Abstract
We have previously characterized a large panel of provirus insertion Notch1 mutant alleles and their products arising in thymomas of MMTVD/myc transgenic mice. Here, we show that these Notch1 mutations represent two clearly distinct classes. In the first class (type I), proviral integrations were clustered just upstream of sequences encoding the transmembrane domain. Type I Notch1 alleles produced two types of mutant Notch1 RNA, one of which encoded the entire Notch1 cytoplasmic domain [N(IC)] and the other of which encoded a soluble ectodomain [N(EC)Mut] which, in contrast to the processed wild-type ectodomain [N(EC)WT], did not reside at the cell surface and became secreted in a temperature-dependent manner. A second, novel class of mutant Notch1 allele (type II) encoded a Notch1 receptor with the C-terminal PEST motif deleted (ΔCT). The type II Notch1ΔCT protein was expressed as a normally processed receptor [N(EC)WT and N(IC)ΔCT] at the cell surface, and its ectodomain was found to be shed into the extracellular medium in a temperature- and calcium-dependent manner. These data suggest that both type I and type II mutations generate two structurally distinct Notch1 N(EC) and N(IC) proteins that may participate in tumor formation, in collaboration with the c-myc oncogene, through distinct mechanisms. Constitutive type I N(IC) and type II N(IC)ΔCT expression may enhance Notch1 intracellular signaling, while secreted or shed type I N(EC)Mut and type II N(EC) proteins may differentially interact in an autocrine or paracrine fashion with ligands of Notch1 and affect their signaling.
Members of the Notch receptor family are transmembrane glycoproteins, which have been implicated in the mechanisms of differentiation, transformation, dementia, and stroke (reviewed in references 1, 6, 9, 18, and 19). In mammals, there are four identified members of this family, which display highly similar structures. The extracellular domain encodes tandem extracellular epidermal growth factor (EGF) repeats and a cysteine-rich region called the Notch/lin-12 repeat. The cytoplasmic domain of each family member harbors six ankyrin repeats, as well as a C-terminal PEST motif. The Notch protein and many of its identified signaling partners are conserved from Drosophila to humans. Genetic studies on Notch activation in Drosophila (25, 34), Caenorhabditis elegans (38), and Xenopus (11) have collectively suggested that removal of the Notch extracellular domain results in a dominant gain-of-function Notch allele. Similar truncated NOTCH1 alleles have been discovered in sporadic human (2, 14), and retrovirally-induced mouse (16, 17) T-cell leukemias. In addition, in vitro transformation of T cells and fibroblasts has been achieved using various engineered forms of cytoplasmic Notch1 (2, 3, 10, 31). Altogether, these data have given rise to the notion that a constitutively active intracytoplasmic Notch1 protein, N(IC), can operate as an oncoprotein, which has been most frequently observed in T cells.
It is important to fully understand the structure of the Notch1 receptor, in order to predict how the receptor will function in its mutated form. Original studies of the Drosophila Notch receptor initially suggested that the mature Notch polypeptide is a 300-kDa glycoprotein, since antisera to the extracellular and intracellular domains of Drosophila Notch recognized a ∼300-kDa protein that had affinity for several lectins (20, 21). However, antisera that recognize the mammalian Notch1 cytoplasmic domain have consistently detected two species of Notch1 proteins by Western blot analysis: a ∼330-kDa protein and smaller polypeptides ranging from 110 to 89 kDa, depending on the source of proteins (2, 7, 16, 21, 31, 37, 43). A pulse-chase analysis of Notch1 and Notch2 posttranslational processing has revealed that the 330-kDa precursor is rapidly cleaved to give rise to the smaller cytoplasmic proteins (2, 16, 43). More recently, Drosophila Notch (30), human Notch2 (7), and murine Notch1 (26) have been shown to be proteolytically processed from a 330-kDa precursor to a 110-kDa membrane-anchored cytoplasmic chain. Several lines of evidence have suggested that the Notch1 precursor becomes cleaved by the convertase furin at a consensus sequence, which occurs just N-terminal of two conserved cysteines (C1675 and C1682) in the juxtamembrane extracellular domain (22, 26). The resulting cleavage products are believed to form a heterodimer comprising an extracellular domain, N(EC), that is tethered to the cell via its association with the 69-amino-acid extracellular stalk preserved in the cytoplasmic subunit, N(IC). The physical nature of the heterodimer association is not well understood, although the conserved cysteine residues in the extracellular stalk of the cytoplasmic subunit are believed to play an essential role (reviewed in reference 18). Moreover, it has been recently shown that ligand-induced activation of Notch1 can induce additional proteolysis of Notch1 on the cytoplasmic face near the plasma membrane, which releases a shorter Notch1 cytoplasmic subunit for interaction with downstream signaling partners (36). The fate of the extracellular cleavage product from the Notch1 precursor, however, has yet to be rigorously analyzed.
Our previous analysis of T-cell tumors arising in MMTVD/myc transgenic (Tg) mice infected with murine leukemia virus (MuLV) revealed the presence of provirus insertional Notch1 mutations in 65 out of 110 characterized tumors (16, 17). These tumors, and derived T-cell lines, had the potential to express a unique array of spontaneously selected and presumably gain-of-function mutant Notch1 proteins. A majority of the integrations occurred in genomic regions coding for sequences between the 34th EGF repeat and the transmembrane (TM) domain, resulting in the production of what we term here “type I” mutant Notch1 alleles (16, 17). The characterized insertion sites seemed to respect a window of integration, implying that specific sequence requirements had to be met for the oncogenic conversion of Notch1. Almost all of the tumors with type I proviral insertions produced elevated levels of two distinct types of truncated transcripts (16, 17). The 3- to 4-kb RNAs initiated at the integration site and terminated at the 3′ end of the gene and thus encoded the TM and cytoplasmic domains [N(IC)]. Another class of transcripts, measuring 6 to 9 kb, appeared to originate at the Notch1 promoter and terminate at the integration site, thus having the capacity to encode only a truncated Notch1 ectodomain [N(EC)Mut] (16, 17). Practically every tumor bearing this type of rearranged Notch1 allele expressed an abundant 280-kDa protein, in addition to a 330-kDa Notch1 precursor (16, 17). This 280-kDa protein was recognized in Western analyses by extracellularly but not intracellularly directed anti-Notch1 antisera, suggesting that p280 could represent the Notch1 ectodomain (16).
Using an array of Notch1-specific RNA probes and domain-specific Notch1 antisera, we have now tested the possibility that the 280-kDa protein expressed in some of the type I tumors was being produced from a truncated Notch1 RNA template. In this analysis, we also report a second type of proviral integration event in Notch1, which we refer to as the type II mutation. This newly described cluster of integrations occurred within an 800-nucleotide span, at the C-terminus-encoding region of Notch1. The resulting mutant alleles encoded all of the Notch1 receptor sequence except for the C terminus, which harbors a PEST domain. These data are the first to suggest that such a mutation in Notch1 could be oncogenic. One Notch1 mutant allele encodes a disassembled Notch1 receptor that is rendered ligand independent (type I), and the other allele retains a receptor structure capable of ligand interaction (type II). Our data suggest that these two distinct Notch1 mutant alleles can nonetheless cooperate with c-myc and accelerate the appearance of thymomas in MMTVD/myc Tg mice.
MATERIALS AND METHODS
Construction of full-length Notch1 and mutant Notch1 derivatives in expression vectors.
