PD-1 on glioblastoma cells promotes brain tumor growth, and therapeutic antibodies cannot suppress its proliferative effects.
Abstract
Brain tumor–initiating cells (BTICs) drive glioblastoma growth through not fully understood mechanisms. Here, we found that about 8% of cells within the human glioblastoma microenvironment coexpress programmed cell death 1 (PD-1) and BTIC marker. Gain- or loss-of-function studies revealed that tumor-intrinsic PD-1 promoted proliferation and self-renewal of BTICs. Phosphorylation of tyrosines within the cytoplasmic tail of PD-1 recruited Src homology 2–containing phosphatase 2 and activated the nuclear factor kB in BTICs. Notably, the tumor-intrinsic promoting effects of PD-1 did not require programmed cell death ligand 1(PD-L1) ligation; thus, the therapeutic antibodies inhibiting PD-1/PD-L1 interaction could not overcome the growth advantage of PD-1 in BTICs. Last, BTIC-intrinsic PD-1 accelerated intracranial tumor growth, and this occurred in mice lacking T and B cells. These findings point to a critical role for PD-1 in BTICs and uncover a nonimmune resistance mechanism of patients with glioblastoma to PD-1– or PD-L1–blocking therapies.
INTRODUCTION
Glioblastoma (GBM) is the most common and fatal primary brain malignancy (1). The current standard of care is surgical debulking, followed by radiotherapy and chemotherapy with temozolomide (2). Despite these treatments and previously unknown approaches such as tumor-treating fields, the median survival of patients remains less than 21 months (3). A notable reason for the dismal prognosis is the existence of brain tumor–initiating cells (BTICs) within the tumor microenvironment (4). BTICs exhibit stem cell–like properties, including the ability to self-renew and differentiate along multiple lineages (4). BTICs drive tumor growth and recurrence because they are more resistant to radio- and chemotherapy than their more differentiated counterparts (5). Thus, BTICs are critical targets for more efficient therapy, and the biology of their growth properties is of high interest.
The GBM tumor microenvironment is infiltrated by immune cells with the intrinsic ability to curb cancer cell growth (6). However, these immune cells are subdued by tumor cells as described by several groups including our own (7, 8). GBM cells use different immunosuppressive mechanisms contributing to the evolution of an immunologically “cold microenvironment” that actively antagonizes antitumor immune responses (9, 10). A wealth of evidence supports the importance of inhibitory receptors, also called “immune checkpoints,” in tumor-mediated immune suppression (11). Of different immune checkpoints, programmed cell death 1 (PD-1) is expressed mainly on T cells and macrophages that, upon engagement by its prominent ligand PD-L1 (programmed cell death ligand 1) on tumor cells, dampens antitumor immune functions (12). Blocking the PD-1 and PD-L1 lock-and-key interaction by monoclonal antibodies thus unleashes antitumor immune responses with a desired therapeutic outcome in different cancers such as melanoma, non–small cell lung cancer (NSCLC), Hodgkin’s lymphoma, and renal cell carcinoma (13–15).
PD-1/PD-L1 checkpoint blockade therapy in GBMs, however, is still under debate (16), and clinical results of immune checkpoint blockade therapies have not shown survival benefits (NCT 02017717). Partly accounting for this limited success is the relatively low infiltration of T cells into GBMs compared to highly immunogenic cancers such as melanoma (9). In addition, the blood-brain barrier is a substantial impediment to the penetration of therapeutic antibodies into the central nervous system (CNS) (17). Nevertheless, blockade of PD-1/PD-L1 by monoclonal antibodies exhibits therapeutic activity in CNS metastases in patients with melanoma or NSCLC (18).
The mechanisms underlying the disappointing response to PD-1/PD-L1 blockade therapies in GBMs remain incompletely understood. To seek further clarity, we examined human GBM specimens for this immune checkpoint axis and observed that about 8% of cells across samples coexpress PD-1 and one of the BTIC markers. As this unexpected finding has not been reported in GBM, we thus characterized PD-1 on BTICs further. This study describes new roles for PD-1 in regulating the proliferation and self-renewal of BTICs through nuclear factor κB (NFκB) without the involvement of PD-L1 ligation.
RESULTS
BTICs express PD-1 in resected GBM brain specimens
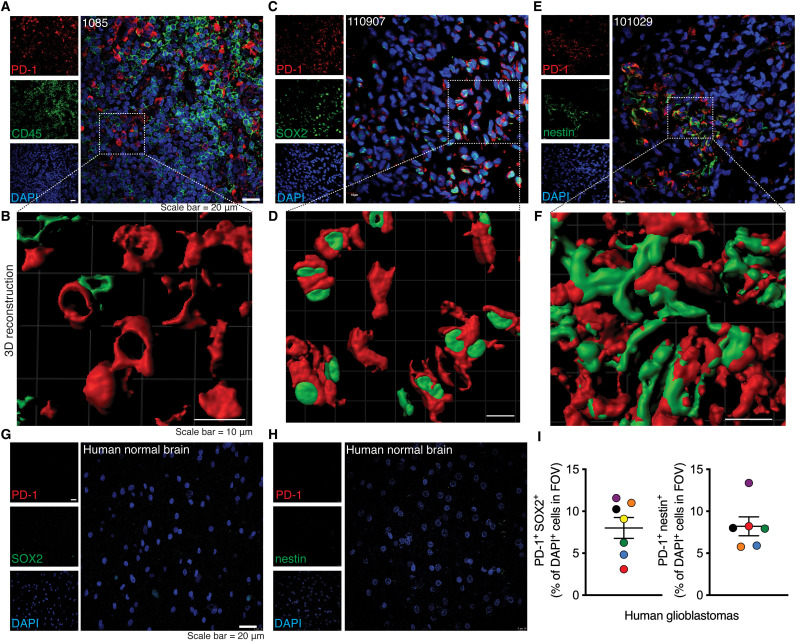
We first assessed the expression of PD-1 across specimens resected from seven patients with GBM (table S1) by immunofluorescence microscopy. While CD45-positive leukocytes express PD-1 (fig. S1A) as commonly seen in the tumor microenvironment, PD-1 was unexpectedly expressed on CD45-negative cells within the GBM tissues (Fig. 1A). Three-dimensional (3D) reconstruction of double labeling corroborated the presence of PD-1 on a nonleukocyte population (Fig. 1B). Although there are no universal BTIC markers, the Sex determining region Y (SRY)-box 2 (SOX2) transcription marker and nestin are commonly used for BTIC identification within the GBM microenvironment (19, 20). Notably, PD-1 was found on cells expressing SOX2 or nestin in the human GBM tissues (Fig. 1, C to F). As controls, no PD-1 or BTIC markers were found in normal brains (Fig. 1, G and H). Expression of PD-1 on BTICs in the human GBMs was affirmed by a different monoclonal antibody made from a distinct clone (fig. S1B). Quantitation of images revealed that about 8% of cells, on average, within the GBM microenvironment coexpressed PD-1 and one of the BTIC markers, SOX2 or nestin (Fig. 1I). The presence of PD-1 on BTICs was found in all seven patients with GBM. We were unable to identify nestin expression in patient 1085, despite the presence of PD-1 and SOX2 double-positive cells.
Fig. 1. The expression of PD-1 on BTICs in resected clinical GBM specimens in situ.
Representative images of immunofluorescence staining and corresponding 3D reconstruction of images of (A and B) CD45 and PD-1, (C and D) PD-1 and BTIC marker SOX2, and (E and F) PD-1 and BTIC marker nestin in sections of human GBM specimens resected from three patients (1085, 110907, and 101029). Representative immunofluorescence images of human normal brains stained for (G) PD-1 and SOX2 or (H) PD-1 and nestin. (I) Quantification of immunofluorescence staining of PD-1+ SOX2+ or PD-1+ nestin+ cells within the tumor microenvironment. Quantification was performed in three to four fields of view (FOVs) of each patient. Each circle is of a different patient with GBM, and the color of the dots in both panels match to the same subject. Green, 026-1; purple, 101220; orange, 110907; blue, 100819; red, 110512; black, 101029; yellow, 1085. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Data are represented as means ± SEM. See also fig. S1 and table S1.
Next, we evaluated the expression of PD-1 on BTICs in a mouse model of GBM. We implanted mouse BT0309 BTIC line, previously generated in C57BL/6 strain and propagated as stem-like cells (21), into the striatum of immunocompetent C57BL/6 mice (fig. S1C). Our previous study confirmed that the mBT0309 BTIC line following intracranial implantation in mice recapitulated the main features of a human World Health Organization grade IV GBM (21). Upon confirmation of tumor growth by T2*-weighted magnetic resonance imaging (MRI) (fig. S1C), brain tissues were collected and subjected to immunofluorescence microscopy. We found PD-1 expression on nestin-positive cells, similar to what we observed in human tissues (fig. S1D). No PD-1 expression was detected in naïve mouse brains (fig. S1E).
GBM patient–derived BTICs and murine BTICs in culture express PD-1
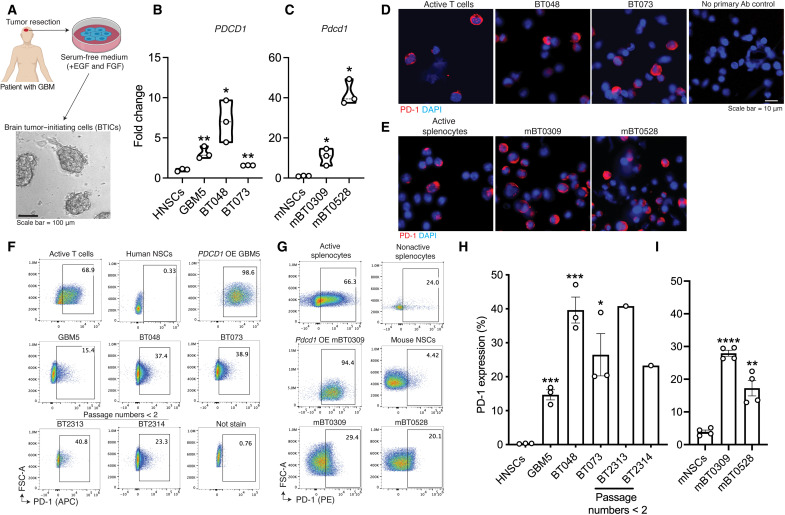
BTICs were generated in-house from resected GBM specimens as described previously (22) and depicted in Fig. 2A. BTICs grew as spheres in the serum-free medium supplemented with epidermal growth factor (EGF) and fibroblast growth factor (FGF) (referred to here as BTIC medium). We previously documented that these BTICs in culture show stem-like properties (23, 24). We assessed the expression of PD-1 on human (PDCD1; Fig. 2B and table S2) and mouse (Pdcd1; Fig. 2C) BTIC lines by reverse transcription quantitative polymerase chain reaction (RT-qPCR). The transcripts of PD-1 were detected in all examined human and mouse BTIC lines; levels were significantly higher than those found in nontransformed neural stem cells isolated from brain tissues of human fetuses or subventricular zone of neonatal mouse brains (Fig. 2, B and C). We also determined the coding sequence (CDS) of PDCD1 in human BTIC GBM5, BT025, and BT073 and blasted the obtained sequence against the National Center for Biotechnology Information (NCBI) database. We found 100% sequence similarity between the transcripts of PDCD1 in BTICs with the NCBI Reference Sequence: NM_005018.3 (fig. S2A), further confirming the mRNA expression of the PDCD1 gene with no mutation or deletion in patient-derived BTICs.
Fig. 2. The expression of PD-1 by patient-derived or murine BTICs in culture.
