Summary
The kinase domain transfers phosphate from ATP to substrates. However, the Legionella effector SidJ adopts a kinase fold yet catalyzes calmodulin (CaM)-dependent glutamylation to inactivate the SidE ubiquitin ligases. The structural and mechanistic basis in which the kinase domain catalyzes protein glutamylation is unknown. Here we present cryo-EM reconstructions of SidJ:CaM:SidE reaction intermediate complexes. We show that the kinase-like active site of SidJ adenylates an active site Glu in SidE resulting in the formation of a stable reaction intermediate complex. An insertion in the catalytic loop of the kinase domain positions the donor Glu near the acyl-adenylate for peptide bond formation. Our structural analysis led us to discover that the SidJ paralog SdjA is a glutamylase that differentially regulates the SidE-ligases during Legionella infection. Our results uncover the structural and mechanistic basis in which the kinase fold catalyzes non-ribosomal amino acid ligations and reveal an unappreciated level of SidE-family regulation.
Keywords: SidJ, SdjA, SdeA, SdeB, SdeC, SidE, glutamylation, pseudokinase, Legionella, effectors
Graphical Abstract
eTOC:
Osinski et al. report cryo-EM reconstructions of the SidJ:CaM:SidE reaction intermediate complex providing structural insights into protein glutamylation by a pseudokinase. Furthermore, they discover that SdjA is also a glutamylase that differentially inactivates the SidE ubiquitin ligases in vitro and during Legionella infection
Introduction
Legionella pneumophila, the causative agent of Legionnaires disease, translocates more than 330 effectors utilizing a Type 4 secretion system to establish a replicative niche known as the Legionella Containing Vacuole (LCV) (Cornejo et al., 2017; Isberg et al., 2009). Within the Legionella effector repertoire are protein domains with recognizable folds that occasionally catalyze unexpected reactions (Black et al., 2019; Mukherjee et al., 2011; Neunuebel et al., 2011; Qiu et al., 2016). For example, the Legionella effector SidJ adopts a kinase-like fold yet catalyzes calmodulin (CaM)-dependent glutamylation to inactivate the SidE ubiquitin (Ub) ligases (Bhogaraju et al., 2019; Black et al., 2019; Gan et al., 2019; Sulpizio et al., 2019). The SidE effectors SdeA, SdeB, SdeC and SidE (collectively referred to as SidE), employ ADP ribosyltransferase (ART) and phosphodiesterase (PDE) activities to catalyze ligation of Ub to proteins independent of E1 and E2 enzymes (Bhogaraju et al., 2016; Kotewicz et al., 2017; Qiu et al., 2016). The SidE ART domain uses NAD+ to ADP-ribosylate Ub on Arg42. ADP ribosylated Ub serves as a substrate for the PDE domain, which hydrolyzes the phosphodiester bond and transfers Ub to Ser residues on proteins forming a Ser-phosphoribosyl (pR)-Ub linkage.
SidE-family Ub ligases have been structurally and mechanistically analyzed (Akturk et al., 2018; Dong et al., 2018; Kalayil et al., 2018; Wang et al., 2018). Co-crystal structures of the SdeA/SidE catalytic core in complexes with Ub/UbR42A and NAD+/NADH provided insights into the molecular mechanism of ADP-ribosylation of Ub by the SidE ART domain (Dong et al., 2018; Wang et al., 2018). The PDE-Ub interactions prior to Ub transfer to substrates were also structurally analyzed, leading to a detailed reaction model (Akturk et al., 2018; Kalayil et al., 2018). All structural studies of SidE-family Ub-ligases to date have focused on interactions with Ub and NAD+ but have not involved interactions with SidJ.
SidE-family ubiquitination promotes infectivity of Legionella pneumophila and is required for proper formation of the LCV (Bardill et al., 2005). However, unrestrained SidE activity is harmful to the host. Therefore, SidE-family ubiquitination is regulated by Legionella deubiquitinases DupA and DupB, which reverse pR ubiquitination (Shin et al., 2020; Wan et al., 2019), and the pseudokinase SidJ, which glutamylates and inactivates the SidE effectors (Bhogaraju et al., 2019; Black et al., 2019; Gan et al., 2019; Sulpizio et al., 2019). Three of the four SidE-family members lie within a contiguous genomic locus that also includes dupA and sidJ (Figure 1A). The dupB gene neighbors the sidJ paralog, sdjA. Despite sharing 52% amino acid sequence identity, SidJ and SdjA are not functionally redundant (Qiu et al., 2017b). As such, the function of SdjA is unknown.
Figure 1. SidJ forms a stable acyl-adenylate intermediate complex with SdeA and SdeC.
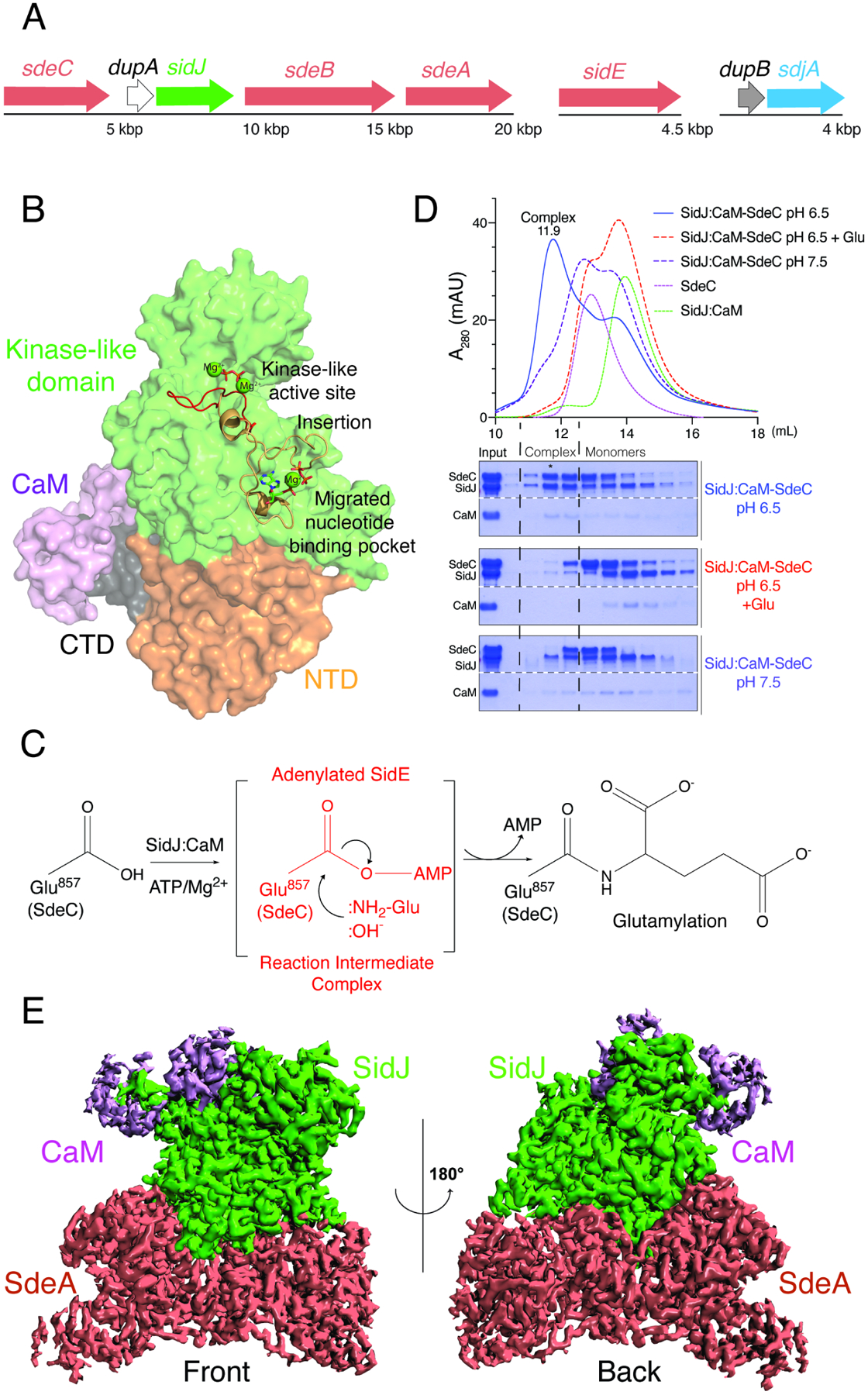
(A) Organization of the sidE family (salmon), sidJ (green) and dupA (white), dupB, (grey) and sdjA (blue) effectors in the genome of L. pneumophila. SdeA, SdeB and SdeC lie in a genomic neighbourhood with SidJ and DupA, while SidE lies in a different locus (upper).
(B) Structure of SidJ (PDB: 6OQQ) depicting the N-terminal domain (NTD; orange), the kinase-like domain (green), the C-terminal domain (CTD; black) and CaM (purple). The kinase-like active site (red) and the migrated nucleotide binding pocket formed by an insertion in the catalytic loop (orange) are highlighted. Mg2+ ions are shown as green spheres.
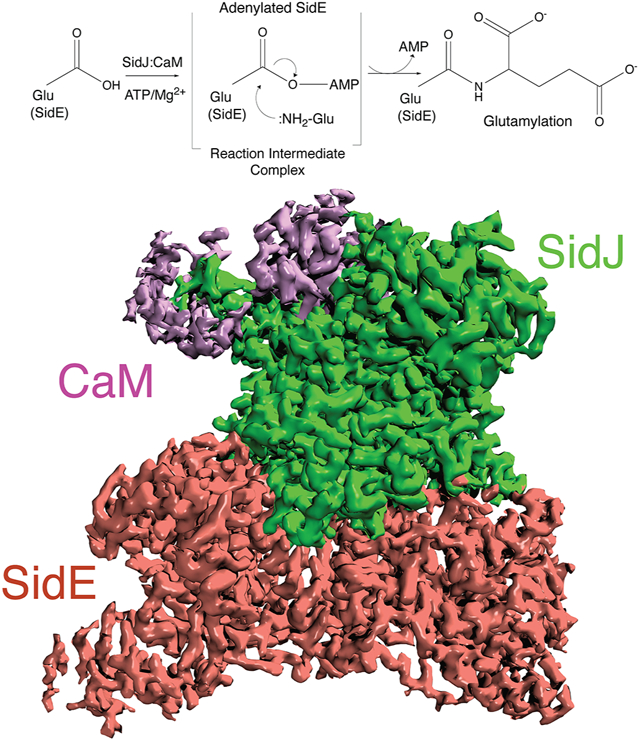
(C) Schematic representation of SidJ-catalyzed glutamylation of the SidE effectors. The SidJ:CaM complex binds ATP/Mg2+, which is used to adenylate the SidE-family active site Glu (SdeC; Glu857). Adenylated SidE forms a stable reaction intermediate complex with SidJ, which facilitates Glu binding, positioning of the NH2 group for attack of the acyl-adenylate and formation of the isopeptide bond. Note that under basic conditions, :OH− can hydrolyze the acyl-adenylate.
(D) SEC trace (upper) and aligned SDS-PAGE and Coomassie staining (lower) of the SidJ:CaM:SdeC complex (blue) and the products resulting from the addition of Glu (dashed red) or increasing the pH to 7.5 (dashed blue). Note that the addition of Glu and increasing the pH dissociates the complex, consistent with the presence of an acyl-adenylate in the complex. Star indicates fraction used for Cryo-EM studies. The white dashed line on the Coomassie stained gel indicates that the samples are on the same gel.
(E) Cryo-EM density map representation of the SidJ:CaM:SdeA complex. SidJ is in green, SdeACore is in salmon and CaM is in purple. Front and back views are shown.
Pseudoenzymes contain a protein domain that resembles a catalytically active counterpart (Kwon et al., 2019; Ribeiro et al., 2019). However, pseudoenzymes lack key catalytic residues believed to be required for activity. Recent work on pseudokinases has revealed that different binding orientations of ATP and active site residue migration can repurpose the kinase scaffold to catalyze novel reactions (Black et al., 2019; Slanina et al., 2021; Sreelatha et al., 2018). These results suggest that the kinase fold is more versatile than previously appreciated and that pseudokinases should be reanalyzed for alternative transferase activities.
Structures of the SidJ:CaM complex uncovered an N-terminal domain (NTD) of unknown function (residues 136–313), a kinase-like domain with two ligand binding pockets (residues 336–758) and a C-terminal domain that binds CaM (residues 759–873) (Bhogaraju et al., 2019; Black et al., 2019; Gan et al., 2019; Sulpizio et al., 2019) (Figure 1B). The canonical kinase-like ATP binding pocket of SidJ contains residues that are required for glutamylation of the SidE ligases, including Lys367 (Lys72 using protein kinase A; PKA nomenclature), N534 (PKA; N171) and D542 (PKA; D184). An insertion in the catalytic loop of the kinase domain (residues 487–532) extends away from the kinase active site and forms a migrated nucleotide binding pocket, which is also required for glutamylation (Bhogaraju et al., 2019; Black et al., 2019; Gan et al., 2019; Sulpizio et al., 2019). From these studies, we proposed a catalytic mechanism, whereby CaM binding allows the kinase–like active site of SidJ to bind ATP and transfer AMP to the active site Glu on SidE forming a high energy acyl-adenylate intermediate. Adenylated SidE binds the migrated nucleotide-binding pocket, positioning the acyl-adenylate and donor Glu for glutamylation and inactivation of the SidE effectors (Black et al., 2019) (Figure 1C). However, others argue that the migrated nucleotide binding pocket is an allosteric site that facilitates glutamylation (Sulpizio et al., 2019).
Here, we present cryo-EM structures of the reaction intermediate complex formed between SidJ and SdeA, and SidJ and SdeC. We propose a model that explains how a pseudokinase catalyzes protein glutamylation. Furthermore, we show that the SidJ paralog, SdjA is an active glutamylase that differentially regulates SidE-family activity in vitro and during Legionella infection.
Results
SidJ and SdeA/C Form a Stable Reaction Intermediate Complex
In several amidoligases, including the ATP-grasp enzymes involved in tubulin glutamylation, a stable reaction intermediate complex facilitates the formation of the peptide bond (Garnham et al., 2015; Mahalingan et al., 2020). We hypothesized that SidJ:CaM would also form a stable reaction intermediate complex with the SidE ligases when Glu was excluded and when the reaction was performed under acidic conditions to prevent base-catalyzed hydrolysis of the acyl-adenylate (Figure 1C). We incubated SidJ97–851:CaM (SidJ:CaM) with SdeA231–1190 (SdeACore) or SdeC231–1222 (SdeCCore), initiated the reactions with ATP/Mg2+, and analyzed the reaction products by SDS-PAGE following size-exclusion chromatography (SEC) in a slightly acidic (pH 6.5) buffer. We detected the formation of stable ternary complexes consisting of SidJ, CaM, and SdeCCore (Figure 1D) and SidJ, CaM and SdeACore (Figure S1A). Upon supplementation of the reaction with Glu, or when the complex was subjected to SEC using a mildly basic buffer (pH 7.5), the SidJ:CaM:SdeCCore complex dissociated (Figure 1D). These results are consistent with the presence of an acyl-adenylate intermediate within the complex.
