Abstract
The speed at which several COVID-19 vaccines went from conception to receiving FDA and EMA approval for emergency use is an achievement unrivaled in the history of vaccine development. Mass vaccination efforts using the highly effective vaccines are currently underway to generate sufficient herd immunity and reduce transmission of the SARS-CoV-2 virus. Despite the most advanced vaccine technology, global recipient coverage, especially in resource-poor areas remains a challenge as genetic drift in naïve population pockets threatens overall vaccine efficacy. In this study, we described the production of insect-cell expressed SARS-CoV-2 spike protein ectodomain constructs and examined their immunogenicity in mice. We demonstrated that, when formulated with CoVaccine HTTM adjuvant, an oil-in-water nanoemulsion compatible with lyophilization, our vaccine candidates elicit a broad-spectrum IgG response, high neutralizing antibody (NtAb) titers against SARS-CoV-2 prototype and variants of concern, specifically B.1.351 (Beta) and P.1. (Gamma), and an antigen-specific IFN-γ secreting response in outbred mice. Of note, different ectodomain constructs yielded variations in NtAb titers against the prototype strain and some VOC. Dose response experiments indicated that NtAb titers increased with antigen dose, but not adjuvant dose, and may be higher with a lower adjuvant dose. Our findings lay the immunological foundation for the development of a dry-thermostabilized vaccine that is deployable without refrigeration.
Keywords: SARS-CoV-2 vaccine, Spike glycoprotein, Recombinant protein, Adjuvant, CoVaccine HT™
Introduction
The emergence and rapid spread of a new infectious respiratory disease, Coronavirus Disease 2019 (COVID-19) has caused an unprecedented public health emergency worldwide since emerging in December 2019. A novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is closely related to SARS-CoV, was identified as the etiologic agent of the new respiratory disease. As of July 2021, there are over 185 million confirmed cases with over 4 million deaths worldwide and over 33 million cases and 606,000 fatalities reported in the U.S. [1], accentuating the continued need for safe and effective, inexpensive and easily deployable vaccines that would rapidly establish global herd immunity. Facilitated by the broad nature of the outbreak, significant SARS-CoV-2 variants have been identified, some of which appear to be more resistant to first-generation vaccines [2]. More than 200 COVID-19 vaccine candidates using various technology platforms are in development [3]. Three vaccines including two of the mRNA platforms, Pfizer/BioNTech BNT162b2 and Moderna mRNA-1273, and the third, a viral-vector platform, Janssen Ad26.COV2.S, with reported interim efficacy rates of 95%, 94.1% and 85% respectively, have been approved by the U.S. Food and Drug Administration (FDA) for emergency use as of July 2021 [4], [5], [6]. These vaccines are now being administered in the U.S. and have also been approved for use in other jurisdictions. As no single vaccine can be produced rapidly in sufficient quantities to satisfy the global demand, diversification using different vaccine platforms would enable worldwide vaccine coverage as well as address the needs of the most vulnerable populations, particularly elderly and immunocompromised individuals or those with other co-morbidities. With mutations emerging even in the absence of strong selective pressure placed on SARS-CoV-2, variants of concern (VOC) will continue to arise under inequitable vaccine availability [2], [7]. Thus, continued research into adaptable and more easily distributed vaccines, compatible with rapid deployment and significant cost efficiencies, must continue unabated.
The recombinant subunit vaccine platform offers a safety advantage over virally vectored vaccines and a distribution advantage relative to many other vaccine platforms. Purified recombinant protein antigens can be engineered to achieve optimal immunogenicity and protective efficacy. Furthermore, a thermostabilized subunit vaccine can be deployed in the field, eliminating stringent cold-chain requirements. Formulation of the vaccine immunogen with a potent adjuvant enhances and focuses immunogenicity while lowering the antigen dose requirement, thereby enabling vaccination of more people with a product carrying significantly more clinical and regulatory precedence compared to nucleic acid-based and viral-vectored approaches.
CoVaccine HT™, is an oil-in-water nanoemulsion adjuvant with an excellent safety profile [8]. In combination with properly selected antigens, CoVaccine HT™ can achieve potent immunogenicity and protective efficacy in rodents and non-human primates (NHPs) [9], [10], [11], [12]. In previous work, we have successfully demonstrated the use of the recombinant protein subunit Drosophila S2 expression system in combination with CoVaccine HT™ to produce vaccines to combat global health threats such as Zika virus (ZIKV) and Ebola virus (EBOV). Immunization with recombinant ZIKV E protein induced potent neutralizing titers in mice [10] and non-human primates [9] and protection against viremia after viral challenge. Similarly, immunization with recombinant subunit formulations consisting of the EBOV glycoprotein and matrix proteins VP40 and VP24 was able to induce potent antibody titers and protection in both mouse [11] and guinea pig models of EBOV disease [12]. In healthy adults, CoVaccine HTTM was shown to be safe and well-tolerated with no risk of severe adverse effects in phase 1 clinical trials (NCT01015703) [8].
The spike (S) glycoprotein, comprised of a receptor binding subunit (S1) and a membrane-fusing subunit (S2) [13] is the main surface protein and present as homotrimers on the viral envelope of SARS-CoV-2. Based on previous preclinical studies of vaccines against the highly pathogenic SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) [14], [15], [16] as well as recent studies of patients with SARS-CoV-2 infections [17], [18], [19], [20], the S protein appears to be the antigenic target of both neutralizing antibody and T cell responses. The majority of current COVID-19 vaccines under preclinical and clinical development use full-length S proteins as antigen targets with further modifications such as removal of the polybasic sites [21], [22], [23], introduction of proline mutations [21], [24], [25], or addition of trimerization domains to preserve the native-like trimeric prefusion structure of S proteins. These antigens have been shown to mimic the native S protein presented on viral particles and preserve neutralization-sensitive epitopes [16], [26]. In a prior study, we evaluated the utility of CoVaccine HT™ adjuvant to induce properly balanced immunity against SARS-CoV-2, when formulated with a commercially available SARS-CoV-2 spike S1 protein [27]. In the current study we produced a native-like trimeric S protein ectodomain with and without stabilizing mutations using the Drosophila S2 cell expression system and assessed the immunogenicity of these S ectodoman trimers formulated with CoVaccine HT™ in mice. The scope of this work demonstrates that CoVaccine HT™ is an effective adjuvant that promotes rapid induction of balanced humoral and cellular immune responses and in combination with spike proteins warrants further development as an effective medical intervention against coronavirus disease.
Materials and methods
Ethics statement
All animal work was conducted in accordance with the Animal Welfare Act and the National Research Council (NRC) Guide for the Care and Use of Laboratory Animals. All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Hawaii at Manoa (UHM) and carried out in the UHM American Association for Accreditation of Laboratory Animal Care (AAALAC) accredited Laboratory Animal Facility.
Recombinant protein expression and purification
Plasmids were generated to express the native-like, trimeric, transmembrane (TM)-deleted spike (S) glycoprotein (SdTM) from SARS-CoV-2 strain Wuhan-Hu-1 (Genbank Accession number NC_045512). The SdTM sequence was designed to encode the SARS-CoV-2 S protein sequence spanning Gln14 to Ser1147. The SdTM gene was produced by de novo synthesis (ATUM, Newark, CA). The gene was also codon-optimized for expression in Drosophila S2 cells, with an altered furin cleavage site (RRAR changed to GSAR) between S1 and S2 domains to prevent cleavage, and contains a trimerization domain of T4 bacteriophage fibritin (foldon) at the C-terminus. Two additional proline substitutions (K986P and V987P) between the heptad repeat 1 and central helix regions and the removal of the S2′ protease cleavage site were introduced by site-directed mutagenesis to generate the stabilized prefusion structure of S protein (SdTM2P). The SARS-CoV-2 receptor binding domain with a foldon trimerization domain at the C-terminus (RBD-F) was also prepared. The RBD-F sequence was designed to encode the SARS-CoV-2 S protein sequence spanning Phe318 to Gly594. The proprietary expression vector pHH202 (Hawaii Biotech Inc., Honolulu, HI) containing S gene variants was transfected into Drosophila S2 cells using the Lipofectamine LTX with PLUS reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Stably transformed cell lines were created by selection with culture medium containing hygromycin B at 300 μg/mL To verify selection, transformants were induced with culture medium containing 200 μM CuSO4. Expression was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
Recombinant S proteins were purified from filtered cell culture supernatants by affinity chromatography (AC) using NHS-activated Sepharose (Cytiva, Marlborough, MA) coupled with 2 mg of a his-tagged human angiotensin I converting enzyme 2 (hACE2), which was also produced using the Drosophila S2 cell expression system and purified by Ni-affinity chromatography in our laboratory. Purified recombinant S proteins were concentrated using Amicon filtration devices (EMD Millipore, Billerica, MA), buffer-exchanged into PBS and analyzed by SDS-PAGE and Western blotting. Antigens were quantified by UV absorbance at 280 nm and stored at −80 °C.
