Abstract
Due to the similar clinical symptoms of influenza (Flu) and coronavirus disease 2019 (COVID-19), there is a looming infection threat of concurrent Flu viruses and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In this work, we introduce a customized isothermal amplification integrated lateral flow strip (LFS) that is capable performing duplex reverse transcription–recombinase polymerase amplification (RT-RPA) and colorimetric LFS in a sequential manner. With customized amplification primer sets targeted to SARS-CoV-2 (opening reading frame 1a/b and nucleoprotein genes) and Flu viruses (Flu A and Flu B), the platform allows the rapid and simultaneous visual screening of SARS-CoV-2 and Flu viruses (Flu A and Flu B) without cross reactivity, false positives, and false negatives. Moreover, it maximally eases the detection, reduces the detection time (1 h), and improves the assay performance to detect as low as 10 copies of the viral RNA. Its clinical application is powerfully demonstrated with 100% accuracy for evaluating 15 SARS-CoV-2-positive clinical samples, 10 Flu viruses-positive clinical samples, and 5 negative clinical samples, which were pre-confirmed by standard qRT-PCR. We envision this portable device can meet the increasing need of online monitoring the serious infectious diseases that substantially affects health care systems worldwide.
Keywords: SARS-CoV-2, Influenza virus, Recombinase polymerase amplification, Lateral flow strip, Simultaneous identification
1. Introduction
Over the past several decades, there have been recurrent large-scale epidemics from emerging viruses such as the pandemic influenza (Flu) viruses and most recently severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Faust and del Rio, 2020). The Flu caused by the spread of the Flu A and Flu B viruses is a common, seasonal, and acute infectious respiratory disease in humans, which causes hundreds of thousands of deaths each year (Krammer et al., 2018). The coronavirus disease 2019 (COVID-19) pandemic caused by the SARS-CoV-2 in late 2019 is a highly infectious respiratory disease with unclear sources (Wang et al., 2020a). Its outbreak in the past one year has caused with over 70 million infected cases and 1.6 million deaths worldwide. More disturbing is that the SARS-CoV-2 mimics the influenza viruses regarding clinical presentation of fever, cough, and fatigue or myalgia, transmission mechanism, and seasonal coincidence (Sheikhzadeh et al., 2020; Wang et al., 2020a). Without timely detection kits, the two respiratory diseases might be misdiagnosed, greatly increasing the risk of epidemic prevention and control, and in turn causing a panic in publics since confluence of COVID-19 and seasonal Flu can result in considerable morbidity and mortality. Some officers, physicians, and researchers have announced the health system and wider society must prepare for the likelihood of co-epidemics of COVID-19 and Flu (Hashemi et al., 2020; Solomon et al., 2020). Accurate and fast diagnostic tests to identify and isolate infected individuals in time seems to be the only way for tracking and controlling the epidemic.
Generally, quantitative RT-PCR (qRT-PCR) is the standard method for virus detection and identification in center laboratories (Ben-Ami et al., 2020; Han et al., 2020). However, it heavily relies on trained technicians and thermal cycling controller, ruling out its application in resource-limited areas. Moreover, the whole detection usually needs 4–6 h, which is difficult to detect bulk samples timely. To simplify and accelerate clinical diagnostic testing, alternative molecular testing strategies such as reverse transcription loop-mediated isothermal amplification (RT-LAMP) (Dao Thi et al., 2020; Ganguli et al., 2020; Wang et al., 2021) and clustered regularly interspaced short palindromic repeats (CRISPR)-assisted diagnosis tests (Abbott et al., 2020; Broughton et al., 2020; Pang et al., 2020) enable equipment-free, rapid, and sensitive detection of viruses. Unfortunately, the requirement of multiple primers of RT-LAMP increases the risk of false positives especially facing with multiple targets, and CRISPR-based methods needs labor-intensive operations. There are still rooms for improvement of the nucleic acids-based detection methods. As a supplement of molecular diagnosis, immunological and serological assays such as chemiluminescent immunoassay (Zhang et al., 2020), enzyme linked immunosorbent assay (ELISA) (Cai et al., 2020; Liu et al., 2020; Xiang et al., 2020) and lateral flow immunoassay (Alves et al., 2020; Feng et al., 2020; Pan et al., 2020) can rapid and specific screening of certain viruses. However, their limited sensitivity hinders their applications for timely diagnosing diseases because it at least takes several days following symptom onset for a patient to mount a detectable antibody response. More than that, when faced with SARS-CoV-2 and Flu viruses, especially Flu A and Flu B, a significant gap in existing detection methods is that these assays were only focus on the detecting of SARS-CoV-2 or Flu viruses. The importance for simultaneous detection of them should be highlighted with the looming infection threat of concurrent Flu viruses and SARS-CoV-2.
