Abstract
Arginine methylation is an influential post-translational modification occurring on histones, RNA binding proteins and many other cellular proteins affecting their function by altering their protein-protein and protein-nucleic acid interactions. Recently, a wealth of information has been gathered implicating the protein arginine methyltransferases (PRMTs), the enzymes that deposit arginine methylation, in transcription, pre-mRNA splicing, DNA damage signaling and immune signaling with major implications for cancer therapy, especially immunotherapy. This review will summarize this recent progress and the current state of PRMT inhibitors, some in clinical trials, as promising drug targets for cancer.
Keywords: Arginine methylation, PRMTs, S-adenosylmethionine, epigenetics, DNA damage signaling, immunotherapy, pre-mRNA splicing, transcription, small molecule inhibitors
I. Protein Arginine Methyltransferases (PRMTs)
In mammals, a family of nine sequence-related protein arginine methyltransferases (PRMT1-9) modifies a wide variety of proteins using the cofactor S-adenosylmethionine (AdoMet) to generate ω-NG,monomethyl-arginines (MMA), ω-NG,NG asymmetric dimethyl-arginines (aDMA) and ω-NG,N′G-symmetric dimethyl-arginines (sDMA) (Figure 1A). Type I PRMTs catalyze the formation of MMA and aDMA (PRMT1, 2, 3, CARM1/PRMT4, 6, and 8). Type II PRMTs (PRMT5 and PRMT9) catalyze the formation of MMA and sDMA, while the type III PRMT (PRMT7) catalyzes only the formation of MMA (Bedford and Clarke, 2009). The core structure of PRMTs is similar forming two domains (Antonysamy et al., 2012; Cheng, 2014), an N-terminal Rossmann-fold where AdoMet binding occurs and a C-terminal β-barrel domain (Tewary et al., 2019). As a prototype of type I, PRMT1, is a ring-shaped head-to-tail homodimer (Zhang and Cheng, 2003). The type II PRMT5 has the Rossmann-fold, a β-barrel domain for dimerization and a TIM (triose phosphate isomerase) barrel for interaction with substrate adaptor proteins (Antonysamy et al., 2012; Ho et al., 2013). The type III PRMT7 monomer adopts a pseudodimer structure reminiscent of type I enzymes (Cura and Troffer-Charlier N, 2014; Hasegawa and Toma-Fukai S, 2014). However, structural data, amino acid substitutions of the PRMT7 active site, and enzymatic analysis point to a restrictive and narrow active site for PRMT7, which is uniquely able to generate MMA (reviewed in (Jain and Clarke, 2019)).
Figure 1. Classification of the types of methyl-arginines and protein arginine methyltransferases.
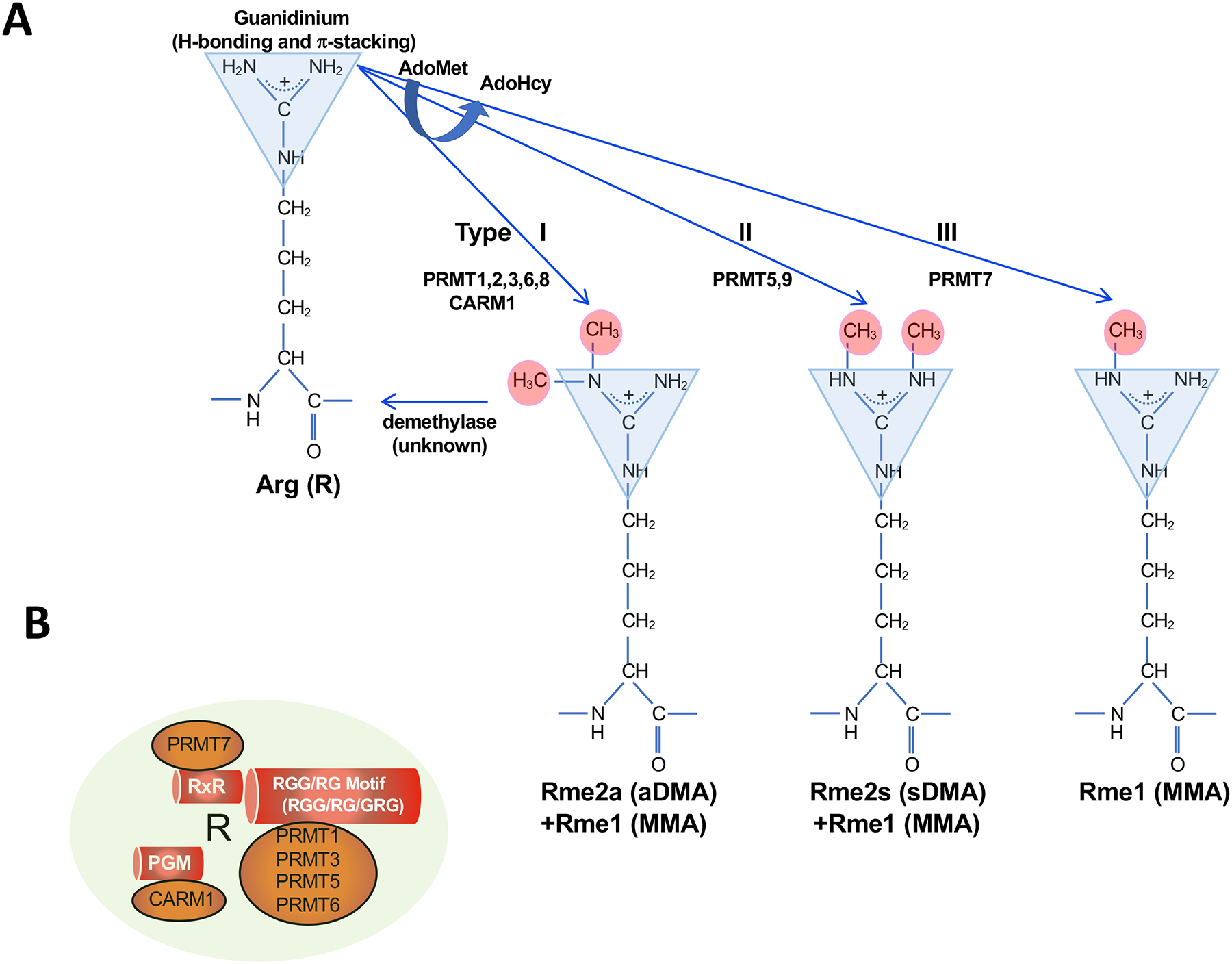
A. Type I and II PRMTs generate monomethyl-arginine (Rme1, MMA) from arginine as a first step, followed by asymmetrical dimethyl-arginine (Rme2a, aDMA; Type I, PRMT1, PRMT2, PRMT3, CARM1, PRMT6 and PRMT8) and symmetrical dimethyl-arginine (Rme2s, sDMA; Type II, PRMT5 and PRMT9) on the guanidino nitrogen atoms using S-adenosylmethionine (AdoMet) converting it to S-adenosylhomocysteine (AdoHcy). The Type III PRMT7 generates only Rme1 (MMA). The guanidinium moiety is shown as a triangle with its H-bonding and π-stacking properties. An arginine demethylase is not known.
B. The known preference of arginine motifs for the PRMTs. PRMT7 has a preference for arginines within the RxR sequences; CARM1 prefers to methylate arginines with neighboring prolines, glycines, and methionines (PGM); PRMT1, PRMT3, PRMT5 and PRMT6 have preference for arginines with neighboring glycines within RGG/RG motifs.
It is important to state that there is no dedicated arginine demethylase, in contrast to lysine demethylases (Pedersen and Helin, 2010). Thus, methylarginines are thought to be a post-translational modification that is long-lasting rather than a rapid ‘ON’ and ‘OFF’ signal generated by phosphorylation, for example. The enzymatic mode of action for arginine demethylation would be similar to lysine demethylation and it seems likely that JmjC proteins may demethylate methylarginine. JmjD6 was wrongly identified as a demethylase for arginine later to be reported as a lysine hydroxylase (Webby et al., 2009). Lysine demethylases (KDM3A, KDM4E, KDM5C) also possess lower methylarginine activity in vitro with unknown physiological importance (Walport et al., 2016). The protein arginine deiminase (PAD) family convert positively charged arginine residue to a neutral citruilline (Thompson and Fast, 2006). Monomethylarginine is a much poorer substrate than unmodified arginine for the PAD enzymes, suggesting it is not of physiological relevance (Thompson and Fast, 2006). The PAD enzymes are therefore antagonistic with the PRMTs for arginines (Guo et al., 2011). We still await the identification of enzyme components necessary to reverse methylated arginines.
II. Biochemical properties of methylated arginines
The guanidinium moiety of arginine is known to carry a positive charge and has the potential to form five hydrogen bonds and π-stacking interactions. Thus, it is not surprising to find arginines proximal to protein-protein, protein-RNA or protein-DNA interfaces. The methylation of the guanidinium moiety increases the level of hydrophobicity of the arginine and provides a bulkier side group without modifying the charge (Evich et al., 2016). The presence of methyl groups can block hydrogen bonds involving a N-H donor, but potentially adds hydrogen bonds formed from carbon hydrogen donors of the methyl group itself (Horowitz and Trievel, 2012). Methyl-arginines are recognized by Tudor domains, although some PHD and WD40 domains also bind methylarginines (reviewed in (Gayatri and Bedford, 2014)). Specifically, methylated arginines interact with the aromatic cage of the Tudor domain where π-stacking interactions occur (Selenko et al., 2001). There are >35 Tudor domain containing proteins but only a subset are currently validated methyl-arginine interactors, including SMN (Survival of motor neuron), SPF30 (Splicing factor 30), and TDRD1/2/3/6/9/11 (Tudor domain-containing protein) (Chen et al., 2011).
III. Overview of the PRMT classes
The methylation of arginines is required to fulfill numerous physiological functions during development, adult homeostasis, and aging. Mouse conditional alleles have been instrumental in defining the tissue-specific requirements of arginine methylation including in the nervous system, skeletal muscle, and the immune system (Blanc and Richard, 2017; Jackson-Weaver et al., 2020). In humans, it is striking to see elevated PRMT expression in cancer (Yang and Bedford, 2013). In addition, PRMT7 has been linked to a genetic disorder: inactivating mutations have been identified for the human PRMT7 gene which results in intellectual disability syndrome termed SBIDDS (Short Stature, Brachydactyly, Intellectual Developmental Disability, and Seizures) (Agolini E, 2018; Poquerusse et al., 2021).