Extensive Interface Interactions Facilitate Complex Formation Between SidJ:CaM and SdeA/CCore
We determined cryo-EM structures of SidJ:CaM:SdeACore and SidJ:CaM:SdeCCore complexes with resolution up to 2.5 Å and 2.8 Å, respectively (Figure 1E, and Figures S1, S2 and Table 1). The density maps reveal details of the interface between the proteins and their active sites, with clear densities for SidJ, CaM and the PDE and ART domains of SdeACore and SdeCCore. Densities for the distal parts of the PDE domain and most of the C-terminal coiled coil are less clear, suggesting a higher degree of flexibility in these regions. We use the SidJ:CaM:SdeACore and the SidJ:CaM:SdeCCore maps for description of the interface and the SidJ:CaM:SdeCCore maps to describe interactions related to catalysis.
Table 1.
Data collection and refinement statistics.
| SidJ:CaM:SdeA (EMD-23862) (PDBID: 7MIR) | SidJ:CaM:SdeC (EMD-23863) (PDBID: 7MIS) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−/Å−2) | 50 | 50 |
| Defocus range (μm) | 0.8 – 2.5 | 0.8 – 2.5 |
| Pixel size (Å) | 1.08 | 1.08 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 3,193,233 | 4,454,035 |
| Final particle images (no.) | 310,154 | 152,589 |
| Map resolution (Å) | 2.5 | 2.8 |
| FSC threshold | 0.143 | 0.143 |
| Refinement | ||
| Initial model used (PDB code) | 5YIM, 6S5T | SidJ-SdeA |
| Model resolution (Å) | 2.6 | 2.9 |
| FSC threshold | 0.5 | 0.5 |
| Map sharpening B factor (Å−2) | −37 | −47 |
| Model composition | ||
| Nonhydrogen atoms | 12,997 | 13,039 |
| Protein residues | 1,603 | 1,616 |
| Ligands | 5 | 6 |
| B factors (Å−2) | ||
| Protein | 48.5 | 54.9 |
| Ligands | 36.9 | 30.8 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.003 |
| Bond angles (°) | 0.485 | 0.468 |
| Validation | ||
| MolProbity score | 1.27 | 1.42 |
| Clashscore | 2.83 | 3.74 |
| Poor rotamers (%) | 0 | 0 |
| Ramachandran plot | ||
| Favored (%) | 96.8 | 96.2 |
| Allowed (%) | 3.2 | 3.8 |
| Disallowed (%) | 0 | 0 |
The overall reaction intermediate complexes are highly similar, with a root mean square deviation (RMSD) of 1.5 Å (Figures S2C and S3). The SidJ/CaM conformation remains similar to those previously determined by X-ray crystallography (Black et al., 2019) and cryo-EM (Bhogaraju et al., 2019). SidJ sits in a V-shaped cleft made up of the PDE and ART domains of SdeA/CCore (Figure 2A). The interfaces between SidJ and SdeA/CCore span ~2,200 Å2 and are predominantly formed by the SidJ NTD (Figure 2B). Within the NTD of SidJ, there are three regions that form extensive contacts with SdeACore. 1) A helix-turn-helix (HTH) motif that forms electrostatic contacts with residues located in the back of the ART domain, 2) a β-hairpin wedged in between the PDE and ART domains and 3) a helical bundle that interacts with the PDE domain. To verify the relevance of these interactions for SidJ-catalyzed glutamylation of SdeA, we mutated D833, K569, D565 and D372 of SdeACore, which lie near the interfaces of the three regions. Reversing the charge on these residues in SdeACore markedly reduced its propensity to be glutamylated by SidJ, and abolished formation of the reaction intermediate complex as judged by SEC (Figure 2C and S2D). Our observation that SidJ interacts with both the SdeA/C ART and PDE domains reconciles previous data showing that SidJ-dependent suppression of SdeA-mediated yeast toxicity requires the PDE and ART domains of SdeA (Havey and Roy, 2015). As expected, SidJ did not glutamylate the isolated ART domain of SdeA (Figure 2D).
Figure 2. SidJ:CaM:SdeA/CCore complex formation is mediated by extensive interface interactions.
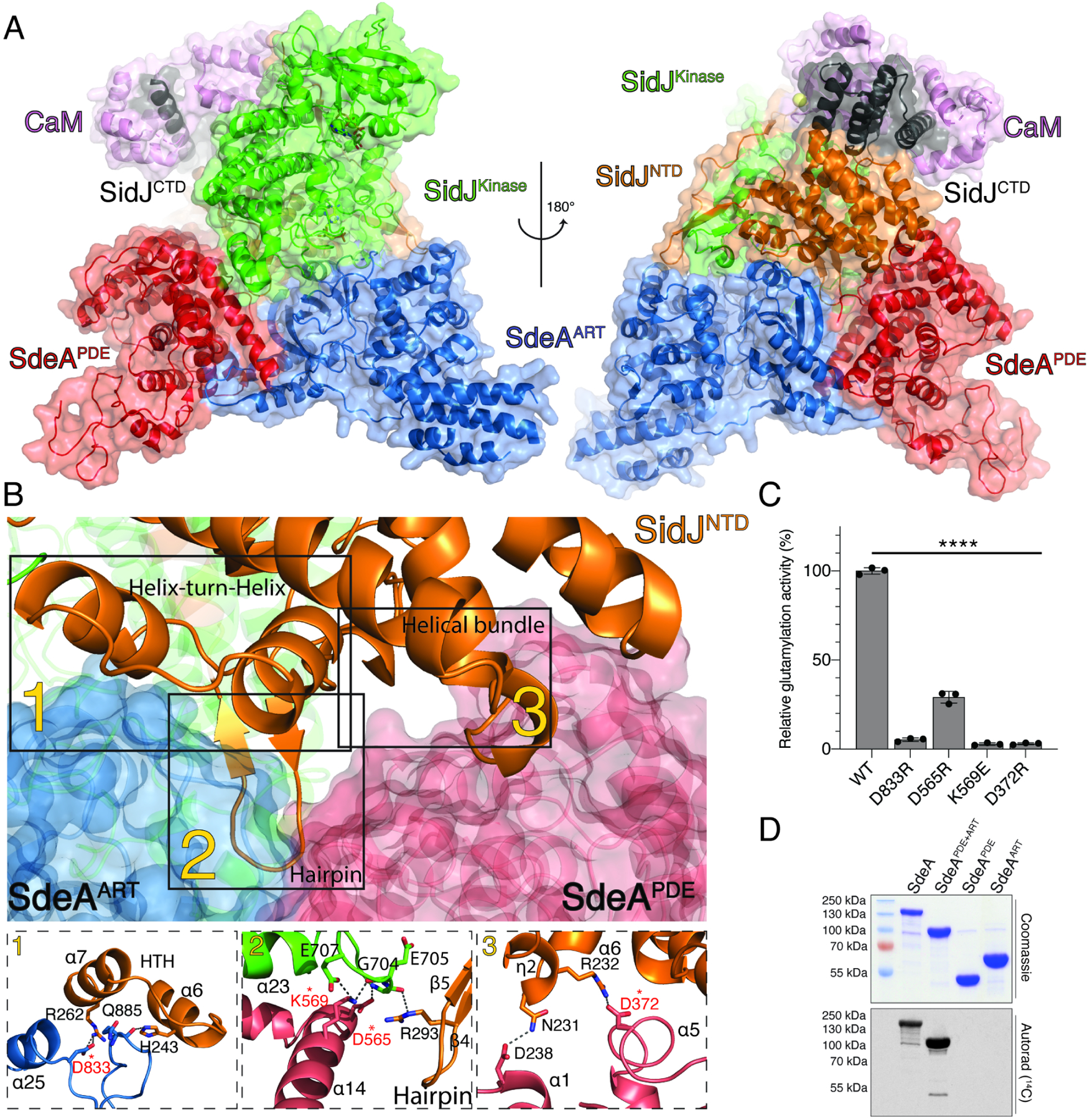
(A) Overview of SidJ:CaM:SdeACore reaction intermediate complex. The SidJ NTD, kinase-like domain and CTD are in orange, green, and black, respectively. The SdeA PDE domain is in red and the ART domain is in blue. CaM is in purple. SidJ lies within a V-shaped cleft between SdeA PDE and ART domains.
(B) Back view of the SidJ:CaM:SdeACore interface. The SidJ NTD forms three distinct regions of interaction with SdeA (labelled 1–3). 1. A loop within the SdeA ART domain interacts with the helix-turn-helix (HTH) motif within the SidJ NTD. 2. A β-hairpin from the SidJ NTD is nestled between the PDE and ART domains of SdeA, forming electrostatic and hydrophobic interactions. A helix from the SidJ kinase-like domain also interacts with the SidJ β-hairpin and the PDE domain of SdeA. 3. A helical bundle from the SidJ NTD interacts with the PDE domain of SdeA. Zoomed in view depicting the residues involved in the interaction interfaces are shown in subpanels (lower) and colored as in A. Residues targeted for mutagenesis are marked with asterisks. Putative hydrogen bonds and salt bridges are depicted as dotted lines.
(C) Glutamylation activity of SidJ using SdeACore and mutations that disrupt interface interactions. Residues mutated are also marked with an asterisk in (B). [3H]Glu was used as a substrate and the reaction products were resolved by SDS PAGE and visualized by Coomassie staining. Radioactive gel bands were excised and 3H incorporation into SdeACore was quantified by scintillation counting, n=3.
(D) Incorporation of [14C]-Glu into full length SdeA, SdeAPDE+ART, SdeAPDE or SdeAART by SidJ. Reaction products were separated by SDS PAGE and visualized by Coomassie staining (upper) and autoradiography (lower).
Data in graphs shown is mean ± SD, ****P <0.0001.
Structural Comparison of the SidJ:SdeA and SidJ:SdeC Interfaces
Although the secondary structural elements that make up the interface between SidJ and SdeACore and SidJ and SdeCCore are conserved, the interacting residues differ between the two complexes (Figure 3). Within the SidJ HTH, Arg262 forms a hydrogen bond with Asp833 within the α-25 helix of SdeA (Figure 3A; left). However, Asp833 is replaced with a Gly in SdeC (Gly830; Figure 3A, right). Nevertheless, Asp882 within the β12-β13 loop in SdeC forms a salt bridge with Arg262 in SidJ thus preserving the integrity of the interface (Figure 3A; right).
Figure 3. SidJ:CaM:SdeACore and SidJ:CaM:SdeCCore complexes are formed by distinct interactions.
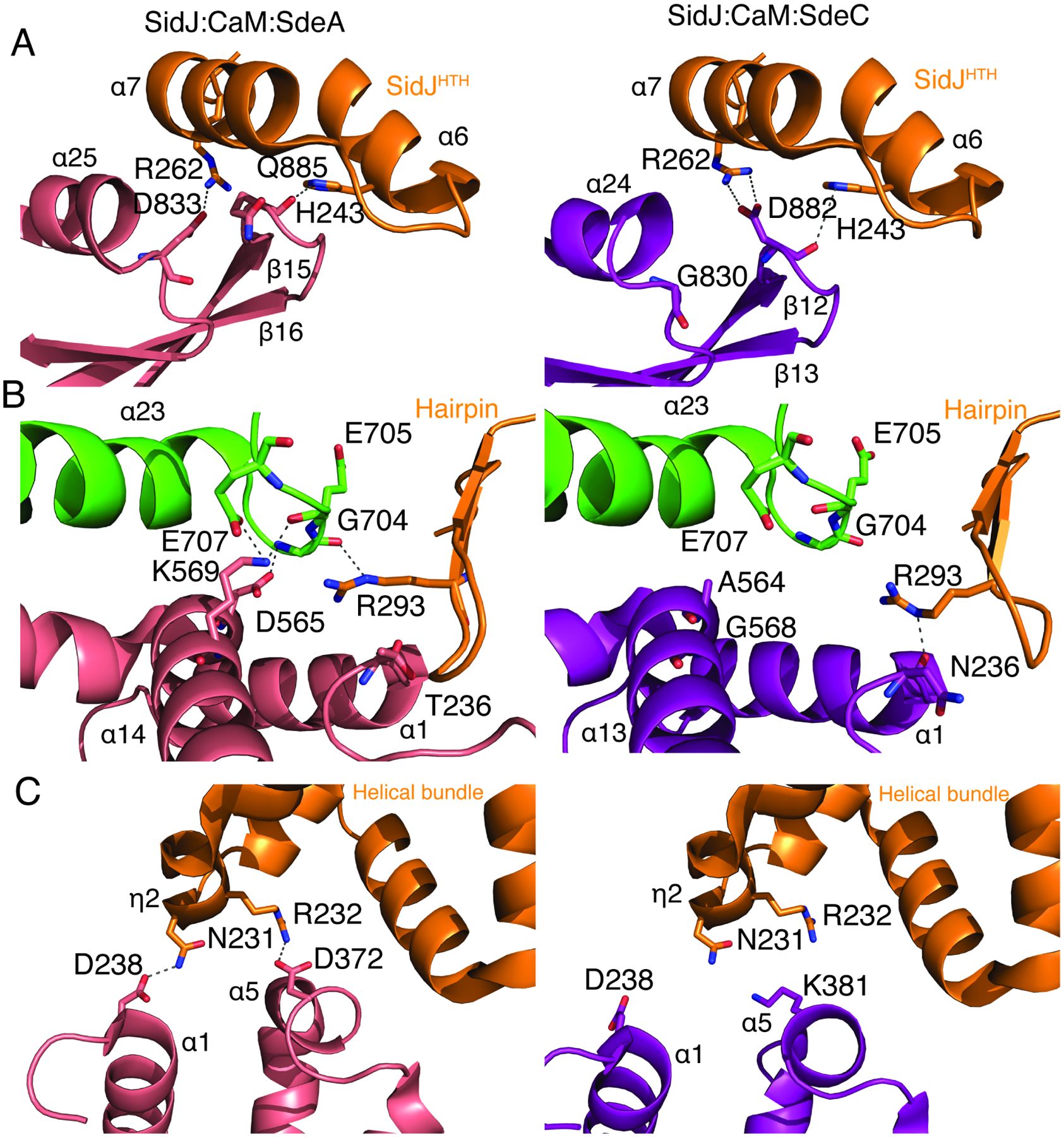
(A) Comparison of interactions between SidJ:CaM:SdeACore (left) and SidJ:CaM:SdeCCore (right) in the Helix-loop-Helix region (orange). SidJ Arg262 forms a hydrogen bond with SdeA Asp833 (left), and SdeC Asp882 (right).
