Abstract
Ovarian cancer is one of the leading causes of cancer related deaths affecting United States women. Early-stage detection of ovarian cancer has been linked to increased survival, however, current screening methods, such as biomarker testing, have proven to be ineffective in doing so. Therefore, further developments are necessary to be able to achieve positive patient prognosis. Ongoing efforts are being made in biomarker discovery towards clinical applications in screening for early-stage ovarian cancer. In this perspective, we discuss and provide examples for several workflows employing mass spectrometry-based proteomics towards protein biomarker discovery and characterization in the context of ovarian cancer; workflows include protein identification and characterization as well as intact protein profiling. We also discuss the opportunities to merge these workflows for a multiplexed approach for biomarkers. Lastly, we provide our insight as to future developments that may serve to enhance biomarker discovery workflows while also considering translational potential.
Introduction
Ovarian cancer is one of the leading causes of cancer deaths affecting women in the United States. Its relatively low five-year survival rate of 48.6% can be attributed to the fact that nearly two-thirds of women are not diagnosed until the cancer has metastasized to other areas of the body[1]. However, early detection of ovarian cancer has been shown to increase the five-year survival rate to over 90%[1]. Therefore, reliable methods of early stage ovarian cancer screening are of the utmost importance in positive patient prognosis.
Several screening methods currently exist for ovarian cancer, with one of the most common methods being biomarker testing[2]. Since the discovery of increased levels of carcinoembryonic antigen (CEA) in ovarian cancer patients, a variety of ovarian cancer biomarkers have been identified and approved for clinical diagnostic usage by the United States Food and Drug Administration (FDA)[3]. These biomarkers include single biomarkers, such as cancer antigen 125 (CA125) and human epididymis protein 4 (HE4), and more recently, biomarker panels such as OVA1® and Overa®[4–8].
Screening using the aforementioned biomarkers has been unable to reach desired levels for both sensitivity and specificity and require orthogonal screening methods and/or physician assessment. Dochez et al. compiled a subset of analyses to compare the diagnostic performance of CA125, HE4, and CA125 + HE4. Benchmarks showed sensitivity ranging from 58%−97% and specificity ranging from 53%−98%; high sensitivity was often accompanied by low specificity or vice versa[9–26]. OVA1® (Vermillion Inc), an FDA approved multiplexed index assay using CA125, transferrin, beta-2-microglobulin, transthyretin, and apolipoprotein A1 biomarkers, was developed as a new screening tool to be used in conjunction with physician assessment. Despite showing promising sensitivity of 82%−100% (dependent on the demographic tested), the assay was plagued by low specificity ranging from 26%−43%[6,27]. Later, a second generation multiplexed index assay was developed under the name Overa®, which replaced beta-2-microglobulin and transthyretin with HE4 and follicle stimulating hormone. With a sensitivity of 90%−96% and specificity of 47%−71%, there were notable improvements[8,28]. However, these tests require higher specificity to see broad, routine implementation for general women’s health screening purposes. These results have even led to news outlets, scientific authorities, and the FDA to caution against excessive use of these tests due to their penchant for false positives[29].
In recent years, efforts have been made to use various “-omics” technologies to identify novel, reliable ovarian cancer biomarkers to be used for early stage ovarian cancer screening purposes from DNA, RNA, proteins, and metabolites[30]. In the case of proteins and metabolites, mass spectrometry has proven to be an invaluable tool towards these efforts. Mass spectrometers are more readily available in clinical laboratories thanks to the growing number of in vitro diagnostic (IVD) assays. Here, we present an overview of different proteomics approaches compatible with mass spectrometry that have been developed over the years for ovarian cancer.
Proteomics Workflows
Instrumentation and workflows for mass spectrometry-based proteomics oftentimes fall into one of two categories: tandem mass spectrometry (MS/MS) based proteomics and intact protein profiling (also commonly referred to as protein fingerprinting and not to be confused with peptide mass fingerprinting). Both workflows can be applied for research use only (RUO) as well as clinical IVD applications. RUO workflows are often focused on biomarker discovery, involving protein identification followed by subsequent characterization to provide insight into protein structure, biological effects, and mechanism of action. Conversely, IVD workflows allow for accessible routine screening based on biomarker assays that have been developed, validated, and quality controlled for a given disease. Figure 1 provides an overview of workflows and usage, and Table 1 outlines several differences between the two. Swiatly et al. have written in-depth reviews on recent mass spectrometry-based proteomics and intact protein profiling approaches in the context of ovarian cancer[31]. Although matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) imaging mass spectrometry is not in the scope of this perspective, Kriegsmann et al. have written an excellent review regarding its use in the context of ovarian cancer[32].
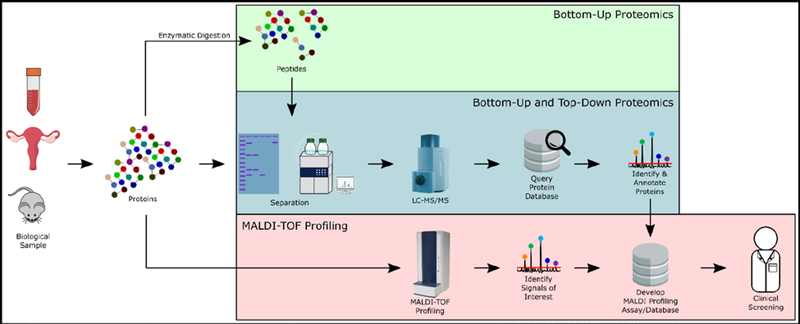
Figure 1:
Workflow from the bench to the clinic. Proteins can be extracted from fluid or tissue samples. Under the protein identification/characterization workflow, researchers have the option of digesting proteins into peptides using various methods prior to separation and MS/MS (bottom-up proteomics; green box). Otherwise, intact proteins can be maintained (top-down proteomics; blue box). In bottom-up and top-down proteomics, separation can be performed using gel electrophoresis or liquid chromatography, and is followed by collection MS/MS spectra. These spectra can be queried against peptide sequencing databases to ascertain their identities (blue box). To use this information for clinical screening purposes, development of a means of detection is still required. Under the intact protein profiling workflow (red), researchers collect protein profiles from intact proteins using MALDI-TOF mass spectrometers. Signals of interest can be putatively identified from these spectra and a database can then be compiled using protein profiles from different disease states, which can then be used in clinical settings to screen patient samples (i.e. benign or malignant).
Table 1:
Comparison of MS/MS based proteomics and MALDI-TOF protein profiling workflows.
| MS/MS Based Proteomics | MALDI-TOF Protein Profiling |
|---|---|
| Allows for analysis of intact proteins or peptides | Used for analysis of intact proteins |
| Requires more extensive sample preparation/cleanup | Minimal sample preparation required |
| Often coupled to online or offline separation/fractionation method | Separation/fractionation is not required |
| Allows for identification and characterization of proteins/peptides | Only provides putative mass value |
| Can perform absolute and/or relative quantitation | Can only perform relative quantitation |
| Sample preparation, cleanup, and separation/fractionation result in lower throughput workflow | Minimal sample preparation results in higher throughput analysis |
MS/MS Based Proteomics
As the name implies, MS/MS based proteomics has traditionally involved the use of a separation method followed by MS/MS. This workflow can be further categorized into bottom-up or top-down approaches. Wither et al. and Donelly et al. provide a more detailed overview of sample preparation and instrumentation used in bottom-up and top-down proteomics, respectively[33,34].