A PCR product corresponding to nucleotides +1 to +565 (using the base pair coordinates of a published complete murine Notch1 cDNA sequence, mmnotcha [GenBank]) was used to screen a λ-ZAP, random-primed murine spleen cDNA library (Stratagene) for Notch1 5′ cDNA sequences, from which clone N1c5 (bp −105 to +406) was isolated after automatic excision (the methods were those specified by Stratagene). Clone N1c27 (bp +256 to +3521) was isolated, using the same PCR probe, from a λ-ZAP murine embryonic kidney 18-day-postcoitum random-primed cDNA library (from J. Pelletier, McGill University). Partial sequencing of each cDNA clone revealed 99% homology to mmnotcha (GenBank), with several silent mutations, except for N1c27, which had a TAT→CAT change (Y→H, codon 75). The full-length murine Notch1 (Notch8.0) cDNA was constructed by ligating the SmaI-DraIII fragment of N1c5 (+50 to +274), the DraIII-ClaI fragment of N1c27 (+274 to +2801), the ClaI-BclI fragment of plasmid pMN4.0 (+2801 to +5553), (33), and the BclI-EcoRI fragment of pKS Motch (+5553 to +8054) (23). The NΔANK plasmid was derived from Notch8.0 by religating a blunt-end NcoI (+5618) to the EcoRV (+6368) site, which deleted amino acids 1848 to 2097. Both Notch8.0 and NΔANK were subcloned into a modified pcDNA3 vector, pcDNA3puro, in which the neomycin resistance gene was replaced with the puromycin resistance gene by exchanging the SmaI-BsmI fragment of pcDNA3 (Invitrogen) with a fragment harboring the puromycin resistance gene from pBabepuro (28) and a simian virus 40 polyadenylation sequence. The N(EC)Nar plasmid was derived from pcDNA3-Notch8.0 by digestion with NarI and religation of a linearized DNA which retained the vector and 1,651 amino acids (aa) of the Notch1 ectodomain. The N(EC)Stu clone was generated by subcloning an EcoRI-StuI fragment from Notch8.0 into pcDNA3(neo), which included 1,468 aa of the 36 Notch1 EGF repeats. The NΔCT plasmid was generated by introducing a stop codon at the HindIII site (aa 2293) of full-length Notch8.0. The integration sites for T28853, L48, T28840, T14418, T14469, T30966, and T3465 were determined by sequencing the chimeric Notch1-proviral PCR product obtained with tumor or cell line genomic DNA, using sense primer 232 (GCTTATGAATTTCACCGTGGGTG at bp 6925 of Notch1 cDNA) and antisense primer 110 from the U3 region of the MuLV LTR, or antisense primer 231 (GAGGAAAGTGGGCTCTGGCAC at bp 7536 of Notch1 cDNA) and sense primer 112 from the U3 region, or sense primer 541 (GAGTGATTGACTACCCGTCAG) from the U5 region of MuLV long terminal repeat (LTR) and antisense primer 540 (GCATCCCACATCTCTGTTTA at bp 7691 of Notch1 cDNA), as previously described (16).
Cell transfection and cell culture.
Isolation and propagation of T-cell lines L42, L45, L46, L48, and L96 have been previously described (16). Lymphocytes were maintained in RPMI 1640 containing 10% fetal bovine serum (Hyclone) and 5 × 10−5 M β-mercaptoethanol at 37°C under 5% CO2. Plasmid DNA was transfected transiently or stably into 293T cells by the calcium phosphate precipitation method, as previously described (5). Individual, stable clones of 293T cells expressing full-length Notch1 (Notch8.0), NΔANK, N(EC)Nar, or pcDNA3puro vector were obtained by selecting colonies after culturing for 2 weeks in 5 μg of puromycin per ml in Dulbecco's minimal essential medium containing 10% fetal calf serum (HyClone). Pools of 293T cells expressing N(EC)Stu or pcDNA3(neo) vector were obtained through selection in 400 μg of G418 per ml for 2 weeks.
Protein extraction and Western blotting.
For Western blot analysis, cells were rinsed twice in 4°C phosphate-buffered saline (PBS) and lysed directly into RIPA buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1.0% sodium desoxycholate) with protease inhibitors (2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 1 μg of pepstatin per ml, 50 μg of 1-chloro-3-tosylamido-7-amino-L-2-heptanone (TLCK) per ml, 100 μg of phenylmethylsulfonyl fluoride per ml). Extracts were cleared by centrifugation at 150,000 × g for 30 min. Protein extracts were mixed with double-strength SDS sample buffer with dithiothreitol or β-mercaptoethanol, boiled for 5 min, subjected to SDS-polyacrylamide gel electrophoresis (PAGE) using 6% acrylamide gels, and transferred to nylon membranes. Protein molecular mass standards including thyroglobulin (∼330 kDa) and myosin (205 kDa) were purchased from Sigma (SDS-6H) and Bio-Rad. Filters were blocked with 5% milk powder in TBST (10 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Tween 20) and then probed with primary antiserum in 0.5% milk powder in TBST. The generation and characterization of Notch1-specific antisera, extra-2, extra-1, intra-1, and intra-2, has been described previously (16). Immunodetection was performed using secondary horseradish peroxidase-conjugated anti-rabbit antiserum (Sigma A0545) followed by chemiluminescent detection (Amersham). After primary Western analyses, blots were incubated in stripping buffer (62.5 mM Tris [pH 6.8], 100 mM β-mercaptoethanol, 2% SDS) for 1 h at 50°C and then reanalyzed with another antiserum. All the resulting autoradiographs were scanned with HP Deskscan II and reproduced for publication using Powerpoint (Microsoft) software.
Trypsinization of cells.
Cells (5 × 107) from each cell line were rinsed in 37°C RPMI, separated into five aliquots for each condition, and incubated for 15 min at 37°C in 5 ml of RPMI containing 0, 0.1, 1, 10, or 100 μg of trypsin per ml. The cells were pelleted and then rinsed twice in 4°C PBS containing phenylmethylsulfonyl fluoride. Final cell pellets were lysed in 250 μl of RIPA buffer containing protease inhibitors. Equal volumes of each lysate were combined with double-strength SDS sample buffer, boiled for 5 min, loaded onto 6% acrylamide–SDS gels, and analyzed by Western blotting with anti-Notch1 antisera.
Notch1 ectodomain dissociation assay.
T lymphocytes (3 × 107 for each condition) were briefly rinsed twice in room temperature PBS and then incubated for 10 min in 250 μl of PBS at 37 or 4°C or 250 μl of PBS with 1 mM EGTA at 4°C. Transiently transfected 293T cells (2 × 106) (48 h posttransfection) were rinsed twice in room temperature PBS and then incubated for 10 min in 100 μl of PBS at 37 or 4°C. After 10 min, the cells were pelleted in a desktop microcentrifuge for 15 s. The supernatant was recovered, and the cell pellet was lysed in RIPA buffer containing protease inhibitors. The supernatant was recentrifuged at 150 × g for 30 min at 4°C, and the pellet was discarded. Equal volumes of each sample were subjected to Western analysis with extra-1 antiserum as described above. The blots were then stripped, reprobed with anti-gp70 followed by horseradish peroxidase-conjugated anti-goat (Sigma) antiserum, and subjected to chemiluminescent detection. The film was scanned and processed for publication using the software mentioned above.
RESULTS
Many type I insertional Notch1 mutant alleles produce truncated Notch1 ectodomain proteins, N(EC)Mut, from truncated RNA and not from processing.
Our analysis of tumors harboring type I Notch1 mutations revealed that some tumors which expressed high levels of truncated 5′ Notch1 RNA and no full-length Notch1 transcripts also produced elevated levels of ∼280-kDa proteins. This suggested that some of the 280-kDa proteins made in these tumors were produced from a truncated Notch1 RNA template and represented a mutated Notch1 ectodomain [N(EC)Mut]. If tumors harboring type I integrations expressed and secreted N(EC)Mut proteins, this could have a potential impact on the transformation mechanism by Notch1. We therefore conducted experiments to test whether N(EC)Mut proteins were expressed, if they resided in a secretory compartment, and whether they became secreted. This analysis was rendered more complicated by the fact that the normal Notch1 receptor, which was retained as an unrearranged allele in most type I tumors, was processed into an ectodomain [N(EC)WT] and an intracytoplasmic domain [N(IC)WT]. N(EC)WT proteins would be expected to have an almost identical structure to N(EC)Mut proteins encoded by the truncated Notch1 RNAs detected in type I tumors. Two tumor cell lines, L96 and L45, were therefore critical in analyzing the putative N(EC)Mut proteins, since these lines harbored distinct type I integrations and abundant, truncated Notch1 5′ RNAs (16) (Fig. 1). Both cell lines were subjected to a Western analysis, using four antisera (16) that have been previously shown to specifically recognize extra- and intracellular epitopes of Notch1 (Fig. 2).