(A) Following surgery of patients with GBM, BTICs were generated from resected specimens by culturing cells in a serum-free medium supplemented with EGF and FGF. RT-qPCR analysis of PD-1 transcripts in (B) human (PDCD1) or (C) mouse (Pdcd1) BTIC lines compared with neural stem cells (NSCs). Fold changes were calculated relative to PD-1 expression in neural stem cells and normalized to GAPDH expression. Immunofluorescence of PD-1 staining on (D) human and (E) mouse BTIC lines in culture. Active T cells or splenocytes were used as controls. Nuclei were counterstained with DAPI. Representative flow cytometry plots of PD-1 expression on (F) human and (G) mouse BTIC lines, with quantitation and comparison to human or mouse neural stem cells shown in (H) and (I), respectively. Human or mouse BTICs (OE) PDCD1 or Pdcd1 and active T cells or splenocytes were used as controls. T cells or splenocytes were activated with anti-CD3/CD28 antibodies. Recently initiated patient-derived BTICs from two resections (BT2313 and BT2314), with less than two passages in culture, were also tested. Human and mouse cells were stained with antibodies conjugated with allophycocyanin (APC) or phycoerythrin (PE) fluorochromes, respectively. Data are representative of two to three independent experiments. Means were compared to neural stem cells with unpaired (two-tailed) t test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are represented as means ± SEM. See also figs. S2 and S3 and table S2. Ab, antibody.
Immunofluorescence staining of fixed cultured cells for PD-1 substantiated the mRNA expression seen in human and mouse BTIC lines (Fig. 2, D and E, and fig. S2B). Flow cytometry affirmed the surface expression of PD-1 on several live human and mouse BTIC lines (Fig. 2, F and G) and on two low-passage patient-derived BTIC lines (Fig. 2, F and H). Seven additional GBM patient–derived BTIC lines expressed surface PD-1, at levels from 12.6 to 56.4% (fig. S2, C and D). In general, the expression levels of surface PD-1 on human and mouse BTICs were significantly higher than those in their respective species neural stem cells (Fig. 2, H and I). We also validated the expression of PD-1 on human BTICs by two monoclonal antibodies made from distinct clones (fig. S2E). The percentage of PD-1–positive BTICs remained relatively stable after multiple passages in culture (fig. S2F). PD-1 was coexpressed with the BTIC markers, musashi1 or nestin, on human and mouse BTICs (fig. S3, A and B). PD-1 and nestin or SOX2 were also codetected in flow cytometry of live single-cell plots (fig. S3, C and D).
We addressed whether PD-1 expression remains stable upon differentiation of BTICs. BTICs differentiate upon treatment with 1% fetal bovine serum–containing medium (22). Differentiation was suggested by the absence of spheres 1 week after treatment (fig. S3E) and reduced stemness marker expression documented in our previous study (21). The mRNA and protein levels of PD-1 were lower in differentiated compared to their undifferentiated counterparts in three of four human BTIC lines; differentiated GBM5 had similar PD-1 mRNA but lower protein levels than undifferentiated cells (fig. S3, F and G).
Tumor-intrinsic PD-1 promotes BTIC growth in culture
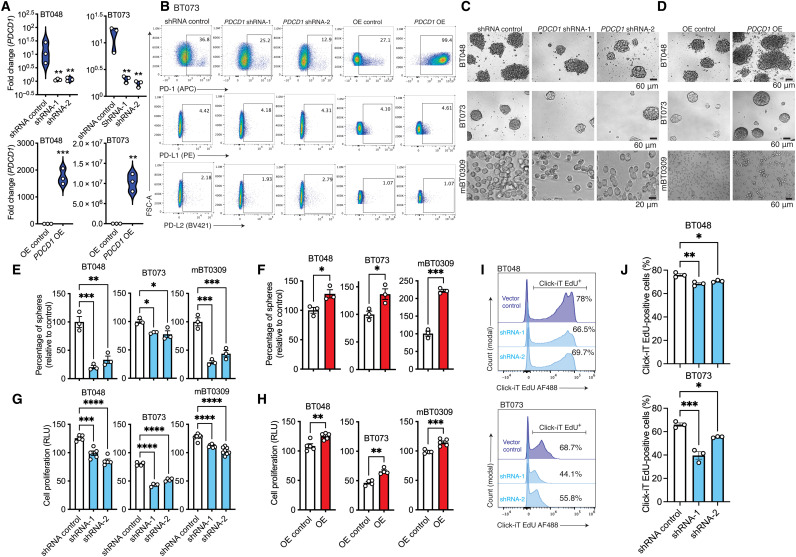
To interrogate intrinsic functions of PD-1 in BTICs, we under- or overexpressed PD-1 in patient-derived and murine BTIC lines as depicted in fig. S4A. While PD-1 expression was detected in all 12 human BTIC lines studied, we conducted most of the subsequent experiments in four patient-derived (BT048, BT073, BT069, and BT100) and two murine (mBT0309 and mBT0528) BTIC lines to address the functions of PD-1. The under- or overexpression of PD-1 was confirmed at mRNA and protein levels by RT-qPCR and flow cytometry, respectively (Fig. 3, A and B, and fig. S4, B to D). The altered expression of PD-1 did not affect the expression of PD-L1 or PD-L2 on the human and mouse BTIC lines (Fig. 3B and fig. S4, C and D). We then investigated the sphere-forming capacity, an indicator of self-renewal, of PD-1–underexpressing or –overexpressing BTICs. The sphere-forming capacity of BTICs was reduced in PD-1–underexpressing human and mouse lines compared to short hairpin–mediated RNA (shRNA) control; conversely, sphere formation was further increased when PD-1 was up-regulated (Fig. 3, C to F). The down-regulation of PD-1 also reduced cell proliferation as monitored through adenosine 5′-triphosphate (ATP) luminescence, while forced expression of PD-1 in BTICs further augmented cell proliferation (Fig. 3, G and H, and fig. S4, E and F). Furthermore, EdU (ethynyl deoxyuridine) labeling found reduced incorporation into DNA of human or mouse BTICs with down-regulated PD-1 expression (Fig. 3, I and J, and fig. S4G), while greater EdU labeling was observed in Pdcd1-overexpressing mouse BTICs than respective vector controls (fig. S4H).
Fig. 3. Tumor-intrinsic PD-1 promotes BTIC growth in culture.
(A) Confirmation of PDCD1 down-regulation or up-regulation in human BTIC lines at mRNA levels by RT-qPCR. Fold changes were calculated relative to PD-1 expression in respective vector controls and normalized to GAPDH expression. (B) Flow cytometry analysis of PD-1, PD-L1, and PD-L2 expression after knocking down or OE PD-1 in BTICs. (C and D) Representative bright-field microscopy images of 72- to 96-hour outcomes of tumor spheres in PD-1 knockdown or overexpression versus respective vector controls of two human (BT048 and BT073) and mouse (mBT0309) BTIC lines. (E and F) Quantification of tumor spheres in PD-1 down-regulated or PD-1 OE versus respective vector controls. (G and H) ATP proliferation assay of human and mouse BTIC with PD-1 down-regulation or up-regulation. (I and J) Representative plots and bar plots of the proliferation of human BTIC lines by measuring incorporation of EdU into DNA following 24-hour treatment. AF488, Alexa Fluor 488; RLU, relative light unit. Data are representative of two to three separate experiments. Means were compared to respective vector controls by unpaired (two-tailed) t test when comparing two groups. For more than two groups, one-way analysis of variance (ANOVA) with Tukey’s post hoc was used: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are represented as means ± SEM. See also fig. S4.
PD-1 as a signaling receptor activates NFκB transcription factor in BTICs
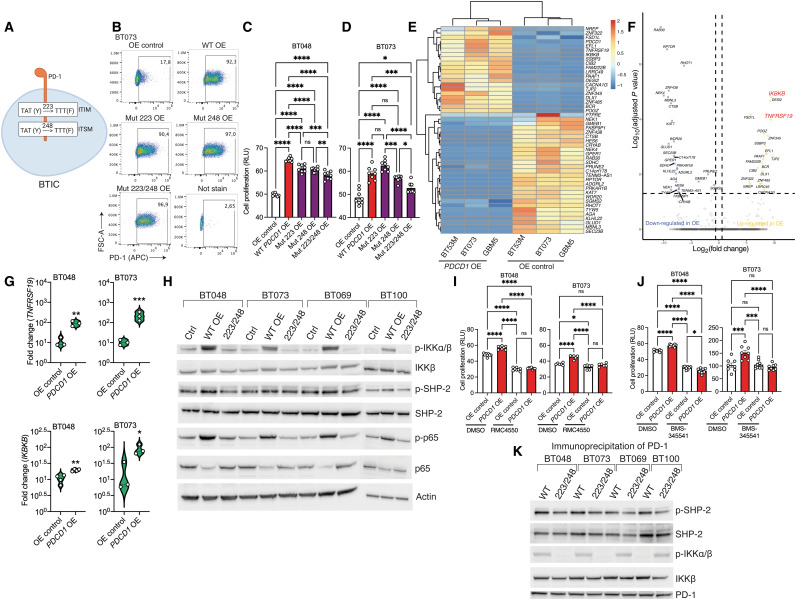
The cytoplasmic tail of PD-1 contains two signaling motifs, an immunoreceptor tyrosine-based inhibitory motif (ITIM) and an immunoreceptor tyrosine-based switch motif (ITSM) (25). To test the involvement of these two sequences in the tumor-intrinsic promoting effects of PD-1, we generated four human BTIC lines (BT048, BT073, BT069, and BT100) overexpressing mutant variants of PD-1 containing tyrosine (Y) to phenylalanine (F) substitutions at ITIM (Y223F mutation) or ITSM (Y248F mutation) motifs as described previously (26). Human BTIC lines with both tyrosine mutations (Y223F/Y248F) were also created (Fig. 4A). These mutations disrupt the signaling functions of ITIM and ITSM motifs in the PD-1 receptor. We confirmed a similar percentage of the expression of mutant or wild-type PD-1 on transfected BT048 and BT073 (Fig. 4B and fig. S5A). While individual ITIM or ITSM mutation decreased cell proliferation in some cell lines when compared to BTICs overexpressing wild-type PD-1, the greatest reduction occurred when both ITIM and ITSM motifs were disrupted (Fig. 4, C and D, and fig. S5B); thus, we proceeded in subsequent experiments with the cells overexpressing double mutation of PD-1.
Fig. 4. PD-1 as a signaling receptor activates NFκB transcription factor in BTICs.
(A) Schematic diagram of site-directed mutagenesis in two signaling motifs (ITIM, Y223F; ITSM, Y248F) of human PD-1 gene. (B) Representative flow cytometry plots of PD-1 expression in BT073 transfected with OE vectors coding wild-type (WT), 223, 248, or 223/248 mutant (Mut) version of PD-1. (C and D) ATP proliferation assay of human BT048 and BT073 cells OE wild-type or mutant versions of PD-1 compared with OE control. ns, not significant. (E) Heatmap of differentially expressed genes between PDCD1–OE cells and OE control of three human BTIC lines. Experiment was conducted once. (F) Volcano plot shows all data points that meet the false discovery rate (FDR) cutoff of 5% and 1.5-fold change criteria. (G) Validation of mRNA expression of TNFRSF19 and IKBKB genes by RT-qPCR in another culture set of BT048 and BT073 cells. Fold changes were calculated relative to PD-1 expression in OE control and normalized to GAPDH expression. (H) The phosphorylation of IKKα/β, SHP-2, and NFκB (p65) was detected in the cell lysates of four human BTIC lines by Western blot. Actin was used as a loading control. Luminescence ATP proliferation test of PDCD1-OE BT048 and BT073 cells compared to OE controls after 72 hours of treatment with (I) SHP-2 inhibitor RMC4550 (3 Mμ) or (J) IKKα/β inhibitor BMS-345541 (1 Mμ). DMSO, dimethyl sulfoxide. (K) After PD-1 immunoprecipitation of cell lysates and SDS–polyacrylamide gel electrophoresis, immunoblot was performed with antibodies for p–SHP-2, SHP-2, p-IKKα/β, IKKβ, and PD-1. Means were compared by unpaired (two-tailed) t test when comparing two groups. For more than two groups, one-way ANOVA with Tukey’s post hoc was used: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are represented as means ± SEM. See also fig. S5.
