Abstract
Introduction
The purpose of this study is to examine the predictive utility of polygenic risk scores (PRSs) for smoking behaviors.
Aims and Methods
Using summary statistics from the Sequencing Consortium of Alcohol and Nicotine use consortium, we generated PRSs of ever smoking, age of smoking initiation, cigarettes smoked per day, and smoking cessation for participants in the population-based Atherosclerosis Risk in Communities (ARIC) study (N = 8638), and the Collaborative Genetic Study of Nicotine Dependence (COGEND) (N = 1935). The outcomes were ever smoking, age of smoking initiation, heaviness of smoking, and smoking cessation.
Results
In the European ancestry cohorts, each PRS was significantly associated with the corresponding smoking behavior outcome. In the ARIC cohort, the PRS z-score for ever smoking predicted smoking (odds ratio [OR]: 1.37; 95% confidence interval [CI]: 1.31, 1.43); the PRS z-score for age of smoking initiation was associated with age of smoking initiation (OR: 0.87; 95% CI: 0.82, 0.92); the PRS z-score for cigarettes per day was associated with heavier smoking (OR: 1.17; 95% CI: 1.11, 1.25); and the PRS z-score for smoking cessation predicted successful cessation (OR: 1.24; 95% CI: 1.17, 1.32). In the African ancestry cohort, the PRSs did not predict smoking behaviors.
Conclusions
Smoking-related PRSs were associated with smoking-related behaviors in European ancestry populations. This improvement in prediction is greatest in the lowest and highest genetic risk categories. The lack of prediction in African ancestry populations highlights the urgent need to increase diversity in research so that scientific advances can be applied to populations other than those of European ancestry.
Implications
This study shows that including both genetic ancestry and PRSs in a single model increases the ability to predict smoking behaviors compared with the model including only demographic characteristics. This finding is observed for every smoking-related outcome. Even though adding genetics is more predictive, the demographics alone confer substantial and meaningful predictive power. However, with increasing work in PRSs, the predictive ability will continue to improve.
Introduction
Cigarette smoking is a complex and multifaceted behavior. Key milestones include smoking initiation, regular use, heavy consumption, and cessation, and both genetic and environmental factors contribute to each step in this behavioral cascade. Though the prevalence of cigarette smoking has decreased in the United States1 through both reduced initiation and increased cessation, it remains a leading cause of preventable death2 and a major contributor to cancer, cardiovascular disease, and lung disease.3 In the United States, approximately 34 million adults smoke,4 despite approximately two-thirds of current smokers reporting a desire to quit.5
Cigarette smoking behaviors are heritable,6,7 and several genetic studies have examined smoking-related phenotypes.8 The largest genome-wide association study (GWAS) of smoking-related phenotypes to date was performed by the GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN) consortium (N up to 1.2 million).9 GSCAN found hundreds of genetic variants, both known and novel, associating with ever smoking, age of smoking initiation, cigarettes smoked per day, and smoking cessation.9 This work has created an extensive catalog of genetic associations for smoking-related behaviors.
These GWAS results now provide the opportunity to develop polygenic risk scores (PRSs) by combining summary single nucleotide polymorphism (SNP) statistics from a GWAS into a single risk variable that can then be tested for its predictive ability for an individual to develop a trait or disease. PRSs have successfully been applied to predict many disorders including breast cancer,10 obesity,11 prostate cancer,12 and schizophrenia.13 A study by Belsky et al. created an early smoking-related PRS based on GWAS results from three meta-analyses of cigarettes smoked per day and examined the association of the PRS with smoking transitions observed in 1037 participants in a 38-year prospective study conducted in New Zealand.14 The authors observed that smokers who had a high smoking-related PRS were more likely to progress to heavy smoking and were less likely to quit.14
With the availability of GWAS results from the large-scale GSCAN consortium and thus more statistical power to capture genetic signals, our goal was to more comprehensively test the utility of genetic variables to predict different steps in smoking behaviors: ie, ever smoking, early versus late smoking initiation, heaviness of smoking, and smoking cessation—in a population-based study and a selected sample.
