A decade of progress in sequencing and in CRISPR/Cas technologies has created a situation without precedent in the history of medicine. Starting with an individual patient, next-generation sequencing can diagnose, in less than 24 h, the genetic basis of a Mendelian disorder.1 Once the causative mutation is found, CRISPR/Cas, in principle, represents a targeted therapy, a first-pass iteration of which can be designed in silico within minutes thanks to straightforward principles of target locus recognition by Cas9.2 The juxtaposition of the two to yield a treatment is not a hypothetical. For Mendelian disorders of hematopoiesis and those that can be treated by editing genes in the liver or the eye, a charted path exists to (1) engineer a CRISPR/Cas-based therapeutic, (2) complete IND-enabling preclinical safety, efficacy, and manufacturing studies, and (3) perform a phase 1/2 clinical trial. Three ongoing such trials have reduced all of this to practice,3,4 with a good safety record in ∼22 (sickle cell disease and transfusion-dependent β-thalassemia), 6 (TTR amyloidosis), and 6 (Leber's congenital amauropathy) subjects dosed to date; in the first case, all subjects for whom data are publicly available have experienced a resolution of major disease symptoms.
This rapid progress illuminates a glaring gap with respect to using CRISPR/Cas to treat N = 1 disease: if current trends continue, the vast majority of patients with clinically tractable monogenic disorders will, in fact, not benefit from a CRISPR/Cas treatment in the next decade. Part of the challenge is the current preclinical framework for advancing editing to the clinic, put in place before any Cas-based IND was filed and before the “rapid and facile design of editing therapeutics” vision became real. Furthermore, >95% of INDs and ongoing trials in the editing space are held by biotechnology companies, and these have a fiduciary responsibility to focus on diseases that represent viable commercial opportunities, such as the hemoglobinopathies or cancer, rather than bona fide N = 1 disorders. The latter do not, at present, appear to represent viable targets in for-profit settings. To wit, a stunning achievement in academic genomic medicine that cured 50 out of 50 pediatric subjects of adenosine deaminase deficiency-severe combined immunodeficiency (ADA-SCID) using lentiviral gene therapy5 was followed, within 3 weeks, by an announcement from a biotechnology company that licensed this program that it was being shelved as not representing a robust commercial opportunity.
This status quo is not acceptable. Patients dying of N = 1 genetic conditions cannot wait for the for-profit sector to work out its business model. Therefore, we must engender change across the entire space in how, where, and with what financial support such treatments are engineered and put to clinical use. Importantly, over three decades of precedent in gene therapy prove that academic/government/nonprofit science and medicine can successfully innovate in developing and administering curative treatments.6
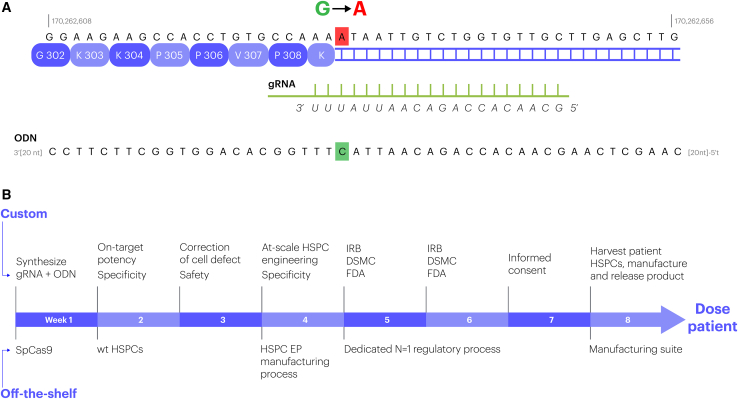
With that said, consider a pediatric patient with a severe inborn disorder of the immune system: a 2-month-old infant who suffered a catastrophic failure of immunity owing to a single G>A SNP in the LCP2 gene. The child died after a failed bone marrow transplant preceded by 8 months of devastating suffering.7 As shown in Figure 1A, genome editing can repair this exact mutation efficiently and precisely in the clinically relevant cell type, hematopoietic stem cells, using the exact same overall approach currently pursued for sickle-cell disease in the clinic (NCT04774536). Experienced contract research organizations (CROs) that manufacture the key reagents—a guide RNA (gRNA) and an oligonucleotide (ODN)—can do so in less than 3 days, and the protein subunit of the editor can be premanufactured in bulk. That Cas9-gRNA-ODN combination can be tested for potency and specificity in engineered proxy cell lines and in healthy hematopoietic stem and progenitor cells (HSPCs) in less than 2 weeks (Figure 1B). There is a clinic-vetted approach3 to (1) harvest the relevant cell type (HSPCs) from the patient, (2) gene-edit them, and (3) transplant them back to the patient. What obstacles exist to using CRISPR to treat severely ill patients such as this in an academic/nonprofit setting?
