Abstract
Lamin A/C provides a nuclear scaffold for compartmentalization of genome function that is important for genome integrity. Lamin A/C dysfunction is associated with cancer, aging, and degenerative diseases. The mechanisms whereby lamin A/C regulates genome stability remain poorly understood. We demonstrate a crucial role for lamin A/C in DNA replication. Lamin A/C binds to nascent DNA, especially during replication stress (RS), ensuring the recruitment of replication fork protective factors RPA and RAD51. These ssDNA-binding proteins, considered the first and second responders to RS respectively, function in the stabilization, remodeling, and repair of the stalled fork to ensure proper restart and genome stability. Reduced recruitment of RPA and RAD51 upon lamin A/C depletion elicits replication fork instability (RFI) characterized by MRE11 nuclease–mediated degradation of nascent DNA, RS-induced DNA damage, and sensitivity to replication inhibitors. Importantly, unlike homologous recombination–deficient cells, RFI in lamin A/C-depleted cells is not linked to replication fork reversal. Thus, the point of entry of nucleases is not the reversed fork but regions of ssDNA generated during RS that are not protected by RPA and RAD51. Consistently, RFI in lamin A/C-depleted cells is rescued by exogenous overexpression of RPA or RAD51. These data unveil involvement of structural nuclear proteins in the protection of ssDNA from nucleases during RS by promoting recruitment of RPA and RAD51 to stalled forks. Supporting this model, we show physical interaction between RPA and lamin A/C. We suggest that RS is a major source of genomic instability in laminopathies and lamin A/C-deficient tumors.
Keywords: nuclear lamina, DNA replication, DNA damage, genomic instability, nuclear envelope
Abbreviations: APH, aphidicolin; ATR, ataxia telangiectasia–mutated and Rad3-related kinase; BLI, biolayer interferometry; BRCA, BReast CAncer gene; BSA, bovine serum albumin; CldU, chlorodeoxyuridine; DAPI, 4′,6-diamidino-2-phenylindole; DSB, double-strand break; EdU, 5-ethynyl-2′-deoxyuridine; HEK-293T, human embryonic kidney 293T cells; HR, homologous recombination; HU, hydroxyurea; IdU, iododeoxyuridine; IF, immunofluorescence; lamin A/C, A-type lamins; MEF, mouse embryonic fibroblast; NA, numerical aperture; ON, overnight; PARP, poly(ADP-ribose) polymerase; PCNA, proliferating cell nuclear antigen; RF, replication fork; RFI, replication fork instability; RS, replication stress; shLuc, cells depleted of lamin A/C, shLmna, or luciferase; Thy, thymidine
The spatial organization of the genome, orchestrated by structural nuclear proteins, such as A-type and B-type lamins, is important for genome function and integrity. Lamins maintain nuclear architecture, a proper response to mechanical stress, chromatin organization, and genome stability (1, 2). Lamin dysfunction is associated with degenerative disorders, premature aging, and cancer, suggesting that these proteins operate as “genome caretakers.” In particular, A-type lamins (lamin A/C) dysfunction causes alterations in telomere biology and DNA repair mechanisms (3, 4, 5, 6, 7, 8, 9). Although DNA replication is the major source of spontaneous DNA damage in dividing cells (10, 11), the role of lamin A/C in replication has been minimally explored. Early studies showed colocalization of lamin A/C with proliferating cell nuclear antigen (PCNA) (12, 13, 14) and a direct association with DNA polymerases pol α, δ, and ε (15), suggesting a role for lamin A/C in replication. Other studies showed that lamin A/C is required for restart of stalled replication forks (RFs) (16) and that mutant forms of lamin A sequester PCNA away from the RF (17, 18, 19). Despite these findings, our mechanistic understanding of lamin A/C function in replication is limited.
During replication, the fork encounters many challenges, including DNA lesions and secondary structures in DNA that cause RF stalling and replication stress (RS) (11). Upon fork stalling, there is uncoupling of DNA polymerases from the minichromosome maintenance helicase complex, which generates regions of ssDNA that are rapidly coated by the heterotrimeric RPA complex (RPA70, RPA32, and RPA14 subunits) (20). The ssDNA–RPA complex plays a key role during RS, activating the ataxia telangiectasia–mutated and Rad3-related kinase (ATR)/Chk1-dependent S-phase checkpoint, which phosphorylates among other factors, chromatin-bound RPA32 on multiple sites including S33 (21, 22, 23). This complex acts as a platform for recruitment of factors, such as BRCA1/2, FANCD2, PALB2, and ultimately RAD51, which replaces RPA on ssDNA and forms a filament that stabilizes and remodels the stalled fork (24, 25). These proteins, classically associated with homologous recombination (HR) repair, protect the stalled fork from degradation by nucleases such as MRE11 (26, 27, 28). RPA and RAD51 have also been shown to facilitate fork reversal, a key protective mechanism that involves the annealing of two newly synthesized strands to form a chicken-foot structure (29) that allows forks with persistent ssDNA gaps to reverse their course and resume replication without chromosomal breakage (30, 31). RPA promotes SMARCAL1 binding to RF, which is required for remodeling forks into reversed structures (32) RAD51 is thought to have two functions in the reversal of stalled RF: (1) promote the initial step of fork regression/reversal in a BReast CAncer gene (BRCA)-independent manner and (2) stabilize the already formed reversed fork from degradation in a BRCA-dependent manner (29, 33, 34). In addition, RAD51 mediates the restart of stalled RFs and repair of DNA damage (35). Interestingly, the protective function of RAD51 from MRE11-mediated degradation and the remodeling of stalled forks do not seem to require its enzymatic recombinase activity (36). In the case of RPA, noncanonical roles include functioning as a sensor of R-loops (RNA:DNA hybrids with ssDNA), which represent a barrier to RF progression. RPA was shown to stimulate the activity of RNaseH1 suppressing R loops (37). Moreover, RPA recruitment to sites of long interspersed nuclear element-1 integration via binding to poly(ADP)-ribose polymerase 2 facilitates retrotransposition (38). These studies showcase the versatility of RPA and RAD51 in the maintenance of genomic stability and their crucial tasks in RF protection during RS. The nuclear pools of RAD51 and RPA are finite and rate limiting for ssDNA protection at stalled forks. Excessive accumulation of ssDNA in cells, for example, by ATR inhibition, TREX1 deficiency, or toxigenic bacterial infection, overwhelms the RPA and/or RAD51 response to DNA damage, leading to exhaustion of RPA and RAD51 nuclear pools and an increase in DNA breakage that stems from RS (39, 40, 41).
Given the roles of structural protein A-type lamins in telomere biology and DNA repair (3, 4, 5, 6, 7, 8, 9), we determined if lamin A/C plays a role in DNA replication. Here, we show for the first time that lamin A/C is essential for the efficient recruitment of RPA and RAD51 to stalled RFs during RS and that their reduced recruitment upon lamin A/C loss causes replication fork instability (RFI) to the same extent as HR-deficient cells. Lamin A/C physically interacts with RPA (this study) and RAD51 (42) and is likely acting to recruit both RPA and RAD51 to RFs. Accordingly, lamin A/C depletion elicits nuclease-mediated degradation of stalled forks, RS-induced genomic instability, and increased sensitivity to drugs that inhibit replication. Overall, our studies indicate that reduced expression of lamin A/C, which is associated with poor prognosis in many cancers, elicits phenotypes of genomic instability because of degradation of ssDNA during replication.
Results
Lamin A/C loss causes nucleolytic degradation of stalled RFs
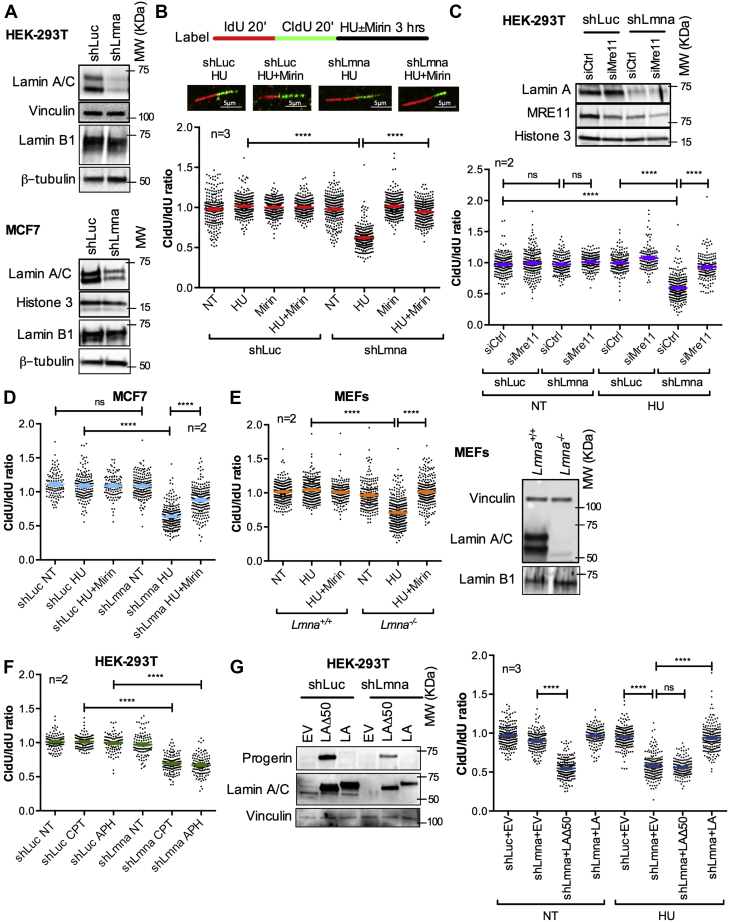
DNA lesions caused by endogenous and exogenous agents, secondary DNA structures, and transcribing RNA polymerases pose a challenge for the RF. Damaged RFs stall and recruit factors that protect them from excessive nucleolytic degradation, while facilitating fork restart to preserve genome stability (11, 25, 31). To determine how lamin A/C loss impacts RF progression and stability upon stalling, we performed DNA fiber assays in human embryonic kidney 293T (HEK-293T) and MCF-7 cells transduced with shRNAs targeting lamin A/C (shLmna) or luciferase (shLuc) as control (Fig. 1A). Replication events were labeled with thymidine (Thy) analogs: IdU (iododeoxyuridine/red tract) followed by CldU (chlorodeoxyuridine/green tract) or vice versa (Fig. S1), for an equal amount of time and detected by immunofluorescence (IF) (43). Progressing forks were identified by continuous IdU–CldU label, and the length of red (IdU) and green (CldU) tracts was measured. We find that lamin A/C depletion does not affect RF progression, as shown by the similar length of red and green tracts (Fig. S1) and an average CldU/IdU ratio of ∼1 (Fig. 1B). However, treatment with 4 mM hydroxyurea (HU) for 3 h to slow down/stall RFs results in shortening of the most recently synthesized DNA filament (CldU tract) and therefore decreased CldU/IdU ratio in lamin A/C-depleted cells (Fig. 1B). Thus, lamin A/C depletion hinders the stability of stalled RFs. Failure to protect stalled RFs causes nucleolytic degradation, a typical phenotype displayed by cells deficient in factors that promote loading of RAD51 on ssDNA like BRCA1/2 and FANCD2, cells lacking RAD51 mediators (RAD51C and XRCC2), and cells with partial inhibition of RAD51 activity or expression (24, 26, 33, 44). In all these contexts, there is inefficient nucleation and/or stabilization of RAD51 protofilaments, and DNA is exposed to the action of nucleases, such as MRE11, EXO1, and DNA2 (36). Since MRE11 initiates end resection, whereas EXO1 and DNA2 are involved in later steps of long-range degradation (45), we sought to determine whether MRE11 is responsible for fork degradation in our cells. We find that green tract shortening upon fork stalling in lamin A/C-depleted cells is due to MRE11-mediated degradation, as treatment with Mirin, an inhibitor of MRE11 nuclease, rescues fork resection (Fig. 1B). Consistently, MRE11 depletion by siRNA restores the average CldU/IdU ratio to ∼1 in lamin A/C-depleted cells (Fig. 1C). Importantly, lamin A/C loss does not affect cell cycle progression, as indicated by the normal cell cycle profile observed by flow cytometry (Fig. S2). Similarly, the 3-h treatment with HU and Mirin does not alter the percentage of cells in S phase (Fig. S2). These control experiments support the validity of our cellular model and experimental conditions to study DNA replication.
