Abstract
Adoptive T cell therapies have shown impressive signals of activity, but their clinical impact could be enhanced by technologies to increase T cell potency and diminish the cost and labor involved in manufacturing these products. Gene editing platforms are under study in this arena to (1) enhance immune cell potency by knocking out molecules that inhibit immune responses; (2) deliver genetic payloads into precise genomic locations and thereby enhance safety and/or improve the gene expression profile by leveraging physiologic promoters, enhancers, and repressors; and (3) enable off-the-shelf therapies by preventing alloreactivity and immune rejection. This review discusses gene editing approaches that have been the best studied in the context of human T cells and adoptive T cell therapies, summarizing their current status and near-term potential for translation.
Introduction
Adoptive T cell therapies, including chimeric antigen receptor (CAR) T cells, tumor-infiltrating lymphocytes, and T cells engineered to express tumor-specific T cell receptors (TCRs) can eradicate large tumor burdens and mediate durable disease control,1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 but most patients treated with such therapies do not benefit.18 Furthermore, adoptive T cell therapies for cancer are expensive and laborious to manufacture, and autologous products manifest wide differences in potency due to interindividual variability. Some think that master cell banks of allogeneic products engineered to mediate antitumor effects, prevent graft-versus-host disease (GVHD), and avoid immune rejection could provide a major advance, as they could be multiply engineered for enhanced potency and manifest greater consistency at a reduced cost. Thus, the field is working vigorously to combine genetic engineering, synthetic biology, and gene editing to deliver more potent adoptive immune cell therapies with greater accessibility and at lower cost.19
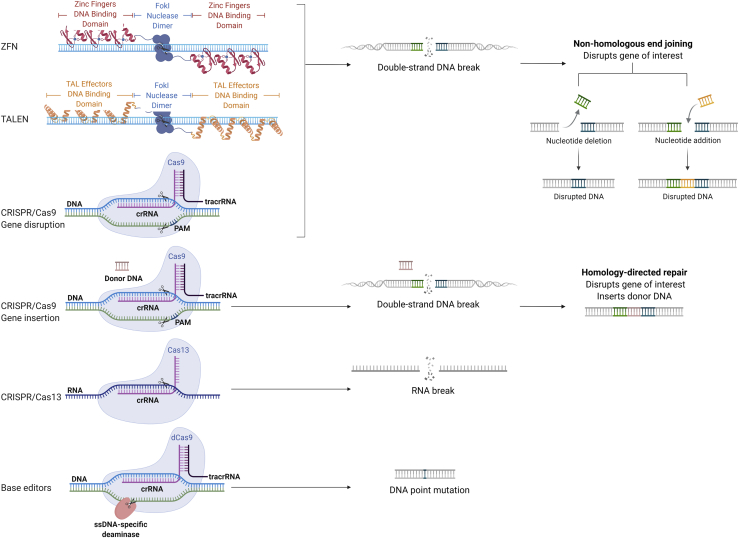
Gene editing changes the DNA of cells in a site-specific manner. Editing of a mammalian cell genome was first demonstrated by expressing an endonuclease that created a double-strand DNA break in murine cells, which was repaired by error-prone non-homologous end joining or homology-directed repair.20 With the goal of enhancing efficiency and translating to therapeutic applications, a variety of nucleases have since been developed and optimized for use in human cells, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), clustered regularly interspaced short palindromic repeats associated with Cas9 or other Cas endonucleases (CRISPR-Cas), homing endonucleases/meganucleases, megaTALs, base editors, and prime editors21, 22, 23, 24 (Figure 1).
Figure 1.
Schematic of a sample of gene editing methods
ZFN and TALEN use FokI nonspecific nuclease in addition to DNA binding domains to induce site-specific cleavage. ZFN, TALEN, and CRISPR-Cas9 create double-strand DNA breaks, which can induce gene knockout by non-homologous end joining. Double-strand breaks can also be repaired by homology-directed repair in the presence of a donor DNA template, resulting in insertion of a transgene (e.g., knock-in). CRISPR-Cas13 cleaves RNA, and base editors introduce site-specific DNA point mutations. Adapted from “CRISPR/Cas9 (crRNA, tracrRNA),” “ZFN and TALEN nucleases for gene editing,” and “CRISPR/Cas9 gene editing,” by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates.