The core and direct clinical proof for detecting viruses still focuses on the nucleic acid detection. As one of the most promising isothermal molecular amplification tools, recombinase polymerase amplification (RPA) shows appealing features of operation simplicity, detection rapidness, and high sensitivity for detecting DNA or RNA from various pathogens at a constant temperate with 30 min (Ahn et al., 2018; Choi et al., 2016; Crannell et al., 2014; Hice et al., 2019; Khater et al., 2019; Lobato and O'Sullivan, 2018; Piepenburg et al., 2006; Yang et al., 2020). Lateral flow strip (LFS) is a paper-based device which permits the performance of simplicity, rapidness, portability, cost-effectiveness, specificity, sensitivity, and low limit of detection for on-site detecting trace analytes (Chen et al., 2020; He et al., 2018; Liu et al., 2017; Qin et al., 2021; Wang et al., 2020b, 2020c; Wu et al., 2019; Yao et al., 2016; You et al., 2017; Zheng et al., 2016). Integration of RPA with LFS can combine their merits together for nucleic acid analysis. Taken above into consideration, here we report the development and initial validation of a customized isothermal amplification integrated lateral flow strip platform, called duplex reverse transcription-RPA based LFS (RT-RPA-LFS), for rapid and simultaneous visual screening of SARS-CoV-2 and Flu viruses. To realize this goal, two pairs of functionalized primer sets were rationally designed for targeting SARS-CoV-2 (ORF1ab and N genes) and Flu viruses (Flu A and Flu B), respectively. We demonstrated that the testing platform can accurately identify SARS-CoV-2, Flu A, and Flu B with no cross-reactivity, no false positives, and no false negatives. To the best of our knowledge, this is the first report in developing such an assay to identify two types of the similar but pathogenic and mortal viruses, which is definitely useful for disease diagnosis, epidemic prevention and control, and in-depth epidemiological analysis.
2. Material and methods
2.1. Chemicals and apparatus
A XYZ 3000 spraying machine and a Guillotine Cutting Module CM4000 were obtained from Bio-Dot Inc. (Irvine, CA, USA). A S1000 Thermal Controller (Bio-Rad, Hercules, CA, USA) was used for the isothermal amplification. A SLA-32 Automatic Nucleic Acid Extractor (TANBead Taiwan dot Nanometer Technology Co., LTD.) was employed to extract total genomic genes in a second-level bio-safety laboratory. The one-step RT-RPA kit was from the GenDx Biotech (Suzhou, China). The fluorescein isothiocyanate (FITC) antibody and goat-anti-mouse secondary antibody were from Sino-pharm Chemical (Wuhan, China). RNase inhibitor (40 U/μL), 4 S Red Plus dye (1000 × ), DNA marker, streptavidin, 20 × PBS buffer (pH 7.4), 5 × TBE buffer, and agarose were from Sangon Biotech (Shanghai, China). N–N-hydroxysuccinimide (NHS) and N-(3-dimethylaminopropyl)-Nethylcarbodiimide hydrochloride (EDC) were from Sigma Chemical Company (St. Louis, MO, USA). Bovine serum albumin (BSA) was from Bio-Dee Biotech. Co. Ltd. (Beijing, China). Red carboxyl modified latex microspheres with a diameter of 100 nm were from the Changsha Made New Biotechnology Co., Ltd. (Changsha, China). Backing pads, nitrocellulose (NC) membranes, sample pads, conjugate pads, and absorbent pads were from the Shanghai Jenin Biotechnology Co., Ltd. (Shanghai, China). All other common chemical reagents were at an analytical grade and supplied by Sinopharm Chemical Reagent Co., Ltd. (Wuhan, China). They can be directly used without further purification. Four kinds of plasmids containing cDNA fragments of ORF1ab gene, N gene, Flu A gene, and Flu B gene, respectively, were prepared by Sangon Biotech (Shanghai, China). Specific primers for SARS-CoV-2 and Flu viruses (Flu A and Flu B) were designed using the software of DNAStarLasergenev7.1. Each primer sets were further pre-confirmed in the Gene Bank. All oligonucleotide sequences (Table S2) were synthesized by General Biosystems (Anhui, China). DEPC treated water (>18 MΩ) was used throughout the research. All clinical samples were collected and stored in the Center for Disease Control and Prevention of Anhui Province (CDC of Anhui). And all clinical verification research have also been approved by the ethics committee of CDC.
2.2. Preparation of FITC antibody labeled latex microsphere conjugates (microlatex-Ab)
To prepare the microlatex-Ab, 10 μL of 2% (w/v) carboxyl modified latex microspheres were activated by reacting with 100 μg EDC and 100 μg NHS for 1 h in 1 mL borate buffer (10 mM H3BO3, pH 8.0). Afterwards, the mixture was added with 4 μL of FITC antibody (1 mg/mL) and gently shaken for 3 h. This is followed by further adding 100 μL of 10% (w/v) BSA solution to block the residual active sites on the surface of latex microspheres. Finally, microlatex-Ab conjugates were obtained by centrifuging above solution at 13,300 rpm for 20 min. The precipitates were resuspended in 100 μL 10% (w/v) BSA solution for further use.
2.3. Fabrication of LFS
The LFS was fabricated according to our previous methods with minor modifications (Qin et al., 2019; Zheng et al., 2018). Typically, the LFS is consists of five components including a sample pad for RT-RPA amplicons loading, a conjugate pad for microlatex-Ab loading, a NC membrane for anchoring two test lines (T1 line and T2 line) and one control (C) line, an absorbent pad for providing the driving force, and a backing pad for supporting all other components. Before assembling, the sample pad and conjugate pad were pre-treated with buffer I (50 mM Tris-HCl, 0.15 mM NaCl and 0.25% Tritonx-100, pH 8.0) and buffer II (10 mM PB, 5% sucrose, 1% trehalose, 0.25% PEG, 20000, and 0.3% Tween 20, pH 8.0), respectively. Then, 5 μL of the microlatex-Ab conjugates were dropped onto the conjugate pad. On the NC membrane, two kinds of biotinylated capture probes (CP1 and CP2) combined with streptavidin were sprayed as T1 line and T2 line, while the secondary antibodies were immobilized as the C line. After drying at 25 °C for 6 h, the sample pad, conjugate pad, NC membrane, and absorbent pad were properly assembled on the backing card.