A conventional immunoaffinity chromatography (IAC) method was also established to purify SdTM2P proteins. For this, the monoclonal antibody (mAb) CR3022 (provided by Mapp Biopharmaceutical, San Diego, CA), was coupled to NHS-activated Sepharose at a concentration of 10 mg/mL and used for IAC in tandem with a HiPrep 26/10 desalting column (Cytiva, Marlborough, MA) equilibrated with PBS allowing quick buffer exchange of the eluted protein from low pH buffer into PBS.
Mouse experiments
Groups (n = 7 or 15 per group) of 7 to 10-week-old Swiss Webster mice of both sexes (bred from original breeding stocks obtained from Taconic Biosciences, Germantown, NY) were immunized intramuscularly (i.m.) at days 0 and 21 with 5 μg of SARS-CoV-2 spike proteins SdTM or SdTM2P (purified by hACE2 affinity chromatography) alone or in combination with 1 mg of CoVaccine HT™ adjuvant (Protherics Medicines Development Ltd, a BTG company, London, United Kingdom). The negative control group received equivalent doses of adjuvant only. Vaccines were prepared by mixing the liquid adjuvant with antigen diluted in 1x PBS to the desired concentration immediately prior to administration to mice. To determine the optimal antigen and adjuvant dosing, mice were also immunized with either 5, 2.5, or 1.25 μg of SdTM2P (purified by mAb CR3022 IAC) and 1 mg of CoVaccine HT™ or 5 μg of SdTM2P formulated with 0.3 mg CoVaccine HT™. Serum samples were collected on days 7, 14, 28, and 35. Three to five mice from each group were euthanized on the seventh day after the first and second vaccinations and splenectomies were performed for preparation of splenocytes. The remaining four to five mice from each group were euthanized 14 days after the second vaccination and terminal bleeds collected by cardiac puncture.
Analysis of antibodies by multiplex microsphere immunoassay (MIA)
The IgG antibody in mouse sera was measured by a multiplex microsphere-based immunoassay as described previously [10], [27], [28]. Briefly, internally dyed, magnetic MagPlex® microspheres (Luminex Corporation, Austin, TX) were coupled to purified receptor binding domain (RBD-F), spike protein (SdTM2P) or bovine serum albumin (BSA) as control [10], [27]. A mixture of RBD, spike SdTM2P, and BSA-coupled beads (approximately 1,250 beads each) was incubated with diluted sera in black-sided 96-well plates for 3 h at 37 °C with gentle agitation in the dark. Following two washes with MIA buffer (1% BSA and 0.02 % Tween 20 in 1x PBS), 50 μl of 1 μg/mL red phycoerythrin (R-PE)-conjugated goat anti-mouse IgG antibodies (Jackson ImmunoResearch, Inc., West Grove, PA) were added and incubated at 37 °C for another 1 h. After washing twice with the MIA buffer, the beads were resuspended in MAGPIX® drive fluid and analyzed on a MAGPIX® Instrument (Luminex Corporation, Austin, TX).
The median fluorescence intensity (MFI) readouts of the experimental samples were converted to antibody concentrations using purified antibody standards prepared from pooled mouse antiserum to SdTM or SdTM2P as described below. For this, IgG was purified from mouse antisera by protein A affinity chromatography and then subjected to immunoaffinity chromatography (IAC) using SdTM2P-coupled NHS-Sepharose (Cytiva, Marlborough, MA) to select only S-reactive IgG. The concentration of purified S-specific IgG was quantified by measuring the UV absorbance of the solution at 280 nm. The purified anti-spike IgG was diluted to concentrations in the range of 4.88–5000 ng/mL and analyzed in the MIA assay. The resulting MFI values were analyzed using a sigmoidal dose–response, variable slope model (GraphPad Prism, San Diego, CA), with antibody concentrations transformed to log10 values. The resulting curves yielded r2 values >0.99 with a well-defined top and bottom and the linear range of the curve was determined. The experimental samples were analyzed side-by-side with the antibody standards at different dilutions (1:50, 1:200, 1:1000, 1:40,000, 1:80,000 or 1:160,000) to obtain MFI values that fall within the linear range of the standard curve. The experimental sample IgG concentrations were interpolated from the standard curves using the same computer program. Finally, the interpolated values were multiplied by the dilution factors and plotted as antibody concentrations (ng/mL).
The IgG subclass profile in serum samples was analyzed using IgG subclass-specific secondary antibodies (Southern Biotech, Birmingham, AL), and the ratios of IgG2a/IgG1 and IgG2b/IgG1 were calculated using the MFI readouts at the serum dilution (1:2000) that is within the linear range of the antibody binding standard curve.
Recombinant vesicular stomatitis virus (rVSV) neutralization assay
Replication-competent rVSV expressing SARS-CoV-2 S protein (Wuhan-Hu-1) with a 21 amino acid deletion in the cytoplasmic tail (rVSV-SARS-CoV-2-S) was generated as described previously [29] and the virus stocks were amplified in Vero E6 cells. For the plaque reduction neutralization test (PRNT) using rVSV- SARS-CoV-2-S, pooled or individual mouse serum samples were heat-inactivated at 56 °C for 30 min. Eight 3-fold serial dilutions of serum samples starting at final 1:10 dilution were prepared and incubated with 100 plaque-forming units (PFU) of rVSV-SARS-CoV-2-S at 37 °C for 1 h. Antibody-virus complexes were added to Vero E6 cell monolayers in 6-well plates and incubated at 37 °C for another hour followed by addition of overlay media mixed with 1% agarose. Three days later, cells were fixed and stained with a solution containing 1% formaldehyde, 1% methanol, and 0.05% crystal violet overnight for plaque enumeration. The neutralization titers (PRNT50) were defined as the highest serum dilution that resulted in 50% reduction in the number of plaques.
Wild-type SARS-CoV-2 virus neutralization assay
PRNT was also performed in a biosafety level 3 facility at BIOQUAL, Inc. (Rockville, MD) using 24-well plates. Mouse sera pooled from individual mice within each group, were diluted to 1:10, and a 1:3 serial dilution series was performed 11 times. Diluted samples were then incubated with 30 plaque-forming units of wild-type SARS-CoV-2 USA-WA1/2020 (BEI NR-52281), B.1.351 (Beta) (BEI NR-55282), or P.1. (Gamma) (BEI NR-54982) variants, in an equal volume of culture medium (DMEM with 10% FBS and gentamicin) for 1 h at 37 °C. The serum-virus mixtures were added to a monolayer of confluent Vero E6 cells and incubated for 1 h at 37 °C in 5% CO2. Each well was then overlaid with 1 ml of culture medium containing 0.5% methylcellulose and incubated for 3 days at 37 °C in 5% CO2. The plates were then fixed with methanol at −20 °C for 30 min and stained with 0.2% crystal violet for 30 min at room temperature. Neutralization titers (PRNT50) were defined as the highest final serum dilution that resulted in 50% reduction in the number of plaques.
