Abstract
Long-read sequencing has the potential to transform variant detection by reaching currently difficult-to-map regions and routinely linking together adjacent variations to enable read based phasing. Third-generation nanopore sequence data has demonstrated a long read length, but current interpretation methods for its novel pore-based signal have unique error profiles, making accurate analysis challenging. Here, we introduce a haplotype-aware variant calling pipeline PEPPER-Margin-DeepVariant that produces state-of-the-art variant calling results with nanopore data. We show that our nanopore-based method outperforms the short-read-based single nucleotide variant identification method at the whole genome-scale and produces high quality single nucleotide variants in segmental duplications and low-mappability regions where short-read based genotyping fails. We show that our pipeline can provide highly-contiguous phase blocks across the genome with nanopore reads, contiguously spanning between 85% to 92% of annotated genes across six samples. We also extend PEPPER-Margin-DeepVariant to PacBio HiFi data, providing an efficient solution with superior performance than the current WhatsHap-DeepVariant standard. Finally, we demonstrate de novo assembly polishing methods that use nanopore and PacBio HiFi reads to produce diploid assemblies with high accuracy (Q35+ nanopore-polished and Q40+ PacBio-HiFi-polished).
Keywords: Nanopore, Variant Calling, Long Reads, PacBio HiFi, Assembly Polishing, Haplotyping, PEPPER, Margin, DeepVariant
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Introduction
Most existing reference-based small variant genotyping methods are tuned to work with short-reads1,2. Short-reads have high base-level accuracy but frequently fail to align unambiguously in repetitive regions3. Short-reads are also generally unable to provide substantial read-based phasing information, and therefore require using haplotype panels for phasing4 that provide limited phasing information for rarer variants.
Third-generation sequencing technologies, like linked-reads5–7 and long-reads8,9, produce sequences that can map more confidently in the repetitive regions of the genome10, overcoming the fundamental limitations of short-reads. Long-reads can generate highly contiguous de novo assemblies11–17, and they are increasingly being used by reference-based analysis methods18–25. The Genome-In-A-Bottle Consortium (GIAB)26 used the additional power of long-reads and linked-reads to expand the small variant benchmarking set to cover more of the genome27. This was essential to the PrecisionFDA challenge V2, which quantified the limitations of short read-based methods to accurately identify small variants in repetitive regions28.
Oxford Nanopore Technologies (ONT) is a commercial nanopore-based high-throughput13 long-read sequencing platform that can generate 100kb+ long reads13,29. Nanopore long-reads can confidently map to repetitive regions of the genome10 including centromeric satellites, acrocentric short arms, and segmental duplications11,30–32. Nanopore sequencing platform promises same-day sequencing and analysis33, but the base-level error characteristics of the nanopore-reads, being both generally higher and systematic, make small variant identification challenging34.
Pacific Biosciences (PacBio) provides a single-molecule real-time (SMRT) sequencing platform that employs circular consensus sequencing (CCS) to generate highly-accurate (99.8%) high-fidelity (PacBio HiFi) reads that are between 15kb-20kb long21. The overall accuracy of PacBio-HiFi-based variant identification is competitive with short-read based methods21. These highly accurate long-reads enabled the small variant benchmarking of major histocompatibility complex (MHC) region35 and difficult-to-map regions27.
In our previous work, we introduced DeepVariant, a universal small variant calling method based on a deep convolutional neural network (CNN)36. We showed that by retraining the neural network of DeepVariant, we can generate highly accurate variant calls for various sequencing platforms36. To limit the computational time, DeepVariant only uses the neural network on candidate sites identified with simple heuristics. However, the higher error-rate of nanopore-reads13,34 causes too many candidate variants to be picked up by the heuristic-based candidate finder of DeepVariant, limiting the extension of DeepVariant to nanopore sequencing platform.
Phasing long reads has been shown to enable or improve methods for small variant calling, structural variant calling, and genome assembly4,21,23,25,37–41. Previously, we trained DeepVariant on PacBio HiFi long-read data and showed highly competitive performance against short-read based methods for small variant identification21. However, the run-time of the haplotype-aware mode of DeepVariant with PacBio HiFi reads remain a bottleneck for production-level scaling.
Here we present a haplotype-aware genotyping pipeline PEPPER-Margin-DeepVariant that produces state-of-the art small variant identification results with nanopore and PacBio HiFi long-reads. PEPPER-Margin-DeepVariant outperforms other existing nanopore-based variant callers like Medaka18, Clair19, and longshot20. For the first time we report that nanopore-based single nucleotide polymorphism (SNP) identification with PEPPER-Margin-DeepVariant outperforms short-read based SNP identification with DeepVariant at whole genome scale. For PacBio HiFi reads, we report PEPPER-Margin-DeepVariant is more accurate, 3× faster, and 1.4× cheaper than the current haplotype-aware pipeline DeepVariant-WhatsHap-DeepVariant. We analyzed our pipeline in the context of GENCODE42 genes and report phasing errors in less than 1.5% of genes, with over 88% of all genes being contiguously phased across six samples. Finally, we extended PEPPER-Margin-DeepVariant to polish nanopore-based de novo assemblies with nanopore and PacBio HiFi reads in a diploid manner. We report Q35+ nanopore-based and Q40+ PacBio-HiFi-polished assemblies with lower switch error rate compared to the unpolished assemblies.
Results
Haplotype-aware variant calling
PEPPER-Margin-DeepVariant is a haplotype-aware pipeline for identifying small variants against a reference genome with long-reads. The pipeline employs several methods to generate highly-accurate variant calls (Figure 1a). Details of these methods are in the online methods section. An overview is presented here:
PEPPER-SNP: PEPPER-SNP finds single nucleotide polymorphisms (SNPs) from the read alignments to the reference using a recurrent neural network (RNN).
Margin: Margin is a phasing and haplotyping method that takes the SNPs reported by PEPPER-SNP and generates a haplotagged alignment file using a hidden Markov Model (HMM).
PEPPER-HP: PEPPER-HP takes the haplotagged alignment file and finds potential SNP, insertion, and deletion (INDEL) candidate variants using a recurrent neural network (RNN).
DeepVariant: DeepVariant takes candidates from PEPPER HP and an haplotagged alignment file from margin and genotypes the proposed candidates in a haplotype-aware manner. DeepVariant has more total parameters than PEPPER, and models with more parameters can generally train to higher accuracy at the cost of increased runtime. By combining PEPPER with DeepVariant in this way, we allow the faster neural network of PEPPER to efficiently scan much more of the genome, and to leverage the larger neural network of DeepVariant to achieve high accuracy on a tractable number of candidates.
Margin: Margin takes the output of DeepVariant and the alignment file to generate a phased VCF file using the same HMM.
Figure 1: Nanopore variant calling results.
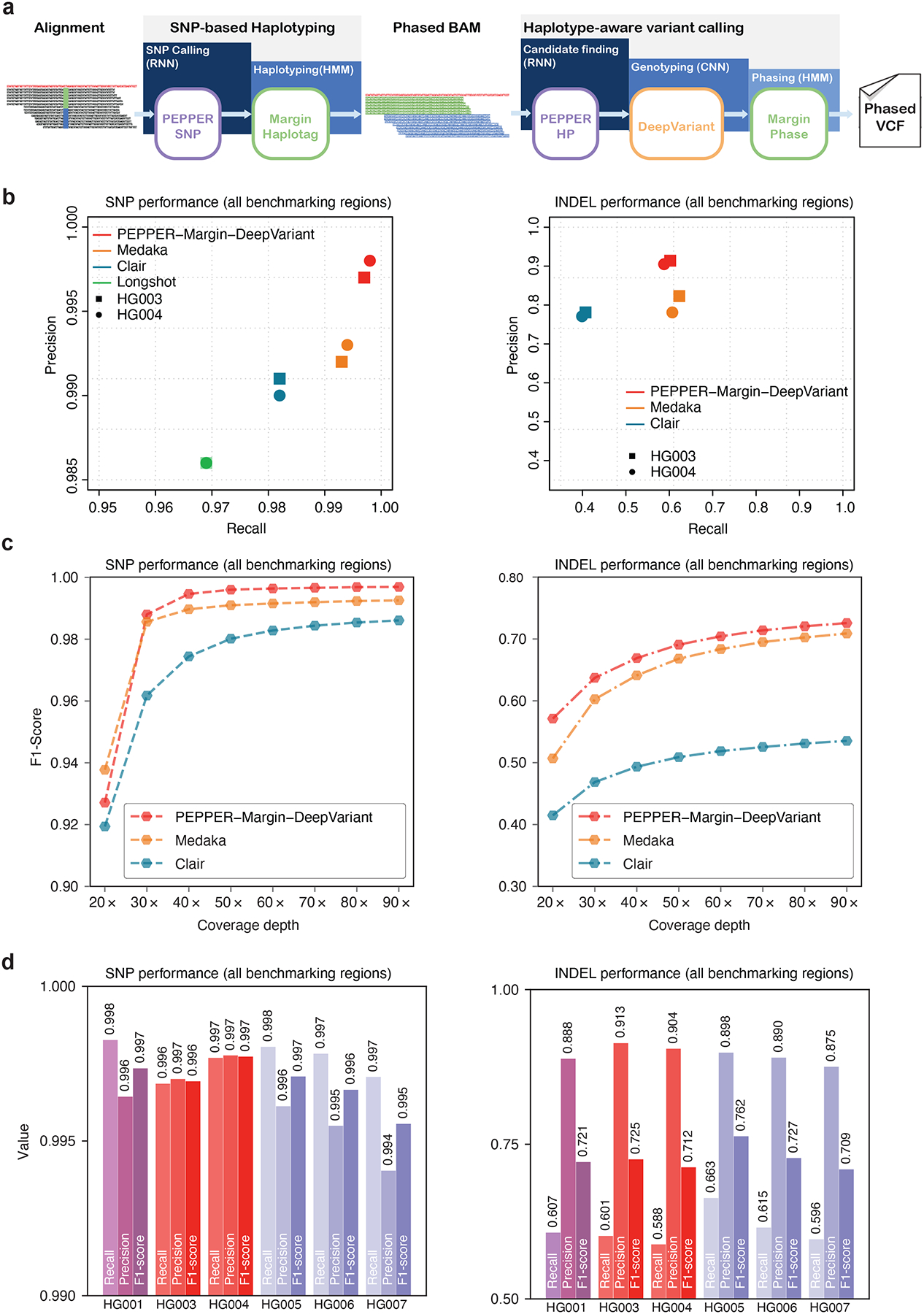
(a) Illustration of haplotype-aware variant calling using PEPPER-Margin-DeepVariant. (b) Nanopore variant calling performance comparison between different nanopore-based variant callers. (c) Evaluating variant calling performance at different coverage of HG003. (d) Variant calling performance of PEPPER-Margin-DeepVariant on six GIAB samples.
It is challenging to identify accurate variants with Oxford nanopore reads due to the error rate. Heuristic-based approaches show robust solutions for highly-accurate sequencing platforms21,36 but fail when introduced with erroneous reads. For example, in 90x HG003 ONT data, at 10% allele frequency, we find 20× more erroneous variants than true variants (Supplementary Figure 2).
Existing variant callers, like Clair19, that use allele frequency to find a set of candidates often need to set the threshold too high, excluding many true variants from being detected. Our pipeline demonstrates an efficient solution using an RNN to find candidates with PEPPER and genotype the candidates accurately with DeepVariant.
The use of haplotype information to get better genotyping results with erroneous reads has been demonstrated before21,22. The schema of the PEPPER-Margin-DeepVariant pipeline follows a similar design of PacBio HiFi-based DeepVariant21 and Medaka18 that use haplotyping to provide better genotyping results. However, Medaka18 is a consensus caller that presents the predicted sequence per position that does not match with reference sequence as variants. In contrast, PEPPER applies the predictions of the RNN to the candidates to find likely candidate variants for DeepVariant to accurately genotype. While maintaining similar candidate sensitivity of the heuristic-based approach, PEPPER reduces the number of erroneous homozygous candidate variants significantly (Supplementary figure 2).
Nanopore variant calling performance
We compared the nanopore variant calling performance of PEPPER-Margin-DeepVariant against Medaka18, Clair19, and Longshot20. We called variants on two samples HG003 and HG004, with 90× coverage. We also compared the performance against Medaka and Clair for the HG003 sample at various coverages ranging from 20× to 90×. Finally, we benchmarked the variant calling performance of PEPPER-Margin-DeepVariant on six Genome-In-A-Bottle (GIAB) samples.
PEPPER-Margin-DeepVariant produces more accurate nanopore-based SNP calls (F1-scores of 0.9969 and 0.9977) for HG003 and HG004 respectively than Medaka (0.9926, 0.9933), Clair (0.9861, 0.9860), and Longshot (0.9775, 0.9776). We also observe higher INDEL performance with PEPPER-Margin-DeepVariant (F1-scores of 0.7257 and 0.7128 for HG003 and HG004) compared to Medaka (0.7089, 0.7128) and Clair (0.5352, 0.5260) (Figure 1b, Supplementary table 1).