2.4. Duplex RT-RPA based LFS detection
The RNA templates of SARS-CoV-2, Flu A, and Flu B were directly extracted using an Automatic Nucleic Acid Extractor. The duplex RT-RPA responsible for SARS-CoV-2 (ORF1ab and N genes) was performed in 48 μL reaction mixture containing 20 μL dissolving buffer, 10 U RNase inhibitor, 200 nM ORF1ab-FP/ORF1ab-RP, 200 nM N-FP/N-R, 2 μL RNA sample, and a certain amount of dry powder (RPA Kit). Then, 2 μL of Mg(OAc)2 solution was added to activate the reaction and the mixture was incubated at 39 °C for 25 min. Of note, the RPA kit enables the steps of reverse transcription and isothermal amplification run in parallel. For the detection of Flu A and Flu B, the amplification primers of SARS-CoV-2 were replaced by 240 nM Flu A-FP/Flu A-RP and 240 nM Flu B-FP/Flu B-RP. Other conditions are identically performed. For LFS detection, 5 μL of the amplification products were diluted with 35 μL 1 × PBS and then dropped onto the sample pad of the LFS. The signals of the T lines and C line were visualized after 3 min and further analyzed with ImageJ (NIH, US) for quantitative analysis.
3. Results and discussion
3.1. Working principle of the duplex RT-RPA-LFS strategy
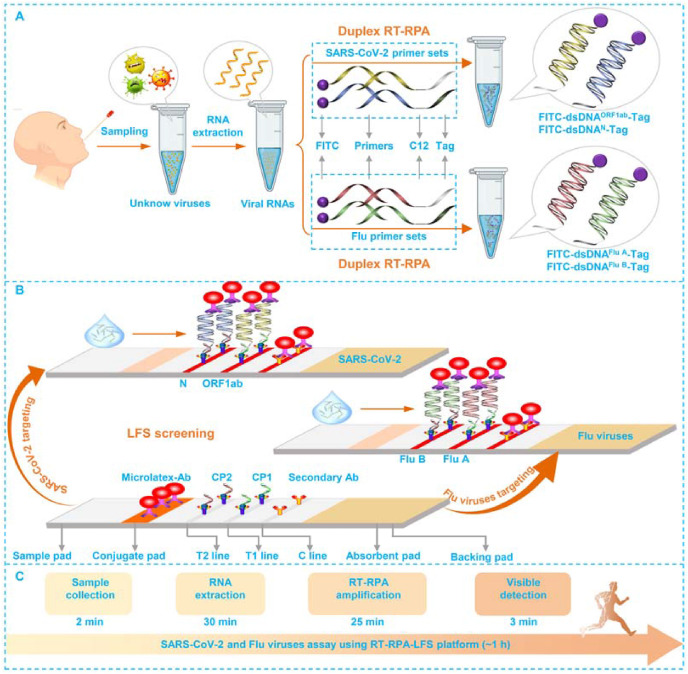
The workflow of this duplex RT-RPA-LFS and the structural analysis of LFS are given in Fig. 1 . For targeting SARS-COV-2, both the 5′ end of ORF1ab-FP and N-FP are modified with FITC, while the 5′-end of ORF1ab-RP and N-RP are labeled with different tag sequences and C12 spacer. The modification of FITC in ORF1ab-FP and N-FP is to facilitate the amplification products to interact with microlatex-Ab on the conjugate pad. The tag sequences of ORF1ab-RP and N-RP are designed to complementary with CP1 and CP2 pre-immobilized on T1 line and T2 line, respectively. The C12 spacer in ORF1ab-RP and N-RP is to block the polymerization on tag sequences. Once presented with SARS-CoV-2 in tested swab samples, the extracted RNA templates are converted to a mass of FITC and Tag dual-labeled double-stranded DNA (dsDNA) amplicons (FITC-dsDNAORF-Tag and FITC-dsDNAN-Tag) via the one-step duplex RT-RPA (Fig. 1A). When loaded onto the LFS, the capillary attraction force would drive the FITC-dsDNAORF-Tag and FITC-dsDNAN-Tag to firstly combine with microlatex-Ab via immune recognition and then be entrapped by CP1 on T1 line and CP2 on T2 line via base pairings. Overused microlatex-Ab conjugates were immobilized by secondary bodies on C line via immune recognition (Fig. 1B).
Fig. 1.
Duplex RT-RPA-LFS based viral assay for SARS-Cov-2 and Flu viruses.
The duplex RT-RPA for targeting of Flu A and Flu B is similarly conducted but with the using another primer couples of Flu A-FP/Flu A-RP and Flu B-FP/Flu B-RP to amplify the genes of Flu A and Flu B, respectively. The labeling of Flu A-FP/Flu A-RP and Flu B-FP/Flu B-RP is referenced to those primers for SARS-CoV-2. The subsequent LFS screenings are identically performed.