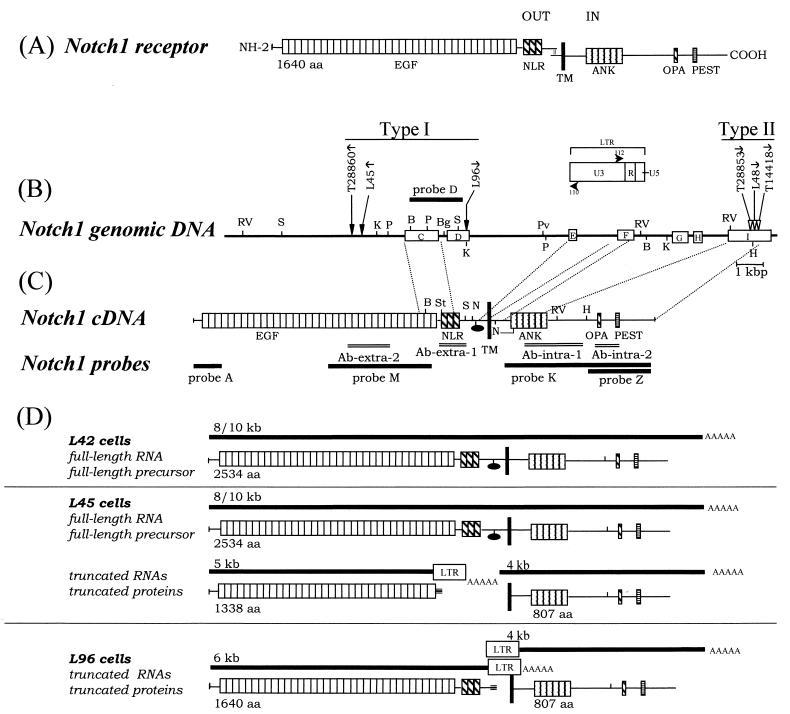
FIG. 1.
Schematic representation of type I and type II provirus insertional Notch1 mutant alleles with their encoded truncated 5′ transcripts and proteins in T-cell tumors arising in MMTVD/myc Tg mice. (A) Simple schematic of the wild-type processed Notch1 receptor at the cell membrane in absence of ligand binding. Cleavage of the molecule has occurred, and the extracellular and intracellular fragments form a complex (vertical double thin lines). Encoded domains are represented: EGF, EGF repeats; NLR, Notch-Lin12 repeats; TM, transmembrane domain; ANK, ankyrin repeats; OPA and PEST, motifs; out, extracellular; in, intracellular. (B) Two distinct clusters of proviral integrations were observed within the middle and C-terminal portions of the Notch1 gene. Mutational insertions were found in tumors (T) and cell lines (L). Type I insertions have been previously mapped (16, 17). Introns are not necessarily drawn to scale, and exons are arbitrarily numbered. PCR and sequencing analysis of exon I between the HindIII site and C terminus revealed no introns. Symbols: thin line, introns; open rectangles, exons; vertical arrows, sites of provirus integration; horizontal arrows, transcriptional orientation of provirus 5′ to 3′. Restriction sites: B, BamHI; Bg, BglII; H, HindIII; K, KpnI; P, PstI; Pv, PvuII; RV, EcoRV; S, SacI. (C) Notch1 cDNA, with encoded domains as in panel A; black oval, furin cleavage consensus sequence. The fragments used for DNA probes (thick lines) and for raising antibodies (double bars) are indicated. Restriction sites are the same as in panel B; also N, NarI; St, StuI. (D) Schematic of truncated Notch1 transcripts (thick lines) detected in distinct T-cell lines with their corresponding primary translational products (16, 17). L42 cells harbor no rearrangement in Notch1, while L45 and L96 cells harbor type I Notch1 mutation. The molecular masses of transcripts (in kilobases) and the number of amino acids in Notch1 are indicated. Symbols: LTR, MuLV LTR; AAAAA, poly(A) tail; triple bar, nonsense coding regions from the LTR region. The normally processed p280 N(EC)WT and p110 N(IC)WT products from the full-length precursor are not illustrated in this section but are shown in panel A.
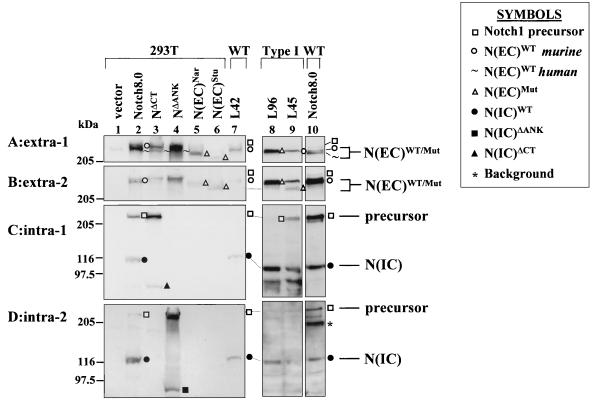
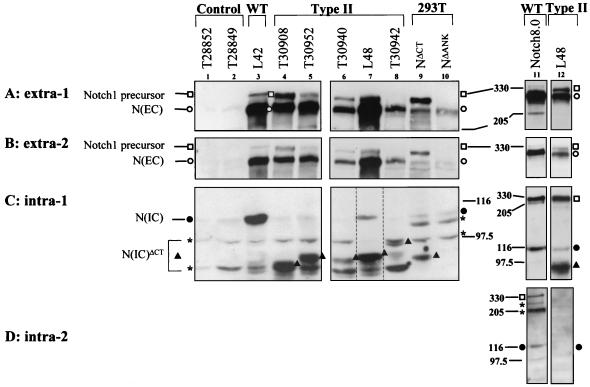
FIG. 2.
Western blot analysis of Notch1 proteins produced by T-cell lines harboring type I Notch1 proviral integrations, compared with full-length and truncated Notch1 receptors ectopically expressed in 293T cells. RIPA cell lysates were subjected to 6% acrylamide SDS-PAGE followed by Western blot analysis using anti-extra-1 (A), anti-extra-2 (B), anti-intra-1 (C), or anti-intra-2 (D) antibodies. The source of protein for individual samples is marked above each lane and includes the following: 293T cells (lane 1) and 293T cells ectopically expressing full-length Notch1 (clone Notch8.0) (lanes 2 and 10) or mutant NΔCT (lane 3), NΔANK (lane 4), N(EC)Nar (lane 5), or N(EC)Stu (lane 6) or T-cell line L42 (lane 7) (wild type [WT] harboring no Notch1 rearrangement), L96 (lane 8), or L45 (lane 9) (both harboring type I proviral insertions in Notch1). The symbols denoting the various Notch1 proteins are indicated in the figure.
L96 cells were previously shown to express a truncated 6-kb Notch1 5′ RNA and no full-length Notch1 transcripts (16) (Fig. 1D). By Western analysis, L96 cells expressed an abundant ∼275-kDa protein that was recognized by extra-1 (Fig. 2A, lane 8; also see Fig. 7), as well as extra-2 (Fig. 2B, lane 8), but by neither of the two antisera recognizing intracellular epitopes (Fig. 2C and D, lanes 8). This p275 protein could therefore have been derived only from the truncated 6-kb Notch1 RNA. L45 cells were previously shown to express both full-length Notch1 8- and 10-kb RNA and, in addition, a 5-kb truncated Notch1 5′ RNA (16) (Fig. 1D). L45 cells expressed a ∼280-kDa protein that was detected with extra-1 and extra-2 (Fig. 2A and B, lanes 9). This 280-kDa protein comigrated with the wild-type processed 280-kDa ectodomain [N(EC)WT] produced by L42 cells, which carry no Notch1 rearrangements (16), and most probably arose from the full-length Notch1 transcripts. In addition, L45 cells expressed a unique ∼250-kDa protein that was detected with extra-2 antiserum (Fig. 2B, lane 9) and, more importantly, undetected with extra-1 antiserum (Fig. 2A, lane 9). The coding potential of the 5-kb Notch1 transcript in L45 cells terminated prior to the extra-1 epitopes (Fig. 1D). A similar ∼250-kDa protein was detected in protein extracts of thymoma T28860, which carried a proviral integration close to that of L45 cells (data not shown). Collectively, these data indicated that N(EC)Mut proteins were stably produced from truncated 5′ Notch1 RNAs in independent T cells or thymomas (see Fig. 7).