PRMT1 is the main type I enzyme with roles in transcription, RNA metabolism, DNA damage signaling, protein stability, protein localization and receptor signaling pathways. PRMT1 has a strong preference for intrinsically disordered sequences, namely the RGG/RG motif commonly found in RNA binding proteins (RBPs) (Thandapani et al., 2013) (Figure 1B). The methylation of RBPs regulates protein-RNA and protein-protein interactions (Guccione and Richard, 2019). Histones represent another major class of PRMT1 substrates and the methyl-mark H4R3me2a located in gene promoter regions serves to activate gene expression (Wang et al., 2001). PRMT1 has many other substrates especially in signaling pathways mediated by DNA damage, growth factors, metabolites, and the immune response. PRMT1 inactivation in cancer cells with type I PRMT inhibitors or genetic depletion causes RNA metabolism defects mainly due to hypomethylation of the RBP RGG/RG motif (Thandapani et al., 2013). Type I PRMT inhibitors also reduce the expression of cell cycle genes because of the loss of the histone activating marks which halts cell proliferation and manifests as hallmarks of DNA damage (Yu et al., 2009). Interestingly, the cellular loss of type I PRMT activity causes a compensatory increase in MMA and sDMA, especially at RGG/RG motifs by type II enzymes (Dhar et al., 2013). Substrate scavenging by PRMT5 after PRMT1 inhibition is thought to be due to PRMT1 being responsible for the majority of arginine methylation in cells, and that PRMT1 inhibition then makes substrates amenable for PRMT5 methylation (Dhar et al., 2013). The outcome of harboring either aDMA or sDMA often has opposing functional implications. For example, histone H4 arginine 3 (H4R3) methylated asymmetrically leads to gene activation, while sDMA causes gene repression (see Figure 2).
Figure 2. Arginine methylation as a regulator of transcriptional activity.
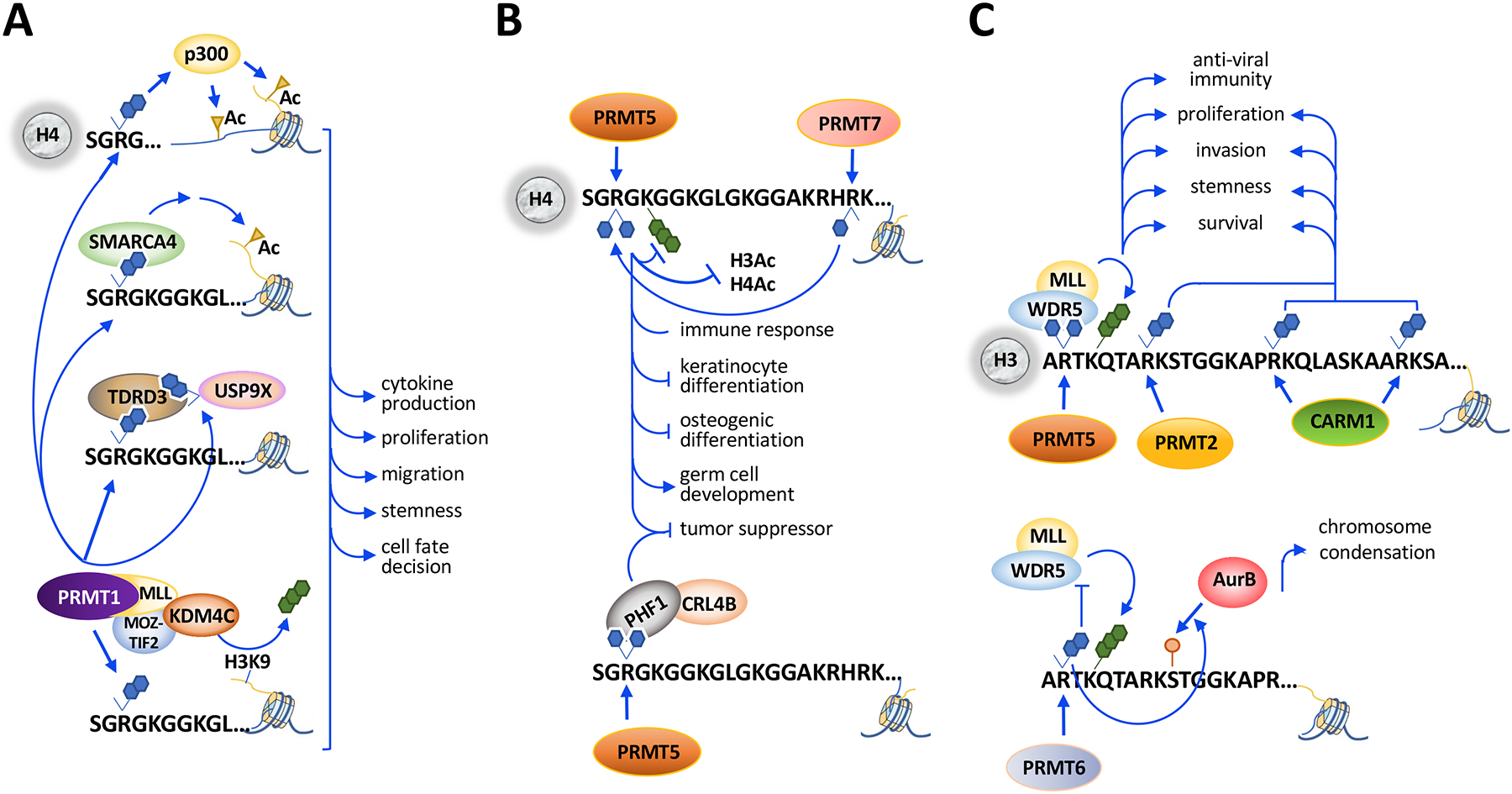
A. PRMT1 methylates H4R3me2a to facilitate p300-mediated acetylation on histones H3 and H4. H4R3me2a is recognized by TDRD3, which recruits methyl-USP9X and binds SMARCA4, which promotes H3 acetylation. PRMT1-MLL (mixed-lineage leukemia) fusion protein deposits H4R3me2a and recruits KDM4C for H3K9me3 demethylation.
B. PRMT5 methylates H4R3me2s which inhibits the deposition of H4K5me3, H4Ac and H3Ac. H4R3me2s is recognized by PHF1 (PHD finger protein 1), which recruits CRL4B (Cullin4B-E3 ligase complex) and transcriptionally represses genes encoding tumor suppressors. PRMT7 methylates H4R17, which enhances H4R3me2a by PRMT5.
C. PRMT5-catalyzed H3R2me2s is recognized by WDR5/MLL complex, which promotes H3K4me3. PRMT5-mediated H3R2me2s promotes transcription activation during anti-viral immunity and cancer progression. PRMT2-catalyzed H3R8me2a and CARM1-catalyzed H3R17me2a/H3R26me2a are also associated with transcriptional activation and cancer progression. PRMT6 methylates H3R2me2a and blocks H3K4-methylwriters including the WDR5/MLL complex and to facilitate AurB (Aurora kinase B)-mediated H3S10 phosphorylation.
PRMT5 is the main type II enzyme that methylates histones and many other substrates including other RBPs, transcription factors, DNA repair proteins, and signal transduction components (for review (Stopa et al., 2015)). Unlike type I PRMTs, PRMT5 forms a complex with MEP50 (WDR77), its obligate binding partner, and one of numerous substrate adaptors which includes pICln, the kinase RioK1, COPR5, Menin, OXR1A, SHARPIN and FAM47E (Chakrapani et al., 2021; Fu et al., 2017; Mulvaney et al., 2021; Stopa et al., 2015; Yang et al., 2020). These PRMT5 substrate adaptors regulate substrate specificity especially at GRG sequences (Musiani et al., 2019) within RGG/RG motifs and this occurs via the PRMT5 TIM barrel (Krzyzanowski et al., 2021; Mulvaney et al., 2021). Note of caution for PRMT5 as it is immunopurified non-specifically with the anti-FLAG M2 antibody (Nishioka and Reinberg, 2003) and thus is falsely found in many proteomic datasets (Mellacheruvu et al., 2013). PRMT5 is also found in many preparations wrongly concluding that different Flag-tagged enzymes have methylation activity when in reality it is PRMT5-mediated activity.
PRMT7, the type III enzyme functions in transcription, DNA damage signaling, stress response and antiviral response (Haghandish et al., 2019; Jain and Clarke, 2019; Szewczyk et al., 2020; Zhu et al., 2021). PRMT7 methylates histones within an RxR consensus (Feng et al., 2013). Interestingly, despite its role in monomethylation, loss of PRMT7 decreases H4R3me2s marks (Blanc et al., 2016), which is explained by allosteric regulation of PRMT5 after the methylation of H4R17me1 by PRMT7 (Jain et al., 2017).
IV. PRMT inhibitors
There are now several clinical-grade small molecule inhibitors for PRMTs and these provide unique tools to study arginine methylation. Specific inhibitors have been identified for PRMT3 (SGC707), CARM1 (TP-064, GSK3359088), PRMT5 (EPZ015666, GSK3326595 (clinical trials NCT02783300, NCT03614728, NCT04676516), JNJ-64619178 (clinical trial NCT03573310), LLY-283, PRT543, PRT811, PF-06939999 (clinical trial NCT03854227)), PRMT6 (EPZ020411, SGC6870) and PRMT7 (SGC3027). Certain inhibitors target multiple PRMTs such as type I PRMTs (MS023, GSK3368715 (clinical trial NCT03666988)), CARM1 and PRMT6 (MS049) and PRMT5 and PRMT7 (DS-437) (for review (Wu et al., 2021)). Using a proteolysis targeting chimera (PROTAC), a PRMT5 degrader was generated confirming this new approach in the PRMT field as another valid therapeutic strategy (Shen et al., 2020). Recently, PRMT5-substrate adaptor interaction inhibitors have been developed, representing a new approach to target PRMT5-mediated methylation events. BRD0639 disrupts the PRMT5-RIOK1 complex and inhibits methylation of some substrates (McKinney et al., 2021).