We sought to understand the downstream pathway of PD-1 more comprehensively by performing RNA sequencing of PDCD1-overexpressing BTICs compared to OE control from three patients. Principal components analysis indicated that the basal (OE control) transcriptome of three BTICs lines is different and PD-1 overexpression resulted in changes unique to each cell line (fig. S5C); however, there were common changes in the expression of genes between PDCD1-overexpressing cells and OE control in all three lines (Fig. 4E). In the differential expression analyses, we applied a 5% FDR (false discovery rate) cutoff and a 1.5-fold change threshold. We identified 48 differentially expressed transcripts in PDCD1-overexpressing versus control across three different BTIC lines (Fig. 4E). Among those genes, expression of IKBKB (inhibitor of NFκB kinase subunit B) and TNFRSF19 [tumor necrosis factor (TNF) receptor superfamily member 19] genes increased 9.49- and 9.39-fold, respectively, in PD-1-overexpressing BTICs compared to controls (Fig. 4F). IKBKB encodes inhibitor of NFκB kinase β (IKKβ) subunit of IKK complex, the core element of the NFκB signaling pathway (27). TNFRSF19 and IKK complex are implicated in the activation of NFκB signaling and gliomagenesis (28, 29). RT-qPCR in another batch of BT048 and BT073 cultures corroborated the enhanced expression of IKBKB and TNFRSF19 genes in PDCD1-overexpressing BTICs compared to OE controls (Fig. 4G).
Western blots of four human BTIC lines indicated that BTICs overexpressing PD-1 had higher levels of phosphorylated IKKα/β and total IKKβ than controls (Fig. 4H). Furthermore, phosphorylation of IKKα/β was lower in BTICs expressing PD-1 with combined ITIM and ITSM mutations compared to cells overexpressing wild-type PD-1, indicating that tyrosine phosphorylation of both ITIM and ITSM motifs was crucial in IKKα/β activation. We also examined the total expression levels of IKKα and IKKβ by flow cytometry and found reduced expression in PD-1 knockdown compared to control BT073 (fig. S5, D and E). Flow cytometry also showed reduced phosphorylation of serine residue 176/180 of IKKα, or serine residue 177/181 of IKKβ (P-IKKα/β), in PDCD1–down-regulated BTICs compared to shRNA control (fig. S5F).
Previous research in T cells revealed that ITIM and ITSM motifs of PD-1 bind the Src homology 2 (SH2) domains of SH2-containing phosphatase 2 (SHP-2) and cause SHP-2 to be phosphorylated at tyrosine residue 542 (p542) (25). In addition, previous findings have implicated the role of SHP-2 in IKKα/β activation; hence, we investigated whether PD-1 also changes the phosphorylation levels of SHP-2 in BTICs. Immunoblots of four human BTIC lines revealed that cells overexpressing wild-type PD-1 had higher levels of p–SHP-2 than controls (Fig. 4H). Consistently, the effects of overexpressing wild-type PDCD1 on p–SHP-2 levels were reversed when cells were transfected with vectors encoding double-mutant PD-1 (Fig. 4H). Phospho-flow cytometry of SHP-2 p542 in two human BTIC lines following PD-1 down-regulation versus control corroborated the involvement of SHP-2 phosphorylation in the downstream signaling of PD-1 (fig. S5, G and H).
Phosphorylation of IKKα/β and SHP-2 has been shown to activate the NFκB pathway (30). Thus, we aimed to explore the activation status of the NFκB pathway in BTICs following up-regulation or down-regulation of PD-1. In all four human BTIC lines studied, forced expression of wild-type PD-1 enhanced phosphorylation levels of p65, a subunit of NFκB transcription complex, compared to controls (Fig. 4H). Overexpression of PD-1 with the double mutation ITIM/ITSM, on the other hand, had no effect on p65 activation (Fig. 4H). Silencing PD-1 significantly reduced phosphorylation of NFκB p65 in two human BTIC lines (fig. S5, I and J). We also affirmed reduced nuclear translocation of p65 in PD-1–down-regulated BTICs versus shRNA control by immunofluorescence staining of cells in culture (fig. S5, K and L).
To affirm the roles of IKKα/β and SHP-2 in tumor-promoting effects of PD-1, we pharmacologically inhibited SHP-2 with RMC4550 and IKKα/β with BMS-345541 in PDCD1-overexpressing BTICs versus controls. The luminescence proliferation assay indicated that RMC4550 and BMS-345541 abrogated the intrinsic growth promoting effects of PD-1 in human BTIC lines (Fig. 4, I and J, and fig. S5, M and N). No toxicity was observed for BMS-345541 on human BTICs in culture (fig. S5O). Because SHP-2 in cancer cells also activates the mitogen-activated protein kinase (MAPK) signaling pathway, we blocked this pathway with the pharmacological inhibitor Skepinone-L and found no inhibitory effects on the proliferation of PD-1–overexpressing BTICs compared to controls (fig. S5P).
Physical interaction between receptors and signaling molecules is required for the initiation of the downstream cascade; we thus performed coimmunoprecipitation (co-IP) to collect protein partners of PD-1 in cell lysates of BTIC lines. Immunoblots of protein partners showed that SHP-2 and IKKα/β interacted with PD-1 in human BTIC lines (Fig. 4K). Notably, mutagenesis of the PD-1 ITIM and ITSM motifs abolished the interaction of SHP-2 and IKKα/β with PD-1 receptor and decreased the interactions of p–SHP-2 and p-IKKα/β with PD-1 (Fig. 4K). These findings imply that physical interactions between ITIM/ITSM of PD-1 and SHP-2 and IKKα/β molecules occurred in the BTICs resulting in the phosphorylation of signaling molecules downstream of PD-1.
Coexpression of PD-1, NFκB activation, and proliferation marker in BTICs in GBM specimens
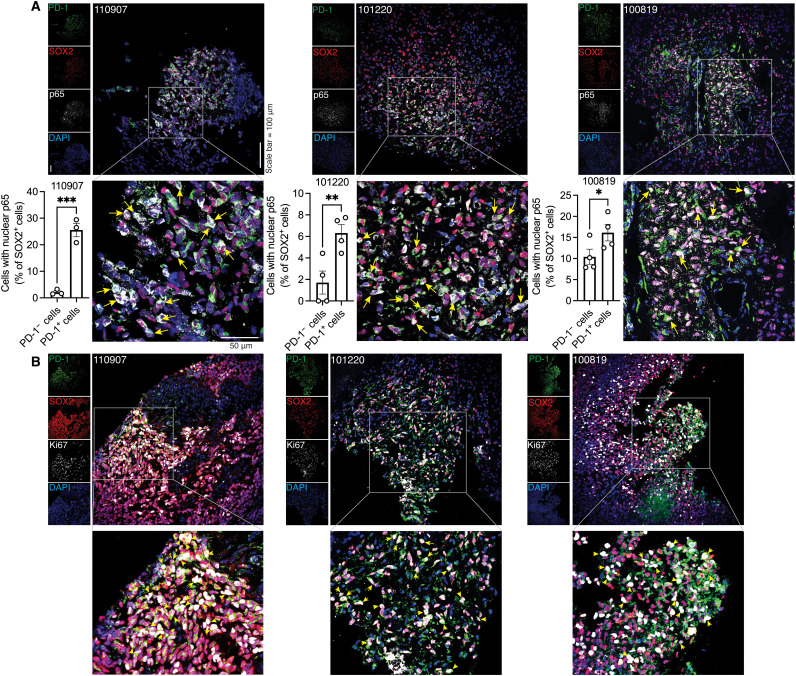
To investigate whether NFκB is activated in PD-1–positive BTICs within the tumor microenvironment, we labeled tumor specimens from three patients with GBM for PD-1, BTIC marker SOX2, and nuclear translocation of p65 as a marker of NFκB activation. We found higher nuclear localization of p65 in PD-1–positive than PD-1–negative BTICs in three of three patients with GBM (Fig. 5A). Moreover, the PD-1 expression corresponded to BTIC proliferation as we observed coexpression of PD-1 and the proliferation marker Ki67 in SOX2-positive BTICs within the GBM microenvironment of three patients (Fig. 5B).
Fig. 5. Coexpression of PD-1, NFκB activation, and proliferation marker in BTICs in GBM specimens.
(A) Representative immunofluorescence laser confocal microscopy images and quantification of PD-1, SOX2, and p65 in sections of human GBM specimens resected from three patients (110907, 101220, and 100819). Quantification was performed in three to four FOVs of each patient. Nuclear translocation (yellow arrows) of p65 in BTICs is shown on the magnified images. Each dot in the bar graphs corresponds to the number of cells per FOV. (B) Representative images of immunofluorescence staining of PD-1, SOX2, and proliferative marker Ki67 in sections of human GBM specimens from three patients, with some triple-positive cells indicated by yellow arrowheads. Nuclei were counterstained with DAPI. Means were compared by unpaired (one-tailed) t test when comparing two groups. *P < 0.05, **P < 0.01, and ***P < 0.001.
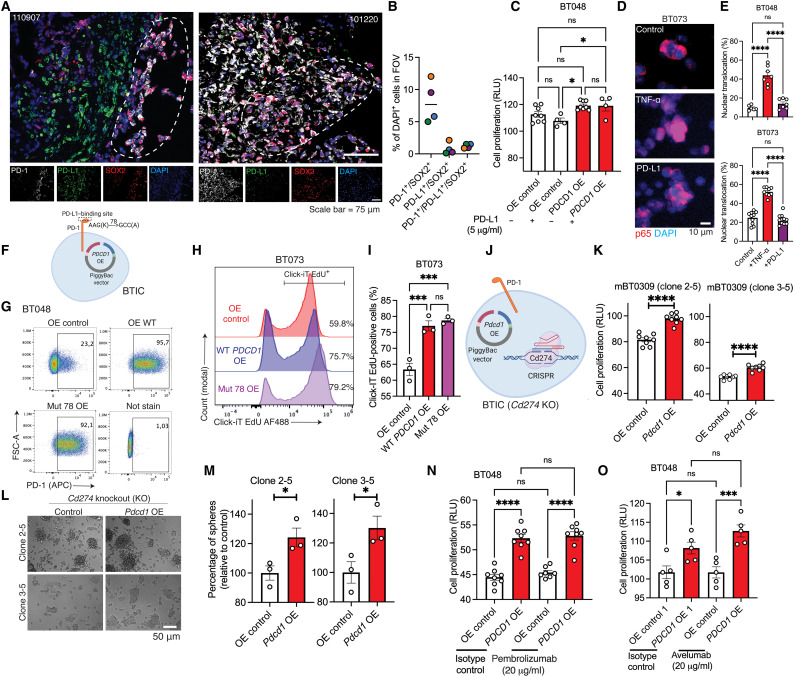
PD-1 promotes BTIC proliferation independent of ligation with PD-L1
Immunofluorescence staining of human GBM specimens revealed the presence of PD-1–positive BTICs in some tumor areas without PD-L1 immunoreactivity (Fig. 6A). In addition, quantification of images show that while around 8% of cells in the tumor microenvironment coexpress PD-1 and SOX2, about 0.8 and 1.2% of cells coexpress PD-L1/SOX2 or PD-1/PD-L1/SOX2, respectively (Fig. 6B). Thus, we asked whether the tumor-intrinsic promoting effects of PD-1 were dependent on PD-L1 ligation. Flow cytometry staining of BTIC lines in culture showed that about 6 to 18% of human and 1% of mouse BTICs expressed PD-L1 (fig. S6, A and B). This PD-L1 expression on human and mouse BTIC lines increased upon treatment of cells with interferon-γ (IFN-γ) (fig. S6, A and B), while there was no change in PD-1 levels on BTICs (fig. S6C). Notably, treatment of either OE control or PDCD1-overexpressing BTICs with IFN-γ did not further increase cell proliferation (fig. S6D).
Fig. 6. Tumor-intrinsic PD-1 does not require PD-L1 ligation to promote BTIC growth.