Materials and Methods
Demographics Data
The Atherosclerosis Risk in Communities Study
The Atherosclerosis Risk in Communities (ARIC) study is a prospective epidemiologic study focusing on understanding atherosclerosis.15 In 1987, approximately 16 000 participants ages 45–64 were recruited from four US communities: Forsyth County, NC; Jackson, MS; Minneapolis, MN; and Washington County, MD.15 A subset of the ARIC participants underwent genetic testing, and the dataset was divided into two samples by ancestry: one dataset consisted of 8638 European American individuals with a mean age of 54.3 ± 6 (Supplementary Table 1); the second dataset consisted of 2412 African American individuals with a mean age of 53.4 ± 6 (Supplementary Table 2). Because the main GSCAN results were generated in those of European ancestry,9 we focused our primary analyses of ARIC to participants of European descent with genetic data.
Smoking history was assessed via self-report.15 An individual was considered an ever smoker (compared with a never smoker) if the individual had smoked. Age of smoking initiation was reported as the age of first regularly smoking cigarettes. Age of smoking initiation was transformed into a binary outcome by stratifying individuals into one of two groups: early initiation (those who initiated smoking at <18 years old) and later initiation (those who initiated smoking at ≥18 years old) based on a median split. Individuals who had ever smoked regularly were also asked the average number of cigarettes smoked per day. The cigarettes smoked per day measure was constructed from the maximum of lifetime number of cigarettes smoked per day. Cigarettes smoked per day was then dichotomized into two levels (heavier (≥21 cigarettes smoked per day) versus lighter (≤20 cigarettes smoker per day) smoking). Finally, of those individuals who had smoked, successful smoking cessation was defined in those who reported no smoking at the last assessment. All smoking-related phenotypes were dichotomized to assist in interpretability of results. For information on the distribution of these key variables, see Supplementary Tables 1 and 2. This study had Institutional Review Board (IRB) approval.
The Collaborative Genetic Study of Nicotine Dependence
The purpose of the Collaborative Genetic Study of Nicotine Dependence (COGEND) was to study genetic contributions to the development of nicotine dependence. The COGEND study included individuals ages 25–44 who were recruited by telephone screening in Detroit, Minneapolis, and St. Louis.16 Extremes in smoking behavior were recruited into the study. Current nicotine dependent cases were defined as having a Fagerström Test for Nicotine Dependence (FTND) score of 4–10 while controls were defined as having an FTND score of 0–1 while having smoked at least 100 cigarettes lifetime.17 The COGEND individuals in this study self-identified as European ancestry, which was confirmed genetically18 (N = 1935) (Supplementary Table 3). All participants were ever smokers. Age of smoking initiation was reported as the age of first regularly smoking cigarettes. Age of smoking initiation was transformed into a binary outcome by stratifying individuals into one of two groups: early initiation (those who initiated smoking at <14 years old) and later initiation (those who initiated smoking at ≥14 years old) based on a median split. Participants were also asked the number of cigarettes smoked per day when smoking the most. Cigarettes smoked per day was then dichotomized into two levels (heavier (≥21 cigarettes smoked per day) versus lighter (≤20 cigarettes smoked per day) smoking). Finally, successful smoking cessation was defined in those who reported no smoking at the last assessment. This study had IRB approval at each data collection site, and participants gave informed consent prior to enrolling.
Genotyping
ARIC participants were genotyped at the Broad Institute on the Affymetrix 6.0 chip. Data for ARIC participants were obtained from the National Center for Biotechnology Information database of Genotypes and Phenotypes (dbGaP) (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) under accession number (phs000090.p5.v1). COGEND data were genotyped using Illumina HumanOmni2.5 from the Genetic Architecture of Smoking and Smoking Cessation (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000404.v1.p1&phv=162951&phd=3684&pha=&pht=2369&phvf=&phdf=&phaf=&phtf=&dssp=1&consent=&temp=1) under accession number (phs000404.v1.p1) and using Illumina 1M from the Study of Addiction: Genetics and Environment (SAGE) (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1) under accession number (phs000092.v1.p1). Additional data on how COGEND individuals were genotyped can be found on the dbGaP repository (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/document.cgi?study_id=phs000092.v1.p1&phd=2274) under accession number (phd002274.1).
GWAS Quality Control and Imputation
Standard GWAS Quality Control (QC) was applied using PLINK software,19 and SNPs were aligned to the + strand of the 1000 Genomes (build 37, 2013). Genotyped SNPs were imputed on the University of Michigan Imputation server using the 1000 Genomes build 37 phase 5 as the reference panel.20 Imputed SNPs were filtered for an info score of ≥0.9 and a minor allele frequency ≥1%. Included SNPs were converted to hard calls.