The availability of PRMT inhibitors has allowed the research community to define tumor type vulnerability (for review (Guccione et al., 2021)). p53 wild type cancer cells are more sensitive to PRMT5 inhibition (Bezzi et al., 2013; Gerhart et al., 2018). Cells with defective splicing machinery have increased sensitivity to type I PRMT and PRMT5 inhibitors (Fedoriw et al., 2019; Fong and al., 2019). The MTAP (methylthioadenosine phosphorylase) gene deficiency creates a vulnerability for cells depleted of PRMT5 protein (Kryukov et al., 2016; Marjon et al., 2016; Mavrakis et al., 2016), inhibited by type I PRMT inhibitors (Dominici et al., 2021; Fedoriw et al., 2019; Gao et al., 2019), and MAT2a inhibitors (Kalev et al., 2021). Inhibition with type I PRMTs increases the MMA/sDMA levels and combined with a PRMT5 inhibitor this brings the methyl-arginine levels below a threshold, causing cytotoxicity (Fedoriw et al., 2019). Cancer cells with high levels of R-loops, as seen in BRCA1 and BRCA2 mutated cells, are sensitive to PRMT5 inhibition (Mersaoui et al., 2019), yet whether there is specificity for certain cancer types, such as breast and ovarian cancer, remains to be defined.
V. Transcriptional Regulation
Epigenetic functions are one of the most well-characterized roles of PRMTs (Figure 2). PRMTs methylate H2AR3(me2a,me2s), H3R2(me2a,me2s), H3R8(me2s), H3R17/26/42(me2a), H4R3(me2a,me2s), and H4R17/19(me1). PRMT1-mediated H4R3me2a is known to facilitate the recruitment of histone acetyltransferases including p300 and subsequent lysine acetylation on H3 and H4 (An et al., 2004). H4R3me2a is also recognized by the Tudor domain-containing protein TDRD3 (Yang et al., 2010), which recruits methyl-USP9X to promote proliferation (Narayanan et al., 2017). Pull-down experiments identified SMARCA4, an ATPase subunit of the SWI/SNF chromatin remodeling complex, as a binder of PRMT1-mediated H4R3me2a for epidermal growth factor receptor expression in colorectal cancer (Yao et al., 2021). PRMT1 and H4R3me2a cooperate with Mixed Lineage Leukemia (MLL) transcriptional complex for hematopoietic cell self-renewal (Cheung et al., 2007; Cheung et al., 2016). PRMT1-mediated H4R3me2a activation mark is critical for cancer cell proliferation, migration, and stemness (reviewed in (Blanc and Richard, 2017)), as well as cell proliferation in multiple developing organs (Gou et al., 2018; Hashimoto et al., 2021) and roles in differentiation such as mature β-cell identity (Kim et al., 2020b) (Figure 2A).
PRMT5-mediated H4R3me2s is found on promoters of tumor suppressor and CDK inhibitor genes to silence their expression and promote cancer cell proliferation (Kaushik et al., 2018; Pal et al., 2004; Yang et al., 2021). One suppression mechanism is the recognition of H4R3me2s by the PHD or ADD domain of DNMT3a to methylate DNA and silence gene expression (Zhao et al., 2009). PRMT5-mediated H4R3me2s also silences epithelial junctional genes to promote cancer cell invasion (Chen et al., 2017). H4R3me2s is also recognized by the PHD finger protein 1, PHF1, which recruits the CUL4B-Ring E3 ligase complex via a PHD finger to silence E-cadherin and FBXW7 expression for cell growth and migration (Liu et al., 2018). PRMT5-mediated histone methylation represses keratinocyte and osteoblasts differentiation genes (Kota et al., 2018; Moena et al., 2020) and regulates the PIWI pathway during germ cell development (Dong et al., 2019; Huang et al., 2021) (Figure 2B). Additionally, PRMT5 deposits activation marks H3R2me1/me2s to recruit the WDR5/MLL complex to promote H3K4me3 deposition for transcriptional activation, a mechanism further demonstrated in SHARPIN-activated cancer cell proliferation and invasion, FOXP1-mediated breast cancer stem cell renewal, the genotoxic stress response and growth hormone production (Cao et al., 2019; Chiang et al., 2017; Lorton et al., 2020; Tamiya et al., 2018; Yang et al., 2020). PRMT7 H4R17me1 enhances allosteric PRMT5-mediated H4R3me2s which represses subsequent H3K4me3, H3Ac and H4Ac (Jain and Clarke, 2019) (Figure 2B).
PRMT2-catalyzed H3R8me2a was identified at promoters (e.g. Bcl2) and enhancers for growth and survival (Dong et al., 2018; Hu et al., 2020). CARM1 functions as a coactivator by methylating H3R17, H3R26 and H3R42, which are recognized by readers including TDRD3 (Yang et al., 2010) (Figure 2C). CARM1 also methylates coactivators and corepressors to regulate transcription (for review (Suresh et al., 2021)). PRMT6 methylates H3R2me2a thus preventing readers of H3K4me3 from binding (reviewed in (Guccione and Richard, 2019)). The H3S10 mediated phosphorylation by Aurora B kinase during mitosis is regulated by PRMT6-mediated H3R2me2a (Kim et al., 2020c) (Figure 2C). PRMT6 also methylates H2AR29 in vitro and accumulates at promoters of PRMT6 repressed genes (Waldmann et al., 2011).
VI. Pre-mRNA splicing
Arginine methylation of RBPs is known to be required for pre-mRNA splicing (reviewed in (Guccione and Richard, 2019)). Type I PRMT inhibitors reduce the methylation of RBPs mainly at RGG/RG sequences (Fedoriw et al., 2019) implying that this RGG/RG disordered region, represents a methyl-arginine reservoir (Thandapani et al., 2013). RNA-seq following type I PRMT inhibition showed profound changes in splicing affecting exon usage and this effect was exacerbated with the treatment of PRMT5 inhibitors (Fedoriw et al., 2019). Inhibition of PRMT5 and type I PRMTs resulted in preferential killing of acute myeloid leukemia (AML) cells harboring mutations within the splicing factor SRSF2 over wild type counterparts (Fong and al., 2019). EZH2 null cells were significantly more resistant to type I PRMT inhibition, suggesting that the effect of the PRMT inhibitor is, in part, mediated by restoration of EZH2 levels (Fong and al., 2019). The differential cassette exon inclusion and intron retention observed in MS023-treated MC38 murine colon adenocarcinoma cells were shown to encode MHC I presented neopeptides that may play a role in influencing tumor immunity (Lu et al., 2021).
The PRMT5/ MEP50/ pICln complex is known to methylate Sm proteins in the cytoplasm for the assembly into mature small nuclear ribonucleoproteins (snRNPs) required for splicing (Guccione and Richard, 2019). The levels of Sm protein sDMA inversely correlates with intron retention and reduction of Sm sDMA is associated with accumulated polyadenylated RNA containing introns in the A549 lung adenocarcinoma cell line (Maron et al., 2020). Cells with reduced PRMT5 activity, either due to genetic manipulation or inhibitor treatment, have many splicing defects. One splicing event that is particularly sensitive to PRMT5 inhibition is MDM4, where a short isoform is generated with a premature stop codon resulting in non-sense mediated decay and reduced protein levels of MDM4, a repressor of p53 (Bezzi et al., 2013) (Figure 3A). SRSF1, a splicing regulator, is a PRMT5 substrate and effector (Cai et al., 2021a; Radzisheuskaya et al., 2019). Knockdown of PRMT5 causes a differential binding of SRSF1 to its alternatively pre-mRNA spliced targets with reduced SRSF1 protein-protein interactions (Radzisheuskaya et al., 2019) (Figure 3A). The methylation of the RBMX RGG/RG motif by PRMT5 regulates higher-order complexes with SRSF1 within nuclear membraneless organelles to generate the short isoform of MDM4 by alternative splicing (Cai et al., 2021a) (Figure 3A). Interestingly, the link between RBMX methylation and p53 activation has clinical significance, as Shashi-XLID syndrome patients have a genetic deletion of 23bp causing a frameshift encoding a truncated RBMXΔRGG/RG protein inducing defects in neuronal differentiation (Cai et al., 2021a). In B cell lymphomas, MYC regulated transcripts are mis-spliced and PRMT5 inhibition induces apoptosis (Koh et al., 2015). CARM1 and PRMT9 also regulate alternative splicing and these roles were reviewed previously (Guccione and Richard, 2019).
Figure 3. Protein arginine methyltransferases regulate the p53 and DNA damage response pathways.
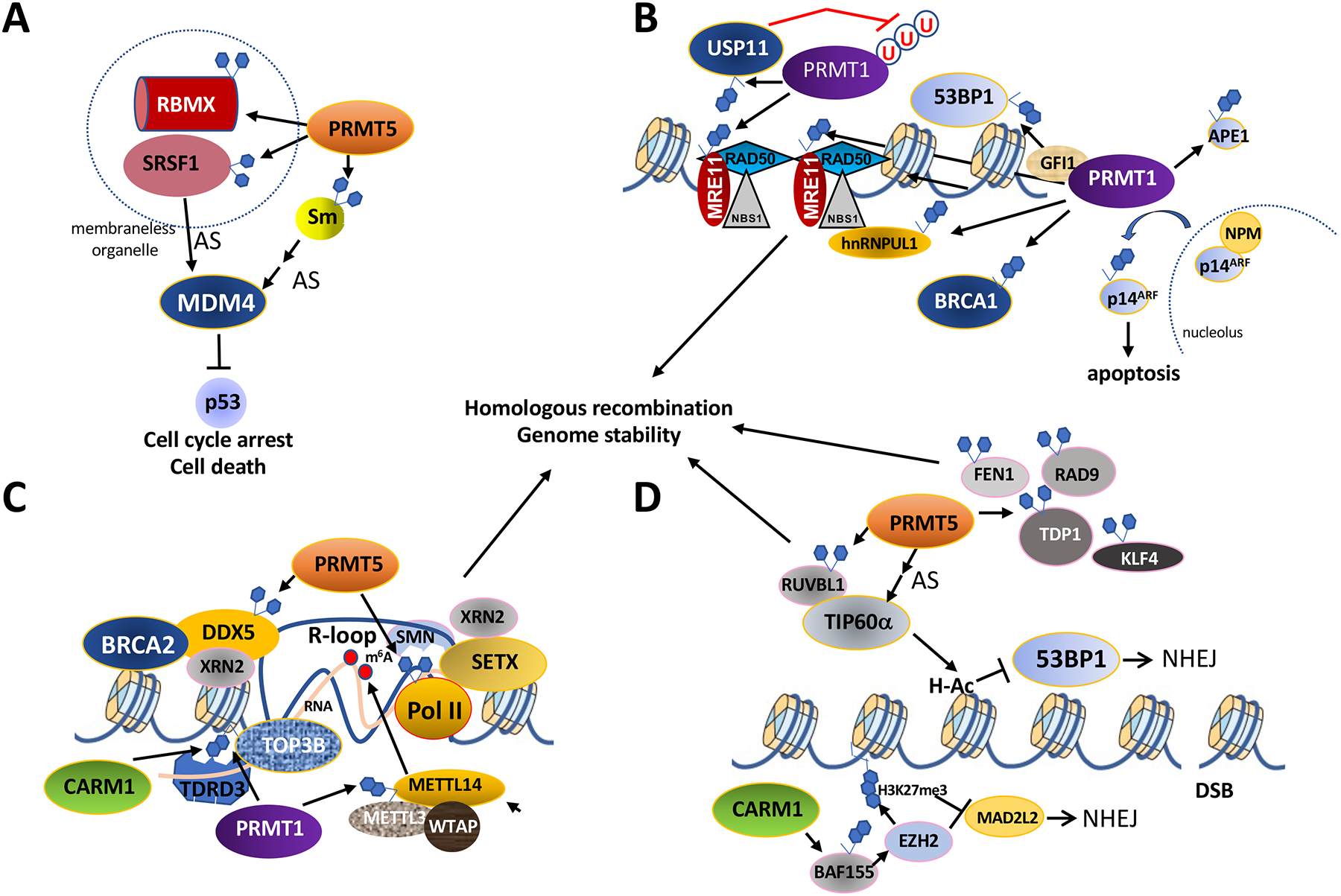
A. PRMT5 methylates Sm proteins, SRSF1 (Serine and arginine rich splicing factor 1), and RBMX (RNA binding motif protein X-linked), to regulate MDM4 alternative splicing (AS) and p53 protein levels. SRSF1 and RBMX can coexist in a membraneless nuclear organelle and this is regulated by the methylation of RBMX by PRMT5.
