ABSTRACT
Autophagy is a widely shared pathway among different eukaryotes, which helps to maintain cellular homeostasis via recycling unwanted cytoplasmic components. Autophagy plays an important role in plant growth, also assists plants in confronting various environmental stresses. Drought stress can activate autophagy pathway in plants to favor their environmental adaptations, however, a direct link to wire drought and autophagy is still missing. We have recently identified a plant-unique COST1 (Constitutively Stressed 1) protein that can negatively regulate plant drought tolerance through direct interaction with an autophagy receptor protein ATG8e (autophagy-related 8e). COST1 thus represents an innovation of plant-specific autophagy regulation, extending our understating of this conserved but complex pathway, as well as underlying its potential in agricultural usage.
KEYWORDS: Drought, autophagy, COST1, stress, signaling
Drought stress can severely affect plant growth and farming, greatly threaten crop yield and food safety, which is compounded by increasing global temperature.1,2 During drought, plants can integrate transcriptional and post-transcriptional signals, as well as coordinate cellular and physiological changes,3–5 for gaining advantages in environmental adaptions. Various biotic and abiotic stresses can trigger activation of autophagy pathway, which is an essential and conserved pathway that can subject unwanted substrates for recycling to achieve cellular homeostasis.6,7 Our recent study of a plant-specific DUF641/COST family protein COST1 indicates that plants evolved a unique route in autophagy regulation for stress responding and consequently better survivals.8
Degradation of COST1 for conferring drought tolerance
cost1 was characterized as a strong drought-tolerant mutant,8 and as known, drought stress can cause gene transcriptional changes.9 However, our qPCR (quantitative polymerase chain reaction) did not detect any increase or decrease of COST1 gene expression during various stresses treatments, including drought, ABA, mannitol, and salt. In consistent, transgenic plants harboring COST1 promoter in fusion with GUS(β-glucuronidase) gene did not show significant difference after biochemical staining of dehydration-treated and -untreated Arabidopsis seedlings. While in stress treated COST1-YFP transgenic seedlings, confocal microscope captured constant moving dots in the leaf epidermal cells, a feature that is shared by autophagy. Immunoblotting study of COST1 at protein level suggests that drought can promote the degradation of COST1, a common phenomenon that can be observed for many post-transcriptionally regulated components in abiotic stress response.5Two well-known pathways are required for getting rid of damaged or unwanted proteins in plants: ubiquitination-mediated 26S-proteasome pathway and autophagy-mediated vacuolar degradation pathway.10,11 Indeed, both MG132 (inhibitor of 26S-proteasome pathway) and Concanamycin A (autophagy-mediated vacuolar degradation pathway inhibitor) can inhibit the degradation of COST1 during dehydration treatment. This indicates that both pathways are involved in the efficient removal of COST1, to release its inhibition of autophagy and thus conferring drought tolerance. It has been well studied that NBR1 can bind to ubiquitinated proteins for assisting in its target degradation,12,13 however, our genetic study by employing drought related water loss assay clearly suggests that NBR1 is not required for COST1-meidated drought regulation. Thus, the role of NBR1 in COST1-regulated pathway or vice versa is still needs to be determined.
COST1 designates an ABA- and H2O2- independent path for drought stress response
