Abstract
Purpose of review
This review is meant to describe the genetic associations with pediatric severe COVID-19 pneumonia and the postinfectious complication of the multisystem inflammatory syndrome in children (MIS-C). Multiple genetic approaches have been carried out, primarily in adults with extrapolation to children, including genome-wide association studies (GWAS), whole exome and whole genome sequencing (WES/WGS), and target gene analyses.
Recent findings
Data from adults with severe COVID-19 have identified genomic regions (human leukocyte antigen locus and 3p21.31) as potential risk factors. Genes related to viral entry into cells (ABO blood group locus, ACE2, TMPRS22) have been linked to severe COVID-19 patients by GWAS and target gene approaches. Type I interferon (e.g. IFNAR2) and antiviral gene (e.g. TLR7) associations have been identified by several genetic approaches in severe COVID-19. WES has noted associations with several immune regulatory genes (e.g. SOCS1). Target gene approaches have identified mutations in perforin-mediated cytolytic pathway genes in children and adults with severe COVID-19 and children with MIS-C.
Summary
Several genetic associations have been identified in individuals with severe COVID-19 and MIS-C via various genetic approaches. Broadly speaking, COVID-19 genetic associations include genes involved with antiviral functions, viral cell entry, immune regulation, chemotaxis of white blood cells, and lymphocyte cytolytic function.
Keywords: COVID-19, cytokine storm syndrome, genetics, multisystem inflammatory syndrome in children, SARS-CoV-2
INTRODUCTION
As of August 2021, the pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus has infected over 198 million individuals worldwide resulting in over 4.2 million deaths yielding a mortality rate of about 2%. Much of the mortality associated with severe infection (COVID-19 pneumonia) is believed to result from proinflammatory hypercytokinemia, including elevated levels of interleukin-6 (IL-6), IL-1, and others [1]. Perhaps, the best evidence for the detrimental role of the immune response in COVID-19 is the benefit afforded by immunosuppressive glucocorticoids, Janus kinase inhibitors, and IL-6 inhibitors. Glucocorticoids specifically, when given during hypoxic phases of the disease, have been demonstrated in randomized, blinded, placebo-controlled trials to improve mortality [2]. Why some infected individuals are asymptomatic, some experience a flu-like illness, and others develop severe and sometimes fatal pneumonia and acute respiratory distress syndrome, however, remains a conundrum.
There have been many identified clinical risk factors for severe COVID-19, including old age, obesity, diabetes, hypertension, and several other chronic conditions. But why some individuals, including younger adults and children, develop severe COVID-19 pneumonia remains unclear. Some have posited initial viral load of infection, but there are likely host genetic factors that contribute to disease severity in COVID-19. Fortunately, children seem to be largely spared of severe acute disease, and many infected children are asymptomatic. However, a small subset of children develops a postinfectious SARS-CoV-2-related multisystem inflammatory syndrome in children (MIS-C) [3]. MIS-C in younger children resembles Kawasaki disease and toxic shock syndrome, whereas in older children MIS-C can present as shock with features of a cytokine storm syndrome (CSS) [4] or macrophage activation syndrome (MAS) [5▪,6▪,7▪]. Unlike most children with severe primary COVID-19, children with MIS-C do not tend to have preexisting medical conditions [8], but do respond to glucocorticoids [9]. Again, the reason for why only some SARS-CoV-2 infected children develop MIS-C but most do not suggest a key role for host genetic susceptibility [10].
Host genetic factors that contribute to severe COVID-19 have been explored largely in adults by genome-wide association studies (GWAS) and whole exome (WES)/genome (WGS) sequencing. Because there is little data on host genetics in children with COVID-19, much is extrapolated from genetic findings in adults. There is even less data on host genetic risk factors for the development of MIS-C, but it shares some features with CSS, such as elevated markers of interferon-gamma (IFNγ), like CXCL9 [11]. IFNγ is classically elevated in hemophagocytic lymphohistiocytosis (HLH) [12], a familial form of CSS, and therefore MIS-C may share genetic contributions that predispose to related CSS. In this review, we will focus on host genetic risk factors for the development of severe primary COVID-19 in children, as well as post-COVID MIS-C.
