ABSTRACT
The control of gynecium development in Arabidopsis thaliana by the auxin response factor ETTIN (ETT) correlates with a reduction in the methylesterification of cell-wall pectins and a decrease in cell-wall stiffness in the valve tissues of the ovary. Here, we determine the list of genes rapidly regulated following the in-vivo activation of an ETT fusion protein, and show these to be significantly enriched in genes encoding cell-wall proteins, including several pectin methylesterases (PMEs) and pectin methylesterase inhibitors (PMEIs). We also perform a genome-wide scan for potential ETT-binding sites, and incorporate the results of this procedure into a comparison of datasets, derived using four distinct methods, to identify genes regulated directly or indirectly by ETT. We conclude from our combined analyses that PMEIs are likely to be key actors that mediate the regulation of gynecium development by ETT, while ETT may simultaneously regulate PMEs to prevent exaggerated developmental effects from the regulation of PMEIs. We also postulate the existence of one or more rapidly-acting intermediate factors in the transcriptional regulation of PMEs and PMEIs by ETT.
KEYWORDS: Arabidopsis thaliana, Gynoecium, Carpel, ETTIN, AUXIN RESPONSE FACTOR, Pectin methylesterase, Plant cell wall
Introduction
The Arabidopsis thaliana gynecium, or female floral whorl, consists of a pistil of two fused carpels. This structure is divided longitudinally into stigma, style and ovary tissues and includes a short stalk termed the gynophore. The ovary consists of two valves, attached to a central replum, along which placental tissues form to generate the ovules.
Loss-of-function mutants of the auxin response transcription factor ETTIN (ETT), also known as AUXIN RESPONSE FACTOR3 (ARF3), show defects in tissue patterning along the longitudinal axis of the gynecium, including an increase in stigma and gynophore tissues and a reduction in the length of the ovary, including the valve tissues.1 These changes are accompanied by defects in abaxial/adaxial patterning, in agreement with the known role of ETT and its paralog ARF4 in the definition of abaxial tissue identity both in floral organs outside the gynecium and in leaves.2 ETT and ARF4 expression is restricted to the abaxial domain of these organs by the presence in the adaxial domain of tasi-RNAs generated from TAS3 by the activity of miRNA390.3
ARF proteins are known to bind DNA motifs termed auxin response elements (AuxREs), which are found in the promoter regions of numerous auxin-responsive genes, as reviewed by Guilfoyle and Hagen.4 To quantitatively characterize ETT’s interactions with these motifs, position-specific scoring matrices (PSSMs) have been generated from Protein Binding Microarray (PBM) data,5 based on variants of the two top-scoring sequences among all possible 10-mers. The recovery of more than one top-scoring matrix is common in analyses of transcription factor binding preferences, and can be caused by the presence of interdependency among nucleotide positions in the binding site.6
The cellulose microfibrils of the plant cell wall are embedded in a mixture of polysaccharides termed pectin. The major component of pectin, homogalacuronan, is synthesized in a highly methyl-esterified form that contributes to cell-wall stiffness. A recent study by Andres-Robin et al.7 showed that the regulation of gynecium patterning by ETT correlated with changes in pectin methylesterase (PME) activity in the cell wall. This study supports a model in which ETT contributes to normal gynecium patterning by causing an increase in PME activity that results in a decrease in the level of methylesterification of cell-wall pectins. This decrease was found to correlate with a reduction in cell-wall stiffness and an increase in valve-length in the ovary. Strikingly, the artificial overexpression of PMEs or PME inhibitors (PMEIs) respectively rescued and phenocopied ett loss-of-function mutants, supporting a possible role for PMEs and/or PMEIs in the mechanism of action of ETT. The findings of Andres Robin et al.7 were in accord with those of previous studies which found PMEs to promote cell wall loosening and developmental processes at the stem apex.8
Here, we use a transcriptomics approach to identify genes that are rapidly regulated following the induction of ETT activity in transgenic plants, and also perform in-silico scans for potential ETT-binding sites in a genome-wide dataset of A. thaliana promoter sequences. Our analyses, combined with those of a study by Simonini et al.9 suggest distinct roles for PMEIs and PMEs in mediating the role of ETT in the gynecium, and indicate the possible presence of rapidly-acting intermediate factors in the transcriptional regulation of these genes.
