Abstract
Motivation
Clinical trials are the essential stage of every drug development program for the treatment to become available to patients. Despite the importance of well-structured clinical trial databases and their tremendous value for drug discovery and development such instances are very rare. Presently large-scale information on clinical trials is stored in clinical trial registers which are relatively structured, but the mappings to external databases of drugs and diseases are increasingly lacking. The precise production of such links would enable us to interrogate richer harmonized datasets for invaluable insights.
Results
We present a neural approach for medical concept normalization of diseases and drugs. Our two-stage approach is based on Bidirectional Encoder Representations from Transformers (BERT). In the training stage, we optimize the relative similarity of mentions and concept names from a terminology via triplet loss. In the inference stage, we obtain the closest concept name representation in a common embedding space to a given mention representation. We performed a set of experiments on a dataset of abstracts and a real-world dataset of trial records with interventions and conditions mapped to drug and disease terminologies. The latter includes mentions associated with one or more concepts (in-KB) or zero (out-of-KB, nil prediction). Experiments show that our approach significantly outperforms baseline and state-of-the-art architectures. Moreover, we demonstrate that our approach is effective in knowledge transfer from the scientific literature to clinical trial data.
Availability and implementation
We make code and data freely available at https://github.com/insilicomedicine/DILBERT.
1 Introduction
The emerging use of neural network architectures in early-stage drug development has recently resulted in several breakthroughs. Later stages of drug development are significantly less amenable to innovation due to the immense infrastructure allocated to the existing establishment of clinical trials. The clinical development of the drug is a long and costly process that typically requires several years and a billion-dollar budget to progress the drug from phase 1 clinical trials to the patients (Dowden and Munro, 2019). The use of state-of-the-art neural network approaches in clinical trials may dramatically speed up the overall drug development process and increase its success rate, thus saving lives.
It is widely recognized that the drug development industry suffers from high attrition rates with less than one in six drugs making it from phase 1 to the market (Hay et al., 2014). ClinicalTrials.gov, the clinical trial repository maintained by the National Institutes of Health (NIH), contains over 284 000 clinical trial entries submitted by various organizations as of January 1, 2021 (see www.clinicaltrials.gov). It is estimated that trained analysts would require tens of thousands of hours of labor to incorporate its full information manually (Wong et al., 2019). Thus it’s critical to develop precise automatic approaches for the clinical trial entry annotation. This work focuses on the harmonization of diseases and interventions presented in the clinical trial entry as free text with the existing centralized standardized taxonomies. The use of automatic natural language processing (NLP) methods is imperative to semantic annotation of a large volume of clinical records, and to linking and standardization of biomedical entity mentions to formal concepts. In biomedical research and healthcare, the entity linking problem is known as medical concept normalization (MCN).
Inspired by metric learning (Hoffer and Ailon, 2015; Huang et al., 2013; Schroff et al., 2015), its usage for multimodal and sentence representation learning (Liu et al., 2017; Reimers and Gurevych, 2019), negative sampling (Mikolov et al., 2013) and Bidirectional Encoder Representations from Transformers (BERT) (Devlin et al., 2019), we present a neural model for Drug and disease Interpretation Learning with Biomedical Entity Representation Transformer (DILBERT). This model directly optimizes the BioBERT representations (Lee et al., 2019) of entity mentions and concept names themselves, rather than classification or ranking layer. We use triplets of free-form entity mention, positive concept names and randomly sampled concept names as negative examples to train our model.
The key contributions of this work are three-fold. First, we develop a simple and effective model that uses metric learning and negative sampling to obtain entity and concept embeddings. Second, we consider the zero-shot scenario with cross-domain evaluation because it is often the case in the biomedical domain, where exist dozens of concept categories and terminologies. Third, we perform extensive experiments of several BERT-based models on a newly annotated dataset of clinical trials in two setups, where each mention is associated with one or more concepts (in-KB) or zero (out-of-KB). A preliminary version of this work has appeared in Miftahutdinov et al. (2021). Compared to the conference version, we have: (i) extended our description of the proposed dataset of clinical trials; (ii) significantly extended the experimental part of this work to assess the performance of DILBERT; in particular, we investigated the impact of dictionary size on the task of disease and drug normalization, adding new experimental results and conclusions; (iii) performed error analysis and discussed the limitations of our model.
The paper is organized as follows. We begin with an overview of existing papers on processing of clinical trials and MCN systems in Section 2. In Section 3, we present our dataset of clinical trials. Description of our novel DILBERT model is presented in Section 4. Descriptions of our experimental setup, academic datasets we used and the results are reported in Section 5. Section 6 contains a discussion of our results and limitations of our approach. Finally, we summarize our contributions and discuss directions for further research in Section 7.
2 Related work
While the majority of biomedical research on information extraction primarily focused on scientific literature (Huang and Lu, 2016), much less work utilizes NLP methods to conduct curation of clinical trial records’ fields (Atal et al., 2016; Boland et al., 2013; Brown and Patel, 2017; Hao et al., 2014; Sen et al., 2018) with most of the work conducted on a subset of clinical trial data severely restricted either by therapeutic indication (Boland et al., 2013; Sen et al., 2018), development status (Brown and Patel, 2017) or terminology (Atal et al., 2016). This restriction ultimately impedes the advancement of crucial downstream tasks concerning drug repurposing (Malas et al., 2019), overall clinical development risk assessment (Lo et al., 2019; Wong et al., 2019) and its aspects such as clinical failure prediction based on safety concerns (Gayvert et al., 2016).
2.1 Downstream applications
Characteristically, Lo et al. (Lo et al., 2019; Wong et al., 2019) leveraged the proprietary dataset of unprecedented size to compute the statistical parameters associated with clinical development phase transitions as well as to build a machine learning model to predict the probability of successful phase transition. As the source data is not available for the independent quality assessment and reproduction of the work there is currently an unmet need in frameworks for the creation of datasets of comparable scale. Gayvert et al. (2016) selected 108 clinical trials of any phase that were annotated as having failed for toxicity reasons. Then intervention names of each trial were manually mapped to DrugBank (Wishart et al., 2018) concepts to collect molecular weight, polar surface area and other compounds’ properties which were then used as an input for decision tree ensemble-based machine learning model predictive of the trial failure due to toxicity.