To assess the robustness of our method, we evaluated the variant calling performance with HG005 sample on GRCh38 and GRCh37 against two GIAB truth versions (v3.3.243 and v4.2.127). In this comparison, we see PEPPER-Margin-DeepVariant perform similarly between GRCh37 GIABv3.3.2 (SNP F1-Score: 0.9971, INDEL F1-Score: 0.7629) and GRCh38 v4.2.1 (SNP F1-Score: 0.9974, INDEL F1-Score: 0.7678) and has higher accuracy compared to Medaka (GRCH37: SNP 0.9938, INDEL 0.7629, GRCh38: SNP 0.9927, INDEL 0.7406), Clair (GRCH37: SNP 0.9789, INDEL 0.5666, GRCh38: SNP 0.9787, INDEL 0.5675) and longshot (GRCh37: SNP 0.9803, GRCh38: SNP 0.9767) (Supplementary table 2). Overall, we see PEPPER-Margin-DeepVariant has consistent performance between different samples, reference sequence and truth sets.
We performed a Mendelian concordance analysis of our method with the HG005/HG006/HG007 trio on GRCh38 inside and outside of the HG005 v4.2.1 high-confidence regions (Supplementary table 3). In the 2.5 Gb high confidence region we observed a paternal and maternal concordance of 99.90%, with overall concordance of 99.75%. In the 315Mb region outside of high confidence excluding centromeres we observed a paternal concordance of 98.20%, maternal concordance of 97.80%, with overall concordance of 95.52%.
To understand performance over realistic coverage ranges, we downsampled the HG003 nanopore sample at coverages varying between 20× and 90× and compared PEPPER Margin-DeepVariant against Medaka and Clair. The INDEL performance of PEPPER-Margin-DeepVariant achieves the highest F1-score at any coverage compared to other tools (Figure 1c, Supplementary table 4). At coverage above 30×, PEPPER-Margin-DeepVariant achieves a higher F1-score than Medaka and Clair (Supplementary table 5). Overall, we observe that PEPPER-Margin-DeepVariant can yield high-quality variant calls at above 40× coverage on Oxford Nanopore data.
We investigated the nanopore variant calling performance of PEPPER-Margin-DeepVariant on six GIAB samples (HG001, HG003-HG007), each sample with various coverage (Supplementary Table 6) and against GRCh37 and GRCh38 reference genomes (Supplementary Table 7). PEPPER-Margin-DeepVariant achieves SNP F1-score 0.995 or higher and INDEL F1-score of 0.709 or higher for each sample, demonstrating the ability to generalize the variant calling across samples and reference genomes (Figure 1d, Supplementary table 8).
We also assessed the ability to use PEPPER-HP as a variant caller if we tune the method for a balanced precision and recall. We find that PEPPER-HP outperforms Medaka in SNP accuracy while having a comparable INDEL accuracy. However, PEPPER-HP in itself is not able to achieve the genotyping accuracy DeepVariant provides. As PEPPER-HP uses a compressed representation of nucleotide bases, it fails to achieve the high genotyping accuracy compared to DeepVariant’s CNN (Supplementary table 9).
Similar to the nanopore-based haplotype-aware pipeline, the PacBio HiFi-based PEPPER-Margin-DeepVariant pipeline produces highly accurate variant calls. We analyzed the PacBio HiFi PEPPER-Margin-DeepVariant variant calling performance on the 35x HG003 and HG004 from precisionFDA28 against DeepVariant-WhatsHap-DeepVariant (current state-of-the-art method21,28) and DeepVariant-Margin-DeepVariant. In this comparison we see that DeepVariant-Margin-DeepVariant produces the best performance (HG003 SNP-F1: 0.9991 INDEL-F1: 0.9945, HG004 SNP-F1: 0.9992, INDEL-F1: 0.9942) compared to DeepVariant-WhatsHap-DeepVariant (HG003 SNP-F1:0.9990 INDEL-F1:0.9942, HG004 SNP-F1: 0.9992, INDEL-F1: 0.9940) and PEPPER-Margin-DeepVariant (HG003 SNP-F1:0.9990, INDEL-F1:0.9944, HG004 SNP-F1: 0.9992, INDEL-F1: 0.9941) (Supplementary table 10).
We compared the run-time and cost of Oxford nanopore-based variant calling pipelines on 50× and 75× HG001 data (Supplementary table 11) using the GCP platform with instance sizes best matching CPU and memory requirements. On a CPU-platform, PEPPER-Margin-DeepVariant is 8× faster than Medaka and 4× faster than Longshot while providing the best variant calling performance at lower cost. Clair is faster and cheaper than PEPPER-Margin-DeepVariant but fails to generate high-quality variant calls. On PacBio HiFi data the PEPPER-Margin-DeepVariant pipeline outperforms DeepVariant-WhatsHap-DeepVariant and is 3× faster and 1.4× cheaper, establishing a faster and more accurate solution to haplotype-aware variant calling with PacBio HiFi data (Supplementary table 12, Supplementary table 13).
Nanopore, Illumina, and PacBio HiFi variant calling
We compared the variant calling performance of Oxford Nanopore and PacBio HiFi long-read based PEPPER-Margin-DeepVariant against Illumina short-read based DeepVariant method44. We used 35x Illumina NovaSeq, 35x PacBio HiFi, and 90x Oxford Nanopore reads basecalled with Guppy v4.2.2 for HG003 and HG004 samples available from PrecisionFDA28. We used GIAB v4.2.1 benchmarking data for HG003 and HG004, which is notable for including difficult-to-map regions. Finally we used GIAB v2.0 stratificiations to compare variant calling performance in difficult-to-map regions and low-complexity regions of the genome.
The SNP F1-score of PacBio HiFi (HG003 SNP-F1: 0.9990, HG004 SNP-F1: 0.9992) is higher than Oxford Nanopore (HG003 SNP-F1: 0.9969, HG004 SNP-F1: 0.9977) and Illumina (HG003 SNP-F1: 0.9963, HG004 SNP-F1: 0.9962) in all benchmarking regions. Notably, both long-read sequencing platforms outperform the short-read based method in accurate SNP identification performance. The INDEL F1-score of Oxford Nanopore (HG003 INDEL-F1: 0.7257, HG004 INDEL-F1: 0.7128) is well below the performance with PacBio HiFi (HG003 INDEL-F1: 0.9945, HG004 INDEL-F1: 0.9941) and Illumina (HG003 INDEL-F1: 0.9959, HG004 INDEL-F1: 0.9958) suggesting further improvement required for nanopore-based methods. On HG003 PacBio-CLR data, we observed a SNP-F1 score of 0.9892 with our method and 0.9755 with Longshot (Supplemental Table 14). Overall, we find that haplotype-aware long-read-based variant calling produces high-quality SNP variant calls comparable to those produced by short-read-based variant identification methods (Figure 2a, Supplementary table 15). This is the first demonstration we are aware of in which SNP variant calls with Oxford Nanopore data achieved similar accuracy to Illumina SNP variant calls.
Figure 2: Comparison between Nanopore, Illumina and PacBio HiFi variant calling performance.
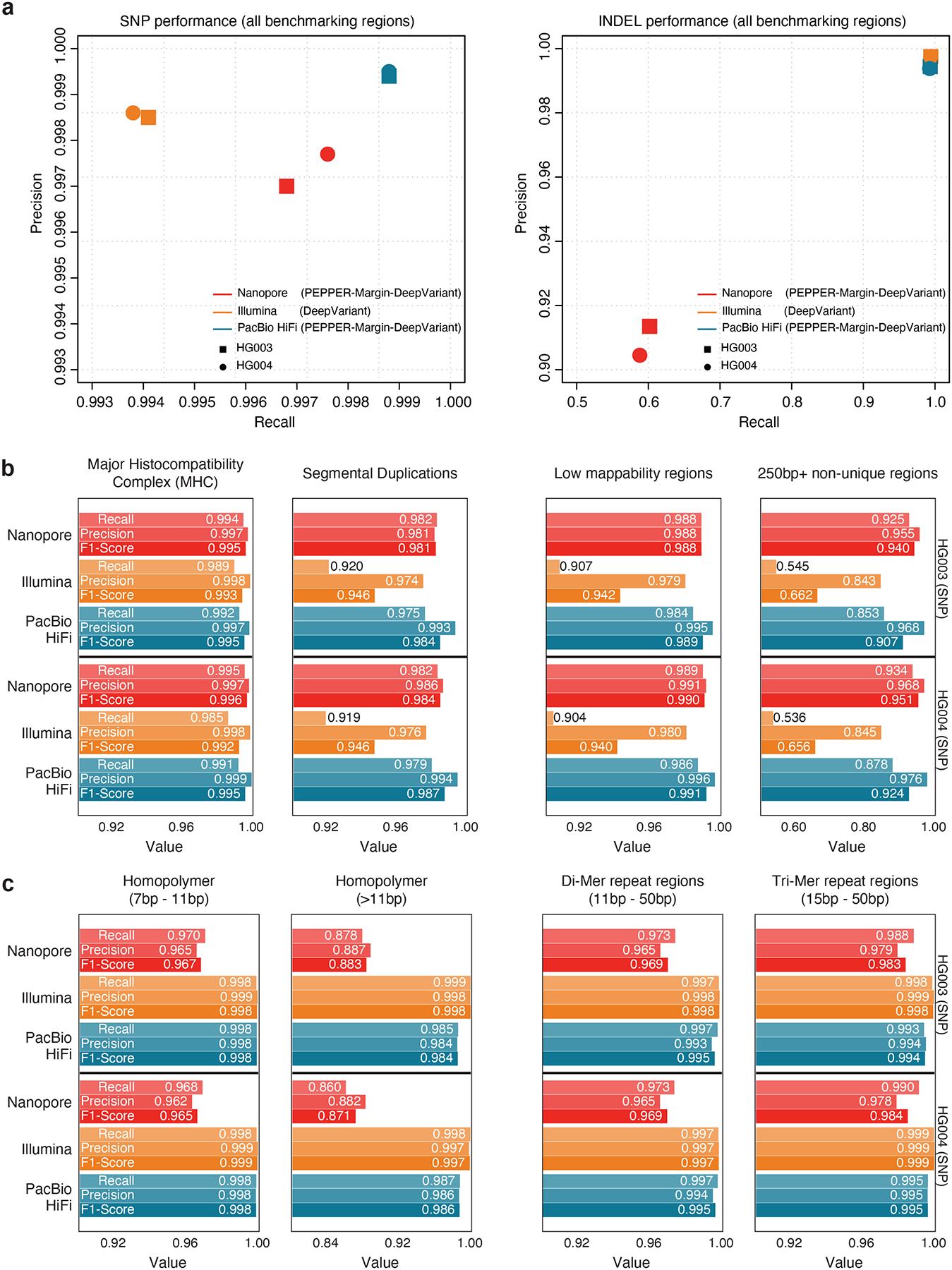
(a) SNP and INDEL performance comparison of Nanopore, Illumina and PacBio HiFi in all benchmarking regions. (b) SNP performance comparison in difficult-to-map regions of the genome. (c) SNP performance comparison in low-complexity regions of the genome.
In segmental duplications, 250bp+ non-unique regions, and low-mappability regions where short-reads have difficulty in mapping, we observe the average SNP F1-scores of Illumina (Seg. Dup. F1-score: 0.94, 250bp+ non-unique:0.66, Low-mappability: 0.94) drop sharply for both HG003 and HG004 samples. Long-read based PacBio HiFi (Seg. Dup. F1-score: 0.99, 250bp+ non-unique:0.90, Low-mappability: 0.99) and Oxford Nanopore (Seg. Dup. F1-score: 0.98, 250bp+ non-unique:0.94, Low-mappability: 0.98) produce more accurate SNP variants. In the major histocompatibility complex (MHC) region, we see Oxford Nanopore (HG003 SNP F1-score: 0.9958, HG004 SNP F1-score: 0.9966) achieve best performance followed by PacBio HiFi (HG003 SNP F1-score: 0.9951, HG004 SNP F1-score: 0.9955) and Illumina HG003 SNP F1-score: 0.9939, HG004 SNP F1-score: 0.9921). In general, the long-read-based haplotype-aware methods outperform short-reads in more repetitive regions of the genome (Figure 2b, Supplementary table 16).
In low-complexity regions like homopolymer, di-mer and tri-mer repeat regions of the genome, the average variant calling performance of Nanopore drops (7bp-11bp homopolymer SNP F1-score: 0.96, 11bp+ homopolymer SNP F1-Score: 0.88) for both HG003 and HG004 samples compared to Illumina (7bp-11bp homopolymer SNP F1-score: 0.998, 11bp+ homopolymer SNP F1-Score: 0.998) and PacBio HiFi (7bp-11bp homopolymer SNP F1-score: 0.998, 11bp+ homopolymer SNP F1-Score: 0.984). In 11bp-50bp di-mer and 15bp-50bp tri-mer repeat regions of the genome, we see the average performance of Oxford Nanopore (di-mer SNP F1-score: 0.969, tri-mer SNP F1-score: 0.984) is lower than PacBio HiFi (di-mer SNP F1-score: 0.995, tri-mer SNP F1-score: 0.995) and Illumina (di-mer SNP F1-score: 0.998, tri-mer SNP F1-score: 0.998). Overall, the Illumina short-read based variant calling method achieves higher accuracy in low-complexity regions of the genome (Figure 2c, Supplementary table 17).