Figure 1.
Lamin A/C loss results in nuclease-mediated degradation of stalled RFs.A, immunoblots showing depletion of lamin A/C in HEK-293T and MCF-7 cells lentivirally transduced with shRNAs directed against LMNA gene (shLmna) or luciferase gene (shLuc) as control. Similar levels of lamin B1 expression were observed. B, single-molecule replication analysis performed in HEK-293T cells generated in (A). Images show DNA fibers in cells labeled with IdU 20 min + CldU 20 min as detected by fluorescence confocal microscopy. Tract lengths are measured using ImageJ. Graph shows the tract length ratio CldU/IdU in untreated cells (NT), cells treated with HU to stall RFs (4 mM HU for 3 h), and cells treated with HU and the MRE11 nuclease inhibitor Mirin (50 μM). Graph shows each individual value of all measurements in three biological repeats (three independent lamin A/C depletions), with ∼300 forks measured per experiment. The bar indicates the average ± SEM of individual values of all three biological repeats. Note how the CldU/IdU ratio <1 in shLmna cells treated with HU is rescued by Mirin. All DNA fiber assays in this figure were performed with the same labeling scheme as in (B). C, immunoblots show depletion levels of MRE11 nuclease in cells generated in (A) and transiently transfected with siRNA directed against MRE11 (siMRE11) or negative control (siCtrl). Histone 3 is the loading control. Graph shows tract length ratio CldU/IdU in cells NT or treated with HU. Graph represents individual values of two biological repeats (two independent MRE11 depletions), with ∼200 forks measured per experiment. The bar represents average ± SEM of individual values of the two biological repeats. D, tract length ratio CldU/IdU in MCF-7 depleted of lamin A/C and treated with HU ± Mirin as in (B). E, immortalized MEFs from Lmna+/+ and Lmna−/− were treated with HU ± Mirin as in (B) and CldU/IdU ratio calculated. Graph represents individual values, and the bar represents the average ± SEM of two different lines of MEFs. Immunoblots show lack of expression of lamin A/C, but not lamin B1, in MEFs from Lmna−/− mice compared with Lmna+/+. Vinculin and histone 3 are loading controls. F, tract length ratio CldU/IdU in HEK-293T generated in (A) and either NT or exposed to camptothecin (100 nM CPT) or aphidicholin (200 nM APH). Graph shows individual values, and the bar represents average ± SEM of two biological repeats, with ∼200 fibers measured per experiment. G, cells generated in (A) were lentivirally transduced with either a mutant lamin A (LAΔ50 or progerin), wildtype lamin A (LA), or an empty vector (EV) control. Immunoblots show levels of expression of progerin and lamin A. Graph shows CldU/IdU ratio in these cells either untreated (NT) or exposed to HU. Average ± SEM from three biological repeats is represented, with ∼100 fibers measured in each experiment. Individual values of all three biological repeats are combined in the graph. In all figures, ∗ denotes p value of statistical significance (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001). CldU, chlorodeoxyuridine; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; IdU, iododeoxyuridine; MEF, mouse embryonic fibroblast; NT, not treated; RF, replication fork.
Interestingly, the decreased CldU/IdU ratio upon RF stalling and the rescue of replication defects by Mirin were also observed in lamin A/C-depleted tumor cells—MCF-7 (Fig. 1D) and U2OS (Fig. S3), and in mouse embryonic fibroblasts (MEFs) from Lmna−/− mice (Fig. 1E). These data indicate that lamin A/C is required for the stability of RFs in multiple cell types. Moreover, compounds such as camptothecin and aphidicolin (APH), which cause RF stalling via different mechanisms, also elicit RFI in lamin A/C-depleted cells (Fig. 1F). Importantly, reconstitution of lamin A in lamin A/C-depleted cells rescues replication defects upon HU treatment (Fig. 1G). In contrast, expression of a truncated mutant form of lamin A (LAΔ50), also known as “progerin,” does not rescue replication defects. Rather, progerin itself imposes a challenge to DNA replication, causing RFI (reduced CldU/IdU ratio) in the absence of any drug. This confirms our published results showing fork stalling and nuclease-mediated degradation of newly replicated DNA upon expression of progerin in normal cells (46). In summary, our data demonstrate that lamin A/C is involved in the protection of the replication intermediates that are formed at stalled RFs, with loss of lamin A/C function mirroring RFI phenotypes of HR-deficient cells.
Lamin A/C loss elicits RS-induced genomic instability
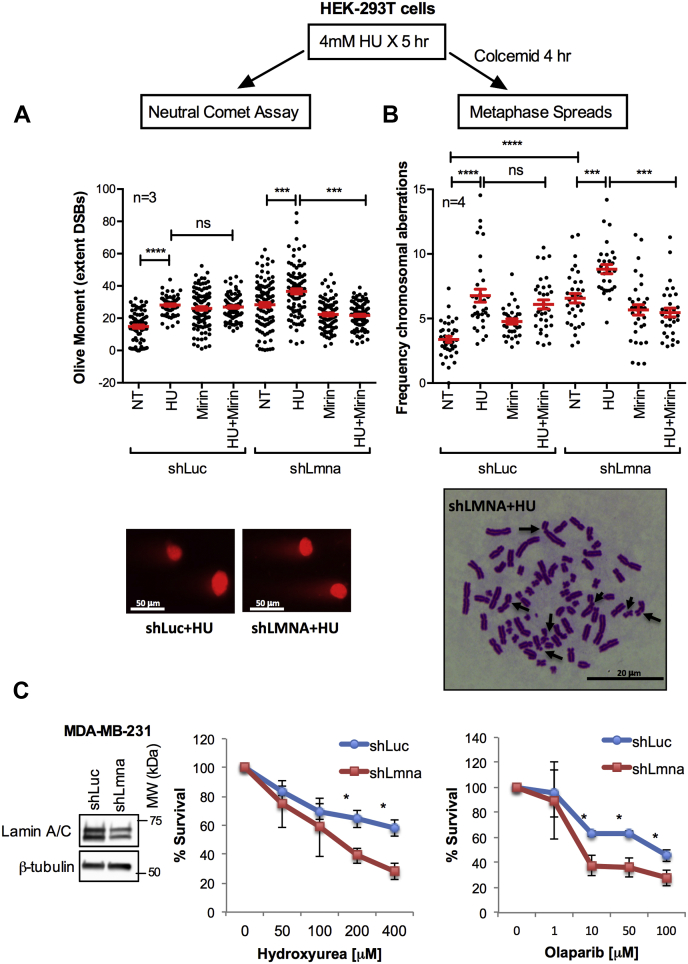
Inability to maintain fork stability in HR-deficient cells results in the accumulation of double-strand breaks (DSBs) and chromosomal aberrations in response to RS (26, 27, 32, 47). To determine if nucleolytic degradation of stalled forks causes genomic instability in lamin A/C-deficient cells, we hindered RF progression by HU treatment and monitored the generation of DSBs by neutral comet assays (Fig. 2A) and chromosomal aberrations in metaphase spreads prepared by colcemid right after induction of RS (Fig. 2B). With this approach, increased DSBs and chromosomal aberrations over basal levels should derive from those RFs that stalled/collapsed during S phase by HU treatment (27). As reported, lamin A/C-deficient cells exhibit higher basal levels of DSBs and chromosomal aberrations than control cells (48). Prolonged exposure to high HU dosage induces fork degradation and accumulation of DNA damage (DSBs) and chromosomal breaks in control and lamin A/C-deficient cells, in agreement with RS-induced DNA damage. Consistent with the notion that DNA damage can derive from nucleolytic degradation of unstable forks, inhibition of MRE11 with Mirin reduces DSBs and chromosomal aberrations in HU-treated lamin A/C-deficient cells, with no effect in control cells (Fig. 2, A and B). Interestingly, we did not observe complex chromosomal aberrations (i.e., triradials, dicentric chromosomes) typical of cells deficient in HR factors such as BRCA1/2 undergoing RS (26, 27, 32, 47). This suggests that in lamin A/C-deficient cells, genomic instability could derive from degradation of replication intermediates that differ from those arising in BRCA-deficient cells. The absence of chromatid-type aberrations in lamin A/C-deficient cells, but their presence in BRCA mutants, likely reflects differences in the role of BRCA genes and lamin A/C in the repair of DNA damage at stalled forks via HR pathways. Overall, our results suggest that excessive remodeling of stalled forks is toxic in lamin A/C-deficient cells and contributes to genomic instability. As evidence for lamin A/C loss hindering the ability of cells to cope with replication challenges, we find that tumor cells (MDA-MB-231) depleted of lamin A/C exhibit increased sensitivity to replication inhibitors, including HU and the poly(ADP-ribose) polymerase 1 (PARP1) inhibitor olaparib (Fig. 2C). Increased sensitivity to olaparib is consistent with lamin A/C-depleted cells exhibiting a replication phenotype that resembles HR-/BRCA-deficient cells.
Figure 2.
RS causes genome instability in lamin A/C-deficient cells. HEK-293T cells depleted of lamin A/C (shLmna) or control cells (shLuc) were either not treated (NT) or exposed to HU (4 mM), Mirin (50 μM), or the combination HU + Mirin for 5 h. A, after the treatments, cells were processed for neutral comet assay to monitor the amount of DNA DSBs generated in response to RS. Graph shows average ± SEM of olive moment (extent DNA DSBs) in three biological repeats. Each individual value from the three biological repeats is combined in the graph. B, after the treatments, cells were incubated in 10 μM colcemid for 4 h, and metaphase spreads were prepared to monitor the frequency of chromosomal aberrations induced by RS (chromosome breaks, chromatid breaks, chromosome end-to-end fusions, and other complex aberrations). Graph shows the average ± SEM of four biological repeats. Each individual value from the four biological repeats is combined in the graph. Note how the increased genomic instability induced by HU is rescued by Mirin in lamin A/C-deficient cells by both assays. C, graphs show survival curves of MDA-MB-231 cells depleted for lamin A and either left untreated or treated with increasing concentrations of HU or the PARP inhibitor Olaparib. Lamin A/C-deficient cells exhibit increased sensitivity to both drugs in three biological repeats. DSB, double-stranded break; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; NT, not treated; PARP, poly(ADP-ribose) polymerase; RS, replication stress.
Lamin A/C is required for RAD51 protective function at stalled RFs
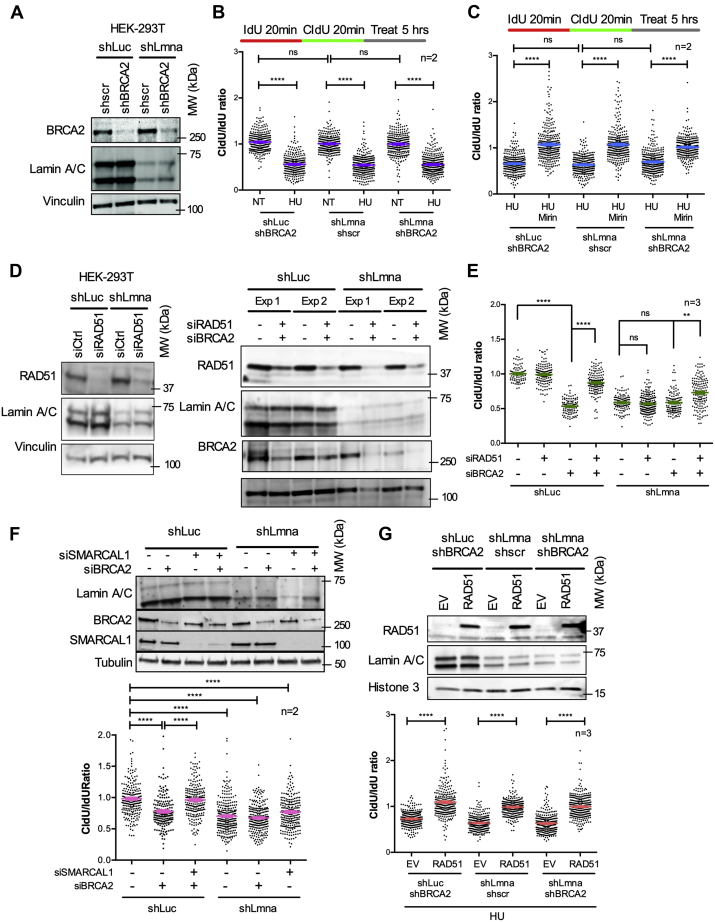
Our results show that lamin A/C is required to stabilize damaged RFs and to prevent genomic instability during RS. Stabilization and protection of stalled forks are essential processes during fork remodeling, a complex mechanism orchestrated by a multitude of factors that cooperate to reinstate DNA replication and prevent damaged forks translating into DSBs (31). BRCA2 plays an important fork protective role by mediating the loading of RAD51 on regressed arms (26, 27). Lamin A/C-depleted cells exhibit similar replication defects as BRCA2-deficient cells (Fig. 1), suggesting that lamin A/C and BRCA2 might be part of the same pathway. To test this hypothesis, we generated HEK-293T cells depleted of both, lamin A/C and BRCA2 (Fig. 3A), and performed DNA fiber assays labeling with Thy analogs for 20, 30, and 40 min to ensure that the length of green tracts was adequate for accurate measurements. BRCA2 depletion did not further exacerbate the RFI phenotype of lamin A/C-depleted cells (Figs. 3B and S4), suggesting that lamin A/C and BRCA2 act on the same RF protective pathway. Consistently, Mirin treatment rescued replication defects to the same extent in all three cell lines (Fig. 3C). Importantly, lamin A/C depletion does not affect BRCA2 expression in these cells, and thus lamin A/C might hinder the RF protective function of BRCA2 or other factors in the pathway.