Box 1.
no caption available
GENOME-WIDE ASSOCIATION STUDIES OF SEVERE COVID-19
The first rigorous insights into the host genetics of severe COVID-19 infection were provided by large GWAS. GWAS is ideal for identifying relatively high-frequency (>5% minor allele frequency) variants linked to specific single-nucleotide polymorphisms (SNPs), and whereas the overall relative risk of these SNPs is generally small, they can highlight specific genes that may have key pathogenic roles in disease [13]. Of note, the GWAS for COVID-19 have examined almost exclusively adult patients, and whereas findings likely have broad applicability to disease pathogenesis they do not address the etiology of rare, severe outcomes in children. The most consistently identified region amongst the several GWAS of severe COVID-19 is in chromosome 3p21.31 spanning numerous genes including SLC6A20, LZTFL1, CCR2, CCR3, CCR9, FYCO1, and CXCR6 (Table 1). This was first identified in a GWAS of 1980 Italian and Spanish patients with severe COVID and respiratory failure, with the risk haplotype conferring a 1.77 odds ratio (OR) for disease [14▪▪]. A second large GWAS utilizing research participants via 23andMe and patient-reported severe disease also identified this genomic region, with the protective haplotype conferring 0.59 OR for severe respiratory disease [15]. This was most pronounced in the European population with an increased frequency of the risk haplotype. This association was also detected in two additional studies that performed transcriptome-wide association studies to link results to tissue-specific gene expression data and found multiple genes within this cluster with significant differences in predicted expression [16,17]. Interestingly, it has recently been proposed that this genomic region is part of a 50 kilobase region of DNA inherited from Neanderthals [18].
Table 1.
Genes implicated in COVID-19 and MIS-C development or severity
| Gene/region | Method of identification | Function | References (PMID#) | Notes |
| Genomic regions | ||||
| 3p21.31 | GWAS | Multiple genes including SLC6A20, LZTFL1, CCR2, CCR3, CCR9, FYCO1, and CXCR6 | 32558485; 34315903; 33888907, 33307546 | |
| HLA locus | Target gene | 33343579, 32717807 | Not replicated by GWAS | |
| Viral entry | ||||
| ABO | GWAS | ABO blood group | 32558485, 34315903, 33888907 | Not replicated in 33307546 |
| ACE2 | GWAS and target gene | Angiotensin-converting enzyme, receptor for SARS-CoV-2 | 33837377, 32681121, 33704002 | Conflicting findings |
| TMPRSS2 | Target gene | Transmembrane serine protease, used for SARS-CoV-2 entry | 33921689, 34075330 | Not replicated by GWAS |
| Type I interferon and antiviral | ||||
| OAS1-3 | GWAS | Degrades viral RNA and inhibits replication | 34315903 | |
| IFNAR1 | WES/WGS | Interferon alpha and beta receptor subunit 1 | 32972995 | Not replicated in 34043590 |
| IFNAR2 | GWAS | Interferon alpha and beta receptor subunit 2 | 34315903, 33307546, 33837377 | |
| TLR7 | WES | Pattern recognition receptor binding viral RNA | 34115965, 32706371 | |
| IRF7 | WES | Transcriptional activation of interferon-induced genes | 32972995 | |
| Immune dysregulation | ||||
| SOCS1 | WES | Negative regulator of cytokine production | 32853638 | MIS-C |
| XIAP | WES | Regulates apoptosis and modulates inflammation | 34224783 | MIS-C |
| CYBB | WES | Component of phagocyte NADPH oxidase | 34224783 | MIS-C |
| TBK1 | WES | Mediates NFκB activation, activates interferon responses | 34210994 | Fatal pediatric COVID-19 |
| Known cytokine storm genes | ||||
| UNC13D | Target genes | Involved in cytolytic vesicle maturation and binding | 33867526 | |
| AP3B1 | Target genes | Involved in vesicle biogenesis | 33867526 | |
| PRF1 | Target gene | Released by cytolytic cells to form pores in target cells | 33256384 | |
| LYST | Target gene | Regulates cytolytic vesicle size and trafficking | 34132389 | |
| STX11 | Target genes | Regulates targeting and membrane fusion of cytolytic vesicles | 33442938 | MIS-C |
GWAS, genome wide association study; HLA, human leukocyte antigen; MIS-C, multisystem inflammatory syndrome in children; NADPH, nicotinamide adenine dinucleotide phosphate; PMID, PUBMED identifier; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WES, whole exome sequencing; WGS, whole genome sequencing.