We further compare the variant calling performance of Illumina, PacBio HiFi and ONT in “easy regions” which cover 76% of the genome27. In this comparison, we see ONT variant calling performance (SNP: 0.9988 INDEL: 0.9719) is comparable to Illumina (SNP: 0.9997, INDEL: 0.9996) and HiFi (SNP: 0.9999, INDEL: 0.9997) showing that in easy regions, all technologies can generate high-quality variants (Supplementary table 18). We further look into regions with no tandem repeats (covering 86% of the genome) and see that ONT performance (SNP: 0.9981, INDEL: 0.97) is comparable to Illumina (SNP: 0.996, INDEL: 0.996). However in tandem repeat and homopolymer regions, the ONT SNP calling performance drops from 0.998 to 0.9748, and the INDEL calling performance drops from 0.97 to 0.54 (Supplementary Table 19) suggesting that ONT variant calling can generate competitive variant calling in the 86% of the genome outside tandem repeat and homopolymer regions, and it suffers only in the 4% of the genome which is highly repetitive.
Phaseset and Haplotagging Accuracy
We compared phaseset accuracy for Margin and WhatsHap on HG001 against GIAB’s phased v3.3.2 variants with 25× nanopore, 50× nanopore, 75× nanopore, and 35× PacBio HiFi data. We generated genotyped variants with PEPPER-Margin-DeepVariant, and used both Margin and WhatsHap to phase the final variant set. The phasesets produced by both tools were analyzed using whatshap stats and whatshap compare against the trio-confirmed truth variants in high-confidence regions.
For all datasets, Margin had a lower switch error rate (0.00875, 0.00857, 0.00816, 0.00895) than WhatsHap (0.00923, 0.00909, 0.00906, 0.00930), but lower phaseset N50 (2.07, 4.21, 6.13, 0.24 Mb) than WhatsHap (2.37, 4.90, 8.27, 0.25 Mb) (Figure 3a, Supplementary Tables 20 and 21).We also compared phaseset accuracy for Margin and WhatsHap on the same data using a novel metric we call “Local Phasing Correctness” (LPC), which is a generalization of the standard switch error and Hamming rate metrics. (see online methods). We plot LPC across various length scale values (Figure 3b). Margin produced more accurate phasing for all length scales for 25× nanopore and 35× CCS. Margin also produced more accurate phasing for 50× nanopore for length scales up to 128kb and for 75× nanopore for length scales up to 242kb.
Figure 3: Margin and WhatsHap phasing results.
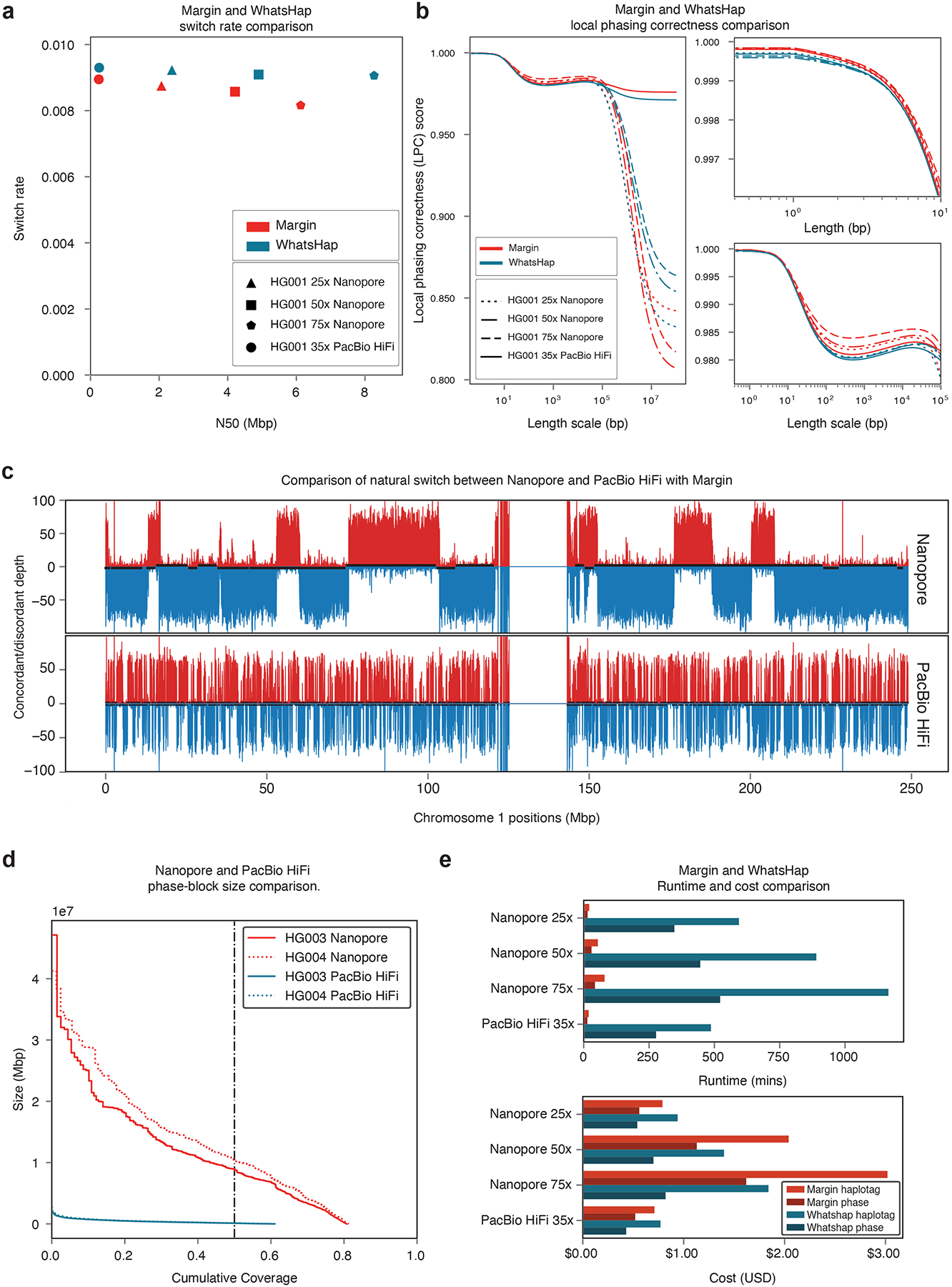
(a) Phaseset switch rate to N50. (b) Novel metric “Local Phasing Correctness” analyzing phaseset accuracy across different length scales. For visibility, we plot the full data (left), short length scales (right top), and medium length scales (right bottom). (c) Novel visualization “Natural Switch Rate” describing haplotagging accuracy for reads. Phasing is consistent across the top/red and bottom/blue blocks, with switch errors occurring at the transitions between these. (d) Phaseset N50 for Nanopore and PacBio HiFi data on HG003 and HG004. (e) Cost and runtime comparison between Margin and Whatshap. Plots (a) and (b) are generated from phased HG001 GIAB v3.3.2 variants; plot (c) is generated from an admixture of reads from HG005 and HG02723.
To analyze haplotagging accuracy, we artificially constructed an admixture sample by trio-binning reads from HG005 and HG02723 and combining an equal amount of maternal reads from each sample, resulting in a 55× nanopore alignment and a 35× PacBio HiFi alignment. We ran PEPPER-SNP, haplotagged each alignment with Margin using these variants, and compared the number of direct-matched reads Rd (truth H1 to tagged H1 or truth H2 to tagged H2) and cross-matched reads Rc (truth H1 to tagged H2 or truth H2 to tagged H1) of the output. In Figure 3c, for each 10kb bucket in chr1 we plot the number of reads that were direct-matched (top, red) and cross-matched (bottom, blue) for both data types, with phasesets plotted in black alternating between top and bottom. This “Natural Switch” plot identifies consistent phasing as regions where the majority of reads are either direct- or cross-matched, and haplotagging switch errors as regions where the majority of reads transition between the two. As the plot shows, nanopore reads allow us to haplotag consistently with phase sets in the range of tens of megabases, whereas PacBio HiFi reads cannot be used for long-range haplotagging. For each bucket we can calculate a local haplotagging accuracy using the ratio: max(Rc,Rd)/(Rc + Rd). On average the haplotagging accuracy is 0.9626 for ONT data and 0.9800 for HiFi data using Margin with reduced performance for WhatsHap (Supplementary Table 22; Supplementary Figures 3, 4, 5, 6). As Margin has higher haplotagging accuracy compared to WhatsHap, we see that the variant calling with Margin exhibits higher accuracy compared to WhatsHap for both Oxford Nanopore and PacBio HiFi data (Supplementary Table 23, Supplementary Table 12).
Lastly we compare the runtime and cost for the haplotag and phase actions on the four HG001 datasets using Margin and WhatsHap. For Margin, we calculate a cost of $1.35 (36m) for 25x ONT, $3.17 (84m) for 50x ONT, $4.64 (123m) for 75x ONT, $1.23 (33m) for 35x PacBio HiFi, and for WhatsHap $1.48 (941m), $2.10 (1336m), $2.66 (1688m), and $1.20 (764m) respectively (Supplementary table 24)
Gene Analysis
We performed an analysis of Margin’s phasing over genic regions to understand its utility for functional studies. With 75× nanopore data from HG001 on GRCh37, we classified each of the GENCODE v35 genes45 (coding and non-coding) as wholly, partially, or not spanned for the GIAB v3.3.2 high confidence regions, the phasesets proposed by Margin, and the switch errors between the two as determined by whatshap compare. In Figure 4a, we first plot the number of gene bodies as spanned by high confidence regions (23712 wholly, 26770 partly, 11372 not), then further compare how many of each division were wholly spanned by Margin’s phasesets (22491, 24877, 9807), and finally how many of these had no detected phasing errors (22191, 24563, unknown). In Figure 4b, we plot the number of genes wholly phased by Margin on GRCh38 (60656) for HG003 and HG004 (53817, 55234) and on GRCh37 (62438) for HG001, HG005, HG006, and HG007 (57175, 53150, 53112, 54116). SNP and INDEL accuracies are largely similar between stratifications of all regions, all genes, and all protein coding genes, with improved performance for protein coding sequence (Supplementary Tables 25, 26).
Figure 4: Gene analysis.
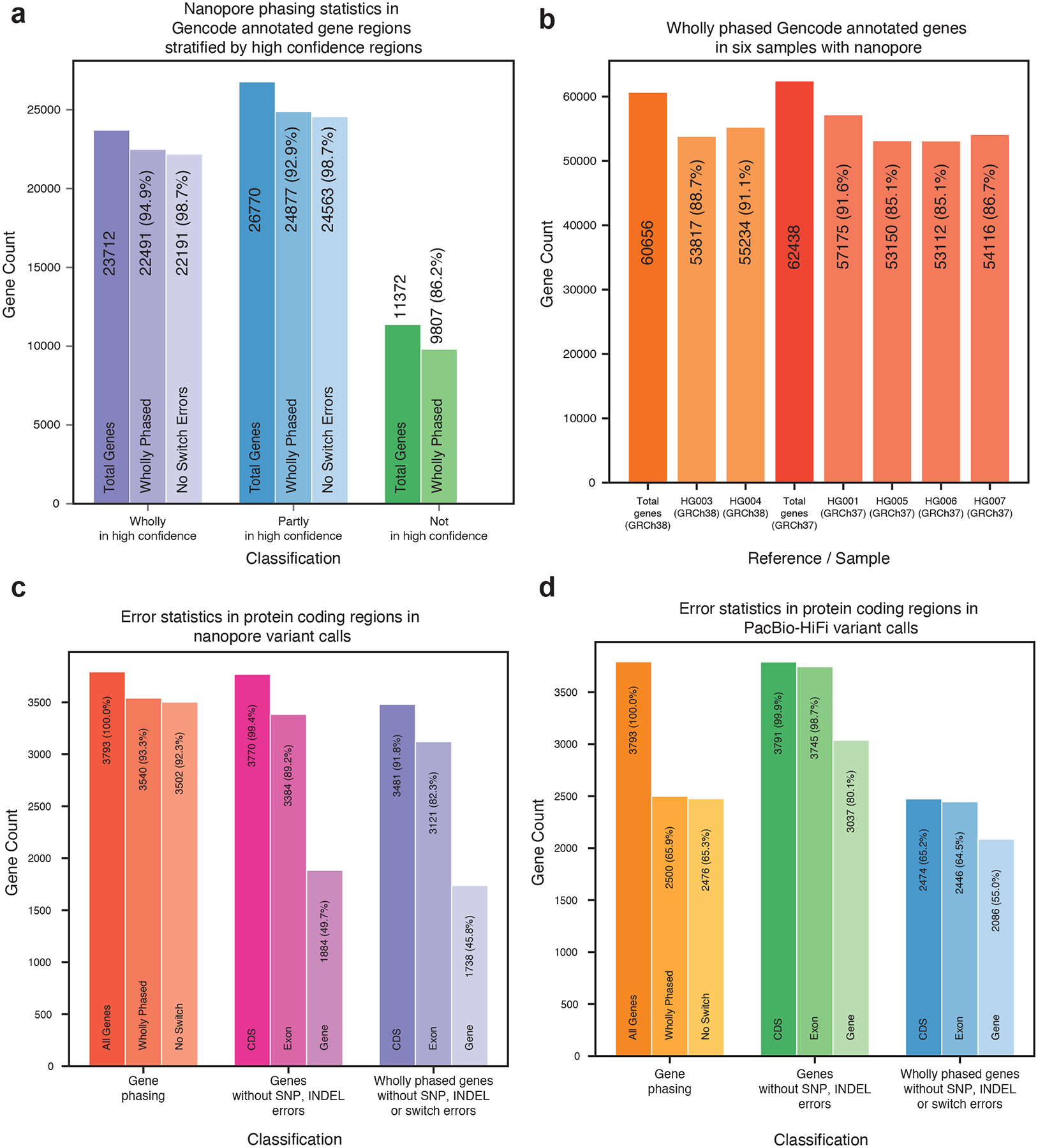
(a) Phasing analysis for HG001 over GENCODE annotated gene regions, stratified by GIAB high confidence coverage. Percentages are relative to their predecessor. (b) Wholly phased GENCODE annotated gene regions. Percentages are relative to the total genes annotated on the reference. (c) Error statistics including wholly phased genes, genes without SNP or INDEL errors, and wholly phased genes without SNP, INDEL, or switch errors over a subset of the protein coding genes, derived on HG001 from 75x nanopore data. Percentages are relative to total genes in subset. (d) The same statistics on HG001 with 35x PacBio HiFi data.