Statistical Analyses
We used Stata/SE (College Station, TX) to summarize demographic and covariate data and to perform regression analyses. Principal components (PCs) of genetic ancestry for ARIC were downloaded from the dbGaP repository (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap). We generated PCs for genetic ancestry for COGEND using EIGENSTRAT4.2.21
Four smoking-related PRSs were generated from the GWAS summary statistics of ever smoking, age of smoking initiation, cigarettes smoked per day, and smoking cessation from GSCAN9 using PRSice software.22 Because GSCAN included individuals from the ARIC and COGEND cohorts, we received updated GSCAN GWAS summary statistics excluding these two studies to preserve independence between generating and applying the PRSs. SNPs were clumped by PRSice.22 PRSs were created by PRSice.22 One of the most documented SNPs of smoking, rs16969968,8 was forced into the creation of each PRS. We included this SNP because rs16969968 confers a change in amino acid within the CHRNA5 gene which alters the nicotinic receptor function and likely is a functional SNP associated with heaviness of smoking.8,23 For each of the smoking-related phenotypes, PRSs were generated for various p value thresholds and for each p value threshold we examined that model’s predictive ability for that phenotype. Consistent with other studies,22,24,25 the percentage of the variance explained generally increased when using PRSs developed from the larger number of SNPs included with the lower p value thresholds (Supplementary Figures 1–4). Because there were only minor differences in the selection of the more generous p value thresholds, we adopted the PRSs generated from the p value threshold of 5.0 × 10−1 for each phenotype for all subsequent analyses for consistency. The final number of independent SNPs compiling each PRS is shown in Supplementary Table 4. Lastly, PRSs were standardized to z-scores for ease of interpretability. The reference group for PRS z-scores was 0.
Logistic regression models were created for the following outcomes: ever smoking, early versus late age of smoking initiation, heaviness of smoking (dichotomized cigarettes smoked per day), and smoking cessation. For each outcome, two main models were compared: the baseline demographics only model—adjusting for age, sex; and the baseline model plus genetic ancestry and the matching smoking-related PRS for the outcome (ie, ever smoking PRS for ever smoking outcome) in addition to the predictors age and sex. Then to more comprehensively assess the contribution of genetic ancestry and PRS, four models were evaluated: the baseline demographics only model—adjusting for age, sex; the baseline model plus genetic ancestry (which added 10 PCs) in addition to the predictors age and sex; the baseline model plus the matching smoking-related PRS for the outcome (ie, ever smoking PRS for ever smoking outcome) in addition to the predictors age and sex; and the baseline model plus genetic ancestry and the matching smoking-related PRS for the outcome (ie, ever smoking PRS for ever smoking outcome) in addition to the predictors age and sex. The pseudo R2 represents the Nagelkerke R2. The effectiveness of each model was compared with each other via the likelihood ratio test. In addition, we created receiver operating characteristic (ROC) curves for outcome, and the area under the ROC curve (AUC) values were estimated. When examining the prevalence of each smoking-related behavior, we assigned each individual a predicted probability of a smoking-related outcome from the corresponding regression analyses. This predicted probability was stratified into deciles, and the prevalence of each outcome was determined within each decile. The prevalence of each outcome within each decile was then plotted for interpretability purposes.
Results
Multiple PRSs and Genetic Ancestry Are Associated With Smoking Behaviors
Smoking-related PRS was significantly associated with each smoking behavior in both the ARIC and COGEND cohorts. For example, in the ARIC cohort, the ever smoking PRS z-score was significantly associated with the ever smoking behavior (odds ratio [OR]: 1.37; 95% confidence interval [CI]: 1.31, 1.43), and the age of smoking initiation PRS z-score was significantly associated with early age of smoking initiation (OR: 0.87; 95% CI: 0.82, 0.92) after adjusting for demographic covariates (age and sex) and genetic ancestry. Adding the PRS z-score corresponding to the outcome substantially increases the predictive ability as measured by pseudo R2 (ie, for ever smoking, demographics only—pseudo R2 = 0.068 vs. demographics plus genetic ancestry PCs and one PRS model—pseudo R2 = 0.107) (Table 1 and Supplementary Table 5). Each of the other smoking-related outcomes showed statistically significant improvement in model fit. Similar results were seen in the COGEND sample (Supplementary Table 6).
Table 1.
