ABSTRACT
As stress-inducible proteins, dehydrins exert functional protective role by alleviating the cell damage when plants are suffering from the stresses. However, the upstream regulatory mechanism of these proteins is not very clear. To unravel the regulatory mechanism of dehydrin, a screen of wheat cDNA library from cold and PEG-treated wheat seedlings was performed and a transcription factor TaERF4a (GenBank NO. AFP49822.1) interacting with wheat dehydrin wzy1-2 gene (Gene ID: 100037544) promoter was identified by yeast one-hybrid assay. The regulator TaERF4a and the wzy1-2 gene can respond to the abiotic stress, the induced transcripts of these two genes exhibit a similar expression trend under adverse environmental conditions. In planta, the dual luciferase transient assay analysis showed that TaERF4a can positively regulate the expression of WZY1-2 dehydrin. Therefore, the obtained results suggest that ERF transcription factor can regulate the expression level of the dehydrin gene in wheat.
KEYWORDS: Dehydrin, transcription factor, abiotic stress, regulatory mechanism
Wheat, as one of the three major gramineous crops, is widely cultivated all over the world. In China, the main producing areas are the north China plain and the northeast plain.1 In these areas, seasonal drought, low temperature, and other abiotic stress have seriously affected the growth and production of wheat. Dehydrins (DHNs), also known as LEA II (late embryogenesis abundant) proteins, are widely distributed in prokaryotic and eukaryotic organisms, including angiosperm, algae plants, and bacteria, fungi, evennematode.2,3 Due to their unique amino acid compositions and structure character, dehydrins exhibit versatile abilities, including heat resistance, reduction of reactive oxygen species and enzyme cryoprotection, etc.4,5 Therefore, they play an important role in protecting cell membrane and protein structure from dehydration and desiccation damage.6 However, despite considerable progress made toward understanding the role of DHNs, the molecular mechanisms by which these proteins perform their functions inside the cells remain elusive.7
In a previous study, we have cloned the dehydrin gene wzy1-2 (Gene ID: 100037544) and wzy1-2 promoter in Zheng yin 1 cultivar of Triticum aestivum and showed its fundamental biological function in plant under adversity stresses.8 Expression pattern analysis indicated that wzy1-2 transcript and WZY1-2 protein accumulation can be induced by osmotic stress, cold, ABA, and MeJA treatments.8 Histochemical analysis of GUS expression demonstrated that wzy1-2 promoter activity could be upregulated by osmotic stress, ABA treatment.8
To elucidate the upstream regulation mechanism of wzy1-2 dehydrin gene, cDNA library from cold and PEG-treated wheat seedlings was screened by using yeast one-hybrid (Y1H) assay with the wzy1-2 promoter as bait. By sequencing the positive clones we isolated a transcription factor encoding 196 amino acids with a predicted molecular weight of 20.77 kDa, named TaERF4a (GenBank NO. AFP49822.1). To further verify the interaction between the TaERF4a and wzy1-2 promoter, the plasmid containing the transcription factor TaERF4a gene was extracted and the cotransformation on selective media were performed. The bait yeast strains (Pwzy1-2 in pAbAi) containing the individual prey vector of the pGADT7-TaERF4a were able to grow on SD/-Leu (synthetic dropout nutrient medium) and SD/-Leu/AbA200 (Aureobasidin, 200 ng/ml) selective medium. However, Y1H Gold yeast cells containing pGADT7-TaERF4a could not grow on SD/-Leu/AbA200 selective medium (Figure 1a). Moreover, a typical AP2/ERF conserved domain was found in the gene sequence, indicating that TaERF4a was a member of the ERF protein family (Figure 1b). Ethylene-response factor (ERF) proteins are members of the second-largest transcription factor family involved in plant growth, development, and environmental stress responses.9 ERF96, a positive regulator of ABA responses, can not only increase ABA response genes RD29A, ABI5, ABF3 expression, but it is also involved in the Se resistance and detoxification.10
Figure 1.

The interaction of TaERF4a with the promoter of wzy1-2 gene in yeast one-hybrid assays (a) and the sequence feature of TaERF4a protein (b). (a) The transformation of pGADT7-TaERF4a with Y1H Gold yeast cells, bait yeast strains (Y1H Gold containing the PAbAi-Pwzy1-2) on selective media respectively to verify interactions. (b) The protein sequence of the TaERF4a.