We identified the 3793 protein coding genes which had at least 80% of their coding sequence covered by the high confidence regions and analyzed the presence of phasing and SNP/INDEL errors on HG001 with 75x nanopore (Figure 4c) and 35x PacBio HiFi (Figure 4d) reads (Supplementary Tables 27, 28). Nanopore data produced 3540 genes wholly spanned by Margin’s phasesets with only 38 exhibiting a switch error (1.07%), as compared to PacBio HiFi with 2500 wholly spanned genes and 24 switch errors (0.96%). We identified the genes with no SNP or INDEL errors in the high confidence region for the entire gene, all annotated exons in the gene, and all coding sequences in the gene; PacBio HiFi produces 3037, 3745, and 3791 respectively with nanopore producing 1884, 3384, and 3770 perfectly called regions. Lastly, we identified how many of these gene regions were perfectly captured (wholly phased with no switch errors and having no SNP or INDEL miscalls) for the entire gene (1738 for nanopore, 2086 for PacBio HiFi), for all annotated exons (3121, 2446), and for all coding sequences (3481, 2471). For nanopore data, we find that for 91.8% of genes the CDS is fully phased and genotyped without error, and for 82.3% of genes all exons are fully phased and genotyped without error.
Diploid polishing of de novo assemblies
Oxford Nanopore-based assemblers like Flye16 and Shasta13 generate haploid assemblies of diploid genomes. By calling and phasing variants against the haploid contigs they produce, it is possible to polish the haploid assembly into a diploid assembly. We implemented such a diploid de novo assembly polishing method with PEPPER-Margin-DeepVariant (Figure 5a). It can polish haploid Oxford Nanopore-based assemblies with either Nanopore or PacBio HiFi reads. A detailed description of this method is presented in the online methods.
Figure 5: Diploid assembly polishing results.
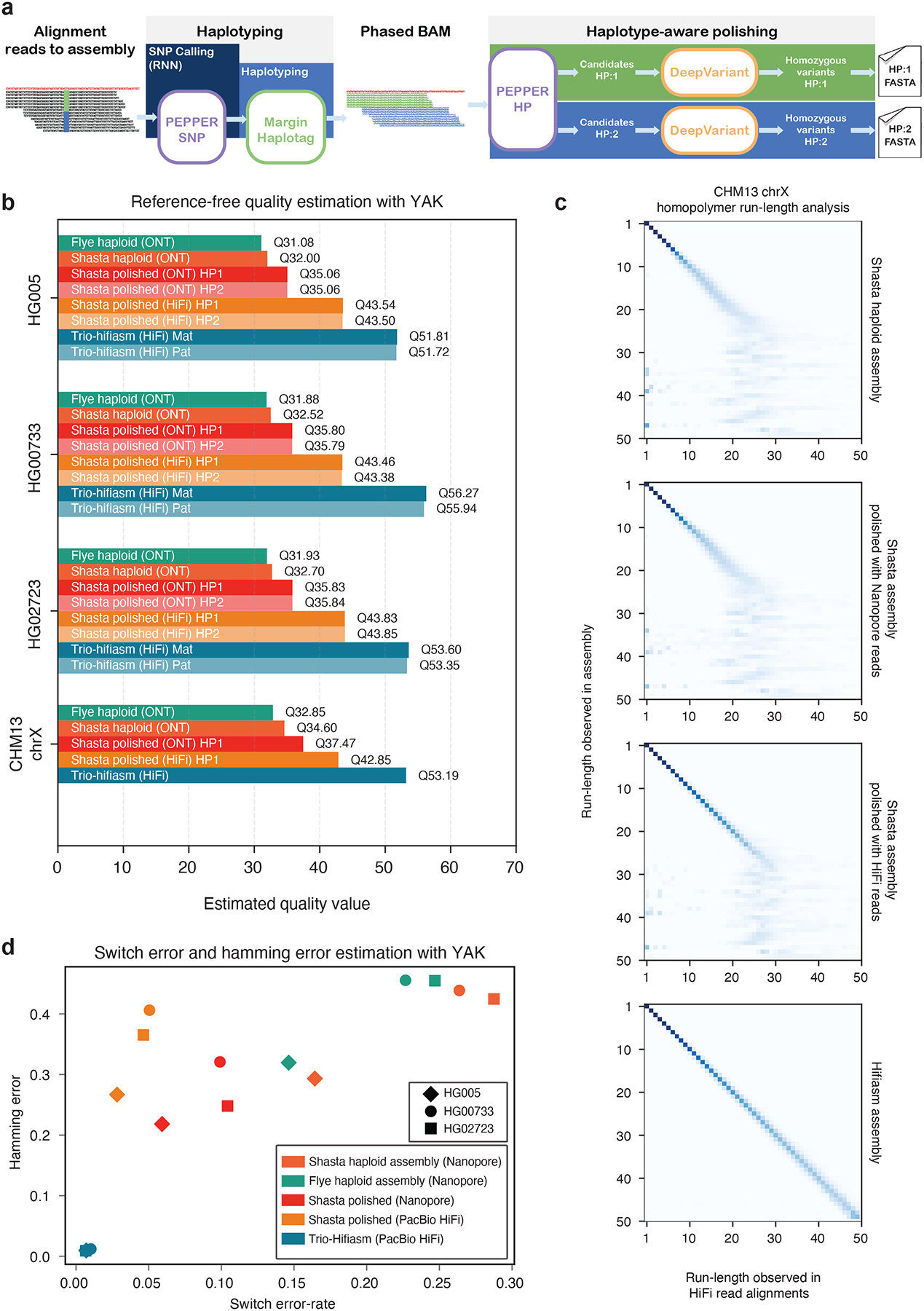
(a) Illustration of the diploid assembly polishing pipeline. (b) Estimated quality values of assemblies using YAK. (c) CHM13-chrX run-length confusion matrix between different assemblies and PacBio HiFi reads aligned to the corresponding assembly. (d) Switch error and hamming error comparison between assemblies.
We generated haploid assemblies using Shasta13 and Flye16 for diploid samples HG005, HG00733, HG02723, and haploid sample CHM13 (chrX) using nanopore reads, and we polished the Shasta assemblies using ONT and PacBio HiFi reads. To evaluate the base-level accuracy of the assemblies we use the kmer-based tool YAK14, which uses Illumina trio data to estimate sequence quality, switch error rates, and hamming error rates. We compare the haploid assemblies, polished diploid assemblies, and trio-aware diploid assemblies generated with hifiasm14. Hifiasm uses parental short-read data to generate maternal and paternal assemblies.
The estimated quality values (QV) of nanopore-based assemblies with Shasta assembler are higher than the nanopore-based Flye assemblies. (Supplementary table 29). As Shasta generated higher quality assemblies compared to Flye, we polished the Shasta assemblies with the PEPPER-Margin-DeepVariant diploid polisher. The nanopore-polished assemblies achieve Q35+ estimated quality and PacBio-HiFi-polished assemblies achieve Q40+ estimated quality for all three diploid samples. Finally, we show that the unpolished CHM13-chrX Shasta assembly (QV34.6) can be improved to QV36.9 with nanopore-based and QV42.7 PacBio-HiFi-based assembly polishing with PEPPER-Margin-DeepVariant. Compared to the nanopore-based Shasta assemblies, the trio-aware PacBio HiFi assembler hifiasm achieves higher quality assemblies with respect to base-level accuracy but the NG50 of the hifiasm assemblies are lower for HG00733 and HG02723 samples. In summary, PEPPER-Margin-DeepVariant achieves Q35+ ONT-based assembly polishing and Q40+ PacBioHiFi-based assembly polishing of ONT assemblies (Figure 5b, Supplementary table 29, Supplementary table 30).
The dominant error modality for ONT data are homopolymers13. In Figure 5c we show the run-length confusion matrix of PacBio HiFi read alignments to four chrX assemblies of CHM13-chrX. The Shasta assembly starts to lose resolution at run-lengths greater than 7 (RL-7) and loses all resolution around RL-25. The nanopore-polished assembly improves homopolymer resolution up to RL-10, but also fails to resolve run-lengths greater than RL-25. The PacBio HiFi polished assembly has fair resolution up to RL-25. The trio-hifiasm assembly shows accurate homopolymer resolution up to and beyond RL-50.
Figure 5d shows the switch error-rate of the assemblies. The switch error –rate and hamming error rate of haploid Shasta assemblies are reduced after polishing with PEPPER-Margin-DeepVariant using ONT data. The trio-hifiasm that use maternal and paternal short-reads to resolve haplotypes have much lower switch error-rate and hamming error-rate (Figure 5d, Supplementary table 30).
In table 1, we compare HG005 assemblies at the small variant level. The analysis shows that the F1-score of unpolished Shasta assembly (INDEL: 0.1203, SNP: 0.4928) improves significantly after polishing with nanopore reads using PEPPER-Margin-DeepVariant (INDEL: 0.3611, SNP: 0.9825). The PacBio-HiFi-polished Shasta assembly achieves similar F1-score (INDEL: 0.9565, SNP: 0.9976) compared to the trio-hifiasm assembly (INDEL: 0.9733, SNP: 0.9988). This analysis provide evidence that PEPPER-Margin-DeepVariant can effectively improve the assembly quality at small variant level.
Table 1:
Small variant accuracy evaluation of HG005 assemblies against GIAB HG005 v3.3.2 benchmarking set. We derive a small variant set against GRCh37 from the assemblies using dipcall51 and compare the variant calls against HG005 GIAB benchmark. We restrict our analysis in regions that are assembled by both Shasta and trio-hifiasm and falls in the high-confidence region defined by GIAB.
| Method | Polished with | Type | True positives | False negatives | False positives | Recall | Precision | F1 score |
|---|---|---|---|---|---|---|---|---|
| Shasta | - | INDEL | 128989 | 253403 | 1629782 | 0.3373 | 0.0732 | 0.1203 |
| SNP | 1279988 | 1696540 | 939228 | 0.4300 | 0.5769 | 0.4928 | ||
| Shasta | Nanopore | INDEL | 279793 | 102605 | 906353 | 0.7317 | 0.2397 | 0.3611 |
| SNP | 2940462 | 36058 | 68863 | 0.9879 | 0.9771 | 0.9825 | ||
| Shasta | PacBio HiFi | INDEL | 367819 | 14575 | 19341 | 0.9619 | 0.9512 | 0.9565 |
| SNP | 2971733 | 4787 | 9689 | 0.9984 | 0.9968 | 0.9976 | ||
| Hifiasm | - | INDEL | 374002 | 8390 | 12451 | 0.9781 | 0.9686 | 0.9733 |
| SNP | 2973193 | 3320 | 3730 | 0.9989 | 0.9987 | 0.9988 |
The current version of the PEPPER-Margin-DeepVariant pipeline does not attempt to polish structural variants (SVs, >50bp in size). The resulting haplotypes preserve all SVs initially contained in the input assembly. Since the input assemblies are haploid, only one (randomly assembled) allele for each heterozygous SV is retained within the pair of output haplotypes. To benchmark SV recall and precision, we first called SVs from the assemblies using svim-asm46 and then validated the reconstructed SV sets using the previously described approach47. Our benchmarks using HG002, HG005, HG0073 and HG02733 genomes show that input Shasta assemblies on average contained signatures of 94.6% and 48.3% of homozygous and heterozygous SVs, respectively. After polishing using PEPPER-Margin-DeepVariant, the average reconstruction rate slightly increased to 95.7% and 50.9% for homozygous and heterozygous SVs, respectively. The average SV precision was 81.6% before and 83.2% after polishing (Supplementary Table 31).