Splenocyte preparation and FluoroSpot assays
Three or five mouse spleens from each group were harvested seven days after the first and second vaccinations, and single cell suspensions were prepared using a gentleMACS Dissociator (Miltenyi Biotec, Auburn, CA). The cells were passed through a cell strainer, resuspended in freezing medium containing 90% FBS and 10% dimethyl sulfoxide (DMSO) after lysis of red blood cells, and cryopreserved in liquid nitrogen. The FluoroSpot assay was performed using the mouse IFN-γ/IL-4 FluoroSpotPLUS kit according to the manufacturer’s instructions (Mabtech, Inc., Cincinnati, OH). Briefly, splenocytes were rested in a 37 °C, 5% CO2 incubator for 3 h after rapidly thawing in a 37 °C water bath followed by slow dilution with culture medium to allow removal of cell debris. A total of 2.5 × 105 cells per well in RPMI-1640 medium supplemented with 10% FBS, penicillin (100 units/mL) and streptomycin (100 μg/mL) were added in a 96 well PVDF membrane plate pre-coated with capture monoclonal antibodies. The cells were stimulated for 24 h with a peptide pool consisting of 17-mer peptides with 10 amino acids overlapping, covering the spike protein of SARS-CoV-2 (BEI Resources NR-52402) at 10 μg/mL or medium containing an equal concentration of DMSO (0.05%) as negative control. Fifty thousand cells were incubated with a cell activation cocktail (BioLegend, San Diego, CA) at 1:500 dilution containing phorbol-12-myristate 13-acetate (PMA, 0.081 μM) and Ionomycin (1.3386 μM) as positive control. Each stimulation condition was set up in triplicate. Plates were developed using specific monoclonal detection antibodies and fluorophore-conjugated secondary reagents. Finally, plates were treated with a Fluorescence enhancer (Mabtech) to optimize detection and then air-dried. The spots were enumerated using the CTL ImmunoSpot® S6 Universal Analyzer (Cellular Technology Limited, CTL, Shaker Heights, OH), and the number of antigen-specific cytokine secreting spot forming cells (SFCs) per million cells for each stimulation condition was calculated by subtracting the number of spots detected in the medium only wells.
Statistical analysis
Statistical analysis was performed using a one-sample t test and Wilcoxon test or Mann-Whitney t test to compare the neutralization titers, and IFN-γ secreting response between adjuvanted and unadjuvanted groups. The difference in the neutralizing antibody titers and the numbers of IFN-γ secreting cells between groups receiving different dosages of vaccines, or IgG subclass antibody profile, anti-S and anti-RBD IgG concentrations between unadjuvanted and adjuvanted groups were determined by one-way, or two-way, ANOVA with Tukey’s multiple comparison test. The correlation between the measured neutralization titers using rVSV-SARS-CoV-2 and authentic SARS-CoV-2 was analyzed by Pearson correlation analysis.
Results
Drosophila S2 cell-expressed recombinant S proteins adjuvanted with CoVaccine HT™ induce robust IgG antibody responses
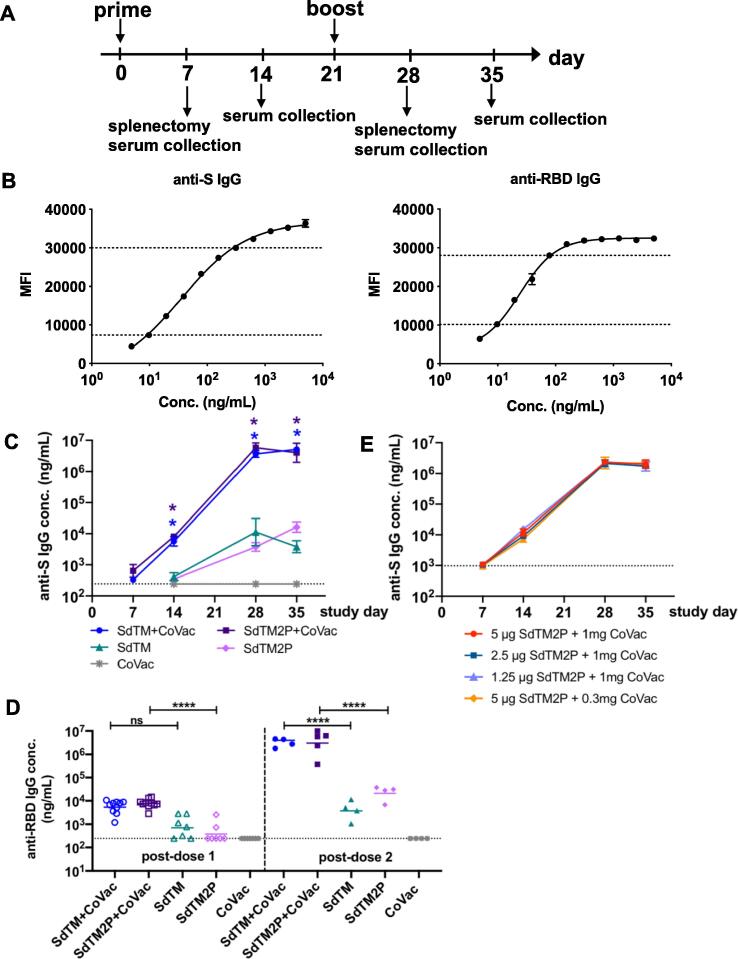
Using the Drosophila S2 cell expression system, we generated trimeric SARS-CoV-2 S protein devoid of transmembrane domain (SdTM) as well as a stabilized prefusion structure of S protein (SdTM2P). To explore different methods for purification of S protein, we employed two methods of purification using hACE2 protein, a known receptor for SARS-CoV-2, for AC, and the SARS-CoV RBD specific mAb CR3022 for IAC. This mAb, originally constructed from an immune svFc phage display library of lymphocytes from a convalescent SARS-CoV patient, was selected due to its cross-reactivity with both SARS-CoV and SARS-CoV 2 receptor binding sites, which contain the majority of neutralizing epitopes [30], [31]. The immunogenicity of recombinant S proteins was evaluated with and without CoVaccine HT™ adjuvant in outbred Swiss Webster mice. Animals were immunized with one or two doses at a 3-week interval by intramuscular injection with 5 μg of SARS-CoV-2 SdTM or SdTM2P (purified by hACE2 AC) alone or formulated with 1 mg CoVaccine HT™ (Fig. 1A). We developed a quantitative IgG assay in which purified anti-S polyclonal antibody was used as a standard and therefore the antigen-specific IgG levels are expressed as concentration (ng/mL) (Fig. 1B). Analysis of IgG antibody in sera obtained on days 7, 14, 28, and 35 indicated that the co-formulation of CoVaccine HT™ with SdTM or SdTM2P significantly increased anti-S IgG antibody levels over those induced by proteins alone (Fig. 1C). In addition, two doses of CoVaccine HT™ adjuvanted SdTM or SdTM2P elicited high levels of IgG antibodies to RBD, which is the target of neutralizing antibodies (Fig. 1D). To further optimize vaccine formulations, we evaluated the IgG antibody responses in mice that received a lower dosage (2.5 or 1.25 μg) of SdTM2P proteins or that received a reduced amount (0.3 mg) of adjuvant. The results demonstrated that vaccination with reduced amounts of antigen or adjuvant did not decrease the levels of antigen-specific IgG antibodies (Fig. 1E).
Fig. 1.
IgG antibody responses to recombinant SARS-CoV-2 S proteins. (A) Groups of Swiss Webster mice (n = 7 or 15) were immunized with one or two doses of recombinant S proteins with or without CoVaccine HT™ (CoVac) adjuvant at a 3-week interval. Sera were collected 1 and 2 weeks after each immunization (days 7, 14, 28, and 35), and spleens were harvested 1 week (day 7 and 28) after each immunization. SARS-CoV-2 S-specific IgG titers were measured by a multiplex microsphere immunoassay (MIA) using SdTM2P and RBD-F coupled beads. (B) The purified anti-S antibody was diluted to concentrations in the range of 4.8 to 5000 ng/mL and analyzed by MIA as a standard (as described in the Materials and Methods). Mouse sera were assayed along with the antibody standard and the IgG concentrations was interpolated from the standard curves using a sigmoidal dose–response computer model (GraphPad Prism). The dotted lines denote the top and bottom of linear range that were used to interpolate antibody concentration. (C) The anti-S and (D) anti-RBD antibody titers in sera from mice immunized with SdTM or SdTM2P (purified by hACE2 AC) with or without adjuvants or (E) anti-S antibody in sera of mice administered different dosages (5, 2.5, or 1.25 μg) of SdTM2P (purified by mAb IAC) with adjuvant (1 or 0.3 mg) are expressed as IgG concentrations (ng/mL). The dotted lines in panels C to E indicate the bottom of the linear range of the standard curve. Statistically significant differences in the concentrations of anti-S IgG (panel C) or anti-RBD IgG (panel D) between groups receiving vaccines with or without adjuvant were determined by two-way ANOVA with Tukey’s multiple comparisons test (*p < 0.05; ****p < 0.0001).