Figure 3.
Replication defects in lamin A/C-deficient cells have similarities with BRCA2-deficient cells.A, HEK-293T cells depleted of lamin A/C (shLmna) or control cells (shLuc) were lentivirally transduced with short hairpin directed against BRCA2 or a scrambled control (shscr). Immunoblots show reduced levels of BRCA2 and lamin A/C proteins. Vinculin serves as loading control. B, single-molecule replication analysis performed in HEK-293T cells generated in (A). Cells were labeled with IdU 20 min + CldU 20 min and left untreated or exposed to 4 mM HU for 5 h. Graph shows average ± SEM of tract length ratio CldU/IdU in cells generated in (A) and either left untreated (NT) or treated with HU. Cells depleted of BRCA2, lamin A/C, or both proteins share similar phenotypes under replication stress (CldU/IdU < 1). Data from two independent experiments from two biological repeats, with ∼200 fibers measured per experiment. Graph shows each individual value of all measurements from the two biological repeats. C, cells generated in (A) and labeled as in (B) are exposed to HU alone or in combination with Mirin and used in single-molecule replication analysis. Graph shows tract length ratio CldU/IdU. Note how inhibition of MRE11 rescues fork degradation similarly in all the three different lines. Data from two independent experiments from two biological repeats, with ∼200 fibers measured per experiment. Graph shows each individual value from the two biological repeats, and the bar indicates the average ± SEM. D, immunoblots showing siRNA-mediated depletion of RAD51 (left panels) and BRCA2 and RAD51 (right panels) in lamin A/C-proficient and lamin A/C-deficient cells. Vinculin is the loading control. Representative image of three biological repeats. E, graph shows tract length ratio CldU/IdU of cells generated in (D) and labeled with IdU 20 min + CldU 20 min prior to exposure to HU for 5 h. Note how RAD51 depletion rescues RFI in BRCA2-deficient cells but not in lamin A-deficient cells. Graph shows each individual value of all measurements from all three replicates. The bar indicates the average of all measurements ± SEM. F, immunoblots showing lamin A/C-proficient and lamin A/C-deficient cells transfected with siRNAs for depletion of BRCA2 and SMARCAL1. Graph shows tract length ratio CldU/IdU of lamin A/C-proficient and lamin A/C-deficient cells depleted of SMARCAL1 and/or BRCA2 via siRNAs. Note how SMARCAL1 depletion rescues resection in BRCA2-deficient cells but not in lamin A/C-deficient cells. Graph shows each individual value of all measurements from two replicates. The bar indicates the average of all measurements ± SEM. G, cell lines generated in (A) were transduced with RAD51 construct or an empty vector (EV) control. Immunoblots show the overexpression of RAD51 in the three cell lines. Graph shows the CldU/IdU ratio in the six different cells. Note how overexpression of RAD51 rescues replication defects in all three lines. Data from three biological repeats and ∼200 fibers measured in each experiment. Graph shows each individual value of all measurements from all three replicates. The bar indicates the average of all individual measurements ± SEM. BRCA, BReast CAncer gene; CldU, chlorodeoxyuridine; dU, iododeoxyuridine; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; NT, not treated; RFI, replication fork instability.
Next, we interrogated the involvement of RAD51, which plays a dual role at stalled RFs: promoting fork reversal independently of BRCA and protecting the regressed arms from nucleolytic degradation in a BRCA-dependent manner (11). Accordingly, depletion of RAD51 and thus inhibition of fork reversal alleviates nucleolytic degradation in BRCA2-deficient cells (28, 33). Paradoxically, overexpression of RAD51 also inhibits nucleolytic degradation in BRCA2-deficient cells (24, 26, 27). This protective function of RAD51 is exerted through the formation of stable nucleoprotein filaments at regressed arms, an ability that is diminished in the absence of BRCA2 and restored by forced expression of RAD51 (34, 36). To define how lamin A/C loss impacts all these RAD51 protective roles, we first tested whether unprotected regressed arms are the entry point for MRE11 in lamin A/C-deficient cells. In lamin A/C-deficient and lamin A/C-proficient cells, we depleted RAD51 (Fig. 3D) and monitored fork stability by DNA fiber assay (Fig. 3E). As expected, depletion of RAD51, BRCA2, or both combined does not affect normal RF progression (Fig. S4D). Consistent with previous studies (28, 33), RAD51 depletion in BRCA2-deficient cells rescues fork resection (Fig. 3E). Unexpectedly, depletion of RAD51 in lamin A/C-deficient cells does not prevent fork resection. Similar results were obtained in two different cell lines depleted of SMARCAL1 (Figs. 3F and S5A), a fork remodeler involved in fork reversal and shown to contribute to RFI in BRCA2-deficient cells (32). The fact that in lamin A/C-deficient cells, knockdown of RAD51 or SMARCAL1 does not rescue fork protection argues against the reversed fork being the point of entry of nucleases, implying that reversed forks are not always required for nucleases to resect nascent DNA. We did find however that overexpression of RAD51 (Fig. 3G) prevents HU-induced fork resection in lamin A/C-deficient cells as well as in BRCA2-deficient cells and in cells depleted of both lamin A/C and BRCA2 (Fig. 3G). As control, we show that RAD51 overexpression does not impact normal RF progression in BRCA2- or lamin A/C-depleted cells (Fig. S5). Altogether, our results suggest that lamin A/C deficiency hinders the protective function of BRCA2 and RAD51 at the RF. The defects however seem to be independent of fork reversal and more related to the inability to properly protect ssDNA generated during RF stalling.
Lamin A/C-deficient cells display delayed replication restart but normal fork reversal
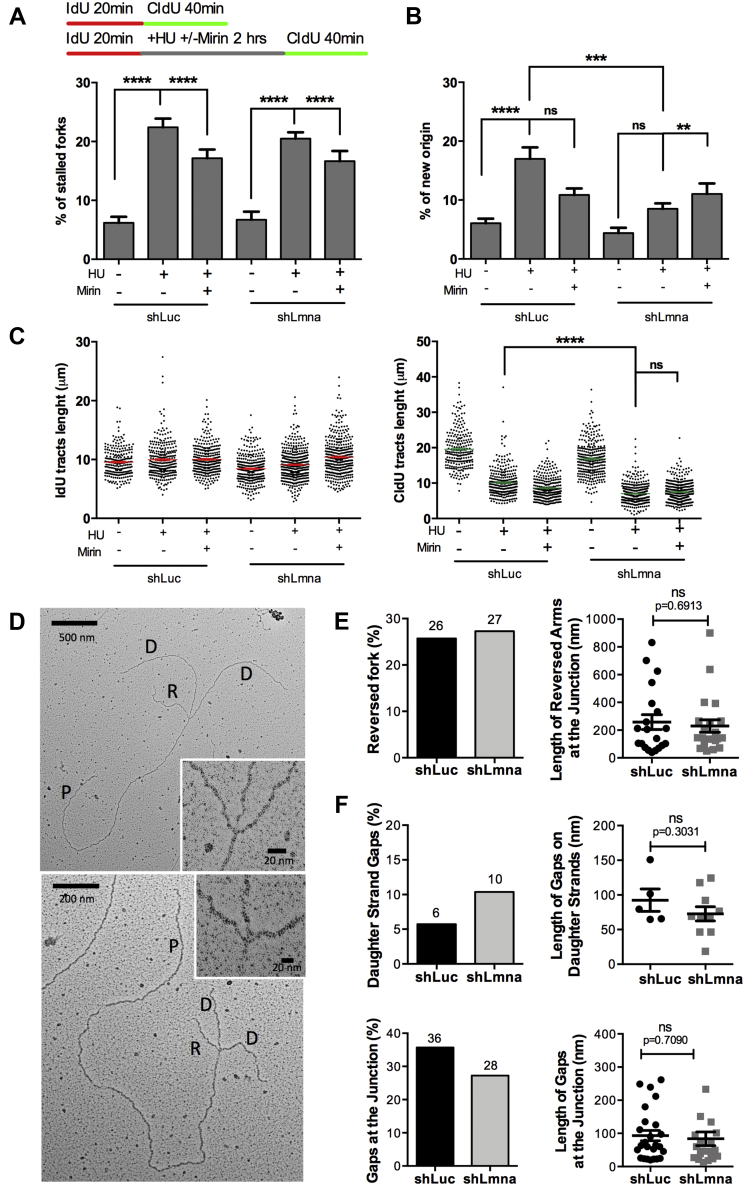
The inability to rescue fork stability in lamin A/C-deficient cells by depletion of factors involved in fork reversal (RAD51 or SMARCAL1) prompted us to investigate RF restart dynamics. Progressing forks are first labeled with IdU (red), then arrested with HU for 2 h, and successively allowed to recover in complete medium supplemented with CldU (green), to label restarting forks (Fig. S1A). This labeling scheme allows to distinguish stalled forks (red only) from restarting forks (red–green) and to identify newly fired origins (green only). Consistent with a normal cell cycle profile (Fig. S2), lamin A/C-depleted cells exhibit a similar percentage of progressing and spontaneously stalled forks as control cells (Fig. 4A). As expected, in both cell lines, there is an increased frequency of stalled forks in response to HU treatment and upon inhibition of MRE11 (HU + Mirin) (Fig. 4A), as controlled nucleolytic resection of regressed arms is required to reinstate DNA replication ((36) and (45)). To survive RS, human cells fire dormant origins within active replicon clusters (49). Interestingly, in lamin A/C-deficient cells, the percentage of new origin fired in response to HU is significantly lower than in control cells (Fig. 4B). This result is difficult to explain, but given the complex nature of DNA replication initiation, we cannot exclude a role of lamin A/C in origin specification/licensing during G1 or suboptimal functioning of S-phase checkpoints. Unlike previous observation (16), we found that in lamin A/C-deficient cells, the proportion of forks restarting replication upon HU treatment is not statistically different from control cells. However, lamin A/C loss results in delayed restart, with shorter CldU tracts compared with control cells (Fig. 4C), similarly to what was previously reported in BRCA2-deficient cells (47). Controlled resection of ssDNA at regressed arms is necessary to restart replication, whereas excessive nucleolytic activity is toxic. Interestingly, inhibition of MRE11 does not rescue timely restart of stalled forks in lamin A/C-deficient cells (Fig. 4C). Our data argue that although lamin A/C is dispensable to reinstate DNA replication, they play a role in ensuring timely restart of RFs.
Figure 4.