(A and B) Representative immunofluorescence images and quantification of GBM specimens stained for PD-1, PD-L1, and SOX2. Dashed outlines correspond to areas where there is minimum PD-L1 around PD-1–positive BTICs. Quantification was performed in three to four FOVs of each patient. Each circle is of a different patient with GBM. Green, 026-1; purple, 101220; orange, 110907; blue, 100819. (C) Proliferation of BT048-OE PD-1 and control in the presence or absence of PD-L1. (D and E) Representative immunofluorescence and quantification of p65 translocation in BTICs after treatment with PD-L1 (5 μg/ml) for 1 hour. For each well of cells, four FOVs were quantified. TNF-α (100 ng/ml) was used as a control to activate p65 translocation. (F) Schematic of human PD-1 K78A version with defective PD-L1 binding. (G) Flow cytometry plots of BTICs OE wild-type or K78A PD-1. (H and I) Representative flow cytometry and bar plots of EdU assay. (J) Schematic of knocking out of mouse PD-L1 (Cd274) gene by CRISPR-Cas9 and overexpressing Pdcd1 in mBT0309. (K) Proliferation assay of Cd274 knockout (KO) clones 2-5 and 3-5 of mBT0309-OE PD-1 compared to control. (L and M) Representative images of 72- to 96-hour outcomes of tumor spheres and quantification in clones 2-5 and 3-5. Proliferation of human BTICs overexpressing PD-1 and control after exposure to (N) PD-1 (pembrolizumab) or (O) PD-L1 (avelumab) antibodies for 72 hours. Results are representative of n = 2 to 3 independent experiments. Unpaired (two-tailed) t test was used to assess significance between two groups. For more than two groups, one-way ANOVA with Tukey’s post hoc was used: *P < 0.05, ***P < 0.001, and ****P < 0.0001. ns, not significant. Data are represented as means ± SEM. See also fig. S6.
To test that PD-1 ligation with PD-L1 promotes BTIC growth, we treated PD-1–overexpressing BT048 and OE control with exogenous recombinant PD-L1 protein known to stimulate PD-1 signaling in T cells (31). We found no promotion of cell proliferation either in PD-1–overexpressing BTICs or OE control (Fig. 6C). Similar results were generated for wild-type BT048 and BT073 cell lines (fig. S6E). Staining of p65 translocation showed that PD-L1 proteins did not promote NFκB activation in two patient-derived BTIC lines opposite to TNF-α, which stimulated p65 translocation in cells (Fig. 6, D and E).
To expand these findings, we generated a mutant version of PD-1 with defective PD-L1/2 binding (K78A mutation) (32) by site-directed mutagenesis (Fig. 6F). We affirmed similar levels of expression of mutant and wild-type PD-1 on transfected BTICs (Fig. 6G). The EdU and ATP proliferation assay show that BTICs expressing the K78A variant were similar in proliferation rate to cells overexpressing native PD-1 (Fig. 6, H and I, and fig. S6F).
We also examined the promoting effects of PD-1 in PD-L1 gene (Cd274) knockout (KO) mouse BT0309 line. We first deleted the Cd274 gene in mBT0309 cells using a CRISPR-Cas9 gene–editing system (Fig. 6J). DNA sequencing showed that the mouse Cd274 gene was knocked out in clone 2-5 mBT0309 (designated as Cd274 KO 2-5) (fig. S6G). Full allelic KO of PD-L1 at the protein level was confirmed by Western blot in clones 1-1, 2-3, 2-5, 3-3, and 3-5 (fig. S6H). We then overexpressed wild-type Pdcd1 in clones 2-5 and 3-5 of Cd274 KO mBT0309 (Fig. 6J) and validated PD-1 overexpression in cells by flow cytometry (fig. S6I). ATP luminescence proliferation assay revealed that forced expression of Pdcd1 in two clones of Cd274 (PD-L1) KO mouse BTICs (clones 2-5 and 3-5) increased cell proliferation compared to OE control (Fig. 6K). There was a significant increase in the sphere numbers of Pdcd1-overexpressing cells compared with OE controls in Cd274 KO mouse (clones 2-5 and 3-5) BTICs (Fig. 6, L and M). Together, these datasets suggest that PD-1 without the requirements of PD-L1 ligation promotes BTIC growth.
Last, we used blocking antibodies for PD-1 or PD-L1 that have been tested in clinical trials for patients with GBM. We hypothesized that these antibodies that inhibit the interaction between PD-1 and PD-L1 (33, 34) would not abrogate the tumor-accelerating effects of PD-1 that are non–PD-L1 dependent as the above data show. PDCD1-overexpressing and control human BTICs were exposed to anti–PD-1 (pembrolizumab) or anti–PD-L1 (avelumab) antibodies, and the proliferation of BTICs was unaltered (Fig. 6, N and O).
PD-1 on BTICs promotes intracranial tumor growth irrespective of immunocompetence or immunodeficiency
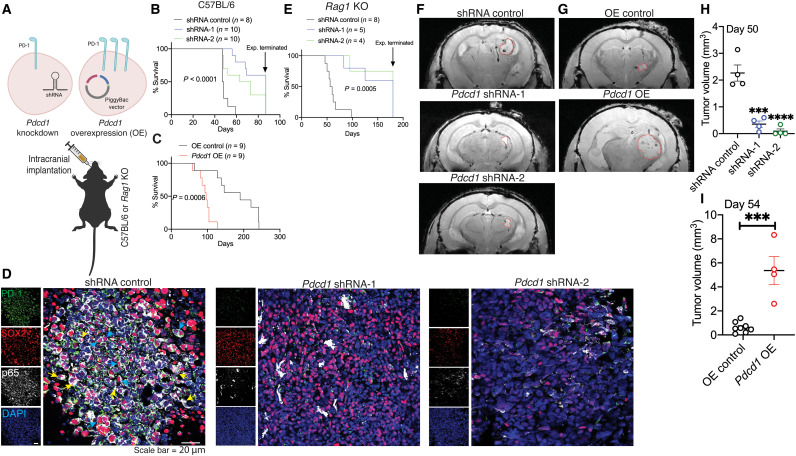
To address in vivo growth benefits of PD-1 on BTICs, we implanted syngeneic mouse BT0309 with stable down- or up-regulated PD-1 into the striatum of immunocompetent C57BL/6 mice (Fig. 7A). We found that the lifespan of mice with down-regulated PD-1 BTICs was longer compared to mice with shRNA control (Fig. 7B). Conversely, lifespan was significantly shortened when Pdcd1-overexpressing cells were implanted compared to mice implanted with OE control (Fig. 7C). Staining for p65 as an index of NFκB activation showed lower immunoreactivity either in cytoplasm or nucleus of SOX2-positive BTICs in mice implanted with Pdcd1–down-regulated cells compared to shRNA control (Fig. 7D).
Fig. 7. PD-1 on BTICs promotes intracranial tumor growth in mice.
(A) Schematic diagram depicts implantation of Pdcd1-knockdown or OE cells into the striatum of mice. Kaplan-Meier analysis to assess the survival of wild-type mice i4mplanted with (B) Pdcd1 knockdown or (C) overexpressing murine BT0309. Cells transduced with lentiviral vectors coding nontarget shRNA or transfected with empty PiggyBac vector were considered as controls for knockdown (shRNA control) or OE (OE control) BTICs, respectively. (D) Representative images of immunofluorescence staining of PD-1, SOX2, and p65 in sections collected from C57BL/6 wild-type mice implanted with Pdcd1-knockdown or shRNA controls. Nuclear translocation (yellow arrowheads) and cytoplasmic localization (blue arrowheads) of p65 in BTICs are shown on the images. (E) Kaplan-Meier analysis of Rag1 KO mice implanted with Pdcd1-knockdown or shRNA controls of mBT0309. Representative T2-weighted MRI images (F) at day 50 of Rag1 KO mice implanted with Pdcd1-knockdown or (G) at day 54 of Rag1 KO mice implanted with Pdcd1 OE mBT0309 (tumor demarcated with red outlines). Quantitation of tumor volume from MRI images of mice implanted with (H) Pdcd1 knockdown or (I) OE BTICs versus respective vector controls. Results are representative of n = 2 to 3 independent experiments. Unpaired (two-tailed) t test was used to assess significance between two groups in (I). For more than two groups in (H), one-way ANOVA with Tukey’s post hoc was used. Kaplan-Meier survival curves were analyzed for statistical differences between groups using the log-rank Mantel-Cox test: ***P < 0.001 and ****P < 0.0001 compared to respective vector control. Data are represented as means ± SEM.
Last, given the functions of PD-1/PD-L1 interaction in antitumor immunity, we determined whether the in vivo tumor-promoting effects of PD-1 on BTICs depended on adaptive immune cells. We implanted stable down- or up-regulated Pdcd1-expressing mouse BT0309 into the striatum of T cell– and B cell–deficient Rag1 KO mice (Fig. 7A). Kaplan-Meier survival plots showed increased longevity of mice implanted with down-regulated PD-1 compared with vector control (Fig. 7E). The tumor size was assessed about 8 weeks after implantation in live asymptomatic animals using T2*-weighted MRI. Images showed significantly reduced tumor volume in mice with down-regulated Pdcd1-expressing BTICs compared with shRNA control mice (Fig. 7, F and H). Conversely, larger tumors were observed in mice implanted with Pdcd1-overexpressing BTICs relative to mice with OE control (Fig. 7, G and I). Thus, PD-1 on BTICs promotes intracranial tumor growth even in the absence of adaptive immune cells.
DISCUSSION
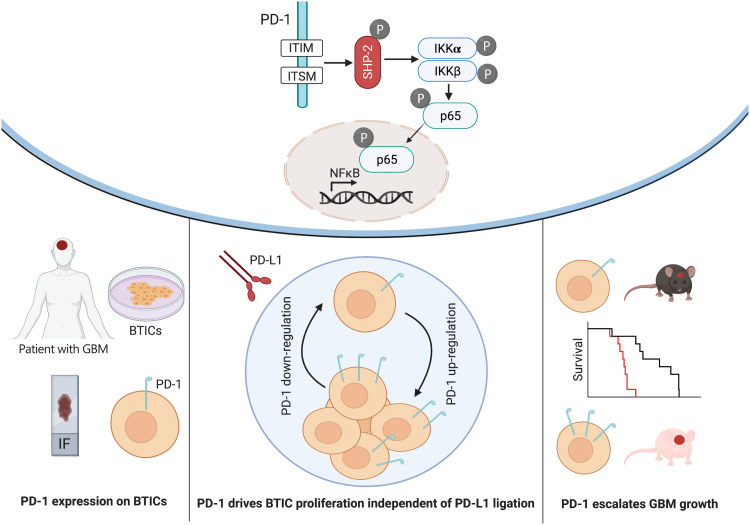
It has become evident that GBM stem cells are the root cause of therapy resistance, relapse, and metastasis through unknown mechanisms. Results from the current study identify a novel regulator of NFκB transcription factor in GBM stem cells and uncover an underlying cause for the resistance to immune checkpoint blockade therapy in patients with GBM (Fig. 8).
Fig. 8. Diagram of PD-1 expression on BTICs and signaling through NFκB.
PD-1 is expressed on BTICs in resected clinical GBM specimens in situ and patient-derived BTICs in culture. Tumor-promoting effects of PD-1 in BTICs did not require interaction with PD-L1; thus, the therapeutic antibodies were unable to overcome the growth advantage of PD-1 in BTICs. Mice with intracranial Pdcd1 over- or underexpressing BTICs had shorter or longer survival, respectively. Mechanistically, phosphorylation of ITIM and ITSM motifs within the cytoplasmic tail of PD-1 recruited SHP-2 phosphatase and activated the NFκB pathway through IKKα/β in BTICs.
PD-1 is mainly expressed on immune cells in patients with cancer that, as an inhibitory receptor, suppresses antitumor functions. There are recent reports showing that a subpopulation of various cancer cells including melanoma, hepatocellular carcinoma, and lung cancer cells express PD-1 (35–37); however, the expression of PD-1 has not been well documented on GBM stem cells. While there are no universal markers for BTICs, stem cell–associated transcription factor SOX2 and nestin are frequently used to identify BTICs (20). By using these BTIC markers, we observed PD-1 expression on BTICs within the GBM microenvironment of seven of seven patients. Notably, there was no nestin immunoreactivity in patient 1085, which might be related to the interpatient heterogeneity of BTIC markers, as previously described (38). The Cancer Genome Atlas project and single-cell RNA sequencing analysis indicate low levels of PD-1 mRNA expression in patients with GBM (39, 40), while protein analysis briefly alluded to PD-1 expression in GBM cell lines (37). This is also the case for melanoma and liver cancer in which expression of PD-1 protein is documented on cancer cells while transcripts are low (40).