Results
Putative immediate targets of ETT are enriched in genes involved in cell-wall remodeling
To identify immediate targets of ETT, we generated transgenic plants in which the translocation to the nucleus of constitutively produced ETT, translationally fused to the hormone-binding domain of the rat glucocorticoid receptor protein (GR), could be induced by exogenous application of the hormone analogue dexamethasone (DEX).10 After verification of the functionality of the ETT-GR fusion protein (Figure S1), inflorescence tissues of transformed plants were treated either with DEX and cycloheximide (CHX) to induce the transcriptional effects of ETT on its target genes while simultaneously blocking protein synthesis, or with CHX alone to block protein synthesis without ETT-induction. The efficiency of the CHX treatments given has been previously demonstrated.11 Apical regions of inflorescences, containing unopened flowers only, were harvested from both groups of plants two hours after induction. RNA was then extracted and processed for expression analyses on CATMA GST microarrays.12
The results of three independent experiments were compared, resulting in a list of 85 putative ETT targets (p-value < .005; Table S1). Among these, 65 genes (76.5%) were repressed, while the remaining 20 (23.5%) were activated. This predominantly repressive activity of ETT is in agreement both with previous experimental data and the presence of a glutamine-rich central region in ETT, shared with other predominantly repressive ARFs.13 Interestingly, 13 (15.3%) of the genes that were rapidly regulated following ETT-activation encoded proteins predicted to be involved in cell-wall structure and/or remodeling, including four PMEs (AT5G07420, AT3G17060, AT2G26450 and AT5G27870), three PMEIs (AT5G50030, AT2G47050 and AT1G10770), one expansin (AT1G69530, also known as Ath.EXPA1), one member of the OLE E 1 allergen and extensin family (AT1G29140), two arabinogalactan proteins (AT3G01700 and AT5G24105, also known as Ath.AGP11 and 41, respectively), and two pectate lyase-like proteins (AT1G14420 and AT3G01270, also known as Ath.PLL8 and 10, respectively).
To compare the frequencies of cell-wall-related genes between putatively ETT-regulated sequences and the entire A. thaliana genome, we used Functional Classification SuperViewer, based on Gene Ontology from TAIR.14 The result of this analysis (Figure S2) shows a highly statistically significant, five-fold over-representation of cell-wall-related genes, among putative immediate ETT targets.
All 13 putative ETT cell-wall targets identified here are expressed in flower buds approaching anthesis, though predominantly in anthers.15 The expansin AT1G69530 additionally shows high expression in the stamen filaments and petals of mature flowers and in siliques and germinating seeds. As all 13 genes are downregulated on induction of ETT activity, their expression in wild-type tissues in which ETT is expressed, including the gynecium, is not necessarily to be expected. However, gene ontogeny terms in TAIR (https://www.arabidopsis.org/), based on comparisons of transcriptomic data, do indicate these 13 genes to be expressed in carpel tissues.
To confirm the repression by ETT of the 13 cell-wall-related genes identified, we performed qRT-PCR analyses on inflorescences from Col-0 plants and from 35S:ETTm transformants16 in which ETT mRNA is stabilized by silent mutations in both of its tasiR-ARF binding sites. All 13 cell wall-related targets identified were significantly down-regulated in 35S:ETTm (Figure 1), consistent with the results of our microarray analyses (Table S1).
Figure 1.