At this point, we would like to highlight the simplicity, rapidness, and robustness of our detection method as follows: First, the isothermal amplification can be operated without needing of professional technicians. The sample loading onto LFS associated with the subsequent signal explanation are much easier. This is what we called simplicity. Second, the clinical sample collection is about 2 min, the RNA extraction takes about 30 min, the duplex RT-RPA-LFS is about 25 min, and the visible detection on a LFS is 3 min (Fig. 1C). This means the whole detection processes is only about 1 h, which is far below than the required detection of common qRT-PCR (4–6 h). Besides, for nucleic acid analysis, this constructed RT-RPA-LFS platform is also comparable to other intensive optimized real time PCR similar protocols with amplification time of 45 min with the help of professional devices (Cepheid, 2012; WHO, 2020). Frankly speaking, compared with other excellent professional rapid COVID-19 diagnosis station (Abbott, 2020; VitaPCR, 2020; Panels BioFire, 2021; Taleghani and Tghipour, 2021) which usually take about 5–45 min for the whole detection of SARS-CoV-2, our constructed RT-RPA-LFS still needs to be further optimized and integrated with other advanced technologies such as microfluidic and isothermal amplification protocols et al. to improve the amplification efficiency and further shorten the consumption of time. Third, unlike reported cases in detecting SARS-CoV-2 or influenza viruses, our study is the first attempt for simultaneously targeting SARS-CoV-2, Flu A, and Flu B, which will play a very useful role for us to tackle with the threat of concurrent influenza and COVID-19 epidemics. This is what we called robustness of the platform.
3.2. Feasibility demonstration for simultaneous identification
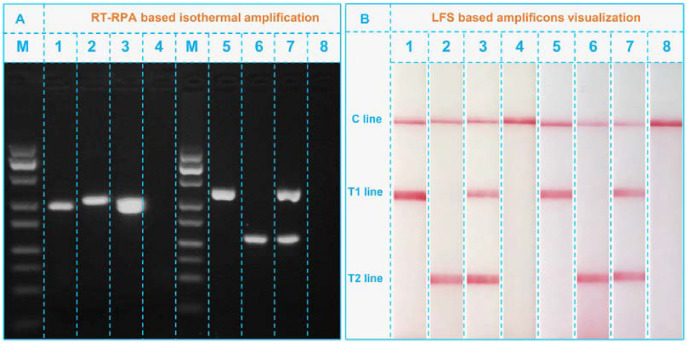
To confirm the feasibility of for simultaneous detecting SARS-CoV-2 and Flu viruses, the amplification products of SARS-CoV-2 and Flu viruses were first visually characterized using gel electrophoresis. As can be seen from Fig. 2 A, the RNA templates of SARS-CoV-2 amplified with ORF1ab-FP/ORF1ab-RP (lane 1) and N-FP/N-RP (lane 2) showed obvious bands of their respective amplicons of FITC-dsDNAORF1ab-Tag (lane 1) and FITC-dsDNAN-Tag (lane 2). Simultaneous using of ORF1ab-FP/ORF1ab-RP and N-FP/N-RP (lane 3) to amplify the SARS-CoV-2 template appeared a wide band with location match well with those bands in lane a and b, indicating the successful duplex RT-RPA. In contrast, the amplifying blank control with ORF1ab-FP/ORF1ab-RP and N-FP/N-RP showed no bands in lane 4, suggesting no false positive results by this amplification. Similarly, the RNA templates from Flu viruses were also successfully amplified with customized Flu A-FP/Flu A-RP, Flu B-FP/Flu B-RP and their mixture in lane 5 (FITC-dsDNAFlu A-Tag), 6 (FITC-dsDNAFlu B-Tag), and 7 (FITC-dsDNAFlu A-Tag and FITC-dsDNAFlu B-Tag), respectively. Lane 8 showed no bands in the absence of Flu virus templates was also evidence of no false positives. One thing needed to be clarified is that the concentrations of the primer set of critical importance for the performance of the constructed RT-RPA-LFS. Firstly, the concentration of the primer set will directly determine the amplification efficiency of the system and also influence the final sensitivity of the platform. Secondly, improper concentration of primer set can induce the presence of dimer of the amplification system, which will further influence the specificity of the lateral flow strip according to our previous research (Qin et al., 2019).
Fig. 2.
(A) Gel electrophoresis analysis of the duplex RT-RPA amplicons. M, DNA marker; lanes 1–3, SARS-CoV-2 RNA templates amplified with ORF1ab-FP/ORF1ab-RP, N-FP/N-RP, and the mixture of ORF1ab-FP/ORF1ab-RP and N-FP/N-RP, respectively; lane 4, blank control amplified with ORF1ab-FP/ORF1ab-RP and N-FP/N-RP; lanes 5–7, Flu RNA templates amplified with Flu A-FP/Flu A-RP, Flu B-FP/Flu B-RP, and the mixture of Flu A-FP/Flu A-RP and Flu B-FP/Flu B-RP, respectively; lane 8, blank control amplified with Flu A-FP/Flu A-RP and Flu B-FP/Flu B-RP. (B) LFS analysis of the RPA amplicons: strips 1–8, FITC-dsDNAORF1ab-Tag, FITC-dsDNAN-Tag, FITC-dsDNAORF1ab-Tag/FITC-dsDNAN-Tag, blank, FITC-dsDNAFlu A-Tag, FITC-dsDNAFlu B-Tag, FITC-dsDNAFlu A-Tag/FITC-dsDNAFlu B-Tag, blank.