Figure 1.
The path from a lethal mutation to a CRISPR therapeutic
(A) The genic context of, and genome editing reagents for, the mutation that killed the infant treated by Lev et al.7 The mutation (mutant allele, shown, is an A; wild-type allele is a G) drives aberrant splicing of LCP2 (genomic coordinates in hg38 are indicated). The gRNA spacer for DSB-driven editing and the cognate ODN repair template,8 are shown below the sequence. The ODN specifies the correct allele (highlighted in red); the ODN does not contain a “PAM-blocking” mutation because the SNP lies in the “seed” region. (B) An accelerated timeline to engineering, derisking, and putting through regulatory review an edited HPSC product to treat terminally ill pediatric subjects such as the one described by Lev et al.7 Every step in the process will require formidable scientific and regulatory innovation; a realistic, actionable path to such innovation exists across the entire problem space.
Under the current preclinical, manufacturing, and regulatory framework, developing a CRISPR/Cas experimental therapeutic of this type is a time-consuming (at minimum 2–3 years) and costly (>US$5 million) process. Considering editing HSPCs as an example, the relevant framework was established in 2007–2011 to support a first-in-human trial for knocking out CCR5 in patients with HIV,9 remains largely intact,3 and is the basis of open or soon-to-begin trials for the hemoglobinopathies sponsored by six different biotechnology and not-for-profit organizations.10 Under this framework, the gRNA and the ODN have to be manufactured to good manufacturing practice (GMP) standards. A required IND-enabling safety experiment is transplantation of a patient dose of edited cells into immunodeficient mice followed by a detailed analysis after 4–6 months. The manufacturing scheme involves three “engineering runs” in the GMP suite with patient lot-sized doses of cells. Each of these workstreams (just several of many that go into an IND!) involves much more than 6 months of work by many individuals, and the total cost for just the above is, at present, >$2 million. All of this is well justified when the plan is to treat chronic severe disease with the intended patient population numbering in the hundreds of thousands. An infant such as that described by Lev et al.7 is unique, however, the window of opportunity to save his life via editing is just a few months, and a cost of $5 million makes this impossible.
The mRNA vaccine and gene therapy fields provide the basis for revision of this process. It took 62 days from design of a SARS-Cov2 vaccine at the NIH to Moderna's first clinical trial. Advancing a new mRNA vaccine for a SARS-Cov2 variant took even less time. In the same vein, over the past 12 years the same delivery platform, lentivirus, and the same manufacturing framework, ex vivo transduction of HSPCs, has been used to cure many rare Mendelian disorders of hematopoiesis (e.g., X-SCID, ADA-SCID, adrenoleukodystrophy), as well as both sickle cell disease and β-thalassemia.6 What changes between diseases is the payload (i.e., the cDNA). This “keep everything the same and change one thing” approach is, in fact, in clinical use for in vivo editing, with a “targeted integration into a safe harbor” platform enabling trials both in hemophilia and lysosomal storage diseases (change of transgene11), and a “nonviral delivery of CRISPR/Cas to the liver” platform enabling trials in TTR amyloidosis and hereditary angioedema (change of gRNA).4
In precise analogy with the above, the vision for a “N = 1 CRISPR cures” approach is to establish a vertically integrated set of indication-agnostic platform capabilities that allow transitioning from one disease to another by changing only a few components. As one considers a patient's journey, and zooming in on the genome-editing step, the relevant capabilities are enumerated below, with key areas for innovation/improvement in italics:
-
1
Designing the mutation-specific editor. The vast, and growing, toolbox of CRISPR-based tools—currently dispersed in the for-profit sector largely for intellectual property reasons—is ready to be consolidated into a platform that can rapidly identify an optimal editing approach for a given genetic condition. Of note, non-DNA double-strand break (DSB)-based approaches to repair point mutations such as base editing and “prime” editing,8 along with identification of novel Cas-like enzymes12 and engineering new ones,13 have finally made every base pair in the human genome editable without reliance on a DSB. Continued innovation in editor design and characterization will be important to fully realize the potential of the CRISPR platform, but ample editing tools already exist.