Recombinant S proteins adjuvanted with CoVaccine HT™ elicit potent neutralizing antibody responses
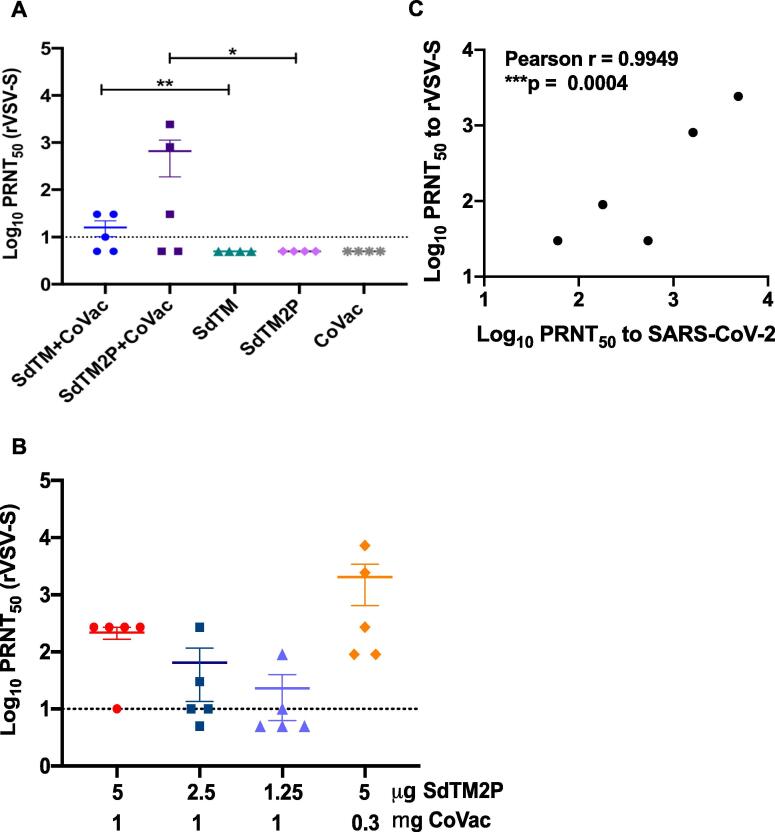
We next examined whether our vaccine candidates induced neutralizing antibody responses. Measurement of SARS-CoV-2 neutralizing antibodies requires biosafety level 3 (BSL3) laboratory facilities, and therefore, alternative assays have been developed, such as one utilizing replication-competent rVSV-ΔG expressing SARS-CoV-2 S glycoprotein (rVSV-SARS-CoV-2-S) [32], [33]. We first compared the neutralizing antibody titers of pooled mouse sera using both SARS-CoV-2 and rVSV-SARS-CoV-2-S (Table 1) and found that the titers are significantly correlated between these two assays (Fig. 2C). To further evaluate the neutralizing antibodies of individual animals, we employed only the PRNT using rVSV-SARS-CoV-2-S. The PRNT50 titers of sera obtained from mice vaccinated with SdTM or SdTM2P in combination with CoVaccine HT™ were significantly higher than those from animals receiving protein alone (Fig. 2A), suggesting that the CoVaccine HT™ adjuvant enhances induction of neutralizing antibody responses. Furthermore, a trend of dose-dependent decreases of neutralization titers was observed when animals were given lower dosages of SdTM2P proteins (Fig. 2B). Of note, the serum PRNT50 titers of mice receiving a lower amount (0.3 mg) of CoVaccine HT™ were comparable or even higher than those of mice given 1 mg of adjuvant indicating that the optimal vaccine formulation may be achieved using lower dosages of adjuvant.
Table 1.
Virus neutralizing antibody titers against SARS-CoV-2 lineage A USA-WA-1/2020, B.1.351, and P.1 variants, and rVSV-SARS-CoV-2-S (Wuhan-Hu-1) in pooled antisera from mice immunized with two doses of S proteins formulated with CoVaccine HT™ adjuvant.
| Immunogena | Adjuvanta | PRNT50b SARS-CoV-2 | PRNT50b rVSV-SARS-CoV-2-S | PRNT50b SARS-CoV-2 | PRNT50b SARS-CoV-2 |
|---|---|---|---|---|---|
| USA-WA1/2020 | Wuhan-Hu-1 | B.1.351 | P.1 | ||
| 5 µg SdTM | 1 mg CoVaccine HT™ | 540 | 30 | 60 | 180 |
| 5 µg SdTM2P | 1 mg CoVaccine HT™ | 1620 | 810 | 180 | 180 |
| 5 µg SdTM | None | <20 | <10 | <20 | <20 |
| 5 µg SdTM2P | None | <20 | <10 | <20 | <20 |
| None | 1 mg CoVaccine HT™ | <20 | <10 | NTc | NTc |
| 5 µg SdTM2P | 1 mg CoVaccine HT™ | NAd | 270 | 180 | 540 |
| 2.5 µg SdTM2P | 1 mg CoVaccine HT™ | 180 | 90 | 60 | 180 |
| 1.25 µg SdTM2P | 1 mg CoVaccine HT™ | 60 | 30 | 60 | 60 |
| 5 µg SdTM2P | 0.3 mg CoVaccine HT™ | 4860 | 2430 | 540 | 1620 |
Two vaccinations with protein with or without adjuvant given intramuscularly.
Pooled antiserum (n = 3 or 5 per group) collected 14 days post-dose 2. Titer equals the highest serum dilution yielding 50% reduction in plaque counts.
NT: Not tested.
NA: Assay results not available.
Fig. 2.
The serum neutralization titers of mice immunized with recombinant subunit SARS-CoV-2 vaccines. The neutralization titers of sera obtained 2 weeks post boosting (day 35) from mice vaccinated with (A) SARS-CoV-2 SdTM or SdTM2P proteins (purified by hACE2 AC) adjuvanted with or without 1 mg of CoVaccine HT™ (CoVac) or (B) different dosages (5, 2.5, or 1.25 μg) of SdTM2P (purified by mAb IAC) with CoVac (1 or 0.3 mg) was measured by a PRNT using rVSV-SARS-CoV-2-S. The data shown are log-transformed PRNT50 values from individual animals, and mean ± SEM of each group is indicated. Horizontal dashed lines represent the limit of detection. For the samples showing low (<50 %) or no neutralizing activity at the starting dilution (1:10) in the assay, a PRNT50 of 5 (half of the limit of detection) was reported when the data were plotted. One sample t and Wilcoxon tests on log-transformed data were used to analyze the significant difference between adjuvanted and protein alone groups in panel A (*p < 0.05, **p < 0.01). There are no significant differences between groups when analyzed by one-way ANOVA with Tukey’s multiple comparisons test in panel B. (C) The correlation between the neutralization titers of pooled serum samples from the same group to rVSV-SARS-CoV-2 and those to WT SARS-SARS-CoV-2 was examined by Pearson correlation analysis. The PRNT50 values are listed in Table 1.
Similar trends were seen with the pooled sera in PRNT using the SARS-CoV-2 wt, lineage B1.351 Beta, and P.1 Gamma VOC (Table 1). Although two doses of 5 μg SdTM or SdTM2P formulated with CoVaccine HTTM yielded PRNT50 titers approximately 3–9 fold-lower than the Lineage A, USA-WA1/2020 strain, both adjuvanted spike antigens were able to generate a humoral response capable of cross-variant neutralization. Interestingly, 2.5 and 1.25 μg of the SdTM2P antigen sustained the same neutralization level across different VOC, suggesting lower concentrations of antigen may generate a minimal threshold of neutralizing antibodies targeting conserved epitopes. Furthermore it appears that the B.1.351 Beta variant was more evasive than the P.1 Gamma variant, an observation consistent with clinical studies using convalescent sera and sera from vaccinated individuals [34].