Discussion
Long-read sequencing technology is allowing gapless human genome assembly11 and enabling investigations in the most repetitive regions of the genome28. In this work, we present PEPPER-Margin-DeepVariant, a state-of-the-art long-read variant calling pipeline for Oxford nanopore data. For the first time, we show that nanopore-based SNP identification outperforms a state-of-the-art short-read based method at whole genome scale. Particularly in segmental duplication and difficult-to-map regions, the nanopore-based method outshines the short-read based method. It seems likely that the anticipated widespread application of long-read variant calling will for the first time accurately illuminate variation in these previously inaccessible regions of the genome.
The genomic contexts where nanopore SNP accuracy suffers for our pipeline are identifiable, meaning that variant calls in these regions can be treated with skepticism while calls outside these contexts can be handled with confidence. The one obvious area that Nanopore variant calling lags is in INDEL accuracy. While the results achieved here are to our knowledge the best shown so far, we believe it is likely that further technological innovations at the platform level will be required to make nanopore INDEL accuracy on par with other technologies in all genomic contexts. However, we find that in the 86% of the genome without tandem repeats or homopolymers, INDEL calls from our method are already of high quality.
PEPPER-Margin-DeepVariant is designed for whole-genome sequencing analysis. Although targeted sequencing with the Oxford Nanopore platform is reasonably popular, several issues may limit the application. For example, read length, read quality, coverage, and heterozygosity of the target region are expected to be different than whole-genome sequencing. Further investigation and benchmarking are required to extend support for variant calling on amplicon sequencing data.
Oxford Nanopore provides a highly-multiplexed sequencing solution with its PromethION device13. With this device and the PEPPER-Margin-DeepVariant pipeline described here it should be comfortably possible to go from biosample collection to complete genome inferences in under half a day. This fast turnaround should enable its use in a medical context, where diagnosis for acute disease situations requires speed.
We have demonstrated our nanopore-based phasing is able to wholly phase 85% of all genes with only 1.3% exhibiting a switch error. This phasing ability could play a useful role in population genetics studies48,49 and clinical genomics50. For clinical applications the accurate identification of compound-heterozygotes should be particularly valuable.
We have extended PEPPER-Margin-DeepVariant to PacBio HiFi reads and demonstrated a more accurate and cheaper solution to the existing WhatsHap-DeepVariant variant calling methods, making cohort-wide variant calling and phasing with PacBio-HiFi more accessible. Currently, we find PacBio-HiFi sequencing analyzed with our method has the best performance, but we expect that improvements to nanopore pore technology and basecalling may close this gap.
We have demonstrated diploid polishing of nanopore-based haploid assemblies with PEPPER-Margin-DeepVariant. We achieve Q35+ nanopore polished assemblies and Q40+ PacBio-HiFi-polished assemblies. We observe that our polishing method can resolve homopolymer errors up to 20bp with PacBio HiFi data. However, our polishing method fails to resolve 25bp+ long homopolymers indicating that they need to be resolved during the consensus generation of the de novo assembly methods. As nanopore assembly methods like Shasta move toward generating fully resolved diploid genome assemblies like trio-hifiasm, our polishing method can enable nanopore-only Q40+ polished diploid assemblies.
Online Methods
Code Availability
The modules of PEPPER-Margin-DeepVariant are publicly available in these repositories:
DeepVariant: https://github.com/google/deepvariant
The PEPPER-Margin-DeepVariant software52 is available under DOI: https://doi.org/10.5281/zenodo.5275510 and we used r0.4 version for the evaluation presented in this manuscript.
For simpler use we have also created a publicly available docker container kishwars/pepper_deepvariant:r0.4 that can run our variant calling and polishing pipelines.
Data availability
Analysis data available
We have made the analysis data available publicly (variant calling outputs, genome assemblies etc.) in: https://console.cloud.google.com/storage/browser/pepper-deepvariant-public/analysis_data.
The source data for the main figures can be found in: https://console.cloud.google.com/storage/browser/pepper-deepvariant-public/figure_source_data/Figure_source_data/
Sequencing data availability
We used several publicly available datasets:
Genome-In-a-Bottle consortium26,28: https://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/
Human Pangenome Reference Consortium (HPRC): https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html
Telomere-to-telomere consortium11,12: https://github.com/nanopore-wgs-consortium/CHM13
Please see supplementary notes to find specific links to the sequencing data that we used for our analysis.
Analysis methods and data pre-processing
Read alignment
We used minimap253 version 2.17-r941 and pbmm2 version 1.4.0 to align reads to a reference genome. The supplementary notes have details on execution parameters.
Subsampling alignment files to different coverages
We used samtools54 version 1.10 to generate alignment files of different coverages. The supplementary notes have details on execution parameters.
Variant calling
We used the following methods to call variants with nanopore data:
PEPPER-Margin-DeepVariant version r0.4.
Medaka version v1.2.1.
Clair version v2.1.1.
Longshot version v0.4.2.
For Illumina short-reads and PacBio HiFi we used DeepVariant version v1.1.0. The details on execution parameters are available in supplementary notes.
Benchmarking variant calls
We used hap.py55 version v0.3.12 to assess the variant calls against GIAB truth set. The hap.py program is available via jmcdani20/hap.py:v0.3.12 docker image. The command used for the assessment is described in the supplementary note.
For HG002, HG003, HG004, HG005 we used GIAB v4.2.1 truth set27 against GRCh38 reference and for HG001, HG005, HG006, HG007 samples, we used v3.3.2 variant benchmarks43 against GRCh37 reference genome. We used GIAB stratification v2.0 files with hap.py to derive stratified variant calling results. The GIAB benchmarking data availability is listed in data availability section of supplementary notes.
Mendelian Analysis
For our Mendelian Analysis we used RTG version 3.1256.
Phasing and haplotagging
We used Margin version v2.0 and WhatsHap22 version v1.0 to haplotag and phase the variants.
Margin is available in https://github.com/UCSC-nanopore-cgl/margin and WhatsHap is available in https://github.com/whatshap/whatshap. The details on how we ran these tools is described in supplementary notes.
Small variant switch error rate and hamming error rate determination
We used a Workflow Description Language (WDL)-based analysis pipeline whatshap.wdl available in https://github.com/tpesout/genomics_scripts to derive the switch error rate and hamming error rate compared to the GIAB truth set. The whatshap.wdl workflow invokes the stats and compare submodules available in whatshap version v1.0.
In our analysis, we compared phased variants against GRCh37 reference against GIAB v3.3.2 truth set to derive switch error rate and Hamming error rate. We only considered variants that have PATMAT annotation in the truth set and that fall in the high-confidence region defined by GIAB benchmarking set. The non-PATMAT annotated variants in the GIAB benchmarking variant set are not trio-confirmed so we did not use those to benchmarking our phasing methods. We used whatshap compare command to generate the whole genome switch error rate and used a custom script defined in whatshap.wdl to derive the hamming error rates.
Local phasing correctness calculation
For the Local Phasing Correctness (LPC) analysis, we used the calcLocalPhasingCorrectness executable found in the https://github.com/UCSC-nanopore-cgl/margin repository. The LPC analysis require a truth variant set and a query variant set. We used GIAB benchmarking set as truth. The calcLocalPhasingCorrectness generates a tsv file describing the results.
We used https://github.com/tpesout/genomics_scripts/plot_haplotagging_lpc.py script to visualize the results. The details of parameters is described in supplementary notes. The methods used in local phasing correctness as a metric is presented separately in the methods description.
Haplotagging accuracy and natural switch determination
We used https://github.com/tpesout/genomics_scripts/haplotagging_stats.py to calculate the haplotagging accuracy. The script calculates average haplotagging accuracy and average tagged reads per 10kb. Details on execution parameters is available in supplementary notes.
We visualized the natural switch error using compare_read_phasing_hapBam.py available in https://github.com/tpesout/genomics_scripts. Details on execution parameters is available in supplementary notes.
Phaseblock N50 calculation
An N50 value is a weighted median; it is the length of the sequence in a set for which all sequences of that length or greater sum to 50% of the set’s total size. We used the ngx_plot.py available from https://github.com/rlorigro/nanopore_assembly_and_polishing_assessment/ to plot phaseblock N50. From a phased VCF file, we extracted the phaseblock name, contig, start position, and end position to create the input file. We used 3272116950 as the size of the genome to maintain consistency with previous work13.
Variant calling and phasing analysis on Gencode annotated regions
We used Gencode v3542 to determine the variant calling and phasing accuracy in gene regions. The Gencode data is publicly available and can be found in data availably section of supplementary notes.
We used https://github.com/tpesout/genomics_scripts/gencode_to_stratification.py script to convert the Gencode regions to a bed file that is acceptable to hap.py. With the newly defined stratified regions from Gencode, we ran hap.py to determine the variant calling accuracy in gene regions.
Nanopore and PacBio HiFi de novo assembly generation
We used Shasta13 with a development build after version 0.7.0 available from https://github.com/chanzuckerberg/shasta (commit 06a639d36d26a4203c0b934d6e63c719750c5398) and Flye16 version 2.8.2 available from https://github.com/fenderglass/Flye to generate nanopore-based de novo assemblies. For PacBio-HiFi-based assemblies we used hifiasm14 version 0.14 available from https://github.com/chhylp123/hifiasm. The commands used to generate the assemblies are provided in execution parameters section of supplementary notes.
Assembly QV and switch error rate analysis
We assessed the assemblies with yak14 version 0.1 available from https://github.com/lh3/yak. YAK is a short-read kmer-based assembly quality estimator. We use short-reads for each sample to estimate the quality of the assembly with k-mer size of 31, With parental short-reads, YAK can also estimate the switch error rate in the assembly. WDL version of the pipeline standard_qc_haploid.wdl is available in https://github.com/human-pangenomics/hpp_production_workflows/.
Homopolymer run-length analysis
We used runLengthMatrix module of margin to derive the homopolymer run-length analysis between assembly and reads. In runLengthMatrix, we convert each read sequence into RLE form and track a map of raw positions to RLE positions. We convert from a raw alignment to RLE alignment by iterating through the matches in raw space and tracking the previous RLE match indices. From this set of matched read and reference RLE positions, we construct a confusion matrix. Details of the command are provided in execution parameters section of supplementary notes.
Small variant accuracy evaluation of assemblies
We used dipcall51 v0.2 to identify the small variants from the assemblies. The dipcall variant identification takes the maternal and paternal haplotypes generated by a phased assembly and a reference genome sequence. It maps the haplotypes to the reference and generates a VCF file containing all small variants identified in the assembly. For the haploid assembly, we provided the haploid assembly as both maternal and paternal haplotypes to dipcall. dipcall also generates a bed file containing regions where the assembly maps to the reference. We intersected the bed files to get regions that are assembled by all assembly methods and intersected with GIAB high-confidence region. Finally, we used hap.py to compare the variant calls derived from the assemblies against GIAB benchmarking VCF to get the accuracy statistics. Please see supplementary notes for dipcall parameters.
Structural variant accuracy evaluation of assemblies
We evaluated SV precision and recall for each assembly as follows. We aligned each set of contigs (either haploid or diploid) against the reference with minimap2 v2.18 using default parameters and the “asm5” preset. We have selected the hg19 reference (instead of the hg38 version) to be able to compare against the curated set of SVs in the HG002 genome47 that was initially produced using the hg19 reference. Given the reference alignments, we used svim-asm 1.0.246 in the respective (haploid or diploid) mode to call SV of size more than 50bp. Given two sets of SV calls, we used the SVbenchmark tool from the SVanalyzer package v0.36 with default parameters to estimate recall and precision. To estimate recall for homozygous and heterozygous SVs separately, we split each truth set into two respective parts. The statistics were evaluated within the GIAB Tier1 high-confidence regions that cover 2.51Gb of the human genome47.
The assemblies produced by hifiasm had high recall (97.8% homozygous 97.0% heterozygous) and precision (93%) against the HG002 curated SVs set (Supplementary Table 31). In addition, the recall and precision of the Shasta and PEPPER-Margin-DeepVariant assemblies measured against the HG002 curated SVs were highly correlated with recall and precision measured against the hifiasm SV calls. This allowed us to estimate the SV recovery accuracy of the Shasta and PEPPER-MarginDeepVariant assemblies for the HG005, HG0073, and HG02733 genomes, for which curated sets of SVs were not available.
Method description PEPPER
PEPPER is a recurrent neural network-based sequence prediction tool. In PEPPER, we use summary statistics derived from reads aligned to a reference to produce base probabilities for each genomic location using a neural network. We translate the position-specific base probabilities to the likelihood values of candidate variants observed from the read alignments. We propose candidate variants with likelihood value above a set threshold to DeepVariant for genotyping. Candidate pre-filtering with PEPPER ensures a balanced classification problem for DeepVariant and achieves high-quality variant calling from erroneous long-reads.