ZFNs and TALENs are chimeric nucleases, composed of programmable, sequence-specific DNA-binding modules linked to a DNA cleavage domain, which induce DNA double-strand breaks that stimulate endogenous DNA repair mechanisms. CRISPR-Cas, derived from archaeal and bacterial antiviral defense systems, induces double-strand DNA breaks at the site targeted by a single guide RNA (sgRNA).25,26 The first human trials using genome editing were conducted using T cells and hematopoietic stem cells (HSCs) collected from patients with HIV/AIDS and targeting CCR5, the gene encoding the CD4+ coreceptor for HIV entry. The CCR5 gene was disrupted via ZFN-mediated double-strand DNA breaks and subsequent non-homologous end joining in one study and was disrupted via CRISPR editing in hematopoietic stem and progenitor cells (HSPCs) in a second study.27,28
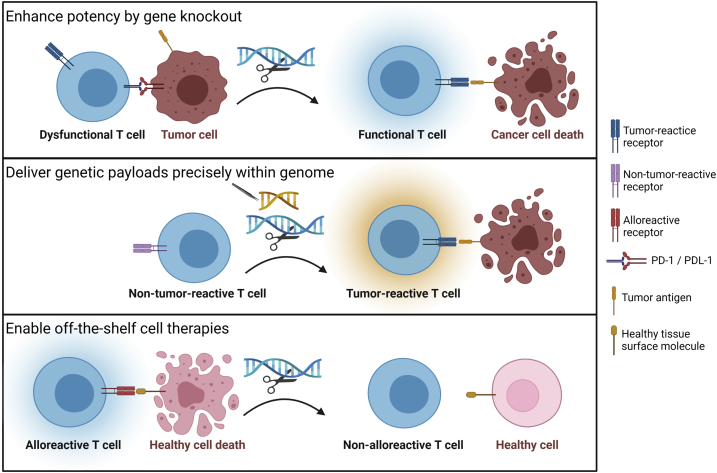
CRISPR-Cas systems are continuing to evolve to incorporate diverse Cas effectors (Figure 1).29, 30, 31, 32 Meganucleases, homing endonucleases derived from I-CreI and I-CeuI, recognize DNA sequences with palindromic sequences and cause DNA double-strand breaks,33 while megaTALs are engineered chimeric proteins fusing homing endonucleases and TAL effector arrays.34 Gene editing can now be accomplished without induction of double-strand DNA breaks: through base editing, wherein a catalytically dead Cas (dCas9) and a base modification deaminase are catalytically fused, thereby editing bases within a small segment of displaced DNA;23 and through prime editing, which relies on dCas9 fused to an engineered reverse transcriptase and prime editing RNA (pegRNA), to target a site in the genome and provide a template for the desired edit36 (Figure 1). In the context of adoptive cell therapy, gene editing platforms are being developed for three major goals: (1) enhancing immune cell potency by knocking out molecules that inhibit immune responses; (2) delivering genetic payloads into precise genomic locations and thereby enhance safety and/or improve the gene expression profile by leveraging physiologic promoters, enhancers, and repressors; and (3) enabling off-the-shelf therapies by preventing alloreactivity and immune rejection (Figure 2). This review focuses on studies employing gene knockout (KO) and/or gene editing in human T cells that have already been or are poised for near-term clinical translation. Note that we do not review approaches underway to enhance therapies using gene overexpression or synthetic circuits, which have been the focus of recent comprehensive reviews.37, 38, 39
Figure 2.
Key goals for gene editing in the context of adoptive cell therapies
Goal 1 seeks to enhance potency by knocking out molecules that inhibit immune responses, such as inhibitory receptors. Goal 2 seeks to deliver genetic payloads into precise genomic locations, such as the locus of the endogenous TCR. Goal 3 seeks to enable off-the-shelf therapies, by preventing alloreactivity and immune rejection. Created with BioRender.com.