We also found that when BTICs were differentiated, the expression of PD-1 on BTICs dropped considerably at the level of protein in all four lines studied. Except for GBM5, all cell lines also showed a reduction in PD-1 mRNA levels. Given the steady expression of PD-1 mRNA in GBM5 after differentiation, the decline in PD-1 protein might be attributed to posttranscriptional mechanisms that regulate the properties and expression of proteins in cancer stem cells (41).
Although PD-1 expression has been observed in a few cancer cell types, research on its tumor-intrinsic activities has shown contradictory results. Unlike its tumor-promoting effects in melanoma and hepatocellular carcinoma (35, 36), PD-1 in lung cancer suppresses tumor cell proliferation (37). In our study, we noted that intrinsic expression of PD-1 within the GBM microenvironment accelerated sphere-forming capacity and proliferation of BTICs, as well as intracranial tumor growth in a mouse GBM model. The specific contribution of tumor-intrinsic PD-1 to tumorigenesis appears to be context dependent, and therefore, studying the tumor-intrinsic functions of PD-1 is of high interest in the field of targeted cancer therapies.
Although PD-1 effects on the proliferation of some cell lines are comparable to those of other recognized factors (42, 43), the effects are small in some of the lines. One explanation might be that we used wild-type BTICs in our experiments as controls that have intrinsic expression of PD-1. Moreover, various mechanisms govern the proliferation of cancer cells, particularly cancer stem cells, implying that cotargeting of several molecules might be necessary to substantially curb tumor cell growth. Furthermore, the reliance of different BTIC lines on PD-1 signaling varied, which could be attributed to the fact that they were derived from patients with diverse genetic and epigenetic backgrounds. We attempted to compare the in vivo growth of PD-1–positive versus PD-1–negative sorted cell; however, because of unstable expression of PD-1 on BTICs, we were unable to implant the cells into the mouse brains.
To date, the downstream signaling pathways of PD-1 have been mainly studied in T cells. Upon binding of PD-1 on T cells to PD-L1, phosphatases SHP-1 and SHP-2 bind to the phosphorylated forms of ITIM/ITSM motifs of PD-1 which leads to the dephosphorylation and subsequent inhibition of phosphatidylinositol 3-kinase (PI3K), Akt, MAPK, and mammalian target of rapamycin (mTOR) pathways (44, 45). While PD-1 signaling in T cells might be more dependent on ITSM phosphorylation (25, 46), the significance of ITIM or ITSM in tumor cells is controversial. In melanoma cells, mutation of either one (ITIM or ITSM) or both (ITIM/ITSM) reduced tumor cell proliferation (35). Our findings showed that inactivating both motifs abolished the proliferative effects of PD-1 at higher levels than individual mutations in BTIC lines. These findings are consistent with those of PD-1 in lung cancer, where double mutations had a larger impact in cancer cells than single ITIM or ITSM mutations (37). In addition, disruption of both tyrosines (Y223F/Y248F) in BT048 and BT073 did not entirely abolish the promoting effects of PD-1 compared to BTICs overexpressing wild-type PD-1, indicating that additional mechanisms are involved in the promoting effects of PD-1 in these lines.
Unlike T cells, PD-1 in melanoma and hepatocellular carcinoma activates PI3K and Akt pathways (35, 36). Contrary to what was observed in melanoma and hepatocellular carcinoma, tumor-intrinsic PD-1 in lung cancer suppresses AKT and extracellular signal–regulated kinase 1/2 pathways (37, 47). Our findings show that PD-1 in GBM cells promoted SHP-2 and IKK phosphorylation, followed by NFκB activation. We noted that SHP-2 along with IKKα/β interacted with PD-1 in BTICs. In addition, mutations of both ITIM and ITSM motifs abrogated phosphorylation of SHP-2 and IKKα/β and their connections with PD-1 in BTICs, implying that the interactions between these molecules are involved in PD-1 signaling in BTICs. This observation is in line with a previous report in other cell lines that a physical interaction between IKKα subunit of IKK complex and SHP-2 triggers NFκB activation (30). Findings from SHP-2 and IKK inhibitors also corroborated the involvement of these two molecules in the proliferative effects of PD-1 in BTICs. Notably, both SHP-2 and IKKα/β inhibitors also constrained the proliferation of control BTICs, which we think might be related to the intrinsic PD-1 expression in wild-type BTICs.
Unlike T cells in which SHP-2 inhibits T cell proliferation, SHP-2 activation by PD-1 in BTICs induced tumor cell growth. This observation is in line with previous reports that identified SHP-2 as a proto-oncogene in GBM cells that activates protumorigenic pathways such as AKT/mTOR (48, 49). A recent study suggested that decreased expression of SHP-2 leads to reduced proliferation, tumorigenicity, and stemness properties of GBM stem cells (49). In addition, NFκB activation contributes to the proliferation and self-renewal properties of BTICs, as well as resistance to conventional therapies (29, 50); therefore, suppression of PD-1 signaling might be a new approach to containing the proliferation and self-renewal properties of BTICs in patients with GBM.
Another unexpected discovery from our investigation was that PD-1 functions in BTICs did not require ligation with PD-L1. An important area for future research is what factors promote PD-1 signaling in the absence of PD-L1. One proposed mechanism is constitutive or ligand-independent PD-1 activation. This mechanism has already been described for other receptors in cancer cells (51, 52). For example, in GBM cells, EGF receptor is constitutively activated regardless of ligand (53). Dimerization of receptor might be a possible mechanism for PD-1 activation independent of PD-L1 in BTICs. However, unlike other CD28 family members, the PD-1 molecule lacks the cysteine residues required for disulfide bond formation and subsequent dimerization (54). Nonetheless, a recent study revealed that SHP-2 connects phosphorylated ITSM residues on two PD-1 molecules, forming a PD-1:PD-1 dimer (46). Thus, it is plausible that RMC4550, a SHP-2 inhibitor, blocks the potential dimerization of PD-1 in BTICs by altering the protein structure of SHP-2. Last, PD-1 may interact with proteins other than PD-L1 or PD-L2 ligands to activate the signaling pathway in BTICs. Thus, interactome studies are required to find proteins in BTICs that may interact with the extracellular domains of PD-1.
The tumor-promoting effects of PD-1 independent of PD-L1 binding are an important consideration in the field of immune checkpoint blockade therapy of patients with GBM. Most commercially available blocking antibodies inhibit interactions between PD-1 and PD-L1 (33, 34). As a result, blocking antibodies tested in therapeutic trials for GBM patients with the goal of reactivating T cells are likely to miss disrupting the critical growth-promoting role of PD-1 intrinsic to BTICs. This is consistent with our findings that two clinical antibodies were ineffective in abrogating the proliferative functions of PD-1 on BTICs. Therefore, investigation of other strategies such as small-molecule inhibitors to block PD-1 downstream signaling or suppression of PD-1 expression by in vivo small interfering RNA or CRISPR-Cas9–based gene therapy might be of high interest to abrogate tumor-promoting effects of PD-1 in GBMs.
While PD-1 is expressed on a small population of cells within the GBM microenvironment, BTICs are critically important in GBM as they drive tumor growth, recurrence, and intratumoral heterogeneity (55). BTICs are responsible for resistance to pharmacology, radiation, and surgery, and thus, PD-1 might be a new molecular target to contain BTIC growth in patients with GBM. In addition, the immunomodulatory effects of tumor-intrinsic PD-1 are areas for further investigation.
Together, identification of PD-1 as a previously unknown driver of NFκB signaling and proliferation in BTICs without the requirement of PD-L1 ligation and understanding of the mechanisms underlying its protumorigenic effects could contribute to the discovery of a new therapeutic approach for patients with GBM.
MATERIALS AND METHODS
Human tissues
Human GBM tissues for immunohistochemistry (n = 7; table S1) were collected in accordance with the protocols approved by the Institutional Review Board of Indiana University. Consent was given by all patients. The fresh tissues surgically removed from patients were snapped frozen and stored in −80°C. The tissues were fixed in formalin and histologically processed for paraffin-embedded blocks for the studies. The hematoxylin and eosin–stained slides were examined by the neuropathologist (C. Hao). All patients provided written informed consent to obtain tumor biopsies. Cerebral cortex of normal human individuals was purchased from AMSBIO (catalog number HP-210).
Animal experimental models
All mice used in this study were on C57BL/6 background. Female wild-type C57BL/6 mice were obtained from Charles River. Rag1 KO mice were a gift of P. Kubes (University of Calgary). Six- to 8-week-old mice (female) were used for all experiments of the GBM preclinical model. Rag1 KO female mice were bred and maintained in our animal facility. All experiments were conducted with ethics approval from the Animal Care Committee at the University of Calgary under regulations of the Canadian Council of Animal Care.
Method details
Generation of GBM patient–derived BTICs and neural stem cells
Human BTIC lines were generated from resected specimens of patients with GBM as described previously (22). We grew cells in a 5% CO2 incubator and in a serum-free NeuroCult NS-A basal medium (STEMCELL Technologies) supplemented with NeuroCult™ Proliferation Supplement (STEMCELL Technologies), EGF (PeproTech Inc.), FGF (R&D Systems), and heparin solution (STEMCELL Technologies); we refer to this as BTIC medium. To propagate the cells, BTIC spheres were dissociated mechanically and plated into T-75 culture flasks. These lines were cultured chronologically, maintained, and authenticated within the University of Calgary BTIC Core. Human and mouse fetal neural stem cells were generated and cultured as previously described (56, 57). Human brain tissues were obtained from 12- to 18-week-old fetuses from therapeutic abortions according to ethical guidelines established by the University of Calgary. The use of these human samples is approved by the Institutional Review Board of the University of Calgary, and consent was obtained from all donors of tissues.
Isolation and characterization of mouse BTICs
Mouse BTIC line mBT0309 and mBT0528 in the C57BL/6 background were isolated, characterized, and maintained in the BTIC medium as described elsewhere (21). These lines have been previously shown to recapitulate key features of a human GBM (21).
Evaluation of BTIC growth
For the sphere formation assay, freshly dissociated cells were plated at 10,000 cells per well in 100 μl of BTIC medium into a 96-well flat bottom plate. The resultant number of spheres above the 60-μm-diameter cutoff, a convenient parameter to describe growth characteristics, was monitored after 3 to 5 days by bright-filed photographing multiple fields per well with subsequent analyzes as previously described (8, 43, 58). For the blockade of signaling pathways, BTICs were treated with the following inhibitors: p38 MAP Kinase Inhibitor Skepinone-l (Millipore, 506174), IKK-α/β inhibitor BMS 345541 (Abcam, ab144822), or RMC-4550 (Chemietek, CT-RMC4550).
Intracranial tumor implantation
After sphere dissociation of BTICs, 20,000 viable cells were resuspended in 2 μl of phosphate-buffered saline (PBS) and implanted stereotactically into the right striatum of mice as described previously (22). After implantation, we daily monitored animals to assess weight loss and physical/neurological abnormalities.
Flow cytometry
Surface staining was performed in PBS for 15 to 20 min at 4°C. For phospho-flow cytometry, cells were fixed in Fix Buffer I (BD, 557870) for 10 min at 37°C, permeabilized in Perm/Wash Buffer (BD, 554723), and then stained for 30 min at room temperature. Cells were subjected to Attune NxT flow cytometry (Thermo Fisher Scientific). Data analysis was performed by FlowJo version 10.7.2 (Treestar). In the flow cytometry dot plots, x axis shows fluorescence intensity detected in each channel for every event, and y axis represents forward scatter area (FSC-A) that detects scatter along the path of the laser and is proportional to cell size. The scale is an arbitrary scale representing increasing intensity of signal. Following antibodies were used for the cell staining: allophycocyanin (APC) anti-human CD279 (BD, 558694), Alexa Fluor 647 anti-human CD279 (BD, 560838), phycoerythrin (PE) anti-human CD274 (BD, 557924), BV421 anti-human CD273 (BD, 563842), PE hamster anti-mouse CD279 (BD, 561788), Brilliant Violet 421 (BV421) anti-NFκB p65 (pS529) (BD, 565446), PE anti–p-IKKα/β (Ser176/180) (Cell Signaling Technology, 14938), Alexa Fluor 488 anti-IKKα/β (Abcam, ab200618), PE anti-SHP2 (pY542) (BD, 560389), PE anti-SOX2 (BioLegend, 656104), and PE anti-nestin (BD, 561230).