Bottom-up proteomics involves the use of digestion enzymes (i.e. trypsin or Lys-C) to digest proteins into peptides for MS/MS analysis. Peptides can be separated via offline (i.e. gel electrophoresis) or online (i.e. high performance liquid chromatography, HPLC; ultra-high performance liquid chromatography, UPLC; ion mobility) methods prior to detection via MS/MS or MSn [35,36]. Common mass spectrometers used in bottom-up proteomics include 3D/linear ion traps, TOF/TOF, orbitrap, hybrid quadrupole-TOF (qTOF), and hybrid quadrupole-orbitrap (Q-Exactive) mass spectrometers. Following MS analysis, peptide sequences can then be queried against protein databases (i.e. MASCOT, UniProt) for peptide annotation[37,38]. Tyanova et al. provides a detailed protocol for protein annotation in MaxQuant using custom databases (including those found on UniProt)[39]. As an example, Kacirova et al. used this workflow to identify cadherin-1 (CDH1), vitronectin (VTN), and basement membrane specific-heparan sulphate proteoglycan core protein (HSPG2) as downregulated proteins in the pathogenesis of endometrial cancer[40]. Malaker et al. also used StcE, a mucin-selective bacterial protease derived from Escherichia coli, to further characterize the structure and function of mucins, which are known to be implicated in various cancers[41]. For example, mucin 16 (MUC16; aka CA125), is a well known protein associated with ovarian cancer. Protocols and instrumentation for this method of proteomics are very mature; however, information about proteoforms and post-translational modifications are not always attainable[42,43].
Top-down proteomics, on the other hand, involves the analysis of intact proteins, which addresses some of the limitations of bottom-up proteomics by allowing the determination of complete protein sequences and post-translational modifications. Separation methods are similar to those used in bottom-up proteomics, but unlike bottom-up proteomics and intact protein profiling, top-down proteomics workflows use mass spectrometers capable of analyzing high molecular weight proteins (tens or hundreds of kilodaltons) such as MALDI-TOF, Fourier-transform ion cyclotron resonance (FT-ICR), or hybrid FT-ICR mass spectrometers. This workflow also uses MS/MS or MSn analysis followed by protein annotation via database searches. Ntai et al. were able to use immunoaffinity enrichment coupled to top-down proteomics on a Q-Exactive mass spectrometer to detect and quantify mutation specific post-translational modifications in the protein KRAS4b, which have been found to promote cancer progression in human cancers[44]. Delcourt et al. also used MALDI-TOF imaging mass spectrometry (IMS) followed by nanoLC-MS/MS on a Q-Exactive mass spectrometer to identify 11 putative biomarkers derived from the serous ovarian cancer microenvironment[45]. Advances in instrumentation and development of novel techniques in recent years have even allowed for analysis of intact protein complexes via native proteomics[46–51]. However, separation of intact proteins has been shown to be more difficult than peptide separation[52–61].
Given sufficient biological and/or technical replicates, quantitative proteomics can be achieved using label-free quantification, which can be performed using software such as Proteome Discoverer (Thermo, Waltham, MA) or MaxQuant[39,62,63]. Alternatively, labeling methods such as the addition of tandem mass tags (TMTs) or isobaric tags for relative and absolute quantitation (iTRAQ) can be leveraged for quantitative proteomics[64,65]. Zhang et al. identified four differentially expressed proteins from epithelial ovarian cancer exosomes using TMTs, while Swiatly et al. identified five proteins using iTRAQ-tagging coupled to mass spectrometry from patient serum that were incorporated into existing diagnostic models to improve performance[66,67]. Relative and/or absolute quantitation can reveal differentially expressed proteins across disease states, allowing for the identification of putative biomarkers. From there, validation via immunohistochemistry and functional characterization of these putative biomarker proteins can provide the function of the protein in the context of the given disease, in this case ovarian cancer[68].
This workflow provides a wealth of information that can lead to an understanding of the biomarker(s) identified and studied. However, proteomics experiments often require a significant amount of sample preparation that varies depending on available equipment and instrumentation. For example, protein digestion in bottom-up proteomics may involve the use of materials that may be incompatible with LC-MS/MS, such as phosphate buffered saline (PBS) or sodium dodecyl sulfate (SDS), requiring additional cleanup steps for removal. Additionally, in a typical proteomics experiment using LC-MS/MS, each single run can take several minutes to hours. Quantitative experiments will also require multiple biological and/or technical replicates. Fractionation of samples, although not required, has also been reported to increase proteome coverage, and may be necessary if the biological sample used was not a rich source of proteins[52–61] can exponentially increase the run time for a single sample[52–61]. Previous studies have shown that sample pooling can be used to alleviate these run times, but caveats associated with doing so must be taken into consideration, as evidenced by Diz et al. and Oberg et al.[69,70]
While MS/MS based proteomics and metabolomics is more commonly used in RUO biomarker discovery applications, targeted IVD assays have also been developed due to the increasing presence of mass spectrometers in clinical laboratories in recent years[71,72]. For example, triple quadrupole (QqQ) and qTOF based LC-MS/MS analysis can be used for small molecule/peptide monitoring and quantitation. Macklin et al. have reviewed recent developments in clinical proteomics and examples of their usage[73]. Traditionally, enzyme-linked immunosorbent assays (ELISA) and biosensors have been used to detect protein biomarkers from clinical samples (i.e. blood, serum, tissue)[74,75]. However, mass spectrometry offers several large advantages over ELISA: higher throughput capabilities, broad applicability towards a large number of biomarkers and assays since compatible antibodies are not required, and the ability to distinguish between compounds that are indistinguishable via immunoassays such as proteoforms. Volmer et al. have noted that the latter capability applies to vitamin D; mass spectrometry is able to distinguish between different vitamin D compounds, whereas immunoassays lack that specificity[76].
Intact Protein Profiling
Intact protein profiling is relatively newer compared to LC-MS/MS based proteomics experiments, vide supra. Profiling is most frequently accomplished using MALDI-TOF mass spectrometry. In the clinic, it comes in the form of benchtop instruments such as the BioTyper (Bruker Daltonics) and VITEK (bioMérieux). MALDI-TOF mass spectrometers are used to obtain a profile of a given biological sample, generally in the microprotein (<= 30 kDa) range. Microproteins are ideal for profiling due to (1) the ease of ionization and subsequent detection compared to larger proteins and (2) the biological importance many microproteins exhibit; in microbiology, these are typically 16S rRNA proteins which in high copy number and have been used to identify microbes to the genus and species level[77–79]. With enough replicates for a given disease state, the profiles are inherently multidimensional and can be mined to select multiple putative biomarkers rather than a single biomarker. These putative biomarkers can then be used to develop models, databases, or scoring algorithms for IVDs to classify the measured profiles (i.e. benign or malignant in the case of tumors)[80–82]. Importantly, one of the main advantages to MALDI-TOF MS intact protein profiling is its high throughput capability (data acquisition for each biological sample only takes several seconds), which allows for large amounts of data to be generated in short order. Therefore, it is an extremely useful technique for data curation.
While MALDI-based profiling has shown promise in the clinic, it is also important to understand the current limitations of the technology. For example, although the microprotein range can be profiled relatively easily, detection of higher mass proteins proves to be more difficult, requiring specialized detectors[83]. Additionally, sample preparation is vitally important and can be the determining factor in accuracy, precision, and reproducibility when profiling for several reasons. Processing protocols for clinical samples need to ensure they are compatible with MALDI-TOF MS, as samples with incompatible materials (i.e. blood) may result in ion suppression and affect screening accuracy[84–86]. Another important factor in sample preparation is matrix selection and application. Like sample processing, matrix selection can easily be standardized once an ideal matrix has been chosen; matrix application, on the other hand, can be prone to variation. Figure 2 shows how differences in matrix application to the same sample can affect reproducibility, which in turn affects downstream statistical analysis.