Models Predicting Each Smoking Phenotype Within European Americans in the ARIC Dataset
| Exposures | Demographics only | Demographics plus genetic ancestry and PRS |
|---|---|---|
| OR [95% CI] | OR [95% CI] | |
| Ever smoking | ||
| Age | 1.00 [0.99, 1.01] | 1.00 [0.99, 1.01] |
| Sex—female | 0.39 [0.35, 0.42] | 0.37 [0.34, 0.41] |
| PRS—ever smoking | 1.37 [1.31, 1.43] | |
| Model prediction | Pseudo R2 = 0.068 | Pseudo R2 = 0.107a |
| Early age of smoking initiation (<18 years) | ||
| Age | 0.99 [0.98, 1.00] | 0.99 [0.98, 1.00] |
| Sex—female | 0.42 [0.38, 0.48] | 0.42 [0.37, 0.47] |
| PRS—age of smoking initiation | 0.87 [0.82, 0.92] | |
| Model prediction | Pseudo R2 = 0.056 | Pseudo R2 = 0.067b |
| Heavier smoking (≥21 cigarettes smoked per day) | ||
| Age | 0.97 [0.96, 0.98] | 0.97 [0.96, 0.98] |
| Sex—female | 0.43 [0.38, 0.48] | 0.42 [0.37, 0.47] |
| PRS—cigarettes smoked per day | 1.17 [1.11, 1.25] | |
| Model prediction | Pseudo R2 = 0.056 | Pseudo R2 = 0.066c |
| Smoking cessation | ||
| Age | 0.97 [0.96, 0.98] | 0.97 [0.96, 0.98] |
| Sex—female | 2.00 [1.79, 2.24] | 2.01 [1.79, 2.25] |
| PRS—smoking cessation | 1.24 [1.17, 1.32] | |
| Model prediction | Pseudo R2 = 0.047 | Pseudo R2 = 0.069d |
ARIC = Atherosclerosis Risk in Communities, CI = confidence interval, OR = odds ratio, PRS = polygenic risk score. The reference for sex is male. The PRSs are in z-scores. The reference group for each PRS is a z-score of 0. Each OR represents a 1 standard deviation increase in PRS z-score.
aThe Χ2 statistic from the likelihood ratio test when comparing to the demographics only model is 264.80 with 11 degrees of freedom and a corresponding p value of <0.0001.
bThe Χ2 statistic from the likelihood ratio test when comparing to the demographics only model is 44.19 with 11 degrees of freedom and a corresponding p value of <0.0001.
cThe Χ2 statistic from the likelihood ratio test when comparing to the demographics only model is 38.11 with 11 degrees of freedom and a corresponding p value of 0.0001.
dThe Χ2 statistic from the likelihood ratio test when comparing to the demographics only model is 85.21 with 11 degrees of freedom and a corresponding p value of <0.0001.
Interestingly, in addition to the increase in predictive ability based on the PRSs, we found that genetic ancestry independently improved the fit of the models. Though all participants were of European ancestry, there were statistically significant genetic differences within European ancestry populations defined by PCs. These PCs were also associated with ever smoking, age of smoking initiation, and smoking cessation. The addition of PCs to the model of heavier smoking did not statistically significantly improve the fit of the model (Supplementary Table 5).
PRSs Modestly Improve Model Prediction Measured by AUC
The baseline model of age and sex has substantial predictive power (Supplementary Figures 5–8). The addition of PCs and PRS further significantly improves model fit for all smoking behaviors (ever smoking—baseline model = 0.617 and full genetics model = 0.665; early vs. late smoking initiation—baseline model = 0.609 and full genetics model = 0.629; heaviness of smoking—baseline model = 0.618 and full genetics model = 0.630; smoking cessation—baseline model = 0.610 and full genetics model = 0.631). Similar results were seen in COGEND (Supplementary Figures 9–12).