(B) Comparison of interactions in the hairpin region (orange). In complex with SdeA, a loop in SidJ extending from the α23 helix of the Kinase domain (green) forms electrostatic interactions with Asp565 and Lys569 in the α14 helix of SdeA (left). In complex with SdeC, the α13 helix of SdeC moves away, bringing the α1 helix closer to the SidJ hairpin, allowing for hydrogen bond formation between SidJ Arg293, and main chain carbonyl of Asn236.
(C) Comparison of interactions with the SidJ helical bundle region (orange). In complex with SdeA (left), SidJ Asn231 and Arg232 form hydrogen bonds with Asp238 and Asp372 on SdeA PDE domain, respectively (right). In complex with SdeC, the SdeC PDE domain moves away from the helical bundle (right).
SidJNTD – orange, SidJKinase – green, SdeA – salmon, SdeC – purple. Dashed lines indicate interactions.
Within the C-lobe of the kinase-like domain of SidJ, a large loop between the α-23 and α-24 helices interacts with the α-14 helix within the PDE domain of SdeACore, adjacent to its interaction with the SidJ NTD β-hairpin (Figure 2B; inset 2 and Figure 3B). Glu707 and Gly704 within the α-23-α24 loop of SidJ interact with Lys569 and Asp565 in SdeACore. Interestingly, in the SidJ:SdeCCore interface, the PDE domain is slightly shifted and the main chain carbonyl of Asn236 within the loop preceding the α-1 helix forms a hydrogen bond with Arg293 within the SidJ β-hairpin (Figure 3B).
Interactions with the helical bundle motif of SidJ are also not conserved between the two complexes. In SdeACore, Asp238 from the α-1 helix and Asp372 from the α-5 helix form electrostatic interactions with residues located within the SidJ helical bundle (Figure 3C, left). However, the SdeC PDE domain is shifted away from the SidJ helical bundle such that SidJ Asn231 is unable to interact with SdeC Asp238 (Asp238 in SdeA). Asp372 in SdeA is replaced by K381 in SdeC, making the interaction with SidJ R232 unfavorable (Figure 3C, right). The shift however is what allows for the interactions around the β-hairpin region of SidJ, bringing SidJ Arg293 and SdeC Asn236 closer, enabling hydrogen bonding (Figure 3B, right). Collectively, the differences between the SidJ:SdeA and SidJ:SdeC interfaces suggest that SidJ, rather than forming interactions with highly conserved residues, recognizes the overall fold of the SidE effectors by a few strong, and multiple weak interactions.
Unique Active Sites in SidJ Catalyze Adenylation and Glutamylation
Interactions also occur within the migrated-nucleotide binding pocket and the ARTT loop within the active site of the ART domain. Remarkably, we observed density that is consistent with a covalent bond between the phosphate of AMP and the γ-carboxyl group of E857 in SdeCCore, which is consistent with an acyl-adenylate (Figure 4A). The acyl-adenylate is stabilized by Arg500, Asn733, His492 and Tyr506 within the SidJ migrated nucleotide binding pocket. Mutation of these residues to Ala markedly reduced glutamylation of SdeA by SidJ (Black et al., 2019). When comparing the active site of SdeA in the reaction intermediate complex to the apo structure of SdeA (Dong et al., 2018), significant rearrangement of the ARTT loop is observed, which facilitates SidJ binding and stabilization of the acyl-adenylate (Figure S4A).
Figure 4. Unique active sites facilitate SidJ-mediated glutamylation of the SidE effectors.
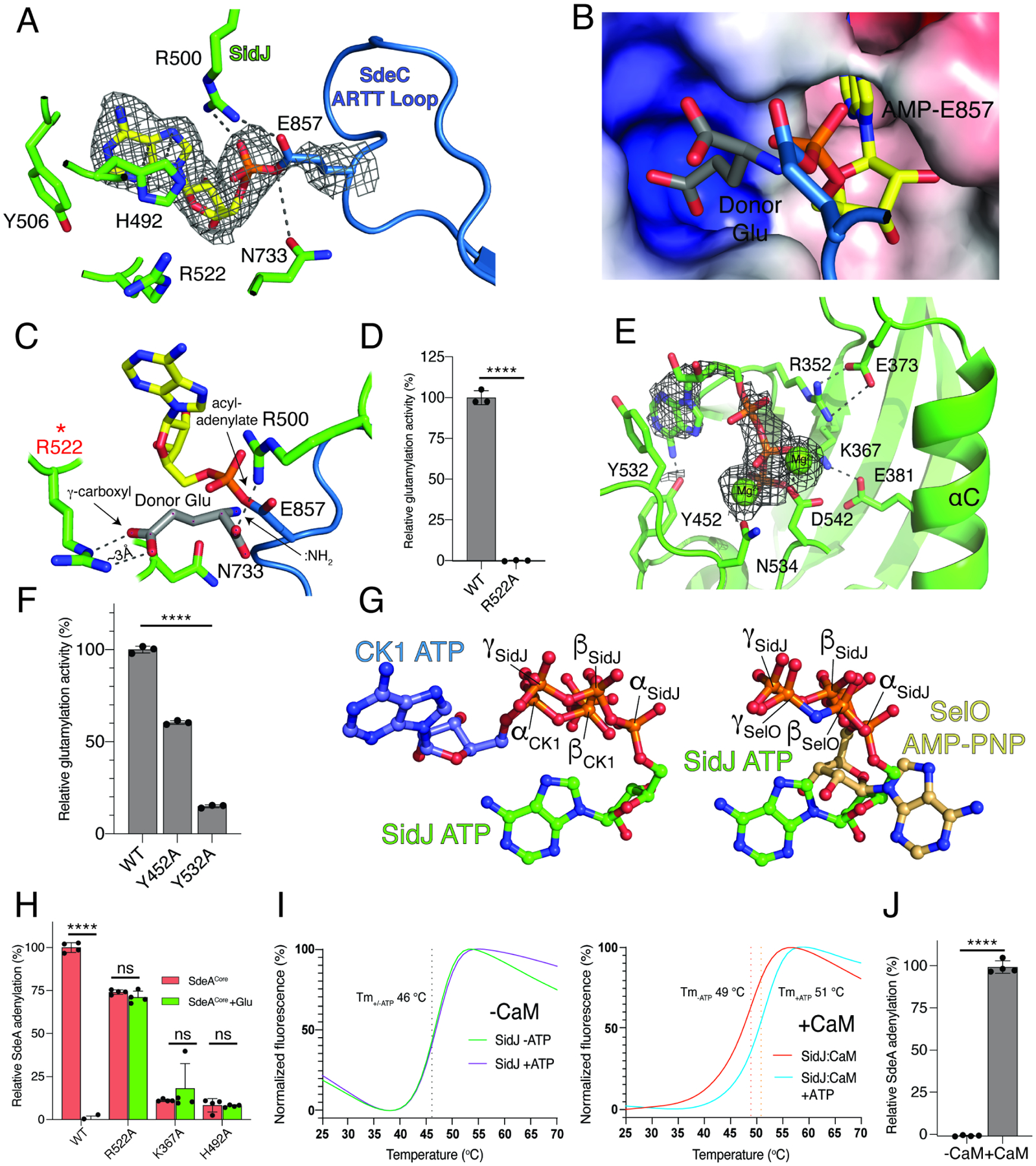
(A) Details of the migrated nucleotide binding pocket of SidJ (green) and the ARTT loop of SdeCCore (blue) showing the interactions (dashed lines) involved in binding the acyl-adenylate reaction intermediate. The AMP is shown as sticks and the Coulomb potential map is shown in mesh. Note the formation of a covalent bond between AMP and the SdeC active site Glu857.
(B, C) Electrostatic surface representation (B) and molecular interactions (C) of the migrated nucleotide binding pocket of SidJ bound to SdeCCore depicting a positively charged cleft adjacent to the acyl-adenylate reaction intermediate. A donor Glu was manually placed inside of the pocket, with its γ-carboxyl group interacting with Arg522 of SidJ. The donor Glu :NH2 group is in position to attack the acyl-adenylate. Electrostatic surface is contoured at 7kT.
(D) Glutamylation activity of SidJ and the Arg522 mutant using SdeACore as substrate. Reaction products were analyzed as in Figure 2C, n=3.
(E) Details of the kinase-like active site of SidJ (green) depicting the molecular interactions (dashed lines) that facilitate ATP/Mg2+ binding. The ATP is in sticks, the Coulomb potential map is shown in mesh and the Mg2+ ions are shown as spheres.
(F) Glutamylation activity of SidJ and kinase-like active site mutants using SdeACore as a substrate. Reaction products were analyzed as in Figure 2C, n=3. Note that the other residues have been mutated and analyzed for glutamylation in our previous work (Black et al., 2019).
(G) Ball-and-stick representation of superimposed nucleotides from SidJ and protein kinase CK1 (left) and SidJ and SelO (right) as a result of superposition of the kinase active sites. The α, β, and γ-phosphates of SidJ, SelO and CK1 are highlighted. Note that the nucleotide orientation of SidJ is similar to SelO, which transfers AMP to proteins (Sreelatha et al., 2018).
(H) Quantification of acyl-adenylate formation following reactions with SidJ59–851 (WT and mutants), SdeACore and [α-32P]ATP. SdeACore E860Q was used to calculate the baseline signal in TCA precipitates. The reactions were terminated by the addition of TCA and the SdeA acyl-adenylate was detected by scintillation counting of the acid-insoluble material (red bars), n=4. Glu was also added to the reactions (green bars), which displaces the 32P-AMP from SdeA into a TCA soluble fraction.
(I) Differential scanning fluorimetry (DSF) depicting the thermal stability profiles of SidJ (left) and SidJ:CaM (right) in the presence or absence of ATP/Mg2+. Protein denaturation was followed by monitoring the fluorescence of SYPRO Orange dye, which binds hydrophobic regions on proteins. The Tm values are shown in the insets. Graphs show a mean of separately normalized triplicates.
(J) Quantification of acyl-adenylate formation following reactions with SidJ59–851, SdeACore, [α-32P]ATP in the presence (+) or absence (−) of CaM. SdeACore E860Q was used to calculate the baseline signal in TCA precipitates. Reaction products were analyzed as in Figure 4H, n=3.
Data in graphs shown as mean ± SD, ****P <0.0001, ns >0.15.
During catalysis, SidJ cannot substitute Asp for Glu, suggesting a high degree of specificity (Black et al., 2019). Analysis of the electrostatic surface in the vicinity of the active site revealed a small positively charged pocket in SidJ that we predict binds Glu (Figure 4B). We modelled a donor Glu into this pocket, positioning the :NH2 group near the acyl-adenylate for nucleophilic attack and formation of the Glu-Glu isopeptide bond. The γ-carboxyl was placed at a distance of ~3 Å from the SidJ Arg522 guanidinium, indicating a potential strong electrostatic interaction (Figure 4C). Mutation of Arg522 abolished SidJ-catalyzed glutamylation of SdeA (Figure 4D). Assuming the carboxyl group of a donor Asp side chain could enter the pocket and interact with Arg522, the distance between the :NH2 group of Asp and the acyl-adenylate would be too long for it to perform a nucleophilic attack. This mode of specificity is analogous to the Glu specificity observed for the tubulin glutamylase, Tubulin-tyrosine ligase-like 6 (TTL6) (Mahalingan et al., 2020) (Figure S4B).
We also observed density in the kinase-like active site that corresponds to ATP and two Mg2+ ions (Figure 4E). The adenine is stabilized by stacking interaction with Ty532 and the phosphates and Mg2+ ions are bound by kinase-like active site residues including Lys367 (PKA; K72), Asn534 (PKA; N171) and Asp542 (PKA; D184). Mutation of these residues reduced glutamylation activity (Black et al., 2019) (Figure 4F). Similar to canonical kinases, the ATP sits in a pocket between the two lobes of the kinase-like domain; however, the nucleotide is positioned in a unique manner. The β- and γ-phosphates of ATP are buried in a positively charged pocket, which is reminiscent of the pseudokinase SelO that binds ATP in a flipped orientation and transfers AMP from ATP to hydroxyl-containing amino acids (Sreelatha et al., 2018) (Figure 4G).
To determine the role of the kinase-like active site in SidJ, we performed adenylation reactions by monitoring 32P incorporation from [α-32P]ATP in the absence of Glu. The reactions were terminated with trichloroacetic acid (TCA) and adenylated SdeACore was detected as 32P-labelled protein in TCA-insoluble material. To account for any auto-modification of SidJ or non-specific modification of SdeA, we utilized a SdeACore E860Q mutant as a control and also added Glu to release the 32P-AMP signal from SdeACore. Although the kinase-like active site residues are required for adenylation, His492 in the migrated nucleotide binding pocket is also required (Figure 4H), likely because it stabilizes the acyl-adenylate (Figure 4A). Notably, although the putative donor Glu-interacting Arg522 within the migrated nucleotide binding pocket is required for glutamylation (Figure 4D), the R522A mutant of SidJ still retains adenylation activity towards SdeACore (Figure 4H). Importantly, the addition of Glu to the R522A mutant reaction failed to decrease the 32P-signal in TCA precipitates. These results provide evidence that Arg522 in SidJ plays a major role in positioning the donor Glu. Taken together, the kinase-like active site of SidJ adenylates the SidE active site Glu, which is stabilized by the migrated nucleotide binding pocket to position the donor Glu for attack of the acyl-adenylate intermediate and the formation of an isopeptide bond.
CaM Binding is Required for SidJ to Bind ATP and Adenylate SdeA
Several bacterial effectors and toxins are activated by eukaryote-specific proteins, such as CaM, to achieve spatial regulation (Guo et al., 2016; Guo et al., 2005; Leppla, 1984). CaM activates SidJ by binding to the IQ helix within the CTD (Black et al., 2019). However, the mechanism by which CaM activates SidJ is unknown. We monitored the thermal stability of SidJ and observed an ATP/Mg2+-dependent thermal shift that occurred only in the presence of CaM (Figure 4I). This thermal shift also occurred in the migrated active site mutant H492A, but not the kinase-like active site D542A mutant (Figure S4C–F). As expected, CaM was required for adenylation of SdeACore (Figure 4J). Thus, CaM binding renders the kinase-like active site of SidJ competent to bind ATP and adenylate the SidE-ligases.