Then, on this basis, all amplicons were used to explore the performance of LFS. As depicted in Fig. 2B, the adding of FITC-dsDNAORF1ab-Tag only led to the appearance of T1 line in strip 1. T2 line cannot be visualized under this condition. Likewise, amplicons of FITC-dsDNAN-Tag can only make T2 line appeared in strip 2. This lay the foundation for duplex tests of ORF1ab and N gene on LFS. As expected, when they were simultaneously loaded onto a LFS (strip 3), T1 line and T2 line co-appeared. Absence of them in strip 4 cannot cause any signals. Strips 5 to 8 are the results of Flu viruses. Since they showed the same phenomenon as that of strips 1 to 4, the LFS is thus able to identify Flu A and Flu B. For the results of strip 4 and strip 8 in Fig. 2B, there is not any observable signal on different T1 and T2 lines, indicating the negative results of both SARS-CoV-2 and common Flu A & B. Taken the results of gel electrophoresis and LFS together, we can confirm the availability of our duplex RT-RPA-LFS platform for simultaneous visual screening of SARS-CoV-2 and common influenza viruses. Meanwhile, the better differentiation properties or the specificity of the detection with this platform can be ascribed to two aspects: (1) the special designed primer sets of RPA can guarantee the specific amplification of the target templates including the SRAS-CoV-2 and Flu A & B; (2) the optimized conditions of LFS can ensure that only the target amplicons can be visually measured while the negative or blank control groups cannot.
3.3. Analytical performance investigation
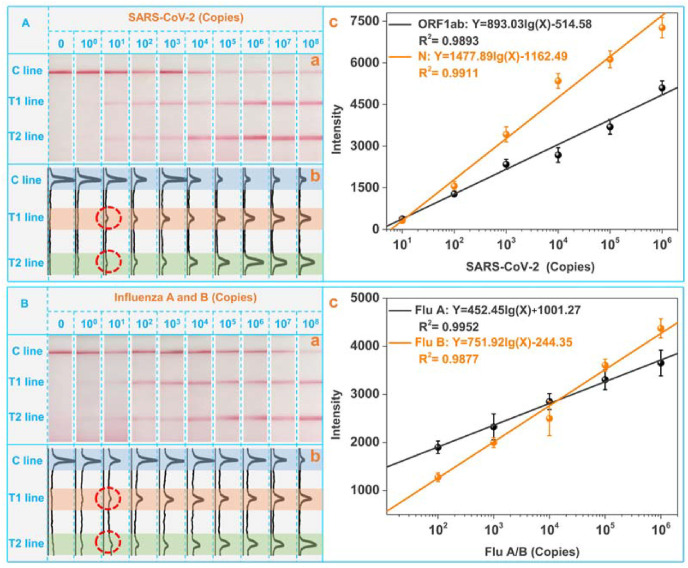
To determine the analytical performance, we tested the duplex RT-RPA-LFS platform with varied amounts of SARS-CoV-2 and influenza viruses templates produced by customized plasmids. As shown in panel a and panel b of Fig. 3 A, both the intensities of T1 line and T2 line were gradually increased with the increasing of SARS-CoV-2 templates from 101 to 108 copies. Panel c plotted the calibration curves for ORF1ab gene and N gene, which are fitted to be Y = 893.03lg(X)-514.58 and Y = 1477.89lg(X)-1162.49 respectively. Y and X represent the T line intensity and virus RNA copy, respectively. One can find that as low as 101 copies of the SRAS-CoV-2 templates can be easily distinguished from the blank control. Fig. 3B displayed the visual detection results of the LFS against Flu viruses with different copies. Intensified T1 line and T2 line also suggest the capabilities to quantify Flu A and Flu B with a lower detection limit of 101 copies. The calibration curves for Flu A and Flu B are Y = 452.45lg(X)+1001.27 and Y = 751.92lg(X)-244.35 respectively. Of note, the quantitative analysis of the detection result is based on the optical intensities of test lines in the images of LFS by the professional software of ImageJ. For the images taken at different batches, the ambient lightness will definitely influence the calibration curves of different targets. In the practical applications, this influence can be well avoided by two strategies: (1) the optical intensities of the test lines can be collected with the developed apps, which can be integrated with the standard color card. The optical signals of color card will be collected together with the LFS. The influence of the ambient lightness to the optical signals of LFS can be modulated based on the optical signals of standard color cards. (2) The optical signals on the LFS can be measured by the portable reader device. All images and optical signals were collected under the consistent conditions of ambient lightness in the portable reader. With these two potential strategies, the standard deviations and errors of the detection can be well guaranteed for the future practical on-site applications.
Fig. 3.
Qualitative analysis of SARS-CoV-2 (A) and Flu A and B (B) at varied copies. From left to right: 0, 100, 101, 102, 103, 104, 105, 106, 107, and 108 copies. Panel a is the LFS results, panel b is the corresponding image obtained by “Image J″, and panel c provides the fitted equations.