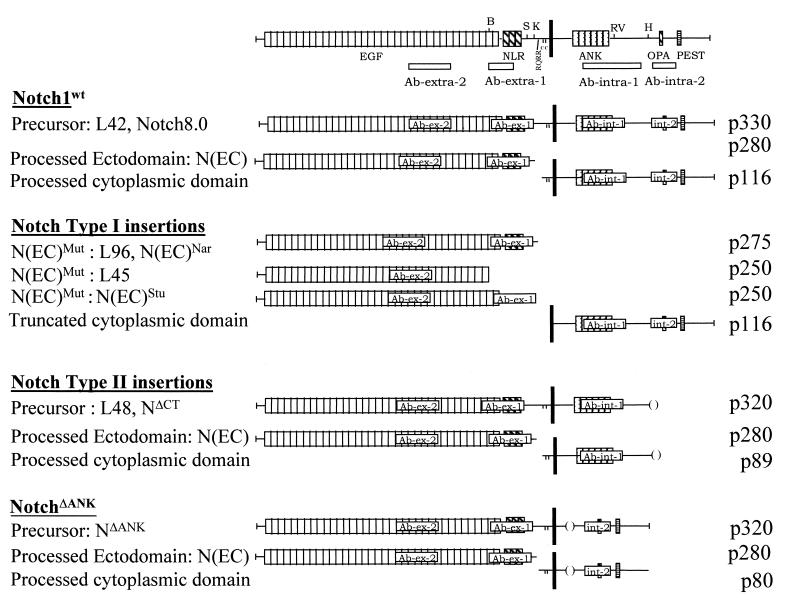
FIG. 7.
Summary of the data on Notch1 proteins studied. A schematic structure for each protein is presented. Individual Notch1 proteins immunodetected in each cell line are listed, along with epitopes present in or absent from each characteristic Notch1 protein, with their distinct molecular masses. The N(EC)WT and N(IC)WT proteins have been reported to associate and to form heterodimers (7, 26). Symbols are the same as in Fig. 1.
The molecular mass (∼250 to 280 kDa) of the truncated N(EC)Mut or processed N(EC)WT ectodomain proteins was much larger than the expected mass of these proteins (∼215 kDa or less, in the absence of posttranslational modifications). Therefore, we constructed two distinct truncated Notch1 cDNAs, N(EC)Nar and N(EC)Stu, which had roughly the same coding potential as the type I mutated Notch1 5′ RNAs to generate an internal molecular weight marker for the Notch1 ectodomain in our SDS-PAGE system. N(EC)Nar encoded the whole 1,651-aa ectodomain preceding the furin cleavage site at aa 1654. N(EC)Stu encoded the 36 EGF repeats (1,468 aa). Each construct was transiently expressed in 293T cells, which express very low levels of the endogenous Notch1 receptor, and the resulting translation products were analyzed by Western blotting (Fig. 2). The protein produced by N(EC)Nar migrated at ∼275 kDa (lane 5), whereas the protein produced by N(EC)Stu (lane 6) migrated faster at ∼250 kDa (Fig. 2A and B). Although the C terminus of the N(EC)Stu protein harbored only part of the extra-1 epitopes, the protein was still detected with extra-1 antiserum (Fig. 2A, lane 6). The molecular masses (∼250 to 280 kDa) of the proteins produced by the mutated alleles in tumors with a type I insertion and by in vitro-constructed Notch1 deletion mutants were in agreement, reinforcing the notion that the ∼250- to 280-kDa N(EC)Mut proteins detected in type I tumors originated from the 5′-truncated RNAs.
To summarize, our Western analysis indicated that the Notch1 ectodomain could be generated by quite different mechanisms: by normal processing of wild-type Notch1 precursor, or from truncated RNAs produced by type I mutant alleles.
In contrast to the processed 280-kDa N(EC)WT proteins, the mutant truncated N(EC)Mut proteins are not located on the cell surface and reside in the secretory compartment.
From the predicted translation products of the truncated Notch1 5′ RNAs, it appeared likely that the N(EC)Mut proteins would be secreted. The predicted coding sequence for these N(EC)Mut proteins included a signal recognition (leader) sequence, which should route these proteins to the secretory pathway. However, these truncated proteins had no TM sequence and would not be expected to lodge in the plasma membrane. We therefore tested whether the N(EC)Mut proteins detected in L96 and L45 cells (harboring the type I Notch1 mutation) resided on the cell surface by treating live cells with increasing amounts of trypsin and then performing a Western analysis. In comparison, we analyzed the trypsin sensitivity of the normally processed Notch1 receptor [N(EC)WT] in L42 cells, which do not harbor an insertional Notch1 mutation. Probing of the resulting immunoblots with extra-1 revealed that p280 N(EC)WT in L42 cells became progressively degraded with increasing amounts of trypsin (Fig. 3A, upper panel). This suggested that nearly all of the processed Notch1 ectodomain detected in L42 cells existed as an assembled receptor at the cell surface. In support of this notion, the p110 N(IC) subunit and Notch1 precursor remained trypsin resistant (Fig. 3A, lower panel). These results were consistent with earlier results (7, 26) showing that extracellular cleavage of the Notch1 precursor occurs before or simultaneously with the export of the processed Notch1 receptor to the cell surface.
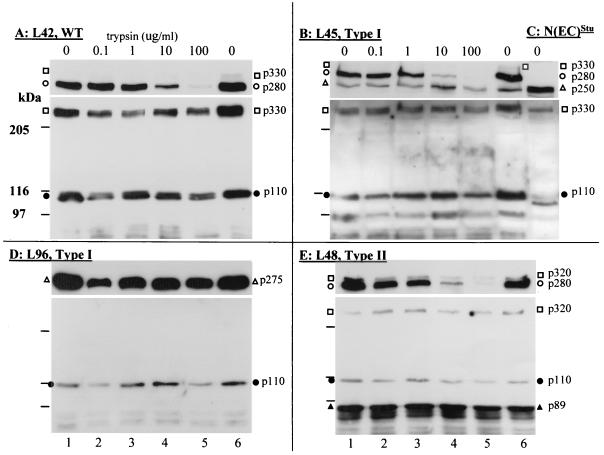
FIG. 3.
Western blot analysis of Notch1 proteins after trypsin treatment of live T cells. The T-cell lines chosen for this analysis include two lines with type I mutation, L45 (B) and L96 (D), and one line with type II mutation, L48 (E). Controls include L42 (A) (wild type [WT], no Notch1 rearrangement) and 293T cells expressing N(EC)Stu as the molecular mass standard (C). The same number of cells (∼107) was used for each trypsin condition. Cells were washed once with RPMI medium without serum at 37°C and then incubated for 15 min at 37°C in RPMI without trypsin (lanes 1 and 6) or with trypsin at 0.1 μg/ml (lane 2), 1 μg/ml (lane 3), 10 μg/ml (lane 4), or 100 μg/ml (lane 5). The cells were washed twice at 4°C in PBS with trypsin inhibitor and immediately lysed in RIPA buffer with protease inhibitors for subsequent Western blot analysis. Immunoblots were first processed with extra-1 (A and D) or extra-2 (B, C, and E) antiserum (upper panel). The blots were then stripped of antisera and reprobed with intra-1 antiserum (lower panel). Symbols for immunoreactive bands are as shown in Fig. 2.
In contrast, the ∼p275 truncated N(EC)Mut in L96 cells appeared to be trypsin resistant (Fig. 3D, upper panel). Furthermore, in L45 cells, the mutant ∼p250 N(EC)Mut was also trypsin resistant, even though the p280 N(EC)WT from the processed Notch1 receptor was trypsin sensitive (Fig. 3B, upper panel). Taken together, these results suggested that the truncated N(EC)Mut proteins are not exposed to the external surface of the plasma membrane and therefore most probably reside in the secretory pathway.
To test whether the truncated ∼p275 N(EC)Mut proteins become secreted, we performed an in vitro assay by incubating live L96 cells in PBS at 37 or 4°C for 15 min and then analyzing the PBS supernatant for the appearance of p275. When live L96 cells were incubated at 37°C but not 4°C, the ∼p275 N(EC)Mut proteins appeared in the PBS medium (Fig. 4C). Reprobing of the Western membrane with intra-1 showed that no N(IC) was released into the PBS medium under any conditions tested (data not shown). Since the truncated N(EC)Mut proteins expressed in L96 cells were not membrane associated (trypsin resistant, Fig. 3D), their presence in the L96-conditioned PBS medium suggests that they have been secreted.
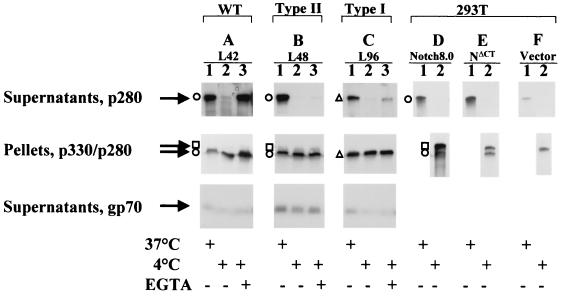
FIG. 4.