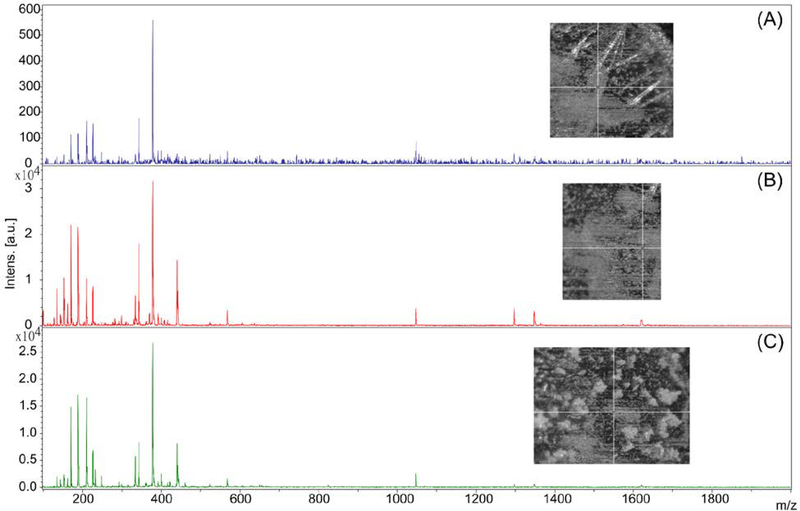
Figure 2:
Example of how differences in matrix application and crystallization can affect MALDI-TOF mass spectra. Three spots of Peptide Calibration Standard (Bruker) were co-crystallized with 50:50 alpha-cyano-4-hydroxy-cinnamic acid: 2,5-dihydroxybenzoic acid in 70:30 acetonitrile:MilliQ water w/ 0.1% trifluoroacetic acid. All spectra were acquired with the same calibrated method (Laser Power: 15%; Detector Gain: 6.2x; Shots: 200; Frequency: 60Hz; Mass Range: 100–2000 Da). (A) The first spot resulted in co-crystallization that produced large, slender crystals. These crystals resulted in weaker ionization producing low intensity spectra. (B) The second spot had much smoother co-crystallization. This resulted in higher intensity ionization across our entire mass range. (C) The third spot produced large, rounder crystals. Smaller mass peaks (< 300 Da) were able to ionize well, but larger mass peaks (> 1200 Da) were not able to ionize well.
Yet another consideration is that MALDI profiling only yields a putative mass-to-charge ratio, with limited information on identity. Therefore, for the purposes of biomarker discovery, intact protein profiling is often used prior to or in conjunction with LC-MS/MS based protein identification in order to generate biomarker leads. Lastly, early protein profiling studies using MALDI-TOF and surface-enhanced laser desorption/ionization TOF (SELDI-TOF) MS introduced doubts to the reproducibility of the technique; however, more recent studies have shown that intact protein profiling is precise enough for classification purposes when standard operating procedures (SOPs) are in place[77,87–97].
Despite the caveats mentioned above, a myriad of research and clinical studies have demonstrated that intact protein profiling has great potential as a widely available screening and diagnostic tool. Profiling is already used in the clinic for identification of pathogenic microorganisms via an FDA approved protocol. In a study by Wilson et al. the BioTyper (Bruker Daltonics) was able to identify 98.4% of bacterial and yeast isolates, with 95.7% of those identification being of high confidence, proving that MALDI-based profiling in this mass rage is a viable, accurate method of classification once the initial database and protocol have been developed and optimized[98]. Another particularly promising example is Mass-Fix[99,100]. Mass-Fix is an assay with accompanying software developed using MALDI-TOF MS designed to detect and quantify monoclonal proteins in plasma cell dyscrasias patients in a high throughput manner; a single technician was able to process hundreds of patient samples in an eight-hour shift. In collaboration with clinical colleagues, it has been used to screen thousands of patients and shows promise as a detection method for M-proteins[101,102]. Furthermore, availability of these assays as IVDs results in the collection of extremely large datasets; retrospective analysis of these intact protein profiling datasets can be used to further characterize putative biomarkers. Mellors et al. used the Mass-Fix assay to identify a post-translational modification in monoclonal proteins by showing that a small percentage of Mass-Fix positive patients displayed light chain N-glycosylation[103]. Eighteen other examples of studies using protein profiling as a diagnostic tool for ovarian cancer can be found in a meta-analysis performed by Li et al[104]. Here, it was shown that based on this subset of studies, the pooled sensitivity and specificity were 77% and 72%, respectively, highlighting the profiling approach holds promise as a diagnostic tool but requires continued development for real world translation in ovarian cancer.
Strategies and Challenges in Merging Identification and Characterization with Profiling Workflows
While protein identification and characterization and intact protein profiling both have their respective strengths and weaknesses, both workflows are not mutually exclusive. As alluded to above, these two workflows are often used as orthogonal methods to provide a wealth of information on disease states. In doing so, comprehensive datasets containing information, such as mechanism of action and progression of disease, and protocols can be generated that pave the way for biomarkers to break into the clinic.
With differentially regulated peaks that are consistently detected and identified by intact protein profiling, further exploration can lead to the identification and characterization of key protein biomarkers. Timms et al. also used this approach when identifying potential diagnostic MALDI-TOF MS peaks that could be combined with CA125 testing to more effectively detect early stage ovarian cancer[81]. Here, patient serum samples were profiled to detect differentially regulated protein peaks, leading to the identification of connective tissue-activating peptide III (CTAPIII) and platelet factor 4 (PE4) using MS/MS on digested peptides and literature searches, respectively. The combination of diagnostic peaks for CTAPIII and PE4 with CA125 allowed for classification of cases 11–15 months earlier than with CA125 alone. Interestingly, CTAPIII had been one of the biomarkers originally considered during OVA1® development, but was presumably dropped due to lack of immunoassay availability[105]. As previously mentioned above, this highlights the utility of mass spectrometry in the clinic as it allows for the development of assays that may otherwise be unavailable via immunoassays alone.
The main concern when following a profiling and identification workflow is to ensure that: (1) samples are properly handled or aliquoted since sample preparation steps for intact protein profiling and protein identification are not always compatible, and (2) there is enough sample to perform both workflows. The first concern simply requires proper experimental planning and design. The latter concern, however, may be harder to address. For example, clinical studies may result in less than ideal sample quantities, and it may not always be feasible to acquire more patient-derived samples. Therefore, it is important to consider these practical limitations when attempting to take advantage of multiple orthogonal discovery workflows.
Conversely, protein biomarkers identified through LC-MS/MS based proteomics can be used to inform classification models, databases, and scoring algorithms. Here, the use of intact protein profiling is pivoted from a discovery role into a detection role. Normally, distinguishing peaks of interest picked from thousands to tens of thousands of signals in an untargeted manner are used to differentiate between conditions. However, it is possible to interrogate the data in a targeted manner from a panel of proteins identified through other proteomics experiments and literature searches, allowing MALDI-TOF MS to act as a means of detection as opposed to a means of discovery. Hernandez et al. used this approach by taking 19 proteins with known relation to ovarian and breast cancer to inform a classification model for data collected via MALDI-TOF imaging mass spectrometry[106].