2.2 Clinical trial record field processing
In Brown and Patel (2017), presented a novel database of approved and failed drugs and their indications for drug repositioning. Pairs of drugs and approved indications were drawn from DrugCentral, which contains UMLS indications mapped from free-text mentions in drug labels. To create a list of failed drugs and their indications, the authors adopted the AACT database and utilized a dictionary-based approach to map interventions to DrugCentral synonyms. Indication information was mapped to UMLS identifiers using the UMLS REST API. In Atal et al. (2016), developed a knowledge-based approach to classify entity mentions to disease categories from a Global Burden of Diseases (GBD) cause list. The proposed method uses MetaMap to extract UMLS concepts from trial fields (health condition, public title and scientific title), link UMLS concepts with ICD10 codes and classify ICD10 codes to candidate GBD categories. The developed classifier identified GBD categories for 78% of the trials. Li and Lu (2012) identified clinical pharmacogenomics (PGx) information from clinical trial records based on dictionaries from a pharmacogenomics knowledge base PharmGKB. First, they applied dictionaries from a pharmacogenomics knowledge base PharmGKB to identify genes, drugs and diseases from clinical trials and assign these entities PharmGKB identifiers. Second, they used a co-occurrence-based method for identifying relationships between three types of entities. 100 identified gene-drug-disease relationships were manually validated and the proposed approach achieves an accuracy of 74%. Given these 26 PGx gene-drug pairs, a total of 240 3-way PGx relationships were found in trial records; 68 relationships are overlapped with 261 results in PharmGKB. Li and Lu noted that their approach failed to identify entity variants not covered by the dictionaries. Summarizing the above, previous studies on clinical trial records, however, have not analyzed the performance of linking of clinical trials to disease and drug concepts, but rather across eligibility criteria (e.g. patient’s demographic, disease category). They also share the common restriction of relying on specific terminologies that cannot be changed in a feasible manner for knowledge transfer purposes (Atal et al., 2016; Boland et al., 2013; Hao et al., 2014; Leveling, 2017; Sen et al., 2018).
Neural architectures have been widely used in recent state-of-the-art models for MCN from scientific texts, user reviews, social media texts and clinical notes (Ji et al., 2020; Leaman and Lu, 2016; Li et al., 2017, 2019; Miftahutdinov and Tutubalina, 2019; Sung et al., 2020; Xu et al., 2020; Zhao et al., 2019; Zhu et al., 2020). Most models share limitations regarding a supervised classification framework: (i) to retrieve concepts from a particular terminology for a given entity mention, models are required re-training, (ii) use additional classification or ranking layer, therefore, during inference compute all similarities between a given mention and all concept names from a dictionary through this layer and sort these scores in descending order. For instance, Ji et al. (2020) fine-tuned BERT with binary classifier layer. Xu et al. (2020) adopted a BERT-based multi-class classifier to generate a list of candidate concepts for each mention, and a BERT-based list-wise classifier to select the most likely candidate. We note that this multi-class candidate generator will require re-training for cross-terminology mapping. In our work, we focus on direct optimization of BERT representations to allow efficient similarity search with a FAISS library (Johnson et al., 2019).
The works that are the closest to ours and use entity and concept representation learning are triplet networks (Mondal et al., 2019), Biomedical Named Encoder (BNE) (Phan et al., 2019) and BioSyn (Sung et al., 2020). Mondal et al. used distances between disease mentions, positive and randomly sampled negative candidates to train a triplet network (Mondal et al., 2019). As encoder, convolutional and pooling layer based on word embeddings was adopted. Sung et al. proposed a BioBERT-based model named BioSyn that maximizes the probability of all synonym representations in the top 20 candidates (Sung et al., 2020). BioSyn uses a combination of two scores, sparse and dense, as a similarity function. Sparse scores are calculated on character-level TF-IDF representations to encode morphological information of given strings. Dense scores are defined by the similarity between CLS tokens of a single vector of input in BioBERT. This model achieves state-of-the-art results in disease and chemical mapping over previous works (Leaman and Lu, 2016; Mondal et al., 2019; Phan et al., 2019; Wright et al., 2019). Phan et al. presented an encoding framework with new context, concept and synonym-based objectives (Phan et al., 2019). Synonym-based objective enforces similar representations between synonymous names, while concept-based objective pulls the name’s representations closer to its concept’s centroid. The word and concept unique identifier (CUI) embeddings are pretrained on 29 million PubMed abstracts annotated with UMLS concepts of diseases and chemicals. However, ranking on these embeddings shows worse results on three sets than supervised models.
Our work differs from the discussed studies in important aspects. First, none of these methods have been applied to free-form descriptions of conditions and interventions from clinical trials. Second, evaluation strategies in the mentioned papers are based on train/test splits provided by datasets’ authors. We follow recent refined evaluation strategy from (Tutubalina et al., 2020) on creation of test sets without duplicates or exact overlaps between train and test sets. Finally, our dataset includes entity mentions for both in-KB and out-of-KB linking.
3 Dataset of clinical trials
NLM maintains a clinical trial registry data bank ClinicalTrials.gov (https://clinicaltrials.gov/) that contains over 284 000 trials from 214 countries. This database includes comprehensive scientific and clinical investigations in biomedicine (Gill et al., 2016). Each trial record provides information about a trial’s title, purpose, description, condition, intervention, eligibility, sponsors, etc. Most information from records is stored in a free text format. In our study, we use publicly available Aggregate Content of ClinicalTrials.gov (AACT) Database (https://www.ctti-clinicaltrials.org/aact-database), v. 20200201.
Since there is no off-the-shelf gold-standard dataset for drug and disease concept normalization for clinical trials, we’ve constructed one through selecting 500 clinical studies from AACT using the following criteria:
The study is interventional, that is the study is a trial. Participants of clinical trials receive intervention/treatment so that researchers can evaluate the effects of the interventions on health-related outcomes.
The study is associated with one or more interventions of the following types: Drug, Biological, Combination Product.