Gene knockout to enhance T cell potency
T cell potency is regulated by a complex array of activating and inhibitory signals evolved to maximize immune competence against viral infection while diminishing risks for autoimmunity and lymphoproliferation. Such physiologic regulators are candidates for knockout to enhance T cell potency, especially in the setting of cancer therapies. For instance, Fas-FasL-dependent activation-induced cell death (AICD) attenuates CAR T cell activity, whereas Fas-ablated CAR T cells generated using CRISPR-Cas9 exhibited decreased AICD and manifested increased lifespan, cytokine secretion, tumor cell killing, and enhanced tumor control.40, 41, 42 CRISPR-Cas9 knockout of diacylglycerol kinase (DGK) was shown to generate CAR T cells resistant to DGK-mediated downregulation of TCR distal molecules (including extracellular signal-related kinases 1 and 2 [ERK1/2]), thus potentiating CAR T cell effector function.43 CRISPR-Cas9-mediated knockout of the adenosine A2A receptor, which is activated by the immunosuppressive factor adenosine, was also found to enhance CAR T cell efficacy.44
Knockout of inhibitory receptors, which are overexpressed in exhausted T cells, have been a focus of numerous studies.19,45,46 Several studies have demonstrated enhanced T cell potency of PD-1-deficient T cells, following PD-1 blockade, and with CRISPR-Cas9-mediated PD-1 knockout.17,47, 48, 49,50, 51, 52, 53 Not surprisingly, therefore, the first-in-human, phase I human clinical trial to test the safety and feasibility of multiplex CRISPR-Cas9 genome editing for a cancer immunotherapy application infused autologous T cells engineered to express the NY-ESO-1 TCR edited at the PDCD1 locus, thereby inducing PD-1 knockout19,54,55 (ClinicalTrials.gov: NCT03399448). In this study, approximately 25% of the T cells expressing the NY-ESO-1 TCR in the infusion product had mutations in the PDCD1 locus, but this decreased to approximately 5% 4 months after infusion, illustrating that PDCD1 knockout actually led to diminished T cell expansion and/or persistence. This may be a result of PD-1 biology, and its known contribution to T cell fitness,56,57 or it could reflect adverse effects of CRISPR-Cas9 editing on T cell fitness.
Measurable levels of off-target genotoxicity, mostly in the form of genomic translocations, were present in the infused product, but no evidence for transformation or lymphoproliferation was observed. Humoral responses to Cas9 did not develop in any of the three patients, which may reflect the low content of Cas9 in the infused product and/or the immunodeficiency in the patients due to their extensive previous treatment histories.40 Safety and feasibility of CRISPR-Cas9-edited T cell therapy is being studied in PD-1 knockout T cells for the treatment of adult solid tumors58 (ClinicalTrials.gov: NCT03545815) and has been shown in a clinical study of the treatment of non-small cell lung cancer using PD-1-edited T cells (ClinicalTrials.gov: NCT02793856).59 These early clinical studies demonstrate feasibility and an early safety signal for CRISPR-Cas9 editing of adoptive T cells but demonstrated diminished fitness of PD-1-edited T cells. Findings of off-target genotoxicity also raise important, but currently unanswered, questions regarding how best to quantify genotoxicity in gene-edited populations and what level of genotoxicity is safe and acceptable in the context of clinical studies.
These studies highlight that while progress in developing technologies to disrupt genes in T cells has advanced significantly, biological understanding has not yet identified the optimal gene, or gene combination, for which knockout will enhance potency and improve patient outcomes. Recent progress in delivering either whole-genome sgRNA libraries or curated sgRNA libraries that target hundreds of genes60,61 into human T cells with high efficiency is enabling comprehensive screens for targets that enhance potency when disrupted. Recently, such efforts in CAR T cells identified increased potency upon deletion of TLE4, a transcriptional corepressor of multiple genes encoding inflammatory cytokines,62 and IKZF2, which is upregulated in exhausted CAR T cells.63, 64, 65 Recently, CRISPR-Cas9 transfection of primary human T cells without TCR stimulation to enable study of genes involved in T cell activation has been shown to result in a knockout efficiency >90%.66 We anticipate significant additional data in the near term identifying additional candidates for knockout to confer enhanced potency in T cells.