Improvement in Prediction in Seen in the Lowest and Highest Deciles of Risk
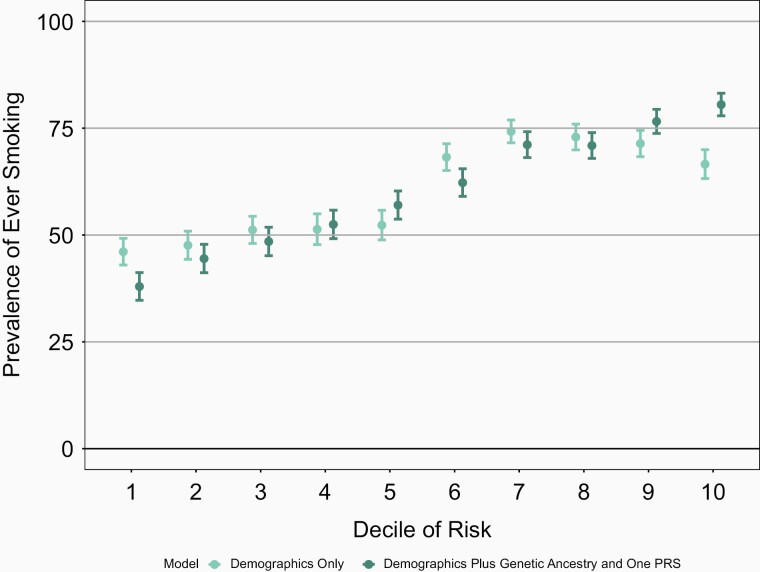
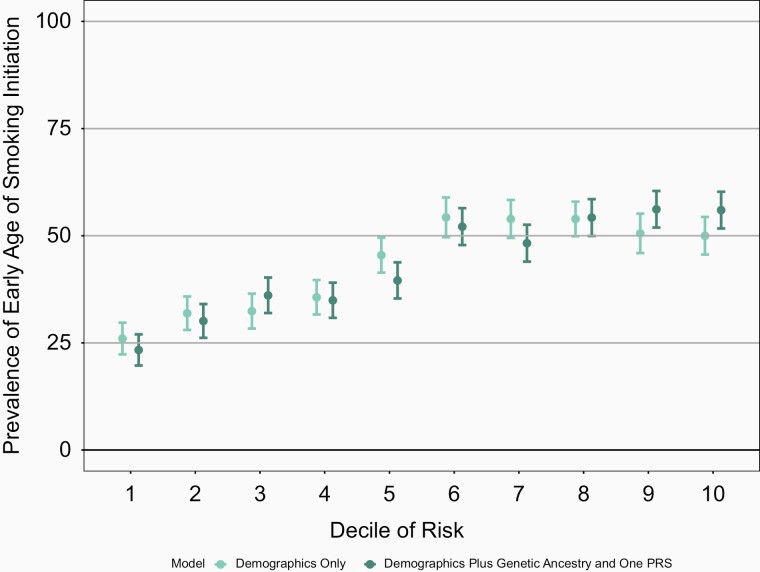
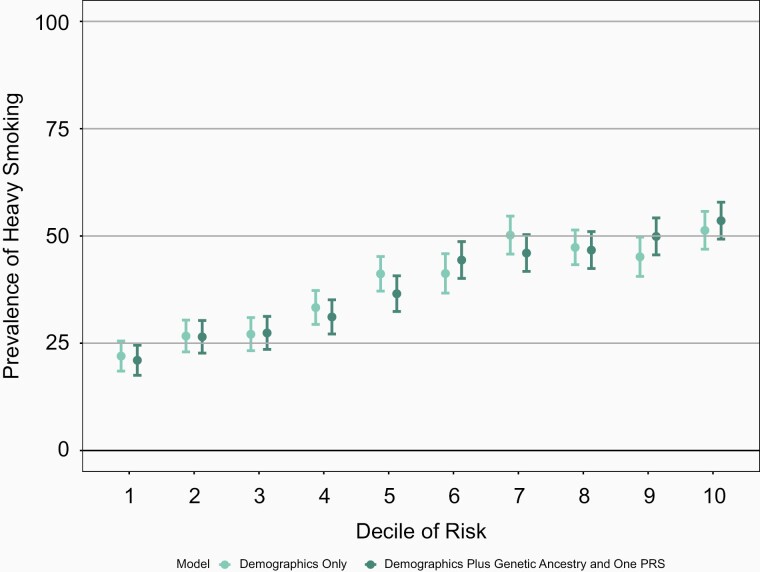
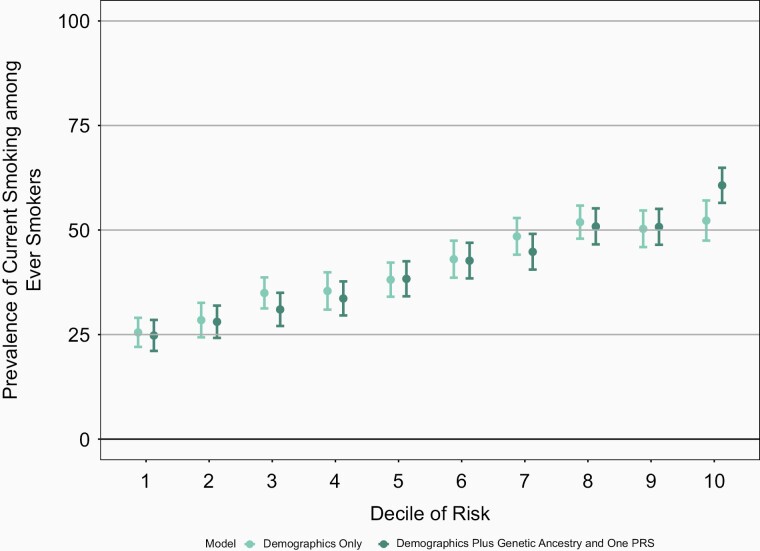
Though the increase in the AUC is modest, the full genetic model which includes genetic ancestry and PRS better differentiates individuals at highest decile and lowest decile risk for a behavior when compared with the baseline model of age and sex only (Figures 1–4 and Supplementary Figures 13–16). For example, the mean prevalence of ever smoking in the highest risk decile as defined by age, sex, genetic ancestry, and PRS is 80.5% compared with 37.9%, the mean prevalence of ever smoking in the lowest decile of risk. In contrast, the highest decile of risk as defined by age and sex alone is 66.6% and the mean prevalence of ever smoking in the lowest decile of risk defined by age and sex is 46.1% (Figure 1). Similarly, the differentiation between models with sex and age only compared with the models which also included genetic ancestry and PRS were greatest in the highest and lowest deciles for the other smoking behaviors. Again, similar results were seen in COGEND (Supplementary Figures 17–20).
Figure 1.
Mean prevalence of ever smoking stratified by deciles of risk within European Americans in the ARIC dataset. The demographics only model included age and sex only. The demographics plus genetic ancestry and one PRS includes age, sex, 10 PCs, and the ever smoking PRS. The decile of risk was estimated for each individual for each model. The prevalence of ever smoking was calculated within each decile independently. The 95% confidence interval was estimated from the prevalence and sample size of each decile. ARIC = Atherosclerosis Risk in Communities, PCs = principal components, PRS = polygenic risk score.
Figure 2.
Mean prevalence of early age of smoking initiation (<18 years) stratified by deciles of risk within European Americans in the ARIC dataset. The demographics only model included age and sex only. The demographics plus genetic ancestry and one PRS includes age, sex, 10 PCs, and the age of smoking initiation PRS. The decile of risk was estimated for each individual for each model. The prevalence of ever smoking was calculated within each decile independently. The 95% confidence interval was estimated from the prevalence and sample size of each decile. ARIC = Atherosclerosis Risk in Communities, PCs = principal components, PRS = polygenic risk score.