The SidJ Paralog SdjA is an Active Glutamylating Enzyme
In L. pneumophila, the T4SS effector SdjA shares ~52% sequence identity to SidJ (Figure 5A); however, the function of SdjA is unknown. When overexpressed in yeast, SidJ, but not SdjA suppresses the growth inhibition phenotype induced by SdeA (Qiu et al., 2017a), suggesting different functions for the two effectors. Moreover, deletion of SidJ in L. pneumophila results in a growth inhibition phenotype that cannot be complemented by endogenous SdjA (Liu and Luo, 2007). Although the predicted kinase-like domains are 66% identical and the catalytic residues and CaM interacting IQ motif are well conserved, the NTDs are significantly different between SidJ and SdjA (Figure S5A and B). Because the majority of the interactions between SidJ and SdeACore lie within the SidJ NTD (Figure 2B), we asked whether SdjA could glutamylate the SidE-ligases. We incubated SdjA with CaM, ATP/Mg2+, [14C]-Glu and full-length SidE-family proteins and observed 14C incorporation into SdeB, SdeC and SidE, but not SdeA (Figure 5B). Consistent with our previous results (Black et al., 2019), SidJ glutamylated all 4 effectors. The SdjA reaction required CaM, ATP/Mg2+ and the residues in both the kinase-like active site and the migrated nucleotide binding pocket (Figure 5C and D). Similar to SidJ, SdjA glutamylated the active site Glu in SdeB, SdeC and SidE (Figure 5E). As expected, SdjA-mediated glutamylation completely abolished Ub ligase activity of SdeB, SdeC and SidE, in vitro (Figure S5C) and in cells (Figure 5F). Thus, SidJ and SdjA are CaM-dependent glutamylases that differentially inactivate the SidE-family of Ub ligases.
Figure 5. SidJ and SdjA differentially regulate the SidE-Ub ligases in vitro.
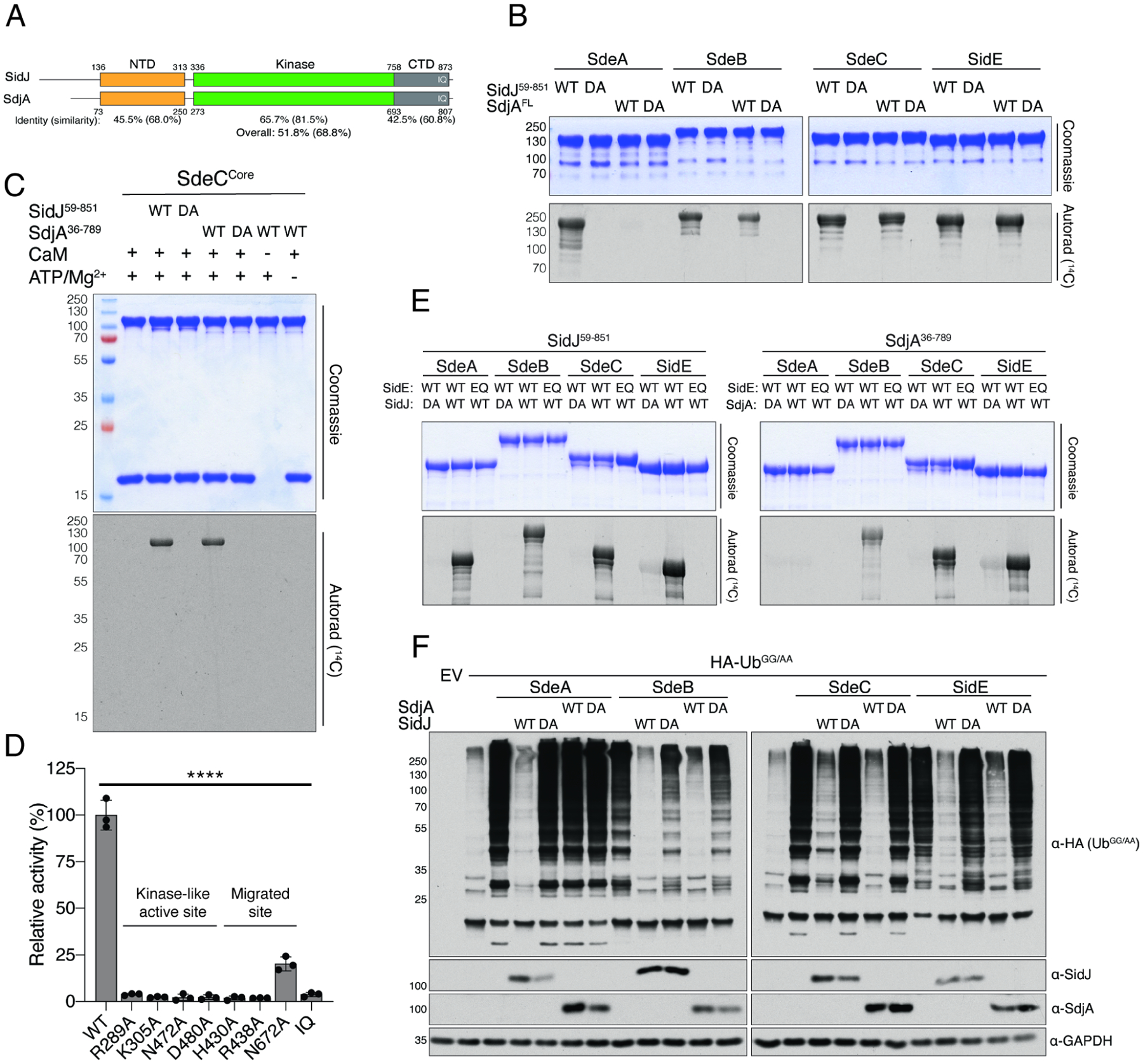
(A) Cartoon representation of SidJ and SdjA depicting the NTD (orange), the kinase-like domain (green), and the CaM binding CTD (grey). The percent sequence identity and similarity are shown for each domain and for the overall protein.
(B) Glutamylation activity of SidJ (SidJ59–851) and SdjA using full length SdeA, SdeB, SdeC and SidE as substrates. Reaction products were analyzed as in Figure 2D. DA denotes SidJD542A or SdjAD480A. Note that SdjA does not glutamylate SdeA.
(C) Glutamylation activity of SidJ59–851 and SdjA36–789 using SdeCCore as substrate. Reaction products were analyzed as in Figure 2D. DA denotes SidJD542A or SdjAD480A. (D) Glutamylation activity of SdjA36–789 and indicated mutants using SdeACore [3H]Glu as substrates. Reaction products were analyzed as in Figure 2C, n=3. The kinase-like active site residues and the residues in the migrated nucleotide binding pocket are highlighted. IQ mutant: SdjAI776D; Q777D; R778E; R781E.
(E) Glutamylation activity of SidJ59–851 and SdjA36–789 using full length SdeA, SdeB, SdeC and SidE as substrates. Reaction products were analyzed as in Figure 2D. DA denotes SidJD542A or SdjAD480A. EQ denotes SdeAE860Q, SdeBE857Q, SdeCE857Q, or SidEE855Q.
(F) Protein immunoblotting of total extracts from HEK293A cells expressing HA-UbGG/AA, Myc-SdeA, Myc-SdeB, Myc-SdeC, or Myc-SidE and SidJ-V5, V5-SdjA or the indicated mutants. GAPDH is shown as a loading control. HA-UbGG/AA was used to specifically interrogate SidE-family activity because it cannot be used by the endogenous Ub machinery. DA denotes SidJD542A or SdjAD480A.
Data in graphs shown as mean ± SD, ****P <0.0001.
The SidJ/SdjA NTDs Determine Specificity For the SidE Effectors
Because most of the interactions between SidJ/CaM with SdeA/CCore come from the NTD, we hypothesized that the NTD could be the determining factor for the differential regulation of the SidE-family by SidJ and SdjA. We generated chimeric SidJ-SdjA proteins, in which the NTD from SidJ was replaced with the NTD from SdjA, and vice versa (Figure 6A). Replacing the HTH motif in SdjA with the corresponding HTH from SidJ resulted in an active chimeric protein that retained its ability to glutamylate SdeB, SdeC and SidE but also glutamylated SdeA (Figure 6B, lane 9). A SidJ chimera containing the SdjA NTD (SidJSdjA-NTD) lost most of its activity towards SdeA (Figure 6B, lane 13). Thus, the variable NTD appears to be the major determinant of SidJ and SdjA specificity for the SidE effectors.
Figure 6. The variable NTD is the major determinant of SidJ and SdjA specificity for the SidE effectors.
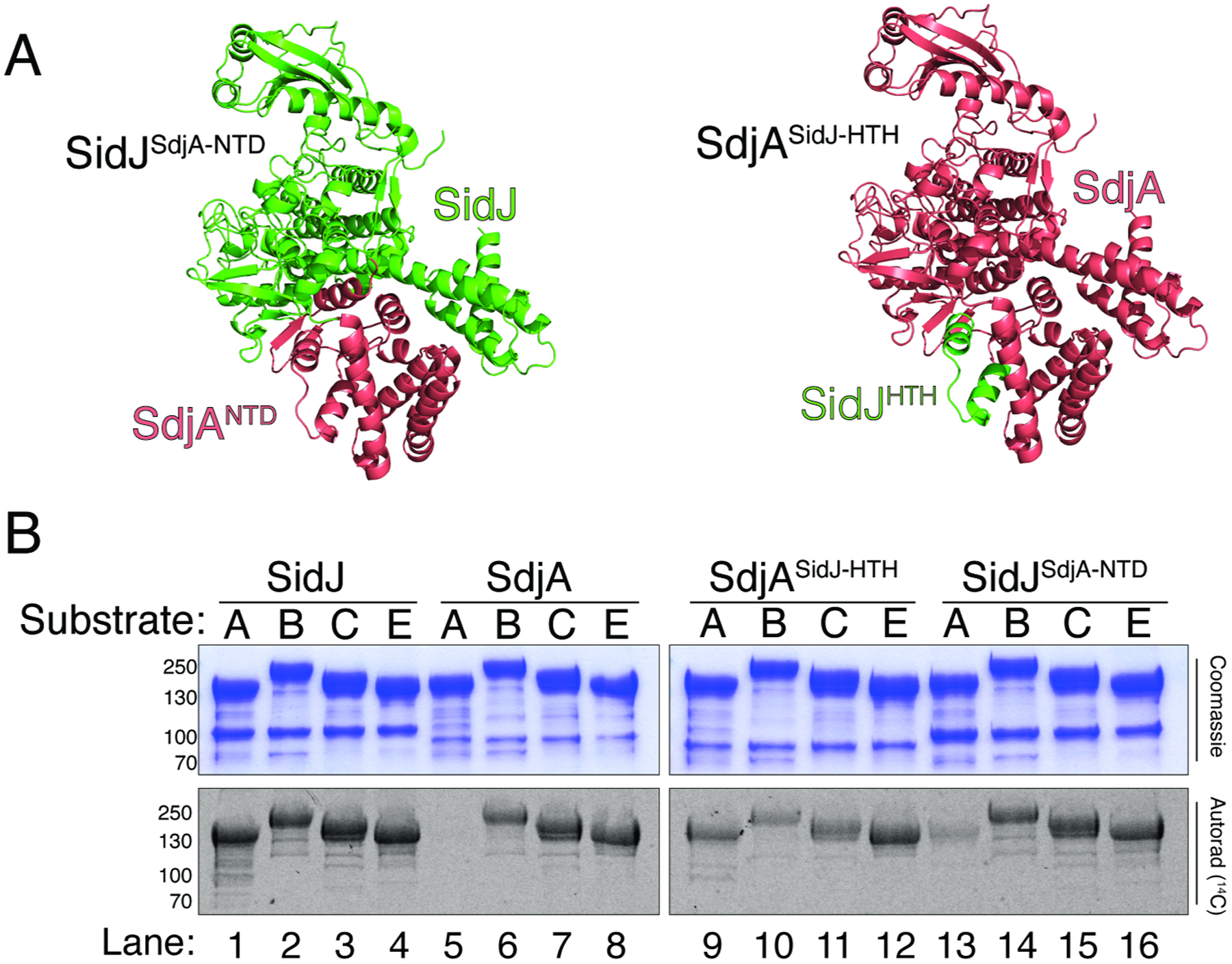
(A) Cartoon representations of SidJSdjA-NTD (left) and SdjASidJ-HTH (right) chimeras depicting the interchanged regions, presented on a crystal structure of SidJ (PDBID: 6OQQ). Portions from SidJ are in green and SdjA in salmon.
(B) Glutamylation activity of SidJ59–851, SdjA36–789, SdjASidJ-HTH and SidJSdjA-NTD using full length SdeA (A), SdeB (B), SdeC (C) and SidE (E) as substrates. Reaction products were analyzed as in Figure 2D. Compare lanes 1 and 13 (SidJ loss of activity), 5 and 9 (SdjA gain of activity).
Differential Regulation of the SidE-effectors by SidJ and SdjA During Legionella Infection.
SidJ is one of only a few T4SS effectors that when deleted from the Legionella genome causes a replication phenotype in infected cells, suggesting that SidJ and SdjA are not redundant (Jeong et al., 2015; Liu and Luo, 2007). We reasoned that in the background of a sdeA deletion, SidJ and SdjA would be functionally redundant (Figure 7A). We generated a ΔsdjAΔsidJΔsdeA Legionella strain and monitored its replication in the environmental host Acanthamoeba castellanii (Figure S6A). Remarkably, both SidJ and SdjA could complement the growth defect in the ΔsdjAΔsidJΔsdeA strain (Figure 7B). In contrast, SidJ, but not SdjA could rescue the growth defect in a ΔsdjAΔsidJ strain (Figure 7C). Collectively, our results suggest that the SidE-Ub ligases are differentially regulated by SidJ and SdjA during Legionella infection.
Figure 7. SidJ and SdjA differentially regulate the SidE-Ub ligases during Legionella infection.