Western blot analysis of the Notch1 ectodomain released in the medium of T-lymphocyte lines and transiently transfected 293T cells. Cells tested include one line with type I mutation, L96 (C), and one line with type II mutation, L48 (B). Controls were cells from line L42 (A) (wild type [WT], no Notch1 rearrangement), as well as 293T cells transiently transfected with an expression vector harboring full-length Notch1 (Notch8.0) (D), NΔCT (E), or expression vector alone (F). Cells (3 × 107) were incubated for 10 min in PBS at 37°C (lanes 1), in PBS at 4°C (lanes 2), or in PBS plus 1 mM EGTA at 4°C (lanes 3). For transiently transfected cells, one 100-mm petri dish of cells was transfected with 5 μg of plasmid vector DNA. Two days after transfection, the cells were divided into two aliquots and incubated for 15 min in PBS at either 4 or 37°C, as indicated. After incubation, cells were separated from the supernatants by low-speed centrifugation (1,000 × g) and the supernatants were cleared with a 30-min high-speed centrifugation (150,000 × g). The supernatants were analyzed by Western blotting with extra-1 antiserum (top panel) or with anti-gp70 envelope protein (bottom panel). The cell pellets were extracted with RIPA and analyzed by Western blotting with extra-1 antiserum as a control for the number of cells (middle panel).
A second class of Notch1 mutant alleles (type II) detected in T-cell tumors carries provirus inserted at the 3′ end of the gene, resulting in truncation of 3′-end sequences.
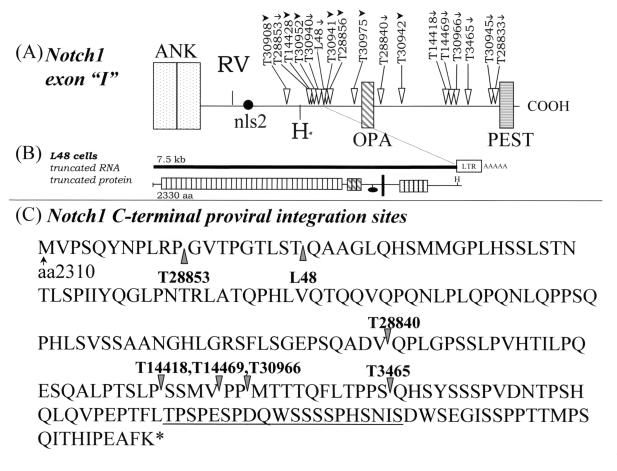
Other provirus integrations were previously detected downstream of the exon encoding the TM domain but were not mapped precisely (16, 17). We have now mapped these 3′-end provirus integrations (type II insertions) by Southern blot analysis with KpnI- or EcoRV-digested tumor DNAs using probe K or Z (Fig. 1C), as previously described (reference 16 and data not shown). The positions and orientations of these newly mapped integration sites are shown in Fig. 5. Further, the precise integration sites in cell line L48 and in tumors T14418, T14469, T30966, T28840, T28853, and T3465 were mapped by PCR and sequence analysis and were found to occur between amino acids 2318 and 2462 (Fig. 5C). The open reading frame of each mutant Notch1 allele was found to terminate at a position corresponding to roughly 20 nonsense amino acids after the integration site, in the U3 region of the MuLV LTR. These provirus insertional mutations formed a distinct cluster, and they represented 29% (19 out of 65) of the total provirus insertional mutations detected in Notch1 (16, 17) (Table 1). These type II mutations always occurred within the exon coding for the fifth and sixth ankyrin repeats, prior to the PEST domain, resulting in a 3′-end truncation of the gene (Fig. 1 and 5). Thus, all of the mutated alleles ultimately conserved the cytoplasmic ankyrin repeats and the second putative nuclear localization signal (nls2) while deleting the C-terminal PEST domain (ΔCT). Intriguingly, all of the type II integrations appeared to be in the sense orientation (Fig. 5A), in contrast to the type I integrations, which occurred in either polarity (16, 17). Nearly all of the tumors harboring the 3′-end type II insertions expressed abundant truncated 6.5- to 7.5-kb transcripts that could have been produced only by the mutated Notch1 allele (Fig. 5B and data not shown). This was specifically verified in cell line L48 by Northern analysis, where a truncated 7.5-kb Notch1 transcript hybridized with probes derived from everywhere along the cDNA (probes A, M, and K) (16) but not with a probe (probe Z) covering the last 1,000 bp (data not shown). Therefore, the 3′-end type II insertional mutations found in the Notch1 gene of a number of T-cell tumors are distinct from the type I insertions previously found near the TM domain and are expected to encode different gene products.
FIG. 5.
Schematic representation of type II proviral insertions in the Notch1 gene 3′ coding region (exon “I”). (A) Horizontal arrows show the orientation of provirus integrations as assessed by Southern blot analysis (arrowheads) or by PCR or sequencing of genomic DNA (arrows). The PCR primers used within the MuLV LTR are shown in Fig. 1B (see Materials and Methods). All other tumors were predicted to have sense proviral integrations based on Southern blot analysis, as previously described (16). (B) Schematic of truncated Notch1 transcript detected in L48 cells harboring the type II Notch1 mutation. (C) Carboxy terminus of type II mutant Notch1 proteins predicted from the site of integration. The actual translational stop signal occurred within 25 nonsense amino acids for each protein, in the U3 region of the MuLV LTR. The PEST motif is underlined. Symbols: ∗, C terminus of Notch1; vertical arrowheads, the point at which a provirus interrupted the Notch1 open reading frame for individual mutated Notch1 alleles.
TABLE 1.
Frequency of Notch1 rearrangement
| Tumor | No. of NDa | No. of NCb | No. of:
|
% type II/all Notch1 insertionsc | |
|---|---|---|---|---|---|
| Type I | Type II | ||||
| First group | |||||
| CD1 Notch1+/+ | 30 | 1 | 24 | 6 | 19 (6/31) |
| Second group | |||||
| CD1 Notch1+/+ | 6 | 1 | 8 | 6 | 40 (6/15) |
| CD1 Notch1+/− | 7 | 2 | 10 | 7 | 37 (7/19) |
| Total tumors | 43 | 4 | 42 | 19 | 29 (19/65) |
ND, no integration detected by Southern analysis of digested tumor DNA with probe D (all tumors tested) or probe K (not all tumors from the first group were tested with probe K; therefore the actual number of type II integrations in the first group may be underestimated).
NC, integration detected that was neither type I nor type II.
Frequency of C-terminal integration of provirus among all of the Notch1 integrations detected for that particular sample group.
The novel type II insertional Notch1 mutant alleles code for C-terminally truncated Notch1 receptors with deletions of the PEST domain, which become normally processed.
Our characterization of the new cluster of type II integrations suggested that the encoded mutant Notch1 protein could be a novel oncogenic form of Notch1. To decipher the mutant Notch1 protein structure, we studied the T-cell line L48 that we previously established (16) and that was subsequently discovered to harbor a type II insertional Notch1 mutation. We conducted a Western analysis of Notch1 proteins expressed in L48 cells. The L48 Notch1 precursor (Fig. 6A and B, lanes 7 and 12) appeared truncated, migrating slightly faster (at ∼320 kDa) than the wild-type receptor (330 kDa) of L42 cells (lanes 3) or than the full-length Notch1 receptor in 293T cells (lanes 11). This ∼320-kDa precursor was not detected with intra-2 (which recognizes the C-terminal OPA domain) (Fig. 6D, lane 12), indicating that the precursor lacked the C terminus.
FIG. 6.
Western blot analysis of Notch1 proteins expressed by thymomas harboring type II proviral integrations. Proteins from frozen tumors or from cells in culture were extracted in RIPA buffer with protease inhibitors and then subjected to Western analysis using extra-1 (A), extra-2 (B), intra-1 (C), and intra-2 (D) antisera. Approximately 80 μg of total extract was loaded in each lane. Extracts were from control tumor cells with no detectable Notch1 integration, namely thymomas T28852 (lane 1) and T28849 (lane 2) and cell line L42 (wild type [WT]) (lane 3) and from thymomas with type II integration, including thymomas T30908 (lane 4), T30952 (lane 5), T30940 (lane 6), and T30942 (lane 8) and one cell line, L48 (lanes 7 and 12). Other controls included 293T cells ectopically expressing NΔCT (lane 9), NΔANK (lane 10), and full-length Notch1 (clone Notch8.0) (lane 11). The type II tumor samples are presented from the most N-terminal (lane 4) to the most C-terminal (lane 8) integration. Lane 7 in panel C (bracketed with dotted lines) had a shorter exposure time. The p320 Notch1 precursor was clearly visible on a longer exposure of this lane (not shown). Symbols are the same as in Fig. 2. N(IC)ΔCT proteins are demarcated by a symbol on the right.