RFI in lamin A/C-depleted cells is independent of fork reversal.A, HEK-293T cells depleted of lamin A/C (shLmna) or control cells (shLuc) were used for DNA fiber assays with a labeling scheme that monitors fork restart. Cells are labeled with IdU (red) for 20 min, then arrested with HU (4 mM for 2 h) in the presence or the absence of Mirin, and released into complete media with CldU (green) for 40 min to label DNA after fork restart. A control sample was included without any drugs (IdU 20 min + CldU 40 min). The percentage of stalled forks (only red label) is quantified in the graph. B, the same labeling scheme as in (A) and new origins (green only) are quantified. C, same labeling scheme as in (A), and measurement of IdU tracts length (left graph), a measure of replication rate, and CldU tracts length (right graph), a measure of fork restart. Note how lamin A/C depletion hinders fork restart. Graph shows each individual value of all measurements from all three biological repeats. The bar indicates the average of all individual measurements ± SEM. D, EM performed in lamin A/C-proficient and lamin A/C-deficient cells to visualize reversed forks and extent of ssDNA gaps at the stalled fork. The cells were treated with HU (4 mM HU for 2 h) to induce RS and RFI in lamin A/C-depleted cells and with Mirin (50 μM for 2 h) to prevent degradation of reversed forks. RF structures are identified as two arms of equal length and reversed forks as four-way junction structures. Two examples of reversed forks in lamin A/C-depleted HEK-293T cells are shown. E, frequency of fork reversal in lamin A/C-proficient and lamin A/C-deficient cells treated with HU and Mirin (left graph) and length (nanometer) of the reversed arms at the junction. Note that there are no differences between control (sLuc) and lamin A/C-deficient (shLmna) cells. F, frequency of ssDNA gaps in daughter strands and at the junction (left graphs) and length of the gaps (right graphs) in lamin A/C-depleted and control cells. CldU, chlorodeoxyuridine; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; IdU, iododeoxyuridine; RFI, replication fork instability; RS, replication stress.
Next, we investigated whether delayed fork restart was caused by an inefficient fork reversal process. To test this directly, we performed EM, the only technique to directly visualize replication intermediates, including RFs that “reverse” their course following RS (28). We crosslinked cells with psolaren to preserve DNA structures and visualized duplex DNA by EM (10 nm fiber) and ssDNA gaps (thickness 5–7 nm). RF structures were identified as two arms of equal length and reversed forks as four-way junction structures (Fig. 4D). We compared lamin A/C-proficient and lamin A/C-deficient cells subjected to HU treatment to stall the fork and to Mirin to prevent degradation of the newly synthesized DNA in the stalled fork. Interestingly, the frequency of fork reversal (Fig. 4E) was indistinguishable between lamin A/C-depleted and control cells. In addition, the frequency and length of ssDNA gaps in the daughter strands and at the junction were similar in lamin A/C-depleted and control cells (Fig. 4F). Thus, lamin A/C loss does not have an impact on the generation of ssDNA at the stalled forks or the ability of forks to reverse as a mechanism of RF protection.
Altogether, our data suggest that the main role of lamin A/C at stalled forks is to mediate the stability of RAD51 nucleoprotein filaments while having a minor role in fork reversal and restart. It is possible that in the absence of lamin A/C, a limited recruitment of RAD51 to stalled forks still allows fork reversal but is not sufficient to protect ssDNA, thus leading to a slower restart. It is also possible that alternative factors such as RAD51 paralogs (RAD51B, RAC51C, RAD51D, XRCC2, and XRCC3) mediate RF reversal in lamin A/C-deficient cells, as shown in other contexts (50, 51).
Lamin A/C is essential for the recruitment of ssDNA-binding proteins to stalled forks
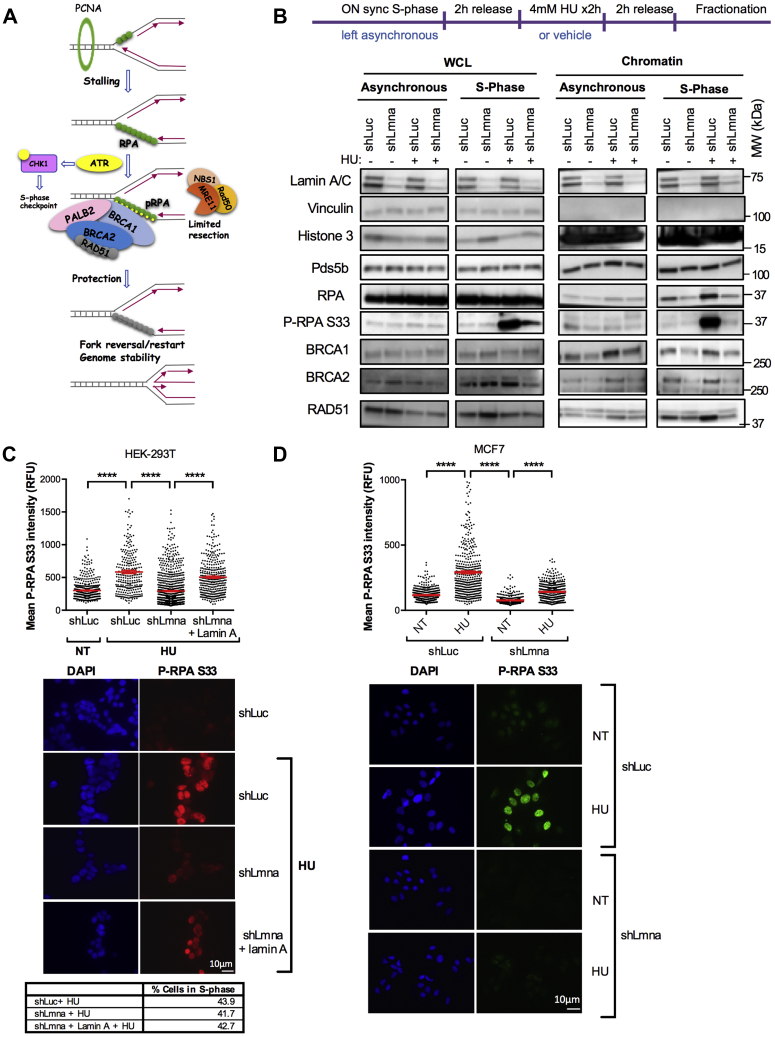
Our data show that lamin A/C-deficient cells are able to reverse stalled forks, yet they are severely impaired in protecting newly synthesized DNA from nucleolytic attack. Degradation of nascent DNA in lamin A/C-deficient cells does not seem to come from regressed arms, given that inhibition of fork reversal does not rescue degradation. Thus, we placed our focus on the initial steps of RF protection during RS, which involve the recruitment of the heterotrimeric RPA complex to ssDNA generated upon RF stalling. RPA, considered the first responder to RS, interacts with a large number of replication and repair proteins, and precedes RAD51 loading on replication intermediates (Fig. 5A). We hypothesized that the fork protective role of lamin A/C could involve binding to the RF and facilitating the recruitment and function of these factors during RS. First, we monitored the association of RPA, RAD51, and BRCA1/2 with asynchronous and S-phase chromatin by subcellular fractionation. Cells were synchronized in S phase by overnight (ON) treatment with APH and released for 2 h in normal media to allow the repair of any DNA damage and the reinitiation of replication (Fig. 5B). Then, we induced RF stalling by HU treatment for 2 h, followed by fractionation. Flow cytometry analysis confirmed efficient S-phase synchronization by this protocol and effective 5-ethynyl-2′-deoxyuridine (EdU) incorporation during release, indicating efficient restart of replication prior to HU treatment (Fig. S6). We did not find changes in global levels of factors involved in RF protection and remodeling upon depletion of lamin A/C in asynchronous or S-phase–synchronized HEK-293T cells, except for the levels of P-RPAS33, a marker of RS, which was markedly increased in control S-phase–synchronized cells under HU treatment, but much less in lamin A/C-deficient cells (Fig. 5B, whole cell lysate). Consistent with RS, we did find a robust increase in RPA, RAD51, and especially P-RPAS33 in S-phase chromatin of control cells treated with HU. In contrast, lamin A/C-depleted cells show markedly reduced levels of all these factors at chromatin upon RS (Fig. 5B; chromatin S phase). These findings indicate that lamin A/C facilitates the recruitment of key HR and fork remodeling factors to chromatin in response to RS. The same robust increase in P-RPAS33 levels was observed by IF in control HEK-293T cells treated with HU (Fig. 5C). In contrast, lamin A/C-depleted cells show markedly reduced levels of P-RPAS33, supporting the idea that lamin A/C loss hinders the proper recruitment of RPA to chromatin during RS, thus preventing phosphorylation. Accordingly, forced expression of lamin A in lamin A/C-depleted cells rescued the levels of P-RPAS33, indicating that this is a direct consequence of lamin A/C loss (Fig. 5C). As control, we calculated the percentage of cells in S phase by fluorescence-activated cell sorting (Fig. 5C, table), which is similar in all conditions, thus implying that the differences in relative fluorescence unit are not because of changes in cell cycle. Importantly, the results are not cell specific, as we confirmed in MCF-7 cells (Fig. 5D). Note how the robust increase in P-RPAS33 levels in control MCF-7 cells treated with HU is prevented by lamin A/C depletion. Altogether, these studies support the idea that lamin A/C loss hinders the proper recruitment of RPA to chromatin during RS, thus preventing phosphorylation and proper RS signaling.
Figure 5.
Lamin A/C deficiency hinders chromatin recruitment and phosphorylation of RPA.A, schematic representation of the events occurring upon stalling of RFs. The uncoupling of the replicative polymerase from the helicase generates ssDNA that is coated by the ssDNA-binding trimeric complex RPA. This activates the ATR/CHK1 intra S-phase checkpoint and ATR-mediated phosphorylation of DNA-bond RPA, which becomes a docking site for a plethora of fork-protective/remodeling factors (BRCA1, PALB2, BRCA2, and RAD51). These factors stabilize the fork and mediate restart of the stalled fork. B, HEK-293T cells proficient (shLuc) and deficient for lamin A/C (shLmna) were either left asynchronous or synchronized in S phase via overnight treatment with 1 μM aphidicholin (APH). Cells were then allowed to recover for 2 h in complete medium, followed by exposure to HU (4 mM) to elicit RS or vehicle as control for 2 h. Subsequently, cells were either lysed directly to prepare whole cell lysates (WCLs) or subjected to subcellular fractionation to isolate the chromatin fraction. Immunoblots show global protein levels from WCL or chromatin-bound proteins from asynchronous and S-phase–synchronized shLuc and shLmna cells. Note how loss of lamin A/C results in a reduction of proteins associated with S-phase chromatin under conditions of RS. Vinculin is the loading control for WCL, whereas Pds5b and H3 are loading controls for the chromatin fraction. Representative immunoblots of three biological repeats. C, immunofluorescence (IF) performed in HEK-293T lamin A/C-proficient (shLuc) and lamin A/C-deficient (shLmna) cells, as well as in cells in which lamin A expression was reconstituted (shLmna + lamin A), and treated with HU to induce RS. Monitored levels of P-RPAS33, a marker of RS that indicates binding of RPA to chromatin and phosphorylation by AKT. Representative fields of cells are shown in the images, and quantification of fluorescence intensity (relative fluorescence units) is shown in the graph (ImageJ program). DAPI staining was used to demarcate nuclei. Results are from two independent experiments, and >200 cells quantified per condition each time. Graph shows each individual value of all measurements from two replicates. The bar indicates the average for those two replicates ± SEM. The table shows percentage of cells in S phase by FACS. D, lamin A/C-proficient and lamin A/C-deficient MCF-7 cells were processed for IF as in (C) to monitor the levels of P-RPAS33 in response to HU treatment. Note the marked decrease in fluorescence intensity in lamin A/C-deficient cells compared with control. Results are from two independent experiments. Graph shows each individual value of all measurements from two replicates. The bar indicates the average for those two replicates ± SEM. ATR, ataxia telangiectasia–mutated and Rad3-related kinase; DAPI, 4′,6-diamidino-2-phenylindole; FACS, fluorescence-activated cell sorting; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; RF, replication fork; RS, replication stress.