Recombinant S proteins in combination with CoVaccine HT™ induce a balanced IgG subtype antibody response
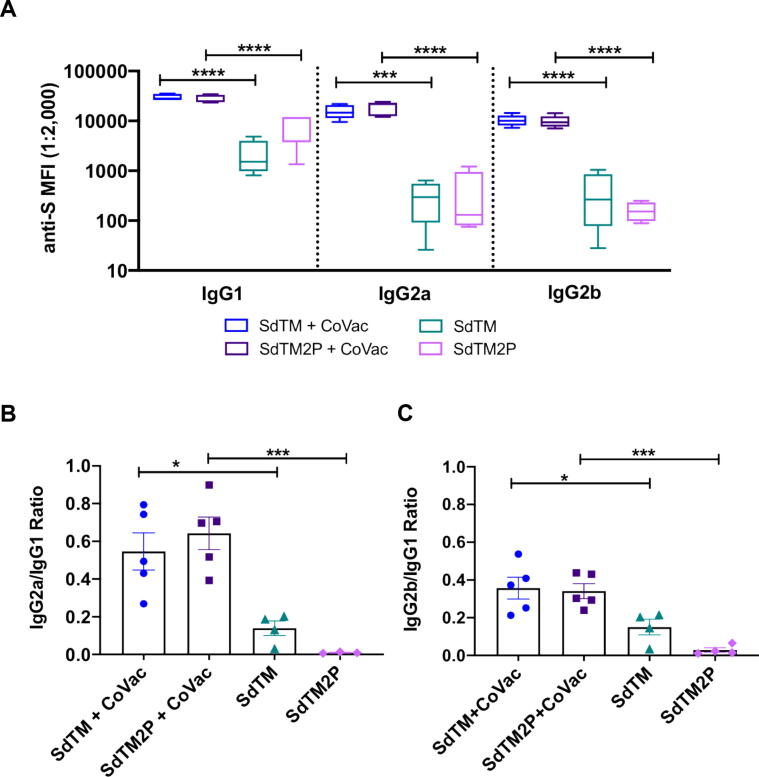
Vaccine-associated enhanced respiratory disease (VARED) has been reported in infants and young children immunized with inactivated whole virus vaccine against respiratory syncytial virus (RSV) and measles virus [35], [36], [37], and associated with Th2-biased immune responses [38]. A similar pulmonary immunopathology was also observed in animals with SARS-CoV vaccines [39], [40], [41]. Thus, we evaluated the balance of Th1 and Th2 by comparing the levels of S-specific IgG2a/b and IgG1, which are indicative of Th1 and Th2 responses, respectively. Both SdTM and SdTM2P adjuvanted with CoVaccine HT™ elicited high anti-S IgG2a/2b and IgG1 subclass antibodies whereas protein alone induced high IgG1 with lower IgG2a and IgG2b (Fig. 3A). To further assess the effect of CoVaccine HT™ adjuvanticity on IgG subclass antibody profiles, the ratios of IgG2a versus IgG1 as well as IgG2b versus IgG1 were calculated. The results indicate that CoVaccine HT™ enhanced the induction of Th1 responses as evidenced by the significantly higher ratios of IgG2a/IgG1 and IgG2b/IgG1 obtained from groups given either SdTM or SdTM2P with adjuvant compared to those without adjuvant (Fig. 3B, 3C).
Fig. 3.
IgG subclass profile induced after vaccination with recombinant S proteins with or without CoVaccine HT™ (CoVac) adjuvant. (A) Mouse sera collected at two weeks post-booster (day 35) were assessed for anti-S specific IgG1, IgG2a, and IgG2b by MIA. The ratios of IgG2a to IgG1 (B) or IgG2b to IgG1 (C) from individual animals were calculated using the MFI values at the serum dilution of 1:2,000. Bars represent mean ± SEM of each group. Statistically significant differences in the anti-S IgG1, IgG2a, and IgG2b antibody titers, or ratios of IgG2a/IgG1 and IgG2b/IgG1 between adjuvanted and protein alone groups were determined by a one-way ANOVA with Tukey’s multiple comparisons test (*p < 0.05; ***p < 0.001; ****p < 0.0001).
CoVaccine HT™ adjuvanted SARS-CoV-2 recombinant S protein vaccine elicits IFN-γ T cell response
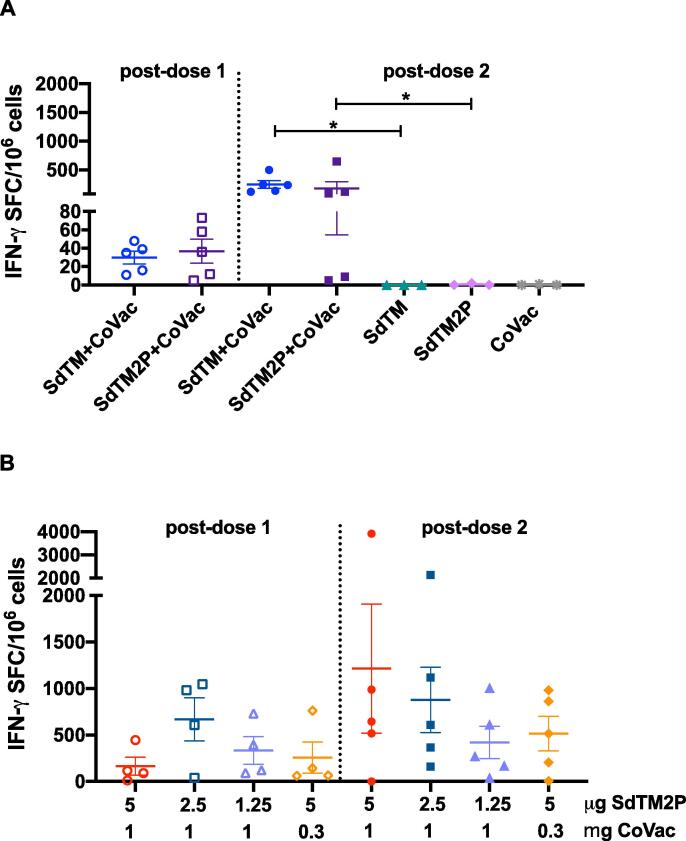
To assess vaccine-induced T cell responses, we analyzed the number of IFN-γ secreting cells after ex vivo stimulation of splenocytes with SARS-CoV-2 spike peptides by a FluoroSpot assay. Stimulation of cells prepared from mice receiving 2 doses of SdTM or SdTM2P with CoVaccine HT™ resulted in robust production of IFN-γ secreting cells (Fig. 4A). Interestingly, one dose of CoVaccine HT™ -adjuvanted S proteins also elicited a rapid IFN-γ secreting response (Fig. 4A). Additionally, a slight decrease in the numbers of IFN-γ secreting cells was observed when mice received a lower amount (2.5 or 1.25 μg) of adjuvanted SdTM2P proteins or a lower amount (0.3 mg) of CoVaccine HT™ (Fig. 4B). However, these differences were not statistically significant. In addition, spike peptide stimulation of immune splenocytes was also analyzed for production of IL-4, but no significant IL-4 production was detected at any timepoint with less than 10 spot forming cells (SFCs) per million cells (data not shown). Altogether, our platform shows great potential towards achieving a rapid, robust, and Th1-focused T cell response even in outbred populations.
Fig. 4.