Two distinct N(IC) cytoplasmic cleavage products were detected in L48 cells with the intra-1 antiserum: p110, which probably arose from the unrearranged allele, and a much more abundant p89, most probably encoded by the mutant allele (Fig. 6C, lanes 7 and 12). The 89-kDa protein was not recognized by intra-2 (Fig. 6D, lane 12), suggesting that this protein was a cytoplasmic Notch1 protein with a C-terminal truncation [N(IC)ΔCT]. The L48 mutant Notch1 allele also expressed a normal 280-kDa ectodomain [N(EC)] (Fig. 6A and B, lanes 7 and 12) that comigrated with wild-type p280 found in the control unrearranged cell line L42 (lanes 3) and in transiently transfected 293T cells expressing full-length Notch1 DNA (lanes 11). These data suggested that the abundant 280- and 89-kDa proteins detected in L48 cells are the processed N(EC) and N(IC)ΔCT domains, respectively, generated from the truncated precursor (Fig. 7). These data have been further supported by immunoprecipitation and Western analysis of L48 cell extracts, where immunoprecipitation with an ankyrin repeat-directed antiserum, intra-1, was capable of coprecipitating N(IC) with a 280-kDa protein (16), the latter of which was recognized by extracellularly directed antisera (results not shown). Together, our data indicated that the C-terminal OPA and PEST sequences are not required for receptor processing.
To determine whether other tumors harboring type II proviral insertion mutations produced a mutant Notch1 receptor protein, a Western analysis was conducted on four thymomas with distinct type II C-terminal insertions (Fig. 5). The results of this analysis (Fig. 6, lanes 4 to 6 and 8) were similar to those obtained for L48 cells. The intra-1 antiserum detected truncated N(IC)ΔCT proteins that ranged in molecular mass from ∼70 to 90 kDa, in good agreement with the mapped integration sites (Fig. 5A). The tumors also produced a 110-kDa cytoplasmic Notch1 protein and a 280-kDa ectodomain, N(EC). The bulk of p280 could be predicted to arise from processing of the truncated Notch1 receptor. The p110 most probably corresponded to the processed N(IC) from the unrearranged allele in the tumor cells. The stromal cells and multiclonal cells within the tumor may also have expressed wild-type Notch1 and thus contributed to the signals observed at p280 and p110. These data showed that the 3′-truncated Notch1 alleles produced fairly abundant levels of processed, mutant Notch1 receptors.
To independently confirm the expression profile arising from the C-terminally truncated Notch1 receptor, we constructed a C-terminal deletion mutant of the murine Notch1 receptor, NΔCT, deleting all sequences beyond the HindIII site (Fig. 1B and 5A), and analyzed the ectopically expressed proteins in 293T cells. Simultaneously, as a control, we also analyzed a completely different Notch1 deletion mutant in which the ankyrin repeats were missing (NΔANK). The NΔANK mutation has been previously characterized in Drosophila as having dominant loss of function (25, 34). High-level expression of each clone was achieved in either transient (Fig. 2, lanes 3 and 4; Fig. 6, lanes 9 and 10) or stable (data not shown) transfection of 293T cells, which expressed very low levels of endogenous Notch1 (Fig. 2A, lane 1). The NΔCT and NΔANK constructs gave rise to a truncated Notch1 precursor that was recognized by both of the extracellularly directed antisera and one of each intracellularly directed antisera (Fig. 2, lanes 3 and 4). Both NΔCT and NΔANK constructs gave rise to a processed 280-kDa N(EC) that was recognized by extra-2 and extra-1 but not by intra-1 or intra-2 (lanes 3 and 4). As observed for ectopically expressed full-length Notch1, p280 appeared as a doublet in the Western blot of 293T cells expressing the deletion mutants NΔCT or NΔANK when analyzed with anti-extra-1 (Fig. 2A, lanes 3 and 4), which can recognize both murine and human Notch1 epitopes (unpublished observations). This p280 doublet was also observed for ectopically expressed full-length Notch1 (lane 2). These data suggested that high-level expression of ectopic processed Notch1 receptor, either full-length or cytoplasmic deletion mutants, was augmenting and stabilizing the expression of the endogenous human Notch1 receptor in 293T cells. Of note, this apparent stabilization was not seen with the ectopically expressed N(EC)Nar or N(EC)Stu proteins (lanes 3 and 4). The processed cytoplasmic domain produced by each deletion mutant was shorter (∼80 to ∼89 kDa) and lacked the deleted epitopes: N(IC)ΔANK lacked intra-1 epitopes, and N(IC)ΔCT lacked intra-2 epitopes (Fig. 2C and D, lanes 3 and 4; Fig. 6C, lanes 9 and 10). Since each construct gave rise to one species of RNA in transfected cells (data not shown), this ruled out the possibility that an artifactual alternative splicing of the transcript produced from the cDNA was somehow giving rise to the truncated proteins. These results strongly indicated that p280 and p80/89 corresponded to the normally processed N(EC) and truncated N(IC) subunits, respectively. Moreover, these data demonstrated that Notch1 receptor processing can occur in the absence of the ankyrin repeat or of the C-terminal domain.
The processed 280-kDa N(EC) proteins generated from C-terminally truncated type II mutant Notch1 precursor are expressed on the cell surface, as are the N(EC)WT proteins.
As shown above, the type II C-terminally truncated Notch1 receptors appeared to become normally processed. To confirm that these receptors were expressed on the cell surface, live L48 cells were treated with increasing amounts of trypsin to degrade extracellular proteins, and the resulting cellular proteins were analyzed by Western blotting with extra-1 or intra-1. The results of this analysis revealed that processed p280 N(EC) protein expressed by L48 cells was progressively degraded by increasing amounts of trypsin (Fig. 3E, upper panel). The C-terminally truncated precursor, ∼p320, and the truncated N(IC) subunit, p89, remained trypsin resistant, as revealed by the stable presence of these proteins in the intra-1 immunoblot (Fig. 3E, lower panel). Iodination of cell surface proteins of L48 cells confirmed that the processed p280 resided on the cell surface, since the p280/N(EC) protein but not the p320, p110, or p89 Notch1 proteins, became strongly iodinated (data not shown). These data showed that the processed N(EC) resides at the cell surface of L48 cells.
The processed Notch1 p280 type II ectodomain N(EC) proteins are shed from transformed T lymphocytes, in a temperature-dependent and calcium-sensitive manner.
It is likely that the proteolytic cleavage that generates the Notch1 ectodomain, which we described as p280 N(EC), occurs at an extracellular, juxtamembrane site of the precursor (26). Our data are consistent with this argument, since the molecular mass of the furin processed N(EC) protein (1,654 aa) was nearly identical to that of the truncated N(EC)Mut produced in L96 cells (1,640 aa) and to that of the N(EC)Nar mutant generated in vitro (1,651 aa). As a strictly extracellular protein, it was possible that the processed Notch1 ectodomain might be able to dissociate from the cell surface under certain circumstances. To test this hypothesis, the supernatants of Notch1-expressing cells were analyzed for shed p280 after incubation under various conditions. Since Notch1 receptor adhesion is calcium dependent (15), we chose to look for ectodomain dissociation in PBS–1 mM EGTA at 4°C to test whether calcium could be mediating the association of N(EC) with the cell surface. Incubation of the T cells at 37°C with 1 mM EGTA provoked a high level of cell death, which precluded testing this condition. The cells were also incubated for 10 min in PBS at 37 or 4°C. The cell supernatants were analyzed by Western blotting for the release of processed N(EC) into the PBS supernatant (Fig. 4).