Figure 3.
Mean prevalence for heavier smoking (≥21 cigarettes smoked per day) stratified by deciles of risk within European Americans in the ARIC dataset. The demographics only model included age and sex only. The demographics plus genetic ancestry and one PRS includes age, sex, 10 PCs, and the cigarettes smoked per day PRS. The decile of risk was estimated for each individual for each model. The prevalence of ever smoking was calculated within each decile independently. The 95% confidence interval was estimated from the prevalence and sample size of each decile. ARIC = Atherosclerosis Risk in Communities, PCs = principal components, PRS = polygenic risk score.
Figure 4.
Mean prevalence for smoking cessation among ever smokers stratified by deciles of risk within European Americans in the ARIC dataset. The demographics only model included age and sex only. The demographics plus genetic ancestry and one PRS includes age, sex, 10 PCs, and the smoking cessation PRS. The decile of risk was estimated for each individual for each model. The prevalence of ever smoking was calculated within each decile independently. The 95% confidence interval was estimated from the prevalence and sample size of each decile. ARIC = Atherosclerosis Risk in Communities, PCs = principal components, PRS = polygenic risk score.
Smoking-Related PRSs From European Populations Did Not Predict Smoking Behaviors Among African American Individuals
As expected, the PRSs generated from European ancestry populations have little predictive value in an African American population. No smoking-related PRSs were significantly associated with any smoking behavior among the African American sample (Supplementary Table 7). While the pseudo R2 improved for each outcome (ever smoking: 0.090–0.101; early age of smoking initiation: 0.038–0.046; heavier smoking: 0.045–0.064; smoking cessation: 0.016–0.034), none reached statistical significance in part because of the reduced predictive ability and also because of reduced power with a smaller sample.