-
2
Manufacturing the editor. A transition away from virus-based platforms has enabled all-nonviral clinic-grade editing modalities ex vivo3 and in vivo.4 This means that the nuclease components of the editing platform can be premanufactured in bulk and the relevant one taken off the shelf. The gRNA and ODN manufacturing process is 72 h. Dramatic innovation in manufacturing schemes and analytical pipelines for such critical reagents should drive a fundamental revision of the regulatory framework to allow the rapid “release” of such a gRNA to treat a terminally ill patient.
-
3
Derisking the editor. On-target potency can be trivially measured in <72 h, and establishing the salutatory effect of repair on the relevant cell biomarker can be accomplished in less than 2–3 weeks. Highly sensitive ex vivo, scalable assays exist to map the genomic landscape of potential off-target sites in <1 week. Innovation in toxicology to eliminate the requirement for animal experiments and, instead, focus on ex vivo, higher-sensitivity, and richer-in-relevant-output assays, will be necessary to derisk the editor and the edited cells in ∼1 month.
-
4
Manufacturing the cell product. Ex vivo manufactured therapies in the editing space all rely on electroporation of purified cells and already use stereotypical equipment. Innovation in manufacturing to transition to closed-loop, and ultimately in vivo, administered editor formulations will be key, but is not limiting given the existence of established, clinic-grade pipelines for T cell, HPSC, and liver editing.
-
5
Putting the editor through regulatory review. Starting with the first genome-editing IND, the US Food and Drug Administration has been a key and enabling partner in the growth of editing-based approaches. It is imperative that both United States and European regulators no longer be treated as a separate authority (which they de jure are) and are made de facto partners: the current framework for regulatory review of experimental therapeutics based on genome editing is incompatible with it benefiting many of the patients who need it the most.
Taken together, the overall ∼2-month timeline from sequencing to treatment (Figure 1B) has no precedent in the genome-editing space and flies in the face of a 12-year established precedent. As someone who contributed to developing that preclinical and regulatory framework and then led its advance to the first genome-editing IND for the hemoglobinopathies, I am acutely aware of what a change from established practice this represents. Our field of genome editing in this regard should learn and benefit from a key example14 set by the acceleration to the clinic of an N = 1 effort using an antisense oligonucleotide as evidenced by recent FDA (FDA-2021-D-0320) guidance on “nonclinical testing of individualized antisense oligonucleotide drug products for severely debilitating or life-threatening diseases.” Ours is a larger challenge: you can stop dosing a subject with an antisense oligonucleotide; in contrast, you cannot “unedit” a subject. An actionable balance must be struck between accelerated derisking and a mindful focus on patient safety.
Large-scale innovation connected to this editing workstream is required across the entire problem space of identifying patients who can benefit from editing. Here, accelerated and expanded whole-genome sequencing capabilities and pipelines must be connected to distinct dedicated pipelines to rapidly functionally identify the causative variant, and thus “feed” the editor engineering pipeline—which, in turn, must be connected with strategic investment in manufacturing capabilities. Dominant in all this must be the community of clinicians in the genomic therapies space. They need programmatic support at every step, from identifying among their patients those who could benefit the most from being this “accelerated N = 1 cures” pipeline all the way through administering the experimental therapeutic.