Several additional genomic regions conferring risk for severe COVID-19 have been identified through GWAS and other approaches. The initial GWAS study also identified a locus in the ABO blood group (Table 1), and particularly that the risk for severe COVID was higher with blood group A versus other blood groups (1.45 OR), whereas blood group O was protective (0.65 OR) [14▪▪]. This association has also been observed in other GWAS [15,17] but not all [16]. Several other putative genome-wide associations have been identified in individual studies with various approaches, including antiviral effectors OAS1, OAS2, OAS3; TYK2; IL10RB; and ACE2[16,17,19] (Table 1). Interestingly, these studies have not identified a significant association in the human leukocyte antigen (HLA) locus, although two targeted analyses of HLA types have reported significant associations with SARS-CoV-2 infection [20,21]. Finally, several studies that also considered gene expression data highlighted low-expression risk alleles of IFNAR2 as conferring risk for severe COVID-19 (see below) [16,17,19] (Table 1).
Finally, there is some evidence that genetic polymorphisms may impact the risk for infection by SARS-CoV-2 through viral entry including ACE2 and TMPRSS2[22,23]. Several studies have utilized a target gene approach to identify functional variants of these proteins associated with COVID-19 disease and severity. For example, two independent studies found that the p.Val160Met variant of TMPRSS2 was associated with decreased disease severity [23,24] (Table 1). Other studies have found more frequent ACE2 allelic variability in uninfected versus SARS-CoV-2 infected individuals in Italy [25], and specific SNPs enriched in health-care works highly exposed but never infected with SARS-CoV-2 [26] (Table 1). However, such target gene associations must be interpreted with caution, as large GWAS above have largely not found associations with these loci to have genome-wide significance [14▪▪,16,19].
POTENTIAL MONOGENIC CAUSES OF SEVERE COVID-19 AND MULTISYSTEM INFLAMMATORY SYNDROME IN CHILDREN
In contrast to GWAS, WES/WGS of cohorts with defined phenotypes has the potential to identify monogenic causes of severe disease. The relatively rare occurrence of life-threatening acute COVID-19 and MIS-C in healthy children indeed suggests the presence of rare, single-gene mutations [27]. Indeed, work from a large international consortium has proposed that rare inborn errors in type I IFN responses may occur in some previously healthy individuals with severe COVID-19 [28▪]. This study performed WES or WGS on 659 previously healthy patients with severe COVID-19 including children as young as 1 month of age and used a targeted analysis of 13 genes related to influenza immunity. Patients with severe COVID-19 showed enrichment in predicted loss of function (pLOF) variants at these loci compared to healthy controls, and 3.5% of severe COVID-19 patients carried variants experimentally found to cause impaired type I IFN responses. This included 4 unrelated patients with biallelic IFNAR1 or IRF7 mutations (Table 1), and dominantly acting variants in 7 other genes in these pathways. This work is further supported by the identification of IFNAR2 in several GWAS discussed above (Table 1), and together with the identification of functional autoantibodies that can inhibit type I IFN responses [29] contribute to a model of COVID-19 pathogenesis where the failure of early interferon responses is central to severe disease progression. However, a recent study has questioned this association by analyzing 4 independent, international cohorts of adult patients with severe COVID-19 [30]. This work found first, very few patients with rare pLOF variants in type I IFN pathway genes, and second, no enrichment for either pLOF or any rare missense variants in cases versus controls. There are a number of methodologic differences between the approaches used, including ancestral matching, different definitions of ‘severe’ COVID-19 on the WHO scale, using pauci-symptomatic or asymptomatic controls versus the general population, stratifying for age, and assessing type I IFN autoantibody presence [31,32]. These conflicting results, though, do raise questions about whether the findings of Zhang et al. are generally applicable, and, as such, the relative contribution of type I IFN pathway mutations as a risk factor for severe COVID-19 remains unsettled.