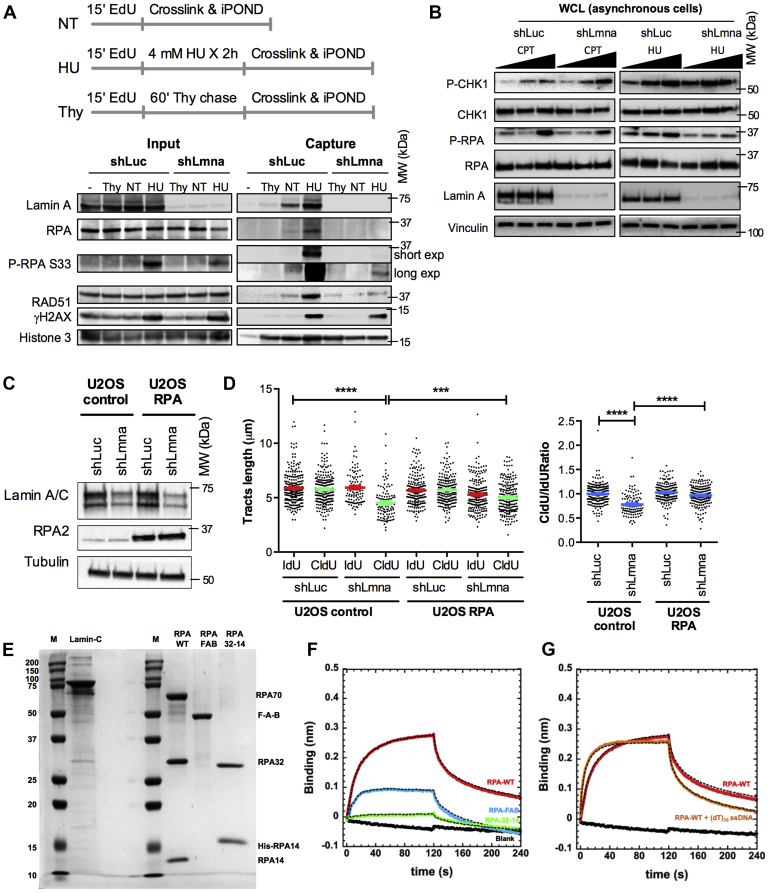
Because subcellular fractionation and IF do not allow distinguishing global chromatin from RFs, we performed isolation of proteins at nascent DNA to directly monitor the binding of proteins to nascent DNA (Fig. 6A). HEK-293T cells were pulse labeled with EdU for 15 min and either left untreated (not treated) as control, subjected to a 60-min Thy chase, or treated for 2 h with HU, which is sufficient to stall RFs without causing DNA DSBs (Fig. S7). “Input” samples show similar levels of proteins in these cells under the different conditions, except for P-RPA and γH2AX, which increase upon HU treatment because of RS and DNA damage (Fig. 6A). This is in line with results in Figure 5B showing that the global levels of proteins are not affected by lamin A/C loss. “Capture” samples show binding of lamin A to nascent DNA (not treated) in control (shLuc) cells consistent with previous observation by nascent chromatin capture proteomics (52). Importantly, we find that lamin A/C association to nascent DNA increases upon fork stalling (HU) supporting the notion that these nuclear proteins assist damaged RFs. As expected, HU treatment results in increased binding of RPA, P-RPA, and RAD51 in control cells, indicating a proper response to fork damage. However, lamin A/C-depleted cells exhibit a robust shortage of binding of RPA, P-RPA, and RAD51 to nascent DNA (Fig. 6A). Histone H3 (loading control) and γH2AX (DNA damage marker) are associated with nascent DNA to the same extent in lamin A/C-proficient and lamin A/C-deficient cells, excluding the possibility that lack of protective factors at stalled forks in lamin A/C-deficient cells is due to differences in loading or to ineffective HU treatment.
Figure 6.
Lamin A/C interacts with RFs and RPA and is required to recruit RPA and RAD51 to stalled forks.A, iPOND of shLmna and shLuc cells under basal conditions (NT) and upon replication stress (HU). HEK-293T shLuc and shLmna cells were EdU labeled and treated with 4 mM HU for 2 h. Thymidine chase experiment is performed to distinguish proteins associated with postreplicative chromatin from those at active RFs. Input (2%) shows global levels of proteins in the starting material. The immunoblot of captured proteins shows that lamin A/C directly associates with RFs, and this interaction increases under replication stress (HU). Note the marked reduction of RF protective factors (RPA, P-RPA, and RAD51) at active forks in lamin A/C-deficient cells. B, immunoblots showing levels of CHK1 and RPA and activation of both proteins by phosphorylation (P-CHK1 and P-RPA) in response to RS induced by HU or camptothecin (CPT) treatment for 2 h. Increasing concentrations of CPT (25 nM, 100 nM, and 1 μM) or HU (0.5 mM, 1 mM, and 2 mM) were used. Note how in the absence of lamin A/C, global levels of P-RPA are reduced. Vinculin is used as loading control. Representative immunoblot of three biological repeats. C, U2OS cells control and super-RPA U2OS cells were transduced with shRNAs for stable depletion of lamin A/C (shLmna), and with shLuc as control. Immunoblots show the reduced levels of lamin A/C and the overexpression of a subunit of the RPA complex (RPA2). D, the graphs show the results of DNA fiber assays performed in the four different cell lines. Note the shortening of the green tracts (left graph) and the CldU/IdU ratio <1 (right graph) in lamin A/C-deficient U2OS control cells, which is not observed in super-RPA U2OS cells. Graph shows each individual value of all measurements from all three replicates. The bar indicates the average for those three replicates with SEM. E, SDS-PAGE analysis of recombinantly purified human lamin C, RPA, RPA-FAB, and RPA32–RPA14. F, biolayer interferometry (BLI) analysis of interactions between RPA or RPA subdomains and lamin C show binding between the two proteins with an estimated Kd of ∼35 μM. The FAB fragment of RPA shows binding to lamin C and very weak binding to RPA32–RPA14 is observed. G, lamin C binds with higher affinity when RPA is prebound to a (dT)35 ssDNA oligonucleotide (Kd ∼1.5 μM). EdU, 5-ethynyl-2′-deoxyuridine; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; iPOND, isolation of proteins at nascent DNA; NT, not treated; RF, replication fork.
These findings strongly suggest that lamin A/C binds to nascent DNA and that facilitates the recruitment of ssDNA-binding proteins during RS, thus explaining the excessive resection phenotype observed in lamin A/C-deficient cells. While the protective role of RAD51 has been extensively studied, only recently it has been shown that P-RPA can directly inhibit DNA resection by limiting the processivity and speed of nucleases (53). We exclude the possibility that low P-RPA observed in lamin A/C-deficient cells is due to a compromised ATR activity, as we found a dose-dependent activation of Chk1, an ATR effector kinase activated in response to RS (54), to increasing concentrations of either camptothecin or HU (Fig. 6B). Our results argue that failure to protect stalled forks is due to faulty recruitment of both RAD51 and RPA to nascent DNA. Being RPA the first responder to RS (45), we propose that limited recruitment of RPA to ssDNA plays a major role in the RFI observed in lamin A/C-depleted cells. To test this model directly, we monitored RF stability by DNA fiber assay in a line of U2OS cells overexpressing the heterotrimeric RPA complex (super-RPA cells) (39) and transduced with shRNAs targeting lamin A/C or control hairpins (Fig. 6C). We show that in U2OS control cells under RS (HU treatment), depletion of lamin A/C results in RFI, as shown by the shortening of the green tract (Fig. 6D, left graph) and by a CldU/IdU ratio <1 (Fig. 6D, right graph). In contrast, RFI is not observed in lamin A/C-depleted cells that overexpress the RPA complex (Fig. 6D). The prevention of RFI upon forced expression of the RPA complex in lamin A/C-deficient cells indicates that suboptimal recruitment of RPA at stalled forks might be the main cause of fork protection failure in lamin A/C-deficient cells. Since RPA binding precedes RAD51 on nascent DNA, reduced amount of this “docking” factor could impact on RAD51 loading and therefore on the ability of cells to remodel stalled forks. Limited recruitment of these factors could also help explain the slow replication restart observed in lamin A/C-deficient cells despite their preserved ability to perform fork reversal. Overall, our study indicates a key role for lamin A/C in the protection of ssDNA from degradation during RS by mediating the recruitment of ssDNA-binding proteins RPA and RAD51 to stalled RFs.
Lamin A/C physically interacts with RPA
Since lamin A/C influences the recruitment of RPA to ssDNA, we next tested whether the proteins interact. Lamin C was recombinantly purified (Fig. 6E), and binding to RPA was tested using biolayer interferometry (BLI). Lamin C was tethered onto the optical probe, and binding to RPA was captured (Kd = 35 μM; Fig. 6F). When DNA is prebound to RPA, the affinity of this complex increases (Kd = 1.5 μM; Fig. 6G). RPA binds to RPA-interacting proteins through two separate protein interaction domains or compositely through interactions with multiple regions in RPA. The F domain in the large RPA70 subunit and the winged helix (wh) domain in the RPA32 subunit are well-established protein interaction regions. To test whether lamin C interaction was coordinated through these domains, we tested lamin C binding to two fragments of RPA. The FAB fragment has the DNA-binding regions A and B and the F domain. The other fragment of RPA had the RPA32 and RPA14 subcomplex (RPA32–RPA14). In BLI experiments, we qualitatively see physical interactions between lamin C and the FAB fragment but poor binding to RPA32–RPA14 (Fig. 6F). Thus, the F domain likely is the major contributor to RPA interactions with lamin C.
Discussion
This study demonstrates a role of A-type lamins in DNA replication in mammalian cells. We propose a model whereby lamin A/C is not essential for RF progression under unperturbed conditions but become critical under RS to protect stalled forks from the action of nucleases (Fig. 1). Loss of lamin A/C hinders the recruitment of RF protective and remodeling factors, especially ssDNA-binding proteins RPA and RAD51 (Figs. 5 and 6), freeing the way for nucleolytic attack by MRE11. This function of lamin A/C prevents genomic instability arising from challenges encountered during DNA replication (Fig. 2).
An important finding is that lamin A/C-depleted cells exhibit markedly reduced levels of P-RPA in response to RS (Figs. 5 and 6). RPA phosphorylation, mediated primarily by ATR (55), is a critical early step in the loading cascade of protecting factors at stalled RFs (56). ATR accumulates at the nuclear envelope during S phase (57). Thus, it is possible that lamin A/C depletion hinders ATR localization at the nuclear periphery and potentially to other ssDNA–RPA entities located away from the nuclear envelope. There is also evidence that RPA directly interacts with MRE11 during unperturbed replication and that P-RPA displaces MRE11 from this interaction (58, 59, 60). Deficiencies in P-RPA in lamin A/C-depleted cells might result in MRE11 being retained at stalled RFs, where it could exert unrestricted nucleolytic activity. In addition, P-RPA is important for the recruitment of BRCA1/2 and PALB2 proteins to stalled forks (27, 56), which in turn are required for the loading of RAD51 that promotes fork reversal and restart (29, 35). Our findings that lamin A/C-depleted cells exhibit deficiencies in RAD51 recruitment to nascent DNA (Figs. 5 and 6) are consistent with lamin A/C loss, hindering the whole cascade of events that protect stalled RFs. Interestingly, direct physical interaction between A-type lamins and BRCA2 has been previously reported (61), which regulates the association of chromatin flanking persistent DSBs along the nuclear envelope. Thus, it is also possible that lamin A/C directly mediates the positioning of BRCA2 at stalled forks. Our data showing that depletion of BRCA2 in lamin-deficient cells does not exacerbate replication defects observed upon dNTP depletion (Fig. 3) support the idea that lamin A/C and BRCA2 cooperate to protect stalled forks. Depletion of either of these proteins would lead to deficiencies in RAD51 loading to stalled RFs.
The mechanisms whereby RAD51 stabilizes the RF in response to RS has received much attention lately. Deficiency in the loading of RAD51 upon lamin A/C loss is especially significant, as RAD51 is essential for the protection of the stalled forks as well as their remodeling via fork reversal, which facilitates proper restart of the RF (29, 33). A direct interaction of lamin A with RAD51 has been reported (42), and thus a direct recruitment of RAD51 by lamin A/C is possible. Interestingly, we find some differences in the effects of BRCA2 deficiency and lamin A/C loss in DNA replication (Fig. 3). The phenotype of BRCA2-deficient cells is rescued by both RAD51 overexpression and RAD51 depletion (Fig. 3), as reported (26, 28). A model to explain this paradoxical effect of RAD51 proposes that overexpression of RAD51 allows the protection of stalled and reversed forks from nucleolytic degradation. The rescue by RAD51 depletion is due to inhibition of fork reversal, the main point of entry of nucleases in BRCA2-deficient cells. In lamin A/C-depleted cells, replication defects are only rescued by RAD51 overexpression and not by RAD51 depletion. This suggests that fork reversal in lamin A/C-deficient cells might be mediated by proteins other than RAD51 and that reversed forks in these cells do not constitute the main point of entry of nucleases.