Gene knockout has also been a focus of numerous studies aimed at preventing T cell fratricide, and this study area is most advanced in the development of CAR T cells for treatment of T cell malignancies. Fratricide-resistant human CAR T cells have been successfully generated using multiple platforms, including base editing of TCR/CD3 and CD7, thus disrupting gene expression via creation of stop codons or elimination of splice sites followed by lentiviral-mediated expression of CARs targeting CD3 or CD7;67 TALEN-mediated disruption of TRAC and lentiviral-mediated CD3-CAR expression;68 CRISPR-Cas9 editing with guides targeting CD7 and TRAC loci followed by lentiviral-mediated CD7-CAR expression;69 CRISPR-Cas9-mediated knockout of CD7 followed by γ-retroviral-mediated CD7-CAR expression;70 as well as by CRISPR-Cas9 editing of the CD5 locus followed by lentiviral-mediated CD5-CAR expression.71 Clinical translation will need to address the expected killing of normal CD3+, CD7+, and CD5+ cells following adoptive transfer and the consequences for T cell-mediated immunity. Highlighting the potential for T cell aplasia with these platforms, a phase I dose escalation study (ClinicalTrials.gov: NCT03081910) of autologous CD5-CAR T cells, which showed minimal fratricide even without CD5 editing, found prolonged cytopenias in two of five patients and reactivation of cytomegalovirus (CMV) and BK virus requiring antiviral therapy in one of five patients.72
Finally, CAR T cell therapy efficacy, as evaluated by progression-free survival and length of overall survival, has been found to decrease with glucocorticoid administration for the treatment of cytokine release syndrome (CRS).73 Based on findings that granulocyte-macrophage colony-stimulating factor (GM-CSF) promotes CRS, TALEN-mediated gene knockout of GM-CSF in CAR T cells suppressed the secretion of CRS biomarkers by monocytes,74 suggesting a potential strategy to mitigate toxicity related to CRS and neuroinflammation,75 and thereby improve outcomes following CAR T cell therapy.
Gene editing to deliver genetic payloads into precise locations in the genome and to enable viral-free gene engineering
When double-strand DNA breaks induced by gene editing platforms discussed herein are repaired by homologous recombination, in the presence of an appropriate DNA template, a genetic payload is inserted into a precise genomic location (e.g., knockin). Viral vectors, such as adeno-associated virus (AAV) vectors, are most commonly used to provide a template for knockin platforms. For instance, in a preclinical model, using CRISPR-Cas9/AAV to insert a CD19-specific CAR into the TRAC locus resulted in enhanced potency compared to CARs delivered via retroviral vectors,76 presumably by modulation of CAR expression levels at baseline and following antigen activation, thereby tuning the CAR signal and preventing CAR T cell exhaustion. Combining gene knockout with knockin of a gene of interest also provides the additional benefit of enabling more precise quantitation of the gene editing efficiency since a knockin gene can function as a marker for tracking and/or selection of gene-edited cells.77 Knockin of selectable markers also provides options for increasing purity and homogeneity of cell products, thereby enhancing therapeutic potency in clinical settings.
Genetic reprogramming of T cells in the context of adoptive T cell therapies has predominantly utilized recombinant viral vectors, including γ-retroviral vectors, lentiviral vectors, and DNA transposons to introduce transgenes. Viral vectors integrate randomly into the genome, which confers risks of insertional mutagenesis and/or diminished transgene expression due to regulatory elements in proximity to the integration. Two clinical cases of lentivirus insertion-mediated CAR T cell clonal expansion have been described: a CD19 CAR T clone with insertion in the TET2 locus, which demonstrated enhanced T cell potency; and a CD22 CAR T clone with insertion in the CBL locus that constituted approximately half of total white blood cells in the second peak of CAR T cell expansion.78,79 Furthermore, two cases of T cell lymphoma have been observed as a result of insertional mutagenesis using the Piggybac transposon platform.80, 81, 82 These results verify that integration into host genes can lead to transformation and leukemogenesis and thereby confirm the risk of random vector integration.
These safety issues, in addition to the expense and laborious manufacturing process involved in generating viral vectors, have led investigators to develop viral-free CRISPR-Cas9 platforms for gene delivery.17,83 Knockin efficiency with viral-free platforms remains lower than with comparable sized AAV templates but could ultimately enhance options for gene delivery by eliminating the need for viral vectors, which may lower cost, increase efficiencies and consistency, and improve safety. Site-specific viral-free gene delivery is an active area of translation for HSCs84 and has been pursued in the context of T cells via CRISPR-Cas9-mediated specific insertion of a BCMA CAR at the TRAC locus, which has demonstrated consistent and high CAR expression.85 Thus, precision payload delivery is likely to be increasingly applied to adoptive T cell therapies for controlled insertion to standardize transgene expression. Furthermore, insertion of a selectable marker provides an opportunity to render a purer product for clinical use.