Similar WES/WGS based studies have also examined individual children and young adult patients with unusually severe COVID-19. One case series of 2 unrelated pairs of previously healthy brothers age 21–32 with severe COVID-19 identified rare loss of function variants of the X-chromosome gene TLR7[33▪] (Table 1). Functionally, these patients’ peripheral blood mononuclear cells showed impaired upregulation of type I IFN-related genes [33▪]. Interestingly, Solanich et al. identified a third pair of previously healthy brothers <30 affected with severe COVID-19 both possessing a missense variant in the TLR7 coding region [34]. It has been suggested that since TLR7 escapes X-inactivation and has higher expression levels in women, TLR7 could play a role in the greater severity of COVID-19 seen in men [33▪]. Indeed a large exome-based study also found an increased burden of rare pLOF variants in TLR7 in patients with severe COVID-19 [35].
Finally, there are several smaller WES studies suggesting potential monogenic causes of severe pediatric COVID-19 as well as MIS-C. Prior to the COVID-19 pandemic, unique heterozygous truncation variants in a suppressor of cytokine signaling 1 (SOCS1) were identified in 2 unrelated boys with immune thrombocytopenia and autoimmune hemolytic anemia in the setting of acute infections [36] (Table 1). One of these patients later developed MIS-C after a documented SARS-CoV-2 infection [36]. This observation inspired a single-center prospective cohort study of 18 MIS-C patients which identified additional candidate variants in XIAP and CYBB[37▪] (Table 1). XIAP has been shown to dysregulate inflammasome activity leading to elevations in IL-1β, IL-18, and CXCL9 [38]. Interestingly, XIAP deficiency has also been associated with CSS, particularly in the context of epstein-barr virus infection (see below) [38]. CYBB encodes the p91phox subunit of the nicotinamide adenine dinucleotide phosphate oxidase necessary for phagocytic oxidative burst, impairing neutrophil cytotoxic responses. Although the full loss of p90phox activity causes chronic granulomatous disease, this patient's variant reduced but did not eliminate the neutrophil respiratory burst [37▪]. Finally, homozygous TBK1 and TNFRSF13B mutations were recently identified in a 3-year-old girl with fatal COVID-19 and a history of autoimmune disease [39]. Notably, TBK1 is a known regulator of the type I IFN pathway, and homozygous variants were recently reported in a child with systemic autoinflammation [40] (Table 1).
CYTOKINE STORM SYNDROME GENES IN COVID-19 AND MULTISYSTEM INFLAMMATORY SYNDROME IN CHILDREN
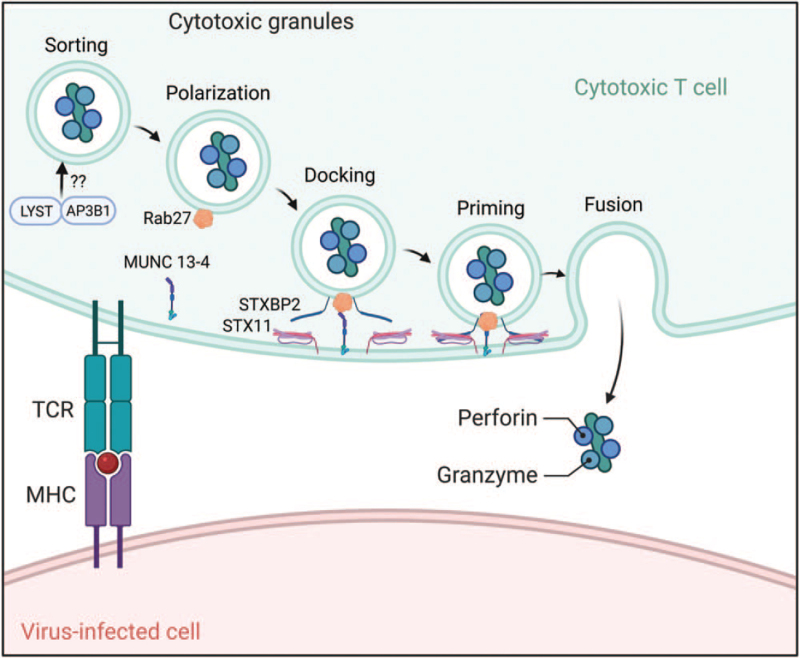
Both severe COVID-19 and MIS-C have been classified under the hyperinflammatory CSS umbrella [1]. CSS are characterized by an overly exuberant immune response to a number of triggers, including many viral pathogens [4]. When CSS occurs, the host often develops multiorgan system failure, including liver dysfunction, cytopenias, and coagulopathies [41]. Although there are multiple pathways leading to CSS [42], the best studied, and perhaps the most common, involves defects in perforin-mediated cytolytic activity of natural killer (NK) cell and cytotoxic CD8 T lymphocytes [12]. There are multiple proteins involved in the delivery of perforin to the target cell (antigen-presenting cell) to trigger apoptotic cell death (Fig. 1). The genes involved in this process are largely known, and bi-allelic defects in perforin-mediated cytolysis result in frequently fatal HLH in infancy [43]. Although familial HLH is rare, secondary forms of HLH or CSS are often associated with heterozygous defects in perforin pathway genes [44]. Indeed, some of these gene defects act as hypomorphs or complete or partial dominant-negatives disrupting lymphocyte-mediated cytolysis [45–49]. Disrupted cytolysis of the target cell results in prolonged engagement of the lytic lymphocyte and the target cell resulting in increased pro-inflammatory cytokines (e.g. IFNγ) believed to contribute to the multiorgan failure seen in CSS [49–51]. This has been reported for other pandemic viruses, such as H1N1 influenza, where fatal cases are associated with heterozygous defects in perforin pathway genes (PRF1, LYST) (Fig. 1) [45].