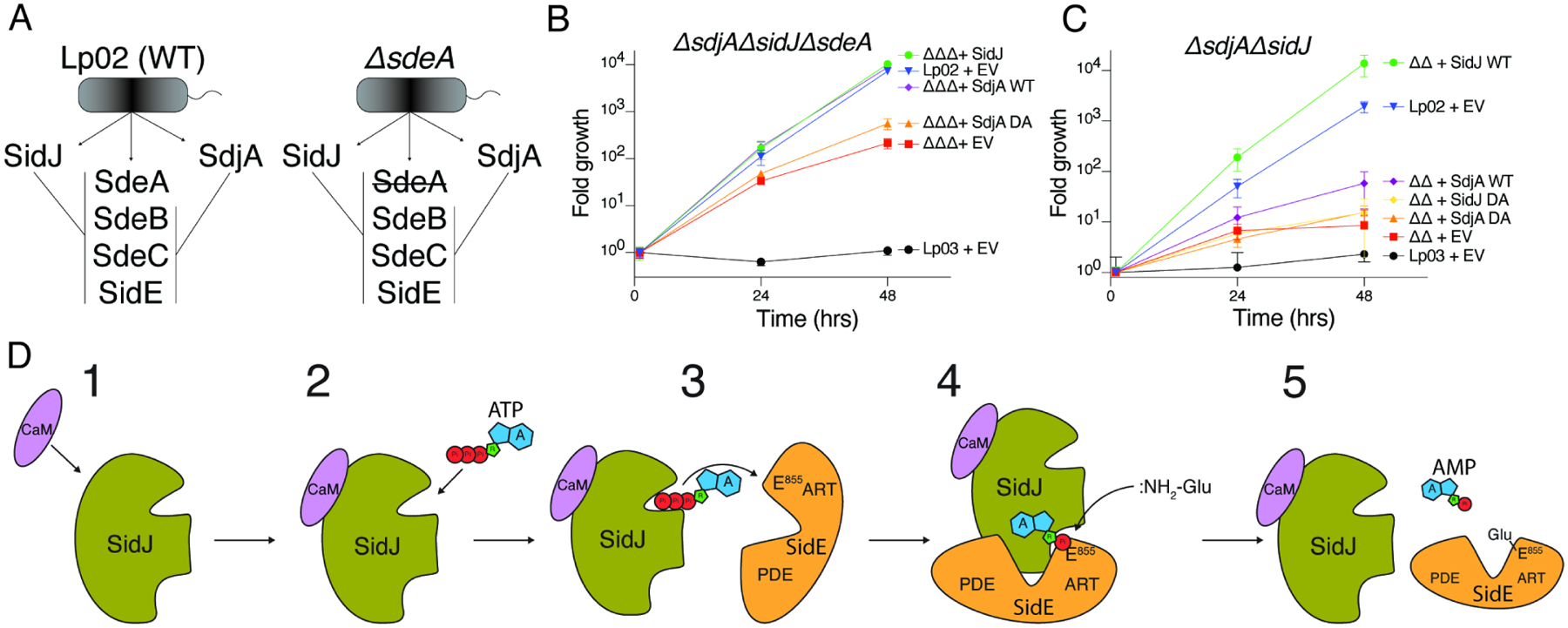
(A) Schematic representation of SidJ/SdjA genetic interaction during Legionella infection. In a wild-type L. pneumophila (Lp02) strain, SdjA is unable to complement SidJ because it is inactive against SdeA (left). In a ΔsdeA background, SdjA is functionally redundant with SidJ (right).
(B, C) Replication of L. pneumophila strains in A. castellanii. Infected amoeba cells were lysed at the indicated timepoints and bacterial replication was quantified by plating serial dilutions of lysates. Results are representative of two independent experiments; error bars denote SD from 1 experiment performed in triplicate. ΔsdjAΔsidJΔsdeA (ΔΔΔ), ΔsidJΔsdeA (ΔΔ), EV; empty vector, DA denotes SidJD542A or SdjAD480A, Lp03; ΔdotA (T4SS deficient). Error bars represent standard deviation.
(D) Model of SidJ-catalyzed glutamylation of the SidE effectors. (1) SidJ is translocated into host cells and binds CaM. (2) CaM binding renders the kinase-like active site competent to bind ATP/Mg2+ and (3) adenylate the active site Glu in the SidE effectors. (4) Adenylated SidE binds the migrated nucleotide binding pocket of SidJ, forming a stable reaction intermediate. Free Glu binds to a positively charged cleft in the migrated nucleotide binding pocket of SidJ which positions the NH2 group for nucleophilic attack of the acyl-adenylate, (5) releasing AMP and forming an isopeptide bond between Glu and SidE.
Discussion
We propose a model for how SidJ glutamylates the SidE-family of Ub ligases (Figure 7D). During infection, SidJ is translocated into the host cell where it binds CaM (1), which allows the kinase-like active site to bind ATP/Mg2+ (2). Notably, because CaM is a eukaryote-specific protein, this mechanism allows for spatial regulation of the SidE-Ub ligases within the host cell and prevents premature inactivation of the SidE-Ub ligases within Legionella. SidJ then adenylates the active site Glu within the ARTT loop in the SidE effectors (3). Adenylated SidE binds the migrated nucleotide binding pocket (4), which positions free Glu near the acyl-adenylate intermediate for nucleophilic attack and subsequent formation of the Glu-Glu isopeptide bond (5).
Our structural and biochemical data suggest that the kinase-like active site performs the adenylation reaction and the migrated nucleotide binding pocket executes the glutamylation reaction. Although His492 in the migrated nucleotide binding pocket is required for adenylation (Figure 4H), the H492A mutant would be unable to stabilize the intermediate and therefore prevent us from detecting the acyl-adenylate in our assays. Consistent with this idea, Arg522 does not contact the acyl-adenylate and thus is not required for adenylation. Three pieces of evidence lead us to believe that Arg522 is required for Glu binding: 1) Arg522 is highly conserved (Figure S6B). 2) Arg522 is required for glutamylation but not adenylation of SidE (Figure 4D and H) and 3) the addition of Glu to adenylation reactions using the SidJ R522A mutant does not liberate AMP from adenylated SidE (Figure 4H).
SidJ homologs are found in taxonomically diverse groups of organisms including archaea and viruses, suggesting that SidJ is being spread by horizontal gene transfer (Figure S6C). Distant SidJ homologs retain strong conservation of residues in both the kinase-like active site and the migrated nucleotide binding pocket (Figure S6B). However, none of these organisms except for Legionella species have SidE homologs or the SidJ CaM-binding IQ motif and they show virtually no conservation within the SidE-interacting NTD (Figure S6B). Thus, their substrates and activation mechanisms will likely differ. Interestingly, SidJ homologs from crocodilepox viruses are located next to a cluster of proteins that differ from SidE but are involved in the modulation of host ubiquitination pathways (Afonso et al., 2006).
In mammals, tubulin glutamylation, tyrosination and glycylation are performed by TTLL enzymes, members of the ATP-grasp superfamily (Song and Brady, 2015; Yu et al., 2015). TTLLs catalyze amino acid ligation via the formation of a high-energy acyl-phosphate intermediate, which is subsequently attacked by the amine group of the donor amino acid (Szyk et al., 2011). Interestingly, both the protein kinase and ATP-grasp fold enzymes share a common topology (Grishin, 1999). In contrast to most protein kinases, SidJ adenylates a carboxyl containing amino acid as an intermediate step in peptide bond formation. We propose that the protein kinase fold can phosphorylate or adenylate carboxylic groups to mediate non-ribosomal amino-acid ligation reactions. Notably, a recent study of “the hidden phosphoproteome” revealed that a vast number of Glu and Asp residues within HeLa cells are phosphorylated (Hardman et al., 2019).
In summary, our work underscores the catalytic versatility of the kinase fold and reveals a previously unappreciated level of regulation of the SidE Ub ligases.
Limitations of the Study
Our biochemical and mutagenesis data suggests that the kinase-like active site performs the adenylation reaction and the migrated nucleotide binding pocket catalyzes the glutamylation reaction. The structures presented here have captured the reaction intermediate complex that facilitates acyl-adenylate stabilization and subsequent glutamylation. However, we lack structural information on the first step of the reaction: adenylation of the SidE active site Glu by the kinase-like active site in SidJ. Likewise, the structural rearrangements that need to occur following the adenylation reaction are unknown at this time.
Furthermore, our discovery that SidJ and SdjA differentially regulate the SidE effectors suggests that SdeB, SdeC and SidE require an additional layer of regulation and may have distinct substrate specificities. Thus, the reason why L. pneumophila has evolved two glutamylases to differentially inactivate the SidE effectors is unknown and warrants further investigation.
STAR METHODS
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vincent S. Tagliabracci (vincent.tagliabracci@utsouthwestern.edu).
Materials Availability
Plasmids, primers, recombinant proteins, experimental strains, and any other research reagents generated by the authors will be distributed upon request to other research investigators under a Material Transfer Agreement.
Data and Code Availability
Cryo-EM maps and final refined coordinates have been deposited in EMDB (The Electron Microscopy Data Bank) and PDB (Protein Data Bank), respectively, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw data for protein immunoblots, SDS-PAGE, and autoradiography have been deposited at Mendeley Data (https://dx.doi.org/10.17632/sgnknvxz3j.1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SidJ | (Black et al., 2019) | N/A |
| Rabbit anti-SdjA | This study | N/A |
| Rabbit anti-SdeA | (Black et al., 2019) | N/A |
| Mouse anti-GAPDH | ThermoFisher | Cat#MA5-15738 |
| Mouse anti-HA antibody | Sigma-Aldrich | Cat# H3663 |
| Donkey anti-Mouse IgG, HRP-Linked Whole Ab | VWR | Cat#95017-554 |
| Donkey anti-Rabbit IgG, HRP-Linked Whole Ab | VWR | Cat# 95017-556 |
| Bacterial Strains | ||
| Rosetta DE3 | Novagen | Cat#70954 |
| Rosetta DH5α | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L. pneumophila SidJ 59-851 (E. coli) | This study | N/A |
| L. pneumophila SidJ 97-851 (E. coli) | This study | N/A |
| L. pneumophila SdjA (E. coli) | This study | N/A |
| L. pneumophila SdjA 36-789 (E. coli) | This study | N/A |
| L. pneumophila SdeA (E. coli) | This study | N/A |
| L. pneumophila SdeB (E. coli) | This study | N/A |
| L. pneumophila SdeC (E. coli) | This study | N/A |
| L. pneumophila SidE (E. coli) | This study | N/A |
| L. pneumophila SdeA 231-1190 (E. coli) | This study | N/A |
| L. pneumophila SdeC 231-1222 (E. coli) | This study | N/A |
| Human CALM2 (E. coli) | This study | N/A |
| Human HA-Ubb (E. coli) | This study | N/A |
| Ulp1 S. cerevisiae SUMO protease | This study | N/A |
| PfuTurbo DNA Polymerase | Thermo | Cat# 50-125-946 |
| Q5 High-Fidelity 2X Master Mix | NEB | M0492L |
| Adenosine 5’-triphosphate disodium salt hydrate | Sigma | Cat# A2383 |
| ATP, [α-32P]- 3000Ci/mmol 10mCi/ml, 250 μCi | PerkinElmer | BLU003H250UC |
| L-[14C(U)]-Glutamic Acid | PerkinElmer | NEC290E050UC |
| L-[3,4-3H]-Glutamic Acid | PerkinElmer | NET490001MC |
| Budget-Solve scintillation cocktail | RPI | Cat# 111167 |
| EN3HANCE Autoradiography Enhancer | PerkinElmer | Cat# 6NE9701 |
| Tris-(2-Carboxyethyl)phosphine, Hydrochloride (TCEP) | Fisher Scientific | Cat# PI20490 |
| Hydrogen peroxide | Sigma | Cat# H1009 |
| L-glutamic acid, Sodium salt | Sigma | Cat# G1626 |
| Sodium Chloride | Fisher Scientific | BP358-10 |
| Tris base | Fisher Scientific | BP152-5 |
| Kanamycin sulfate | Millipore Sigma | K1377 |
| Ampicillin sodium salt | Millipore Sigma | A9518 |
| Chloramphenicol | Millipore Sigma | C0378 |
| PolyJet In Vitro DNA Transfection Reagent | SignaGen Laboratories | SL100688 |
| Protease Inhibitor Cocktail Tablets (Complete) | Millipore Sigma | 11697498001 |
| Dithiothreitol (DTT) | Millipore Sigma | D0632 |
| DMEM, High Glucose | Thermo Fisher Scientific | 11-965-118 |
| Fetal bovine serum | Millipore Sigma | F2442 |
| Penicillin-Streptomycin | Millipore Sigma | P0781 |
| Magnesium Chloride hexahydrate | Millipore Sigma | M2670 |
| Deposited Data | ||
| SidJ:CALM2:SdeA | This study | EMDB: EMD-23862 PDB: 7MIR |
| SidJ:CALM2:SdeC | This study | EMDB: EMD-23863 PDB: 7MIS |
| Experimental Models: Organisms/Strains | ||
| Legionella pneumophila Philadelphia-1 (Lp02) | Dr. Ralph Isberg | N/A |
| Legionella pneumophila Philadelphia-1 ΔdotA (Lp03) | Dr. Ralph Isberg | N/A |
| Lp02 ΔsidJ | (Black et al., 2019) | N/A |
| Lp02 ΔsdjA | This study | N/A |
| Lp02 ΔsdeA | This study | N/A |
| Lp02 ΔsidJΔsdjA | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sidJ) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sidJ D542A) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sdjA) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sdjA D480A) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sidJ) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sidJ D542A) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sdjA) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sdjA D480A) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908) | This study | N/A |
| HEK293a | Invitrogen | R70507 |
| Acanthamoeba castellanii | Dr. Kim Orth | N/A |
| Oligonucleotides | ||
| See Table S1 for primers used in this study | ||
| Recombinant DNA | ||
| ppSUMO L. pneumophila SidJ 59-851 | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SidJ 97-851 | This study | N/A |
| ppSUMO L. pneumophila SdjA 36-789 | This study | N/A |
| ppSUMO CALM2 | (Black et al., 2019) | |
| ppSUMO L. pneumophila SdeA | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SdeB | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SdeC | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SidE | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SdeA 231-1190 | This study | N/A |
| ppSUMO L. pneumophila SdeC 231-1222 | This study | N/A |
| ppSUMO L. pneumophila SidJSdjA-NTD | This study | N/A |
| ppSUMO L. pneumophila SdjASidJ-HTH | This study | N/A |
| pETDuet1 L. pneumophila SdjA CALM2 | This study | N/A |
| pProEx2 L. pneumophila SdeA 231-1190 | This study | N/A |
| pProEx2 L. pneumophila SdeC 231-1222 | This study | N/A |
| pSR47s | Dr. Shaeri Mukherjee | N/A |
| pSR47s sdeA flanks | This study | N/A |
| pSR47s sdjA flanks | This study | N/A |
| pSR47s sidJ flanks | (Black et al., 2019) | N/A |
| pJB908 sidJ | (Black et al., 2019) | N/A |
| pJB908 sidJ D542A | (Black et al., 2019) | N/A |
| pJB908 SdjA | This study | N/A |
| pJB908 SdjA D480A | This study | N/A |
| pJB908 | Dr. Ralph Isberg | N/A |
| pcDNA-HA-Ubb | Dr. Jenna Jewell | N/A |
| pcDNA-HA-Ubb GG/AA | (Black et al., 2019) | N/A |
| pcDNA-SidJ-V5 | (Black et al., 2019) | N/A |
| pcDNA-SidJ-V5 D542A | (Black et al., 2019) | N/A |
| pCCF-V5-SdjA | This study | N/A |
| pCCF-V5-SdjA D480A | This study | N/A |
| pCDNA-Myc-SdeA (c. opt) | (Black et al., 2019) | N/A |
| pCCF-Myc-SdeB | (Black et al., 2019) | N/A |
| pCCF-Myc-SdeC (c. opt) | (Black et al., 2019) | N/A |
| pCCF-Myc-SidE | (Black et al., 2019) | N/A |
| Software and Algorithms | ||
| Pymol | Schrödinger, Inc. | N/A |
| WebLogo server | (Crooks et al., 2004) | http://weblogo.threeplusone.com/ |
| Dalilite server | (Holm and Rosenstrom, 2010) | http://ekhidna2.biocenter.helsinki.fi/dali/ |
| Jackhmmer server | (Finn et al., 2015) | https://www.ebi.ac.uk/Tools/hmmer/search/jackhmmer |
| BLAST server | (Altschul et al., 1990) | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| MAFFT server | (Katoh and Standley, 2013) | https://mafft.cbrc.jp/alignment/software/ |
| Pfam database | (Finn et al., 2016) | http://pfam.xfam.org/ |
| RELION | (Zivanov et al., 2018) | https://relion.readthedocs.io/en/latest/index.html |
| crYOLO | (Wagner et al., 2019) | https://cryolo.readthedocs.io/en/stable/ |
| Gctf | (Zhang, 2016) | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| DeepEMhancer | (Sanchez-Garcia et al., 2020) | https://github.com/rsanchezgarc/deepEMhancer |
| Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| Phenix | (Adams et al., 2010) | https://www.phenix-online.org |
| Coot | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| CCP4 7.1 | (Winn et al., 2011) | https://www.ccp4.ac.uk/?page_id=1417 |
| GraphPad Prism | N/A | https://www.graphpad.com/scientific-software/prism/ |
| ESPript | (Robert and Gouet, 2014) | http://espript.ibcp.frESPript/ESPript/ |
Experimental Model and Subject Details
Legionella pneumophila
Legionella pneumophila subsp. pneumophila strain Philadelphia 1 derived strain Lp02 and Lp03 and mutants were cultured at 37°C in AYE liquid medium, or on CYA solid medium, prepared according to ATCC: Medium 1099 CYE, i.e. ACES [N-(2-acetamido)-2- aminoethanesulfonic acid]-buffered charcoal yeast extract (CYE) agar plates or grown in ACES buffered yeast extract (AYE) liquid cultures supplemented immediately before use with 100x concentrated sterile filtered solutions of ferric nitrate (final 0.135 g/L) and cysteine (final 0.4 g/L). Thymidine was added to a final concentration of 100 μg/mL for maintenance of the thymidine auxotrophic strains. Additional supplements used were used where applicable: kanamycin (30–50 μg/mL), sucrose (5%). Protein knockouts were confirmed by PCR and immunoblotting.