We use PEPPER in two steps in the variant calling pipeline. Initially, we use the PEPPER-SNP submodule to find single nucleotide polymorphisms (SNPs) from the initial unphased alignment file. In this setup, we tune PEPPER-SNP to have high precision so Margin can use the SNPs confidently to phase the genome. To this end, we also exclude insertions and deletions (INDELs) from the callset as they have notably worse performance for nanopore reads. Margin can then tag reads in the alignment file with predicted haplotypes.
After Margin, we use the PEPPER-HP submodule on the phased alignment file to generate haplotypespecific likelihoods for each candidate variant observed from the read alignments. In PEPPER-HP, we consider SNPs, insertions, and deletions as potential candidate variants. We propose the candidate variants with likelihood values higher than a set threshold to DeepVariant for genotyping with a more extensive convolutional neural network (CNN). We tune PEPPER-HP to achieve high-sensitivity but low-precision during candidate finding. The PEPPER-Margin-DeepVariant suite can identify small variants with high-quality from erroneous reads.
PEPPER-SNP
PEPPER-SNP is a submodule of PEPPER used to identify single nucleotide polymorphisms from reads aligned to a reference sequence. PEPPER-SNP works in three steps: image generation, inference, and candidate finding. First, we generate summary statistics from reads aligned to a reference sequence. We encode basic alignment statistics at each genomic location in an image-like tensor format. Second, we apply a recurrent neural network to predict the two most likely bases at each genomic location. Finally, we use the base predictions from each genomic location to compute the likelihoods of SNPs we observe from the reads. We filter candidate variants with a likelihood value below a set threshold to find a set of SNPs. Likelihood thresholds for PEPPER-SNP were determined by training the model on HG002 chr1-19, selecting an appropriate threshold, and verifying on chr20. The SNP set we get from PEPPER-SNP is used by Margin to phase the alignment file.
PEPPER-SNP: Image generation
In the image generation step of PEPPER-SNP, we generate summary statistics of base-level information per genomic location. The summary provides weighted observation of bases from all reads divided into nucleotide and orientation.
In PEPPER-SNP, we do not encode insert bases observed in reads to reference alignment as we only look for SNPs. We use a position value to represent a location in the reference sequence. For each genomic location, we iterate over all reads that align to that genomic location and encode ten features to encode base-level information: {A,C,G,T,Gap(*)} divided into two read orientations: {forward,reverse}. Finally, we normalize the weights of each genomic position based on the read coverage.
Supplemental figure 7 describes the feature encoding scheme we use in the image generation step of PEPPER-SNP. The top row of the image, annotated as REF, describes the reference base observed at each genomic location. The colors describing the bases are {A : Blue,C : Red,G : Green,T : Yellow,Gap(*) : White}. Each row after REF describes a feature; each feature encodes an observation of nucleotide bases from a forward, or a reverse strand read. We use ten features to encode base-level information: {A,C,G,T,Gap(*)} divided into two read orientations: {forward,reverse}. For example, AF encodes the observations of base A from forward-strand reads, and AR encodes observations of base A from reverse-strand reads. The columns describe genomic locations to the reference sequence.
In each column, we encode each observation as weights, which we show as alpha of each base. The grey weights are zero weights. At position (23), the weight distribution of {C,G} bases indicates a potential heterozygous variant at that position. The REF row is shown in the figure to describe the scheme; in practice, we do not encode the REF row.
In the inference step of PEPPER-SNP, we use a recurrent neural network for sequence prediction. The network architecture consists of two bidirectional gated recurrent unit (GRU) layers and a linear transformation layer. The linear transformation layer produces a prediction of two bases for each genomic location present in the summary image. To identify potential variants, we use 15 class-labels for base prediction: {AA,AC,AG,AT,A*,CC,CG,CT,C*,GG,GT,G*,TT,T*,**}. We do not use co-linear classes like CA and AC as two separate classes, as it is not possible to differentiate between these two classes from the summary observations.
From the image-generation module of PEPPER-SNP, we get summary images in 1kb chunks. We use a sliding window method during inference and chunk the 1kb segments into multiple overlapping windows of 100bp segments. We first run inference on the leftmost window and go to the next window with 50bp overlapping bases. We pass the hidden state output from the left window to the next window and keep a global inference counter to record base-predictions.
PEPPER-SNP: Inference model
Supplemental figure 8 describes the neural network-based inference scheme. The two dotted boxes indicate two adjacent windows with overlapping sequences. The top panel shows the inference scheme. We start with the first window and produce base predictions for each genomic location present in that window; then, we slide the window to the right. We do this from left to right on the entire genomic sequence. We record the base predictions from all windows in a global counter and report them to the candidate finder to calculate candidate likelihoods.
We trained the inference model using a gradient descent method. We use adaptive moment estimation (Adam57) to compute gradients based on a cross-entropy loss function. The loss function is defined to calculate the prediction performance at each genomic location against a labeled set of expected base observations derived from the Genome-In-A-Bottle (GIAB) truth set. The gradient optimization attempts to minimize the loss function by tuning the parameters of the neural network.
We trained PEPPER-SNP with 100× coverage of HG002 data subsampled at different coverage values (20x–100x). We split the training sets into three sets: train, test, and holdout. We use chromosome 1 to 19 for training, chromosome 20 for testing, and we keep 21 and 22 as holdout sets. We train the models for several epochs and test after each epoch. Finally, we pick a model that performs the best on the holdout dataset.
PEPPER-SNP: Candidate finding
In the candidate finding step of PEPPER-SNP, we take the base-predictions and derive likelihoods of SNPs we observe from the read alignments. If the likelihood value of a variant is above a set threshold, we pick that allele to be a real variant.
In PEPPER-SNP, we derive a allele probability (AP) and a non-reference observation likelihood (NR) for each observed SNP. First, at each genomic location (pos) we use the prediction vector of baseclasses {AA,AC,AG,AT,A*,CC,CG,CT,C*,GG,GT,G*,TT,T*,**} to derive two prediction vectors V1[pos] = [A1,C1,G1,T1,Gap(*)1] and V2[pos] = [A2,C2,G2,T2,Gap(*)2]. For example, the predicted value of class {GT} contributes to V1[pos][G1] and V2[pos][T2] values. Then, we iterate over all the reads to find potential SNPs by recording each read base that does not match the reference base. We calculate the likelihood of candidate base b, observed at position pos, by taking the maximum likelihood from the prediction vectors V1[pos] and V2[pos] as shown in equation 1 where we denote allele likelihood as AP.
| (1) |
We also calculate the likelihood of non-reference base observation (NR) to estimate the likelihood of observing any allele other than the reference allele at a location. We derive non-reference base observation likelihood from prediction vectors V1 and V2 independently and take the maximum value between two values. For each prediction vector, we take the sum of the values, subtract the observation likelihood of the reference base, and divide by the sum of the prediction vector. Equation 2 describes the calculation NR in PEPPER-SNP.
| (2) |
Finally, we derive a genotype for the variant from the prediction vector V1 and V2. If a variant has a likelihood above a set threshold observed in both V1 and V2, we set the genotype as a homozygous alternate (1/1). If the likelihood is above the threshold in one vector but not in the other, we call it a heterozygous variant (0/1). For each variant, we use NR value of that position to be the genotyping quality.
PEPPER-SNP: Code availability
PEPPER-SNP is available at https://github.com/kishwarshafin/pepper.
PEPPER-HP: Haplotype-aware sequence prediction.
PEPPER-HP is a haplotype-aware sequence prediction tool designed to find candidate variants from read alignments. In PEPPER-HP, we take a phased alignment file as input where each read has a haplotag of {0,1,2} indicating which haplotype the read represents (or lack of haplotype information), and we output a set of SNP, insertion, and deletion candidates for genotyping using DeepVariant.
Similar to the PEPPER-SNP submodule, PEPPER-HP has three steps, image generation, inference, and candidate finding. In the image generation step, we generate two sets of summary statistics, one per haplotype, and save them as image-like tensors. We use a recurrent neural network to predict bases on each haplotype for each genomic location in the inference step. Finally, we calculate likelihood values for SNP and INDEL candidates based on base-predictions of each haplotype. We consider candidates with likelihood values over a certain threshold to be candidate variants and propose them to DeepVariant for genotyping. Similarly, likelihood thresholds for PEPPER-HP were determined by training the model on HG002 chr1-19, selecting an appropriate threshold, and verifying on chr20.
PEPPER-HP: Image generation
In the image generation step of PEPPER-HP, the input is an alignment file with phased reads, and we generate image-like summary statistics of base-level information per genomic location for each haplotype {0,1}. The summary of haplotype 1 provides weighted observation of bases from reads with haplotag 1 divided into nucleotide and orientation. Similarly, the haplotype 2 summary provides weighted observations of bases from reads with haplotag 2. Reads that are unphased or have haplotag 0 contribute to summary statistics for both haplotypes.
In PEPPER-HP, we represent position in reference sequence using two values: position and index. The position value indicates a location in the reference sequence, and we use the index to accommodate insert alleles anchored to a position. All reference sequence positions have an index of 0. On each haplotype {0,1}, we iterate over all haplotype associated reads that align a genomic location and encode ten features to encode base-level information: {A,C,G,T,Gap(*)} divided into two read orientations: {forward,reverse}. The weights depend on the mapping quality and base quality of the associated reads. Finally, we normalize the weights of each genomic position based on the haplotype associated read coverage.
PEPPER-HP: Inference model
In Supplemental figure 9, we describe the feature encoding scheme of PEPPER-HP. We derive two summary statistics based on the haplotype association of the reads. The top row of the image, annotated as REF, describes the reference base observed at each genomic location. The colors describing the bases are {A : Blue,C : Red,G : Green,T : Yellow,Gap(*) : White}. Each column represents a reference position with two values (pos,index). For example, pos (14,0) is the reference reference position 14 and (14,1) is the insert base anchored in position (14,0). For each haplotype, we use ten features to encode base-level information: {A,C,G,T,Gap(*)} divided into two read orientations: {forward,reverse}. From the summary, we can see that at location (23,0) HP-1 observers C bases where the reference is T but in HP-2 the reads observe C bases that match with the reference, denoting a heterozygous variant present in haplotype-1 sequence.
We use a recurrent neural network for sequence prediction on haplotype-specific images of PEPPERHP. The network architecture consists of two bidirectional gated recurrent unit (GRU) layers and a linear transformation layer. For each haplotype, the linear transformation layer predicts a base for each genomic location present in the image-like tensor. We use five class-labels for base prediction: {A,C,G,T,Gap(*)}.
The haplotype-specific images in 1kb chunks, and we use a sliding window method to go over the 1kb chunk in overlapping 100bp segments on both haplotype images simultaneously. We start from the leftmost window of 100bp and slide the window to 50bp to the right for the next step. We use two global counters from the base predictions, one per haplotype, to record the haplotype-specific base predictions.
The inference scheme of PEPPER-HP is shown in Supplemental figure 10. We have two haplotypespecific image for each genomic region representing two haplotypes. For each haplotype, we start from the leftmost window and generate haplotype-specific base predictions. The base predictions are recorded in two global counters.
We train the PEPPER-HP inference model using a gradient descent method. We use adaptive moment estimation (Adam) to compute gradients based on a cross-entropy loss function. The loss function is defined to calculate the prediction performance at each genomic location against a labeled set of expected base observations derived from the Genome-In-A-Bottle (GIAB) truth set. We use GIAB v3.3.2 truth set as the variants in v3.3.2 are phased. We use phase-specific base predictions to optimize PEPPER-HP model for each haplotype.
We train PEPPER-HP with three sets of HG002 data with 50×, 80×, and 100× coverage. We further generate multiple train sets by arbitrarily downsampling the three training sets at different fractions. We split the training sets into three sets: train, test, and holdout. We use chromosome 1 to 19 for training, chromosome 20 for testing, and we keep 21 and 22 as holdout sets. We train the models for several epochs and test after each epoch. Finally, we pick the model that performs the best on the holdout dataset.
PEPPER-HP: Candidate finding
In the candidate finding step of PEPPER-HP, we evaluate variants observed from the read alignments. We use the base-predictions from the neural network to calculate the likelihood of an observed allele. If the likelihood value of a candidate variant is above a set threshold, we pick that variant as a potential candidate for DeepVariant to assess.