As a drug terminology source, we use a Drugbank v. 5.1.8 that contains 14 325 concept unique identifiers (CUIs). As a condition terminology source, we use MeSH v. 20200101. 500 selected trials contain 1075 and 819 entries in the ‘Intervention’ and ‘Condition’ fields respectively. Each entry was manually annotated by the two annotation experts with a background in biomedical data curation. The calculated inter-annotator agreement (IAA) using Kappa was 92.32% for the entire dataset. Disagreement was resolved through mutual consent.
Phase of the clinical study is defined by the FDA. There were five phases included: Early Phase 1, Phase 1, Phase 2, Phase 3 and Phase 4. As shown in Table 1, the selected trials cover various phases evenly. Statistics of annotated texts are summarized in Table 2. 749 out of 1075 non-unique mentions (69.6%) were mapped to one or more drug concepts. 838 (80%) of lower-cased interventions are unique. 804 out of 819 non-unique mentions (98.2%) were mapped to one or more concepts, while there are 638 (78%) lower-cased unique mentions. Interestingly, MeSH concepts linked to conditions belong to several MeSH categories including Diseases (C), Psychiatry and Psychology (F) and Analytical, Diagnostic and Therapeutic Techniques and Equipment (E). We note that NLM provided automatically assigned MeSH terms to trials’ interventions. 716 out of 1075 entries (66.6%) were mapped to MeSH terms. Our analysis revealed that MeSH terminology does not include investigational drugs, the data on which is crucial for the downstream tasks. Table 3 contains a sample of annotated texts.
Table 1.
Statistics of trials’ phases
| Phase | No. of trials in our dataset | No. of trials in clinicaltrials.gov |
|---|---|---|
| Phase 2 | 128 | 47 398 |
| Phase 3 | 116 | 32 707 |
| Phase 1 | 108 | 34 121 |
| Phase 4 | 88 | 27 726 |
| Phase 1/Phase 2 | 0 | 11 582 |
| Phase 2/Phase 3 | 0 | 5632 |
| Early Phase 1 | 0 | 3500 |
| Total | 500 | 162 666 |
Table 2.
Statistics of annotated texts
| Mention | No. of texts | No. of texts with CUIs | No. of unique texts | No. of unique texts with CUIs |
|---|---|---|---|---|
| Intervention types | ||||
| Drug | 850 | 657 | 671 | 584 |
| Biological | 118 | 58 | 102 | 55 |
| Other | 57 | 21 | 27 | 21 |
| Procedure | 19 | 1 | 16 | 1 |
| Radiation | 11 | 3 | 9 | 3 |
| Device | 11 | 4 | 11 | 4 |
| Combination product | 5 | 3 | 5 | 3 |
| Dietary supplement | 2 | 2 | 2 | 2 |
| Diagnostic test | 1 | 0 | 1 | 0 |
| Behavioral | 1 | 0 | 1 | 0 |
| Total | ||||
| Intervention | 1075 | 749 | 838 | 661 |
| Condition | 819 | 804 | 638 | 638 |
Table 3.
Sample of manually annotated trials’ texts
| NCT | Type | Text | Concept |
|---|---|---|---|
| Intervention (with DrugBank CUIs) | |||
| NCT00559975 | Biological | Adjuvanted influenza vaccine combine with CpG7909 | Agatolimod sodium (DB15018) |
| NCT01575756 | Biological | Haemocomplettan® P or RiaSTAPTM | Fibrinogen human (DB09222) |
| NCT00081484 | Drug | epoetin alfa or beta | Erythropoietin (DB00016) |
| NCT03375593 | Drug | Ibuprofen 600 mg tab | Ibuprofen (DB01050) |
| NCT01170442 | Drug | vitamin D3 5000 IU | Calcitriol (DB00136) |
| NCT02493335 | Drug | Placebo orodispersible tablet twice daily | nil (no concept) |
| Condition (with MeSH CUIs) | |||
| NCT02009605 | Condition | Squamous Cell Carcinoma of Lung | Carcinoma, Non-Small-Cell Lung (D002289) |
| NCT04169763 | Condition | Stage IIIC Vulvar Cancer AJCC v8 | Vulvar Neoplasms (D014846) |
4 Model
In this section, we present a neural model for Drug and disease Interpretation Learning with Biomedical Entity Representation Transformer (DILBERT). We address MCN as a retrieval task by fine-tuning the BERT-based network using metric learning (Hoffer and Ailon, 2015; Huang et al., 2013; Schroff et al., 2015), negative sampling (Mikolov et al., 2013), specifically, triplet constraints. This idea was successfully applied to learn multimodal embeddings (Liu et al., 2017; Wu et al., 2013) and recent sentence embeddings via a sentence-BERT model (Reimers and Gurevych, 2019). Compared to a pair of independent sentences or images, two concept names can have relationships as synonyms, hypernyms, hyponyms, etc., that we consider during the training phase to facilitate the concept ranking task at the retrieval phase.
Architecture Following denotations proposed by Humeau et al. (2019), we encode both entity mention m and candidate concept name c into vectors:
| (1) |
where T is the transformer that is allowed to update during fine-tuning, is a function that reduces that sequence of vectors into one vector. There are two main ways of reducing the output into one representation via : choose the first output of T (corresponding to the token CLS) or compute the elementwise average over all output vectors to obtain a fixed-size vector. As a pretrained transformer model, we use BioBERT base v1.1 (Lee et al., 2019).
Scoring The score of a candidate ci for a entity mention m is given by a distance metric, e.g. Euclidean distance:
| (2) |
A noteworthy aspect of the proposed model is its scope: by design, it aims at the cross-terminology mapping of entity mentions to a given lexicon without additional re-training. This approach allows for fast, real-time inference, as all concept names from a terminology can be cached. This is a requirement for processing biomedical documents of different subdomains such as clinical trials, scientific literature, drug labels.
Optimization The network is trained using a triplet objective function. Given an user-generated entity mention m, a positive concept name cg and a negative concept name cn, triplet loss tunes the network such that the distance between m and cg is smaller than the distance between m and cn. Mathematically, we minimize the following loss function:
| (3) |
where ϵ is margin that ensures that cg is at least ϵ closer to m than cn. As scoring metric, we use Euclidean distance or cosine similarity and we set ϵ = 1 in our experiments.