Acanthamoeba castellanii
Acanthamoeba castellanii was maintained as a monolayer culture in ATCC 712 PYG medium (20 g/L protease peptone, 1 g/L yeast extract, 100 mM glucose, 4 mM Mg2SO4, 0.4 mM CaCl2, 0.1% (w/v) sodium citrate dihydrate, 0.05 mM Fe(NH4)2(SO4)2·6H2O, 2.5 mM NaH2PO4, 2.5 mM K2HPO4, final pH adjusted to 6.5) in tissue culture flasks at 23°C.
HEK293A
HEK293A cells were grown in DMEM containing 10% (vol/vol) FBS with 100 μg/mL penicillin/streptomycin (GIBCO) at 37 °C with 5% CO2. Cells were routinely tested for Mycoplasma contamination by PCR based methods.
Method Details
Antibodies
Mouse anti-GAPDH antibodies (CB1001) were from EMD Millipore. Mouse anti-HA antibodies were from Sigma (H3663) and anti-Legionella pneumophila antibodies (ab20943) were from Abcam. Rabbit anti-SdeA, SidJ and SdjA were raised by Cocalico and purified in-house as previously described in (Sreelatha et al., 2018), and in “Production of SdjA Antibodies” section.
Generation of Plasmids and Strains
SidJ, SdjA, SdeA, SdeB, SdeC and SidE coding sequences (CDS) were amplified by PCR using Legionella pneumophila Philadelphia-1 strain genomic DNA (gDNA) as a template. SidJ, SdjA, SdeA, SdeC CDS were also codon-optimized for mammalian expression and synthesized as gBlocks (SidJ, SdjA, SdeC; Integrative DNA Technologies, SdeA; Genscript). CALM2 and FcγRIIa were amplified from the Ultimate™ ORF Lite human cDNA collection (Life Technologies) and cloned into mammalian or bacterial expression vectors. Amino acid mutations were introduced via Quick Change site-directed mutagenesis. Briefly, primers were designed manually, or using the Agilent Quick Change Primer design tool: https://www.genomics.agilent.com and used in PCR reactions to generate the desired mutation using PfuTurbo DNA polymerase. Reaction products were digested with Dpn1 restriction endonuclease and mutations were confirmed by Sanger sequencing. For mammalian cell expression in HEK293A cells, codon-optimized SidJ, SdjA, SdeA and SdeC were amplified from gBlocks by PCR and cloned in frame with a C-terminal V5 tag (SidJ), N-terminal V5 tag (SdjA) or amplified with an N-terminal Myc tag (SdeA, SdeC) into the CMV promoter-driven vector pcDNA (Invitrogen). pcDNA-HA-tagged Ubiquitin B (UbB) was a generous gift from Dr. Jenna Jewell and used as a template to generate the UbGG/AA mutant.
L. pneumophila strains Lp02, Lp03 (Lp02 ΔdotA), and thymidine auxotrophic derivatives used in this study were derived from Legionella pneumophila Philadelphia-1 strain and were generous gifts from Dr. Ralph Isberg. Legionella bacteria were maintained on ACES [N-(2-acetamido)-2- aminoethanesulfonic acid]-buffered charcoal yeast extract (CYE) agar plates or grown in ACES buffered yeast extract (AYE) liquid cultures supplemented with ferric nitrate (0.135 g/L) and cysteine (0.4 g/L). Thymidine was added to a final concentration of 100 μg/mL for maintenance of the thymidine auxotrophic strains.
SidJ, SdjA and SdeA knockout strains were generated using the R6K suicide vector pSR47s (KanR, sacB), a generous gift from Dr. Shaeri Mukherjee, UCSF. Briefly, ~800bp regions flanking the SidJ ORF were amplified and cloned using Gibson assembly into pSR47s to generate pSR47s-ΔsdeA, pSR47s-ΔsidJ, and pSR47s-ΔsdjA, which was maintained in S17-1 λpir E. coli. pSR47s vectors with flanks were introduced by electroporation into Thy− strain Lp02, or ΔsidJ, or ΔsidJ/ΔsdeA, and colonies having undergone homologous recombination were selected with kanamycin (30 μg/mL). Bacteria were streaked on CYA Thy+ plate. Merodiploids were resolved on 5% sucrose, and the resulting colonies were screened for loss of respective genes by PCR and protein immunoblotting. Complementing strains were generated using the RSF1010 cloning vector pJB908 (AmpR tdΔi), a gift from Dr. Ralph Isberg. Mutants were cloned into pJB908 and transformed by electroporation into the appropriate parental strain. Transformants were selected for on CYE medium without thymidine and complementation was verified by PCR and protein immunoblotting. For intracellular replication assays and HEK293 cell challenge, Legionella were scraped from a 2/3-day heavy patch to inoculate 2 mL AYE broth. Serial dilutions of the inoculum were grown 24–48 hours at 37°C with shaking to an OD600 of 2.5 to 4, at which time the cultures acquired a brown pigmentation.
Intracellular replication in amoeba
18 hours prior to infection, confluent amoeba monolayers were washed with A. castellani buffer ACB (4 mM Mg2SO4, 0.4 mM CaCl2, 0.1% (w/v) sodium citrate dihydrate, 0.05 mM Fe(NH4)2 (SO4)2·6H2O, 2.5 mM NaH2PO4, 2.5 mM K2HPO4, final pH adjusted to 6.5), to remove non-adherent cells, and collected by scraping. Resuspended in fresh ACB, amoeba were counted, and 6×105 cells were seeded into individual wells of 24-well plates 1–2 h prior to infection. Plates were equilibrated to reach 37°C. All subsequent incubations were performed at 37°C. Legionella cultures at post-exponential phase were diluted in A. castellanii buffer and ~6×104 bacteria were added to each well for a multiplicity of infection (MOI) of 0.1 (assuming 1600OD=109 Legionella CFU). Infections were synchronized by centrifugation at 230 × g for 5 minutes. Infections were allowed to proceed for 1 hour, then extracellular bacteria were removed by washing each well 3 times in ACB, before adding ACB buffer to a final volume of 0.5 mL/well. Timepoint zero (1 HPI) was lysed immediately after washing, and subsequent timepoints were taken after 24h, and 48h. Amoeba cells were lysed by addition of saponin to a final concentration of 0.05%, and scraped off the plate with a mini cell scraper. Wells were subsequently washed with 500 μL of Milli-Q H2O, and combined with saponin lysates. Serial dilutions of the infectious inoculum and the amoeba lysates were plated on CYE plates to confirm the MOI, quantify infection efficiency, and assess bacterial growth. All solutions used during the infection were filtered using sterile 0.22 μm syringe filters.
Protein purification
L. pneumophila SidJ residues 59–851 or 97–851, SdjA FL and 36–789, human CaM (CALM2), and the SidE effectors (and indicated truncations) were cloned into a modified pET28a bacterial expression vector (ppSumo), containing an N-terminal 6X-His tag followed by the yeast Sumo (smt3) CDS. Ubiquitin, CaM, SdeA 231–1190 and SdeC 231–1222 were also cloned into pProEX2 containing an N-terminal 6X-His tag followed by a TEV protease cleavage site. For purification from E. coli, plasmids were transformed into Rosetta DE3 cells. Cells were grown in Miller modification of Luria Bertani (Miller LB) broth with 100 mg/L Ampicillin or 50 mg/L Kanamycin, in presence of 34 mg/L of Chloramphenicol. At the OD600 of 0.6–1.0, protein expression was induced with 0.4 mM IPTG for 16–18 hours at 18 °C. Cells were harvested by centrifugation and lysed in 50 mM Tris-HCl pH 8, 300 mM NaCl, 15 mM Imidazole, 1 mM PMSF, (containing 5 mM β-ME) by sonication. Cell lysates were cleared by centrifugation at 35,000 × g for 30 minutes. The cleared lysate was incubated with washed Ni-NTA beads for a minimum of one hour at 4°C. Beads were passed over a gravity column and washed with 20 column volumes of 50 mM Tris-HCl pH 8, 300 mM NaCl, 25 mM imidazole, 5 mM β-ME. Proteins were eluted with 50 mM Tris-HCl pH 8, 300 mM NaCl, 300 mM imidazole, 1 mM DTT. SdjA purifications were performed in 500 mM NaCl to aid stability. Full-length SdjA cloned into ppSumo was coexpressed with human 6X-His-CALM2 in pProEX2 and grown under triple selection (Kan, Amp, Cm), and was purified in one step (Ni-NTA). Proteins were cut overnight at 4 °C with 6X-His tagged Ulp1 Sumo protease or TEV protease, followed by gel filtration chromatography using a Superdex 200 gel filtration column attached to an AKTA Pure FPLC chromatography system (GE Healthcare) in 25 mM Tris 7.5 or 8.0, 150 mM or 300 mM of NaCl, 1 mM DTT. Proteins were concentrated (~15–30 mg/mL SidJ, ~30 mg/mL CALM2, ~30 mg/mL SdeACore, 60 mg/mL SdeCCore, ~10 mg/mL full-length SidE effectors), aliquoted, flash frozen in liquid N2 and stored at −80 °C until use.
Size-exclusion chromatography for SidJ:CaM:SdeC and SdeA complexes
For preparation of the reaction intermediate complex sample, tag-less proteins were preincubated for 15 minutes at 37 °C: 2.5 mg SidJ 97–851, 0.83 mg CALM2 and 2.5 mg SdeA/CCore. Conditions within the preincubation were: 50 mM Tris 7.5, 50–75 mM NaCl, 10 mM MgCl2, 4 mM ATP, 1 mM DTT, 300 μL. The reaction was spun at 22,000 × g for 10 minutes at 4 °C, diluted to 400 μL with size-exclusion buffer (25 mM Bis-tris pH 6.5, 100 mM NaCl, 1 mM TCEP, 2 mM MgCl2, 1 mM ATP), and subjected to SEC on a Superdex S200 increase 10/300 column.
For SEC experiments, the same conditions were used, with less protein: 0.5 mg of each SidJ and SdeC and + 0.17 mg CALM2. ATP was omitted from the SEC buffer in SdeC samples. Preincubation was supplemented with 5 mM L-Glu for experiment with Glu treatment. Tris pH 7.5 was used for basic condition size-exclusion experiment shown in Figure 1C.
Cryo-EM grid preparation
The reaction intermediate complex sample was prepared as above, in size-exclusion buffer. All proteins were tag-less. Concentration of proteins within the complex fractions were assessed according to Bradford reagent (Bio-rad protein assay), and were ~1–1.5 mg/mL. The peak fraction was diluted to 0.35 mg/mL using size-exclusion buffer, and spun at 22,000 × g for 5–10 minutes at 4 °C. The sample used for grid preparation was never frozen and used within 1 hour after preparation. Grids were prepared using a FEI Vitrobot mark IV (ThermoFisher Scientific) at 4 °C with 95% relative humidity and plunged into liquid ethane. 3.5 μL of sample was applied to a Quantifoil 300-mesh R1.2/1.3 grid (Quantifoil, EMS), which was glow discharged using Pelco EasiGlo. The sample was preincubated with grid for 10 seconds before blotting.
Cryo-EM Data collection and processing
Prior to data collection, sample grids were screened on a Talos Artica microscope, and a pilot dataset of SidJ-SdeC-CaM was collected on a Titan Krios microscope at the Cryo Electron Microscopy Facility at UT Southwestern. Datasets of both SidJ-SdeA-CaM and SidJ-SdeC-CaM complexes used for final data processing were collected at the Pacific Northwest Cryo-EM Center (PNCC) on a Titan Krios microscope operating at 300 kV, with the post-column energy filter (Gatan) and a K3 direct detection camera (Gatan), using SerialEM(Mastronarde, 2005). Movies were acquired at a pixel size of 0.54 Å; in super-resolution counting mode, with an accumulated total dose of 50 e-/Å2 over 78 frames. The defocus range of the images was set to be −0.8 to −2.5 μm. 11,694 and 9,409 movies were collected for SidJ-SdeA-CaM complex and SidJ-SdeC-CaM complex, respectively.