Detection of IFN-γ secreting cells from mice immunized with SARS-CoV-2 vaccines. The splenocytes were obtained from mice (n = 3 or 5 per group) immunized with one dose (open symbols) or two doses (closed symbols) of (A) either 5 μg of SARS-CoV-2 SdTM or SdTM2P proteins (purified by hACE2 AC) adjuvanted with 1 mg of CoVaccine HT™ (CoVac) or (B) different dosages (5, 2.5, or 1.25 μg) of SdTM2P (purified by mAb IAC) with CoVac (1 or 0.3 mg). The cells were incubated for 24 h with medium or a peptide pool covering the S protein of SARS-CoV-2 (10 μg/mL), and the IFN-γ secreting cells were enumerated by FluoroSpot. The data are shown as the mean values of triplicate assays from individual animals using the number of spot forming cells (SFC) per 106 splenocytes after subtraction of the number of spots formed by cells in medium control wells (<5 spots). Bars represent mean ± SEM of each group. Significant differences in numbers of IFN-γ secreting cells between groups receiving 2 doses of vaccines with or without adjuvant in panel A was determined by Mann-Whitney t test (*p < 0.05). There are no significant differences between groups given either one or two doses of vaccines when analyzed by one-way ANOVA with Tukey’s multiple comparisons test in panel B.
Discussion
An ideal COVID-19 vaccine is expected to induce both humoral and cellular immunity, high titers of neutralizing antibodies and Th1-biased response to reduce potential risk of vaccine-associated enhancement of disease [42], [43], [44]. Using a well-established insect cell expression system, we have generated two versions of the SARS-CoV-2 S protein ectodomain and formulated them with a potent adjuvant, CoVaccine HT™. The vaccine candidates elicit both neutralizing antibody and cellular immunity with a balanced Th1/Th2 response in an outbred mouse model. The results obtained in this model may thus inform future vaccine development in animal models more closely related to humans [45]. These data support further preclinical and clinical development of CoVaccine HT™ adjuvanted SARS-CoV-2 vaccines to mitigate the ongoing COVID-19 pandemic.
Stabilization of S proteins in the prefusion trimeric conformation results in increased expression, conformational homogeneity, and production of potent neutralizing antibody responses [16], [25]. Current SARS-CoV-2 vaccines under development use either RBD or full-length S protein with or without modifications for stabilization of prefusion conformation as the major antigen targets [46], [47], [48], [49]. Although RBD is a primary target for potent neutralizing antibodies, it lacks other neutralizing epitopes present on full-length S. This might suggest that full-length S-based vaccines would broaden the neutralizing repertoire and cross-neutralize circulating spike variants of concern. In the current work, we produced trimeric S ectodomains in which the furin cleavage site was mutated (SdTM) and that were further stabilized in the prefusion S form by removing the S2′ protease cleavage site and introducing two proline substitutions (SdTM2P). The introduction of two prolines in the S2 subunit resulted in significantly greater production yield (∼3-fold increase) in Drosophila S2 cells (unpublished data). Vaccination with adjuvanted SdTM or SdTM2P elicits comparable levels of IgG antibody and IFN-γ cellular responses; however, the SdTM2P vaccine generated a slightly higher level of neutralizing antibodies than SdTM, which was also reported in studies of other SARS-CoV-2 and MERS-CoV vaccines [16], [22].
CoVaccine HT™ is a novel adjuvant that consists of a sucrose fatty acid sulfate ester (SFASE) immobilized on the oil droplets of a submicrometer emulsion of squalane in water (oil-in-water emulsion) [50]. It has been used for influenza virus and malaria vaccines and shown to enhance humoral and cellular protective immunity, in particular antibody response [51], [52], [53], [54], [55]. In addition, we have successfully utilized CoVaccine HT™ in our previous recombinant subunit EBOV, ZIKV and preliminary SARS-CoV-2 vaccine studies and have shown this adjuvant to elicit robust antibody responses [9], [10], [11], [27]. Use of CoVaccine HT™ with SdTM and SdTM2P yielded significantly enhanced total IgG and neutralizing antibody responses after both the first and second dose as compared to protein alone which reached similar IgG concentrations after two doses as did a single dose of the adjuvanted formulations. Although not directly comparable, the elicitation of S- and RBD-specific IgG and neutralizing antibody titers, along with IFN-γ secreting cells, after each dose followed similar kinetics as other 2P-S subunit vaccine candidates formulated with saponin or CpG based adjuvants [56], [57], while similarities in IgG and neutralizing antibody titers were observed compared with alum only-formulated candidates [58], [59]. Interestingly antibody levels (total IgG or neutralizing) did not decrease with a decreased dose of adjuvant; instead, a pronounced increase in neutralizing antibody titers was observed, indicating further opportunity for formulation optimization.
The decrease in neutralization capacity of the humoral response against the B.1.351 Beta and P.1 Gamma VOC is consistent with clinical data of vaccines using the pre-fusion stabilized spike [34], [60] that are still protective with only a marginal decrease in efficacy [61], [62], [63], [64]. Specifically, NVX-CoV2373 vaccine, a subunit vaccine in a Phase 1/2 clinical trial where the most common circulating strain is the B.1.351 Beta VOC showed sufficient protection against COVID-19 [65]. It is likely that a similar vaccine formulated with CoVaccine HTTM, based on our sustained neutralization results, may yield similar or better results in an appropriate challenge model.
In addition to improved kinetics, the addition of CoVaccine HTTM modulated the humoral response more towards Th1 type relative to protein alone, as indicated by higher levels of IgG2a and IgG2b. Antigen-specific IFN-γ production after splenocyte restimulation ex vivo was more variable, but also increased with the use of adjuvant, particularly after two doses, and was not strongly dependent on adjuvant concentration. These robust responses in outbred mice observed with relatively low antigen and adjuvant doses are particularly encouraging in light of our previous work which has demonstrated the successful thermostabilization of this adjuvant in combination with other glycoprotein antigens [66]. More recently, we have demonstrated that these recombinant subunits, in combination with CoVaccine HT™, can be formulated as a glassy solid using lyophilization. In this format, lyophilized vaccine formulations consisting of filovirus glycoprotein and adjuvant were able to maintain native quaternary structure and immunogenicity for at least 12 weeks at 40 °C [67], [68]. Other vaccines, using a similar lyophilization process, demonstrated potency after at least 1 year at 40 °C [69]. The similarity of the COVID-19 vaccine candidate formulation to the formulation of other vaccine candidates which were successfully thermostabilized suggests it also may be thermostabilized in a single vial format. This would allow easier vaccine stockpiling and distribution in regions of the world where maintaining cold-chain logistics necessary for transporting and storing vaccines from other platforms may be difficult.
The urgent need to rapidly vaccinate the human population worldwide both to slow the increase in fatalities and to prevent the emergence of escape mutants sharply highlights the limitations of vaccines requiring stringent cold chains as even under the best conditions, doses are lost due to inadequate storage or handling. The use of protein subunit vaccines not only offers additional vaccines to be used in parallel to rapidly immunize the global population, but also offers an easier opportunity to develop thermostabilized vaccines which would simplify rapid and far-flung delivery. Using recombinant spike protein without the transmembrane domain and with pre-fusion complex stabilization, we have demonstrated robust immune responses with the CoVaccine HT™ adjuvant in an outbred mouse model. This immune response is observed within 7 days of the first dose and peaks within 14 days after the second dose, showing robust humoral and cell mediated immunity. The immune response was potent with as little as 2.5 µg of protein and 0.3 mg of adjuvant, indicating an economical dose-sparing format should be feasible and further development should include testing of thermostabilized formulations in rodents and other species.
Funding
This work was supported in part by the National Institute of Allergy and Infectious Diseases (grant number R01AI132323), the Centers of Biomedical Research Excellence, the National Institute of General Medical Sciences (grant number P30GM114737), the Division of Intramural Research, NIAID, NIH and institutional funds. The funding sources had no involvement in study design; in the collection, analysis and interpretation of data; in writing of the report; and in the decision to submit the article for publication.