ABA can be accumulated to high levels during drought stress, which plays a critical role in orchestrating gene expression as well as in regulating protein modifications. With the discovery of ABA receptors in 2009,14,15 signaling transduction pathway mediated by ABA is finally unveiled.16 In addition to the ABA-mediated drought stress regulation, there also exists an ABA-independent pathway working in parallel for plant responding to drought.9 cost1 mutant is strongly drought tolerant, and genetic studies by crossing cost1 mutant with both ABA signaling and ABA biosynthesis deficient mutants (relevant mutants used were abi1-1C, ost1, and aba3), suggests that COST1 works independently of ABA signaling pathway. Constantly, cost1 does not show significantly difference in ABA-mediated seed germination assay when compared with WT (wild-type). But, it’s worth noting that drought stress can also induce the transcription of some ATG-responsive genes like ATG18,17 underlying there is a cross talk between autophagy-dependent and autophagy-independent pathways. In addition, H2O2 is another signaling molecule that can act both independently and coordinately with ABA in plant drought stress response, and our genetic study by employing a ghr1 mutant also clearly indicated that COST1 functions independently of H2O2.18
Negative feedback regulation of autophagy by COST1
Plant autophagy pathway components are very much overlapped with the findings in yeast and human, and the vast majority of those factors tend to be essential for the formation of autophagosome, a double membrane-bound structure that can engulf substrates for delivering to lysosome or vacuole for degradation. To data, TOR (target of rapamycin), a highly conserved central energy sensor, is the only negative regulator that can directly modulate autophagy in plants.19 In addition to its quick turnover through the autophagy pathway, COST1 can directly interact with an autophagy adaptor protein ATG8e and inhibit autophagy, which featured itself as a negative regulator in autophagy like TOR but with plant-specific innovation. There are nine ATG8 proteins in Arabidopsis;20 interactions between COST1 and other ATG8 isoforms remain unknown, raising the possibility of specific interaction of certain ATG8 isoforms with COST1 or not. Unclear also is the interaction between COST1 and other ATG pathway proteins and adaptors. More, COST1 interacts with ATG8e through which domain, AIM (ATG8-interacting motif) or UIM (ubiquitin-interacting motif)21,22or a novel unidentified motif, is still an open question. By immunoblotting assay, constitutively activated autophagy was observed in cost1 mutant background.23 While in COST1 overexpression plants, autophagy is inhibited and the relevant adaptor ATG8e protein seems to be significantly reduced especially during drought.8
Perspectives
The past decades have achieved great progress in understanding drought stress response as well as in knowing plant autophagy formation and the pathway regulation.5–7 The finding of COST1 eventually links drought stress and autophagy directly with each other. In addition to TOR, COST1 is to date the first direct negative regulator of autophagy pathway discovered in plants. Clear answer of COST1 in drought stress sensing is still lacking, and the relevant E3 ligase that mediates the degradation of COST1 remains to be identified; nor do we know if there is a COST1-independent pathway in stress-mediated autophagy activation (Figure 1). Moreover, besides the important function of COST1 in drought stress response, plant growth is severely retarded in cost1 mutant, which renders a critical role of COST1 in balancing stress tolerance and plant growth.24 Phylogenetic study suggests that COST1-like proteins are highly conserved and broadly distributed in all higher plants,8 studies of COST1 thus have both values in expanding our understanding of plant autophagy regulation, and in engineering more stress tolerant crops with a balance of yield.
Figure 1.
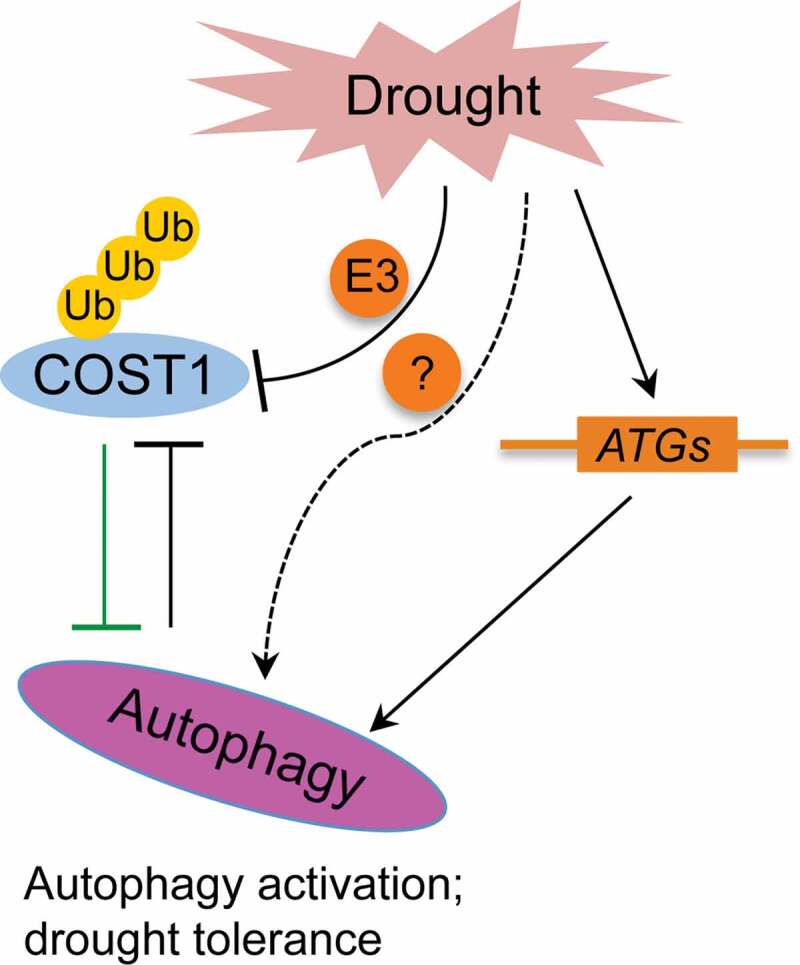
A proposed working model of COST1 in linking between drought stress and autophagy in plants. Drought stress can promote degradation of COST1 through 26S-proteasome pathway and autophagy pathway, while the relevant E3 ligase that mediates the ploy ubiquitination (Ub denotes ubiquitin) of COST1 is yet to be identified. Dysfunction of COST1 would release its inhibition of autophagy and activate this pathway, confers plant drought tolerance. Drought can also induce the transcription of some stress-responsive ATG genes expression and thus add in autophagy induction. There may also exist a COST1-independent pathway that can sense drought and activate autophagy directly or indirectly, which is denoted as a dashed line with a question mark on it. Arrow-headed and bar-headed lines denote activation and inhibition. The green line represents a function of COST1 in promoting plant growth under normal condition.