We evaluate the SNPs and INDELs observed in read alignments to find potential candidate variants. First, at each genomic location (pos,index) we use the haplotype-specific prediction values of baseclasses VH1[pos,index] = [A1,C1,G1,T1,Gap(*)1] and VH2[pos,index] = [A2,C2,G2,T2,Gap(*)2]. We iterate over all the reads to find potential variants by recording the read base that does not match the reference base. We calculate the allele likelihood of a SNP candidate APSNP with base observation b, observed at position (pos,index), by taking the maximum likelihood from the prediction vectors VH1[(pos,index)] and VH2[(pos,index)] as described in equation 3. For inserts and deletes we extend the likelihood calculation to cover the length of the allele.
| (3) |
We also calculate the likelihood of observing a variant other than the reference allele at any location. In equation 4, let R[(pos,index)] be the reference base at location (pos,index) and N be the maximum index value observed in position pos. The reference base at any position with index > 0 is gap(*). We take the maximum value of observing a non-reference base between (pos,0) and (pos,max_index[pos]). For each index, we calculate the total value of the prediction vector, subtract the observation likelihood of the reference base and divide by the sum of the prediction vector. The non-reference observation likelihood NR is the maximum value we observe across index values. For insertion and deletion alleles we cover the allele length and take the maximum value as the NR for those candidates.
| (4) |
Based on the allele likelihood and non-reference observation likelihood, we calculate a likelihood value for each type of candidate SNP, insert and delete. Then for each type, we set a threshold value and if a candidate passes the threshold value, we propose the candidate to DeepVariant for genotyping.
PEPPER-HP: Code availability
PEPPER-HP is available at https://github.com/kishwarshafin/pepper as a submodule.
Margin
Margin is a suite of tools employing Hidden Markov Models (HMMs) to perform genomic analysis with long reads. MarginPhase (the first module) was introduced alongside WhatsHap as a tool performing joint genotyping and phasing22. MarginPolish (the second module) was introduced as a graph-based assembly polisher which can do standalone polishing and is the first step in a two-part polishing framework MarginPolish-HELEN13. Release 2.0 of Margin incorporates both tools into one suite, including diploid-aware polishing in MarginPolish which has informed improvements in a new iteration of MarginPhase.
In this paper we focus exclusively on improvements made to the phasing submodule. The core partitioning algorithm is described in our previous work22, but we provide a summary here of the previous methodology, followed by a description of the modifications made in the current iteration. First, we give a high-level overview of the phasing workflow.
Overview
Margin takes as input an alignment (BAM), reference (FASTA), and variant set (VCF). It determines regions to run on from the alignment and variant set, and it breaks the input into chunks to enable multiprocessing. For each chunk, it extracts reference substrings around each variant site, and read substrings aligning around each variant site. For each variant site, it calculates the alignment likelihood between all substrings and all variant alleles. These likelihoods are used in the core phasing algorithm which bipartitions reads and assigns alleles to haplotypes. After all chunks have been analyzed, we stitch the chunks together to produce results across whole contigs. Last, we output a copy of the input BAM with haplotagged reads and a copy of the input VCF with phased variants.
Parameterization.
Margin is parameterized with a configuration file. For each configuration, parameters are grouped into polish and phase sections. There is overlap in the parameters used by both submodules, so some parameters used by the phasing algorithm fall under the polish heading and vice versa. When referenced in this document, we specify the full path of the parameter and the default value associated with the intended configuration. Default parameter values and thresholds were determined by experimentation on the HG002 sample.
Core Phasing Algorithm
For the core phasing algorithm, we construct a graph G = (VG,EG) describing all possible bipartitions of reads with positions as variant sites, vertices as a combination of position and read bipartitions, and edges as possible transitions between bipartitions for adjacent positions. For example, at position Pi with aligned reads R1 and R2, we have the possible vertices Vi0 with haplotypes H1 = {R1,R2} and H2 = {} (R1,R2/.), Vi1 = R1/R2, Vi2 = R2/R1, and Vi3 = ./R1,R2. At position Pj with the same aligned reads R1, R2 and a new read R3, vertex Vj0 = R1,R2,R3/. and Vj1 = R1,R2/R3 are both connected to vertex Vi0 because all reads shared between vertices are have the same haplotype assignment, but Vj2 = R1/R2,R3 is not connected to Vi0 because read R2 has different haplotype assignments in the two vertices. We extend each vertex as described above to additionally represent all possible genotypes. After running the forward-backward algorithm on this graph, at each position the posterior distribution over states describing read bipartitions and genotypes can be marginalized to determine the most likely genotype.
The state space for this algorithm increases exponentially with the number of reads at each position. To account for this, Margin implements a pruning and merging heuristic where the input is divided into smaller pieces, unlikely states are pruned, and the resulting graphs are merged before running the full forward-backward algorithm.
Improved Functionality
One of the most significant changes to the Margin workflow is that we now only analyze sites proposed by the input VCF. Previously we considered any reference position where less than 80% of the nucleotides agreed with the reference base as a candidate variant site. To determine which proposed variants are considered, we read the input VCF and remove all INDEL variants (phase.onlyUseSNPVCFEntries = false), all homozygous variants (phase.includeHomozygousVCFEntries = false), all non-PASS variants (phase.onlyUsePassVCFEntries = true), and all variants with a quality score below phase.minVariantQuality = 10. For each chunk, we perform an adaptive sampling of variants (phase.useVariantSelectionAdaptiveSampling = true) where we start by taking all variants with a quality score above phase.variantSelectionAdaptiveSamplingPrimaryThreshold = 20. If the average distance between these variants for the chunk is greater than phase. variantSelectionAdaptiveSamplingDesiredBasepairsPerVariant = 2000, we take variants ordered by quality score descending until we achieve the desired number of variants. These values were determined after experimentation on HG002.
Instead of considering only the nucleotide aligned directly to the variant position as the first iteration of Margin had done, we now extract substrings from the reference and reads and perform an alignment to determine which allele the read most likely originated from. In theory, this allows Margin to use INDELs during phasing, although for our current evaluations we do not test this functionality. When extracting reference bases, we take phase.columnAnchorTrim = 12 bp from the reference before the variant position, and the same amount upstream from the end of the variant position. All alleles (including the reference allele) in the VCF are recreated by substituting the replaced reference base with the allelic sequence from the VCF. Note that there is no further modification of the reference sequences for cases where multiple variants fall within the extracted region. From the reads, we extract all sequence between the first and last position matched to the reference over the extracted reference region. Reads substrings for which the average base-level quality score over the substring is less than polish.minAvgBaseQuality = 10 are excluded at these positions.
The original Margin phasing HMM used an emission model based off of a read error model (empirically determined) and a mutation likelihood model (based on likelihood of specific mutations). In the new implementation, we replace this with a likelihood generated by aligning the read substrings to the allelic substrings.
As in the previous iteration, after running the forward-backward algorithm on the HMM we marginalize over possible genotypes at each variant site to determine final predicted haplotypes. Given these haplotypes and the read-to-allele alignment likelihoods, we compute from which haplotype the read most likely originated using the joint probability of the read substrings aligning to the alleles for each haplotype. This haplotyping step is performed for all reads used for phasing, as well as other reads filtered out before phasing (as described below in the Chunking section). If a read does not span any variants or has equal likelihood of aligning to both haplotypes, no prediction is made regarding the haplotype from which it originated.
Chunking
Margin divides input into polish.chunkSize = 100000 bp chunks with polish.chunkBoundary = 10000 bp boundary on both ends, resulting in 2x polish.chunkBoundary overlap between chunks. Once the boundaries have been determined, the ordering of the chunks is mutated (polish.shuffleChunks = true) by default based on descending order of size (polish.shuffleChunksMethod = size_desc), with random ordering (random) also as a configurable option. While the ordering does not have a large effect on the runtime during phasing, we found that deep chunks would take drastically longer to complete for polishing, and ensuring that they were completed first would reduce overall runtime for the submodule.
When operating on a chunk, Margin first extracts all the reads that have an alignment between the start and end of the chunk, tracking all alignments falling between the start and end of the extended boundary. Margin then collects a set of reads to run the main algorithm on, first by removing reads that have a MQ score below polish.filterAlignmentsWithMapQBelowThisThreshold = 10, are secondary alignments (polish.includeSecondaryAlignments = false), or are supplementary alignments (polish.includeSupplementaryAlignments = false). Given this set of reads, we downsample to an expected depth of polish.maxDepth = 64 for haplotagging and polish.maxDepth = 32 for variant phasing. The downsampling is biased to maximize coverage over heterozygous variants given the constraint on expected coverage. To accomplish this, we compute the sampling probabilities for each read according to the following linear program, which we solve using the LP Solve library (http://lpsolve.sourceforge.net/).
| (5) |
where pi is the probability of selecting read i, vi is the number of heterozygous variants on read i, li is the length of read i, C is the desired coverage, and L is the length of the chunk.
The algorithm runs on the chunk region with the primary set of reads (all reads kept after filtering and downsampling) and assigns reads and variants to haplotypes. Then it takes any removed reads (either through filtering or downsampling) and assigns them to haplotypes as described above. Margin tracks the assignment of variants to haplotypes within the chunk (not including the boundaries), and of reads to haplotypes for the whole region (including the boundaries).
To stitch two chunks together, we need to determine whether the two previous haplotypes (P1,P2) are oriented with the two current haplotypes (C1,C2) in cis (P1C1,P2C2) or in trans (P1C2,P2C1). To do this, we compare the number of reads that are in cis and in trans between the two chunks; if there are more reads in trans, we switch the haplotypes of the current chunk’s reads and variants. To mulithread this process, Margin separates all the chunks in a contig into numThreads contiguous groups. The chunks in each of the groups are stitched together by a single thread, and then the same stitching process is used to stitch each of these groups together to complete the whole contig.
The final assignment of a read to a haplotype is determined by the haplotype it was assigned to in the first chunk for the contig. The final assignment of an allele to a haplotype is determined by the chunk it falls within (boundary region excluded).
Phaseset Determination
When writing the output VCF, Margin makes predictions about which sets of variants are confidently inherited together and annotates the output with phaseset (PS) tags. Margin will assign a phaseset to a variant if it is heterozygous, a SNP (phase.onlyUseSNPVCFEntries = true), and if it agrees with the genotype from the input VCF (phase.updateAllOutputVCFFormatFields = false). For variants meeting this criteria, as Margin iterates through the VCF it will extend the current phaseset unless (a) the variant is the first in the contig, (b) there are no reads spanning between the current variant and the previous variant, (c) there is an unlikely division of reads for the variant (explained below), or (d) the reads spanning the current variant and the previous variant are discordant above some threshold (explained below). The values described below were determined after experimentation on HG002.
To identify unlikely divisions of reads (which we take as potential evidence there is an error in the phasing), we take the number of primary reads assigned to each haplotype and find the binomial p-value for that division of reads. If that probability is less than the threshold phase.phasesetMinBinomialReadSplitLikelihood = 0.0000001, we create a new phase set for this variant.
Within each chunk region (boundary region excluded) and after determining haplotype assignment for the reads, we track which primary reads were used for phasing and to which haplotype they were assigned in the chunk. This serves as a check against poorly-phased or poorly-stitched chunks. To determine discordancy in the phasing between variants, we compare the number of reads which are in cis or “concordant” (Cc) given the read assignment to adjacent variants, and the number in trans or “discordant” (Cd) between variants. If the discordancy ratio Cd/(Cc + Cd) is greater than phase.phasesetMaxDiscordantRatio = 0.5, we create a new phase set for this variant.
Margin: Code availability
Margin is available at https://github.com/UCSC-nanopore-cgl/margin.
Local Phasing Correctness
The local phasing correctness (LPC) is a novel metric for measuring phasing accuracy that we developed for this study. More precisely, the LPC is a family of metrics parameterized by a varying parameter ρ ∈ [0,1], which controls the degree of locality. The LPC can be seen as a generalization of the two most common metrics used to evaluate phasing accuracy: the switch error rate and the Hamming rate. The switch error rate corresponds to the LPC with ρ = 0 (fully local), and the Hamming rate is closely related to the LPC with ρ = 1 (fully global). With intermediate values of ρ, the LPC can measure meso-level phasing accuracy that the two existing metrics cannot quantify.
The LPC consists of a sum over all pairs of heterozygous variants where each pair contributes an amount that decays with greater genomic distance. If the variants are incorrectly phased relative to each other, the pair contributes 0. This sum is normalized by its maximum value so that the LPC is always takes a value between 0 and 1. In mathematical notation,
where d(i,j) is the distance between variants i and j, and δ(i,j) is an indicator for whether variants i and j are correctly phased relative to each other. In the case that ρ = 0 the above formula is undefined, and we instead take the limit of LPCρ as ρ → 0.
If we take d(i,j) to be |i−j|, then it can be shown that LPC0 is equivalent to the complement of the switch error rate (i.e. the “switch correctness” rate). LPC1 is not equivalent to the Hamming rate, but they are monotonic functions of each other. Thus, LPC1 and Hamming rate always produce the same relative ranking but not the same numerical value. Alternatively, we can take d(i,j) to be the genomic distance between the variants, measured in base pairs. In doing so, the LPC no longer has the provable relationships to switch error rate and Hamming rate, neither of which has any mechanism to incorporate genomic distance. However, we consider the genomic distance to be a more relevant measurement of distance for phasing accuracy than the variants’ ordinal numbers. Accordingly, we believe that this amounts to a further strength of the LPC over existing metrics, and all LPC values reported in this work use this definition of distance.