Discussion
Genetic predictors using smoking-related PRSs and markers of genetic ancestry statistically improve the prediction of multiple smoking behaviors compared with predictive models that include age and sex only. This result was observed for every outcome studied: ever smoking, early age of smoking initiation, heaviness of smoking, and smoking cessation. This finding is seen in both a population-based cohort and replicated in a sample selected for smoking behaviors.
This project builds on the ongoing efforts to predict risk for adverse health outcomes. Recent work has shown the potential impact that PRSs can have. For example, Khera et al. examined the impact that genetics has on body mass index (BMI).26 The authors observed that adults in the top PRS decile had an average BMI of 30 kg/m2, while adults in the bottom PRS decile had an average BMI of 25.2 kg/m2.26 The difference between the top and bottom PRS deciles was 13.0 kg in average weight.26 In another study by Maas et al., the authors generated a PRS examining breast cancer risk and presented the combined environmental and genetic risks.10 The authors observed that women in the highest decile for breast cancer, which included genetic risk, had a 23.5% mean risk of breast cancer compared with a mean risk of 4.4% in the lowest decile.10 The authors also found that if women in the highest decile of risk for breast cancer reduced their modifiable risk factors (ie, obtained low BMI and reduced alcohol consumption), these women’s mean risk would drop to the risk level of an average woman in the general population.10 The authors observed that epidemiologic risk factors had an AUC value of 0.588 when predicting breast cancer. When incorporating the breast cancer PRS with the epidemiologic risk factors into one model, the AUC value increased to 0.648,10 a 0.06 difference which is in the range of what we have seen with the improvement of AUC with the different smoking behaviors.
This work also highlights the potential power of using the upper and lower deciles in stratifying risk groups for smoking-related behaviors. For instance, using the PRS predicting ever smoking, genetic ancestry, and demographic variables of age and sex, only 37.9% of the lowest decile of predicted risk smoked compared with 80.5% of those in the highest decile of predicted risk. Across the other smoking-related phenotypes there is also large differentiation between the lowest and highest deciles of risk. With the use of demographic factors along with genetic risk factors, groups at the highest risk can be identified and potentially targeted for more intensive interventions to most efficiently utilize public health resources. For example, increased prevention efforts may be focused on those at highest risk of initiating smoking. Earlier and more aggressive smoking cessation interventions may be targeted for those at highest risk of heavier smoking and failed smoking cessation.
However, we must interpret this differentiation using PRSs with caution. Even though models utilizing PRSs to predict smoking-related behaviors were statistically more predictive than models without adjusting for PRS, the utility of adding PRSs for these outcomes was modest when defined by the measures such as AUC. For example, the addition of genetics increased the predictive ability from 0.617 to 0.665 with the ever smoking phenotype. Graphically, we can see that there is little differentiation of those with intermediate risk whether using demographic predictors alone or with the addition of genetic risk. The greatest change in risk prediction is at the lowest and highest deciles of risk.
Lastly, these smoking-related PRSs were tailored from individuals of European ancestry.9 When we applied these smoking-related PRS to individuals of African American ancestry, the PRS did not predict any smoking behavior. This finding is consistent with other work that notes that European-derived PRSs lose their usefulness when applied to non-European populations.27 These results once again highlight the urgent need to develop new methods to improve PRSs so that they can be applied to other populations. To address this disparity, we need to engage populations of non-European ancestry so that the upcoming benefits of genomic studies can be applied to all regardless of each person’s ancestry. In the future, we hope that PRSs can be predictive and personally tailored for each individual, regardless of their ancestral background.
With the growing efforts in genomics, we anticipate that the power of genetics through the use of PRSs and genetic ancestry will continue to increase and individuals will be genetically screened in the future to determine their propensity to multiple illnesses. Those individuals at highest risk could then be counseled on their predispositions to diseases and be advised about behavioral and environmental changes that can reduce this risk. As with most interventions, those at greatest risk will likely benefit the most. In the context of predicting smoking behaviors, we can improve the predictive ability of the model in terms of statistical significance with PRS, but the utility of this improvement remains modest at this time. However, with the increasing amount of GWAS summary statistics available from larger cohort sizes and focusing on genetic variation that results in the biologic changes that alters risk of disease, these models will improve, and we should prepare for the near future when genetic predictors along with demographic predictors will impart clinically meaningful prediction of risk for smoking behaviors and we can then better tailor prevention and treatment interventions for individuals.