However, the use of intact protein profiling as a detection strategy is dependent on the ability of MALDI-TOF MS to detect these proteins; if these peaks are not detectable via MALDI-TOF MS, they cannot be reliably used in intact protein profiling for screening purposes. Furthermore, it must be known whether the chosen panel of proteins is up or downregulated in diseased vs healthy conditions and to what degree. While some biomarkers show similar expression across multiple cancers (i.e. BRCA1/2 downregulation in ovarian and breast cancer), others can display differential expression in different cancers (i.e. cystatin A)[107–109]. Although quantitation has traditionally been performed using LC-MS/MS workflows, Källback et al. has shown MALDI-TOF and MALDI-FT-ICR MS can provide comparable results[110]. With that being said, given these criteria are met, panels can be compiled from identified potential biomarkers and narrowed while optimizing data acquisition parameters to ensure high quality data can be collected on these select peaks.
The Future of Biomarker Discovery and Clinical Screening
As the use of mass spectrometry-based proteomics continues to increase and knowledge on biomarkers continues to grow, it is important to consider the ultimate goals of each study regardless of the workflow(s) used. Discovery workflows continue to benefit from advances in instrumentation. Technical improvements such as the emergence of dual source instruments show promise by allowing labs to perform both LC-MS/MS based proteomics and MALDI-TOF MS based intact protein profiling on single instruments as opposed to purchasing two separate instruments. Considerations must also be given as to whether a study can be translated to the clinic, and if so, how that may be done. Unfortuantely, clinical implementation of mass spectrometry-based instrumentation has been slow, though great strides have been made in recent years. Assays must be developed, validated, and in certain cases approved by regulating bodies (i.e. FDA) in order to be used in the clinic, and proper instrumentation to perform these assays must be available. However, availability of mass spectrometers in clinical laboratories is limited due to the high amounts of capital required for their acquisition. Genzen has noted that the cost associated with analytical instruments causes clinical laboratories to favor those that are compatible with a large number of their laboratory developed tests (LDTs), which could number in the dozens to hundreds[111]. This results in instruments with more niche uses, such as mass spectrometers, having lower purchasing priority compared to other more commonly used instruments. At present, MALDI-TOF MS based intact protein profiling has three main barriers to mainstream adoption as a screening method for early stage ovarian cancer: (1) a lack of standardized sample preparation protocols, (2) low performance instrumentation in comparison to their research laboratory counterparts, and (3) obtaining regulatory approval. Before standardized sample preparation protocols can be developed, easily accessible methods of sample collection must first be standardized. Costas et al. have previously reviewed potential sample collection methodologies to be adopted for screening purposes in the context of early-stage ovarian cancer[112]. Boylan et al. have used LC-MS/MS based bottom-up proteomics to compare one of these methodologies, Pap tests, to more traditional sample collection methods (i.e. tissue collection) in a patient diagnosed with high grade serous ovarian cancer.[113] Here, 4934 proteins were identified from tumor tissue, Pap test fluid, or cervical swabs, with 2293 proteins being found in samples from all three collection methods (i.e. CA125, HE4, leucine rich alpha 2 glycoprotein). While MALDI-TOF MS can be found in clinical laboratory settings, compared to RUO instruments, clinical laboratory based instrumentation is often less advanced; features such as increased mass resolving power or tandem mass spectrometry capabilities via TOF/TOF mass analyzers are absent. But despite the basic feature set in clinical MALDI-TOF MS, previous efforts in intact protein profiling (i.e. clinical microbial identification, Mass-Fix) has shown that they are more than capable. Currently, clinical laboratory instrumentation can only be used for applications that have obtained regulatory approval. Interestingly, proposed legislation such as the Verifying Accurate Leading-edge IVCT Development (VALID) Act of 2020/2021 could change the way IVD assays are regulated, which could allow mass spectrometry-based tests and assays to be more easily approved. Genzen and Johnson and Marchant have discussed the VALID Act in more detail and its implications in the future of in vitro clinical tests[111][114]. Lastly, as evidenced by Mass-Fix, significant amounts of collaboration between academic and clinical institutions is necessary for the development of clinical assays utilizing MALDI-TOF protein profiling. It also had the benefit of a large serum database from which biological material could be sourced to build a spectral database for analysis. Unfortunately, ovarian cancer currently lacks the large collaborations and resources that were available for the development of Mass-Fix, and reliable identification and detection of early stage biomarkers has not been established. With that being said, Mass-Fix has demonstrated a promising example of a pipeline for mass spectrometry-based screening. Therefore, we believe that with the establishment of standardized sample collection methods and discovery of reliable biomarkers, a similar pipeline can be developed for ovarian cancer with a large, diverse sample size that can be used to develop spectral databases and appropriate collaborations in place. Overall, we have seen exciting developments in mass spectrometry-based biomarker discovery and screening in recent years, and we await further discoveries in the coming years.
Acknowledgements
This publication was supported by the National Institute of General Medical Sciences Award Number R01GM125943 (LMS), the National Cancer Institute Award Number R01CA240423 (LMS) of the National Institutes of Health, the Research Corporation for Science Advancement Scialog Award #26222 (LMS), and by the National Science Foundation grant 2128044 (LMS).
References
- [1].SEER Cancer Stat Facts: Ovarian Cancer, (n.d.). https://paperpile.com/app/p/9ce8d25d-d68a-084a-9108-4bd5f9c55caa (accessed February 11, 2021).
- [2].Can Ovarian Cancer Be Found Early?, (n.d.). https://paperpile.com/app/p/53075792-e0f8-0ba1-ac3b-2cebb6a8e926 (accessed February 11, 2021).
- [3].Khoo SK, MacKay EV, Carcinoembryonic antigen (CEA) in ovarian cancer: factors influencing its incidence and changes which occur in response to cytotoxic drugs, Br. J. Obstet. Gynaecol 83 (1976) 753–759. [DOI] [PubMed] [Google Scholar]