Cell proliferation analysis by ATP luminescence and Click-iT EdU-labeling assays
Cell viability was assessed using the ATP CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega, G7572) based on the manufacturer’s protocol using 100 μl of CellTiter-Glo Reagent per well. After 10 min of incubation at room temperature, 100 μl of the solution was transferred to a white 96-well plate, and the luminescence signal was measured with a luminometer (Labsystems). For Click-iT EdU proliferation assay, cells were incubated with 1 μM EdU for 24 to 48 hours. After cell permeabilization, EdU staining was performed with the Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Cells were subjected to Attune NxT flow cytometry (Thermo Fisher Scientific). Data analysis was performed by FlowJo version 10.7.2 (Treestar).
Sequencing of CDS of PDCD1 gene in human BTICs
Briefly, frozen cell pellets were gently thawed on ice and lysed using QIAGEN lysis buffer and 2-min centrifugation at 14,000 rpm with QIAshredder spin columns (QIAGEN, 79654). RNA was purified from the resulting lysate using the RNeasy Mini Kit (QIAGEN, 74104) according to the manufacturer’s instructions. RNA was reverse-transcribed to complementary DNA (cDNA) using a SuperScript III First-Strand synthesis system (Thermo Fisher Scientific, 18080051). The human PD-1 cDNA was amplified using the primers human PD-1 forward (ATGCAGATCCCACAGGCGC) and human PD-1 reverse (GATCGCAGATCCTTCGCGGCCGC) using KAPA HiFi HotStart ReadyMix (Roche, KK2601). The resulting PCR products were cloned into pBluescript SK(–), and the DNA insert was sequenced.
Quantitative real-time PCR
Cells were lysed in 1 ml of TRIzol (Thermo Fisher Scientific). RNA was purified with the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Concentration and quality of RNA were assessed via measuring optical density at wavelengths of 260 and 280 nm by a NanoDrop spectrophotometer (Thermo Fisher Scientific). Samples were subjected to deoxyribonuclease (DNase) digestion by Ribonuclease-Free DNase Set (QIAGEN, 79254). RNA was reverse-transcribed to cDNA using the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, 11754050). Forward and reverse primers to detect transcripts were purchased from QIAGEN: PDCD1 (QT01005746), Pdcd1 (QT00111111), SOX2 (QT00237601), NESTIN (QT00235781), MSI1 (QT00025389), MKI67 (QT00014203), IKBKB (QT00062482), TNFRSF19 (QT00024829), and CDKN1A (QT00062090) with GAPDH (QT00079247) or Gapdh (QT01658692) as housekeeping controls. Transcripts were quantified by RT-qPCR on the QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) using the QuantiFast SYBR Green PCR Kit (QIAGEN, 204054). The relative expression levels between genes were calculated using a comparative cycle threshold method, with expression levels normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Reactions were run in triplicate or quadruplicate.
Confocal immunofluorescence microscopy
We evaluated formalin-fixed paraffin-embedded specimens from patients with GBM. The sections of 10 μm in thickness were subjected to deparaffinization and antigen retrieval using heat-mediated antigen retrieval with tris/EDTA buffer (pH 9). Samples were permeabilized with 0.25% Triton X-100 in PBS 10 min for intracellular markers. Tissues were blocked by incubation for 1 hour at room temperature with horse-blocking solution [PBS, 10% horse serum, 1% bovine serum albumin (BSA), 0.1% cold fish stain gelation, 0.1% Triton X-100, and 0.05% Tween 20]. Tissues were then incubated overnight at 4°C with following antibodies resuspended in antibody dilution buffer (PBS, 1% BSA, 0.1% cold fish stain gelation, and 0.1% Triton X-100): human CD45 (1:100; Dako, M0701), mouse CD45 (1:100; BD, 550539), PD-1 (1:1000; Cell Signaling Technology, 3501), PD-1 (1:1000; Thermo Fisher Scientific, UM800091CF), SOX2 (1:200; Thermo Fisher Scientific, 14-9811-82), human nestin (1:200; Millipore, MAB5326), mouse nestin (1:1000; Novus Biologicals, NB100-1604), p65 (1:400; Cell Signaling Technology, 6956), Ki67 (1:400; Cell Signaling Technology, 9449), human PD-L1 (1:100; Abcam, ab210931), and mouse PD-L1 (1:200; Thermo Fisher Scientific, 14-5983-82). The corresponding fluorophore-conjugated secondary antibodies (1:500; Jackson ImmunoResearch Laboratories or Thermo Fisher Scientific) were then added with 4′,6-diamidino-2-phenylindole (DAPI) (1:1000). Fluoromount-G (SouthernBiotech, 0100-01) was used to adhere coverslips to the slides. Laser confocal immunofluorescence images were obtained at room temperature using the Leica TCS SP8 laser confocal microscope with a 25/0.5 numerical aperture (NA) water objective. Samples were excited by 405-, 488-, 552-, and 640-nm lasers and detected by two low dark current Hamamatsu photomultiplier tube detectors and two high-sensitivity hybrid detectors on the SP8. The following settings were used to capture images of samples: 8 bits, bidirectional scanning in a z-stack, 4× frame averaging, 1 airy unit pinhole, 0.75 zoom, and 2048 by 2048 pixels x-y resolution. For all samples in each series of trials, the same laser, gain, and offset settings were used to optimize contrast and reduce saturation. A sample slide stained with only the secondary antibodies and DAPI was always included for each experiment as a control. Three and four fields of view (FOVs) were obtained from each sample of human and mouse brain slices. Image capture was performed using Leica Application Suite X, image threshold and particle analysis were performed with ImageJ, and 3D rendering was performed with Imaris (Bitplane).
Immunofluorescence staining of cells
BTIC was seeded in a 96-well Black/Clear Flat Bottom TC–treated Imaging Microplate (Falcon, 353219) at a density of 10,000 cells per well in BTIC medium. After incubation at 37°C incubator, cells were washed with PBS and then fixed in 4% paraformaldehyde for 10 min.
For immunocytochemistry, cells were permeabilized using 0.5% Triton X-100 and 5% BSA for 1 to 2 hours at room temperature. Primary antibodies for PD-1 (1:100; Cell Signaling Technology, 3501), SOX2 (1:100; Thermo Fisher Scientific, 14-9811-82), human nestin (1:200; Millipore, MAB5326), mouse nestin (1:500; Novus Biologicals, NB100-1604), Ki67 (1:1000; Cell Signaling Technology, 9449), Musashi1 (1:500; Thermo Fisher Scientific, 14-9896-82), and p65 (1:800; Cell Signaling Technology, 6956) were incubated overnight at 4°C. Cells were subsequently incubated with corresponding fluorophore-conjugated secondary antibodies (1:500 Jackson ImmunoResearch Laboratories or Thermo Fisher Scientific) at room temperature for 1 hour and washed, and nuclei were counterstained with Hoechst (1:1000; Sigma-Aldrich) and imaged using ImageXpress (Molecular Devices). The number of cells in four FOVs per well was obtained and then normalized to obtain a percentage value for each well. This value was averaged across three to four replicates per condition. Nuclear translocation HT analysis in the MetaXpress High-Content Image Acquisition and Analysis software (Molecular Devices) was used to quantify p65 nuclear translocation from the fluorescence microscopy images.
RNA sequencing and analysis
BTICs were plated in 12-well plates at a density of 106 cells per well in 1 ml of BTIC medium. Cells were incubated for 3 hours at 37°C with 5% CO2, after which total RNA was isolated using the RNeasy Mini Kit (QIAGEN) from cell pellets. Total RNA samples were verified for integrity using RNA assays on an Agilent TapeStation instrument. Satisfactory samples [RIN (RNA integrity number) scores were >9.3] were then subjected to enrichment of polyadenylated [poly(A)] mRNA, cDNA synthesis, and Illumina compatible library preparation using the NEBNext Poly(A) mRNA Magnetic Isolation Module, the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, and NEBNext Multiplex Index Primers, all according to the New England Biolabs–recommended protocols. Libraries were then quantitated by both Agilent TapeStation High Sensitivity assays and a KAPA qPCR Library quantification assay before pooling. Sequencing was performed on an Illumina NextSeq 500 sequencer with 75-cycle high-output sequencing kits, as per standard Illumina methods, to yield an average of 70 million reads per sample. Differential expression analysis was performed between BTIC lines overexpressing PD-1 (n = 3) and lines transfected with OE control vector (n = 3). The R package “DESeq2” was used for differential expression analysis. Visualization of the analysis results was performed using the R packages “EnhancedVolcano” and “pheatmap.” The adjusted P value cutoff was set at 0.05 and log2(fold change) at +0.58 for filtering of the final gene list. The gene list was also evaluated via literature review.
Generation of Cd274 KO mouse BTIC cells by CRISPR gene-editing system
CRISPR gene KO of PD-L1 gene (Cd274) in mBT0309 was carried out using the pSpCas9(BB)-2A–green fluorescent protein (GFP; pX458) vector and a short-guide sequence targeting exon three, 5′-GTGACCACCAACCCGTGAGT-3′. PX458 vector was a gift from F. Zhang (Addgene plasmid #48138; http://n2t.net/addgene:48138) (59). Short-guide sequence oligonucleotides were synthesized, annealed, cloned into pX458 vector, and confirmed by DNA sequencing at DNA laboratory, University of Calgary. Three micrograms of DNA4 plasmid (pX458 containing Cd274 short guide RNA sequence) was transfected into cells using the Mouse Embryonic Stem Cell Nucleofector Kit (Lonza, VPH-1001) according to the manufacturer’s recommendation. After transfection, cells were harvested, and genomic DNA was isolated using the KAPA Express Extract Kit (KAPA Biosystems, KK7100) according to the manufacturer’s instructions. Genomic DNA fragments of Cd274 around the short-guide RNA site were amplified by PCR. Wild-type Cd274 DNA fragment was also amplified as control. The primers used to amplify the genomic DNA region of Cd274 exon number 3 were as follows: 5′-AATAGCCAGCCCGAGTTGAT-3′ (forward) and 5′-TCACCTGGGATGATCAGCTC-3′ (reverse). PCR products were used for nuclease mutation detection using the SURVEYOR Mutation Detection Kit (Transgenomic) according to the manufacturer’s protocol. After transfection, GFP-positive cells were fluorescence-activated cell sorting–sorted into a 96-well plate, at the Flow Cytometry Facility, University of Calgary. Single cells were then expanded for further analysis (i.e., Western blot and DNA sequencing). For DNA sequencing of Cd274 KO clones, DNA fragments of cells around the short guide RNA site were amplified by PCR. PCR products were subcloned into pEGFP-C2 vector (Clontech), and plasmid DNA from individual clones were sent for Sanger DNA sequencing at DNA laboratory, University of Calgary to confirm all indel.
Immunoprecipitation and immunoblotting
Whole-cell extracts were generated by NP40 Cell Lysis Buffer (Thermo Fisher Scientific, FNN0021) supplemented with 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (Sigma-Aldrich, P2714-1BTL). Protein concentrations were determined using a BCA (bicinchoninic acid) assay kit (Sigma-Aldrich) with BSA as standard. Equal amounts (50 μg per lane) of total protein and 10 μl of PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific) were then run on NuPAGE 10% Bis-Tris Protein Gels (Thermo Fisher Scientific) with Mops SDS Running Buffer (Thermo Fisher Scientific) and using the Invitrogen NuPage Novex Gel System. After transferring into Hybond 0.2-μm polyvinylidene difluoride (Amersham) using a wet transfer system (Bio-Rad), membranes were blocked with 5% BSA and then probed overnight with the following primary antibodies: PD-1 (1:1000; Cell Signaling Technology, 3501), p–SHP-2 (Y542) (1:1000; Cell Signaling Technology, 3751), SHP-2 (1:1000; Cell Signaling Technology, 3752), p-IKKα/β (Ser176/180) (1:1000; Cell Signaling Technology, 2697), IKKβ (1:1000; Cell Signaling Technology, 2370), p-NFκB p65 (Ser536) (1:1000; Cell Signaling Technology, 3033), NFκB p65 (1:1000; Cell Signaling Technology, 8242), PD-L1 (1:250; R&D System, MAB90781), Mre11 (1:8000; Novus Biologicals, NB100-142), and actin (1:5000; Abcam, ab20272). The following day, membranes were washed with 0.1% 1× tris-buffered saline with Tween 20, incubated with the corresponding horseradish peroxidase–linked secondary antibody (1:50,000; Abcam), then developed using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific), and imaged with the ChemiDoc Touch Imaging System (Bio-Rad). co-IP of PD-1 was performed with PD-1 mouse antibody clone EH12 (BioLegend, 329902) covalently conjugated to Dynabeads protein G (Thermo Fisher Scientific, 10003D) according to the kit’s instructions.