Soluble p280 was released into the medium after cells were incubated at 37°C in PBS, regardless of whether the cells expressed a full-length Notch1 receptor (control L42 cells) (Fig. 4A, lane 1) or a C-terminally truncated receptor (L48 cells) (Fig. 4B, lane 1). Neither the Notch1 precursor nor the Notch1 cytoplasmic domain was detected in the media under any conditions, showing that the cells were not simply becoming lysed and releasing cellular contents into the PBS (data not shown). We showed above that practically all of the processed Notch1 p280 expressed in L42 and L48 cells was trypsin sensitive and thus present at the cell surface (Fig. 3A and E). Therefore, the p280 found in the PBS medium from L42 and L48 cells was likely to have been shed, not secreted. Very little p280 was released from the cells during a 4°C incubation (Fig. 4A and B, lanes 2), indicating that release of p280 from the cells was temperature dependent. However, the addition of EGTA to the 4°C incubation resulted in a measurable release of p280 from both L42 and L48 cells (lanes 3). These data suggested that a portion of the p280 detected on the cell surface was bound via a calcium-sensitive interaction. Under all the conditions tested, a large portion of the immunoreactive p280 remained associated with the cell pellet (Fig. 4, middle panel). The differential amounts of p280 released by the cells under the various conditions were not seen for a distinct viral protein that is constitutively shed (42), the MuLV envelope gp70 protein (Fig. 4, bottom panel).
The same assay was conducted on 293T cells transiently transfected with full-length Notch1 (Notch8.0), C-terminally-truncated Notch1 (NΔCT), or expression vector alone. In all three groups of cells incubated at 37°C, but not at 4°C, the presence of p280 in the medium could be detected (Fig. 4D to F). Significantly, the ectodomain of the human Notch1 protein expressed endogenously by 293T cells was also observed to be released at 37°C (Fig. 4F, lane 1).
In summary, our data revealed that the processed Notch1 p280 N(EC) generated from the wild-type ∼p330 precursor or from the type II C-terminally truncated mutant ∼p320 precursor behave similarly and are both released into the extracellular medium under physiological conditions.
DISCUSSION
Two distinct Notch1 proviral insertional mutant alleles are associated with the development of murine T-cell leukemia.
We report here a comparative analysis of two distinct clusters of proviral insertional mutations of the Notch1 gene separated by approximately 20 kbp. One cluster, previously reported (16), occurred within a 3-kbp domain in the extracellular-encoding region prior to the TM domain (type I), whereas the second newly identified cluster occurred within an 800-bp segment of the exon coding for the Notch1 C terminus (type II). Several features distinguish these two types of insertional Notch1 mutants. First, in type I mutants, the proviruses are inserted in both sense and antisense orientations around the TM-coding region, whereas in all 16 type II C-terminal mutants analyzed, the provirus was integrated in the sense orientation. This indicates a strong selection for such a sense orientation of the provirus and suggests that the LTR may be required to function as a promoter (promoter insertion), although the putative 1.3-kb 3′-end RNAs which could be synthesized from the LTR promoter have not yet been detected. Second, the truncated cytoplasmic N(IC) type I mutant proteins are synthesized from a truncated 3.5- to 4.0-kb RNA, while the N(IC)ΔCT type II mutant proteins are generated by receptor processing. Third, before any cytoplasmic processing, the N(IC) mutant proteins produced from the type I mutant alleles encode the TM domain and the complete cytoplasmic domain while the N(IC)ΔCT type II mutant proteins harbor the extracellular stalk, the TM domain, and the cytoplasmic domain with the PEST motif deleted. Fourth, N(IC) type I mutant proteins are produced constitutively under the regulation of a surrogate promoter (LTR or cryptic, respectively, for sense and antisense provirus orientations), while N(IC)ΔCT type II C-terminally truncated mutant proteins are made under the regulation of the Notch1 promoter itself. Finally, the N(EC)Mut truncated proteins produced by type I mutant alleles are trypsin resistant and appear to reside not at the cell surface but, rather, in the secretory pathway. In contrast, the N(EC) proteins produced from the C-terminally truncated type II mutant receptor appear to be normally processed and expressed at the cell surface (trypsin sensitive). Therefore, type I and type II Notch1 mutant proteins generated in this biological system have very different structures.
Removal of PEST as a novel mechanism to activate Notch1 oncogenically.
C-terminal truncating mutations in Notch1 or in other members of the mammalian Notch family have not been previously reported. However, the type II C-terminal truncations arising in our thymomas closely resemble the Drosophila mutants N60g11 and l(l)N3, both of which carry a deletion of the Drosophila Notch OPA and PEST sequences (8, 27). Excision of just the OPA domain, but not the PEST domain, from the full-length Drosophila Notch receptor generates a protein that behaves similarly to the wild-type receptor (25, 34). The existing data would thus seem to indicate that removal of the PEST domain, and not the OPA domain, is what gives rise to mutant Notch1 receptor activity. Controversy exists whether l(l)N3 and N60g11 alleles behave as gain-of-function mutations (4, 27) or not (8). If NΔCT behaved as a hypomorphic allele in our system, a higher incidence of type II than type I mutations might have been expected in a Notch1+/−-deficient background; however, we did not observe such bias (Table 1) (17). Moreover, the wild-type Notch1 allele is retained and expressed by tumors harboring type II C-terminal insertions, suggesting that the NΔCT mutations behave as dominant alleles. In further support of this argument, the evidence implicating Notch1 in T-cell tumor formation is heavily biased toward the notion that constitutive activation of Notch1 can give rise to an oncogenic protein (2, 10, 13, 31). Based on the high frequency of proviral integration within the Notch1 C terminus in tumors arising in MMTVD/myc Tg mice, it is arguable that type II NΔCT alleles, like the truncated N(IC) Notch1 mutants (2, 3, 14, 16, 17), are gain-of-function mutations involved in tumor formation in collaboration with the c-myc transgene.
There are several possible mechanisms by which C-terminally truncated Notch1 mutants may participate in T-cell transformation. It seems logical to presume that deletion of the PEST sequences could contribute to the generation of an oncogenic form of Notch1. Removal of the PEST domain from the c-fos proto-oncogene contributes to its oncogenic activation (40). Deletion of PEST sequences from various proteins is known to stabilize and to augment their levels (reviewed in reference 35). This also appears to be the case with the Notch1 intracytoplasmic domain [N(IC)ΔCT], since high levels of the C-terminally truncated N(IC)ΔCT (89 kDa) were detected in L48 cells and in other tumors harboring C-terminal provirus integrations. Furthermore, a previous pulse-chase analysis of the mutant Notch1 receptor expressed in L48 cells showed that the truncated N(IC)ΔCT, p89, had a longer half-life than did the coexpressed wild-type p110 N(IC) protein (16). From the analysis of the various Notch1 insertional mutants, it appears that constitutively high levels of N(IC) proteins are required for transformation. This could be achieved by overexpression of a truncated Notch1 3′ end RNA (type I mutations) or by deletion of the PEST domain (type II mutations). Thus, by seemingly different mechanisms, two structurally different mutant Notch1 alleles may have the same final impact on cytoplasmic signaling.
It is also possible that deletion of the Notch1 C terminus could influence Notch1 signaling apart from simply augmenting N(IC) protein levels. The region C-terminal of the ankyrin repeats is known to contain a binding site for the products of the dishevelled (dsh) and the numb genes. Dsh is a downstream component of the wingless signaling pathway, and Numb is an asymmetrically localized cell fate determinant. Both gene products inhibit Notch signaling by binding to its C-terminal domain (4), more specifically its PEST domain in the case of Numb (41). Deletion of its Dsh binding domain or its PEST domain (binding Numb) allows Notch to escape suppression by Dsh (4) or Numb (41), respectively. Moreover, ectopic expression of the C-terminal Notch domain alone in transgenic flies was sufficient to suppress Dsh-dependent bristle production, presumably by competitively binding the inhibitory Dsh protein from the wild-type Notch receptor (4). Since 100% of the type II C-terminal integrations analyzed here occurred in the sense orientation, this opens the possibility that 3′ transcripts are made from the LTR promoter and actively produce C-terminal Notch1 proteins. Such proteins would be expected to bind to and titrate out Dsh-like proteins and inhibit their function, as reported for Drosophila (4). However, we have so far been unable to detect such transcripts or proteins in tumors or cells producing type II C-terminally truncated Notch1 receptors. Such short PEST-containing Notch1 proteins could be active at relatively low levels. Future work should be performed to determine whether this pathway is involved in cell transformation.
Fate of the wild-type and mutant Notch1 ectodomains.