The for-profit sector will not undertake such an across-the-board initiative in the next 5 years. In the United States this leaves the federal, state, and nonprofit sectors. Importantly, strategic initiatives such as the NIH Somatic Cell Genome Editing Consortium,15 the newly emerged ARPA-H, and California Institute of Regenerative Medicine already exist and have the explicit mandate of accelerating paths to innovative cures. Philanthropy-supported research and clinical institutions with a strong track record of innovation in clinic-directed genome editing will also have a key role to play. Academic university-based medical centers of clinical innovation in genomic therapies are the logical venue as hubs of administering such N = 1 cures. Industry-public partnerships will also be essential (the relevant CROs will benefit from their pro bono support by having clinical data on how their reagents perform). In broad strokes, therefore, if there has ever been a use-case scenario for the urgent necessity of the e pluribus unum mindset, the entirely tractable cause of patients with terminal N = 1 genetic disease is the proverbial and literal poster child.
The song that inspired the title of this piece was envisaged by its author as something to which a child can relate. A formidable fraction of severe genetic disease manifests in the pediatric population. As the foregoing has argued, a first-of-its-kind initiative to deliver the extraordinary medical promise of CRISPR/Cas must no longer remain in the realm of our imagination.
Acknowledgments
I am indebted to colleagues at the Innovative Genomics Institute—Alexander Marson, Jennifer Doudna, Ross Wilson, Matthew Kan, and Brad Ringeisen— as well as Mark Walters (UCSF), Donald Kohn (UCLA), and Timothy Yu (Boston Childrens) for their essential role in inspiring and informing the narrative above. None of these individuals or their institutions bears any responsibility for any mistakes or misalignments with reality in the above, which are mine alone. I am grateful to Benton Cheung (IGI) for drawing the figure.
References
- 1.Owen M.J., Niemi A.-K., Dimmock D.P., Speziale M., Nespeca M., Chau K.K., Van Der Kraan L., Wright M.S., Hansen C., Veeraraghavan N. Rapid sequencing-based diagnosis of thiamine metabolism dysfunction syndrome. N. Engl. J. Med. 2021;384:2159–2161. doi: 10.1056/NEJMc2100365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.-S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 4.Gillmore J.D., Gane E., Taubel J., Kao J., Fontana M., Maitland M.L., Seitzer J., O’Connell D., Walsh K.R., Wood K. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 5.Kohn D.B., Booth C., Shaw K.L., Xu-Bayford J., Garabedian E., Trevisan V., Carbonaro-Sarracino D.A., Soni K., Terrazas D., Snell K. Autologous ex vivo lentiviral gene therapy for adenosine deaminase deficiency. N. Engl. J. Med. 2021;384:2002–2013. doi: 10.1056/NEJMoa2027675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M. Gene therapy comes of age. Science. 2018;359:eaan4672. doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- 7.Lev A., Lee Y.N., Sun G., Hallumi E., Simon A.J., Zrihen K.S., Levy S., Beit Halevi T., Papazian M., Shwartz N. Inherited SLP76 deficiency in humans causes severe combined immunodeficiency, neutrophil and platelet defects. J. Exp. Med. 2021;218:e20201062. doi: 10.1084/jem.20201062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 9.Holt N., Wang J., Kim K., Friedman G., Wang X., Taupin V., Crooks G.M., Kohn D.B., Gregory P.D., Holmes M.C. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urnov F.D. The Cas9 hammer-and sickle: a challenge for genome editors. CRISPR J. 2021;4:6–13. doi: 10.1089/crispr.2021.29120.fur. [DOI] [PubMed] [Google Scholar]
- 11.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pausch P., Al-Shayeb B., Bisom-Rapp E., Tsuchida C.A., Li Z., Cress B.F., Knott G.J., Jacobsen S.E., Banfield J.F., Doudna J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369:333–337. doi: 10.1126/science.abb1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J., Hu C., Moufawad El Achkar C., Black L.E., Douville J., Larson A., Pendergast M.K., Goldkind S.F., Lee E.A., Kuniholm A. Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 2019;381:1644–1652. doi: 10.1056/NEJMoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha K., Sontheimer E.J., Brooks P.J., Dwinell M.R., Gersbach C.A., Liu D.R., Murray S.A., Tsai S.Q., Wilson R.C., Anderson D.G. The NIH somatic cell genome editing program. Nature. 2021;592:195–204. doi: 10.1038/s41586-021-03191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]