FIGURE 1.
Genes involved in the perforin-mediated cytolytic pathway of cytotoxic CD8 T lymphocytes and natural killer cells. Mutations in perforin or genes involved in delivering and releasing perforin containing cytotoxic granules to the immunologic synapse (e.g. STX11) may contribute to a CSS-like hyperinflammatory state in children with severe COVID-19 and MIS-C. CSS, cytokine storm syndrome; MIS-C, multisystem inflammatory syndrome in children.
Like H1N1 influenza, SARS-CoV-2 triggers a range of host immune responses. Similarly, severe cases of COVID-19 were found to be enriched for heterozygous missense and splice-site variants in familial HLH genes (UNC13D, AP3B1) (Fig. 1 and Table 1) among those with higher serum cytokine levels [52▪]. In another small cohort of severe COVID-19 patients, excluding the elderly and those with preexisting conditions, 2 of 22 adults were noted to have the familial HLH-associated missense mutation, PRF1 p.A91V (Fig. 1) [53▪], similar to 2 of 14 with fatal H1N1 influenza [45]. In terms of children or infants, one 6-week old with COVID-19 was identified as having familial HLH (homozygous nonsense LYST mutation, p.Gly1675) (Fig. 1) (Table 1) triggered by SARS-CoV-2 [54]. Likewise, a toddler with COVID-19 was noted to have familial HLH (homozygous frameshift mutation in STX11, p.Gln230Alafs∗125) (Fig. 1) (Table 1), and months later developed MIS-C, both episodes responsive to IL-1 blockade [55]. Intriguingly, familial HLH genes are significantly enriched when exploring the SARS-CoV-2 host protein interactome [56▪▪]. Thus, like other infectious CSS, mutations in known familial HLH genes (Fig. 1) may serve as risk factors for both children and adults with COVID-19, and possibly for children with MIS-C as well.
A knowledge of the genetics contributing to pediatric COVID-19 and MIS-C may benefit clinicians in terms of choosing therapeutics. Those children with evidence of HLH-associated gene mutations may derive the most benefit from therapies targeting excessive inflammation used in other CSS, including glucocorticoids, cytokine blockers (e.g. IL-1, IL-6), and lymphocyte targeted treatments (e.g. calcineurin inhibitors) [1]. In contrast, SARS-CoV-2-infected children with genetic defects in innate immune responses, including type I IFN, may benefit from treatment with recombinant interferon [57] to help control viral replication in advance of a potential CSS. Genetics may thus tailor therapy for children with COVID-19 and MIS-C, but patient selection and timing of treatment will be critical.
CONCLUSION
Only a small subset of SARS-CoV-2 infected children and young adults develop severe COVID-19, suggesting a susceptibility to cellular infection and/or inability to control the virus. Similarly, small numbers of SARS-CoV-2 infected children develop a postinfectious life-threatening hyperinflammatory episode, MIS-C, with CSS features. For COVID-19 pneumonia, genes identified in younger adults and children converge on an inadequate antiviral, particularly type I interferon, response. In contrast, studies of children with MIS-C identified mutations in CSS genes. The potential to identify children at risk for severe complications of SARS-CoV-2 infection has implications for targeted therapy directed toward diminished or excessive inflammatory responses.