Therefore, TaERF4a as the potential regulator of wzy1-2 gene was selected for further research. Next, we aimed to determine the subcellular localization of TaERF4a using GFP translational fusions. The p35 S::GFP and p35 S::TaERF4a-GFP fusion constructs were used for transient expression analysis. The GFP fluorescent of the control was shown in the nucleus and the cytoplasm of the Nicotiana benthamiana protoplasts, but the TaERF4a-GFP signal was only detected in the nucleus (Figure 2a). In addition, tissue-specific analysis indicated that the expression of the TaERF4a was highest in leaves during wheat seedling stage (Figure 2b). Moreover, we examined the expression patterns of the TaERF4a and WZY1-2 at different time points in response to abiotic stress. Under PEG treatment, the expression level of TaERF4a was gradually increased, reached a high level at 6 h and then declined. Interestingly, the expression of wzy1-2 gene showed the similar tendency with this transcription factor (Figure 2c). To characterize the ability of TaERF4a to activate wzy1-2 gene in plant, A. tumefaciens containing the p35 S::ERF4a (effector vector) together with the wzy1-2 promoter::Rluc (reporter vector) were co-infiltrated into the tobacco leaves. The luciferase activity (Rluc/Fluc ratio) of the infected plants with TaERF4a protein was significantly increased (by 5.21 fold) (Figure 2d). In addition, the luciferase activity was also significantly increased after 24 h 20% PEG treatment compared with that of untreated (Figure 2e).
Figure 2.
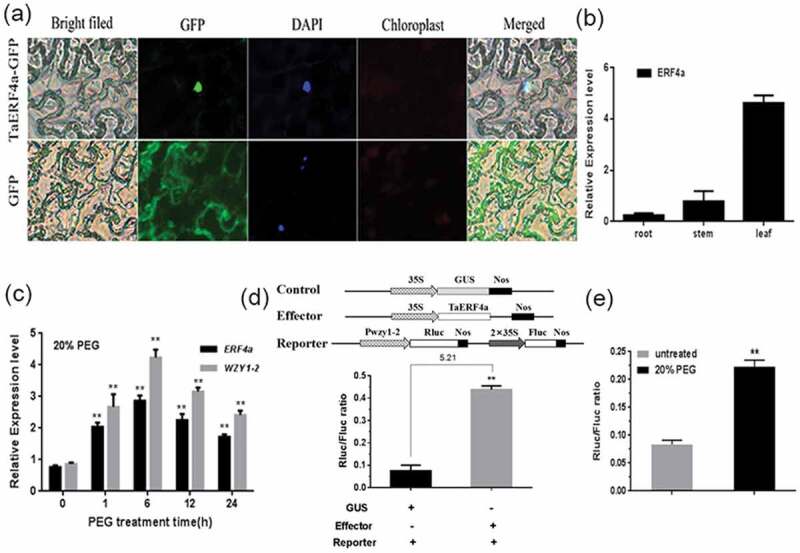
Identification of upstream transcription factor TaERF4a of dehydrin wzy1-2 gene. (a). Subcellular localization of TaERF4a protein in N. benthamiana leaves performed by fluorescent microscope (Leica, DM5000 B, magnificant 20×). Tobacco leaf epidermal cells agro-infiltrated with p35 S::GFP and p35 S::TaERF4a-GFP vectors were used for transient expression of fusion proteins. The fluorescence images were visualized after 48 h infiltration. (b) Transcript level of TaERF4a in different tissues collected at 14 DAP seedlings. (c) The expression of TaERF4a, wzy1-2 genes in response to PEG treatment at 14 DAP seedlings. Total RNA was extracted from leaves at the indicated times. Wheat actin gene (Accession No. AB181991) was used as an internal reference. (d) Transient luciferase activity assay of TaERF4a in N. benthamiana leaves. (e) The relative luciferase enzyme activity of TaERF4a after the PEG treatment. The experiments were performed with at least three biological repeats. Significance analysis was determined by Student’s t-test (* = P < .05, ** = P < .01).