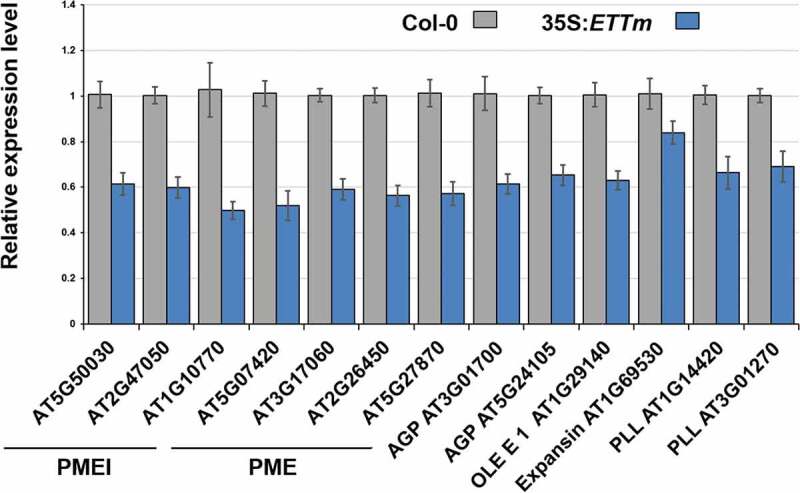
qRT-PCR analyses of 13 cell-wall-related genes. All genes studied are significantly lower expressed (p ≤ 0.05) in plants overexpressing a stabilized ETTm transgene compared to wild-type. Standard errors of the mean are shown.
Comparison of datasets produced using four distinct methods discriminates between possible direct and indirect targets of ETT
We scanned a genome-wide database of presumptive A. thaliana promoter sequences using both of the published PSSMs for ETT.5 The highest affinity sites corresponded to a weight score17 of 8.1 in these scans. A weight-score cutoff value of 7.0 produced a list of 808 promoters containing at least one potential ETT-binding site, though only ~8% of genes identified to bind ETT in ChIP-seq analyses9 were present in this list (data not shown). We therefore reduced the weight-score cutoff to 6.0, which produced a list of 6695 genes (Table S2) that included ~37% of those previously identified by ChIP-seq.
We compared datasets of ETT-regulated genes produced by four distinct methods: in vivo induction of ETT-GR (Table S1), RNA-seq comparison of wild-type and ett-3 inflorescences without exogenous indole acetic acid (IAA),9 ChIP-seq of ETT-GFP transformants without exogenous IAA,9 and in-silico scans of A. thaliana promoters (Table S2), the results of which are shown in Figure 2. Twenty three of the genes identified in the present work by the in-vivo activation of ETT-GR were furthermore identified as potentially direct ETT-targets by ChIP-seq and/or promoter-scanning procedures. These genes included four cell-wall-related proteins: PMEI AT5G50030, PME AT3G17060, expansin AT1G69530 and pectolyase-like protein AT1G14420. A further 26 putative immediate ETT-targets showed differential expression between wild-type and ett-3 inflorescences, though were not predicted as potential direct targets of ETT in ChIP-seq or promoter-scanning procedures. The majority of the cell-wall proteins identified through the in-vivo activation of ETT-GR were present in this group, including PMEs AT2G26450 and AT5G07420, PMEIs AT1G10770 and AT2G47050, OLE E 1 protein AT1G29140, pectolyase-like protein AT3G01270, and arabinogalactan protein AT3G01700 (Figure 2). Only two cell wall proteins identified by the in-vivo activation of ETT-GR (Table S1) and subsequent qPCR studies (Figure 1) failed to be identified in one or more of the other three analyses used for comparison: arabinogalactan protein AT5G24105 and PME AT5G27870.
Figure 2.
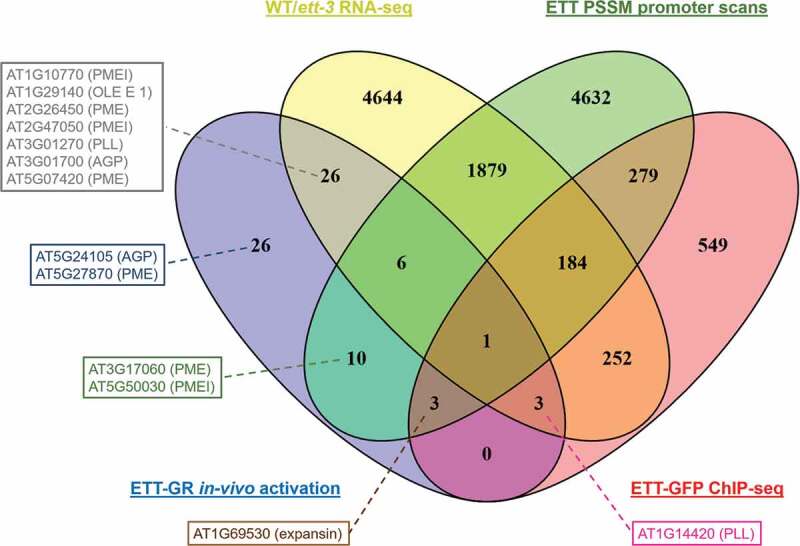
Comparison of four datasets to confirm putative immediate ETT targets. Total numbers of genes occurring in each combination of datasets are indicated. Ten chloroplast genes that were rapidly regulated following ETT-GR induction (Table S1) were excluded from the analysis. The positions of 13 cell-wall-related genes (Table S1, Figure S1) are indicated in boxes.