B. The MRE11-RAD50-NBS1 sensor complex at DSBs (double stranded breaks) activates the ATM (Ataxia telangiectasia mutated) kinase to phosphorylate the histone variant H2AX (γH2AX) and many other proteins to trigger DSB repair. PRMT1 methylation of MRE11 regulates its resection activities. The methylation of RNA binding protein hnRNPUL1 regulates interaction with NBS1. GFI1 is a transcription factor that enhances the PRMT1-mediated methylation of MRE11 in T cells and USP11 regulates the activity of PRMT1 by deubiquination. Circled ‘U’ denote ubiquitin. The methylation of the DSB repair proteins 53BP1, BRCA1, APE1 (apurinic/apyrimidinic endonuclease 1) is mediated by PRMT1, but the role of these methylation events remain unknown. Methylation of p14ARF (alternative reading frame tumor suppressor product of the CDKN2A locus) allows its nucleolar export from NPM (nucleophosmin) to promoter apoptosis.
C. DDX5 (DEAD-box helicase 5) is methylated by PRMT5 and this regulates association with XRN2 (5’−3’ exonuclease 2) for R-loop (RNA/DNA hybrid) resolution. DDX5 recruitment to DNA breaks is regulated by BRCA2 (Breast cancer gene 2). PRMT5 also methylates the RNA polymerase II C-terminal domain which lead to the recruitment of the helicase SETX (Senataxin) and XRN2 by SMN to resolve R-loops. CARM1 and PRMT1 methylates histones which attract the methyl-reader protein TDRD3 via its Tudor domain to recruit topoisomerase TOP3B (DNA topoisomerase III beta) to resolve R-loops. The resolution of R-loops prevents endogenous DNA damage and maintains genome stability. The RNA of the RNA/DNA hybrid of R-loops harbors m6A (N6-methyladenosine) and this event is regulated by PRMT1 methylation of the m6A methyltransferase (WTAP, METTL3, METTL14) component METTL14.
D. PRMT5 methylates RUVBL1 (RuvB like AAA ATPase) activating TIP60α which acetylates H4K16 to block 53BP1 recruitment favoring HR (homologous recombination) over NHEJ (non-homologous end-joining). The alternative splicing (AS) of TIP60α is regulated by PRMT5 to promote HR. CARM1 methylation of the SWI/SNF complex subunit BAF155 at R1064 increases EZH2 (Enhancer of zeste 2 polycomb repressive complex 2 subunit) H3K27me3 methylation repressing NHEJ Shieldin component MAD2L2. The upregulation of NHEJ in HR-proficient high-grade serous ovarian cancers by EZH2 inhibition creates sensitivity to poly (ADP-ribose) polymerase inhibitors. PRMT5 also methylates FEN1, RAD9, TDP1, and KLF4 to maintain genomic stability.
VII. DNA damage repair
Arginine methylation of DNA damage proteins by PRMT1 including MRE11, 53BP1, hnRNPUL1 and BRCA1 is essential to maintain genomic stability and to ensure DNA repair by homologous recombination repair (HR) and non-homologous end joining (NHEJ) (Figure 3B–D). The double stranded break (DSB) sensor protein MRE11 is methylated by PRMT1 at its RGG/RG motif and this methylation event is required for association with its DNA template for processive exonuclease activity during resection required for HR (Boisvert et al., 2005) (Figure 3B). This methylation event is enhanced in T cells by the GFI1 chaperone protein (Vadnais et al., 2018) and by USP11 in epithelial cells with deubiquitylation of PRMT1 regulating its ability to bind and methylate MRE11 (Sanchez-Bailon et al., 2021). Converting the MRE11 RGG to KGG in mice (R-K) causes them to be hypersensitive to IR and their cells exhibit genomic instability due to failure to activate the ATR-CHK1 pathway (Yu et al., 2012). The methylation of MRE11 is a conserved event, as Drosophila MRE11 methylation by DART1 is required to maintain genomic stability (Yuan et al., 2018). DNA damage by ultraviolet (UV) light increases the association and methylation of p14ARF by PRMT1 favoring its nucleolar export to fulfill its proapoptotic role as a tumor suppressor (Repenning et al., 2021) (Figure 3B). APE1 (Apurinic/apyrimidinic endonuclease 1) is a major enzyme of the base excision repair (BER) pathway and its methylation by PRMT1 influences its mitochondrial localization (Zhang et al., 2020b).
PRMT1 methylates components of the N6-methyladenosine (m6A) methyltransferase complex to regulate UV irradiation DNA repair. The m6A RNA modification is an epitranscriptomic mark that regulates DNA repair (Xiang et al., 2017). m6A accumulates on RNA at DSBs forming RNA/DNA hybrids or R-loops recruiting RAD51 and BRCA1 for HR (Abakir et al., 2020; Zhang et al., 2020a). m6A is deposited by a methyltransferase complex consisting of METTL3, METTL14, and WTAP and METTL14 has a C-terminal RGG/RG motif that is methylated by PRMT1 (Wang et al., 2021) (Figure 3C). Genetic knockout and inhibition of PRMT1, which dampens METTL14 arginine methylation, sensitizes mouse embryonic stem cells to mitomycin C and cisplatin-induced cell death (Wang et al., 2021). In another study, METTL14 was shown to be monomethylated on R255 affecting interaction with WTAP (Liu et al., 2021).
CARM1 and PRMT1 limits accumulation of R-loops by recruiting topoisomerase TOP3B to them by methylating histones which are then bound by the TDRD3/TOP3B complex (Yang et al., 2014) (Figure 3C). Recently, it was shown that elevated CARM1 expression causes an increase in NHEJ repair by increasing the expression of MAD2L2, a component of the Shieldin complex, a repressor of HR (Karakashev et al., 2020) (Figure 3D). CARM1 methylation of the SWI/SNF complex BAF155 at R1064 creates a vulnerability to EZH2 inhibition. The upregulation of NHEJ in HR-proficient high-grade serous ovarian cancers by EZH2 inhibition creates sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitors (Karakashev et al., 2020). Notably, CARM1 has been shown to localize at the DNA replication fork and function in replication fork speed regulation in a methyltransferase-independent manner. It functions to regulate PARylation by PARP1/2 and prevent ssDNA gaps from triggering ATR activation, thereby increasing replication stress tolerance (Genois et al., 2021). Thus, CARM1 is emerging as an attractive cancer therapeutic target.
PRMT5-deficient HeLa cells are hypersensitive to ionizing radiation (IR) and accumulate DNA damage (Clarke et al., 2017; Hamard et al., 2018; Mersaoui et al., 2019; Pastore et al., 2020; Wei et al., 2020). PRMT5 methylation of the TIP60 subunit, RUVBL1, a AAA+ ATPase, on R205 affects the activity of TIP60 (Clarke et al., 2017) (Figure 3D). In hematopoietic progenitor cells, the alternative splicing of TIP60 by PRMT5 affects the levels of full-length TIP60 (TIP60α) (Hamard et al., 2018). Genome-wide assessment of R-loops shows that methylation of DDX5 by PRMT5 facilitates the resolution of the DNA/RNA hybrids at specific genomic loci (Villarreal et al., 2020; Yu et al., 2020). DDX5 recruitment and activity at DSBs is regulated by BRCA2 (Sessa et al., 2021) (Figure 3C).
A CRISPR/Cas9 screen identified sgRNAs targeting PRMT5 as creating conditional lethality with the nucleoside analog Gemcitabine in pancreatic ductal adenocarcinoma (PDAC) (Wei et al., 2020). PRMT5-deficient PDAC cells have replicative catastrophes and HR defects with reduced replicative protein A (RPA) protein levels (Wei et al., 2020). In the prostate cancer cell line LNCaP, it was shown that PRMT5 and pICln epigenetically upregulate genes involved in the DDR pathway and the targeting of either PRMT5 or pICln sensitizes these cells to IR (Owens et al., 2020). In hematopoietic stem cells, PRMT5 regulates the splicing of DNA damage genes involved in HR (RAD52) and interstrand crosslink (FANCA, FANCG, RTEL1) repair to maintain genomic stability (Tan et al., 2019). Depletion of PRMT5 activates the p53 pathway and sensitizes stem cell factor-dependent lympho-hematopoietic progenitor cells to the DNA crosslinking agent, mitomycin C (Tan et al., 2019). PRMT5 also epigenetically regulates the expression of the E3 ligase RNF168, a key component of the DDR pathway, to regulate H2AX proteostasis (Du et al., 2019).
Myeloproliferative neoplasms with the common mutation, JAK2V617F, express high levels of PRMT5 (Liu et al., 2011). Synergistic efficacy was obtained by inhibiting PRMT5 and the JAK1/2 inhibitor Ruxolitinib (Pastore et al., 2020). PRMT5 inhibition reduced methylation of E2F1 increasing the affinity for the tumor suppressor Rb and leading to decreased expression of E2F1 downstream targets including DDR genes (Pastore et al., 2020). PRMT5 also methylates FEN1 (flap endonuclease 1) (Guo et al., 2010), RAD9 (DNA repair protein 9) (He et al., 2011), TDP1 (tyrosyl-DNA-phosphodiesterase 1) (Rehman et al., 2018), and KLF4 (Krüppel-like factor 4) (Hu et al., 2015) required for DNA repair.