MRI of mouse brain tumors
We used a 9.4T horizontal bore magnet and a Bruker console with Paravision 5.1 to monitor intracranial tumor growth at the Experimental Imaging Center, University of Calgary. All imaging was performed with a helium cooled cryocoil. A T2*-weighted Fast Low Angle Shot sequence was used to determine the tumor volume [repetition time (TR) = 1000 ms, echo time (TE) = 6.5 ms, flip angle = 60, NA = 4, voxel size = 0.075 mm by 0.075 mm by 0.25 mm]. Tumor volume was calculated in Fiji software (ImageJ, National Institutes of Health) by manually drawing a region of interest around the tumor area.
Stable knockdown and overexpress PD-1 in human and mouse BTICs
To stably knockdown PD-1 gene in BTIC lines, pLKO.1 lentiviral vectors containing human shRNAs specific for 3′ untranslated region (3′UTR; Sigma-Aldrich, TRCN0000083508) or CDS (Sigma-Aldrich, TRCN0000417811) of PDCD1 or mouse shRNAs specific for 3′UTR (Sigma-Aldrich, TRCN0000097670) or CDS (Sigma-Aldrich, TRCN0000097674) of Pdcd1 gene were packaged to Lentiviral particles. MISSION pLKO.1-puro Nontarget shRNA Control Plasmid DNA (Sigma-Aldrich, SHC016-1EA) was used as shRNA control. For generation-stable PD-1–overexpressing cells, human (Origene, SC117011) or mouse (Origene, MC209155) PD-1 sequence was inserted into PB-CMV-MCS-EF1α-Puro PiggyBac vector (System Biosciences, PB510B-1).
Site-directed mutagenesis and overexpression of PD-1 in BTICs
To abrogate PD-1 signaling in BTICs, we mutated tyrosine (Y) to phenylalanine (F) residues in ITIM (Y223F mutation) and ITSM (Y248F mutation). In addition, to generate a mutant version of PD-1 with defective PD-L1 binding, we substituted lysine (K) to alanine (A) residues (K78A mutation). The site-directed mutagenesis was performed using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB E5520S) according to the manufacturer’s instructions. The following mutagenic primers were used to mutate the human PD-1 (PDCD1): Y223F mutation, 5′-GTGTTCTCTGTGGACTTTGGGGAGCTGGATTTC-3′ (forward) and 5′-GAAATCCAGCTCCCCAAAGTCCACAGAGAACAC-3′ (reverse); Y248F mutation, 5′-CCTGAGCAGACGGAGTTTGCCACCATTGTCTTTC-3′ (forward) and 5′-GAAAGACAATGGTGGCAAACTCCGTCTGCTCAGG-3′ (reverse); K78A mutation, 5′-CAGCAACCAGACGGACGCCCTGGCCGCCTTCCCCG-3′ (forward) and 5′-CGGGGAAGGCGGCCAGGGCGTCCGTCTGGTTGCTG-3′ (reverse).
Then, the mutant human PDCD1 constructs were ligated into PiggyBac transposon-based vector (System Biosciences, PB510B-1). The ligated constructs were transformed into competent Escherichia coli cells (NEB 5-alpha, C2987) according to the manufacturer’s instructions. Fidelity of constructs was validated by DNA sequencing and restriction enzyme digestion. BTICs were transfected with the generated constructs by Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, 11668019) based on the company protocol. Transfected cells were selected by treating the cells with puromycin (1 μg/ml; Sigma-Aldrich, P8833). Overexpression of mutant or wild-type PD-1 was confirmed by RT-qPCR and flow cytometry for all BTIC lines.
Statistics analysis
Statistical significance (P < 0.05) was determined using Student’s t tests (comparing two variables) or one-way analysis of variance (ANOVA) (comparing three or more variables) with Tukey’s post hoc. Survival curves were calculated according to the Kaplan-Meier method; survival analysis was performed using the log-rank Mantel-Cox test. Data were analyzed using Prism 9.2.0 (GraphPad Software) unless otherwise noted. In violin plots, center lines represent median and two quartile lines. All data were presented as means ± SEM. We have indicated the n values used for each analysis in the figure captions. The results were reproducible and conducted with established controls. All data shown are representative of two to three independent experiments with similar results, unless otherwise indicated. Cell cultures were routinely screened for mycoplasma.
Acknowledgments
We appreciate the generous support by F. Visser at the Hotchkiss Brain Institute Molecular Core Facility. We acknowledge the Hotchkiss Brain Institute Advanced Microscopy Platform and the Cumming School of Medicine for support and use of Leica TCS SP8 and ImageXpress Micro XLS High-Content Analysis System. We also thank the flow cytometry core, the UCDNA sequencing, and genetic analysis laboratory at the University of Calgary. We also thank the Genome Engineering Center at the Cumming School of Medicine, University of Calgary for generating CRISPR Cd274 gene KO cell line; the BTIC Core headed by S. Weiss and G. Cairncross for isolating BTIC lines from patient-resected specimens; and C. C. Poon and J. Kelly for providing resected samples for the low-passage BTIC lines. This work was enabled by the bioinformatics services of the International Microbiome Centre (IMC), which is supported by the University of Calgary’s Cumming School of Medicine and the province of Alberta. We also wish to acknowledge Pfizer and Merck KGaA, Darmstadt, Germany for providing avelumab antibodies. Graphical abstract and schematic diagrams were created with BioRender.com.
Funding: This study was supported by grants from the Canadian Institutes of Health Research and the Canadian Cancer Society. R.M. was supported by a fellowship from the University of Calgary’s Eyes High program.
Author contributions: Conceptualization: R.M. and V.W.Y. Methodology: R.M. and V.W.Y. Validation: R.M. and V.W.Y. Formal analysis: R.M., A.G., M.K., and P.B. Investigation: R.M., A.G., and F.J.Z. Resources: V.W.Y., H.A.L., A.C.B., C.H., D.J.M., and J.F.D. Writing (original draft): R.M. Writing (review and editing): V.W.Y., R.M., D.J.M., F.J.Z., H.A.L., J.F.D., S.S., M.K., and P.B. Visualization: R.M. and A.G. Supervision: V.W.Y. Project administration: V.W.Y. Funding acquisition: V.W.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE160417. The GenBank accession numbers for sequences of human PD-1 in BTIC lines reported in this paper are MW051354 (GBM5), MW051355 (BT025), and MW051356 (BT073). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Tables S1 and S2
Legend for data file S1
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Louis D. N., Perry A., Wesseling P., Brat D. J., Cree I. A., Figarella-Branger D., Hawkins C., Ng H. K., Pfister S. M., Reifenberger G., Soffietti R., von Deimling A., Ellison D. W., The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro-Oncology 23, 1231–1251 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O.; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Stupp R., Taillibert S., Kanner A., Read W., Steinberg D., Lhermitte B., Toms S., Idbaih A., Ahluwalia M. S., Fink K., Di Meco F., Lieberman F., Zhu J. J., Stragliotto G., Tran D., Brem S., Hottinger A., Kirson E. D., Lavy-Shahaf G., Weinberg U., Kim C. Y., Paek S. H., Nicholas G., Bruna J., Hirte H., Weller M., Palti Y., Hegi M. E., Ram Z., Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA 318, 2306–2316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B., Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Osuka S., Van Meir E. G., Overcoming therapeutic resistance in glioblastoma: The way forward. J. Clin. Invest. 127, 415–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klemm F., Maas R. R., Bowman R. L., Kornete M., Soukup K., Nassiri S., Brouland J.-P., Iacobuzio-Donahue C. A., Brennan C., Tabar V., Gutin P. H., Daniel R. T., Hegi M. E., Joyce J. A., Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 181, 1643–1660.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirzaei R., Sarkar S., Dzikowski L., Rawji K. S., Khan L., Faissner A., Bose P., Yong V. W., Brain tumor-initiating cells export tenascin-C associated with exosomes to suppress T cell activity. Oncoimmunology 7, e1478647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar S., Yang R., Mirzaei R., Rawji K., Poon C., Mishra M. K., Zemp F. J., Bose P., Kelly J., Dunn J. F., Yong V. W., Control of brain tumor growth by reactivating myeloid cells with niacin. Sci. Transl. Med. 12, eaay9924 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Gangoso E., Southgate B., Bradley L., Rus S., Galvez-Cancino F., McGivern N., Güç E., Kapourani C. A., Byron A., Ferguson K. M., Alfazema N., Morrison G., Grant V., Blin C., Sou I., Marques-Torrejon M. A., Conde L., Parrinello S., Herrero J., Beck S., Brandner S., Brennan P. M., Bertone P., Pollard J. W., Quezada S. A., Sproul D., Frame M. C., Serrels A., Pollard S. M., Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell 184, 2454–2470.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antunes A. R. P., Scheyltjens I., Lodi F., Messiaen J., Antoranz A., Duerinck J., Kancheva D., Martens L., De Vlaminck K., Van Hove H., Hansen S. S. K., Bosisio F. M., Van der Borght K., De Vleeschouwer S., Sciot R., Bouwens L., Verfaillie M., Vandamme N., Vandenbroucke R. E., De Wever O., Saeys Y., Guilliams M., Gysemans C., Neyns B., De Smet F., Lambrechts D., Van Ginderachter J. A., Movahedi K., Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 24, 595–610 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Egen J. G., Ouyang W., Wu L. C., Human anti-tumor immunity: Insights from immunotherapy clinical trials. Immunity 52, 36–54 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J., Chiarion-Sileni V., Gonzalez R., Grob J. J., Rutkowski P., Lao C. D., Cowey C. L., Schadendorf D., Wagstaff J., Dummer R., Ferrucci P. F., Smylie M., Hogg D., Hill A., Márquez-Rodas I., Haanen J., Guidoboni M., Maio M., Schöffski P., Carlino M. S., Lebbé C., McArthur G., Ascierto P. A., Daniels G. A., Long G. V., Bastholt L., Rizzo J. I., Balogh A., Moshyk A., Hodi F. S., Wolchok J. D., Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 381, 1535–1546 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Ansell S. M., Lesokhin A. M., Borrello I., Halwani A., Scott E. C., Gutierrez M., Schuster S. J., Millenson M. M., Cattry D., Freeman G. J., Rodig S. J., Chapuy B., Ligon A. H., Zhu L., Grosso J. F., Kim S. Y., Timmerman J. M., Shipp M. A., Armand P., PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 372, 311–319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian S. L., Hodi F. S., Brahmer J. R., Gettinger S. N., Smith D. C., McDermott D. F., Powderly J. D., Carvajal R. D., Sosman J. A., Atkins M. B., Leming P. D., Spigel D. R., Antonia S. J., Horn L., Drake C. G., Pardoll D. M., Chen L., Sharfman W. H., Anders R. A., Taube J. M., McMiller T. L., Xu H., Korman A. J., Jure-Kunkel M., Agrawal S., McDonald D., Kollia G. D., Gupta A., Wigginton J. M., Sznol M., Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirzaei R., Sarkar S., Yong V. W., T cell exhaustion in glioblastoma: Intricacies of immune checkpoints. Trends Immunol. 38, 104–115 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Arvanitis C. D., Ferraro G. B., Jain R. K., The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer 20, 26–41 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg S. B., Gettinger S. N., Mahajan A., Chiang A. C., Herbst R. S., Sznol M., Tsiouris A. J., Cohen J., Vortmeyer A., Jilaveanu L., Yu J., Hegde U., Speaker S., Madura M., Ralabate A., Rivera A., Rowen E., Gerrish H., Yao X., Chiang V., Kluger H. M., Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: Early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol. 17, 976–983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z., Mesci P., Bernatchez J. A., Gimple R. C., Wang X., Schafer S. T., Wettersten H. I., Beck S., Clark A. E., Wu Q., Prager B. C., Kim L. J. Y., Dhanwani R., Sharma S., Garancher A., Weis S. M., Mack S. C., Negraes P. D., Trujillo C. A., Penalva L. O., Feng J., Lan Z., Zhang R., Wessel A. W., Dhawan S., Diamond M. S., Chen C. C., Wechsler-Reya R. J., Gage F. H., Hu H., Siqueira-Neto J. L., Muotri A. R., Cheresh D. A., Rich J. N., Zika virus targets glioblastoma stem cells through a SOX2-integrin α. Cell Stem Cell 26, 187–204.