- [4].Bast RC Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC, Reactivity of a monoclonal antibody with human ovarian carcinoma, J. Clin. Invest 68 (1981) 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hellström I, Raycraft J, Hayden-Ledbetter M, Ledbetter JA, Schummer M, McIntosh M, Drescher C, Urban N, Hellström KE, The HE4 (WFDC2) protein is a biomarker for ovarian carcinoma, Cancer Res. 63 (2003) 3695–3700. [PubMed] [Google Scholar]
- [6].Ueland FR, Desimone CP, Seamon LG, Miller RA, Goodrich S, Podzielinski I, Sokoll L, Smith A, van Nagell JR Jr, Zhang Z, Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors, Obstet. Gynecol 117 (2011) 1289–1297. [DOI] [PubMed] [Google Scholar]
- [7].Bristow RE, Smith A, Zhang Z, Chan DW, Crutcher G, Fung ET, Munroe DG, Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay, Gynecol. Oncol 128 (2013) 252–259. [DOI] [PubMed] [Google Scholar]
- [8].Coleman RL, Herzog TJ, Chan DW, Munroe DG, Pappas TC, Smith A, Zhang Z, Wolf J, Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses, Am. J. Obstet. Gynecol 215 (2016) 82.e1–82.e11. [DOI] [PubMed] [Google Scholar]
- [9].Dochez V, Caillon H, Vaucel E, Dimet J, Winer N, Ducarme G, Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review, J. Ovarian Res. 12 (2019) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ferraro S, Braga F, Lanzoni M, Boracchi P, Biganzoli EM, Panteghini M, Serum human epididymis protein 4 vs carbohydrate antigen 125 for ovarian cancer diagnosis: a systematic review, J. Clin. Pathol 66 (2013) 273–281. [DOI] [PubMed] [Google Scholar]
- [11].Dikmen ZG, Colak A, Dogan P, Tuncer S, Akbiyik F, Diagnostic performances of CA125, HE4, and ROMA index in ovarian cancer, Eur. J. Gynaecol. Oncol 36 (2015) 457–462. [PubMed] [Google Scholar]
- [12].Chen X, Zhou H, Chen R, He J, Wang Y, Huang L, Sun L, Duan C, Luo X, Yan H, Development of a multimarker assay for differential diagnosis of benign and malignant pelvic masses, Clin. Chim. Acta. 440 (2015) 57–63. [DOI] [PubMed] [Google Scholar]
- [13].Yanaranop M, Anakrat V, Siricharoenthai S, Nakrangsee S, Thinkhamrop B, Is the Risk of Ovarian Malignancy Algorithm Better Than Other Tests for Predicting Ovarian Malignancy in Women with Pelvic Masses?, Gynecol. Obstet. Invest 82 (2017) 47–53. [DOI] [PubMed] [Google Scholar]
- [14].Wilailak S, Chan KKL, Chen CA, Nam JH, Ochiai K, Aw TC, Sabaratnam S, Hebbar S, Sickan J, Schodin BA, Charakorn C, Sumpaico WW, Distinguishing benign from malignant pelvic mass utilizing an algorithm with HE4, menopausal status, and ultrasound findings, J. Gynecol. Oncol 26 (2015) 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang J, Gao J, Yao H, Wu Z, Wang M, Qi J, Diagnostic accuracy of serum HE4, CA125 and ROMA in patients with ovarian cancer: a meta-analysis, Tumour Biol. 35 (2014) 6127–6138. [DOI] [PubMed] [Google Scholar]
- [16].Zhen S, Bian L-H, Chang L-L, Gao X, Comparison of serum human epididymis protein 4 and carbohydrate antigen 125 as markers in ovarian cancer: A meta-analysis, Mol Clin Oncol 2 (2014) 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Abdel-Azeez HA, Labib HA, Sharaf SM, Refai AN, HE4 and mesothelin: novel biomarkers of ovarian carcinoma in patients with pelvic masses, Asian Pac. J. Cancer Prev 11 (2010) 111–116. [PubMed] [Google Scholar]
- [18].Holcomb K, Vucetic Z, Miller MC, Knapp RC, Human epididymis protein 4 offers superior specificity in the differentiation of benign and malignant adnexal masses in premenopausal women, Am. J. Obstet. Gynecol 205 (2011) 358.e1–6. [DOI] [PubMed] [Google Scholar]
- [19].Moore RG, Brown AK, Miller MC, Skates S, Allard WJ, Verch T, Steinhoff M, Messerlian G, DiSilvestro P, Granai CO, Bast RC Jr, The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass, Gynecol. Oncol 108 (2008) 402–408. [DOI] [PubMed] [Google Scholar]
- [20].Goff BA, Agnew K, Neradilek MB, Gray HJ, Liao JB, Urban RR, Combining a symptom index, CA125 and HE4 (triple screen) to detect ovarian cancer in women with a pelvic mass, Gynecol. Oncol 147 (2017) 291–295. [DOI] [PubMed] [Google Scholar]
- [21].Meys EMJ, Kaijser J, Kruitwagen RFPM, Slangen BFM, Van Calster B, Aertgeerts B, Verbakel JY, Timmerman D, Van Gorp T, Subjective assessment versus ultrasound models to diagnose ovarian cancer: A systematic review and meta-analysis, Eur. J. Cancer 58 (2016) 17–29. [DOI] [PubMed] [Google Scholar]
- [22].Al Musalhi K, Al Kindi M, Al Aisary F, Ramadhan F, Al Rawahi T, Al Hatali K, Mula-Abed W-A, Evaluation of HE4, CA-125, Risk of Ovarian Malignancy Algorithm (ROMA) and Risk of Malignancy Index (RMI) in the Preoperative Assessment of Patients with Adnexal Mass, Oman Med. J 31 (2016) 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, Gajewski W, Kurman R, Bast RC Jr, Skates SJ, A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass, Gynecol. Oncol 112 (2009) 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li F, Tie R, Chang K, Wang F, Deng S, Lu W, Yu L, Chen M, Does risk for ovarian malignancy algorithm excel human epididymis protein 4 and CA125 in predicting epithelial ovarian cancer: a meta-analysis, BMC Cancer. 12 (2012) 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wei SU, Li H, Zhang B, The diagnostic value of serum HE4 and CA-125 and ROMA index in ovarian cancer, Biomed Rep 5 (2016) 41–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sandri MT, Bottari F, Franchi D, Boveri S, Candiani M, Ronzoni S, Peiretti M, Radice D, Passerini R, Sideri M, Comparison of HE4, CA125 and ROMA algorithm in women with a pelvic mass: correlation with pathological outcome, Gynecol. Oncol 128 (2013) 233–238. [DOI] [PubMed] [Google Scholar]
- [27].Miller RW, Smith A, DeSimone CP, Seamon L, Goodrich S, Podzielinski I, Sokoll L, van Nagell JR Jr, Zhang Z, Ueland FR, Performance of the American College of Obstetricians and Gynecologists’ ovarian tumor referral guidelines with a multivariate index assay, Obstet. Gynecol 117 (2011) 1298–1306. [DOI] [PubMed] [Google Scholar]
- [28].Urban RR, Pappas TC, Bullock RG, Munroe DG, Bonato V, Agnew K, Goff BA, Combined symptom index and second-generation multivariate biomarker test for prediction of ovarian cancer in patients with an adnexal mass, Gynecol. Oncol 150 (2018) 318–323. [DOI] [PubMed] [Google Scholar]
- [29].Simon S, FDA Warns Against Ovarian Cancer Screening Tests, American Cancer Society. (2016). https://www.google.com/url?q=https://www.cancer.org/latest-news/fda-warns-against-ovarian-cancer-screening-tests.html&sa=D&source=editors&ust=1614275357165000&usg=AOvVaw2Fh3LueHRjyXHlrS4EbNXU (accessed February 25, 2021). [Google Scholar]
- [30].Clifford C, Vitkin N, Nersesian S, Reid-Schachter G, Francis J-A, Koti M, Multi-omics in high-grade serous ovarian cancer: Biomarkers from genome to the immunome, Am. J. Reprod. Immunol 80 (2018) e12975. [DOI] [PubMed] [Google Scholar]
- [31].Swiatly A, Plewa S, Matysiak J, Kokot ZJ, Mass spectrometry-based proteomics techniques and their application in ovarian cancer research, J. Ovarian Res 11 (2018) 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kriegsmann J, Kriegsmann M, Casadonte R, MALDI TOF imaging mass spectrometry in clinical pathology: a valuable tool for cancer diagnostics (review), Int. J. Oncol 46 (2015) 893–906. [DOI] [PubMed] [Google Scholar]