Acknowledgements
We thank the dedicated and steadfast researchers and clinicians, as well as the participating patients and their families, for their heroic efforts to better understand the ongoing SARS-CoV-2 pandemic.
Financial support and sponsorship
G.S.S. was supported by NIH K08-AR072075. R.Q.C. was supported by the Arthritis Foundation, Alabama Chapter Endowed Chair in Pediatric Rheumatology.
Conflicts of interest
R.Q.C. has served as a consultant to Sobi, Novartis, Pfizer, and Sironax. R.Q.C. has received grant support from Sobi for investigator-initiated clinical trials. G.S.S. has received consulting fees from Novartis and Sobi.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1.Henderson LA, Canna SW, Schulert GS, et al. On the alert for cytokine storm: immunopathology in COVID-19. Arthritis Rheumatol 2020; 72:1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Group TWREAfC-TRW. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19 a meta-analysis. JAMA 2020; 324:1330–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feldstein LR, Tenforde MW, Friedman KG, et al. Characteristics and outcomes of US children and adolescents with Multisystem Inflammatory Syndrome in Children (MIS-C) compared with severe acute COVID-19. JAMA 2021; 325:1074–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med 2020; 383:2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5▪.Diorio C, Henrickson SE, Vella LA, et al. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J Clin Investig 2020; 130:5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]; These 3 papars are some of the first reports exploring the pathophysiology of the host immune responses to pediatric COVID-19 and MIS-C. Lymphopenia and hypercytokinemia are notable.
- 6▪. Diorio C, Shraim R, Vella LA, et al. Proteomic profiling of MIS-C patients reveals heterogeneity relating to interferon gamma dysregulation and vascular endothelial dysfunction. medRxiv 2021. doi: 10.1101/2021.04.13.21255439. [DOI] [PMC free article] [PubMed] [Google Scholar]; These 3 papars are some of the first reports exploring the pathophysiology of the host immune responses to pediatric COVID-19 and MIS-C. Lymphopenia and hypercytokinemia are notable.
- 7▪.Lee PY, Day-Lewis M, Henderson LA, et al. Distinct clinical and immunological features of SARS-CoV-2-induced multisystem inflammatory syndrome in children. J Clin Investig 2020; 130:5942–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]; These 3 papars are some of the first reports exploring the pathophysiology of the host immune responses to pediatric COVID-19 and MIS-C. Lymphopenia and hypercytokinemia are notable.
- 8.Reiff DD, Mannion ML, Samuy N, et al. Distinguishing active pediatric COVID-19 pneumonia from MIS-C. Pediatr Rheumatol Online J 2021; 19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Son MBF, Murray N, Friedman K, et al. Multisystem inflammatory syndrome in children – initial therapy and outcomes. N Engl J Med 2021; 385:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang SY, Zhang Q, Casanova JL, et al. Severe COVID-19 in the young and healthy: monogenic inborn errors of immunity? Nat Rev Immunol 2020; 20:455–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Smith JJ, Verweyen EL, Clay GM, et al. Inflammatory biomarkers in COVID-19-associated multisystem inflammatory syndrome in children, Kawasaki disease, and macrophage activation syndrome: a cohort study. Lancet Rheumatol 2021; 3:e574–e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crayne CB, Albeituni S, Nichols KE, Cron RQ. The immunology of macrophage activation syndrome. Front Immunol 2019; 10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009; 461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14▪▪.Severe Covid GG, Ellinghaus D, Degenhardt F, et al. Genomewide association study of severe covid-19 with respiratory failure. N Engl J Med 2020; 383:1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study was the first GWAS performed on patients with severe COVID-19. It was the first study which identified putative region in Ch3p21.31 linked to risk of severe disease.