Discussion
Transcriptomics experiments suggest a key role for PMEIs in the regulation of tissue patterning by ETT
Andres-Robin et al.7 showed a correlation between the regulation by ETT of tissue-patterning in the A. thaliana gynecium and enzymatic and biochemical characteristics that pointed to a link with the methylesterification status of cell-wall pectins. A model emerged in which ETT acts to promote PME activity in the cell wall. Here, we provide evidence that ETT acts as a negative regulator of four PMEs and three PMEIs (Table S1). The regulation of all seven of these genes was confirmed by qRT-PCR (Figure 1), and in six cases by comparisons with one or more of three further datasets (Figure 2).
From the overexpression experiments in mutant and wild-type backgrounds performed by Andres-Robin et al.7 the negative regulation by ETT of PMEIs would be consistent with the known effects of ETT on the methylesterification of pectins, cell-wall stiffness and valve length, whereas the negative regulation by ETT of PMEs would be predicted to have the opposite effect. We therefore postulate that the regulation of valve-length by ETT is controlled predominantly through the negative regulation of PMEIs. The significance of the simultaneous negative regulation by ETT of several PMEs is unclear, though may be an example of a “gas and brake mechanism”,18 in which an upstream regulator acts in the same direction on mutually antagonistic classes of downstream components. Such mechanisms are known in the control of shade-avoidance in plants and are postulated to prevent exaggerated downstream effects.
The above conclusions on the possible developmental significance of ETT-targets remain, however, speculative as the biochemical properties and developmental effects of these molecules have yet to be directly investigated. For example, certain PMEs are known to show distinct enzymatic activities depending on the prior pattern of pectin methylesterification encountered,19 while certain PMEIs are known to show differential interaction specificities among PMEs.20,21 In addition, both PME activities and certain PMEI-PMI interactions are known to be pH-dependent.19,21 We do not, therefore, know at present whether the PMEIs identified here as immediate ETT-targets would be active against the PMEs also shown to be regulated by ETT. Neither do we yet know whether those PMEs, if over-expressed in the gynecium, would show similar effects on pectin demethylesterification and valve-length to the effects previously demonstrated for PME5.7
Comparison of datasets suggests the presence of rapidly acting intermediate components in the regulation of PMEs and PMEIs by ETT
Though several PMEs and PMEIs, among other cell wall-related genes, appear to be rapidly downregulated as a response to ETT, it is currently unclear whether ETT interacts directly with the promoters of these genes. Comparisons between datasets confirm the regulatory interactions identified for three PMEIs and three PMEs (Figure 2). However, the presumptive promoters of only two of these genes, PMEI AT5G50030 and PME AT3G17060, contain potential ETT-binding sites, and neither gene emerged as a putative direct target of ETT in the ChIP-seq analysis of Simonini et al.9 Taken together, the data described here suggest the involvement of PMEs and PMEIs in ETT’s mode of action in the gynecium, but suggest the operation of one or more rapidly-acting intermediate components, downstream of ETT, in the transcriptional regulation of these genes.