Image processing and 3D reconstruction
Unless described otherwise, all datasets were processed with Relion (Scheres, 2012). For the pilot dataset, the motion corrected micrographs from Relion were preprocessed with MicAssess (Li et al., 2020). Particles were picked with crYOLO (Wagner et al., 2019). A 4.5 Å map of SidJ:CaM:SdeC was obtained from the pilot dataset. This map was low pass filtered and used as the initial model for data processing as described below. For both datasets collected at PNCC, movies were aligned and summed with MotionCor2 (Zheng et al., 2017), with a downsampled pixel size of 1.08 Å. MicAssess (Li et al., 2020) was used to preprocess all motion corrected images. The CTF parameters were calculated using Gctf (Zhang, 2016), and images with estimated CTF max resolution better than 5 Å were selected for further processing. For the SidJ:CaM:SdeA dataset, 3,193,233 particles were picked using crYOLO (Wagner et al., 2019) from 9,839 images. 2D and 3D classifications identified two subclasses, which were combined with 386,460 particles. After further 3D refinement, CTF refinement and particle polishing, an additional step of alignment-free 3D classification with eight classes was performed. 310,154 particles from three subclasses with more complete maps for SdeA were combined to calculate the final map at 2.5 Å resolution. For the SidJ:CaM:SdeC dataset, 4,454,035 particles were picked using crYOLO5 from 8,432 images. 212,768 particles were selected after 2D and 3D classifications for subsequent 3D refinement, CTF refinement and particle polishing, followed by alignment-free 3D classification. 152,589 particles from the two best classes were used to calculate the final map at 2.8 Å resolution. Map resolution values were calculated by Relion with the gold standard FSC method.
Model building and refinement
For initial model building, the existing crystal structure of SdeA (PDBID: 5YIM), and cryo-EM structure of SidJ:CALM2 (6S5T) were fit into experimental density of SidJ:CaM:SdeA using Chimera. Models were manually adjusted in Coot to account for main chain rearrangements, compared to the crystal structure. Fit was further enhanced by rigid fit and morph routines in Phenix real space refine. Resultant model was split into SidJ:CaM and SdeA, and fit into SidJ:CaM:SdeC map using chimera. Sequence of SdeA was mutated in coot, to match SdeC sequence. Rearranged areas were manually adjusted. AceDRG from CCP4 7.1 suite (Winn et al., 2011) was used to generate AMPGlu link restraints. DeepEMhancer (Sanchez-Garcia et al., 2020) was used to generate post-processed maps, with the two unfiltered half maps from Relion, for further manual model building and inspection in Coot(Emsley et al., 2010). The models were then subjected to real space refinement in Phenix(Adams et al., 2010), with secondary structure restraints and non-crystallographic symmetry restraints. Model geometries were assessed with MolProbity(Chen et al., 2010) as a part of the Phenix validation tools (Table 1). Figures were rendered in PyMOL (The PyMOL Molecular Graphics System, Schrödinger), Coot or Chimera (Pettersen et al., 2004).
Production of SdjA antibodies
L. pneumophila SdjA 36–789 D480A was purified as above as a 6X-His-SUMO fusion and was cleaved off of Ni-NTA resin with Ulp1 protease (on-column cleavage). The protein was further purified by Superdex 200 16/600 gel filtration chromatography and used to inoculate rabbits for generation of rabbit anti-SdjA anti-serum (Cocalico Biologicals). Total IgG was partially purified by ammonium sulfate precipitation as described (Kent, 1999). In short, 1 mL NHS-activated HP columns (Cytiva, 17071601) were washed with 1 mM HCl, and crosslinked with the antigen by repeatedly passing 2 mL of 5 mg/mL protein in coupling buffer (0.2 M NaHCO3, 0.5 M NaCl, pH 8.3) for 30 minutes, at ~1 ml/min. 15 mL of rabbit serum was centrifugated at 10,000 × g for 30 minutes. To 15 mL of serum, 35 mL of ice cold, saturated Ammonium Sulfate solution in BBS (Borate Buffered Saline) was added to precipitate the antibodies for 2 hrs at 4°C (400 g of Ammonium Sulfate dissolved in 500 mL of hot BBS; BBS: 0.2 M Sodium Borate, 160 mM NaCl, pH 8.0). The precipitate was spun at 10,000 × g for 30 min, and resuspended in 15 mL of BBS, and spun for 10 minutes at 10,000 × g to remove insoluble particles. Antibodies from solution were affinity-purified by passing it through a Hi Trap NHS-activated HP column, crosslinked with WT untagged SdjA 36–789, or SdeA 519–1100 (SdeAART domain).
Antibodies were eluted with 9 mL of 100 mM Glycine pH 2.5, into a tube with 1 mL of 1 M Tris 8.0 for neutralization, concentrated to ~1 mg/mL, aliquoted and stored at −20°C until use.
Glutamylation assays
Glutamylation reactions were performed in a reaction mixture containing 50 mM Tris-HCl pH 7.5, 50 or 75 mM NaCl, 1 mM DTT, 1 mM MgCl2, 0.3 mM ATP, and 100 μM [U-14C] or [3H] L-Glu (specific radioactivity 200 cpm/pmol for 14C, 1500 cpm/pmol for 3H). For standard endpoint reactions, a typical 20 μL reaction contained 10 μg SidE effector, 1 ug SidJ/SdjA, and 2 μg CaM. Reactions were initiated by adding ATP/Mg2+, Glu and CaM, and were allowed to proceed at 37 °C for 60 minutes, after which they were terminated by addition of 7 uL of STOP mix (0.5 M EDTA, 5X SDS-PAGE loading dye; 2:5 ratio). Reaction products were resolved by SDS-PAGE and visualized by Coomassie staining. Gels were then soaked in EN3HANCE reagent for 30 minutes, followed by a 30-minute soak in H2O, then dried. 14C incorporation was detected by autoradiography.
For kinetic assays, 0.1 ug of SidJ was incubated with 10 ug SdeAFL for 15 minutes at room temperature (For SdjA, 0.6 ug of protein was incubated for 20 min with 5 ug of SdeCCore). 100 μM [3H] L-Glu (specific activity of 1500 cpm/pmol). Triplicate 20 uL reactions were incubated for 15 min at room temperature and terminated as above. After SDS-PAGE separation, Coomassie stained SidE bands were excised, placed in glass scintillation vials and digested in 1 mL of 30% H2O2 at 60 – 75 °C overnight. After digestion was complete, vials were cooled down to ambient temperature, mixed with 12 mL of Budget-Solve scintillation cocktail (RPI, 111167) and 3H incorporation quantified by scintillation counting.
Adenylation assays
SdeA adenylation end-point assays using [α32P]ATP were performed in a 20 uL reaction volume for 1 h at room temperature using 15 μg SidJ, 15 μg SdeA and 4.5 μg CALM2. Proteins were prediluted in dilution buffer (50 mM Bis-tris 6.5, 50 mM NaCl, 1 mM DTT). Reactions were initiated with ATP/Mg2+. The final reaction conditions were: 50 mM Bis-tris pH 6.5, 50 mM NaCl, 1 mM DTT, 150 μM [α32P]ATP (SA: 1000 cpm/pmol), 1 mM MgCl2, 1 mg/mL fatty acid-free BSA, −/+ 1 mM Glu. Reactions were stopped by addition of 500 μL of 30 mM ATP in 20% Trichloroacetic acid (TCA). Reactions were incubated on ice for 40 minutes, spun at 20,000 × g for 10 minutes at 4 °C and washed twice with 250 μL of 20% TCA solution. Radioactivity in the pellets was quantified by scintillation counting.
Differential Scanning Fluorimetry/thermal shift assay
Protein thermal stability in solution was monitored by measurement of fluorescence of SYPRO Orange dye (Thermo). Triplicate 25 μL reactions contained 5.5 μM SidJ59–851 and 5.5 μM CALM2. Reaction conditions were: 10 mM Tris 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM ATP and 625x dilution of SYPRO Orange dye (Invitrogen, S6650).
Reactions were performed in domed PCR tubes in a BioRad CFX96 thermocycler. Samples were subjected to a gradient of temperature 25–85 °C at a rate of +1 °C/minute. Fluorescent signal was read using FRET mode of the thermocycler. Melting curves were normalized within each replicate, averaged and plotted in Prism 9. The temperature at the derivative peak as reported by BioRad CFX manager, was used as the melting temperature.
SidE Ubiquitination Assay in HEK293A Cells
HEK293A cells were collected and plated into individual wells of a 24-well dish at near confluency. The following day, individual wells were transfected with pcDNA-HA-Ub, pcDNA-HA-UbGG/AA, pcDNA-SidJ-V5 (or mutants), pcDNA-Myc-SdeA -SdeB, -SdeC, or -SidE (or mutants), or empty vector using PolyJet transfection reagent (SignaGen) with a total of ~1 μg plasmid DNA per well: HA-Ub (0.5 μg): SidJ-V5 or V5-SdjA (0.5μg): myc-SdeA (0.02 μg), myc-SdeB/C/SidE (0.04 μg). DNA was premixed separately for each well and diluted with 25 μL of serum free (S.F.), antibiotic free DMEM. 25 μL of PolyJet solution in S.F. DMEM containing ~3.1 μL of PolyJet transfection reagent per well was mixed with DNA solutions and added dropwise to cells. The medium was replaced 5 hours after transfection with DMEM containing 10% FBS and Pen/Strep. ~16–20 hrs after transfection, cells were washed twice with ice cold PBS and lysed directly on the plate with 2X SDS-PAGE loading buffer (25 mM Tris-PO4 pH 6.8, 20% (w/v) glycerol, 2.5% (w/v) SDS, 0.04% (w/v) bromophenol blue and 2% β-mercaptoethanol (β-ME). Samples were boiled, resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with the indicated antibodies.
SidE effector in-vitro Ubiquitin laddering and inactivation assays
Full-length SdeA, SdeB, SdeC and SidE were glutamylated in 20μL reactions containing 0.2 mg/mL SidE effector, 0.2 mg/mL, SidJ 59–851 (WT or D542A, D545A) or SdjA 36–789 (WT or D480A) and 0.1 mg/mL CALM2 in 50 mM Tris HCl pH 7.5, 50 mM NaCl, 1 mM DTT, containing 1 mM glutamic acid. Some reactions contained no SidJ/SdjA or contained catalytic mutants of each SidE effector to inactive ART activity (SdeAE860Q, SdeBE857Q, SdeCE857Q and SidEE855Q). Reactions were initiated by addition of ATP to 1 mM and MgCl2 to 5 mM and incubated for 60 min at 37°C. Reactions were then diluted 1:10 (for a final concentration of 0.02 mg/mL SidE effector) into fresh tubes containing 50 mM Tris HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 5 mM EDTA and 0.15 mg/mL HA-UbB and 100 μM NAD+ or 0.15 mg/mL ADPR-HA-UbB. The phosphoribosyl-ubiquitination assay was conducted at 37°C for 10 min (for SdeA, SdeB, and SdeC) or 15 min (for SidE). Reactions were terminated by addition of SDS loading buffer with β-ME and boiling.
Bioinformatics
For exploration of sequence diversity among SidJ homologs, a homolog set was collected using five iterations of PSI-Blast on the NR database. In the resulting sequence set, redundancy was removed at 70% sequence identity threshold using CD-hit (Huang et al., 2010). The sequences were aligned with Mafft (Katoh et al., 2019), and in-house script removed columns with gaps in the original SidJ (L. pneumophila) sequence. The sequence logos were built using the Weblogo server (Crooks et al., 2004). A phylogenetic tree was built for selected SidJ-like kinase domains, using the PhyML Maximum Likelihood method (Dereeper et al., 2008) with an Approximate Likelihood Ratio Test for branch support, and visualized with iToL (Letunic and Bork, 2016).
Quantification and Statistical Analysis
Quantification of biochemical data was performed by scintillation counting of triplicate or quadruplicate samples, as described in individual methods for respective assays. GraphPad Prism 9 was used to calculate the means, normalize the data, plot the graphs, and to conduct T-tests. All error bars represent standard deviation. Value of n (number of replicates) for each experiment is indicated in the figure legends. No methods were used to verify statistical assumptions.
Supplementary Material
Table S1. List of primers and DNA oligonucleotides used in the study (Related to STAR Methods).
Highlights:
SidJ forms a stable reaction intermediate complex with the SidE effectors
Structures of two reaction intermediate complexes were determined by cryo-EM
Two active sites in SidJ facilitate adenylation and glutamylation
SdjA is a glutamylase that differentially regulates the SidE effectors.