Our study also shows that although RS does not compromise the viability of lamin A/C-deficient cells, it results in accumulation of DSBs and chromosomal aberrations (Fig. 2). This genomic instability might increase the susceptibility to acquire mutations in the genome that promote malignancy. For instance, decreased expression of A-type lamins has been observed in more than 30 cases of high-grade premalignant cervical epithelium lesions (62). Consistently, depletion of lamin A/C in primary breast epithelial cells leads to cancer-like altered morphology and aneuploidy (63). Importantly, different subtypes of advanced breast cancers, including triple-negative breast cancers, feature low expression of A-type lamins, whereas high levels of these proteins are observed in normal mammary tissues (64, 65). Perhaps the more complete study is one performed on a large well-characterized series of early stage operable breast cancers (938) with long-term follow-up (146 months on average), which showed that reduced or loss of expression of lamin A/C is associated with high histological grade, larger tumor size, poor Nottingham Prognostic Index (used to determine the prognosis following breast cancer surgery via pathological parameters, such as size of the lesion, number of involved lymph nodes, and grade of the tumor), more distant metastasis, and shorter breast cancer–specific survival (66). Further studies are needed to determine the extent to which RFI upon lamin A/C loss contributes to malignancy and cancer progression.
We propose that the role exerted by lamin A/C in preventing the degradation of stalled forks (Fig. 1) and in the resultant genome instability (Fig. 2) has important implications for therapy. Depletion of A-type lamins in the triple-negative breast cancer cell line MDA-MB-231, harboring wildtype BRCA1/2 proteins, increases sensitivity to HU and PARP inhibitors (Fig. 2), which exploit defects in DNA replication and DSB repair. The increased sensitivity to PARP inhibitors is particularly interesting, as these compounds are at the forefront for the treatment of breast cancers that exhibit BRCA1/2 mutations. These results suggest that expression levels of lamin A/C might serve as a prognostic marker, similar to BRCA deficiency, for breast cancer cells. Additional studies are also needed to determine whether the phenotypes observed in lamin A/C-depleted cells are recapitulated in cells deficient in well-known lamin-binding proteins such as LAP2 isoforms (67) and Retinoblastoma family (68) and whether other disease-associated LMNA gene mutations cause RS.
Collectively, these results advance our understanding of the role exerted by A-type lamins during DNA replication. In particular, we provide strong evidence for loss of lamin A/C shifting the balance between protection and resection at stalled forks, resulting in genomic instability. These findings and the role of progerin eliciting RS suggest that special attention should be paid to the contribution of RS to genomic instability in different laminopathies. In a broader view, one could envision that while an intact nuclear lamina protects the genome of normal cells from the onset of mutations that trigger malignancy, disruption of this nuclear skeleton in cancer cells could increase the vulnerability of cancer cells to therapeutic compounds.
Experimental procedures
Cell culture
HEK-293 cells and primary MEFs from Lmna+/+ and Lmna−/− mice were immortalized with SV40LT. All these cells and cancer cells—U2OS, MCF-7, and MDA-MB-231—were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum + 1% antibiotics/antimycotics.
Constructs and viral transduction
Retroviral and lentiviral transductions were performed as previously described (5). Viral envelope and packaging plasmids were gifts from Sheila Stewart (WUSM), shRNAs targeting lamin A/C and BRCA2 were purchased from Sigma–Aldrich, lamin A- and progerin-expressing plasmids were a gift from Brian Kennedy (Buck Institute and National University Singapore), and RAD51 construct from Simon Powell (Memorial Sloan Kettering). Constructs for expression of the heterotrimeric RPA complex (super-RPA) were provided by Luis Toledo (University of Copenhagen).
RNA interference
Transient transfection of siRNAs was performed using Lipofectamine RNAiMax transfection reagent (Life Technologies). SMARTpool siRNA Dharmacon was used to deplete BRCA2 (L-003462-00; 50 nM; 48 h), MRE11 (L-009271-00; 50 nM, 48 h), SMARCAL1 (D-013058-02-0002; 50 nM, 48 h), whereas RAD51 was depleted by employing siRNA Ambion (4390827; 50 nM, 48 h) as described (28). In each experiment, silencer select negative siRNA (4390843; Ambion) was used as transfection control.
DNA fiber assays
Fiber assays were performed as previously described (69), with some modifications. Briefly, asynchronous cells were labeled with Thy analogs: 20 μM IdU followed by 200 μM CldU. The specific labeling scheme and treatments for each experiment are shown in the legends to the figures. Cells were collected by trypsinization, washed, and resuspended in 100 μl of PBS. Then, 2 μl of cell suspension was dropped on a polarized slide (Denville Ultraclear), and cell lysis was performed in situ by adding 8 μl lysis buffer (200 mM Tris–HCl [pH 7.5]; 50 mM EDTA; and 0.5% SDS). Stretching of high-molecular weight DNA was achieved by tilting the slides at 15 to 45°. The resulting DNA spreads were air dried and fixed for 5 min in 3:1 methanol:acetic acid and refrigerated ON. For immunostaining, stretched DNA fibers were denatured with 2.5 N HCl for 60 min, washed 3× for 5 min in PBS, then blocked with 5% bovine serum albumin (BSA) in PBS for 30 min at 37 °C. Rat anti-CldU/BrdU (Abcam; ab6326) (1:100), chicken antirat Alexa 488 (Invitrogen; A21470) (1:100), mouse anti-IdU/BrdU (BD Biosciences; 347580) (1:20), and goat antimouse IgG1 Alexa 547 (Invitrogen; A21123) (1:100) antibodies were used to reveal CldU-labeled and IdU-labeled tracts, respectively. Labeled tracts were visualized under a Leica SP5X confocal microscope using 63× oil objective lens (numerical aperture [NA] of 1.40, with LAS AF software), and tract lengths were measured using ImageJ (the National Institutes of Health). A total of 100 to 300 fibers were analyzed for each condition in each experiment. All analyses were performed blinding the samples. Statistical analysis of the tract length was performed using GraphPad Prism (GraphPad Software, Inc).
Labeling schemes and treatments
To monitor DNA end resection, cells are labeled with Thy analogs for equal amounts of time: 20 min IdU followed by 20 min CldU and then exposed for 3 to 5 h to either 4 mM HU, 50 μM Mirin, 4 mM HU + 50 μM Mirin, 200 nM aphidicolin, 100 nM camptothecin, or vehicle. To exclude that end resection was an artifact of the used labeling scheme, progressive forks have been first labeled for 20 min with IdU and then exposed for 2 h to 4 mM HU + CldU or 4 mM HU + 50 μM Mirin + CldU.
Analysis of cell cycle profile
Asynchronous and S-phase–synchronized cells pulse labeled with the Thy analog EdU for 20 min were trypsinized, washed once with PBS, and then suspended in 500 μl of PBS prior to the dropwise addition of 500 μl of formaldehyde (7.4%) and incubated at room temperature for 10 min. Fixed cells were washed once with PBS, blocked in 1% BSA/PBS for 10 min, pelleted at 2000 rpm, and permeabilized in 0.5% saponin/1% BSA/PBS for 30 min at room temperature. Cells were then pelleted at 2000 rpm, washed with 1% BSA/PBS once, pelleted at 2000 rpm, and incubated in the Click-it reaction solution prepared according to the manufacturer (Click-it EdU Alexa Fluor 488 Imaging Kit; Life Technologies; #C10337) for 30 min at room temperature in the dark. Cells were washed with 1% BSA/PBS once, pelleted at 2000 rpm, and incubated with 4′,6-diamidino-2-phenylindole (DAPI) staining solution for 30 min. Stained cells were analyzed for fluorescent DNA content and EdU content using the BD Biosciences LSR II flow cytometer, and the cell cycle profiles were created by the program FlowJo. DNA staining solution consists of 1% BSA/PBS, 0.1 mg/ml RNase (Thermo-Scientific; 12091-039), and 2 μg/ml DAPI.
Subcellular fractionation
Subcellular fractionation was performed using “subcellular protein fractionation kit for cultured cells” from Fisher Thermo Scientific according to the manufacturer's instructions. The day before, 5 × 106 cells were seeded in 150 mm dishes and cultured ON in complete medium. Cells were left asynchronous (control) or synchronized in S phase by ON exposure to 1 μM APH. The next day, synchronized cells were released for 2 h in complete media and exposed to either 4 mM HU or vehicle for 2 h prior to subcellular fractionation.
Immunoblotting
Immunoblotting was carried out by lysing cells in UREA buffer (8 M urea, 40 mM Tris [pH 7.5], and 1% NP-40), for 10 min on ice. Lysates were centrifuged at 14,000 rpm for 12 min at 4 °C, and the DNA pellet was removed from the sample. About 80 μg of total protein was separated by SDS-PAGE on a 4 to 15% Criterion TGX Gel (Bio-Rad) and transferred to a nitrocellulose membrane using the Trans-Blot Turbo system (Bio-Rad). Membranes were blocked using 5% BSA in PBS + 0.1% Tween-20 for 1 h at room temperature and then incubated ON at 4 °C with the appropriate antibody diluted in blocking solution. Membranes were washed three times using PBS + 0.1% Tween-20 after both primary and secondary antibody incubations. Membranes were developed using Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore).
Antibodies used were BRCA1 (Calbiochem; OP93), RAD51 (H-92: sc-8349 SCBT; AB3756; Millipore), 53BP1 (H-300: sc-22760; SCBT), vinculin (N-19; sc-7649; SCBT), γH2AX (cs-2577; Cell Signaling), lamin A/C (H-110; sc-20681; SCBT), RPA (Ab-2; Calbiochem; #NA18), P-RPA32 on S4/S8 (Bethyl; A300-245A), P-RPA on S33 (Bethyl; A300-246A), BRCA2 (Abcam; Ab-1; OP95; Ab90451), MRE11 (Novus Biological; NB100-142), PCNA (PC-10; sc-56; SCBT), CHK1 (sc8408; SCBT), P-CHK1 (Cell Signaling; 2348s), histone 3 (Ab1791; AbCam), and SMARCAL1 (sc376377; SCBT).
IF
Approximately 5 × 105 cells were treated with HU (4 mM for 4 h) or vehicle. After washing, cells were incubated 10 min on ice with an extraction buffer (25 mM Hepes [pH 7.4], 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM Sucrose, 0.5% TX-100 in double-distilled water). After three washes in 1× PBS, cells were fixed in 4% formaldehyde in PBS for 10 min, permeabilized in 0.5% TX-100 for 30 min at room temperature and blocked 1 h at 37 °C in 1% BSA/PBS. Incubation with primary antibody was performed ON at 4 °C and secondary antibody for 1 h at 37 °C, both in a humid chamber. After washes in PBS, cells were counterstained with DAPI in Vectashield. Microscopy and photo capture were performed on a Leica DM5000B microscope using 40× or 63× oil objective lenses with a Leica DFC350FX digital camera and the Leica Application Suite. Antibody used was P-RPA on S33 (Bethyl; A300-246A).
Isolation of proteins at nascent DNA
Isolation of proteins at nascent DNA was performed as originally described (70) with minor modifications. HEK-293T cells were pulse labeled with 10 μM EdU (Life Technologies) for 15 min and left untreated or treated with 4 mM HU for 2 h. For the Thy chase experiments, cells were washed 3× wit× complete medium and incubated for 60 min in medium supplemented with 10 μM (Thy) (Sigma–Aldrich). Protein–DNA crosslinking was performed in 1% formaldehyde for 15 min at RT, quenched with 0.125 M glycine for 5 min, and washed 3× with PBS. Cells were then permeabilized with 0.25% Triton X-100/PBS for 30 min, washed once with 0.5% BSA/PBS and once with PBS, and incubated in click reaction buffer (10 mM sodium-l-ascorbate, 20 μM biotin azide [Life Technologies], and 2 mM CuSO4) for 2 h at room temperature. In the “no-click” control sample, dimethyl sulfoxide was used in place of biotin azide in the click reaction buffer. Cells were washed once with 0.5% BSA/PBS and once with PBS prior to be resuspended in lysis buffer (50 mM Tris–HCl, pH 8.0, and 1% SDS) supplemented with protease/phosphatase inhibitor (Cell Signaling), and chromatin was solubilized by sonication in a Bioruptor (Diagenode) at 4 °C at the highest setting for 30 to 45 cycles (30 s on and 30 s off). Samples were centrifuged for 10 min at 5000g, and supernatants were diluted 1:1 with PBS (v/v) containing protease/phosphatase inhibitors and incubated ON at +4 °C with streptavidin-agarose beads (EMD Millipore). Beads were washed twice with lysis buffer, once with 1 M NaCl, twice with lysis buffer, and captured proteins were eluted by boiling beads in 2× NuPAGE LDS Sample Buffer (Life Technologies) containing 200 mM DTT for 40 min at 95 °C. Proteins were resolved by electrophoresis using BioRad 4 to 12% Bis–Tris gels and detected by Western blotting.
