., Guilak F., Meulenbelt I., Ramos Y. F. M., Cartilage from human-induced pluripotent stem cells: Comparison with neo-cartilage from chondrocytes and bone marrow mesenchymal stromal cells. Cell Tissue Res. 10.1007/s00441-021-03498-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meulenbelt I., Min J. L., van Duijn C. M., Kloppenburg M., Breedveld F. C., Slagboom P. E., Strong linkage on 2q33.3 to familial early-onset generalized osteoarthritis and a consideration of two positional candidate genes. Eur. J. Hum. Genet. 14, 1280–1287 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Ramos Y. F., Bos S. D., van der Breggen R., Kloppenburg M., Ye K., Lameijer E. W., Nelissen R. G., Slagboom P. E., Meulenbelt I., A gain of function mutation in TNFRSF11B encoding osteoprotegerin causes osteoarthritis with chondrocalcinosis. Ann. Rheum. Dis. 74, 1756–1762 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Kiezun A., Pulit S. L., Francioli L. C., van Dijk F., Swertz M., Boomsma D. I., van Duijn C. M., Slagboom P. E., van Ommen G. J., Wijmenga C.; Genome of the Netherlands Consortium, de Bakker P. I., Sunyaev S. R., Deleterious alleles in the human genome are on average younger than neutral alleles of the same frequency. PLOS Genet. 9, e1003301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coutinho de Almeida R., Ramos Y. F. M., Mahfouz A., den Hollander W., Lakenberg N., Houtman E., van Hoolwerff M., Suchiman H. E. D., Rodriguez Ruiz A., Slagboom P. E., Mei H., Kielbasa S. M., Nelissen R., Reinders M., Meulenbelt I., RNA sequencing data integration reveals an miRNA interactome of osteoarthritis cartilage. Ann. Rheum. Dis. 78, 270–277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tuerlings M., van Hoolwerff M., Houtman E., Suchiman E., Lakenberg N., Mei H., van der Linden E., Nelissen R., Ramos Y., Coutinho de Almeida R., Meulenbelt I., RNA sequencing reveals interacting key determinants of osteoarthritis acting in subchondral bone and articular cartilage. Arthritis Rheumatol., (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoffels J. M., Zhao C., Baron W., Fibronectin in tissue regeneration: Timely disassembly of the scaffold is necessary to complete the build. Cell. Mol. Life Sci. 70, 4243–4253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pankov R., Yamada K. M., Fibronectin at a glance. J. Cell Sci. 115, 3861–3863 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Vinod E., Boopalan P., Sathishkumar S., Reserve or resident progenitors in cartilage? Comparative analysis of chondrocytes versus chondroprogenitors and their role in cartilage repair. Cartilage 9, 171–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dicks A., Wu C. L., Steward N., Adkar S. S., Gersbach C. A., Guilak F., Prospective isolation of chondroprogenitors from human iPSCs based on cell surface markers identified using a CRISPR-Cas9-generated reporter. Stem Cell Res. Ther. 11, 66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su X., Zuo W., Wu Z., Chen J., Wu N., Ma P., Xia Z., Jiang C., Ye Z., Liu S., Liu J., Zhou G., Wan C., Qiu G., CD146 as a new marker for an increased chondroprogenitor cell sub-population in the later stages of osteoarthritis. J. Orthop. Res. 33, 84–91 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Li W., Liu S., Xu J., RaptorX-Property: A web server for protein structure property prediction. Nucleic Acids Res. 44, W430–W435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hadler N. M., Dourmashkin R. R., Nermut M. V., Williams L. D., Ultrastructure of a hyaluronic acid matrix. Proc. Natl. Acad. Sci. U.S.A. 79, 307–309 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thyberg J., Electron microscopy of cartilage proteoglycans. Histochem. J. 9, 259–266 (1977). [DOI] [PubMed] [Google Scholar]
- 26.Loeser R. F., Integrins and chondrocyte-matrix interactions in articular cartilage. Matrix Biol. 39, 11–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeser R. F., Carlson C. S., McGee M. P., Expression of beta 1 integrins by cultured articular chondrocytes and in osteoarthritic cartilage. Exp. Cell Res. 217, 248–257 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Homandberg G. A., Meyers R., Williams J. M., Intraarticular injection of fibronectin fragments causes severe depletion of cartilage proteoglycans in vivo. J. Rheumatol. 20, 1378–1382 (1993). [PubMed] [Google Scholar]
- 29.Zack M. D., Arner E. C., Anglin C. P., Alston J. T., Malfait A. M., Tortorella M. D., Identification of fibronectin neoepitopes present in human osteoarthritic cartilage. Arthritis Rheum. 54, 2912–2922 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Homandberg G. A., Wen C., Hui F., Cartilage damaging activities of fibronectin fragments derived from cartilage and synovial fluid. Osteoarthr. Cartil. 6, 231–244 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Reed K. S. M., Ulici V., Kim C., Chubinskaya S., Loeser R. F., Phanstiel D. H., Transcriptional response of human articular chondrocytes treated with fibronectin fragments: An in vitro model of the osteoarthritis phenotype. Osteoarthr. Cartil. 