Acknowledgments
We thank Diana Tomchick and members of the Tagliabracci laboratory for discussions. Results shown are derived from work performed at the Pacific Northwest Center for Cryo-EM. This work was funded by NIH Grants DP2GM137419 (V.S.T.), F30HL143859 (M.H.B.), a W. M. Keck Foundation grant (V.S.T and K.P.), Welch Foundation Grant I-1911 (V.S.T.), Polish National Agency for Scientific Exchange scholarship PPN/BEK/2018/1/00431 (K.P.). We thank the Structural Biology Laboratory and the Cryo Electron Microscopy Facility at UT Southwestern Medical Center which are partially supported by grant RP170644 from the Cancer Prevention & Research Institute of Texas (CPRIT) for cryo-EM studies. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. V.S.T. is a Michael L. Rosenberg Scholar in Medical Research, a CPRIT Scholar (RR150033) and a Searle Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Tulman ER, Delhon G, Lu Z, Viljoen GJ, Wallace DB, Kutish GF, and Rock DL (2006). Genome of crocodilepox virus. J Virol 80, 4978–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akturk A, Wasilko DJ, Wu X, Liu Y, Zhang Y, Qiu J, Luo ZQ, Reiter KH, Brzovic PS, Klevit RE, et al. (2018). Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector. Nature 557, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bardill JP, Miller JL, and Vogel JP (2005). IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 56, 90–103. [DOI] [PubMed] [Google Scholar]
- Bhogaraju S, Bonn F, Mukherjee R, Adams M, Pfleiderer MM, Galej WP, Matkovic V, Lopez-Mosqueda J, Kalayil S, Shin D, et al. (2019). Inhibition of bacterial ubiquitin ligases by SidJ-calmodulin catalysed glutamylation. Nature 572, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Kalayil S, Liu Y, Bonn F, Colby T, Matic I, and Dikic I (2016). Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 167, 1636–1649 e1613. [DOI] [PubMed] [Google Scholar]
- Black MH, Osinski A, Gradowski M, Servage KA, Pawlowski K, Tomchick DR, and Tagliabracci VS (2019). Bacterial pseudokinase catalyzes protein polyglutamylation to inhibit the SidE-family ubiquitin ligases. Science 364, 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo E, Schlaermann P, and Mukherjee S (2017). How to rewire the host cell: A home improvement guide for intracellular bacteria. J Cell Biol 216, 3931–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Mu Y, Xie Y, Zhang Y, Han Y, Zhou Y, Wang W, Liu Z, Wu M, Wang H, et al. (2018). Structural basis of ubiquitin modification by the Legionella effector SdeA. Nature 557, 674–678. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, and Eddy SR (2015). HMMER web server: 2015 update. Nucleic Acids Res 43, W30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44, D279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan N, Zhen X, Liu Y, Xu X, He C, Qiu J, Liu Y, Fujimoto GM, Nakayasu ES, Zhou B, et al. (2019). Regulation of phosphoribosyl ubiquitination by a calmodulin-dependent glutamylase. Nature 572, 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC, Milligan RA, and Roll-Mecak A (2015). Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases. Cell 161, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin NV (1999). Phosphatidylinositol phosphate kinase: a link between protein kinase and glutathione synthase folds. J Mol Biol 291, 239–247. [DOI] [PubMed] [Google Scholar]
- Guo M, Kim P, Li G, Elowsky CG, and Alfano JR (2016). A Bacterial Effector Co-opts Calmodulin to Target the Plant Microtubule Network. Cell Host Microbe 19, 67–78. [DOI] [PubMed] [Google Scholar]
- Guo Q, Shen Y, Lee YS, Gibbs CS, Mrksich M, and Tang WJ (2005). Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin. EMBO J 24, 3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman G, Perkins S, Brownridge PJ, Clarke CJ, Byrne DP, Campbell AE, Kalyuzhnyy A, Myall A, Eyers PA, Jones AR, et al. (2019). Strong anion exchange-mediated phosphoproteomics reveals extensive human non-canonical phosphorylation. EMBO J 38, e100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havey JC, and Roy CR (2015). Toxicity and SidJ-Mediated Suppression of Toxicity Require Distinct Regions in the SidE Family of Legionella pneumophila Effectors. Infect Immun 83, 3506–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, and Rosenstrom P (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res 38, W545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, and Li W (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, O’Connor TJ, and Heidtman M (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong KC, Sexton JA, and Vogel JP (2015). Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathog 11, e1004695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalayil S, Bhogaraju S, Bonn F, Shin D, Liu Y, Gan N, Basquin J, Grumati P, Luo ZQ, and Dikic I (2018). Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature 557, 734–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, and Yamada KD (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20, 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent UM (1999). Purification of antibodies using ammonium sulfate fractionation or gel filtration. Methods Mol Biol 115, 11–18. [DOI] [PubMed] [Google Scholar]
- Kotewicz KM, Ramabhadran V, Sjoblom N, Vogel JP, Haenssler E, Zhang M, Behringer J, Scheck RA, and Isberg RR (2017). A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host Microbe 21, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon A, Scott S, Taujale R, Yeung W, Kochut KJ, Eyers PA, and Kannan N (2019). Tracing the origin and evolution of pseudokinases across the tree of life. Sci Signal 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppla SH (1984). Bacillus anthracis calmodulin-dependent adenylate cyclase: chemical and enzymatic properties and interactions with eucaryotic cells. Adv Cyclic Nucleotide Protein Phosphorylation Res 17, 189–198. [PubMed] [Google Scholar]
- Letunic I, and Bork P (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cash JN, Tesmer JJG, and Cianfrocco MA (2020). High-Throughput Cryo-EM Enabled by User-Free Preprocessing Routines. Structure 28, 858–869 e853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, and Luo ZQ (2007). The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75, 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingan KK, Keith Keenan E, Strickland M, Li Y, Liu Y, Ball HL, Tanner ME, Tjandra N, and Roll-Mecak A (2020). Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes. Nat Struct Mol Biol 27, 802–813. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Liu X, Arasaki K, McDonough J, Galan JE, and Roy CR (2011). Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr., Yergey A, and Machner MP (2011). De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333, 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, and Luo ZQ (2016). Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Yu K, Fei X, Liu Y, Nakayasu ES, Piehowski PD, Shaw JB, Puvar K, Das C, Liu X, et al. (2017a). A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Research 27, 865–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Yu K, Fei X, Liu Y, Nakayasu ES, Piehowski PD, Shaw JB, Puvar K, Das C, Liu X, et al. (2017b). A unique deubiquitinase that deconjugates phosphoribosyl-linked protein ubiquitination. Cell Res 27, 865–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AJM, Das S, Dawson N, Zaru R, Orchard S, Thornton JM, Orengo C, Zeqiraj E, Murphy JM, and Eyers PA (2019). Emerging concepts in pseudoenzyme classification, evolution, and signaling. Sci Signal 12. [DOI] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo J, Sorzano C, and Vargas J (2020). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. bioRxiv, 2020.2006.2012.148296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Mukherjee R, Liu Y, Gonzalez A, Bonn F, Liu Y, Rogov VV, Heinz M, Stolz A, Hummer G, et al. (2020). Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB. Mol Cell 77, 164–179 e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanina H, Madhugiri R, Bylapudi G, Schultheiss K, Karl N, Gulyaeva A, Gorbalenya AE, Linne U, and Ziebuhr J (2021). Coronavirus replication-transcription complex: Vital and selective NMPylation of a conserved site in nsp9 by the NiRAN-RdRp subunit. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, and Brady ST (2015). Post-translational modifications of tubulin: pathways to functional diversity of microtubules. Trends Cell Biol 25, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha A, Yee SS, Lopez VA, Park BC, Kinch LN, Pilch S, Servage KA, Zhang J, Jiou J, Karasiewicz-Urbanska M, et al. (2018). Protein AMPylation by an Evolutionarily Conserved Pseudokinase. Cell 175, 809–821 e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpizio A, Minelli ME, Wan M, Burrowes PD, Wu X, Sanford EJ, Shin JH, Williams BC, Goldberg ML, Smolka MB, et al. (2019). Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Piszczek G, and Roll-Mecak A (2011). Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol 18, 1250–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, et al. (2019). SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Sulpizio AG, Akturk A, Beck WHJ, Lanz M, Faca VM, Smolka MB, Vogel JP, and Mao Y (2019). Deubiquitination of phosphoribosyl-ubiquitin conjugates by phosphodiesterase-domain-containing Legionella effectors. Proc Natl Acad Sci U S A 116, 23518–23526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi M, Feng H, Zhu Y, Liu S, Gao A, and Gao P (2018). Structural Insights into Non-canonical Ubiquitination Catalyzed by SidE. Cell 173, 1231–1243 e1216. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I, Garnham CP, and Roll-Mecak A (2015). Writing and Reading the Tubulin Code. J Biol Chem 290, 17163–17172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of primers and DNA oligonucleotides used in the study (Related to STAR Methods).
Data Availability Statement
Cryo-EM maps and final refined coordinates have been deposited in EMDB (The Electron Microscopy Data Bank) and PDB (Protein Data Bank), respectively, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Raw data for protein immunoblots, SDS-PAGE, and autoradiography have been deposited at Mendeley Data (https://dx.doi.org/10.17632/sgnknvxz3j.1).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SidJ | (Black et al., 2019) | N/A |
| Rabbit anti-SdjA | This study | N/A |
| Rabbit anti-SdeA | (Black et al., 2019) | N/A |
| Mouse anti-GAPDH | ThermoFisher | Cat#MA5-15738 |
| Mouse anti-HA antibody | Sigma-Aldrich | Cat# H3663 |
| Donkey anti-Mouse IgG, HRP-Linked Whole Ab | VWR | Cat#95017-554 |
| Donkey anti-Rabbit IgG, HRP-Linked Whole Ab | VWR | Cat# 95017-556 |
| Bacterial Strains | ||
| Rosetta DE3 | Novagen | Cat#70954 |
| Rosetta DH5α | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L. pneumophila SidJ 59-851 (E. coli) | This study | N/A |
| L. pneumophila SidJ 97-851 (E. coli) | This study | N/A |
| L. pneumophila SdjA (E. coli) | This study | N/A |
| L. pneumophila SdjA 36-789 (E. coli) | This study | N/A |
| L. pneumophila SdeA (E. coli) | This study | N/A |
| L. pneumophila SdeB (E. coli) | This study | N/A |
| L. pneumophila SdeC (E. coli) | This study | N/A |
| L. pneumophila SidE (E. coli) | This study | N/A |
| L. pneumophila SdeA 231-1190 (E. coli) | This study | N/A |
| L. pneumophila SdeC 231-1222 (E. coli) | This study | N/A |
| Human CALM2 (E. coli) | This study | N/A |
| Human HA-Ubb (E. coli) | This study | N/A |
| Ulp1 S. cerevisiae SUMO protease | This study | N/A |
| PfuTurbo DNA Polymerase | Thermo | Cat# 50-125-946 |
| Q5 High-Fidelity 2X Master Mix | NEB | M0492L |
| Adenosine 5’-triphosphate disodium salt hydrate | Sigma | Cat# A2383 |
| ATP, [α-32P]- 3000Ci/mmol 10mCi/ml, 250 μCi | PerkinElmer | BLU003H250UC |
| L-[14C(U)]-Glutamic Acid | PerkinElmer | NEC290E050UC |
| L-[3,4-3H]-Glutamic Acid | PerkinElmer | NET490001MC |
| Budget-Solve scintillation cocktail | RPI | Cat# 111167 |
| EN3HANCE Autoradiography Enhancer | PerkinElmer | Cat# 6NE9701 |
| Tris-(2-Carboxyethyl)phosphine, Hydrochloride (TCEP) | Fisher Scientific | Cat# PI20490 |
| Hydrogen peroxide | Sigma | Cat# H1009 |
| L-glutamic acid, Sodium salt | Sigma | Cat# G1626 |
| Sodium Chloride | Fisher Scientific | BP358-10 |
| Tris base | Fisher Scientific | BP152-5 |
| Kanamycin sulfate | Millipore Sigma | K1377 |
| Ampicillin sodium salt | Millipore Sigma | A9518 |
| Chloramphenicol | Millipore Sigma | C0378 |
| PolyJet In Vitro DNA Transfection Reagent | SignaGen Laboratories | SL100688 |
| Protease Inhibitor Cocktail Tablets (Complete) | Millipore Sigma | 11697498001 |
| Dithiothreitol (DTT) | Millipore Sigma | D0632 |
| DMEM, High Glucose | Thermo Fisher Scientific | 11-965-118 |
| Fetal bovine serum | Millipore Sigma | F2442 |
| Penicillin-Streptomycin | Millipore Sigma | P0781 |
| Magnesium Chloride hexahydrate | Millipore Sigma | M2670 |
| Deposited Data | ||
| SidJ:CALM2:SdeA | This study | EMDB: EMD-23862 PDB: 7MIR |
| SidJ:CALM2:SdeC | This study | EMDB: EMD-23863 PDB: 7MIS |
| Experimental Models: Organisms/Strains | ||
| Legionella pneumophila Philadelphia-1 (Lp02) | Dr. Ralph Isberg | N/A |
| Legionella pneumophila Philadelphia-1 ΔdotA (Lp03) | Dr. Ralph Isberg | N/A |
| Lp02 ΔsidJ | (Black et al., 2019) | N/A |
| Lp02 ΔsdjA | This study | N/A |
| Lp02 ΔsdeA | This study | N/A |
| Lp02 ΔsidJΔsdjA | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sidJ) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sidJ D542A) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sdjA) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908 sdjA D480A) | This study | N/A |
| Lp02 ΔsidJΔsdjA (pJB908) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sidJ) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sidJ D542A) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sdjA) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908 sdjA D480A) | This study | N/A |
| Lp02 ΔsidJΔsdjAΔsdeA (pJB908) | This study | N/A |
| HEK293a | Invitrogen | R70507 |
| Acanthamoeba castellanii | Dr. Kim Orth | N/A |
| Oligonucleotides | ||
| See Table S1 for primers used in this study | ||
| Recombinant DNA | ||
| ppSUMO L. pneumophila SidJ 59-851 | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SidJ 97-851 | This study | N/A |
| ppSUMO L. pneumophila SdjA 36-789 | This study | N/A |
| ppSUMO CALM2 | (Black et al., 2019) | |
| ppSUMO L. pneumophila SdeA | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SdeB | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SdeC | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SidE | (Black et al., 2019) | N/A |
| ppSUMO L. pneumophila SdeA 231-1190 | This study | N/A |
| ppSUMO L. pneumophila SdeC 231-1222 | This study | N/A |
| ppSUMO L. pneumophila SidJSdjA-NTD | This study | N/A |
| ppSUMO L. pneumophila SdjASidJ-HTH | This study | N/A |
| pETDuet1 L. pneumophila SdjA CALM2 | This study | N/A |
| pProEx2 L. pneumophila SdeA 231-1190 | This study | N/A |
| pProEx2 L. pneumophila SdeC 231-1222 | This study | N/A |
| pSR47s | Dr. Shaeri Mukherjee | N/A |
| pSR47s sdeA flanks | This study | N/A |
| pSR47s sdjA flanks | This study | N/A |
| pSR47s sidJ flanks | (Black et al., 2019) | N/A |
| pJB908 sidJ | (Black et al., 2019) | N/A |
| pJB908 sidJ D542A | (Black et al., 2019) | N/A |
| pJB908 SdjA | This study | N/A |
| pJB908 SdjA D480A | This study | N/A |
| pJB908 | Dr. Ralph Isberg | N/A |
| pcDNA-HA-Ubb | Dr. Jenna Jewell | N/A |
| pcDNA-HA-Ubb GG/AA | (Black et al., 2019) | N/A |
| pcDNA-SidJ-V5 | (Black et al., 2019) | N/A |
| pcDNA-SidJ-V5 D542A | (Black et al., 2019) | N/A |
| pCCF-V5-SdjA | This study | N/A |
| pCCF-V5-SdjA D480A | This study | N/A |
| pCDNA-Myc-SdeA (c. opt) | (Black et al., 2019) | N/A |
| pCCF-Myc-SdeB | (Black et al., 2019) | N/A |
| pCCF-Myc-SdeC (c. opt) | (Black et al., 2019) | N/A |
| pCCF-Myc-SidE | (Black et al., 2019) | N/A |
| Software and Algorithms | ||
| Pymol | Schrödinger, Inc. | N/A |
| WebLogo server | (Crooks et al., 2004) | http://weblogo.threeplusone.com/ |
| Dalilite server | (Holm and Rosenstrom, 2010) | http://ekhidna2.biocenter.helsinki.fi/dali/ |
| Jackhmmer server | (Finn et al., 2015) | https://www.ebi.ac.uk/Tools/hmmer/search/jackhmmer |
| BLAST server | (Altschul et al., 1990) | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| MAFFT server | (Katoh and Standley, 2013) | https://mafft.cbrc.jp/alignment/software/ |
| Pfam database | (Finn et al., 2016) | http://pfam.xfam.org/ |
| RELION | (Zivanov et al., 2018) | https://relion.readthedocs.io/en/latest/index.html |
| crYOLO | (Wagner et al., 2019) | https://cryolo.readthedocs.io/en/stable/ |
| Gctf | (Zhang, 2016) | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| DeepEMhancer | (Sanchez-Garcia et al., 2020) | https://github.com/rsanchezgarc/deepEMhancer |
| Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| Phenix | (Adams et al., 2010) | https://www.phenix-online.org |
| Coot | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot |
| CCP4 7.1 | (Winn et al., 2011) | https://www.ccp4.ac.uk/?page_id=1417 |
| GraphPad Prism | N/A | https://www.graphpad.com/scientific-software/prism/ |
| ESPript | (Robert and Gouet, 2014) | http://espript.ibcp.frESPript/ESPript/ |