For example, ETT might repress one or more transcriptional activators, whose activity would thereby be reduced in the gynecium, leading to lower transcription rates of their own direct targets, possibly including some of the likely indirect targets of ETT identified here. Our experiments (Table S1) and those of Simonini et al.9 have indicated a number of transcription factors as possible direct targets of ETT, so that observation could represent a starting point to investigate this hypothesis further.
Material and methods
Plant growth conditions
Wild-type, transgenic and/or mutant A. thaliana lines were grown on peat-based compost at 20°C under short-day conditions (8 h light/16 h dark cycles) for 4 weeks before being transferred in long-day conditions (16 h light/8 h dark cycles) to induce flowering for genetic transformation, induction experiments and/or gene expression analyses.
Transgene constructs, induction experiments and microarray analyses
The ETT coding sequence was inserted into pG0229-35S::GR10 between the CaMV-35S promoter and a sequence encoding the hormone-binding domain of the rat glucocorticoid receptor, so as to conserve the reading frame of the latter element. The resulting plasmid was transferred to Agrobacterium tumefaciens C58/pMP90, a culture of which was then used to transform plants of the A. thaliana Col-0 ecotype by standard “floral dip” procedures. A homozygous individual, stably expressing the transgene from a single insertion event, was selected from the ensuing T2 generation.
The ETT-GR transgene construct was then introduced into an ett-22 mutant background by cross pollination. The functionality of the ETT-GR fusion protein was demonstrated on homozygous plants from which all flowers later than Stage 822 had been removed. Inflorescences were dipped for 2 min, once per day for five days, into aqueous solutions of DEX (10 µg/ml) containing Silwet L-77 surfactant (0.01% v/v), or mock solutions lacking DEX. Gynoecia were observed three days after final treatments using a Leica MZ12 stereomicroscope coupled to a DFC320 digital camera. Fifty measurements were made using ImageJ for each treatment-group of the ett-22 35S:ETT-GR line, and 30 measurements for each treatment-group of the 35S:ETT-GR and ett-22 lines.
For microarray analyses, two populations, each composed of ten T3 descendants of the original T2 35S:ETT-GR transformant in a wild-type Col-0 genetic background, were grown to maturity. CHX and DEX treatments, RNA extraction and microarray analyses were performed as described by Reymond et al.,11 and the entire experiment was repeated twice to provide three biological replicates. Statistical analysis of microarray data was also performed as previously described.11 The complete microarray dataset is available at CATdb (http://tools.ips2.u-psud.fr/CATdb) as Project: GEN45-Carpel_development. The list of putative ETT-targets obtained was functionally analyzed using Classification Superviewer14 at http://bar.utoronto.ca.
Quantitative RT-PCR
Total RNA was extracted from inflorescences using Spectrum Plant Total RNA Kit (Sigma). Any contaminating DNA was eliminated by treatments with Turbo DNAase (Ambion) and 4-µg aliquots of total RNA were then reverse transcribed using RevertAid Reverse Transcriptase (Fermentas). Reverse transcriptase reactions of 20 μl volume were diluted to 1 ml, and 5-μl aliquots of these dilutions were subjected to qPCR using SYBR GREEN (Roche) in a StepOne Plus apparatus (Applied). Primer efficiencies were determined from standard curves using serial cDNA dilutions and PCR amplification was performed using a three-step protocol, incorporating a melting curve. Results were normalized to the expression of GAPDH (AT3G26650), chosen from a list of potential control genes using BESTKEEPER,23 and analyzed using the 2−(ΔΔCt) method. Each expression value was determined from the mean of a minimum of three plants, and each experiment was repeated twice independently. The primers used are given in the Table S3.
In-silico promoter analyses and global data comparisons
A genome-wide set of A. thaliana promoters (http://arabidopsis.med.ohio-state.edu/)24 was scanned with PSSMs for ETT5 using the Matrix-Scan program in RSAT.17 Scans were combined and filtered on the basis of score-values. Gene lists produced using various different methods were compared in Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/).
Supplementary Material
Acknowledgments
We are grateful to R. Scott Poethig for seeds of 35S:ETTm and acknowledge funding from Génoplante (G-45) and the French National Research Agency (ANR-BLAN-0211-01) to CPS. IPS2 is supported by LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). AMR and MCR received doctoral studentship from the French Ministry of Higher Education, Research and Innovation and from the Rhone-Alps Region, respectively.