Positive and negative sampling Suppose that a pair of the entity mention with the corresponding CUI is given as well as the vocabulary. For positive examples, vocabulary is restricted to the concepts that have the same CUI as a mention. Multiple positive concept names could be explained by the presence of synonyms in the vocabulary. Negative sampling (Mikolov et al., 2013) uses the rest part of the vocabulary. We explore several strategies to select positive and negative samples for a training pair (entity mention, CUI):
random sampling: we sample several concept names with the same CUI as positive examples and random negatives from the rest of the vocabulary;
random + parents: we sample k concept names from the concept’s parents in addition to positive and negative names gathered with the random sampling strategy;
re-sampling: using a model trained with random sampling, we identify positives and hard negatives via the following steps: (i) encode all mentions and concept names found in training pairs using the current model, (ii) select positives with the same CUI, which are closest to a mention, (iii) for each mention, retrieve the most similar k concept names (i.e. its nearest neighbors) and select all names that are ranked above the correct one for the mention as negative examples. We follow this strategy from (Gillick et al., 2019);
re-sampling + siblings: we modify the re-sampling strategy by using k concept names from the concept’s siblings as negatives.
Inference At inference time, the representation for all concept names can be precomputed and cached. The inference task is then reduced to finding the closest concept name representation to entity mention representation in a common embedding space.
Out-of-KB prediction To deal with nil prediction in clinical trials, we apply three different strategies for selection of a threshold value. Namely, the intervention or condition mention is considered out of KB if the nearest candidate has a larger distance than a threshold value. Our first strategy is to set the threshold equal to the minimum distance of false-positive (FP) cases. In this case, we consider a mention mapped to a concept by our model but having no appearance in the terminology as FP. Thus in the list of mentions ordered by increasing the distance to the nearest concept, threshold will be equal to the distance of first nill appearance. This strategy has high precision and low recall. Our second strategy set the threshold to the maximum distance of true-positive (TP) cases. Which is opposite to the first strategy and in ordered by distances list threshold will be equal to the last appearance of the TP case. Thus the second strategy has low precision and high recall. The third strategy uses a weighted average of the first two threshold values. The proportion of TP cases used as a weight for the first strategy’s threshold, the proportion of TP cases used as a weight for the second strategy’s threshold. The third strategy is optimal and balances the low recall of first strategy by the high recall of second and similar for the precision metric.
5 Experiments
For the experimental evaluation of DILBERT, we have posed and attempted to answer the following research questions:
RQ1: how does DILBERT perform on clinical trials with both in-KB and out-of-KB cases?
RQ2: can DILBERT outperform state-of-the-art MCN models on existing benchmarks of scientific benchmarks?
RQ3: how dictionary size at prediction time affect the overall performance?
Below we describe the results of our experimental evaluation on the questions above.
5.1 Datasets
We have conducted our experimental evaluation of the proposed model on two datasets: a publicly available benchmark BioCreative V CDR Disease & Chemical (Li et al., 2016), (ii) our dataset of clinical trials named CT Condition & Intervention. The BioCreative V CDR Disease & Chemical is a part of a Biomedical Language Understanding & Reasoning Benchmark (BLURB) (Gu et al., 2020).
Statistics of two datasets are summarized in Table 4. BioCreative V CDR Disease & Chemical consists of chemical & disease mentions in Pubmed abstracts with spans of text annotated as concepts. The format is as follows: PMID <tab> START OFFSET <tab> END OFFSET<tab>text MENTION <tab> mention TYPE (e.g. Disease) <tab> database IDENTIFIER <tab> Individual mentions (e.g. 3403780 29 47 metabolic acidosis Disease D000138). The main difference between BioCreative V and our dataset consists in documents annotated and subsequent particularities of the annotations such as in clinical trials interventions and conditions are organized in fields whereas in abstracts presented in a free-form text as well as differences in vocabularies. Statistics of terminologies synonyms represented in Table 5.
Table 4.
Statistics of the datasets used in the experiments
| CDR Disease | CDR Chem | CT Condition | CT Intervention | |
|---|---|---|---|---|
| Domain | Abstracts | Abstracts | Clinical trials | Clinical trials |
| Entity type | Disease | Chemicals | Conditions | Drugs |
| Terminology | MEDIC | CTD Chemicals | MeSH | Drugbank |
| Entity level statistics | ||||
| % numerals | 0.11% | 7.32% | 7.69% | 25.3% |
| % punctuation | 1.21% | 0.07% | 14.28% | 24.83% |
| Avg. len | 14.88 | 11.27 | 17.92 | 21.68 |
| Number of pre-processed entity mentions | ||||
| Train set | 4182 | 5203 | – | – |
| Dev set | 4244 | 5347 | 100 | 100 |
| Test set | 4424 | 5385 | 719 | 975 |
| Filtered test | 1240 (28.02%) | 826 (15.38%) | 642 (78.4%) | 846 (78.7%) |
Note: Two sets of annotated clinical trials’ fields are marked with ‘CT’.
Table 5.
Synonyms statistics in terminologies
| Vocabulary Name | No. of concepts | No. of concept names | Avg synonyms per concept |
|---|---|---|---|
| MEDIC | 12513 | 73178 | 5.85 |
| CTD Chem | 171285 | 407615 | 2.38 |
| Drugbank | 14325 | 141617 | 9.89 |
BioCreative V CDR (Li et al., 2016) introduces a challenging task for the extraction of chemical-disease relations (CDR) from PubMed abstracts. Disease and chemical mentions are linked to the MEDIC (Davis et al., 2012) and CTD (Davis et al., 2019) dictionaries, respectively. We utilize the CTD chemical dictionary (v. November 4, 2019) that consists of 171 203 CUIs and 407 247 synonyms and the MEDIC lexicon (v. July 6, 2012) that contains 11 915 CUIs and 71 923 synonyms.
According to BioCreative V CDR annotation guidelines, annotators used two MeSH branches to annotate entities: (i) ‘Diseases’ [C], including signs and symptoms, (ii) ‘Drugs and Chemicals’ [D]. The terms ‘drugs’ and ‘chemicals’ are often used interchangeably. Annotators annotated chemical nouns convertible to single atoms, ions, isotopes, pure elements and molecules (e.g. calcium, lithium), class names (e.g. steroids, fatty acids), small biochemicals, synthetic polymers.