- 15.Shelton JF, Shastri AJ, Ye C, et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet 2021; 53:801–808. [DOI] [PubMed] [Google Scholar]
- 16.Pairo-Castineira E, Clohisey S, Klaric L, et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021; 591:92–98. [DOI] [PubMed] [Google Scholar]
- 17.Pathak GA, Singh K, Miller-Fleming TW, et al. Integrative genomic analyses identify susceptibility genes underlying COVID-19 hospitalization. Nat Commun 2021; 12:4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeberg H, Paabo S. A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc Natl Acad Sci USA 2021; 118:e2026309118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaziano L, Giambartolomei C, Pereira AC, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med 2021; 27:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Correale P, Mutti L, Pentimalli F, et al. HLA-B∗44 and C∗01 prevalence correlates with Covid19 spreading across Italy. Int J Mol Sci 2020; 21:5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Littera R, Campagna M, Deidda S, et al. Human leukocyte antigen complex and other immunogenetic and clinical factors influence susceptibility or protection to SARS-CoV-2 infection and severity of the disease course. The Sardinian Experience. Front Immunol 2020; 11:605688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fricke-Galindo I, Falfan-Valencia R. Genetics Insight for COVID-19 susceptibility and severity: a review. Front Immunol 2021; 12:622176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monticelli M, Hay Mele B, Benetti E, et al. Protective role of a TMPRSS2 variant on severe COVID-19 outcome in young males and elderly women. Genes 2021; 12:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravikanth V, Sasikala M, Naveen V, et al. A variant in TMPRSS2 is associated with decreased disease severity in COVID-19. Meta Gene 2021; 29:100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benetti E, Tita R, Spiga O, et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet 2020; 28:1602–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Sanz J, Jimenez D, Martinez-Campelo L, et al. Role of ACE2 genetic polymorphisms in susceptibility to SARS-CoV-2 among highly exposed but non infected healthcare workers. Emerg Microbes Infect 2021; 10:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancho-Shimizu V, Brodin P, Cobat A, et al. SARS-CoV-2-related MIS-C: a key to the viral and genetic causes of Kawasaki disease? J Exp Med 2021; 218:e20210446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28▪.Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020; 370:eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]; Performing WES/WGS on 659 patients with life-threatening COVID-19, found that at least 3.5% had functional errors in type I interferon responses.
- 29.Bastard P, Orlova E, Sozaeva L, et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med 2021; 218:e20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Povysil G, Butler-Laporte G, Shang N, et al. Rare loss-of-function variants in type I IFN immunity genes are not associated with severe COVID-19. J Clin Investig 2021; 131:e147834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Povysil G, Butler-Laporte G, Gharavi AG, et al. Association of rare predicted loss-of-function variants of influenza-related type I IFN genes with critical COVID-19 pneumonia. Reply. J Clin Investig 2021; 131:e152475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Cobat A, Bastard P, et al. Association of rare predicted loss-of-function variants of influenza-related type I IFN genes with critical COVID-19 pneumonia. J Clin Investig 2021; 131:e152474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33▪.van der Made CI, Simons A, Schuurs-Hoeijmakers J, et al. Presence of genetic variants among young men with severe COVID-19. JAMA 2020; 324:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]; This case series of brothers with severe COVID-19 identified rare pLOF variants in TRL7.
- 34. Solanich X, Vargas-Parra G, van der Made CI, et al. Genetic screening for TLR7 variants in young and previously healthy men with severe COVID-19: a case series. Front Immunol 2021; 12:719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosmicki JA, Horowitz JE, Banerjee N, et al. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am J Hum Genet 2021; 108:1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee PY, Platt CD, Weeks S, et al. Immune dysregulation and multisystem inflammatory syndrome in children (MIS-C) in individuals with haploinsufficiency of SOCS1. J Allergy Clin Immunol 2020; 146:1194–1200. e1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37▪.Chou J, Platt CD, Habiballah S, et al. Mechanisms underlying genetic susceptibility to multisystem inflammatory syndrome in children (MIS-C). J Allergy Clin Immunol 2021; 148:723–738. [DOI] [PMC free article] [PubMed] [Google Scholar]; This prospective cohort study performed WES on children with MIS-C, and found that 3/18 had potentially causative genetic variants that could contribute to hyperinflammation.