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silver D. J., Roversi G. A., Bithi N., Wang S. Z., Troike K. M., Neumann C. K., Ahuja G. K., Reizes O., Brown J. M., Hine C., Lathia J. D., Severe consequences of a high-lipid diet include hydrogen sulfide dysfunction and enhanced aggression in glioblastoma. J. Clin. Invest. 131, e138276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisklakova A., McKenzie B., Zemp F., Lun X., Kenchappa R. S., Etame A. B., Rahman M. M., Reilly K., Pilon-Thomas S., McFadden G., Kurz E., Forsyth P. A., M011L-deficient oncolytic myxoma virus induces apoptosis in brain tumor-initiating cells and enhances survival in a novel immunocompetent mouse model of glioblastoma. Neuro-Oncology 18, 1088–1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly J. J. P., Stechishin O., Chojnacki A., Lun X., Sun B., Senger D. L., Forsyth P., Auer R. N., Dunn J. F., Cairncross J. G., Parney I. F., Weiss S., Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells 27, 1722–1733 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Jensen K. V., Cseh O., Aman A., Weiss S., Luchman H. A., The JAK2/STAT3 inhibitor pacritinib effectively inhibits patient-derived GBM brain tumor initiating cells in vitro and when used in combination with temozolomide increases survival in an orthotopic xenograft model. PLOS ONE 12, e0189670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y., Grisdale C. J., Islam S. A., Bose P., Lever J., Zhao E. Y., Grinshtein N., Ma Y., Mungall A. J., Moore R. A., Lun X., Senger D. L., Robbins S. M., Wang A. Y., MacIsaac J. L., Kobor M. S., Luchman H. A., Weiss S., Chan J. A., Blough M. D., Kaplan D. R., Cairncross J. G., Marra M. A., Jones S. J. M., Comprehensive genomic profiling of glioblastoma tumors, BTICs, and xenografts reveals stability and adaptation to growth environments. Proc. Natl. Acad. Sci. U.S.A. 116, 19098–19108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marasco M., Berteotti A., Weyershaeuser J., Thorausch N., Sikorska J., Krausze J., Brandt H. J., Kirkpatrick J., Rios P., Schamel W. W., Köhn M., Carlomagno T., Molecular mechanism of SHP2 activation by PD-1 stimulation. Sci. Adv. 6, eaay4458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okazaki T., Maeda A., Nishimura H., Kurosaki T., Honjo T., PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc. Natl. Acad. Sci. U.S.A. 98, 13866–13871 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q., Lenardo M. J., Baltimore D., 30 Years of NF-κB: A blossoming of relevance to human pathobiology. Cell 168, 37–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Z., Kloss J. M., Tuncali S., Tran N. L., Loftus J. C., TROY signals through JAK1-STAT3 to promote glioblastoma cell migration and resistance. Neoplasia 22, 352–364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rinkenbaugh A. L., Cogswell P. C., Calamini B., Dunn D. E., Persson A. I., Weiss W. A., Lo D. C., Baldwin A. S., IKK/NF-κB signaling contributes to glioblastoma stem cell maintenance. Oncotarget 7, 69173–69187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You M., Flick L. M., Yu D., Feng G. S., Modulation of the nuclear factor kappa B pathway by Shp-2 tyrosine phosphatase in mediating the induction of interleukin (IL)-6 by IL-1 or tumor necrosis factor. J. Exp. Med. 193, 101–110 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirahara K., Ghoreschi K., Yang X.-P., Takahashi H., Laurence A., Vahedi G., Sciume G., Hall A. O. H., Dupont C. D., Francisco L. M., Chen Q., Tanaka M., Kanno Y., Sun H.-W., Sharpe A. H., Hunter C. A., O’Shea J. J., Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity 36, 1017–1030 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lázár-Molnár E., Yan Q., Cao E., Ramagopal U., Nathenson S. G., Almo S. C., Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc. Natl. Acad. Sci. U.S.A. 105, 10483–10488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H. T., Lee S. H., Heo Y.-S., Molecular interactions of antibody drugs targeting PD-1, PD-L1, and CTLA-4 in immuno-oncology. Molecules 24, 1190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu K., Tan S., Chai Y., Chen D., Song H., Zhang C. W., Shi Y., Liu J., Tan W., Lyu J., Gao S., Yan J., Qi J., Gao G. F., Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy. Cell Res. 27, 151–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleffel S., Posch C., Barthel S. R., Mueller H., Schlapbach C., Guenova E., Elco C. P., Lee N., Juneja V. R., Zhan Q., Lian C. G., Thomi R., Hoetzenecker W., Cozzio A., Dummer R., Mihm M. C. Jr., Flaherty K. T., Frank M. H., Murphy G. F., Sharpe A. H., Kupper T. S., Schatton T., Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell 162, 1242–1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Li X., Liu S., Guo L., Zhang B., Zhang J., Ye Q., Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology 66, 1920–1933 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Yang X., Zhang C., Wang Y., Cheng T., Duan L., Tong Z., Tan S., Zhang H., Saw P. E., Gu Y., Wang J., Zhang Y., Shang L., Liu Y., Jiang S., Yan B., Li R., Yang Y., Yu J., Chen Y., Gao G. F., Ye Q., Gao S., Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc. Natl. Acad. Sci. U.S.A. 117, 6640–6650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dirkse A., Golebiewska A., Buder T., Nazarov P. V., Muller A., Poovathingal S., Brons N. H. C., Leite S., Sauvageot N., Sarkisjan D., Seyfrid M., Fritah S., Stieber D., Michelucci A., Hertel F., Herold-Mende C., Azuaje F., Skupin A., Bjerkvig R., Deutsch A., Voss-Böhme A., Niclou S. P., Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 10, 1787 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirosh I., Venteicher A. S., Hebert C., Escalante L. E., Patel A. P., Yizhak K., Fisher J. M., Rodman C., Mount C., Filbin M. G., Neftel C., Desai N., Nyman J., Izar B., Luo C. C., Francis J. M., Patel A. A., Onozato M. L., Riggi N., Livak K. J., Gennert D., Satija R., Nahed B. V., Curry W. T., Martuza R. L., Mylvaganam R., Iafrate A. J., Frosch M. P., Golub T. R., Rivera M. N., Getz G., Rozenblatt-Rosen O., Cahill D. P., Monje M., Bernstein B. E., Louis D. N., Regev A., Suva M. L., Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao H., Wang H., Li C., Fang J. Y., Xu J., Cancer cell-intrinsic PD-1 and implications in combinatorial immunotherapy. Front. Immunol. 9, 1774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chua B. A., Van Der Werf I., Jamieson C., Signer R. A. J., Post-transcriptional regulation of homeostatic, stressed, and malignant stem cells. Cell Stem Cell 26, 138–159 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H., Yu X., Han X., Hao J., Zhao J., Bebek G., Bao S., Prayson R. A., Khalil A. M., Jankowsky E., Yu J. S., Piwil1 regulates glioma stem cell maintenance and glioblastoma progression. Cell Rep. 34, 108522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar S., Mirzaei R., Zemp F. J., Wei W., Senger D. L., Robbins S. M., Yong V. W., Activation of NOTCH signaling by tenascin-C promotes growth of human brain tumor-initiating cells. Cancer Res. 77, 3231–3243 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Wei F., Zhong S., Ma Z., Kong H., Medvec A., Ahmed R., Freeman G. J., Krogsgaard M., Riley J. L., Strength of PD-1 signaling differentially affects T-cell effector functions. Proc. Natl. Acad. Sci. U.S.A. 110, E2480–E2489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn E., Araki K., Hashimoto M., Li W., Riley J. L., Cheung J., Sharpe A. H., Freeman G. J., Irving B. A., Ahmed R., Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, 4749–4754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patsoukis N., Duke-Cohan J. S., Chaudhri A., Aksoylar H. I., Wang Q.; A. Council, Berg A., Freeman G. J., Boussiotis V. A., Interaction of SHP-2 SH2 domains with PD-1 ITSM induces PD-1 dimerization and SHP-2 activation. Commun. Biol. 3, 128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du S., McCall N., Park K., Guan Q., Fontina P., Ertel A., Zhan T., Dicker A. P., Lu B., Blockade of tumor-expressed PD-1 promotes lung cancer growth. Onco. Targets. Ther. 7, e1408747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sang Y., Hou Y., Cheng R., Zheng L., Alvarez A. A., Hu B., Cheng S. Y., Zhang W., Li Y., Feng H., Targeting PDGFRα-activated glioblastoma through specific inhibition of SHP-2-mediated signaling. Neuro-Oncology 21, 1423–1435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roccograndi L., Binder Z. A., Zhang L., Aceto N., Zhang Z., Bentires-Alj M., Nakano I., Dahmane N., O’Rourke D. M., SHP2 regulates proliferation and tumorigenicity of glioma stem cells. J. Neuro-Oncol. 135, 487–496 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Liu R., Qu X., Yu H., Chu H., Zhang Y., Zhu W., Wu X., Gao H., Tao B., Li W., Liang J., Li G., Yang W., α-ketoglutarate-activated NF-κB signaling promotes compensatory glucose uptake and brain tumor development. Mol. Cell 76, 148–162.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Ueda T., Mawji N. R., Bruchovsky N., Sadar M. D., Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J. Biol. Chem. 277, 38087–38094 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Daniel A. R., Lange C. A., Protein kinases mediate ligand-independent derepression of sumoylated progesterone receptors in breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 14287–14292 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakraborty S., Li L., Puliyappadamba V. T., Guo G., Hatanpaa K. J., Mickey B., Souza R. F., Vo P., Herz J., Chen M. R., Boothman D. A., Pandita T. K., Wang D. H., Sen G. C., Habib A. A., Constitutive and ligand-induced EGFR signalling triggers distinct and mutually exclusive downstream signalling networks. Nat. Commun. 5, 5811 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X., Schwartz J. C., Guo X., Bhatia S., Cao E., Lorenz M., Cammer M., Chen L., Zhang Z. Y., Edidin M. A., Nathenson S. G., Almo S. C., Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity 20, 337–347 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Prager B. C., Bhargava S., Mahadev V., Hubert C. G., Rich J. N., Glioblastoma stem cells: Driving resilience through chaos. Trends Cancer 6, 223–235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chojnacki A., Kelly J. J., Hader W., Weiss S., Distinctions between fetal and adult human platelet-derived growth factor-responsive neural precursors. Ann. Neurol. 64, 127–142 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Chojnacki A., Weiss S., Production of neurons, astrocytes and oligodendrocytes from mammalian CNS stem cells. Nat. Protoc. 3, 935–940 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Sarkar S., Döring A., Zemp F. J., Silva C., Lun X., Wang X., Kelly J., Hader W., Hamilton M., Mercier P., Dunn J. F., Kinniburgh D., van Rooijen N., Robbins S., Forsyth P., Cairncross G., Weiss S., Yong V. W., Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat. Neurosci. 17, 46–55 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., Zhang F., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Tables S1 and S2
Legend for data file S1
Data file S1