Supplementary Material
A Contributorship Form detailing each author’s specific involvement with this content, as well as any supplementary data, are available online at https://academic.oup.com/ntr.
Funding
The study was funded by the National Institutes of Health (NIH) training grant 5T32MH014677 to Michael J. Bray. This study was also funded by the National Institute on Drug Abuse grant R01DA036583 to Laura J. Bierut, R01DA042090 to Dana B. Hancock, and R01DA042195 to Richard A. Grucza. In addition, Laura J. Bierut was also supported by National Institute on Aging grant R56AG058726 and National Cancer Institute grants U19CA203654 and P30CA091842. Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I. The authors thank the staff and participants of the ARIC study for their important contributions. Funding for GENEVA was provided by National Human Genome Research Institute grant U01HG004402 (E. Boerwinkle).
Declaration of Interests
Laura J. Bierut and the spouse of Nancy L. Saccone are listed as inventors on Issued U.S. Patent 8,080,371,“Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. There are no other conflicts of interest to disclose.
Acknowledgments
We gratefully acknowledge Scott Vrieze and his group for providing the GSCAN data. International Agency for Research on Cancer Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
References
- 1. Wang TW, Asman K, Gentzke AS, et al. Tobacco product use among adults—United States, 2017. MMWR Morb Mortal Wkly Rep. 2018;67(44):1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. WHO Report on the Global Tobacco Epidemic, 2017: Monitoring Tobacco Use and Prevention Policies: Executive Summary. Geneva: World Health Organization; 2017. [Google Scholar]
- 3. Surgeon General. The health consequences of smoking—50 years of progress: a report of the surgeon general. Paper presented at US Department of Health and Human Services; 2014.
- 4. Cornelius ME, Wang TW, Jamal A, Loretan CG, Neff LJ. Tobacco product use among adults—United States, 2019. MMWR Morb Mortal Wkly Rep. 2020;69(46):1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Babb S, Malarcher A, Schauer G, Asman K, Jamal A. Quitting smoking among adults—United States, 2000–2015. MMWR Morb Mortal Wkly Rep. 2017;65(52):1457–1464. [DOI] [PubMed] [Google Scholar]
- 6. Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res. 1999;1(suppl 2):S51–S57. doi: 10.1080/14622299050011811. [DOI] [PubMed] [Google Scholar]
- 7. Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35(4):397–406. [DOI] [PubMed] [Google Scholar]
- 8. Hancock DB, Markunas CA, Bierut LJ, Johnson EO. Human genetics of addiction: new insights and future directions. Curr Psychiatry Rep. 2018;20(2):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu M, Jiang Y, Wedow R, et al. ; 23andMe Research Team; HUNT All-In Psychiatry . Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maas P, Barrdahl M, Joshi AD, et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol. 2016;2(10):1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belsky DW, Moffitt TE, Houts R, et al. Polygenic risk, rapid childhood growth, and the development of obesity: evidence from a 4-decade longitudinal study. Arch Pediatr Adolesc Med. 2012;166(6):515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aly M, Wiklund F, Xu J, et al. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belsky DW, Moffitt TE, Baker TB, et al. Polygenic risk and the developmental progression to heavy, persistent smoking and nicotine dependence: evidence from a 4-decade longitudinal study. JAMA Psychiatry. 2013;70(5):534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Howard G, Wagenknecht LE, Burke GL, et al. Cigarette smoking and progression of atherosclerosis: the Atherosclerosis Risk in Communities (ARIC) Study. JAMA. 1998;279(2):119–124. [DOI] [PubMed] [Google Scholar]
- 16. Chen LS, Baker TB, Grucza R, et al. Dissection of the phenotypic and genotypic associations with nicotinic dependence. Nicotine Tob Res. 2012;14(4):425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–1127. [DOI] [PubMed] [Google Scholar]
- 18. Saccone NL, Schwantes-An TH, Wang JC, et al. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav. 2010;9(7):741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 22. Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jansen AG, Dieleman GC, Jansen PR, Verhulst FC, Posthuma D, Polderman TJC. Psychiatric polygenic risk scores as predictor for attention deficit/hyperactivity disorder and autism spectrum disorder in a clinical child and adolescent sample. Behav Genet. 2020;50(4):203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamri A, Mao S, Desai D, Gupta M, Paré G, Anand SS. Fine-tuning of genome-wide polygenic risk scores and prediction of gestational diabetes in South Asian women. Sci Rep. 2020;10(1):8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khera AV, Chaffin M, Wade KH, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(3):587–596.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.