Acknowledgments
I would like to thank Dr. Frantisek Baluska for this invitation, and Dr. Diane Bassham for her suggestionsin writing this mini-review paper.
Funding Statement
This work has been supported by grant no. IOS 1353867 to DCB from the US National Science Foundation [IOS 1353867].
Disclosure statement
No potential conflict of interest was reported by the author.
References
- 1.Gornall J, Betts R, Burke E, Clark R, Camp J, Willett K, Wiltshire A.. Implications of climate change for agricultural productivity in the early twenty-first century. Philos Trans R Soc Lond B Biol Sci. 2010;365(1554):1–3. doi: 10.1098/rstb.2010.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamaoui M, Jemo M, Datla R, Bekkaoui F.. Heat and drought stresses in crops and approaches for their mitigation. Front Chem. 2018;6:26. doi: 10.3389/fchem.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tardieu F, Simonneau T, Muller B. The physiological basis of drought tolerance in crop plants: a scenario-dependent probabilistic approach. Annu Rev Plant Biol. 2018;69:733–759. doi: 10.1146/annurev-arplant-042817-040218. [DOI] [PubMed] [Google Scholar]
- 5.Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Signorelli S, Tarkowski Ł, Van den Ende W, Bassham DC. Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 2019;24(5):413–430. doi: 10.1016/j.tplants.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z, Wang X, Bassham DC, Walley J, Yin Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 2017;41(1):33–46.e7. doi: 10.1016/j.devcel.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao Y, Song WM, Wang P, Yu X, Li B, Jiang C, Shiu SH, Zhang H, Bassham DC. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci U S A. 2020;117(13):7482–7493. doi: 10.1073/pnas.1918539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Su T, Li X, Yang M, Shao Q, Zhao Y, Ma C, Wang P. Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Front Plant Sci. 2020;11:164. doi: 10.3389/fpls.2020.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 12.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung H, Lee HN, Marshall RS, Lomax AW, Yoon MJ, Kim J, Kim JH, Vierstra RD, Chung T. NBR1 mediates selective autophagy of defective proteins in arabidopsis. J Exp Bot. 2019;71(1):73-89. doi: 10.1093/jxb/erz404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S, Fung P, Nishimura N, Jensen D, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow T, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 16.Cutler S, Rodriguez P, Finkelstein R, Abrams S, Merchant S, Briggs W, Ort D. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5(7):954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 18.Hua D, Wang C, He J, Liao H, Duan Y, Zhu Z, Guo Y, Chen Z, Gong Z. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in arabidopsis. Plant Cell. 2012;24(6):2546–2561. doi: 10.1105/tpc.112.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Bassham DC, Schumacher K. TOR is a negative regulator of autophagy in arabidopsis thaliana. PLoS One. 2010;5(7):e11883. doi: 10.1371/journal.pone.0011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16(11):2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall RS, Hua Z, Mali S, McLoughlin F, Vierstra RD. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177(3):766–781.e24. doi: 10.1016/j.cell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR Motifs and Cargo receptors. J Mol Biol. 2019;432(1):80-103. doi: 10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Bao Y, Mugume Y, Bassham DC. Biochemical methods to monitor autophagic responses in plants. Methods Enzymol. 2017;588:497–513. [DOI] [PubMed] [Google Scholar]
- 24.Bao Y, Bassham DC. COST1 balances plant growth and stress tolerance via attenuation of autophagy. Autophagy. 2020;16(6):1157–1158. doi: 10.1080/15548627.2020.1752981. [DOI] [PMC free article] [PubMed] [Google Scholar]