Comparison of the current study with reported methods for SARS-CoV-2 and Flu viruses detection is summarized in Table S1. From the table, we find that most of the reported works are merely detect SARS-CoV-2 or Flu viruses. Our work is the first one to simultaneously identify SARS-CoV-2 and Flu viruses using a same platform. Furthermore, our work is superior in terms of the items including detection speed, operation simplicity, and detection range.
3.4. Application of the strategy in clinical samples
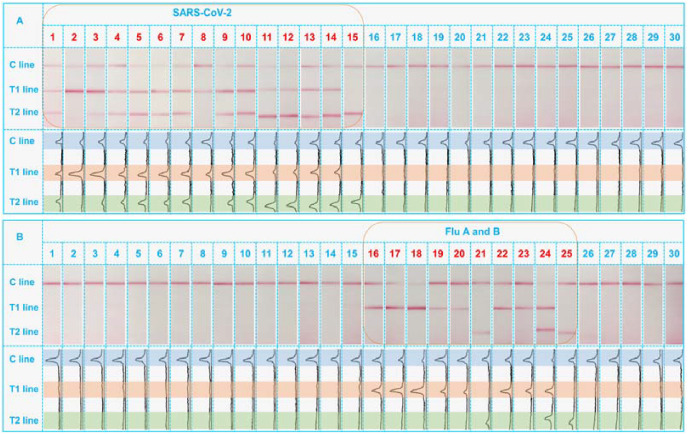
After verified with desirable performance, the practicability of the platform was finally considered by detecting 30 clinical samples, which were initially analyzed using standard qRT-PCR in our center (Anhui Provincial Center for Disease Control and Prevention) to confirm they are consist of 15 SRAS-CoV-2 positive samples, 10 Flu viruses positive samples, and 5 negative samples. The corresponding identification results by the duplex RT-RPA-LFS are collected in Fig. 4 . One can see in Fig. 4A that the use of primer sets of ORF1ab-FP/ORF1ab-RP and N-FR/N-RP to amplify the SARS-CoV-2 specimens resulted in either the co-appearance of two lines (12 samples) or appearance one of the T line (3 samples) on strips 1 to 15. No visible signals can be seen from influenza virus positive samples (strips 16 to 25) and negative samples (strips 26 to 30). In Fig. 4B, instead of giving positive signals in strips 1 to 15, the use of Flu A-FP/Flu A-RP and Flu B-FP/Flu B-RP primers made the visualization of Flu A and Flu B on strips 16 to 25. Among the Flu virus positive samples, 7 samples are Flu A positive, 2 samples are Flu B positive, and 1 sample are co-infected with Flu A and Flu B. There are not any signals on both T1 and T2 lines for other negative samples. The different comparison results about these 30 clinical samples demonstrate a 100% consistency with the RT-PCR for practical simultaneous identification of SRAS-CoV-2 and Flu viruses. Besides, more clinical samples (104 samples) including the negative samples, positive sample of Flu A, Flu B and COVID-19 had also been adopted and tested for the accuracy analysis. Results demonstrated that the accuracy of the detection can be as high as 99.04% (only 1 false negative of RT-RPA-LFS in 89 RT-PCR positive samples and 15 negative samples) with the false positive rate of 0% and the false negative rate of 1.11%. The stability of the constructed RT-RPA-LFS platform can be considered from the two aspects of RPA and LFS, respectively. The RPA reagents were freeze-dried and can be stored with comparable performance as new kits for at least one year. The stability of LFS have also be studies in our lab, which can maintain the good detection results after stored for half a year under ambient conditions (Wu et al., 2020). Therefore, the stability of this RT-RPA-LFS can be ensured for at least for half a year.
Fig. 4.
Real clinical samples analysis towards SARS-CoV-2 positive, Flu viruses positive, and negative samples.
4. Conclusions
In conclusion, we have presented a proof of concept and diagnostic performance of a duplex RT-RPA-LFS assay for simultaneously detection and identification of SARS-CoV-2 and Flu viruses from clinical patient samples. This platform skillfully employs duplex RT-RPA to amplify the virus RNA templates and use LFS to give visible results. This user-friendly model thus strengthens the operation simplicity, detection speed, and performance robustness. As low as 101 copy of each of tested virus gene can be visualized within 1 h. The inspiring result is that using this platform, we checked 30 infected samples from SARS-CoV-2, Flu A, and Flu B positive clinical patients with 100% accuracy. Moreover, it can be also explored for rapid diagnosis of other infectious diseases with simply modification of the amplification primer sets. As such, these will benefit public health and prosperity by fast diseases diagnosing to reduce the burden of viral disease.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by the grants of the NSFC (32172295, 21804028), Ministry of Science and Technology of China (2018YFC1603606), the key R&D program of Anhui (202004a07020002, 202004a07020004, 201904d07020016), the Anhui Provincial NSF (1908085QC121), the China Postdoctoral Science Foundation (2019M652167), the Fund of State Key Lab of Chemo/Biosensing and Chemometrics (Hunan University), the postdoc grant of Anhui (2020B412) the Young and Middle-aged Leading Scientists, Engineers and Innovators of the XPCC (No. 2019CB017) and China Agriculture Research System-48 (CARS-48).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113771.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abbott I.D. 2020. NOWTM COVID-19.https://www.globalpointofcare.abbott/en/product-details/id-now-covid-19.html [Google Scholar]
- Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., Pande T., Endy D., La Russa M.F., Lewis D.B., Qi L.S. Cell. 2020;181:865–876.e812. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn H., Batule B.S., Seok Y., Kim M.G. Anal. Chem. 2018;90:10211–10216. doi: 10.1021/acs.analchem.8b01309. [DOI] [PubMed] [Google Scholar]