As shown in Tutubalina et al. (2020), the CDR dataset contains a high amount of mention duplicates and overlaps between official sets. To obtain more realistic results, we evaluate models on preprocessed official and refined CDR test sets from Tutubalina et al. (2020).
For preprocessing of clinical trials, we use heuristic rules to split composite mentions into separate mentions (e.g. combination of ribociclib + capecitabine into ribociclib and capecitabine) by considering each mention containing ‘combination’, ‘combine’, ‘combined’, ‘plus’, ‘vs’ or ‘+’ as composite. We process all characters to lowercase forms and remove the punctuation for both mentions and synonyms.
5.2 Baselines
BioBERT ranking This is a baseline model that used the BioBERT model for encoding mention and concept representations. Each entity mention or concept name is passed first through BioBERT and then through a mean pooling layer to yield a fixed-sized vector. The inference task is then reduced to finding the closest concept name representation to entity mention representation in a common embedding space. We use the Euclidean distance as the distance metric. The nearest concept names are chosen as top-k concepts for entities. We use the publicly available code provided by Tutubalina et al. (2020) at https://github.com/insilicomedicine/Fair-Evaluation-BERT.
BioSyn BioSyn (Sung et al., 2020) is a recent state-of-the-art model that utilizes the synonym marginalization technique and the iterative candidate retrieval. The model uses two similarity functions based on sparse and dense representations, respectively. The sparse representation encodes the morphological information of given strings via TF-IDF, the dense representation encodes the semantic information gathered from BioBERT. For reproducibility, we use the publicly available code provided by the authors at https://github.com/dmis-lab/BioSyn. We follow the default parameters of BioSyn as in Sung et al. (2020): the number of top candidates k is 20, the mini-batch size is 16, the learning rate is 1e-5, the dense ratio for the candidate retrieval is 0.5, 20 epochs for training.
5.3 Evaluation strategies
Recent study (Tutubalina et al., 2020) on evaluation in concept normalization summarized that there are several evaluation setups depending on two modalities such as domain and terminology:
Stratified and zero-shot evaluation
In- and out-of-domain evaluation (or cross-domain setup)
Single- and out-of-terminology evaluation (or cross-terminology setup)
In general, terminology is homogeneous and include concept names and corresponding CUIs of a specific entity type (e.g. diseases, drugs). Most MCN methods are trained and evaluated on sets of widely differing sizes, entity types, domains and a narrow subsample of concepts from a target terminology. This clearly does not correspond to real-world applications.
The first evaluation setup focuses on the intersection between CUIs in train and test/dev sets. Here stratified, proposed in Tutubalina et al. (2018), is intended to show how well a model recognizes known concepts with different surface forms of entity mentions: each concept appearing in the test/dev sets, appears at least once in the training set. In contrast, zero-shot evaluation shows how well a model links entity mentions to novel concepts: dev and test sets contain novel concepts only for which there are no training mentions. The same terminology is used during training and evaluation.
The second evaluation setup focuses on domain change, i.e. differences between train and test/dev corpora. This is the case of biomedicine, a field with a lot of sub-domains and information sources (e.g. abstracts, patent documents, clinical records, drug labels, social media). All sub-domains differ substantially in their structure, language, writing style which is reflected in surface forms of entity mentions. In-domain evaluation is intended to how well a model maps entity mentions from the seen domain under the implicit hypothesis that the training data and test data come from the same underlying distribution. In contrast, out-of-domain strategy is designed to train and evaluate on concepts from the seen target terminology and mentions of the same type from another domain. For example, models trained to map disease mentions to MeSH terminology from PubMed abstracts (source domain) and evaluated on disease mentions with MeSH CUIs from clinical records (target domain). Adapting trained models to different language varieties would be desirable to enable better generalizability.
The third evaluation setup is more complicated due to shift in terminology which entails changes in entity type and surface form mentions. Single-terminology evaluation is the most popular type of evaluation: concept names and CUIs in the test/dev sets are seen during training. Out-of-terminology strategy is a sophisticated version of zero-shot: dev and test sets contain novel concepts from a target terminology, while another terminology is used during training. For example, models trained to map disease mentions to MeSH concepts and evaluated on drug mentions with DrugBank CUIs. Tutubalina et al. (2020) provided the first cross-terminology evaluation study and showed that knowledge transfer can be effective between diseases, chemicals and genes from abstracts.
In our study, we focus on in-domain and out-of-domain evaluation. We investigate the effectiveness of transferring concept normalization from the general biomedical domain to the clinical trial domain. We trained neural models on the CDR Disease and CDR Chemical train sets for linking clinical conditions and interventions, respectively.
5.4 Experimental setup
We experiment with BioBERTbase v1.1 and PubMedBERTbase both with 12 heads, 12 layers, 768 hidden units per layer and a total of 110 M parameters. Empirical results showed that PubMedBERT achieves 1-2% lower results for all training settings. Due to that fact, we reported only BioBERT results. Epsilon, the number of positive and negative examples, and distance metric were chosen optimally on dev sets. We choose to be the average over all outputs of BERT. We have evaluated different epsilons starting from 0.5 up to 4.0 with 0.5 step for Euclidean distance metric, for cosine distance from 0.05 up to 0.3 with 0.05 step. These experiments have quite similar results. We have evaluated a number of positive and negative examples. For positives we iterated over values from 15 to 35, for negatives from 5 to 15. We found that the optimal is to sample 30 positive examples and 5 negative examples per mention. For the random + parents strategy, we evaluated the number of names of concept’s parents from 1 to 5. Similar, we evaluated the number of names of concept’s siblings from 1 to 5. We found that hard negative sampling (with siblings) achieves the same optima as random negative sampling. The highest metrics are achieved at 5 concept names of the concept’s parents on the CT Condition and CDR Chemical sets. The highest accuracy is achieved at 2 names of the concept’s parents on other sets. As a result, we trained the DILBERT model with Euclidean distance and the following parameters: batch size is equal to 48, learning rate was set to 1e-5, epsilon to 1.0.