Metaphase spreads
Cells were cultured in 10 mm dishes to 70% confluence, and asynchronous cells were exposed to dimethyl sulfoxide, 4 mM HU, 50 μM Mirin, or 4 mM HU + 50 μM Mirin for 5 h. Cells were allowed to complete S phase in complete medium supplemented with 10 μM Colcemid (Sigma; #D1925) in order to arrest them in metaphase. After collecting the culture media, cells were washed with 1× PBS (which was also collected), and the trypsinized cells were collected. All the fractions were combined and centrifuged to pellet the cells. Supernatant was then aspirated to leave ∼2 ml of media + cell pellet, which was resuspended by gentle flicking. About 9 ml of preheated (37 °C) hypotonic buffer (0.56% KCl) was added then to the cell suspension, which was incubated in a 37 °C water bath to allow hypotonic swelling of the cells. A small amount (∼500 μl) of fixing solution (3:1 methanol:acetic acid) was added, and the cells were pelleted at 4 °C by centrifugation. Cells were kept on ice from this point onward. After aspirating the supernatant to leave ∼2 ml, the cells were resuspended by gentle flicking, and 9 ml fixing solution was added dropwise. The suspension was centrifuged once more to pellet the cells, and fixing solution was added in a similar manner. This mixture was stored at −20 °C ON. Cells were then pelleted, resuspended in ∼500 μl of fresh fixing solution, dropped onto microscope slides, and allowed to air dry at room temperature ON. Metaphase spreads were stained by dipping the microscope slides into a Coplin jar containing a solution of Giemsa (Sigma; #GS500) diluted in milliQ water 1:20, for 20 min. The excess staining was washed off with tap water, and slides were air dried at room temperature. Metaphase spreads were visualized under a Leica DM5000B microscope using 100× oil objective lens (numerical aperture of 1.3) with a Leica DFC350FX digital camera and the Leica Application Suite (version 4.1.0).
Comet assay
Neutral comet assays were performed using CometSlide assay kits (Trevigen). Cells were treated with either vehicle (DMSO), 4 mM HU, 50 μM Mirin, or HU + Mirin for 5 h. Cells were embedded in agarose, lysed, and subjected to neutral electrophoresis. Before image analysis, cells were stained with ethidium bromide and visualized under a fluorescence Leica DM5000B microscope using 20× objective lens (numerical aperture of 0.5) with a Leica DFC350FX digital camera and the Leica Application Suite (version 4.1.0). Single-cell electrophoresis results in a comet-shaped distribution of DNA. The comet head contains high molecular weight and intact DNA, and the tail contains the leading ends of migrating fragments. Olive comet moment was calculated by multiplying the percentage of DNA in the tail by the displacement between the means of the head and tail distributions, as described (71). We utilized the program CometScore, version 1.5 (TriTek) to calculate Olive Comet Moment. A total of 60 to 100 comets were analyzed per sample in each experiment.
XTT assay
Cell viability is monitored using “Cell proliferation kit II (XTT)” from Sigma according to the manufacturer's instructions. Cells were plated in 96-well dishes at low confluency and cultured for 6 days in complete medium supplemented with different concentrations of the replication inhibitor HU (50, 100, 200, and 400 μM) or the PARP inhibitor Olaparib (1, 10, 50, and 100 μM). Drugs were freshly replaced every other day.
EM analysis
The procedure was performed as described (72). Lamin A/C-proficient and lamin A/C-deficient cells were treated with HU and Mirin for the indicated times. In vivo psoralen crosslinking of the DNA was achieved by a repetitive exposure of living cells to 4,5′,8-trimethylpsoralen (10 μg/ml final concentration) followed by irradiation pulses with UV 365-nm monochromatic light (UV Stratalinker 1800; Agilent Technologies). The cells were then lysed with cell lysis buffer (buffer C1: 1.28 M sucrose, 40 mM Tris–Cl, pH 7.5, 20 mM MgCl2, and 4% Triton X-100; QIAGEN) and then digested by digestion buffer (QIAGEN; buffer G2: 800 mM guanidine–HCl, 30 mM Tris–HCl, pH 8.0, 30 mM EDTA, pH 8.0, 5% Tween-20, and 0.5% Triton X-100) and 1 mg/ml proteinase K at 50 °C for 2 h. Chloroform/isoamyl alcohol (24:1) was used to collect DNA via phase separation (centrifugation at 8000 rpm for 20 min) followed by DNA precipitation by adding 0.7× volume of isopropanol. The DNA was then washed with 70% ethanol, air dried, and resuspended in 200 μl Tris–EDTA buffer. About 100 U restriction enzyme high-fidelity PvuII was used for 12 μg mammalian genomic DNA digestion (4–5 h of incubation). Poly-Prep chromatography columns were used for RI enrichment. Benzoylated naphthoylated diethylaminoethyl cellulose granules were resuspended in 10 mM Tris–HCl, pH 8.0, and 300 mM NaCl to a final concentration of 0.1 g/ml. The columns were washed and equilibrated with 10 mM Tris–HCl, pH 8.0, and 1 M NaCl and 10 mM Tris–HCl, pH 8.0, and 300 mM NaCl, respectively. The sample DNA was then loaded and incubated for 0.5 h. After washing the columns (10 mM Tris–HCl, pH 8.0, and 1 M NaCl), the DNA was eluted in caffeine solution (10 mM Tris–HCl, pH 8.0, 1 M NaCl, and 1.8% [w/v] caffeine) for 10 min followed by sample collection. DNA is then purified and concentrated, using an Amicon size-exclusion column and resuspended in Tris–EDTA. With DNA spreading by the “BAC method,” the DNA was loaded on carbon-coated 400-mesh copper grids. The DNA was then coated with platinum by platinum-carbon rotary shadowing (High Vacuum Evaporator MED 020; Bal-Tec). Microscopy was performed with a transmission electron microscope (Tecnai G2 Spirit; FEI; LaB6 filament; high tension ≤120 kV) and acquired with a side mount charge-coupled device camera (2600 × 4000 pixels; Orius 1000; Gatan, Inc). The images were processed with DigitalMicrograph, version 1.83.842 (Gatan, Inc) and analyzed with ImageJ.
Statistical analysis
All datasets from each DNA fiber assay experiment have been first subjected to normality test. Most of the dataset does not meet the criteria of a normal distribution. Since each dataset includes more than 100 values to calculate statistical significance of the observed differences, we have used nonparametric tests, such as Wilcoxon matched-pairs signed rank test for paired samples and Mann–Whitney test for unpaired samples; one-way ANOVA was used for all the experiments in which more than two comparisons were performed. GraphPad Prism 6 was used for the calculations. In all cases, differences were considered statistically significant when p < 0.05.
RPA binding to lamin
Human RPA, RPA-FAB, and RPA-32–RPA14 proteins were purified as described (73). Lamin C was overproduced and purified from codon-optimized Lamin C (amino acid residues 1–152) engineered into pRSF-Duet plasmid (GenScript, Inc). The construct carries a tandem N-terminal Strep tag, polyhistidine tag (6× His), and a SUMO tag along with a SUMO protease cleavage site engineered after the tags. The plasmid was transformed into BL21-pLysS cells, and a 10 ml ON culture from a single transformant was grown in Luria Broth media. Larger 1 l cultures were grown from the starter culture and at 37 °C until absorbance at 600 nm reached 0.6 and lamin C overproduction was induced with 0.5 mM IPTG. Induction was carried out at 37 °C for 4 h. Harvested cells were resuspended in 120 ml cell resuspension buffer (50 mM Hepes, pH 8.0, 750 mM NaCl, 5 mM β-mercaptoethanol, 1.5× protease inhibitor cocktail, 1 mM PMSF, 10% [v/v] glycerol, and 10 mM imidazole). Cells were lysed using 0.4 g/ml lysozyme followed by sonication. Clarified lysates were fractionated on a Ni2+–NTA agarose column. Bound proteins were eluted using a step gradient in cell resuspension buffer containing 0 to 500 mM imidazole. Fractions containing lamin C were pooled and concentrated using an Amicon spin concentrator (30 kDa molecular weight cutoff). Concentrated lamin C was dialyzed against lamin C storage buffer (50 mM Hepes, pH 8.0, 750 mM NaCl, 5 mM β-mercaptoethanol, 1.5× protease inhibitor cocktail, 10% [v/v] glycerol). Lamin C was flash frozen using liquid nitrogen and stored at −80 °C. RPA concentration was measured spectroscopically using ε280 = 87,210 M−1 cm−1 (hRPA-WT), 42,400 M−1 cm−1 (hRPA-FAB), or 24,410 M−1 cm−1 (hRPA-32-14). The concentration of lamin C was calculated using the Bradford method with BSA as reference.
Lamin C binding to RPA was captured using BLI. Experiments were performed using a single channel BLItz instrument (Sartorius, Inc), and binding sensograms were collected in advanced kinetics mode at room temperature with shaking at 2200 rpm. Strep-tagged lamin C was bound onto streptavidin biosensors (Sartorious, Inc), which were prehydrated by incubating the tips in BLI buffer (30 mM Hepes, pH 7.8, 100 mM KCl, 5 mM MgCl2, 6% [v/v] glycerol) for 10 min. Binding and dissociation of RPA, RPA-FAB, or RPA-32-14 to the lamin C-bound tip was measured. Sensograms were fitted using the associated BLItz Pro software to obtain Kd values.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge Denisse Carvajal and Annabel Quinet de Andrade for their help setting up the DNA fibers technology in our laboratory.
Author contributions
S. Graziano, A. V., E. A., and S. Gonzalo conceptualization; S. Graziano methodology; S. Graziano validation; S. Graziano and N. C.-B. formal analysis; S. Graziano, N. C.-B., B. T.-C., S. K., J. J., E. S., and U. M. investigation; A. V. and E. A. resources; S. Graziano data curation; S. Graziano writing–original draft; E. A. and S. Gonzalo writing–review and editing; A. V., E. A., and S. Gonzalo supervision; S. Gonzalo project administration; S. Gonzalo funding acquisition.
Funding and additional information
Research in the S. Gonzalo laboratory is supported by the National Institute on Aging, National Institutes of Health (NIH) grant RO1AG058714. The work in the A. V. laboratory is supported by the NIH grant R01GM108648 and the Department of Defense Breast Cancer Research Program breakthrough award BC151728. Work in the E. A. laboratory is supported by grants from the NIH GM130746 and GM133967. S. Graziano was recipient of a predoctoral fellowship from Siteman Cancer Center. N. C.-B. is recipient of a postdoctoral fellowship from the American Heart Association. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Patrick Sung
Footnotes
Present address for Simona Graziano: Cell Therapy Analytics and Formulation, Cell Therapy CMC, Novo Nordisk A/S, Denmark.
Present address for Alessandro Vindigni and Jessica Jackson: Department of Internal Medicine, Washington University School of Medicine, St Louis, MO, USA.