Funding Statement
This work was supported by the Agence Nationale de la Recherche [BLAN-0211-01]; Agence Nationale de la Recherche [ANR-10-LABX-0040-SPS]; and Genoplante [G45].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed Publisher’s website.
References
- 1.Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC.. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:1–5. [DOI] [PubMed] [Google Scholar]
- 2.Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 4.Guilfoyle TJ, Hagen G. Auxin response factors. J Plant Growth Regul. 2001;20:281–291. doi: 10.1007/s003440010026. [DOI] [Google Scholar]
- 5.Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulyk ML, Johnson PLF, Church GM. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andres-Robin A, Reymond MC, Dupire A, Battu V, Dubrulle N, Mouille G, Lefebvre V, Pelloux J, Boudaoud A, Traas J. Evidence for the regulation of gynoecium morphogenesis by ETTIN via cell wall dynamics. Plant Physiol. 2018;178(3):1222–1232. doi: 10.1104/pp.18.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peaucelle A, Louvet R, Johansen JN, Höfte H, Laufs P, Pelloux J, Mouille G. Arabidopsis phyllotaxis is controlled by the methyl-esterification status of cell-wall pectins. Curr Biol. 2008;18:1943–1948. doi: 10.1016/j.cub.2008.10.065. [DOI] [PubMed] [Google Scholar]
- 9.Simonini S, Bencivenga S, Trick M, Ostergaard L. Auxin-induced modulation of ETTIN activity orchestrates gene expression in Arabidopsis. Plant Cell. 2017;29:1864–1882. doi: 10.1105/tpc.17.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Ito T, Wellmer F, Meyerowitz EM. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet. 2004;36:157–161. doi: 10.1038/ng1286. [DOI] [PubMed] [Google Scholar]
- 11.Reymond MC, Brunoud G, Chauvet A, Martínez-Garcia JF, Martin-Magniette M-L, Monéger F, Scutt CP. A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell. 2012;24(7):2812–2825. doi: 10.1105/tpc.112.097915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowe ML, Serizet C, Thareau V, Aubourg S, Rouze P, Hilson P, Beynon J, Weisbeek P, Van Hummelen P, Reymond P, et al. CATMA: a complete Arabidopsis GST database. Nucleic Acids Res. 2003;31:156–158. doi: 10.1093/nar/gkg071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Provart NJ, Zhu T. A browser-based functional classification SuperViewer for Arabidopsis genomics. Curr Comput Mol Biol. 2003:271–272. [Google Scholar]
- 15.Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- 16.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutiérrez-nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turatsinze J-V, Thomas-Chollier M, Defrance M, van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 18.Crocco CD, Holm M, Yanovsky MJ, Botto JF. Function of B-BOX under shade. Plant Signal Behav. 2011;6:101–104. doi: 10.4161/psb.6.1.14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senechal F, L’Enfant M, Domon J-M, Rosiau E, Crépeau M-J, Surcouf O, Esquivel-Rodriguez J, Marcelo P, Mareck A, Guérineau F. Pectin methylesterase inhibitor 7 modulates the processive activity of co-expressed pectin methylesterase3 in a pH-dependent manner. J Biol Chem. 2015;290:23320–23335. doi: 10.1074/jbc.M115.639534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senechal F, Mareck A, Marcelo P, Lerouge P, Pelloux J. Arabidopsis PME17 activity can be controlled by pectin methylesterase inhibitor 4. Plant Signal Behav. 2015;10:e983351. UNSP. doi: 10.4161/15592324.2014.983351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hocq L, Sénéchal F, Lefebvre V, Lehner A, Domon J-M, Mollet J-C, Dehors J, Pageau K, Marcelo P, Guérineau F. Combined experimental and computational approaches reveal distinct pH dependence of pectin methylesterase inhibitors. Plant Physiol. 2017;173(2):1075–1093. doi: 10.1104/pp.16.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth D, Bowman J, Meyerowitz E. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 24.Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 2011;39:D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