We evaluate this solution in information retrieval (IR) scenario, where the goal is to find within a dictionary of concept names and their identifiers the top-k concepts for every entity mention in texts. In particular, we use the top-k accuracy as an evaluation metric, following the previous works (Phan et al., 2019; Pradhan et al., 2014; Sung et al., 2020; Suominen et al., 2013; Tutubalina et al., 2020; Wright et al., 2019). Let Acc@k be 1 if a right CUI is retrieved at rank k, otherwise 0. All models are evaluated with Acc@1. For composite entities, we define Acc@k as 1 if each prediction for a single mention is correct.
5.5 Results
5.5.1 Concept normalization on clinical trials
RQ1 In contrast with the CDR sets, 30.4% and 1.8% of intervention and condition mentions in the CT dataset are not appeared in terminologies, respectively. We investigate different strategies for the out-of-KB prediction (i.e. nil prediction) on clinical trials’ texts.
We tested three strategies for nil prediction on the dev set which containing 100 randomly selected mentions and evaluated the selected threshold values on the test set. This procedure was repeated 20 times. For intervention normalization, the first strategy showed an average accuracy of 79.41 with std of 3.5; second—accuracy of 71.77 and std of 3.5; third—accuracy of 85.73, std of 1.3.
In the first set of experiments, we evaluate the performance of neural models on clinical trials in cross-domain setup.
Table 6 presents the performance of the DILBERT models compared to BioSyn and BioBERT ranking on the datasets of clinical trials. We test the DILBERT model’s transferability on two sets of interventions and conditions where each mention is associated with one concept only (see ‘single concept’ columns). We evaluate the model on test sets with all mentions, including single concepts, composite mentions and out-of-KB cases (see ‘full set’ columns). Several observations can be made based on Table 6. First, DILBERT outperformed BioSyn and BioBERT ranking. Adding randomly sampled positive examples from parent-child relationships gives a statistically significant improvement in 1-2% on the CT Condition set while staying on par with random sampling on interventions. Third, DILBERT models obtained higher results on test sets with single concepts. Models achieve much higher performance for the normalization of interventions rather than conditions.
Table 6.
Out-of-domain performance of the proposed DILBERT model and baselines in terms of Acc@1 on the filtered test set of clinical trials (CT)
| Model | CT condition |
CT intervention |
||
|---|---|---|---|---|
| Single concept | Full set | Single concept | Full set | |
| BioBERT ranking | 72.60 | 71.74 | 77.83 | 56.97 |
| BioSyn | 86.36 | – | 79.58 | – |
| DILBERT with different sampling strategies | ||||
| Random sampling | 85.73 | 84.85 | 82.54 | 81.16 |
| Random + 2 parents | 86.74 | 86.36 | 81.84 | 79.14 |
| Random + 5 parents | 87.12 | 86.74 | 81.67 | 79.14 |
| Resampling | 85.22 | 84.63 | 81.67 | 80.21 |
| Resampling + 5 siblings | 84.84 | 84.26 | 80.62 | 76.16 |
Note: Highest score in a column is marked as bold.
5.5.2 Concept normalization on scientific benchmarks
RQ2 In Table 7, we present in-domain results of models evaluated on the CDR data. In all our experiments when comparing DILBERT and BioSyn models, we use paired McNemar’s test (McNemar, 1947) with a confidence level at 0.05 to measure statistical significance. Table 7 shows that DILBERT outperformed BioSyn on the CDR Disease test set staying on par with BioSyn on the CDR Chemical test set. We compare results on refined test sets with results on the CDR corpus’s official test set. We observe the significant decrease of Acc@1 from 93.6% to 75.8% and from 95.8% to 83.8% for DILBERT on disease and chemical mentions, respectively. Similar to the CT dataset, models achieve much higher performance for the normalization of chemicals rather than diseases.
Table 7.
In-domain performance of the proposed DILBERT model in terms of Acc@1 on the refined test set of the Biocreative V CDR corpus
| Model | CDR disease | CDR chemical |
|---|---|---|
| BioBERT ranking | 66.4 | 80.7 |
| BioSyn | 74.1 | 83.8 |
| DILBERT, random sampling | 75.5 | 81.4 |
| DILBERT, random + 2 parents | 75.0 | 81.2 |
| DILBERT, random + 5 parents | 73.5 | 81.4 |
| DILBERT, resampling | 75.8 | 83.3 |
| DILBERT, resampling + 5 siblings | 75.3 | 82.1 |
Note: Highest score in a column is marked as bold.
5.5.3 Effect of dictionary size at the prediction time
RQ3 Both BioSyn and DILBERT models compute similarities between mentions and concept names at the subword- and word-levels. This can help in linking mentions that look like existing terms like ‘visual defects’ and ‘visual disorder’. However, these models would link mentions to KB incorrectly in two major cases: (i) surface forms of mentions and concept names are similar, yet have a different meaning [e.g. ‘chlorfenac’ (C041190) and ‘chlorferon’ (C305311)], (ii) both expressions share the same meaning, yet are different in surface form [‘metindol’ and ‘indomethacin’ are the same anti-inflammatory drug (D007213)]. Moreover, KBs are could be outdated, and their coverage of synonyms can be very incomplete.
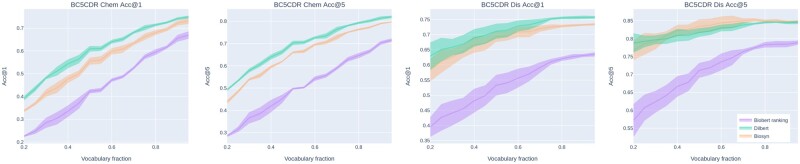
We performed a set of experiments with models trained on CDR Disease & Chemical sets and fragmentary dictionaries at the prediction time. This experiment aims to test how well the model remembered concepts from the training dictionary. To create fragmentary dictionaries, we grouped the initial versions of the dictionaries by CUI and employed a random sample of items using a given fraction of axis items to return. We note that if the number of concept names after sampling turned out to be fractional, then we round the number down of concept names to the smallest integer. For instance, 95% of 10 concept names is 9. The number of fraction ranges from 0.95 to 0.20 with a -0.05 step. We carried out the procedure for reducing the vocabulary, conducted experiments four times and averaged the results. The results are shown in Figure 1. First, These results in terms of Acc@1 demonstrate that degradation in the metrics from the full dictionary to a 30% of the dictionary is significant. The models learned the similarities between mentions and most similar concept names that may be missing at the time of predictions. It is expected that the performance of drug normalization models decreased more than disease normalization models since drug names are highly heterogeneous (there are active compound names, brand names, proprietary identifiers, etc). Second, in terms of Acc@5, models show degradation is smaller. Finally, DILBERT handled modification of the target dictionary without re-training slightly better than other models.