Our results with trypsinized T cells are consistent with a proposed model suggesting that the Notch1 receptor precursor resides in a subcellular compartment and that upon processing, the receptor transits to the cell surface (7, 26, 30). It is furthermore possible that the mutated N(IC) proteins have undergone additional processing events on their cytoplasmic face, as has been observed by Kopan's group (12, 22, 36). We have found that the truncated N(EC)Mut proteins remain trypsin resistant, even though their structures closely resemble the processed p280 N(EC)WT subunit. These data suggest that stable expression of the wild-type ectodomain at the cell surface can possibly be achieved only with normal processing and subsequent association with the N(IC) cytoplasmic subunit. Our coimmunoprecipitation analysis has shown that a portion of the processed p280 N(EC)WT remains associated with the intracellular N(IC) or N(IC)ΔCT fragment, as a heterodimer (16). Since the Notch1 receptor with a deletion in the ankyrin repeat domain also produced the processed N(EC) and N(IC)ΔANK subunits, it is likely that this receptor also resides at the cell surface as an assembled heterodimer. Our data therefore agree with the results obtained by others with the wild-type Notch2 (7) and Notch1 (26) proteins and extend them to Notch1 receptors carrying cytoplasmic deletions.
Another unusual property of the N(EC) proteins was observed in the course of our dissociation assays. We found that under physiological conditions, the N(EC) proteins processed from wild-type or C-terminally truncated type II mutant precursors were shed while N(EC)Mut generated from type I mutants were secreted at a significant rate in the medium of expressing cells (Fig. 4). Shedding and secretion occurred in a temperature-sensitive manner, and at least a portion of the ectodomain released could be enhanced by the chelation of calcium ions from the medium. Shedding of N(EC) from the type II C-terminally truncated Notch1 receptor could give rise to an uncoupled cytoplasmic domain that would resemble previously identified oncogenic versions of Notch1 (2, 10, 14, 16, 31). It is interesting that the truncated Notch1 proteins produced by type I alleles, N(EC)Mut and N(IC), are in fact the individual components of a decoupled Notch1 receptor as well.
The notion that both Notch1 (Fig. 4) and its ligand Delta (32) may naturally shed their ectodomains in different physiological contexts adds another intriguing but complex facet to the Notch signaling cascade. It has recently been reported that soluble forms of Drosophila Delta are generated by cleavage at the cell surface and that these forms act as an agonist of Notch activity (32). In contrast, artificially truncated and secreted ectodomain from the Notch ligands Delta and Serrate were found to antagonize Notch signaling (39). Similar truncation mutations of the human homologue of Serrate, Jagged 1, found in patients with Alagille syndrome, are likely to yield extracellular secreted forms and to antagonize Notch1 signaling (24, 29). Together, these data suggest that precise processing of Delta is essential to producing an agonist soluble form. Since Notch and its ligand have the same general structure, it is of interest that the soluble forms of the Notch ligands previously studied are very similar to the soluble forms of Notch1 ectodomain generated by type I and type II mutations in our system. If the same paradigm applies for Notch as for its ligands, the observed extracellular shedding of the processed N(EC) from type II tumors would be expected to stimulate signaling of the ligand(s) of Notch1, whereas secretion of the truncated N(EC)Mut ectodomain from type I tumors would be expected to antagonize the signaling of the ligand(s) of Notch1. Such an enhanced or decreased signaling of possibly more than one ligand of Notch1 distributed on the tumor cell itself (autocrine effect) or on other stromal cells (paracrine effect) could somehow contribute to the tumor development. This would represent a new role for the soluble Notch1 ectodomain in the complex mechanism of tumor formation.
ACKNOWLEDGMENTS
This work was supported by grants to P.J. from the Medical Research Council of Canada and from the National Cancer Institute of Canada.
We thank Benoît Laganière and Ginette Massé for excellent technical assistance. We are grateful to Jerry Pelletier (McGill University) for providing the kidney cDNA library and to Rita Gingras for typing the manuscript.
REFERENCES
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the tan-1 gene, a human homolog of drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 4.Axelrod J D, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between wingless and notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- 5.Balsalobre A, Jolicoeur P. Fos proteins can act as negative regulators of cell growth independently of the fos transforming potential. Oncogene. 1995;11:455–465. [PubMed] [Google Scholar]
- 6.Blaumueller C M, Artavanis-Tsakonas S. Comparative aspects of Notch signaling in lower and higher eukaryotes. Perspect Dev Neurobiol. 1997;4:325–343. [PubMed] [Google Scholar]
- 7.Blaumueller C M, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 8.Brennan K, Tateson R, Lewis K, Martinez Arias A. A functional analysis of Notch mutations in Drosophila. Genetics. 1997;147:177–188. doi: 10.1093/genetics/147.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos-Ortega J A. Numb diverts notch pathway off the tramtrack. Neuron. 1996;17:1–4. doi: 10.1016/s0896-6273(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 10.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffman C R, Skoglund P, Harris W A, Kintner C R. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- 12.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 13.Diévart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 14.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 15.Fehon R G, Kooh P J, Rebay I, Regan C L, Xu T, Muskavitch M A T, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 16.Girard L, Hanna Z, Beaulieu N, Hoemann C D, Simard C, Kozak C A, Jolicoeur P. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 17.Girard L, Jolicoeur P. A full-length Notch1 allele is dispensable for transformation associated with a provirally activated truncated Notch1 allele in Moloney MuLV-infected MMTV(D)/myc transgenic mice. Oncogene. 1998;16:517–522. doi: 10.1038/sj.onc.1201562. [DOI] [PubMed] [Google Scholar]
- 18.Greenwald I. Structure/function studies of lin-12/Notch proteins. Curr Opin Genet Dev. 1994;4:556–562. doi: 10.1016/0959-437x(94)90072-b. [DOI] [PubMed] [Google Scholar]
- 19.Gridley T. Human genetics. Notch, stroke and dementia. Nature. 1996;383:673. doi: 10.1038/383673a0. [DOI] [PubMed] [Google Scholar]
- 20.Johansen K M, Fehon R G, Artavanis-Tsakonas S. The notch gene product is a glycoprotein expressed on the cell surface of both epidermal and neuronal precursor cells during Drosophila development. J Cell Biol. 1989;109:2427–2440. doi: 10.1083/jcb.109.5.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd S, Baylies M K, Gasic G P, Young M W. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev. 1989;3:1113–1129. doi: 10.1101/gad.3.8.1113. . (Erratum, 3:2020.) [DOI] [PubMed] [Google Scholar]
- 22.Kopan R, Schroeter E H, Weintraub H, Nye J S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopan R, Weintraub H. Mouse notch: expression in hair follicles correlates with cell fate determination. J Cell Biol. 1993;121:631–641. doi: 10.1083/jcb.121.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Krantz I D, Deng Y, Genin A, Banta A B, Collins C C, Qi M, Trask B J, Kuo W L, Cochran J, Costa T, Pierpont M E, Rand E B, Piccoli D A, Hood L, Spinner N B. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 25.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 26.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah N G, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyman D, Young M W. Further evidence for function of the Drosophila Notch protein as a transmembrane receptor. Proc Natl Acad Sci USA. 1993;90:10395–10399. doi: 10.1073/pnas.90.21.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgenstern J P, Land H. A series of mammalian expression vectors and characterisation of their expression of a reporter gene in stably and transiently transfected cells. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.4.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oda T, Elkahloun A G, Pike B L, Okajima K, Krantz I D, Genin A, Piccoli D A, Meltzer P S, Spinner N B, Collins F S, Chandrasekharappa S C. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 30.Pan D, Rubin G M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 31.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of t cell neoplasms in mice transplanted with bone marrow expressing activated notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi H, Rand M D, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the Notch ligand Delta by the metalloprotease kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 33.Reaume A G, Conlon R A, Zirngibl R, Yamaguchi T P, Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- 34.Rebay I, Fehon R G, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 35.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 36.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 37.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 38.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 39.Sun X, Artavanis-Tsakonas S. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development. 1997;124:3439–3448. doi: 10.1242/dev.124.17.3439. [DOI] [PubMed] [Google Scholar]
- 40.Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E, Kishimoto T, Inagaki M, Okazaki K, Sagata N, et al. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol Cell Biol. 1995;15:5682–5687. doi: 10.1128/mcb.15.10.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakamatsu Y, Maynard T M, Jones S U, Weston J A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to Notch-1. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 42.Yu Y, Wong P K. Studies on compartmentation and turnover of murine retrovirus envelope proteins. Virology. 1992;188:477–485. doi: 10.1016/0042-6822(92)90501-f. [DOI] [PubMed] [Google Scholar]
- 43.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]