- [33].Wither MJ, Hansen KC, Reisz JA, Mass spectrometry-based bottom-up proteomics: Sample preparation, LC-MS/MS analysis, and database query strategies, Curr. Protoc. Protein Sci 86 (2016) 16.4.1–16.4.20. [DOI] [PubMed] [Google Scholar]
- [34].Donnelly DP, Rawlins CM, DeHart CJ, Fornelli L, Schachner LF, Lin Z, Lippens JL, Aluri KC, Sarin R, Chen B, Lantz C, Jung W, Johnson KR, Koller A, Wolff JJ, Campuzano IDG, Auclair JR, Ivanov AR, Whitelegge JP, Paša-Tolić L, Chamot-Rooke J, Danis PO, Smith LM, Tsybin YO, Loo JA, Ge Y, Kelleher NL, Agar JN, Best practices and benchmarks for intact protein analysis for top-down mass spectrometry, Nat. Methods 16 (2019) 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Eng JK, McCormack AL, Yates JR, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database, J. Am. Soc. Mass Spectrom 5 (1994) 976–989. [DOI] [PubMed] [Google Scholar]
- [36].Schweppe DK, Eng JK, Yu Q, Bailey D, Rad R, Navarrete-Perea J, Huttlin EL, Erickson BK, Paulo JA, Gygi SP, Full-Featured, Real-Time Database Searching Platform Enables Fast and Accurate Multiplexed Quantitative Proteomics, J. Proteome Res 19 (2020) 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Perkins DN, Pappin DJ, Creasy DM, Cottrell JS, Probability-based protein identification by searching sequence databases using mass spectrometry data, Electrophoresis. 20 (1999) 3551–3567. [DOI] [PubMed] [Google Scholar]
- [38].Consortium UniProt, UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res. 49 (2021) D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tyanova S, Temu T, Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics, Nat. Protoc 11 (2016) 2301–2319. [DOI] [PubMed] [Google Scholar]
- [40].Kacírová M, Bober P, Alexovič M, Tomková Z, Tkáčiková S, Talian I, Mederová L, Bérešová D, Tóth R, Andrašina I, Kožlejová Z, Kilík R, Divín R, Sabo J, Differential Urinary Proteomic Analysis of Endometrial Cancer, Physiol. Res 68 (2019) S483–S490. [DOI] [PubMed] [Google Scholar]
- [41].Malaker SA, Pedram K, Ferracane MJ, Bensing BA, Krishnan V, Pett C, Yu J, Woods EC, Kramer JR, Westerlind U, Dorigo O, Bertozzi CR, The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins, Proc. Natl. Acad. Sci. U. S. A. 116 (2019) 7278–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nesvizhskii AI, Aebersold R, Interpretation of shotgun proteomic data: the protein inference problem, Mol. Cell. Proteomics 4 (2005) 1419–1440. [DOI] [PubMed] [Google Scholar]
- [43].Smith LM, Kelleher NL, Proteoforms as the next proteomics currency, Science. 359 (2018) 1106–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ntai I, Fornelli L, DeHart CJ, Hutton JE, Doubleday PF, LeDuc RD, van Nispen AJ, Fellers RT, Whiteley G, Boja ES, Rodriguez H, Kelleher NL, Precise characterization of KRAS4b proteoforms in human colorectal cells and tumors reveals mutation/modification cross-talk, Proceedings of the National Academy of Sciences. 115 (2018) 4140–4145. 10.1073/pnas.1716122115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Delcourt V, Franck J, Leblanc E, Narducci F, Robin Y-M, Gimeno J-P, Quanico J, Wisztorski M, Kobeissy F, Jacques J-F, Roucou X, Salzet M, Fournier I, Combined Mass Spectrometry Imaging and Top-down Microproteomics Reveals Evidence of a Hidden Proteome in Ovarian Cancer, EBioMedicine. 21 (2017) 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Skinner OS, Haverland NA, Fornelli L, Melani RD, Do Vale LHF, Seckler HS, Doubleday PF, Schachner LF, Srzentić K, Kelleher NL, Compton PD, Top-down characterization of endogenous protein complexes with native proteomics, Nat. Chem. Biol 14 (2018) 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Robinson CV, Chung EW, Kragelund BB, Knudsen J, Aplin RT, Poulsen FM, Dobson CM, Probing the nature of noncovalent interactions by mass spectrometry. A study of Protein−CoA ligand binding and assembly, J. Am. Chem. Soc 118 (1996) 8646–8653. [Google Scholar]
- [48].Rostom AA, Robinson CV, Detection of the intact GroEL chaperonin assembly by mass spectrometry, J. Am. Chem. Soc 121 (1999) 4718–4719. [Google Scholar]
- [49].Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV, Evidence for macromolecular protein rings in the absence of bulk water, Science. 310 (2005) 1658–1661. [DOI] [PubMed] [Google Scholar]
- [50].Barrera NP, Di Bartolo N, Booth PJ, Robinson CV, Micelles protect membrane complexes from solution to vacuum, Science. 321 (2008) 243–246. [DOI] [PubMed] [Google Scholar]
- [51].Chorev DS, Baker LA, Wu D, Beilsten-Edmands V, Rouse SL, Zeev-Ben-Mordehai T, Jiko C, Samsudin F, Gerle C, Khalid S, Stewart AG, Matthews SJ, Grünewald K, Robinson CV, Protein assemblies ejected directly from native membranes yield complexes for mass spectrometry, Science. 362 (2018) 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dufresne J, Bowden P, Thavarajah T, Florentinus-Mefailoski A, Chen ZZ, Tucholska M, Norzin T, Ho MT, Phan M, Mohamed N, Ravandi A, Stanton E, Slutsky AS, Dos Santos CC, Romaschin A, Marshall JC, Addison C, Malone S, Heyland D, Scheltens P, Killestein J, Teunissen CE, Diamandis EP, Michael Siu KW, Marshall JG, The plasma peptides of ovarian cancer, Clin. Proteomics 15 (2018) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dufresne J, Florentinus-Mefailoski A, Bowden P, Marshall JG, A method for the extraction of the endogenous tryptic peptides (peptidome) from human EDTA plasma, Anal. Biochem 549 (2018) 188–196. [DOI] [PubMed] [Google Scholar]
- [54].Marshall J, Jankowski A, Furesz S, Kireeva I, Barker L, Dombrovsky M, Zhu W, Jacks K, Ingratta L, Bruin J, Kristensen E, Zhang R, Stanton E, Takahashi M, Jackowski G, Human serum proteins preseparated by electrophoresis or chromatography followed by tandem mass spectrometry, J. Proteome Res 3 (2004) 364–382. [DOI] [PubMed] [Google Scholar]
- [55].Tucholska M, Bowden P, Jacks K, Zhu P, Furesz S, Dumbrovsky M, Marshall J, Human serum proteins fractionated by preparative partition chromatography prior to LC-ESI-MS/MS, J. Proteome Res 8 (2009) 1143–1155. [DOI] [PubMed] [Google Scholar]
- [56].Tucholska M, Scozzaro S, Williams D, Ackloo S, Lock C, Siu KWM, Evans KR, Marshall JG, Endogenous peptides from biophysical and biochemical fractionation of serum analyzed by matrix-assisted laser desorption/ionization and electrospray ionization hybrid quadrupole time-of-flight, Anal. Biochem 370 (2007) 228–245. [DOI] [PubMed] [Google Scholar]
- [57].Zhu P, Bowden P, Zhang D, Marshall JG, Mass spectrometry of peptides and proteins from human blood, Mass Spectrom. Rev 30 (2011) 685–732. [DOI] [PubMed] [Google Scholar]
- [58].Manadas B, Mendes VM, English J, Dunn MJ, Peptide fractionation in proteomics approaches, Expert Rev. Proteomics 7 (2010) 655–663. [DOI] [PubMed] [Google Scholar]