In conclusion, we identified an ERF transcription factor TaERF4a which presumably binds to the promoter of the dehydrin wzy1-2 gene and regulate its expression. Some reports have shown that ERF can recognize and bind specifically to the GCC box, dehydration responsive elements (DREs)/C-repeat elements (CRTs) in the promoter of stress-responsive genes.11,12 The DRE as the cis-acting element also exist in the promoter of the dehydrin wzy1-2 gene,8 so we speculate that TaERF4a may bind to this motif to regulate the wzy1-2 gene expression. Therefore, we further explore the binding site of the TaERF4a in the wzy1-2 gene promoter and develop the transgenic plant to further confirm regulatory relationship between TaERF4a and wzy1-2 gene and identify whether this regulator can improve the plant stress resistance.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No. 31671608) and Weinan basic research program in shaanxi province (2019ZDYF-JCYJ-133).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Ren YJ, Gao C, Han HF, Li QQ.. Response of water use efficiency and carbon emission to no-tillage and winter wheat genotypes in the North China Plain. Sci Total Environ. 2018;635:1–3. doi: 10.1016/j.scitotenv.2018.04.204. [DOI] [PubMed] [Google Scholar]
- 2.Gasulla F, de Nova PG, Esteban-Carrasco A, Zapata JM, Barreno E, Guera A.. Dehydration rate and time of desiccation affect recovery of the lichen alga Trebouxia erici: alternative and classical protective mechanisms. Planta. 2009;231:195–208. doi: 10.1007/s00425-009-1019-y. [DOI] [PubMed] [Google Scholar]
- 3.Rorat T. Plant dehydrins–tissue location, structure and function. Cell Mol Biol Lett. 2006;11:536–556. doi: 10.2478/s11658-006-0044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara M, Fujinaga M, Kuboi T. Metal binding by citrus dehydrin with histidine-rich domains. J Exp Bot. 2005;56:2695–2703. doi: 10.1093/jxb/eri262. [DOI] [PubMed] [Google Scholar]
- 5.Hara M, Kondo M, Kato T. A KS-type dehydrin and its related domains reduce Cu-promoted radical generation and the histidine residues contribute to the radical-reducing activities. J Exp Bot. 2013;64:1615–1624. doi: 10.1093/jxb/ert016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halder T, Agarwal T, Ray S. Isolation, cloning, and characterization of a novel Sorghum dehydrin (SbDhn2) protein. Protoplasma. 2016;253:1475–1488. doi: 10.1007/s00709-015-0901-7. [DOI] [PubMed] [Google Scholar]
- 7.Hanin M, Brini F, Ebel C, Toda Y, Takeda S, Masmoudi K. Plant dehydrins and stress tolerance: versatile proteins for complex mechanisms. Plant Signal Behav. 2011;6:1503–1509. doi: 10.4161/psb.6.10.17088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu WN, Zhang DP, Lu XX, Zhang LS, Yu ZY, Lv H, Zhang H. Characterisation of an SKn-type dehydrin promoter from wheat and its responsiveness to various abiotic and biotic stresses. Plant Mol Biol Rep. 2014;32:664–678. doi: 10.1007/s11105-013-0681-1. [DOI] [Google Scholar]
- 9.Dong W, Ai X, Xu F, Quan T, Liu S, Xia G. Isolation and characterization of a bread wheat salinity responsive ERF transcription factor. Gene. 2012;511:38–45. doi: 10.1016/j.gene.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 10.Jiang L, Yang J, Liu C, Chen Z, Yao Z, Cao S. Overexpression of ethylene response factor ERF96 gene enhances selenium tolerance in Arabidopsis. Plant Physiol Biochem. 2020;149:294–300. doi: 10.1016/j.plaphy.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 11.Catinot J, Huang JB, Huang PY, Tseng MY, Chen YL, Gu SY, Lo W-S, Wang L-C, Chen Y-R, Zimmerli L, et al. ETHYLENE RESPONSE FACTOR 96 positively regulates A rabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate - and ethylene-responsive defence genes. Plant Cell Environ. 2015;38:2721–2734. doi: 10.1111/pce.12583. [DOI] [PubMed] [Google Scholar]
- 12.Cheng MC, Liao PM, Kuo WW, Lin TP. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