- Alves D., Curvello R., Henderson E., Kesarwani V., Walker J.A., Leguizamon S.C., McLiesh H., Raghuwanshi V.S., Samadian H., Wood E.M., McQuilten Z.K., Graham M., Wieringa M., Korman T.M., Scott T.F., Banaszak Holl M.M., Garnier G., Corrie S.R. ACS Sens. 2020;5:2596–2603. doi: 10.1021/acssensors.0c01050. [DOI] [PubMed] [Google Scholar]
- Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M., Meshorer E., Benedek G., Fogel I., Oiknine-Djian E., Gertler A., Rotstein Z., Lavi B., Dor Y., Wolf D.G., Salton M., Drier Y., Klochendler A., Eden A., Klar A., Geldman A., Arbel A., Peretz A., Shalom B., Ochana B.L., Avrahami-Tzfati D., Neiman D., Steinberg D., Ben Zvi D., Shpigel E., Atlan G., Klein H., Chekroun H., Shani H., Hazan I., Ansari I., Magenheim I., Moss J., Magenheim J., Peretz L., Feigin L., Saraby M., Sherman M., Bentata M., Avital M., Kott M., Peyser M., Weitz M., Shacham M., Grunewald M., Sasson N., Wallis N., Azazmeh N., Tzarum N., Fridlich O., Sher R., Condiotti R., Refaeli R., Ben Ami R., Zaken-Gallili R., Helman R., Ofek S., Tzaban S., Piyanzin S., Anzi S., Dagan S., Lilenthal S., Sido T., Licht T., Friehmann T., Kaufman Y., Pery A., Saada A., Dekel A., Yeffet A., Shaag A., Michael-Gayego A., Shay E., Arbib E., Onallah H., Ben-Meir K., Levinzon L., Cohen-Daniel L., Natan L., Hamdan M., Rivkin M., Shwieki M., Vorontsov O., Barsuk R., Abramovitch R., Gutorov R., Sirhan S., Abdeen S., Yachnin Y., Daitch Y. Clin. Microbiol. Infect. 2020;26:1248–1253. doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.F., Chen J., li Hu J., Long Q.X., Deng H.J., Liu P., Fan K., Liao P., Liu B.Z., Wu G.C., Chen Y.K., Li Z.J., Wang K., Zhang X.L., Tian W.G., Xiang J.L., Du H.X., Wang J., Hu Y., Tang N., Lin Y., Ren J.H., Huang L.Y., Wei J., Gan C.Y., Chen Y.M., Gao Q.Z., Chen A.M., He C.L., Wang D.X., Hu P., Zhou F.C., Huang A.L., Wang D.Q. J. Infect. Dis. 2020;222:189–193. doi: 10.1093/infdis/jiaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cephid | GeneXpert family of system, https://www. cepheid.com/en_US/systems/GeneXpert-Family-of-Systems, (accessed January 29, 2021).
- Chen W., Cai F., Wu Q., Wu Y., Yao B., Xu J. Food Chem. 2020;328:127081. doi: 10.1016/j.foodchem.2020.127081. [DOI] [PubMed] [Google Scholar]
- Choi G., Jung J.H., Park B.H., Oh S.J., Seo J.H., Choi J.S., Kim D.H., Seo T.S. Lab Chip. 2016;16:2309–2316. doi: 10.1039/c6lc00329j. [DOI] [PubMed] [Google Scholar]
- Crannell Z.A., Rohrman B., Richards-Kortum R. Anal. Chem. 2014;86:5615–5619. doi: 10.1021/ac5011298. [DOI] [PubMed] [Google Scholar]
- Dao Thi V.L., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Barreto Miranda I., Schnitzler P., Kräusslich H.G., Knop M., Anders S. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J.S., del Rio C. JAMA Intern. Med. 2020;180:1045–1046. doi: 10.1001/jamainternmed.2020.2306. [DOI] [PubMed] [Google Scholar]
- Feng M., Chen J., Xun J., Dai R., Zhao W., Lu H., Xu J., Chen L., Sui G., Cheng X. ACS Sens. 2020;5:2331–2337. doi: 10.1021/acssensors.0c00927. [DOI] [PubMed] [Google Scholar]
- Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., Ramirez S.A.S.D., Valera E., Cunningham B.T., King W.P., Bashir R. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Byun J.H., Cho Y., Rim J.H. Lancet Infect. Dis. 2020;21:165. doi: 10.1016/S1473-3099(20)30424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi S.A., Safamanesh S., Ghasemzadeh-moghaddam H., Ghafouri M., Azimian A. J. Med. Virol. 2021;93:1008–1012. doi: 10.1002/jmv.26364. [DOI] [PubMed] [Google Scholar]
- He H., Liu B., Wen S., Liao J., Lin G., Zhou J., Jin D. Anal. Chem. 2018;90:12356–12360. doi: 10.1021/acs.analchem.8b04330. [DOI] [PubMed] [Google Scholar]
- Hice S.A., Clark K.D., Anderson J.L., Brehm-Stecher B.F. Anal. Chem. 2019;91:1113–1120. doi: 10.1021/acs.analchem.8b04751. [DOI] [PubMed] [Google Scholar]
- Khater M., Escosura-Muñiz A.d.l., Altet L., Merkoçi A. Anal. Chem. 2019;91:4790–4796. doi: 10.1021/acs.analchem.9b00340. [DOI] [PubMed] [Google Scholar]
- Krammer F., Smith G.J.D., Fouchier R.A.M., Peiris M., Kedzierska K., Doherty P.C., Palese P., Shaw M.L., Treanor J., Webster R.G., García-Sastre A. Nat. Rev. Dis. Primers. 2018;4:3. doi: 10.1038/s41572-018-0002-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zhang M., Liu X., Sharma A., Ding X. Biosens. Bioelectron. 2017;96:213–219. doi: 10.1016/j.bios.2017.04.047. [DOI] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461-00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato I.M., O'Sullivan C.K. TrAC Trends Anal. Chem. (Reference Ed.) 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., Long X., Guo S., Zhao Z., Liu Y., Hu H., Xue H., Li Y. J. Infect. 2020;81:e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panels BioFire® FilmArray® Respiratory Panels, 2021. https://www.biofiredx.com/products/the-filmarray-panels/filmarrayrp/, (accessed January 29, 2021).
- Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., Wu J., Xiao H., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H., Le X.C. Anal. Chem. 2020;92:16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Qiao D., Xu J., Song Q., Yao L., Lu J., Chen W. Food Chem. 2019;294:224–230. doi: 10.1016/j.foodchem.2019.05.030. [DOI] [PubMed] [Google Scholar]
- Qin P., Xu J., Yao L., Wu Q., Yan C., Lu J., Yao B., Liu G., Chen W. Food Chem. 2021;339:127891. doi: 10.1016/j.foodchem.2020.127891. [DOI] [PubMed] [Google Scholar]
- Sheikhzadeh E., Eissa S., Ismail A., Zourob M. Talanta. 2020;220:121392. doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon D.A., Sherman A.C., Kanjilal S. J. Am. Med. Assoc. 2020;324:1342–1343. doi: 10.1001/jama.2020.14661. [DOI] [PubMed] [Google Scholar]
- Taleghani N., Taghipour F. Biosens. Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VitaPCR VitaPCR Platform Features, 2020. ht t p s ://w w w. menarinidiagnostics.com/en-us/Home/Laboratory-products/COVID-19/VitaPCRTM-platform/Features, (accessed October 28, 2020).
- Wang C., Horby P.W., Hayden F.G., Gao G.F. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L., Zhang J., Zhang L., Hu X., Zheng X., Bai J., Zhang Y., Zhang Y., Song M., Tang Y. Nat. Biomed. Eng. 2020;4:1150–1158. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- Wang L., Shen X., Wang T., Chen P., Qi N., Yin B.C., Ye B.C. Biosens. Bioelectron. 2020;165:112364. doi: 10.1016/j.bios.2020.112364. [DOI] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. Biosens. Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020. https://extranet.who.int/pqweb/sites/default/files/documents/200724_final_pqpr_eul_0511_070_00_xpert_xpress%20%281%29.pdf.
- Wu J., Liu Y., Cui Y., Zhao X., Dong D. Biosens. Bioelectron. 2019;142:111508. doi: 10.1016/j.bios.2019.111508. [DOI] [PubMed] [Google Scholar]
- Wu Q., Song Q., Wang X., Yao L., Xu J., Lu J., Liu G., Chen W. Anal. Sci. 2020;36:653–658. doi: 10.2116/analsci.19P218. [DOI] [PubMed] [Google Scholar]
- Xiang J., Yan M., Li H., Liu T., Lin C., Huang S., Shen C. medRxiv. 2020 2020.2002.2027.20028787. [Google Scholar]
- Yang X., Xie J., Hu S., Zhan W., Duan L., Chen K., Zhang C., Yin A., Luo M. Sensor. Actuator. B Chem. 2020;311:127903. [Google Scholar]
- Yao L., Teng J., Zhu M., Zheng L., Zhong Y., Liu G., Xue F., Chen W. Biosens. Bioelectron. 2016;85:331–336. doi: 10.1016/j.bios.2016.05.031. [DOI] [PubMed] [Google Scholar]
- You M., Lin M., Gong Y., Wang S., Li A., Ji L., Zhao H., Ling K., Wen T., Huang Y., Gao D., Ma Q., Wang T., Ma A., Li X., Xu F. ACS Nano. 2017;11:6261–6270. doi: 10.1021/acsnano.7b02466. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang X., Liu J., Ban Y., Li N., Wu Y., Liu Y., Ye R., Liu J., Li X., Li L., Qin X., Zheng R. Int. Immunopharm. 2020;88:106861. doi: 10.1016/j.intimp.2020.106861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Teng J., Cheng L., Ye Y., Pan D., Wu J., Xue F., Liu G., Chen W. Biosens. Bioelectron. 2016;80:574–581. doi: 10.1016/j.bios.2016.01.091. [DOI] [PubMed] [Google Scholar]
- Zheng W., Yao L., Teng J., Yan C., Qin P., Liu G., Chen W. Actuator. B: Inside Chem. 2018;264:320–326. doi: 10.1016/j.snb.2018.02.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.