- [59].Mostovenko E, Hassan C, Rattke J, Deelder AM, van Veelen PA, Palmblad M, Comparison of peptide and protein fractionation methods in proteomics, EuPA Open Proteomics. 1 (2013) 30–37. [Google Scholar]
- [60].Yeung D, Mizero B, Gussakovsky D, Klaassen N, Lao Y, Spicer V, Krokhin OV, Separation Orthogonality in Liquid Chromatography-Mass Spectrometry for Proteomic Applications: Comparison of 16 Different Two-Dimensional Combinations, Anal. Chem 92 (2020) 3904–3912. [DOI] [PubMed] [Google Scholar]
- [61].Camerini S, Mauri P, The role of protein and peptide separation before mass spectrometry analysis in clinical proteomics, J. Chromatogr. A 1381 (2015) 1–12. [DOI] [PubMed] [Google Scholar]
- [62].Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol 26 (2008) 1367–1372. [DOI] [PubMed] [Google Scholar]
- [63].Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M, Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ, Mol. Cell. Proteomics 13 (2014) 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AKA, Hamon C, Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS, Anal. Chem 75 (2003) 1895–1904. [DOI] [PubMed] [Google Scholar]
- [65].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ, Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents, Mol. Cell. Proteomics 3 (2004) 1154–1169. [DOI] [PubMed] [Google Scholar]
- [66].Zhang W, Ou X, Wu X, Proteomics profiling of plasma exosomes in epithelial ovarian cancer: A potential role in the coagulation cascade, diagnosis and prognosis, Int. J. Oncol 54 (2019) 1719–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Swiatly A, Horala A, Matysiak J, Hajduk J, Nowak-Markwitz E, Kokot ZJ, Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery, Int. J. Mol. Sci 19 (2018). 10.3390/ijms19082240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Scopes RK, Overview of protein purification and characterization, Curr. Protoc. Protein Sci Chapter 1 (2001) Unit 1.1. [DOI] [PubMed] [Google Scholar]
- [69].Diz AP, Truebano M, Skibinski DOF, The consequences of sample pooling in proteomics: an empirical study, Electrophoresis. 30 (2009) 2967–2975. [DOI] [PubMed] [Google Scholar]
- [70].Oberg AL, Vitek O, Statistical design of quantitative mass spectrometry-based proteomic experiments, J. Proteome Res 8 (2009) 2144–2156. [DOI] [PubMed] [Google Scholar]
- [71].Jannetto PJ, Fitzgerald RL, Effective Use of Mass Spectrometry in the Clinical Laboratory, Clinical Chemistry. 62 (2016) 92–98. 10.1373/clinchem.2015.248146. [DOI] [PubMed] [Google Scholar]
- [72].Banerjee S, Empowering Clinical Diagnostics with Mass Spectrometry, ACS Omega. 5 (2020) 2041–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Macklin A, Khan S, Kislinger T, Recent advances in mass spectrometry based clinical proteomics: applications to cancer research, Clin. Proteomics 17 (2020) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Engvall E, Perlmann P, Enzyme-linked immunosorbent assay, Elisa. 3. Quantitation of specific antibodies by enzyme-labeled anti-immunoglobulin in antigen-coated tubes, J. Immunol 109 (1972) 129–135. [PubMed] [Google Scholar]
- [75].Franier BDL, Thompson M, Early stage detection and screening of ovarian cancer: A research opportunity and significant challenge for biosensor technology, Biosens. Bioelectron 135 (2019) 71–81. [DOI] [PubMed] [Google Scholar]
- [76].Volmer DA, Mendes LRB, Stokes CS, Analysis of vitamin D metabolic markers by mass spectrometry: Current techniques, limitations of the “gold standard” method, and anticipated future directions, Mass Spectrometry Reviews. 34 (2015) 2–23. 10.1002/mas.21408. [DOI] [PubMed] [Google Scholar]
- [77].Williams TL, Andrzejewski D, Lay JO, Musser SM, Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells, J. Am. Soc. Mass Spectrom 14 (2003) 342–351. [DOI] [PubMed] [Google Scholar]
- [78].Eguen T, Straub D, Graeff M, Wenkel S, MicroProteins: small size-big impact, Trends Plant Sci. 20 (2015) 477–482. [DOI] [PubMed] [Google Scholar]
- [79].Staudt A-C, Wenkel S, Regulation of protein function by “microProteins,” EMBO Rep. 12 (2011) 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kim J-H, Park CW, Um D, Baek KH, Jo Y, An H, Kim Y, Kim TJ, Mass spectrometric screening of ovarian cancer with serum glycans, Dis. Markers. 2014 (2014) 634289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Timms JF, Menon U, Devetyarov D, Tiss A, Camuzeaux S, McCurrie K, Nouretdinov I, Burford B, Smith C, Gentry-Maharaj A, Hallett R, Ford J, Luo Z, Vovk V, Gammerman A, Cramer R, Jacobs I, Early detection of ovarian cancer in samples pre-diagnosis using CA125 and MALDI-MS peaks, Cancer Genomics Proteomics. 8 (2011) 289–305. [PubMed] [Google Scholar]
- [82].Kawakami E, Tabata J, Yanaihara N, Ishikawa T, Koseki K, Iida Y, Saito M, Komazaki H, Shapiro JS, Goto C, Akiyama Y, Saito R, Saito M, Takano H, Yamada K, Okamoto A, Application of Artificial Intelligence for Preoperative Diagnostic and Prognostic Prediction in Epithelial Ovarian Cancer Based on Blood Biomarkers, Clin. Cancer Res. 25 (2019) 3006–3015. [DOI] [PubMed] [Google Scholar]
- [83].van Remoortere A, van Zeijl RJM, van den Oever N, Franck J, Longuespée R, Wisztorski M, Salzet M, Deelder AM, Fournier I, McDonnell LA, MALDI imaging and profiling MS of higher mass proteins from tissue, J. Am. Soc. Mass Spectrom 21 (2010) 1922–1929. [DOI] [PubMed] [Google Scholar]
- [84].Börnsen KO, Influence of salts, buffers, detergents, solvents, and matrices on MALDI-MS protein analysis in complex mixtures, Methods Mol. Biol 146 (2000) 387–404. [DOI] [PubMed] [Google Scholar]
- [85].Chandler J, Haslam C, Hardy N, Leveridge M, Marshall P, A Systematic Investigation of the Best Buffers for Use in Screening by MALDI-Mass Spectrometry, SLAS Discov. 22 (2017) 1262–1269. [DOI] [PubMed] [Google Scholar]
- [86].Taylor AJ, Dexter A, Bunch J, Exploring Ion Suppression in Mass Spectrometry Imaging of a Heterogeneous Tissue, Anal. Chem 90 (2018) 5637–5645. [DOI] [PubMed] [Google Scholar]
- [87].Munteanu B, Hopf C, Emergence of whole-cell MALDI-MS biotyping for high-throughput bioanalysis of mammalian cells?, Bioanalysis. 5 (2013) 885–893. [DOI] [PubMed] [Google Scholar]
- [88].Albrethsen J, The first decade of MALDI protein profiling: a lesson in translational biomarker research, J. Proteomics. 74 (2011) 765–773. [DOI] [PubMed] [Google Scholar]
- [89].Engwegen JYMN, Mehra N, Haanen JBAG, Bonfrer JMG, Schellens JHM, Voest EE, Beijnen JH, Validation of SELDI-TOF MS serum protein profiles for renal cell carcinoma in new populations, Lab. Invest 87 (2007) 161–172. [DOI] [PubMed] [Google Scholar]
- [90].Rogers MA, Clarke P, Noble J, Munro NP, Paul A, Selby PJ, Banks RE, Proteomic profiling of urinary proteins in renal cancer by surface enhanced laser desorption ionization and neural-network analysis: identification of key issues affecting potential clinical utility, Cancer Res. 63 (2003) 6971–6983. [PubMed] [Google Scholar]