Fig. 1.
In-domain performance of the proposed DILBERT model in terms of Acc@1 on the refined test set of the Biocreative V CDR corpus using reduced dictionaries
5.6 Error analysis
The error analysis on the Clinical Trials set showed that the DILBERT model incorrectly maps 98 mentions: 6 of them are linked to the correct concept’s parent and 7 to the correct concept’s child. In many cases, the correct concept is a broader version of the predicted concept. In particular, the entity ‘Brainstem Glioma’ is linked to the concept D020339 (Optic glioma) whereas the correct concept is D005910 (Gliomas). Moreover, the entity ‘Unspecified Adult Solid Tumor, Protocol Specific’ is linked to D009382 (the unknown primary tumor) but the correct concept is D009369 (neoplasms). It should be noted that the mean length of incorrectly mapped entities is 30.8 chars while the length of correctly mapped entities is 19.9. The reason is that most of the mislinked entities contain some extra information that doesn’t allow map the mention to the correct concept (as in the ‘Brainstem Glioma’ example). Some errors occur due to the model was not trained on structural information on biomedical concepts. For instance ‘Advanced Urothelial Carcinoma’ is linked to D014571 (cancer of the urinary tract) but the actual concept is D001749 (urinary bladder cancer). Here the model correctly recognizes that the concept is related to the cancer disease, but couldn’t relate it to the urinary bladder.
6 Discussion and limitations
DILBERT is a novel model that performs medical concept normalization via deep neural networks, metric learning and negative sampling. In our work, we evaluate how well the neural model recognizes new concepts from a clinical domain that were not present in the dictionary the model was trained with. Our cross-domain experiments demonstrate that the proposed model performs reasonably well and transfers knowledge from a scientific domain to a clinical domain. We have shown how DILBERT improves upon the current state of the art and analyzed the influence of dictionary size, nil prediction cases on the results. We provide a tool for the precise large-scale annotation of clinical records. However, there are still interesting problems and limitations.
First, the model is dependent on concept names in a dictionary used at the prediction time. The continuous flow of new molecular entities coming in clinical trials creates the necessity of timely dictionary updates for the model to be able to improve upon its performance or at least preserve it. We intend to conduct further research on concept discovery and dictionary completion which we consider to be a valuable add-on to the model described.
Second, the model does not take into consideration parent-child concept relations inherent to biomedical ontologies and in particular to the disease ontologies. This leads to the mislabeling of the entities with semantically similar concepts. In future work, it might be interesting to incorporate information on an ontology hierarchy or term co-occurrence graph based on a large collection of texts into the neural models to explicitly introduce multimodal connections between the concepts.
Third, zero-shot evaluation is still an open research direction. Here we present a cross-terminology evaluation that is a more complicated version of zero-shot evaluation due to a shift in domain and dictionary. It would be interesting to investigate how joint training on several entity types and dictionaries affects the MCN performance.
7 Conclusion
In this article, we studied the task of drug and disease normalization in clinical trials. We designed a triplet-based metric learning model named DILBERT that optimizes to pull pairs of mention and concept BioBERT representations closer than negative samples. We precomputed concept name representation for a given terminology to allow fast inference. The model computed a Euclidean distance metric between a given mention and concepts in a target dictionary to retrieve the nearest concept name. The advantage of this architecture is the ability to search for the closest concept in a different terminology without retraining the model. In particular, we trained a model on the CDR Chemical dataset with the CTD chemical dictionary and used it to predict on our drug dictionary. We perform a detailed analysis of our architecture that studies in-domain and cross-domain performance across two corpora as well as the performance on reduced disease and drug dictionaries. Extensive experiments show the competitiveness of the proposed DILBERT model. Moreover, we present an error analysis and discuss limitations. This work suggests several interesting directions for future research. We could train out models jointly on several entity types. The most common entity types are disease, drugs, genes, adverse drug reactions. Moreover, we could leverage an ontology hierarchy or term co-occurrence graph to improve our model.
Financial Support: none declared.
Conflict of Interest: none declared.
Contributor Information
Zulfat Miftahutdinov, R&D department, Insilico Medicine Hong Kong, 999077 Pak Shek Kok, Hong Kong.
Artur Kadurin, R&D department, Insilico Medicine Hong Kong, 999077 Pak Shek Kok, Hong Kong.
Roman Kudrin, R&D department, Insilico Medicine Hong Kong, 999077 Pak Shek Kok, Hong Kong.
Elena Tutubalina, R&D department, Insilico Medicine Hong Kong, 999077 Pak Shek Kok, Hong Kong.
References
- Atal I. et al. (2016) Automatic classification of registered clinical trials towards the global burden of diseases taxonomy of diseases and injuries. BMC Bioinformatics, 17, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M. et al. (2013) Feasibility of feature-based indexing, clustering, and search of clinical trials. Methods Inf. Med., 52, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.S., Patel C.J. (2017) A standard database for drug repositioning. Sci. Data, 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. et al. (2012) Medic: a practical disease vocabulary used at the comparative toxicogenomics database. Database, 2012, bar065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A. et al. (2019) The comparative toxicogenomics database: update 2019. Nucleic Acids Res., 47, D948–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J. et al. (2019) Bert: pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Minneapolis, USA, pp. 4171–4186.
- Dowden H., Munro J. (2019) Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov., 18, 495–496. [DOI] [PubMed] [Google Scholar]
- Gayvert K. et al. (2016) A data-driven approach to predicting successes and failures of clinical trials. Cell Chem. Biol., 23, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. et al. (2016) Emerging role of bioinformatics tools and software in evolution of clinical research. Perspect. Clin. Res., 7, 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillick D. et al. (2019) Learning dense representations for entity retrieval. In Proceedings of the 23rd Conference on Computational Natural Language Learning (CoNLL), Hong Kong, China, pp. 528–537.