Supporting information
References
- 1.Denais C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Steensel B., Belmont A.S. Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redwood A.B., Perkins S.M., Vanderwaal R.P., Feng Z., Biehl K.J., Gonzalez-Suarez I., Morgado-Palacin L., Shi W., Sage J., Roti-Roti J.L., Stewart C.L., Zhang J., Gonzalo S. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle. 2011;10:2549–2560. doi: 10.4161/cc.10.15.16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Suarez I., Redwood A.B., Grotsky D.A., Neumann M.A., Cheng E.H., Stewart C.L., Dusso A., Gonzalo S. A new pathway that regulates 53BP1 stability implicates cathepsin L and vitamin D in DNA repair. EMBO J. 2011;30:3383–3396. doi: 10.1038/emboj.2011.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez-Suarez I., Redwood A.B., Perkins S.M., Vermolen B., Lichtensztejin D., Grotsky D.A., Morgado-Palacin L., Gapud E.J., Sleckman B.P., Sullivan T., Sage J., Stewart C.L., Mai S., Gonzalo S. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009;28:2414–2427. doi: 10.1038/emboj.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B., Wang J., Chan K.M., Tjia W.M., Deng W., Guan X., Huang J.D., Li K.M., Chau P.Y., Chen D.J., Pei D., Pendas A.M., Cadiñanos J., López-Otín C., Tse H.F. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S., Liu B., Wang Y., Hao Q., Zhou Z. Lamin A is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Rep. 2015;13:1396–1406. doi: 10.1016/j.celrep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Liu B., Wang Z., Zhang L., Ghosh S., Zheng H., Zhou Z. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat. Commun. 2013;4:1868. doi: 10.1038/ncomms2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao K., Blair C.D., Faddah D.A., Kieckhaefer J.E., Olive M., Erdos M.R., Nabel E.G., Collins F.S. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J. Clin. Invest. 2011;121:2833–2844. doi: 10.1172/JCI43578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berti M., Vindigni A. Replication stress: Getting back on track. Nat. Struct. Mol. Biol. 2016;23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spann T.P., Moir R.D., Goldman A.E., Stick R., Goldman R.D. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moir R.D., Spann T.P., Herrmann H., Goldman R.D. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 2000;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy B.K., Barbie D.A., Classon M., Dyson N., Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaara M., Itkonen H., Hillukkala T., Liu Z., Nasheuer H.-P., Schaarschmidt D., Pospiech H., Syväoja J.E. Segregation of Replicative DNA Polymerases during S Phase. J. Biol. Chem. 2012;287:33327–33338. doi: 10.1074/jbc.M112.357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh M., Hunt C.R., Pandita R.K., Kumar R., Yang C.R., Horikoshi N., Bachoo R., Serag S., Story M.D., Shay J.W., Powell S.N., Gupta A., Jeffery J., Pandita S., Chen B.P. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell Biol. 2013;33:1210–1222. doi: 10.1128/MCB.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilton B.A., Liu J., Cartwright B.M., Liu Y., Breitman M., Wang Y., Jones R., Tang H., Rusinol A., Musich P.R., Zou Y. Progerin sequestration of PCNA promotes replication fork collapse and mislocalization of XPA in laminopathy-related progeroid syndromes. FASEB J. 2017;31:3882–3893. doi: 10.1096/fj.201700014R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheaton K., Campuzano D., Ma W., Sheinis M., Ho B., Brown G.W., Benchimol S. Progerin-induced replication stress facilitates premature senescence in Hutchinson-Gilford progeria syndrome. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobb A.M., Murray T.V., Warren D.T., Liu Y., Shanahan C.M. Disruption of PCNA-lamins A/C interactions by prelamin A induces DNA replication fork stalling. Nucleus. 2016;7:498–511. doi: 10.1080/19491034.2016.1239685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson E., Nievera C.J., Klimovich V., Fanning E., Wu X. RPA2 is a direct downstream target for ATR to regulate the S-phase checkpoint. J. Biol. Chem. 2006;281:39517–39533. doi: 10.1074/jbc.M605121200. [DOI] [PubMed] [Google Scholar]
- 22.Marechal A., Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi W., Feng Z., Zhang J., Gonzalez-Suarez I., Vanderwaal R.P., Wu X., Powell S.N., Roti Roti J.L., Gonzalo S., Zhang J. The role of RPA2 phosphorylation in homologous recombination in response to replication arrest. Carcinogenesis. 2010;31:994–1002. doi: 10.1093/carcin/bgq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhat K.P., Cortez D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018;25:446–453. doi: 10.1038/s41594-018-0075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemacon D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., Vindigni A. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 2017;8:860. doi: 10.1038/s41467-017-01180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zellweger R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neelsen K.J., Lopes M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 31.Quinet A., Lemacon D., Vindigni A. Replication fork reversal: Players and guardians. Mol. Cell. 2017;68:830–833. doi: 10.1016/j.molcel.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taglialatela A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R., Cejka P., Costanzo V., Ciccia A. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell. 2017;68:414–430.e8. doi: 10.1016/j.molcel.2017.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., Lopes M. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017;8:859. doi: 10.1038/s41467-017-01164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolinjivadi A.M., Sannino V., de Antoni A., Técher H., Baldi G., Costanzo V. Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017;591:1083–1100. doi: 10.1002/1873-3468.12556. [DOI] [PubMed] [Google Scholar]
- 35.Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mason J.M., Chan Y.L., Weichselbaum R.W., Bishop D.K. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019;10:4410. doi: 10.1038/s41467-019-12297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angst E., Dawson D.W., Nguyen A., Park J., Go V.L., Reber H.A., Hines O.J., Eibl G. Epigenetic regulation affects N-myc downstream-regulated gene 1 expression indirectly in pancreatic cancer cells. Pancreas. 2010;39:675–679. doi: 10.1097/MPA.0b013e3181c8b476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi T., Makino T., Moran J.V. Poly(ADP-Ribose) polymerase 2 recruits replication protein A to sites of LINE-1 integration to facilitate retrotransposition. Mol. Cell. 2019;75:1286–1298.e12. doi: 10.1016/j.molcel.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 40.Ibler A.E.M., ElGhazaly M., Naylor K.L., Bulgakova N.A., F El-Khamisy S., Humphreys D. Typhoid toxin exhausts the RPA response to DNA replication stress driving senescence and Salmonella infection. Nat. Commun. 2019;10:4040. doi: 10.1038/s41467-019-12064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf C., Rapp A., Berndt N., Staroske W., Schuster M., Dobrick-Mattheuer M., Kretschmer S., König N., Kurth T., Wieczorek D., Kast K., Cardoso M.C., Günther C., Lee-Kirsch M.A. RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat. Commun. 2016;7:11752. doi: 10.1038/ncomms11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B.X., Chen J., Chao B., Zheng Y., Xiao X. A lamin-binding ligand inhibits homologous recombination repair of DNA double-strand breaks. ACS Cent. Sci. 2018;4:1201–1210. doi: 10.1021/acscentsci.8b00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieminuszczy J., Schwab R.A., Niedzwiedz W. The DNA fibre technique - tracking helicases at work. Methods. 2016;108:92–98. doi: 10.1016/j.ymeth.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 44.Ying S., Hamdy F.C., Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- 45.Delamarre A., Barthe A., de la Roche Saint-André C., Luciano P., Forey R., Padioleau I., Skrzypczak M., Ginalski K., Géli V., Pasero P., Lengronne A. MRX increases chromatin accessibility at stalled replication forks to promote nascent DNA resection and Cohesin loading. Mol. Cell. 2020;77:395–410. doi: 10.1016/j.molcel.2019.10.029. [DOI] [PubMed] [Google Scholar]
- 46.Kreienkamp R., Graziano S., Coll-Bonfill N., Bedia-Diaz G., Cybulla E., Vindigni A., Dorsett D., Kubben N., Batista L.F.Z., Gonzalo S. A cell-intrinsic interferon-like response links replication stress to cellular aging caused by progerin. Cell Rep. 2018;22:2006–2015. doi: 10.1016/j.celrep.2018.01.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., John S., Day A., Crespo A.V., Shen B., Starnes L.M. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalo S., Kreienkamp R. DNA repair defects and genome instability in Hutchinson-Gilford progeria Syndrome. Curr. Opin. Cell Biol. 2015;34:75–83. doi: 10.1016/j.ceb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge X.Q., Jackson D.A., Blow J.J. Dormant origins licensed by excess Mcm2 7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor M.R.G., Špírek M., Chaurasiya K.R., Ward J.D., Carzaniga R., Yu X., Egelman E.H., Collinson L.M., Rueda D., Krejci L., Boulton S.J. Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell. 2015;162:271–286. doi: 10.1016/j.cell.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor M.R.G., Špírek M., Jian Ma C., Carzaniga R., Takaki T., Collinson L.M., Greene E.C., Krejci L., Boulton S.J. A polar and nucleotide-dependent mechanism of action for RAD51 paralogs in RAD51 filament remodeling. Mol. Cell. 2016;64:926–939. doi: 10.1016/j.molcel.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soniat M.M., Myler L.R., Kuo H.C., Paull T.T., Finkelstein I.J. RPA phosphorylation inhibits DNA resection. Mol. Cell. 2019;75:145–153. doi: 10.1016/j.molcel.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buisson R., Boisvert J.L., Benes C.H., Zou L. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol. Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S., Opiyo S.O., Manthey K., Glanzer J.G., Ashley A.K., Amerin C., Troksa K., Shrivastav M., Nickoloff J.A., Oakley G.G. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012;40:10780–10794. doi: 10.1093/nar/gks849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy A.K., Fitzgerald M., Ro T., Kim J.H., Rabinowitsch A.I., Chowdhury D., Schildkraut C.L., Borowiec J.A. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 2014;206:493–507. doi: 10.1083/jcb.201404111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar A., Mazzanti M., Mistrik M., Kosar M., Beznoussenko G.V., Mironov A.A., Garrè M., Parazzoli D., Shivashankar G.V., Scita G., Bartek J., Foiani M. ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell. 2014;158:633–646. doi: 10.1016/j.cell.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robison J.G., Elliott J., Dixon K., Oakley G.G. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- 59.Olson E., Nievera C.J., Liu E., Lee A.Y., Chen L., Wu X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol. Cell Biol. 2007;27:6053–6067. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oakley G.G., Tillison K., Opiyo S.A., Glanzer J.G., Horn J.M., Patrick S.M. Physical interaction between replication protein A (RPA) and MRN: Involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry. 2009;48:7473–7481. doi: 10.1021/bi900694p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kusch T. Brca2-Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J. Cell Sci. 2015;128:717–727. doi: 10.1242/jcs.159988. [DOI] [PubMed] [Google Scholar]
- 62.Broers J.L., Ramaekers F.C. The role of the nuclear lamina in cancer and apoptosis. Adv. Exp. Med. Biol. 2014;773:27–48. doi: 10.1007/978-1-4899-8032-8_2. [DOI] [PubMed] [Google Scholar]
- 63.Capo-chichi C.D., Cai K.Q., Simpkins F., Ganjei-Azar P., Godwin A.K., Xu X.X. Nuclear envelope structural defects cause chromosomal numerical instability and aneuploidy in ovarian cancer. BMC Med. 2011;9:28. doi: 10.1186/1741-7015-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wazir U., Ahmed M.H., Bridger J.M., Harvey A., Jiang W.G., Sharma A.K., Mokbel K. The clinicopathological significance of lamin A/C, lamin B1 and lamin B receptor mRNA expression in human breast cancer. Cell Mol. Biol. Lett. 2013;18:595–611. doi: 10.2478/s11658-013-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsumoto A., Hieda M., Yokoyama Y., Nishioka Y., Yoshidome K., Tsujimoto M., Matsuura N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN1, SUN2, and nesprin-2 in breast cancer. Cancer Med. 2015;4:1547–1557. doi: 10.1002/cam4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alhudiri I.M., Nolan C.C., Ellis I.O., Elzagheid A., Rakha E.A., Green A.R., Chapman C.J. Expression of Lamin A/C in early-stage breast cancer and its prognostic value. Breast Cancer Res. Treat. 2019;174:661–668. doi: 10.1007/s10549-018-05092-w. [DOI] [PubMed] [Google Scholar]
- 67.Gesson K., Rescheneder P., Skoruppa M.P., von Haeseler A., Dechat T., Foisner R. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 2016;26:462–473. doi: 10.1101/gr.196220.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson B.R., Nitta R.T., Frock R.L., Mounkes L., Barbie D.A., Stewart C.L., Harlow E., Kennedy B.K. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9677–9682. doi: 10.1073/pnas.0403250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., Marino F., Lucic B., Biasin V., Gstaiger M., Aebersold R. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sirbu B.M., Couch F.B., Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olive P.L., Banath J.P., Durand R.E. Detection of etoposide resistance by measuring DNA damage in individual Chinese hamster cells. J. Natl. Cancer Inst. 1990;82:779–783. doi: 10.1093/jnci/82.9.779. [DOI] [PubMed] [Google Scholar]
- 72.Neelsen K.J., Chaudhuri A.R., Follonier C., Herrador R., Lopes M. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol. Biol. 2014;1094:177–208. doi: 10.1007/978-1-62703-706-8_15. [DOI] [PubMed] [Google Scholar]
- 73.Binz S.K., Dickson A.M., Haring S.J., Wold M.S. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.