- [91].van Winden AWJ, Gast M-CW, Beijnen JH, Rutgers EJT, Grobbee DE, Peeters PHM, van Gils CH, Validation of previously identified serum biomarkers for breast cancer with SELDI-TOF MS: a case control study, BMC Med. Genomics 2 (2009) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Albrethsen J, Kaas A, Schönle E, Swift P, Kocova M, Gammeltoft S, Hansen L, Mortensen HB, Evaluation of a type 1 diabetes serum cohort by SELDI-TOF MS protein profiling, Proteomics Clin. Appl 3 (2009) 383–393. [DOI] [PubMed] [Google Scholar]
- [93].Baggerly KA, Morris JS, Coombes KR, Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments, Bioinformatics. 20 (2004) 777–785. [DOI] [PubMed] [Google Scholar]
- [94].Albrethsen J, Reproducibility in protein profiling by MALDI-TOF mass spectrometry, Clin. Chem 53 (2007) 852–858. [DOI] [PubMed] [Google Scholar]
- [95].Oberle M, Wohlwend N, Jonas D, Maurer FP, Jost G, Tschudin-Sutter S, Vranckx K, Egli A, The Technical and Biological Reproducibility of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Based Typing: Employment of Bioinformatics in a Multicenter Study, PLoS One. 11 (2016) e0164260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Petukhova VZ, Young AN, Wang J, Wang M, Ladanyi A, Kothari R, Burdette JE, Sanchez LM, Whole Cell MALDI Fingerprinting Is a Robust Tool for Differential Profiling of Two-Component Mammalian Cell Mixtures, J. Am. Soc. Mass Spectrom 30 (2019) 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Galey MM, Young AN, Petukhova VZ, Wang M, Wang J, Salvi A, Russo A, Burdette JE, Sanchez LM, Detection of Ovarian Cancer Using Samples Sourced from the Vaginal Microenvironment, J. Proteome Res 19 (2020) 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wilson DA, Young S, Timm K, Novak-Weekley S, Marlowe EM, Madisen N, Lillie JL, Ledeboer NA, Smith R, Hyke J, Griego-Fullbright C, Jim P, Granato PA, Faron ML, Cumpio J, Buchan BW, Procop GW, Multicenter Evaluation of the Bruker MALDI Biotyper CA System for the Identification of Clinically Important Bacteria and Yeasts, Am. J. Clin. Pathol 147 (2017) 623–631. [DOI] [PubMed] [Google Scholar]
- [99].Milani P, Murray DL, Barnidge DR, Kohlhagen MC, Mills JR, Merlini G, Dasari S, Dispenzieri A, The utility of MASS-FIX to detect and monitor monoclonal proteins in the clinic, Am. J. Hematol 92 (2017) 772–779. [DOI] [PubMed] [Google Scholar]
- [100].Kohlhagen M, Dasari S, Willrich M, Hetrick M, Netzel B, Dispenzieri A, Murray DL, Automation and validation of a MALDI-TOF MS (Mass-Fix) replacement of immunofixation electrophoresis in the clinical lab, Clin. Chem. Lab. Med 59 (2020) 155–163. [DOI] [PubMed] [Google Scholar]
- [101].Murray DL, Puig N, Kristinsson S, Usmani SZ, Dispenzieri A, Bianchi G, Kumar S, Chng WJ, Hajek R, Paiva B, Waage A, Vincent Rajkumar S, Durie B, Mass spectrometry for the evaluation of monoclonal proteins in multiple myeloma and related disorders: an International Myeloma Working Group Mass Spectrometry Committee Report, Blood Cancer Journal. 11 (2021). 10.1038/s41408-021-00408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Mellors P, Dasari S, Kohlhagen M, Arendt BK, Gertz MA, Kumar SK, Buadi FK, Willrich MAV, Lust JA, Kapoor P, Lacy MQ, Dingli D, Hwa YL, Fonder A, Hobbs MA, Hayman SR, Kourelis T, Warsame RM, Muchtar E, Leung N, Go RS, Lin Y, Gonsalves WI, Siddiqui M, Kyle RA, Rajkumar SV, Murray DL, Dispenzieri A, MASS-FIX for the diagnosis of plasma cell disorders: A single institution experience of 4118 patients, Blood. 136 (2020) 48–49. [Google Scholar]
- [103].Mellors PW, Dasari S, Kohlhagen MC, Kourelis T, Go RS, Muchtar E, Gertz MA, Kumar SK, Buadi FK, Willrich MAV, Lust JA, Kapoor P, Lacy MQ, Dingli D, Hwa Y, Fonder A, Hobbs M, Hayman S, Warsame R, Leung NR, Lin Y, Gonsalves W, Siddiqui M, Kyle RA, Rajkumar SV, Murray DL, Dispenzieri A, MASS-FIX for the detection of monoclonal proteins and light chain N-glycosylation in routine clinical practice: a cross-sectional study of 6315 patients, Blood Cancer J. 11 (2021) 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li K, Pei Y, Wu Y, Guo Y, Cui W, Performance of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) in diagnosis of ovarian cancer: a systematic review and meta-analysis, J. Ovarian Res 13 (2020) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fung ET, A recipe for proteomics diagnostic test development: the OVA1 test, from biomarker discovery to FDA clearance, Clin. Chem 56 (2010) 327–329. [DOI] [PubMed] [Google Scholar]
- [106].Cordero Hernandez Y, Boskamp T, Casadonte R, Hauberg-Lotte L, Oetjen J, Lachmund D, Peter A, Trede D, Kriegsmann K, Kriegsmann M, Kriegsmann J, Maass P, Targeted Feature Extraction in MALDI Mass Spectrometry Imaging to Discriminate Proteomic Profiles of Breast and Ovarian Cancer, Proteomics Clin. Appl 13 (2019) e1700168. [DOI] [PubMed] [Google Scholar]
- [107].Kastelic L, Turk B, Kopitar-Jerala N, Stolfa A, Rainer S, Turk V, Lah TT, Stefin B, the major low molecular weight inhibitor in ovarian carcinoma, Cancer Lett. 82 (1994) 81–88. [DOI] [PubMed] [Google Scholar]
- [108].Kos J, Krasovec M, Cimerman N, Nielsen HJ, Christensen IJ, Brünner N, Cysteine proteinase inhibitors stefin A, stefin B, and cystatin C in sera from patients with colorectal cancer: relation to prognosis, Clin. Cancer Res 6 (2000) 505–511. [PubMed] [Google Scholar]
- [109].Duncan JA, Reeves JR, Cooke TG, BRCA1 and BRCA2 proteins: roles in health and disease, Mol. Pathol 51 (1998) 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Källback P, Vallianatou T, Nilsson A, Shariatgorji R, Schintu N, Pereira M, Barré F, Wadensten H, Svenningsson P, Andrén PE, Cross-validated Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging Quantitation Protocol for a Pharmaceutical Drug and Its Drug-Target Effects in the Brain Using Time-of-Flight and Fourier Transform Ion Cyclotron Resonance Analyzers, Anal. Chem 92 (2020) 14676–14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Genzen JR, Regulation of Laboratory-Developed Tests, Am. J. Clin. Pathol 152 (2019) 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Costas L, Frias-Gomez J, Guardiola M, Benavente Y, Pineda M, Pavón MÁ, Martínez JM, Climent M, Barahona M, Canet J, Paytubi S, Salinas M, Palomero L, Bianchi I, Reventós J, Capellà G, Diaz M, Vidal A, Piulats JM, Aytés Á, Ponce J, Brunet J, Bosch FX, Matias-Guiu X, Alemany L, de Sanjosé S, Screenwide Team, New perspectives on screening and early detection of endometrial cancer, Int. J. Cancer 145 (2019) 3194–3206. [DOI] [PubMed] [Google Scholar]
- [113].Boylan KLM, Afiuni-Zadeh S, Geller MA, Argenta PA, Griffin TJ, Skubitz APN, Evaluation of the potential of Pap test fluid and cervical swabs to serve as clinical diagnostic biospecimens for the detection of ovarian cancer by mass spectrometry-based proteomics, Clin. Proteomics 18 (2021) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Johnson WG, Marchant GE, Legislating in the time of a pandemic: window of opportunity or invitation for recklessness?, J Law Biosci. 7 (2020) lsaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]