VIII. Tumor immunity and anti-viral responses
The immune system uses toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) to activate the TBK1/IRF3 pathway inducing type I interferons (IFNs) and pro-inflammatory cytokines in response to pathogens. PRMTs regulate the activation of TLR and IFN at multiple levels to modulate immune responses (Kim et al., 2016; Sengupta et al., 2020). Herein we discuss recent advances involving PRMT function in anti-tumor and anti-viral immunity.
PRMTs have been identified as regulators of cancer immunity. CRISPR/Cas9 knockout of PRMT1 in MC38, a murine colon adenocarcinoma cell line, improved anti-PD-1 immunotherapy effectiveness when inoculated in syngeneic C57BL/6 mice (Hou et al., 2021). In another study, a type I PRMT inhibitor (PT1001B) combined with anti-PD-L1 inhibition was most effective in reducing tumor growth in pancreatic ductal adenocarcinoma (Panc2)-derived tumors in syngeneic C57BL/6 mice (Zheng et al., 2020). MS023 treatment significantly improved immunotherapy to anti-PD1 therapy in MC38-injected C57BL/6 mice and the long-term survivors had enhanced immune memory and improved tumor killing response when rechallenged with MC38 cells 6 months later (Lu et al., 2021). Interestingly, a PRMT1 polymorphism, rs975484, regulates the levels of PD-L1 and PD-L2 and may serve as a predictor of immune checkpoint blockade efficiency in hepatocellular carcinoma (Schonfeld et al., 2020). A CRISPR/Cas9 screen delivered in CD8+ T cells identified CARM1 as a negative regulator of tumor-specific T cells using B16F10 melanoma-bearing C57BL/6 mice (Kumar et al., 2021).
PRMT5 inhibition sensitized unresponsive melanomas to anti-PD1 immunotherapy in melanoma (Kim et al., 2020a). PRMT5 contributes to the anti-tumor immune response by methylating interferon-γ-inducible protein 16 (IFI16), a repressor of the DNA sensor cGAS/STING pathway, and also transcriptionally downregulates NLRC5 (nucleotide-binding oligomerization domain-like receptor family caspase recruitment domain containing), a regulator of the major histocompatibility complex class I (MHCI) antigen presentation pathway (Kim et al., 2020a) (Figure 4A). PRMT5 also suppresses the cGAS/ STING pathway by directly associating with and methylating cGAS on R124, inhibiting its DNA binding activity (Ma et al., 2021) (Figure 4A). A CRISPR/Cas9 genetic screen identified PRMT7 as enhancing anti-tumor responsiveness to immune checkpoint blockade (Manguso et al., 2017). PRMT7 inhibition combined with immune checkpoint inhibitors triggers a strong anti-tumor T cell immunity with reduced B16F10 melanoma growth in syngeneic C57BL/6 mice (Srour et al., 2021). PRMT7-deficiency in B16F10 melanomas increased dsRNA repetitive element expression accumulation or ‘viral mimicry’ which led to an increase in IFN genes (Srour et al., 2021) (Figure 4B). Taken together, inhibition of PRMT1-, CARM1-, PRMT5- and PRMT7-regulated cancer immunity is gaining traction and thus PRMT inhibitors provide exciting new options for enhancing the effectiveness of immune checkpoint inhibitors.
Figure 4. Arginine methylation negatively regulates the anti-viral response.
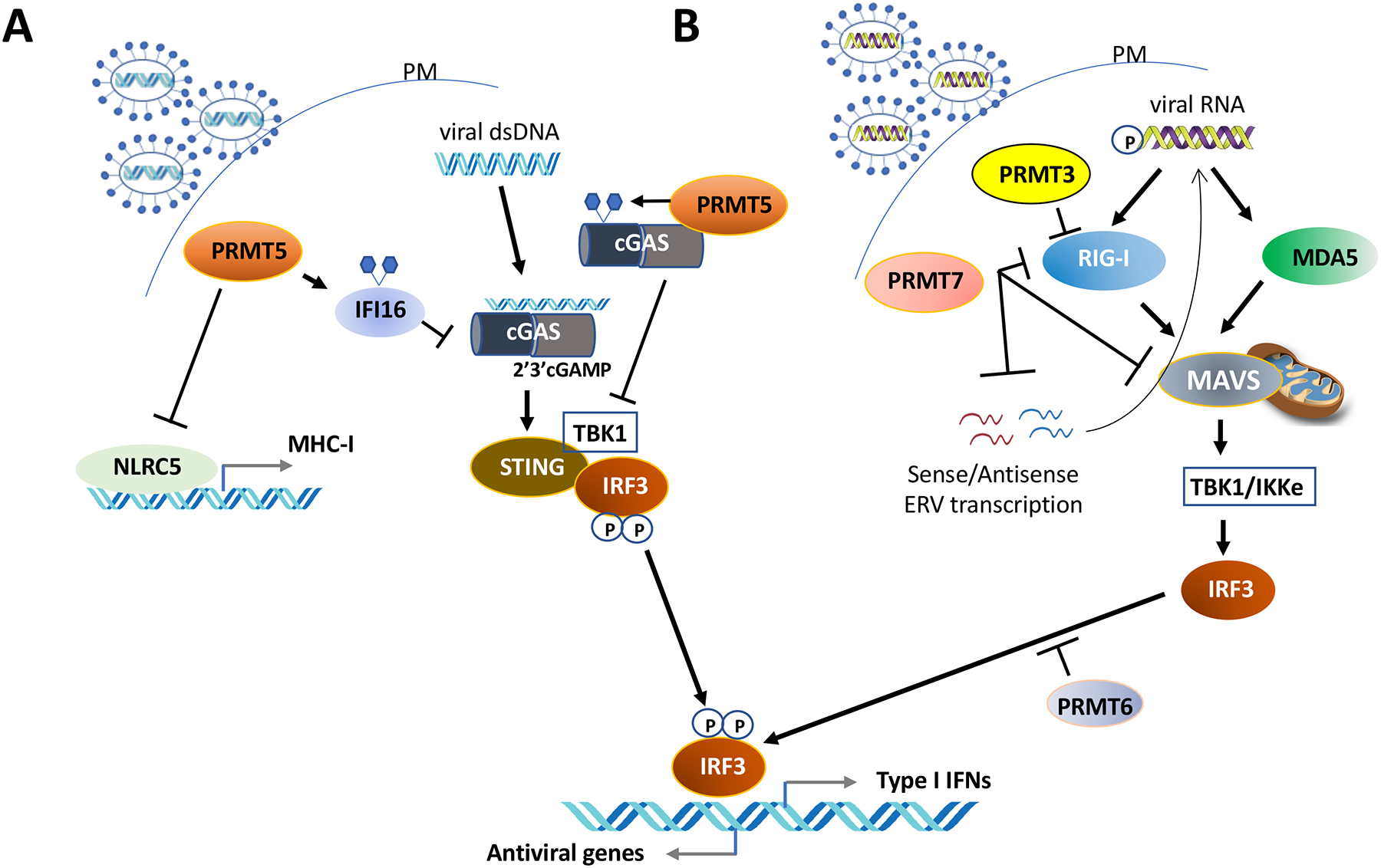
A. Methylation of IFI16 (interferon gamma inducible protein 16) and cGAS (Cyclic GMP-AMP synthase) by PRMT5 inhibits the cGAS/STING (Stimulator of Interferon genes) pathway. PRMT5 also negatively regulates the transcription of NLRC5 (NLR family CARD domain containing 5), a crucial transactivator of MHC (major histocompatibility complex) class genes.
B. PRMT7 inhibits RIG-I-like receptors (retinoic acid-inducible gene-I-like receptors, RLRs), MDA5 (melanoma differentiation-associated gene 5), MAVS (mitochondrial antiviral signaling protein) and dsRNA ERV (Endogenous retrovirus) repetitive sequences to inhibit RIG-I signaling. PRMT3 and PRMT6 also inhibit this pathway preventing IRF-3 (Interferon regulatory factor 3) induced type I interferons.
Further roles of PRMTs in the antiviral response have been documented. In zebrafish, PRMT3 or PRMT7 deficiency/inhibition activates IRF-3-mediated IFN production, and the mutant fish are more resistant to viral infections (Zhu et al., 2020a; Zhu et al., 2020b). Moreover, in mammals PRMT7-mediated monomethylation of MAVS negatively regulates the antiviral response (Zhu et al., 2021) (Figure 4B). PRMT6 prevents activation of IRF3-mediated IFN production by associating with IRF3 and sequestering it (Jiang et al., 2019; Zhang et al., 2019) (Figure 4B). Additionally, PRMTs methylate viral proteins necessary for their life cycle. Recently, PRMT1 was shown to methylate SARS-CoV-2 nucleocapsid (N) protein at R95 and R177 to facilitate N-mediated suppression of stress granules and viral replication. Type I PRMT inhibitors reduced viral titers of SARS-CoV-2 and may represent a new therapeutic application for these inhibitors (Cai et al., 2021b).
IX. Challenges Ahead
Notably in this review, we highlight many cellular processes regulated by PRMT1 and PRMT5, as these PRMTs are at the forefront of the field. This has been fueled by 1) their major contributions to the levels of cellular methyl-arginines and, 2) the availability of specific inhibitors of type I PRMTs and PRMT5 as well as the striking cellular phenotypes induced by these inhibitors. CARM1 is another well-characterized methyltransferase with new specific inhibitors positioning it as an attractive therapeutic target. We know very little about certain methyltransferases such as PRMT7 and PRMT9. Why is the activity of PRMT7 important when type I and type II PRMTs also generate MMA? Does the function of PRMT7-mediated RxR MMA differ from PRMT5 RGRG MMA, for example? We know that the major role for PRMT7 is not to prime for dimethylation by type I and type II PRMTs (Jain and Clarke, 2019). This is further highlighted by differences in mouse phenotypes: whole body knockout of PRMT1 or PRMT5 leads to early embryonic lethality, while PRMT7 null mice are viable (Blanc and Richard, 2017). Also, the roles of type II PRMT9 and its substrates remain to be further defined. PRMT9 is not redundant with PRMT5, the major type II enzyme. PRMT9 displays tissue and subcellular distribution that is distinct from PRMT5, and has specific functions, as observed in splicing with the methylation of spliceosome associated protein SAP145 (Yang et al., 2015).