CRediT authorship contribution statement
Chih-Yun Lai: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Supervision, Project administration. Albert To: Conceptualization, Methodology, Validation, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing. Teri Ann S. Wong: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing – review & editing. Michael M. Lieberman: Conceptualization, Methodology, Formal analysis, Writing – review & editing. David E. Clements: Methodology, Resources, Writing – review & editing. James T. Senda: Methodology, Resources, Writing – review & editing. Aquena H. Ball: Investigation, Writing – original draft. Laurent Pessaint: Investigation. Hanne Andersen: Investigation. Wakako Furuyama: Resources. Andrea Marzi: Resources. Oreola Donini: Writing – original draft, Writing – review & editing. Axel T. Lehrer: Conceptualization, Methodology, Validation, Resources, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
Axel T. Lehrer and Oreola Donini are named inventors on a patent application covering a recombinant subunit vaccine for SARS-CoV-2. David E. Clements and James T. Senda, are current employees of Hawaii Biotech Inc. Laurent Pessaint and Hanne Andersen are current employees of BIOQUAL, Inc. Oreola Donini is a current employee of Soligenix Inc. We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Please adjust the text as necessary, but if your standard text shown in the proof is typically applied in such cases, this will be fine for us.
Acknowledgements
The authors thank Protherics Medicines Development Ltd (London, UK.) for the gift of CoVaccine HT™ adjuvant, and Mapp Biopharmaceutical for the monoclonal antibody CR3022. The authors also acknowledge the following reagent was obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Health (NIH): Peptide Array, SARS-Related Coronavirus 2 Spike Glycoprotein, NR-52402.
References
- 1.Center JHUCR. COVID-19 case tracker.
- 2.Lazarevic I., Pravica V., Miljanovic D., Cupic M. Immune Evasion of SARS-CoV-2 Emerging Variants: What Have We Learnt So Far? Viruses. 2021;13(7):1192. doi: 10.3390/v13071192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Draft landscape of COVID-19 candidate vaccines. https://wwwwhoint/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. 13 July 2021.
- 4.Pfizer-BioNTech COVID-19 Vaccine. Food and Drug Administration; 2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/pfizer-biontech-covid-19-vaccine.
- 5.Moderna COVID-19 Vaccine. U.S, Food and Drug Adminitration; 2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine.
- 6.Oliver Sara E., Gargano Julia W., Scobie Heather, Wallace Megan, Hadler Stephen C., Leung Jessica, et al. The Advisory Committee on Immunization Practices' Interim Recommendation for Use of Janssen COVID-19 Vaccine - United States, February 2021. MMWR Morb Mortal Wkly Rep. 2021;70(9):329–332. doi: 10.15585/mmwr.mm7009e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winger Anna, Caspari Thomas. The Spike of Concern-The Novel Variants of SARS-CoV-2. Viruses. 2021;13(6):1002. doi: 10.3390/v13061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inc. BI. Open-label Safety and Tolerability Study of CoVaccine HT™ in Healthy Volunteers. clinicaltrials.gov; 2010.
- 9.Medina Liana O., To Albert, Lieberman Michael M., Wong Teri Ann S., Namekar Madhuri, Nakano Eileen, et al. A Recombinant Subunit Based Zika Virus Vaccine Is Efficacious in Non-human Primates. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.0246410.3389/fimmu.2018.02464.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.To A., Medina L.O., Mfuh K.O., Lieberman M.M., Wong T.A.S., Namekar M., et al. Recombinant Zika Virus Subunits Are Immunogenic and Efficacious in Mice. mSphere. 2018;3 doi: 10.1128/mSphere.00576-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehrer Axel T., Wong Teri-Ann S., Lieberman Michael M., Humphreys Tom, Clements David E., Bakken Russell R., et al. Recombinant proteins of Zaire ebolavirus induce potent humoral and cellular immune responses and protect against live virus infection in mice. Vaccine. 2018;36(22):3090–3100. doi: 10.1016/j.vaccine.2017.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehrer Axel T., Wong Teri-Ann S., Lieberman Michael M., Johns Lisa, Medina Liana, Feldmann Friederike, et al. Recombinant subunit vaccines protect guinea pigs from lethal Ebola virus challenge. Vaccine. 2019;37(47):6942–6950. doi: 10.1016/j.vaccine.2019.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent Advances in the Vaccine Development Against Middle East Respiratory Syndrome-Coronavirus. Front Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham Rachel L., Donaldson Eric F., Baric Ralph S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallesen Jesper, Wang Nianshuang, Corbett Kizzmekia S., Wrapp Daniel, Kirchdoerfer Robert N., Turner Hannah L., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. PNAS. 2017;114(35):E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okba Nisreen M.A., Müller Marcel A., Li Wentao, Wang Chunyan, GeurtsvanKessel Corine H., Corman Victor M., et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wajnberg Ania, Amanat Fatima, Firpo Adolfo, Altman Deena R., Bailey Mark J., Mansour Mayce, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science (New York, NY) 2020;370(6521):1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Peng, Yang Xing-Lou, Wang Xian-Guang, Hu Ben, Zhang Lei, Zhang Wei, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grifoni Alba, Weiskopf Daniela, Ramirez Sydney I., Mateus Jose, Dan Jennifer M., Moderbacher Carolyn Rydyznski, et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amanat F., Strohmeier S., Rathnasinghe R., Schotsaert M., Coughlan L., García-Sastre A., et al. Introduction of two prolines and removal of the polybasic cleavage site leads to optimal efficacy of a recombinant spike based SARS-CoV-2 vaccine in the mouse model. bioRxiv Prep Server Biol. 2020 doi: 10.1128/mBio.02648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercado Noe B., Zahn Roland, Wegmann Frank, Loos Carolin, Chandrashekar Abishek, Yu Jingyou, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keech Cheryl, Albert Gary, Cho Iksung, Robertson Andreana, Reed Patricia, Neal Susan, et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbett Kizzmekia S., Edwards Darin K., Leist Sarah R., Abiona Olubukola M., Boyoglu-Barnum Seyhan, Gillespie Rebecca A., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh Ching-Lin, Goldsmith Jory A., Schaub Jeffrey M., DiVenere Andrea M., Kuo Hung-Che, Javanmardi Kamyab, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science (New York, NY) 2020;369(6510):1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham Barney S., Gilman Morgan S.A., McLellan Jason S. Structure-Based Vaccine Antigen Design. Annu Rev Med. 2019;70(1):91–104. doi: 10.1146/annurev-med-121217-094234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haun Brien K., Lai Chih-Yun, Williams Caitlin A., Wong Teri Ann S., Lieberman Michael M., Pessaint Laurent, et al. CoVaccine HT™ Adjuvant Potentiates Robust Immune Responses to Recombinant SARS-CoV-2 Spike S1 Immunization. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.599587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namekar Madhuri, Kumar Mukesh, O'Connell Maile, Nerurkar Vivek R., Klein Robyn. Effect of serum heat-inactivation and dilution on detection of anti-WNV antibodies in mice by West Nile virus E-protein microsphere immunoassay. PLoS ONE. 2012;7(9):e45851. doi: 10.1371/journal.pone.004585110.1371/journal.pone.0045851.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furuyama W., Shifflett K., Pinski A.N., Griffin A.J., Feldmann F., Okumura A., et al. Rapid protection from COVID-19 in nonhuman primates vaccinated intramuscularly but not intranasally with a single dose of a recombinant vaccine. bioRxiv Prep Server Biol. 2021 doi: 10.1128/mbio.03379-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ter Meulen Jan, van den Brink Edward N., Poon Leo L.M., Marissen Wilfred E., Leung Cynthia S.W., Cox Freek, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3(7):e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan Meng, Wu Nicholas C., Zhu Xueyong, Lee Chang-Chun D., So Ray T.Y., Lv Huibin, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science (New York, NY) 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Experim Med. 2020;217 doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case James Brett, Rothlauf Paul W., Chen Rita E., Liu Zhuoming, Zhao Haiyan, Kim Arthur S., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28(3):475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann Markus, Arora Prerna, Groß Rüdiger, Seidel Alina, Hörnich Bojan F., Hahn Alexander S., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184(9):2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 36.Chin J., Magoffin R.L., Shearer L.A., Schieble J.H., Lennette E.H. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am J Epidemiol. 1969;89:449–463. doi: 10.1093/oxfordjournals.aje.a120957. [DOI] [PubMed] [Google Scholar]