- 38.Mudde ACA, Booth C, Marsh RA. Evolution of our understanding of XIAP deficiency. Front Pediatr 2021; 9:660520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt A, Peters S, Knaus A, et al. TBK1 and TNFRSF13B mutations and an autoinflammatory disease in a child with lethal COVID-19. NPJ Genom Med 2021; 6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosukcu C, Taskiran EZ, Batu ED, et al. Whole exome sequencing in unclassified autoinflammatory diseases: more monogenic diseases in the pipeline? Rheumatology 2021; 60:607–616. [DOI] [PubMed] [Google Scholar]
- 41.Crayne C, Cron RQ. Pediatric macrophage activation syndrome, recognizing the tip of the Iceberg. Eur J Rheumatol 2019; 7: (Suppl): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canna SW, Cron RQ. Highways to hell: mechanism based management of Cytokine Storm Syndromes. J Allergy Clin Immunol 2020; 146:949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janka GE. Familial and acquired hemophagocytic lymphohistiocytosis. Annu Rev Med 2012; 63:233–246. [DOI] [PubMed] [Google Scholar]
- 44.Schulert GS, Cron RQ. The genetics of macrophage activation syndrome. Genes Immun 2020; 21:169–181. [DOI] [PubMed] [Google Scholar]
- 45.Schulert GS, Zhang M, Fall N, et al. Whole-exome sequencing reveals mutations in genes linked to hemophagocytic lymphohistiocytosis and macrophage activation syndrome in fatal cases of H1N1 Influenza. J Infect Dis 2016; 213:1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spessott WA, Sanmillan ML, McCormick ME, et al. Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion. Blood 2015; 125:1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang K, Jordan MB, Marsh RA, et al. Hypomorphic mutations in PRF1, MUNC13-4, and STXBP2 are associated with adult-onset familial hemophagocytic lymphohistiocytosis. Blood 2011; 118:5794–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang M, Behrens EM, Atkinson TP, et al. Genetic defects in cytolysis in macrophage activation syndrome. Curr Rheumatol Rep 2014; 16:439–446. [DOI] [PubMed] [Google Scholar]
- 49.Zhang M, Bracaglia C, Prencipe G, et al. A heterozygous RAB27A mutation associated with delayed cytolytic granule polarization and hemophagocytic lymphohistiocytosis. J Immunol 2016; 196:2492–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anft M, Netter P, Urlaub D, et al. NK cell detachment from target cells is regulated by successful cytotoxicity and influences cytokine production. Cell Mol Immunol 2020; 17:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins MR, Rudd-Schmidt JA, Lopez JA, et al. Failed CTL/NK cell killing and cytokine hypersecretion are directly linked through prolonged synapse time. J Exp Med 2015; 212:307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52▪.Luo H, Liu D, Liu W, et al. Germline variants in UNC13D and AP3B1 are enriched in COVID-19 patients experiencing severe cytokine storms. Eur J Hum Genet 2021; 29:1312–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two studies reported mutations in CSS-associated genes among cohorts of severe COVID-19 patients.
- 53▪.Cabrera-Marante O, Rodriguez de Frias E, Pleguezuelo DE, et al. Perforin gene variant A91 V in young patients with severe COVID-19. Haematologica 2020; 105:2844–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two studies reported mutations in CSS-associated genes among cohorts of severe COVID-19 patients.
- 54.Lange M, Linden T, Muller HL, et al. Primary Haemophagocytic Lymphohistiocytosis (Chédiak-Higashi Syndrome) triggered by acute SARS-CoV-2 infection in a 6-week-old infant. Br J Haematol 2021; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vagrecha A, Patel HB, Mamdouhi T, et al. Effect of COVID-19 on anakinra-induced remission in homozygous STX11 hemophagocytosis lymphohistiocytosis. Pediatr Blood Cancer 2021; 68:e28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56▪▪.Ding J, Hostallero DE, El Khili MR, et al. A network-informed analysis of SARS-CoV-2 and hemophagocytic lymphohistiocytosis genes’ interactions points to Neutrophil extracellular traps as mediators of thrombosis in COVID-19. PLoS Comput Biol 2021; 17:e1008810. [DOI] [PMC free article] [PubMed] [Google Scholar]; CSS genes were identified in a network-informed analysis of interactions with SARS-CoV-2 proteins. CSS genes were found to be enriched within the SARS-CoV-2 host protein interactome.
- 57.Sosa JP, Ferreira Caceres MM, et al. Effects of Interferon Beta in COVID-19 adult patients: systematic review. Infect Chemother 2021; 53:247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]