29, 235–247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh P., Schwarzbauer J. E., Fibronectin matrix assembly is essential for cell condensation during chondrogenesis. J. Cell Sci. 127, 4420–4428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh P., Schwarzbauer J. E., Fibronectin and stem cell differentiation—Lessons from chondrogenesis. J. Cell Sci. 125, 3703–3712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almonte-Becerril M., Gimeno L. I., Villarroya O., Benito-Jardon M., Kouri J. B., Costell M., Genetic abrogation of the fibronectin-α5β1 integrin interaction in articular cartilage aggravates osteoarthritis in mice. PLOS ONE 13, e0198559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C. S., Fu H., Baratang N., Rousseau J., Kumra H., Sutton V. R., Niceta M., Ciolfi A., Yamamoto G., Bertola D., Marcelis C. L., Lugtenberg D., Bartuli A., Kim C., Hoover-Fong J., Sobreira N., Pauli R., Bacino C., Krakow D., Parboosingh J., Yap P., Kariminejad A., McDonald M. T., Aracena M. I., Lausch E., Unger S., Superti-Furga A., Lu J. T.; Baylor-Hopkins Center for Mendelian Genomics, Cohn D. H., Tartaglia M., Lee B. H., Reinhardt D. P., Campeau P. M., Mutations in fibronectin cause a subtype of spondylometaphyseal dysplasia with “Corner Fractures”. Am. J. Hum. Genet. 101, 815–823 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castelletti F., Donadelli R., Banterla F., Hildebrandt F., Zipfel P. F., Bresin E., Otto E., Skerka C., Renieri A., Todeschini M., Caprioli J., Caruso R. M., Artuso R., Remuzzi G., Noris M., Mutations in FN1 cause glomerulopathy with fibronectin deposits. Proc. Natl. Acad. Sci. U.S.A. 105, 2538–2543 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Min J. L., Meulenbelt I., Kloppenburg M., van Duijn C. M., Slagboom P. E., Mutation analysis of candidate genes within the 2q33.3 linkage area for familial early-onset generalised osteoarthritis. Eur. J. Hum. Genet. 15, 791–799 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Deelen J., Beekman M., Uh H. W., Helmer Q., Kuningas M., Christiansen L., Kremer D., van der Breggen R., Suchiman H. E., Lakenberg N., van den Akker E. B., Passtoors W. M., Tiemeier H., van Heemst D., de Craen A. J., Rivadeneira F., de Geus E. J., Perola M., van der Ouderaa F. J., Gunn D. A., Boomsma D. I., Uitterlinden A. G., Christensen K., van Duijn C. M., Heijmans B. T., Houwing-Duistermaat J. J., Westendorp R. G., Slagboom P. E., Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 10, 686–698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riyazi N., Meulenbelt I., Kroon H. M., Ronday K. H., Hellio le Graverand M. P., Rosendaal F. R., Breedveld F. C., Slagboom P. E., Kloppenburg M., Evidence for familial aggregation of hand, hip, and spine but not knee osteoarthritis in siblings with multiple joint involvement: The GARP study. Ann. Rheum. Dis. 64, 438–443 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keurentjes J. C., Fiocco M., So-Osman C., Onstenk R., Koopman-Van Gemert A. W., Poll R. G., Kroon H. M., Vliet Vlieland T. P., Nelissen R. G., Patients with severe radiographic osteoarthritis have a better prognosis in physical functioning after hip and knee replacement: A cohort-study. PLOS ONE 8, e59500 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos Y. F., den Hollander W., Bovee J. V., Bomer N., van der Breggen R., Lakenberg N., Keurentjes J. C., Goeman J. J., Slagboom P. E., Nelissen R. G., Bos S. D., Meulenbelt I., Genes involved in the osteoarthritis process identified through genome wide expression analysis in articular cartilage; the RAAK study. PLOS ONE 9, e103056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meulenbelt I., Bos S. D., Chapman K., van der Breggen R., Houwing-Duistermaat J. J., Kremer D., Kloppenburg M., Carr A., Tsezou A., Gonzalez A., Loughlin J., Slagboom P. E., Meta-analyses of genes modulating intracellular T3 bio-availability reveal a possible role for the DIO3 gene in osteoarthritis susceptibility. Ann. Rheum. Dis. 70, 164–167 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Dambrot C., van de Pas S., van Zijl L., Brandl B., Wang J. W., Schalij M. J., Hoeben R. C., Atsma D. E., Mikkers H. M., Mummery C. L., Freund C., Polycistronic lentivirus induced pluripotent stem cells from skin biopsies after long term storage, blood outgrowth endothelial cells and cells from milk teeth. Differentiation 85, 101–109 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Bae S., Park J., Kim J. S., Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farndale R. W., Buttle D. J., Barrett A. J., Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim. Biophys. Acta 883, 173–177 (1986). [DOI] [PubMed] [Google Scholar]
- 46.Bomer N., den Hollander W., Ramos Y. F., Bos S. D., van der Breggen R., Lakenberg N., Pepers B. A., van Eeden A. E., Darvishan A., Tobi E. W., Duijnisveld B. J., van den Akker E. B., Heijmans B. T., van Roon-Mom W. M., Verbeek F. J., van Osch G. J., Nelissen R. G., Slagboom P. E., Meulenbelt I., Underlying molecular mechanisms of DIO2 susceptibility in symptomatic osteoarthritis. Ann. Rheum. Dis. 74, 1571–1579 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faas F. G., Avramut M. C., van den Berg B. M., Mommaas A. M., Koster A. J., Ravelli R. B., Virtual nanoscopy: Generation of ultra-large high resolution electron microscopy maps. J. Cell Biol. 198, 457–469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 to S6