The interplay between different PRMT types (I, II, III) and their substrate ‘sharing’ or ‘scavenging’ observed especially during PRMT inhibition needs to be better understood. For example, with the large number of substrates already known for most of the PRMTs, especially PRMT1 and PRMT5, how might the regulation of these enzymes affect the myriad activities of the methyl-accepting proteins. Specifically, how are the roles of arginine methylation in transcriptional activation/repression, pre-mRNA splicing, DNA damage repair, and tumor immunity and anti-viral responses coordinated? Although some PRMTs are thought to be constitutive enzymes which methylate a wide range of substrates, the activity of some PRMTs has been shown to be regulated in response to precise cellular responses and other types of post-translational modifications (Blanc and Richard, 2017; Wu et al., 2021).
Also, it will be pivotal to comprehend if a single arginine within an RGG/RG motif, for example, is always available to both type I and II of PRMTs or if this only occurs in the absence/inhibition or the overexpression of a PRMT. What is known, by studying methylation at H4R3 in the triple negative breast cancer cell line MDA-MB-468, is that the stoichiometry is 0.2% MMA and 0.015% DMA with only aDMA being present (Zappacosta et al., 2021). In this system, sDMA on H4R3 was only observed when PRMT5 and MEP50 were overexpressed (Zappacosta et al., 2021). Thus, perturbation of PRMT expression or inhibition is required to have H4R3me2s and H4R3me2a coexist in MDA-MB-468. In addition to the regulation of arginine methylation by several types of PRMTs, the possibility of the existence of a true dedicated arginine demethylase influences our understanding of the dynamics of methylarginines.
The greatest advancement in recent years has been the use of small molecules to define cancer vulnerabilities to PRMT inhibition, and this has led to exciting clinical trials for cancer therapy. The combination of PRMT inhibitors with DNA damaging agents, and the vulnerability of cells with splicing defects, are valuable discoveries for therapeutic efficacy. This is in line with previous reports that DNA damage signaling and pre-mRNA splicing are the major pathways regulated by PRMT1 and PRMT5. However, the stratification based on p53 levels, is surprising and raises the interesting question ‘why is the p53 pathway via MDM4 splicing a strong indication of PRMT inhibition?’ i.e. why does this specific splicing event prevail functionally over the other splicing events in some cells? Another unexpected finding is the anti-viral and anti-tumor effects of ablation or inhibition of type I PRMTs, CARM1, PRMT5, and PRMT7, and their synergistic effect with immune checkpoint inhibitors for immunotherapy. The generation of neoantigens, the regulation of immune ligands such as PD-L1 and the modulation of interferons and cytokines with PRMT depletion or inhibition is promises that exciting times are ahead in the field, as light is shed on the molecular details.
Acknowledgements
We apologize to colleagues whose work could not be cited or was only referred to by citing review articles due to space restrictions. J.X. is funded by R01DE026468, R01DE030928 and R21DE028617 by the National Institute of Dental and Craniofacial Research, USA. S.R. is funded by FDN-154303 by the Canadian Institutes of Health Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, Rubio-Roldan A, Li M, Eleftheriou M, Crutchley J, et al. (2020). N(6)-methyladenosine regulates the stability of RNA:DNA hybrids in human cells. Nat Genet 52, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agolini E, D. M, Bellacchio E, Alesi V, Radio FC, Torella A, Musacchia F, Tartaglia M, Dallapiccola B, Nigro V, Digilio MC, Novelli A. (2018). Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review. Clin Genet 93, 675–681. [DOI] [PubMed] [Google Scholar]
- An W, Kim J, and Roeder RG (2004). Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748. [DOI] [PubMed] [Google Scholar]
- Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, et al. (2012). Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A 109, 17960–17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, and Clarke SG (2009). Protein arginine methylation in mammals: who, what, and why. Mol Cell 33, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, and Guccione E (2013). Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev 27, 1903–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc RS, and Richard S (2017). Arginine methylation: the coming of Age. Molecular Cell 65, 8–24. [DOI] [PubMed] [Google Scholar]
- Blanc RS, Vogel G, Chen T, Crist C, and Richard S (2016). PRMT7 Preserves Satellite Cell Regenerative Capacity. Cell Rep 14, 1528–1539. [DOI] [PubMed] [Google Scholar]
- Boisvert F-M, Déry U, Masson J-Y, and Richard S (2005). Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes & Dev 19, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Cinkornpumin JK, Yu Z, Villarreal OD, Pastor WA, and Richard S (2021a). Deletion of RBMX RGG/RG motif in Shashi-XLID syndrome leads to aberrant p53 activation and neuronal differentiation defects. Cell Reports 36, 109337. [DOI] [PubMed] [Google Scholar]
- Cai T, Yu Z, Wang Z, Liang C, and Richard S (2021b). Arginine Methylation of SARS-Cov-2 Nucleocapsid Protein Regulates RNA Binding, its Ability to Suppress Stress Granule Formation and Viral Replication. J Biol Chem 297, 100821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Wu G, Zhu J, Tan Z, Shi D, Wu X, Tang M, Li Z, Hu Y, Zhang S, et al. (2019). Genotoxic stress-triggered β-catenin/JDP2/PRMT5 complex facilitates reestablishing glutathione homeostasis. Nat Commun 10, 3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrapani B, Khan MIK, Kadumuri RV, Gupta S, Verma M, Awasthi S, Govindaraju G, Mahesh A, Rajavelu A, Chavali S, et al. (2021). The uncharacterized protein FAM47E interacts with PRMT5 and regulates its functions. Life Sci Alliance 4, e202000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, and Pawson T (2011). Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 12, 629–642. [DOI] [PubMed] [Google Scholar]
- Chen H, Lorton B, Gupta V, and Shechter D (2017). A TGFbeta-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene 36, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X (2014). Structural and Functional Coordination of DNA and Histone Methylation. Cold Spring Harb Perspect Biol 6, a0187747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N, Chan LC, Thompson A, Cleary ML, and So CW (2007). Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol 9, 1208–1215. [DOI] [PubMed] [Google Scholar]
- Cheung N, Fung TK, Zeisig BB, Holmes K, Rane JK, Mowen KA, Finn MG, Lenhard B, Chan LC, and So CW (2016). Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 29, 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K, Zielinska AE, Shaaban AM, Sanchez-Bailon MP, Jarrold J, Clarke TL, Zhang J, Francis A, Jones LJ, Smith S, et al. (2017). PRMT5 Is a Critical Regulator of Breast Cancer Stem Cell Function via Histone Methylation and FOXP1 Expression. Cell Rep 21, 3498–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TL, Sanchez-Bailon MP, Chiang K, Reynolds JJ, Herrero-Ruiz J, Bandeiras TM, Matias PM, Maslen SL, Skehel JM, Stewart GS, et al. (2017). PRMT5-Dependent Methylation of the TIP60 Coactivator RUVBL1 Is a Key Regulator of Homologous Recombination. Mol Cell 65, 900–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cura V, and Troffer-Charlier N, L. M, Bonnefond L, Cavarelli J. (2014). Cloning, expression, purification and preliminary X-ray crystallographic analysis of mouse protein arginine methyltransferase 7. Acta Crystallogr F Struct Biol Commun 70, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Vemulapalli V, Patananan AN, Huang GL, Di Lorenzo A, Richard S, Comb MJ, Guo A, Clarke SG, and Bedford MT (2013). Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci Rep 3, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici C, Sgarioto N, Yu Z, Sesma-Sanz L, Masson JY, Richard S, and Raynal NJ (2021). Synergistic effects of type I PRMT and PARP inhibitors against non-small cell lung cancer cells. Clin Epigenetics 13, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Li Q, Yang C, Huo D, Wang X, Ai C, Kong Y, Sun X, Wang W, Zhou Y, et al. (2018). PRMT2 links histone H3R8 asymmetric dimethylation to oncogenic activation and tumorigenesis of glioblastoma. Nat Commun 9, 4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Wang X, Cao C, Wen Y, Sakashita A, Chen S, Zhang J, Zhang Y, Zhou L, Luo M, et al. (2019). UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat Commun 10, 4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Hansen LJ, Singh SX, Wang F, Sun R, Moure CJ, Roso K, Greer PK, Yan H, and He Y (2019). A PRMT5-RNF168-SMURF2 Axis Controls H2AX Proteostasis. Cell Rep 28, 3199–3211 e3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evich M, Stroeva E, Zheng YG, and Germann MW (2016). Effect of methylation on the side-chain pKa value of arginine. Protein Sci 25, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw A, Rajapurkar SR, O’Brien S, Gerhart SV, Mitchell LH, Adams ND, Rioux N, Lingaraj T, Ribich SA, Pappalardi MB, et al. (2019). Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss. Cancer Cell 36, 100–114 e125. [DOI] [PubMed] [Google Scholar]
- Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT, Bedford MT, Masson JY, et al. (2013). Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem 288, 37010–37025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JY, and al., e. (2019). Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell 36, 194–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T, Lv X, Kong Q, and Yuan C (2017). A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget 8, 54809–54820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Zhang L, Villarreal OD, He W, Su D, Bedford E, Moh P, Shen J, Shi X, Bedford MT, et al. (2019). PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic Acids Res 47, 5038–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayatri S, and Bedford MT (2014). Readers of histone methylarginine marks. Biochim Biophys Acta 1839, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genois MM, Gagne JP, Yasuhara T, Jackson J, Saxena S, Langelier MF, Ahel I, Bedford MT, Pascal JM, Vindigni A, et al. (2021). CARM1 regulates replication fork speed and stress response by stimulating PARP1. Mol Cell 81, 784–800 e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart SV, Kellner WA, Thompson C, Pappalardi MB, Zhang XP, Montes de Oca R, Penebre E, Duncan K, Boriack-Sjodin A, Le B, M. C., et al. (2018). Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep 8, 9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou Y, Li J, Jackson-Weaver O, Wu J, Zhang T, Gupta R, Cho I, Ho TV, Chen Y, Li M, et al. (2018). Protein Arginine Methyltransferase PRMT1 Is Essential for Palatogenesis. J Dent Res 97, 1510–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, and Richard S (2019). The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol 20, 642–657. [DOI] [PubMed] [Google Scholar]