- 37.Fulginiti V.A., Eller J.J., Downie A.W., Kempe C.H. Altered reactivity to measles virus. Atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202:1075–1080. doi: 10.1001/jama.202.12.1075. [DOI] [PubMed] [Google Scholar]
- 38.Browne Sarah K., Beeler Judy A., Roberts Jeffrey N. Summary of the Vaccines and Related Biological Products Advisory Committee meeting held to consider evaluation of vaccine candidates for the prevention of respiratory syncytial virus disease in RSV-naïve infants. Vaccine. 2020;38(2):101–106. doi: 10.1016/j.vaccine.2019.10.048. [DOI] [PubMed] [Google Scholar]
- 39.Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23(17-18):2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng Chien-Te, Sbrana Elena, Iwata-Yoshikawa Naoko, Newman Patrick C., Garron Tania, Atmar Robert L., et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE. 2012;7(4):e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeyanathan Mangalakumari, Afkhami Sam, Smaill Fiona, Miller Matthew S., Lichty Brian D., Xing Zhou. Immunological considerations for COVID-19 vaccine strategies. Nat Rev Immunol. 2020;20(10):615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krammer Florian. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 44.Graham Barney S. Rapid COVID-19 vaccine development. Science (New York, NY) 2020;368(6494):945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 45.Tuttle A.H., Philip V.M., Chesler E.J., Mogil J.S. Author Correction: Comparing phenotypic variation between inbred and outbred mice. Nat Meth. 2020;17:947. doi: 10.1038/s41592-020-0932-7. [DOI] [PubMed] [Google Scholar]
- 46.Nanishi E., Borriello F., O'Meara T.R., McGrath M.E., Saito Y., Haupt R.E., et al. Alum:CpG adjuvant enables SARS-CoV-2 RBD-induced protection in aged mice and synergistic activation of human elder type 1 immunity. bioRxiv. 2021 [Google Scholar]
- 47.Dalvie N.C., Rodriguez-Aponte S.A., Hartwell B.L., Tostanoski L.H., Biedermann A.M., Crowell L.E., et al. Engineered SARS-CoV-2 receptor binding domain improves immunogenicity in mice and elicits protective immunity in hamsters. bioRxiv Prep Server Biol. 2021 [Google Scholar]
- 48.Chen Wen-Hsiang, Tao Xinrong, Agrawal Anurodh Shankar, Algaissi Abdullah, Peng Bi-Hung, Pollet Jeroen, et al. Yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1) formulated with aluminum hydroxide induces protective immunity and reduces immune enhancement. Vaccine. 2020;38(47):7533–7541. doi: 10.1016/j.vaccine.2020.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Yunfei, Wang Lichun, Cao Han, Liu Cunbao. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J Med Virol. 2021;93(2):892–898. doi: 10.1002/jmv.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blom Anneke G., Hilgers Luuk A. Th. Sucrose fatty acid sulphate esters as novel vaccine adjuvants: effect of the chemical composition. Vaccine. 2004;23(6):743–754. doi: 10.1016/j.vaccine.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 51.Bodewes R., Kreijtz J.H.C.M., van Amerongen G., Geelhoed-Mieras M.M., Verburgh R.J., Heldens J.G.M., et al. A single immunization with CoVaccine HT-adjuvanted H5N1 influenza virus vaccine induces protective cellular and humoral immune responses in ferrets. J Virol. 2010;84(16):7943–7952. doi: 10.1128/JVI.00549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heldens J.G.M., Glansbeek H.L., Hilgers L.A.T., Haenen B., Stittelaar K.J., Osterhaus A.D.M.E., et al. Feasibility of single-shot H5N1 influenza vaccine in ferrets, macaques and rabbits. Vaccine. 2010;28(51):8125–8131. doi: 10.1016/j.vaccine.2010.09.097. [DOI] [PubMed] [Google Scholar]
- 53.Mahdi Abdel Hamid M., Remarque E.J., van Duivenvoorde L.M., van der Werff N., Walraven V., Faber B.W., et al. Vaccination with Plasmodium knowlesi AMA1 formulated in the novel adjuvant co-vaccine HT™ protects against blood-stage challenge in rhesus macaques. PloS one. 2011;6:e20547. doi: 10.1371/journal.pone.0020547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kusi Kwadwo A, Remarque Edmond J, Riasat Vanessa, Walraven Vanessa, Thomas Alan W, Faber Bart W, et al. Safety and immunogenicity of multi-antigen AMA1-based vaccines formulated with CoVaccine HT™ and Montanide ISA 51 in rhesus macaques. Malar J. 2011;10(1) doi: 10.1186/1475-2875-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lakhan Nerissa, Stevens Natalie E., Diener Kerrilyn R., Hayball John D. CoVaccine HT™ adjuvant is superior to Freund's adjuvants in eliciting antibodies against the endogenous alarmin HMGB1. J Immunol Methods. 2016;439:37–43. doi: 10.1016/j.jim.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 56.Tian Jing-Hui, Patel Nita, Haupt Robert, Zhou Haixia, Weston Stuart, Hammond Holly, et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat Commun. 2021;12(1) doi: 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuo Tsun-Yung, Lin Meei-Yun, Coffman Robert L., Campbell John D., Traquina Paula, Lin Yi-Jiun, et al. Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-77077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Tingting, Zheng Qingbing, Yu Hai, Wu Dinghui, Xue Wenhui, Xiong Hualong, et al. SARS-CoV-2 spike produced in insect cells elicits high neutralization titres in non-human primates. Emerg Microbes Infect. 2020;9(1):2076–2090. doi: 10.1080/22221751.2020.1821583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seephetdee C., Buasri N., Bhukhai K., Srisanga K., Manopwisedjaroen S., Lertjintanakit S., et al. Mice Immunized with the Vaccine Candidate HexaPro Spike Produce Neutralizing Antibodies against SARS-CoV-2. Vaccines (Basel) 2021;9 doi: 10.3390/vaccines9050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Pengfei, Nair Manoj S., Liu Lihong, Iketani Sho, Luo Yang, Guo Yicheng, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 61.Alter Galit, Yu Jingyou, Liu Jinyan, Chandrashekar Abishek, Borducchi Erica N., Tostanoski Lisa H., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596(7871):268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sadoff Jerald, Gray Glenda, Vandebosch An, Cárdenas Vicky, Shukarev Georgi, Grinsztejn Beatriz, et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. New Engl J Med. 2021;384(23):2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Daming, Dejnirattisai Wanwisa, Supasa Piyada, Liu Chang, Mentzer Alexander J., Ginn Helen M., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv Prep Server Biol. 2021 [Google Scholar]
- 65.Shinde Vivek, Bhikha Sutika, Hoosain Zaheer, Archary Moherndran, Bhorat Qasim, Fairlie Lee, et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. New Engl J Med. 2021;384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Preston Kendall B., Wong Teri Ann S., To Albert, Tashiro Taylor E., Lieberman Michael M., Granados Alex, et al. Single-vial filovirus glycoprotein vaccines: Biophysical characteristics and immunogenicity after co-lyophilization with adjuvant. Vaccine. 2021;39(39):5650–5657. doi: 10.1016/j.vaccine.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chisholm Carly Fleagle, Kang Taek Jin, Dong Miao, Lewis Kasey, Namekar Madhuri, Lehrer Axel T., et al. Thermostable Ebola virus vaccine formulations lyophilized in the presence of aluminum hydroxide. Eur J Pharmaceut Biopharmaceut Off J Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik eV. 2019;136:213–220. doi: 10.1016/j.ejpb.2019.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Preston Kendall B., Monticello Connor R., Wong Teri Ann S., To Albert, Donini Oreola, Lehrer Axel T., et al. Preservation of Quaternary Structure in Thermostable, Lyophilized Filovirus Glycoprotein Vaccines: A Search for Stability-Indicating Assays. J Pharm Sci. 2020;109(12):3716–3727. doi: 10.1016/j.xphs.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westfall Jennifer, Yates Jennifer L., Van Slyke Greta, Ehrbar Dylan, Measey Thomas, Straube Richard, et al. Thermal stability and epitope integrity of a lyophilized ricin toxin subunit vaccine. Vaccine. 2018;36(40):5967–5976. doi: 10.1016/j.vaccine.2018.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]