The ρ parameter is mathematically convenient but difficult to interpret. To improve interpretability, we can reparameterize the LPC with λ = − log2/logρ, which we call the “length scale”. This is the distance (measured either in base pairs or number of variants) at which a pair of variants has 1/2 of the maximum weight. The length scale gives an approximate sense of the scale of distances that the LPC incorporates, although it is worth stressing that pairs that are closer together always receive more weight than pairs that are further apart.
Local Phasing Correctness: Code availability
The code used to calculate LPC is available at https://github.com/UCSC-nanopore-cgl/margin.
DeepVariant
DeepVariant is a small variant identification method based on deep neural networks that achieve high performance on different short-read and long-read sequencing platforms36. DeepVariant has released models for different datatypes and a details of the methods implemented in DeepVariant can be found in associated releases21,36. Here we present an overview of the methods we implemented to achieve high performance with nanopore data.
Adapting DeepVariant to Oxford Nanopore reads
DeepVariant performs variant calling in three stages, “make examples”, “call variants”, and “postprocess variants”. In “make examples”, potential variant positions are identified by applying a minimal threshold for evidence. In positions meeting the candidate generation criteria, the reads overlapping the position are converted into a pileup of a 221-bp window centered at the variant. Multiple features of the reads are represented as different dimensions in the pileup, including the read base, base quality, mapping quality, strand, whether the read supports the variant, and whether the base matches the reference. Prior to this work, the heuristics for candidate generation were simple (at least 2 reads supporting a variant allele and a variant allele fraction at least 0.12). However, the higher error rate of Oxford Nanopore data generated far too many candidates for DeepVariant to call variants in a genome in a reasonable time. To integrate DeepVariant with PEPPER, we developed the ability to import candidates directly from a VCF to replace the logic in “make examples”. This allows DeepVariant to read the output of PEPPER, and in theory, can be used in a similar manner with the outputs of other methods.
DeepVariant has released models for different datatypes21,36. In order to achieve high performance on Oxford Nanopore data, a new DeepVariant model trained for this datatype was required. For this, we modified the training process for DeepVariant. The training process for DeepVariant is very similar to the variant calling process (generating candidates from make examples in a similar manner), but with the addition of a step which reconciles a candidate variant with the truth label from Genome-in-a-Bottle. We adapted this process to use the VcfCandidateImporter to propose PEPPER candidates. Because the representations of variants can diverge from the representation in GIAB (even for the same event), modifying the process for training required multiple rounds of iteration to identify mislabeled edge cases and to modify the proposed candidate representation for training.
With these modifications, training of DeepVariant models using the existing machinery could proceed.
These modifications also allowed existing logic for the stages “call variants” and “postprocess variants” to work directly with the trained Oxford Nanopore model.
DeepVariant: Code availability
All code in the DeepVariant repository incorporates the improvements made for this paper (https://github.com/google/deepvariant). This repository also contains a model retraining tutorial (https://github.com/google/deepvariant/blob/r1.1/docs/deepvariant-training-case-study.md).
Training PEPPER-DeepVariant for ONT basecallers
It is necessary to train new models once the underlying read quality of Oxford Nanopore changes (i.e. basecaller update). It is possible to take the existing PEPPER model and re-train it with 100x HG002 on one gpu for few hours to get a model for the newer chemistry. Similarly, it is possible to take the existing DeepVariant model and train the model for few hours on one GPU to get a model for the new basecaller. A case study of re-training an Illumina model for BGISEQ is available here (https://github.com/google/deepvariant/blob/r1.1/docs/deepvariant-training-case-study.md). As the basecaller updates of ONT are incremental, we believe retraining to newer basecall sets will not have resource bottleneck.
Assembly polishing with PEPPER-Margin-DeepVariant
The assembly polishing method of PEPPER-Margin-DeepVariant is described below:
PEPPER-SNP: PEPPER-SNP finds single nucleotide polymorphisms (SNPs) from the read alignments to a haploid assembly using a RNN. For assembly polishing, we use the same infrastructure described in the haplotype-aware variant calling section.
Margin: Margin takes the SNPs reported by PEPPER-SNP and generates a haplotagged alignment file using a HMM. For assembly polishing, we use the same infrastructure described in the haplotype-aware variant calling section.
- PEPPER-HP: PEPPER-HP takes the haplotagged alignment file and evaluates each haplotype independently and produces haplotype-specific candidate SNP and INDEL-like errors present in the assembly.
- Haplotype-1 and Haplotype-2 candidate finding: For haplotype-1, we take reads with HP-1 and HP-0 and generate base-level summary statistics. Then we use a RNN to produce nucleotide base predictions for each location of the genome. Finally, we find all the observed SNP or INDEL-like candidates in the reads with HP-1 and HP-0 tag and calculate a likelihood of each candidate to be a potential error in the assembly. If the likelihood of the candidate is above a set threshold, we propose that candidate as a potential edit for haplotype-1(HP-1) of the assembly. For haplotype-2, we take reads with HP-2 and HP-0 and find SNP and INDEL-like candidate errors following the same process described in Haplotype-1 candidate finding.
- DeepVariant: DeepVariant takes the haplotype-specific candidate set from PEPPER-HP and the haplotagged alignment from Margin to identify errors present in the assembly using a convolutional neural network (CNN).
-
Haplotype-1 and haplotype-2 polishing: For each candidate error from haplotype-1 set proposed by PEPPER-HP, we generate a feature set representing base, base-quality, mapping quality etc. in a tensor like format. In haplotype-specific polishing setup, reads are sorted by their haplotags in the order {HP-1, HP-0, HP-2}. Then we use a pre-trained inception_v3 CNN to generate a prediction of {0/0, 1/1} for each candidate. Candidates in haplotype-1 that are classified as {1/1} are considered as missing heterozygous variants from the haploid assembly or an error present in the assembly. We apply the candidates of haplotype-1 classified as {1/1} to the haploid assembly using bcftools consensus to get a polished assembly haplotype_1.fasta representing one haplotype of the sample.For haplotype-2, we sort the reads by their haplotags in the order {HP-2, HP-0, HP-1}. Candidates in haplotype-2 that are classified as {1/1} are similarly considered to be a missing heterozygous variant or an error present in the assembly, and are applied to the haploid assembly to get the second polished haplotype (haplotype_2.fasta).
-
We trained the PEPPER-Margin-DeepVariant assembly polishing method with HG002 assembly generated by the Shasta assembler. We first aligned the HG002 Shasta assembly to GRCh38 reference with dipcall51 to associate assembly contigs with GRCh38 reference. Then we aligned HG002 GIAB v4.2.1 small variant benchmarking set to the assembly and marked the high-confidence regions to restrict training only in regions that GIAB notes as high-quality. The alignment between HG002 GIAB v4.2.1 to the assembly produced the training set. We trained PEPPER and DeepVariant on contigs that associate to chr1-chr19 and used chr20 as a holdout set. We used the same approach for Oxford Nanopore and PacBio HiFi based models.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Human Genome Research Institute of the National Institutes of Health under Award Numbers U41HG010972, R01HG010485, U01HG010961, and OT2OD026682 (K.S., T.P., M.K., J.M.E, K.H.M, M.J., B.P.). We thank Circulomics Inc. for sharing HG001 Nanopore data. We thank Justin Zook and Justin Wagner from the National Institute of Standards and Technology (NIST) for providing a draft version of the HG005 v4.2.1 benchmark. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
K.S. has performed paid internships at NVIDIA Corp and Google LLC. P.C., M.N., A.K., S.G., G.B., A.C. are employees of Google and own Alphabet stock as part of the standard compensation package. M.J. has received reimbursement for travel, accommodation, and conference fees to speak at events organized by ONT. The remaining authors declare no competing interests.
References
- 1.Altshuler DM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W & Freudenberg J Mappability and read length. Front. Genet 5, 381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaisson MJP et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belton JM et al. Hi-C: A comprehensive technique to capture the conformation of genomes. Methods 58, 268–276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falconer E & Lansdorp PM Strand-seq: A unifying tool for studies of chromosome segregation. Seminars in Cell and Developmental Biology vol. 24 643–652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisenfeld NI, Kumar V, Shah P, Church DM & Jaffe DB Direct determination of diploid genome sequences. Genome Res. 27, 757–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain M et al. Improved data analysis for the MinION nanopore sequencer. Nat. Methods 12, 351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eid J et al. Real-time DNA sequencing from single polymerase molecules. Science (80-.) 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Jain C, Rhie A, Hansen N, Koren S & Phillippy AM A long read mapping method for highly repetitive reference sequences. bioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miga KH et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Logsdon Glennis A. et al. “The structure, function and evolution of a complete human chromosome 8.” Nature 593, no. 7857 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shafin K et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H, Concepcion GT, Feng X, Zhang H & Li H Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 1–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurk S et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 30, 1291–1305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolmogorov M, Yuan J, Lin Y & Pevzner PA Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol 37, 540–546 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Ruan J & Li H Fast and accurate long-read assembly with wtdbg2. Nat. Methods 17, 155–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.nanoporetech/medaka: Sequence correction provided by ONT Research. https://github.com/nanoporetech/medaka.
- 19.Luo R et al. Exploring the limit of using a deep neural network on pileup data for germline variant calling. Nat. Mach. Intell 2, 220–227 (2020). [Google Scholar]
- 20.Edge P & Bansal V Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat. Commun 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wenger AM et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebler J, Haukness M, Pesout T, Marschall T & Paten B Haplotype-aware diplotyping from noisy long reads. Genome Biol. 20, 116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huddleston J et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sedlazeck FJ et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson MD et al. WhatsHap: Weighted Haplotype Assembly for Future-Generation Sequencing Reads. J. Comput. Biol 22, 498–509 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Zook JM et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. data 3, 160025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner J et al. Benchmarking challenging small variants with linked and long reads. BioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson ND et al. precisionFDA Truth Challenge V2: Calling variants from short-and long-reads in difficult-to-map regions. bioRxiv (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain M et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol 36, 338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain M et al. Linear assembly of a human centromere on the Y chromosome. Nat. Biotechnol 36, 321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiddes IT et al. Comparative Annotation Toolkit (CAT)—simultaneous clade and personal genome annotation. Genome Res. 28, 1029–1038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eichler EE, Clark RA & She X An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet 5, 345 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Euskirchen P et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 134, 691–703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rang FJ, Kloosterman WP & de Ridder J From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 19, 90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin C-S et al. A diploid assembly-based benchmark for variants in the major histocompatibility complex. Nat. Commun 11, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poplin R et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol 36, 983 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Edge P, Bafna V & Bansal V HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 27, 801–812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez OL et al. A novel framework for characterizing genomic haplotype diversity in the human immunoglobulin heavy chain locus. Front. Immunol 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin C-S et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koren S et al. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol 36, 1174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porubsky D et al. Fully phased human genome assembly without parental data using single-cell strand sequencing and long reads. Nat. Biotechnol 39, 302–308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrow J et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zook JM et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol 37, 561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baid G et al. An Extensive Sequence Dataset of Gold-Standard Samples for Benchmarking and Development. bioRxiv (2020). [Google Scholar]
- 45.Frankish A et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heller David, and Vingron Martin. “SVIM-asm: Structural variant detection from haploid and diploid genome assemblies.” Bioinformatics 36.22–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zook JM et al. A robust benchmark for detection of germline large deletions and insertions. Nat. Biotechnol 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tewhey R, Bansal V, Torkamani A, Topol EJ & Schork NJ The importance of phase information for human genomics. Nat. Rev. Genet 12, 215–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Browning SR & Browning BL Haplotype phasing: existing methods and new developments. Nat. Rev. Genet 12, 703–714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glusman G, Cox HC & Roach JC Whole-genome haplotyping approaches and genomic medicine. Genome Med. 6, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat. Methods 15, 595–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.K. S, T. P, P. C, N. M, A. K, S. G, G. B, M. K, J.M. E, K.H. M, P. C, M. J, A. C, & B. P (2021). PEPPER-Margin-DeepVariant (Version r0.4) [Computer software] Zenodo, 10.5281/zenodo.5275510. [DOI] [Google Scholar]
- 53.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krusche P et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol 37, 555–560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cleary JG et al. Joint variant and de novo mutation identification on pedigrees from high-throughput sequencing data. J. Comput. Biol 21, 405–419 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Newey WK Adaptive estimation of regression models via moment restrictions. J. Econom 38, 301–339 (1988). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Analysis data available
We have made the analysis data available publicly (variant calling outputs, genome assemblies etc.) in: https://console.cloud.google.com/storage/browser/pepper-deepvariant-public/analysis_data.
The source data for the main figures can be found in: https://console.cloud.google.com/storage/browser/pepper-deepvariant-public/figure_source_data/Figure_source_data/
Sequencing data availability
We used several publicly available datasets:
Genome-In-a-Bottle consortium26,28: https://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/
Human Pangenome Reference Consortium (HPRC): https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html
Telomere-to-telomere consortium11,12: https://github.com/nanopore-wgs-consortium/CHM13
Please see supplementary notes to find specific links to the sequencing data that we used for our analysis.