- Guccione E, Schwarz M, Di Tullio F, and Mzoughi S (2021). Cancer synthetic vulnerabilities to protein arginine methyltransferase inhibitors. Curr Opin Pharmacol 59, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Bedford MT, and Fast W (2011). Discovery of peptidylarginine deiminase-4 substrates by protein array: antagonistic citrullination and methylation of human ribosomal protein S2. Mol Biosyst 7, 2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, and Shen B (2010). Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. . Nat Chem Biol 6, 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghandish N, Baldwin RM, Morettin A, Dawit HT, Adhikary H, Masson JY, Mazroui R, Trinkle-Mulcahy L, and Côté J (2019). PRMT7 methylates eukaryotic translation initiation factor 2α and regulates its role in stress granule formation. Mol Biol Cell 30, 778–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard PJ, Santiago GE, Liu F, Karl DL, Martinez C, Man N, Mookhtiar AK, Duffort S, Greenblatt S, Verdun RE, et al. (2018). PRMT5 Regulates DNA Repair by Controlling the Alternative Splicing of Histone-Modifying Enzymes. Cell Rep 24, 2643–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, and Toma-Fukai S, K. J, Fukamizu A, Shimizu T. (2014). Protein arginine methyltransferase 7 has a novel homodimer-like structure formed by tandem repeats. FEBS Lett 588, 1942–1948. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Kumabe A, Kim JD, Murata K, Sekizar S, Williams A, Lu W, Ishida J, Nakagawa T, Endo M, et al. (2021). Loss of PRMT1 in the central nervous system (CNS) induces reactive astrocytes and microglia during postnatal brain development. J Neurochem 156, 834–847. [DOI] [PubMed] [Google Scholar]
- He W, Ma X, Yang X, Zhao Y, Qiu J, and Hang H (2011). A role for the arginine methylation of Rad9 in checkpoint control and cellular sensitivity to DNA damage. Nucleic Acids Res 39, 4719–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MC, Wilczek C, Bonanno JB, Xing L, Seznec J, Matsui T, Carter LG, Onikubo T, Kumar PR, Chan MK, et al. (2013). Structure of the Arginine Methyltransferase PRMT5-MEP50 Reveals a Mechanism for Substrate Specificity. PLoS ONE 8, e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, and Trievel RC (2012). Carbon-oxygen hydrogen bonding in biological structure and function. J Biol Chem 287, 41576–41582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Wang Y, Shi L, Chen Y, Xu C, Saeedi A, Pan K, Bohat R, Egan NA, McKenzie JA, et al. (2021). Integrating genome-wide CRISPR immune screen with multi-omic clinical data reveals distinct classes of tumor intrinsic immune regulators. J Immunother Cancer 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N, Lan L, Bahar I, and Wan Y (2015). Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat Commun 6, 8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Yan C, Xie P, Cao Y, Shao J, and Ge J (2020). PRMT2 accelerates tumorigenesis of hepatocellular carcinoma by activating Bcl2 via histone H3R8 methylation. Exp Cell Res 394, 112152. [DOI] [PubMed] [Google Scholar]
- Huang X, Hu H, Webster A, Zou F, Du J, Patel DJ, Sachidanandam R, Toth KF, Aravin AA, and Li S (2021). Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex. Nat Commun 12, 4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Weaver O, Ungvijanpunya N, Yuan Y, Qian J, Gou Y, Wu J, Shen H, Chen Y, Li M, Richard S, et al. (2020). PRMT1-p53 Pathway Controls Epicardial EMT and Invasion. Cell Rep 31, 107739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, and Clarke SG (2019). PRMT7 as a unique member of the protein arginine methyltransferase family: A review. Arch Biochem Biophys 665, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K, Jin CY, and Clarke SG (2017). Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc Natl Acad Sci U S A 114, 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liu L, Yang S, Cao Y, Song X, Xiao J, and Feng H (2019). Black carp PRMT6 inhibits TBK1-IRF3/7 signaling during the antiviral innate immune activation. Fish Shellfish Immunol 93, 108–115. [DOI] [PubMed] [Google Scholar]
- Kalev P, Hyer ML, Gross S, Konteatis Z, Chen CC, Fletcher M, Lein M, Aguado-Fraile E, Frank V, Barnett A, et al. (2021). MAT2A Inhibition Blocks the Growth of MTAP-Deleted Cancer Cells by Reducing PRMT5-Dependent mRNA Splicing and Inducing DNA Damage. Cancer Cell 39, 209–224 e211. [DOI] [PubMed] [Google Scholar]
- Karakashev S, Fukumoto T, Zhao B, Lin J, Wu S, Fatkhutdinov N, Park PH, Semenova G, Jean S, Cadungog MG, et al. (2020). EZH2 Inhibition Sensitizes CARM1-High, Homologous Recombination Proficient Ovarian Cancers to PARP Inhibition. Cancer Cell 37, 157–167 e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Liu F, Veazey KJ, Gao G, Das P, Neves LF, Lin K, Zhong Y, Lu Y, Giuliani V, et al. (2018). Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia 32, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim H, Feng Y, Li Y, Tamiya H, Tocci S, and Ronai ZA (2020a). PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci Transl Med 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yoon BH, Oh CM, Lee J, Lee K, Song H, Kim E, Yi K, Kim MY, Kim YK, et al. (2020b). PRMT1 Is Required for the Maintenance of Mature β-Cell Identity. Diabetes 69, 355–368. [DOI] [PubMed] [Google Scholar]
- Kim JH, Yoo BC, Yang WS, Kim E, Hong S, and Cho JY (2016). The Role of Protein Arginine Methyltransferases in Inflammatory Responses. Mediators Inflamm 2016, 4028353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim NH, Park JE, Hwang JW, Myung N, Hwang KT, Kim YA, Jang CY, and Kim YK (2020c). PRMT6-mediated H3R2me2a guides Aurora B to chromosome arms for proper chromosome segregation. Nat Commun 11, 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, Al-Haddawi M, Tan SY, Osato M, Sabò A, et al. (2015). MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100. [DOI] [PubMed] [Google Scholar]
- Kota SK, Roening C, Patel N, Kota SB, and Baron R (2018). PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone 117, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, et al. (2016). MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzanowski A, Gasper R, Adihou H, Hart P, and Waldmann H (2021). Biochemical Investigation of the Interaction of pICln, RioK1 and COPR5 with the PRMT5-MEP50 Complex. Chembiochem 22, 1908–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Zeng Z, Bagati A, Tay RE, Sanz LA, Hartono SR, Ito Y, Abderazzaq F, Hatchi E, Jiang P, et al. (2021). CARM1 Inhibition Enables Immunotherapy of Resistant Tumors by Dual Action on Tumor cells and T cells. Cancer Discov 11, 2050–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Zhao X, Perna F, Wang L, Koppikar P, Abdel-Wahab O, Harr MW, Levine RL, Xu H, Tefferi A, et al. (2011). JAK2V617F-mediated phosphorylation of PRMT5 downregulates its methyltransferase activity and promotes myeloproliferation. Cancer Cell 19, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Gao J, Yang Y, Qiu R, Zheng Y, Huang W, Zeng Y, Hou Y, Wang S, Leng S, et al. (2018). PHD finger protein 1 (PHF1) is a novel reader for histone H4R3 symmetric dimethylation and coordinates with PRMT5-WDR77/CRL4B complex to promote tumorigenesis. Nucleic Acids Res 46, 6608–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang H, Zhao X, Luo Q, Wang Q, Tan K, Wang Z, Jiang J, Cui J, Du E, et al. (2021). Arginine methylation of METTL14 promotes RNA N6-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat Commun 12, 3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorton BM, Harijan RK, Burgos ES, Bonanno JB, Almo SC, and Shechter D (2020). A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5. Biochemistry 59, 3696–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, De Neef E, Thomas JD, Sabio E, Rousseau B, Gigoux M, Knorr DA, Greenbaum B, Elhanati Y, Hogg SJ, et al. (2021). Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell 184, 4032–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Yang M, Wang Q, Sun C, Shi H, Jing W, Bi Y, Shen X, Ma X, Qin Z, et al. (2021). Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Sci Adv 7, eabc1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, and al., e. (2017). In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, et al. (2016). MTAP Deletions in Cancer Create Vulnerability to Targeting of the MAT2A/PRMT5/RIOK1 Axis. Cell Rep 15, 574–587. [DOI] [PubMed] [Google Scholar]
- Maron MI, Burgos ES, Gupta V, Casill AD, Kosmyna B, Chen H, Gamble MJ, Query CC, and Shechter D (2020). Type I PRMTs and PRMT5 Independently Regulate Both snRNP Arginine Methylation and Post-Transcriptional Splicing. bioRxiv, 2020.2011.2018.389288. [Google Scholar]
- Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, et al. (2016). Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213. [DOI] [PubMed] [Google Scholar]
- McKinney DC, McMillan BJ, Ranaghan M, Moroco JA, Brousseau M, Mullin-Bernstein Z, O’Keefe M, McCarren P, Mesleh MF, Mulvaney KM, et al. (2021). Discovery of a first-in-class inhibitor of the PRMT5-substrate adaptor interaction. bioRxiv, 2021.2002.2003.429644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. (2013). The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersaoui SY, Yu Z, Coulombe Y, Karam M, Busatto FF, Masson JY, and Richard S (2019). Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J 38, e100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moena D, Merino P, Lian JB, Stein GS, Stein JL, and Montecino M (2020). Switches in histone modifications epigenetically control vitamin D3-dependent transcriptional upregulation of the CYP24A1 gene in osteoblastic cells. J Cell Physiol 235, 5328–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney KM, Blomquist C, Acharya N, Li R, Ranaghan MJ, O’Keefe M, Rodriguez DJ, Young MJ, Kesar D, Pal D, et al. (2021). Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol Cell July 28:S1097–2765(21)00588–8, doi: 10.1016/j.molcel.2021.1007.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiani D, Bok J, Massignani E, Wu L, Tabaglio T, Ippolito MR, Cuomo A, Ozbek U, Zorgati H, Ghoshdastider U, et al. (2019). Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci Signal 12. [DOI] [PubMed] [Google Scholar]
- Narayanan N, Wang Z, Li L, and Yang Y (2017). Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells. Cell Discov 3, 16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, and Reinberg D (2003). Methods and tips for the purification of human histone methyltransferases. Methods 31, 49–58. [DOI] [PubMed] [Google Scholar]