- Gu Y. et al. (2020) Domain-specific language model pretraining for biomedical natural language processing. arXiv, preprint arXiv:2007.15779.
- Hao T. et al. (2014) Clustering clinical trials with similar eligibility criteria features. J. Biomed. Inf., 52, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay M. et al. (2014) Clinical development success rates for investigational drugs. Nat. Biotechnol., 32, 40–51. [DOI] [PubMed] [Google Scholar]
- Hoffer E., Ailon N. (2015) Deep metric learning using triplet network. In: International Workshop on Similarity-Based Pattern Recognition, Copenhagen, Denmark. Springer, pp. 84–92.
- Huang C.-C., Lu Z. (2016) Community challenges in biomedical text mining over 10 years: success, failure and the future. Brief. Bioinf., 17, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-S. et al. (2013) Learning deep structured semantic models for web search using clickthrough data. In Proceedings of the 22nd ACM International Conference on Information & Knowledge Management, San Francisco, USA, pp. 2333–2338.
- Humeau S. et al. (2019) Poly-encoders: transformer architectures and pre-training strategies for fast and accurate multi-sentence scoring. CoRR, 2, 2–2. [Google Scholar]
- Ji Z. et al. (2020) Bert-based ranking for biomedical entity normalization. AMIA Summits Transl. Sci. Proc., 2020, 269. [PMC free article] [PubMed] [Google Scholar]
- Johnson J. et al. (2019) Billion-scale similarity search with GPUs. IEEE Trans. Big Data, 7, 535–547. [Google Scholar]
- Leaman R., Lu Z. (2016) Taggerone: joint named entity recognition and normalization with semi-Markov models. Bioinformatics, 32, 2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. et al. (2019) Biobert: pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveling J. (2017) Patient selection for clinical trials based on concept-based retrieval and result filtering and ranking. In: TREC, Gaithersburg, USA.
- Li F. et al. (2019) Fine-tuning bidirectional encoder representations from transformers (BERT)-based models on large-scale electronic health record notes: an empirical study. JMIR Med. Inf., 7, e14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2017) Cnn-based ranking for biomedical entity normalization. BMC Bioinformatics, 18, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu Z. (2012) Systematic identification of pharmacogenomics information from clinical trials. J. Biomed. Inf., 45, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. (2016) Biocreative V CDR task corpus: a resource for chemical disease relation extraction. Database, 2016, baw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. (2017) Learning a recurrent residual fusion network for multimodal matching. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, pp. 4107–4116.
- Lo A. et al. (2019) Machine learning with statistical imputation for predicting drug approvals. Harvard Data Sci. Rev., 1, doi: 10.1162/99608f92.5c5f0525. [Google Scholar]
- Malas T. et al. (2019) Drug prioritization using the semantic properties of a knowledge graph. Sci. Rep., 9, 6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNemar Q. (1947) Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika, 12, 153–157. [DOI] [PubMed] [Google Scholar]
- Miftahutdinov Z., Tutubalina E. (2019) Deep neural models for medical concept normalization in user-generated texts. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics: Student Research Workshop, Florence, Italy, pp. 393–399.
- Miftahutdinov Z. et al. (2021) Drug and disease interpretation learning with biomedical entity representation transformer. In Proceedings of the 43rd European Conference on Information Retrieval, Lucca, Italy.
- Mikolov T. et al. (2013) Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems, Lake Tahoe, USA, pp. 3111–3119.
- Mondal I. et al. (2019) Medical entity linking using triplet network . In: Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, USA, pp. 95–100..
- Phan M. et al. (2019) Robust representation learning of biomedical names. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, pp. 3275–3285.
- Pradhan S. et al. (2014) SemEval-2014 task 7: analysis of clinical text. In: SemEval@ COLING, Dublin, Ireland, pp. 54–62.
- Reimers N., Gurevych I. (2019) Sentence-bert: sentence embeddings using siamese bert-networks. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China, pp. 3973–3983.
- Schroff F. et al. (2015) Facenet: a unified embedding for face recognition and clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, USA, pp. 815–823.
- Sen A. et al. (2018) The representativeness of eligible patients in type 2 diabetes trials: a case study using gist 2.0. J. Am. Med. Inf. Assoc., 25, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M. et al. (2020) Biomedical entity representations with synonym marginalization. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Seattle, USA, pp. 3641–3650.
- Suominen H. et al. (2013) Overview of the share/clef ehealth evaluation lab 2013. In: International Conference of the Cross-Language Evaluation Forum for European Languages, Valencia, Spain, Springer, pp. 212–231. [Google Scholar]
- Tutubalina E. et al. (2018) Medical concept normalization in social media posts with recurrent neural networks. J. Biomed. Inf., 84, 93–102. [DOI] [PubMed] [Google Scholar]
- Tutubalina E. et al. (2020) Fair evaluation in concept normalization: a large-scale comparative analysis for BERT-based models. In Proceedings of the 28th International Conference on Computational Linguistics, Barcelona, Spain, pp. 6710–6716.
- Wishart D. et al. (2018) Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Research, 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. et al. (2019) Estimation of clinical trial success rates and related parameters. Biostatistics, 20, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. et al. (2019) Normco: deep disease normalization for biomedical knowledge base construction. In: Automated Knowledge Base Construction.
- Wu P. et al. (2013) Online multimodal deep similarity learning with application to image retrieval. In Proceedings of the 21st ACM international conference on Multimedia, pp. 153–162.
- Xu D. et al. (2020) A generate-and-rank framework with semantic type regularization for biomedical concept normalization. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Seattle, USA, Association for Computational Linguistics, pp. 8452–8464. https://www.aclweb.org/anthology/2020.acl-main.748.
- Zhao S. et al. (2019) A neural multi-task learning framework to jointly model medical named entity recognition and normalization. Proc. AAAI Conference Artif. Intell., 33, 817–824. [Google Scholar]
- Zhu M. et al. (2020) Latte: latent type modeling for biomedical entity linking. In: AAAI Conference on Artificial Intelligence (AAAI), New York, USA, Vol. 34, pp. 9757–9764. [Google Scholar]