- Owens JL, Beketova E, Liu S, Tinsley SL, Asberry AM, Deng X, Huang J, Li C, Wan J, and Hu CD (2020). PRMT5 Cooperates with pICln to Function as a Master Epigenetic Activator of DNA Double-Strand Break Repair Genes. iScience 23, 100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, and Sif S (2004). Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24, 9630–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore F, Bhagwat N, Pastore A, Radzisheuskaya A, Karzai A, Krishnan A, Li B, Bowman RL, Xiao W, Viny AD, et al. (2020). PRMT5 Inhibition Modulates E2F1 Methylation and Gene-Regulatory Networks Leading to Therapeutic Efficacy in JAK2(V617F)-Mutant MPN. Cancer Discov 10, 1742–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen MT, and Helin K (2010). Histone demethylases in development and disease. Trends Cell Biol 20, 662–671. [DOI] [PubMed] [Google Scholar]
- Poquerusse J, Whitford W, Taylor J, Alburaiky S, Snell RG, Lehnert K, and Jacobsen JC (2021). Novel PRMT7 mutation in a rare case of dysmorphism and intellectual disability. J Hum Genet 10.1038/s10038-021-00955-5. [DOI] [PubMed] [Google Scholar]
- Radzisheuskaya A, Shliaha PV, Grinev V, Lorenzini E, Kovalchuk S, Shlyueva D, Gorshkov V, Hendrickson RC, Jensen ON, and Helin K (2019). PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat Struct Mol Biol 26, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman I, Basu SM, Das SK, Bhattacharjee S, Ghosh A, Pommier Y, and Das BB (2018). PRMT5-mediated arginine methylation of TDP1 for the repair of topoisomerase I covalent complexes. Nucleic Acids Res 46, 5601–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repenning A, Happel D, Bouchard C, Meixner M, Verel-Yilmaz Y, Raifer H, Holembowski L, Krause E, Kremmer E, Feederle R, et al. (2021). PRMT1 promotes the tumor suppressor function of p14(ARF) and is indicative for pancreatic cancer prognosis. EMBO J, e106777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Bailon MP, Choi SY, Durfficy ER, Sharma K, McNee GS, Gunnell E, Chiang K, Sahay D, Maslen S, Stewart GS, et al. (2021). Arginine methylation and ubiquitylation crosstalk controls DNA end-resection and homologous recombination repair. Nat Commun In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfeld M, Zhao J, Komatz A, Weinman SA, and Tikhanovich I (2020). The polymorphism rs975484 in the protein arginine methyltransferase 1 gene modulates expression of immune checkpoint genes in hepatocellular carcinoma. J Biol Chem 295, 7126–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, and Sattler M (2001). SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol 8, 27–31. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Kennemer A, Patrick K, Tsichlis P, and Guerau-de-Arellano M (2020). Protein Arginine Methyltransferase 5 in T Lymphocyte Biology. Trends Immunol 41, 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Gomez-Gonzalez B, Silva S, Perez-Calero C, Beaurepere R, Barroso S, Martineau S, Martin C, Ehlen A, Martinez JS, et al. (2021). BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repairdouble dagger. EMBO J 40, e106018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Gao G, Yu X, Kim H, Wang L, Xie L, Schwarz M, Chen X, Guccione E, Liu J, et al. (2020). Discovery of First-in-Class Protein Arginine Methyltransferase 5 (PRMT5) Degraders. J Med Chem 63, 9977–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srour N, Villarreal OD, Yu Z, Preston S, Miller WH Jr, Szewczyk MM, Barsyte-Lovejoy D, Xu H, del Rincon SV, and Richard S (2021). PRMT7 ablation stimulates anti-tumor immunity and sensitizes melanoma to immune checkpoint blockade. bioRxiv 10.1101/2021.07.28.454202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopa N, Krebs JE, and Shechter D (2015). The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci 72, 2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S, Huard S, and Dubois T (2021). CARM1/PRMT4: Making Its Mark beyond Its Function as a Transcriptional Coactivator. Trends Cell Biol 31, 402–417. [DOI] [PubMed] [Google Scholar]
- Szewczyk MM, Ishikawa Y, Organ S, Sakai N, Li F, Ackloo S, Eram MS, dilworth D, Fukushi H, Harding R, et al. (2020). Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Nat Commun 11, 2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamiya H, Kim H, Klymenko O, Feng Y, Zhang T, Han JY, Murao A, Snipas SJ, Jilaveanu L, Brown K, et al. (2018). SHARPIN-mediated regulation of protein arginine methyltransferase 5 controls melanoma growth. J Clin Invest 128, 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DQ, Li Y, Yang C, Li J, Tan SH, Chin DWL, Nakamura-Ishizu A, Yang H, and Suda T (2019). PRMT5 Modulates Splicing for Genome Integrity and Preserves Proteostasis of Hematopoietic Stem Cells. Cell Rep 26, 2316–2328. [DOI] [PubMed] [Google Scholar]
- Tewary SK, Zheng YG, and Ho MC (2019). Protein arginine methyltransferases: insights into the enzyme structure and mechanism at the atomic level. Cell Mol Life Sci 76, 2917–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thandapani P, O’Connor TR, Bailey TL, and Richard S (2013). Defining the RGG/RG motif. Mol Cell 50, 613–623. [DOI] [PubMed] [Google Scholar]
- Thompson PR, and Fast W (2006). Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol 1, 433–441. [DOI] [PubMed] [Google Scholar]
- Vadnais C, Chen R, Fraszczak J, Yu Z, Boulais J, Pinder J, Frank D, Khandanpour C, Hébert J, Dellaire G, et al. (2018). GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat Commun 9, 1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal OD, Mersaoui SY, Yu Z, Masson JY, and Richard S (2020). Genome-wide R-loop analysis defines unique roles for DDX5, XRN2, and PRMT5 in DNA/RNA hybrid resolution. Life Sci Alliance 3, e202000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann T, Izzo A, Kamieniarz K, Richter F, Vogler C, Sarg B, Lindner H, Young NL, Mittler G, Garcia BA, et al. (2011). Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Chowdhury R, Schiller R, Ge W, Kawamura A, and Schofield CJ (2016). Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun 7, 11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huang Z-Q, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. (2001). Methylation of histone H4 at arginine 3 facilitates transcriptional activation by nuclear hormone receptor. Science 293, 853–857. [DOI] [PubMed] [Google Scholar]
- Wang Z, Pan Z, Adhikari S, Harada BT, Shen L, Yuan W, Abeywardana T, Al-Hadid Q, Stark JM, He C, et al. (2021). m(6) A deposition is regulated by PRMT1-mediated arginine methylation of METTL14 in its disordered C-terminal region. EMBO J 40, e106309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates J.R.r., et al. (2009). Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93. [DOI] [PubMed] [Google Scholar]
- Wei X, Yang J, Adair SJ, Ozturk H, Kuscu C, Lee KY, Kane WJ, O’Hara PE, Liu D, Demirlenk YM, et al. (2020). Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc Natl Acad Sci U S A 117, 28068–28079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Schapira M, Arrowsmith CH, and Barsyte-Lovejoy D (2021). Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat Rev Drug Discov 20, 509–530. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. (2017). RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ma DW, Cao YP, Li DZ, Zhou X, Feng JF, and Bao J (2021). PRMT5 functionally associates with EZH2 to promote colorectal cancer progression through epigenetically repressing CDKN2B expression. Theranostics 11, 3742–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lin X, Segers F, Suganthan R, Hildrestrand GA, Rinholm JE, Aas PA, Sousa MML, Holm S, Bolstad N, et al. (2020). OXR1A, a Coactivator of PRMT5 Regulating Histone Arginine Methylation. Cell Rep 30, 4165–4178 e4167. [DOI] [PubMed] [Google Scholar]
- Yang Y, and Bedford MT (2013). Protein arginine methyltransferases and cancer. Nat Rev Cancer 13, 37–50. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, et al. (2015). PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun 6, 6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, and Bedford MT (2010). TDRD3 Is an Effector Molecule for Arginine-Methylated Histone Marks. Mol Cell 40, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, and Bedford MT (2014). Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell 53, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Gui T, Zeng X, Deng Y, Wang Z, Wang Y, Yang D, Li Q, Xu P, Hu R, et al. (2021). PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med 13, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Chen T, Hébert J, Li E, and Richard S (2009). A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation. Mol Cell Biol 29, 2982–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Mersaoui SY, Guitton-Sert L, Coulombe Y, Song J, Masson JY, and Richard S (2020). DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer 2, zcaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Vogel G, Coulombe Y, Dubeau D, Spehalski E, Hebert J, Ferguson DO, Masson JY, and Richard S (2012). The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res 22, 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Tian R, Zhao H, Li L, and Bi X (2018). Multiple Arginine Residues Are Methylated in Drosophila Mre11 and Required for Survival Following Ionizing Radiation. G3 (Bethesda) 8, 2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappacosta F, Wagner CD, Della Pietra A 3rd, Gerhart SV, Keenan K, Korenchuck S, Quinn CJ, Barbash O, McCabe MT, and Annan RS (2021). A Chemical Acetylation-Based Mass Spectrometry Platform for Histone Methylation Profiling. Mol Cell Proteomics 20, 100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al. (2020a). METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol Cell 79, 425–442 e427. [DOI] [PubMed] [Google Scholar]
- Zhang H, Han C, Li T, Li N, and Cao X (2019). The methyltransferase PRMT6 attenuates antiviral innate immunity by blocking TBK1-IRF3 signaling. Cell Mol Immunol 16, 800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, and Cheng X (2003). Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Q, Li L, Mu D, Hua K, Ci S, Shen L, Zheng L, Shen B, and Guo Z (2020b). Arginine methylation of APE1 promotes its mitochondrial translocation to protect cells from oxidative damage. Free Radic Biol Med 158, 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al. (2009). PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol 16, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng NN, Zhou M, Sun F, Huai MX, Zhang Y, Qu CY, Shen F, and Xu LM (2020). Combining protein arginine methyltransferase inhibitor and anti-programmed death-ligand-1 inhibits pancreatic cancer progression. World J Gastroenterol 26, 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Li X, Cai X, Zha H, Zhou Z, Sun X, Rong F, Tang J, Zhu C, Liu X, et al. (2021). Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. Mol Cell 81, 3171–3186. [DOI] [PubMed] [Google Scholar]
- Zhu J, Liu X, Cai X, Ouyang G, Fan S, Wang J, and Xiao W (2020a). Zebrafish prmt7 negatively regulates antiviral responses by suppressing the retinoic acid-inducible gene-I-like receptor signaling. FASEB J 34, 988–1000. [DOI] [PubMed] [Google Scholar]
- Zhu J, Liu X, Cai X, Ouyang G, Zha H, Zhou Z, Liao Q, Wang J, and Xiao W (2020b). Zebrafish prmt3 negatively regulates antiviral responses. FASEB J 34, 10212–10227. [DOI] [PubMed] [Google Scholar]