Gene editing to create allogeneic, off-the-shelf T cell therapies
Most clinical trials of adoptive cell therapy for cancer employ autologous T cells. Manufacturing of such products is expensive, labor intensive, requires long timelines, and results in significant product heterogeneity. Allogeneic T cell banks could provide off-the-shelf T cell therapies that circumvent these limitations. However, numerous challenges must be overcome if allogeneic T cells are to deliver potent and effective antitumor responses. First, the endogenous TCR must be knocked out to prevent GVHD. Conversely, mismatched human leukocyte antigen (HLA) alleles on the surface of allogeneic T cells must be knocked out to prevent rejection by host T cells, while approaches must be incorporated to prevent natural killer cell-mediated graft rejection, which increases in the absence of self HLA molecules.
T cell editing to introduce an exogenous, high-avidity, tumor-specific TCR is complicated by the presence of the endogenous TCR due to risk associated with inappropriate pairing of the exogenous and endogenous TCR chains. Such mispairing not only decreases transgenic TCR activity via competition for CD3 binding but also can contribute to GVHD or autoimmunity based on unpredictable specificities of new hybrid heterodimers. Addressing this risk, engineered meganuclease technology and TALEN technology have each been applied to target the TCR α- chain as a means of preventing GVHD and creating a universal, allogeneic T cell product.86, 87, 88 In addition, TCR-edited T cells have been generated through ZFN-mediated disruption of endogenous TCR α and β chain genes followed by lentiviral transfer of a tumor antigen-specific TCR. ZFN-edited cells, as compared to unedited T cells, exhibited increased specificity in antigen recognition.89, 90, 91
The first clinical trials of allogeneic CAR T cell products utilized the UCART19 platform. Patients with refractory leukemia were treated with CD19 CAR T cells generated from a healthy donor and subjected to TALEN-mediated disruption of the TCR and CD52 loci. To further minimize the risk of GVHD, T cells that retained their TCR because of inefficient TALEN gene editing were depleted by immune selection prior to infusion. To prevent rejection, patients received a potently immunosuppressive anti-CD52 monoclonal antibody (mAb) (alemtuzumab),92 and ultimately underwent allogeneic stem cell transplant.93 In the first trial, UCART19 was administered to two infants with relapsed CD19/CD52-acute lymphocytic leukemia (ALL), one of which developed grade 2 GVHD in the skin. In subsequent trials (ClinicalTrials.gov: NCT02808442 and NCT02746952), 7 children and 14 adults received UCART19. During the 9-month follow-up period after infusion, grade 1 acute skin GVHD occurred in 10%. CRS occurred in 91%, of which 14% had grade 3–4 CRS, and grade 1 or 2 neurotoxicity occurred in 38%. 67% of patients had a complete response or complete response with incomplete hematological recovery 28 days after infusion, and 10 of 14 responders proceeded to subsequent allogeneic stem cell transplant.94,95 These studies were the first to demonstrate feasibility of using allogeneic cells, and additional clinical testing of CD19/CD22 CAR T cells with CRISPR-Cas9-edited TRAC and CD52 loci demonstrated safety and anti-leukemia efficacy (ClinicalTrials.gov: NCT04227015).96
Additional universal CAR T cells have been generated and tested clinically by TALEN editing of a CD123 CAR T cell for blastic plasmacytoid dendritic cell neoplasm (ClinicalTrials.gov: NCT03203369) and by TALEN editing of a CD123 CAR T cell for acute myeloid leukemia (ClinicalTrials.gov: NCT03190278). Furthermore, there are ongoing phase 1 and phase 1/2a trials of allogeneic CD19 CAR T cell therapies with TRAC-specific insertion of the CD19 CAR (ClinicalTrials.gov: NCT04649112 and NCT03666000). Gene-edited CAR T cells have induced antitumor effects in leukemia, although it remains unknown whether the responses observed are durable and how long the allogeneic cells persisted following adoptive transfer, which is being explored in part through long-term follow-up of patients exposed to UCART19 (ClinicalTrials.gov: NCT02735083).
To evade donor HLA-mediated immune rejection, CRISPR-Cas9 T cell editing has been applied to β2-microglobulin (B2M) and HLA-II α chain genes (HLA-DRA, DQA, and DPA), showing decreased alloreactivity in vitro.97 HLA class I, HLA class II, and TCR triple-knockout T cells by CRISPR-Cas9 editing of B2M, class II major histocompatibility complex transactivator (CIITA), and TRAC loci did not induce GVHD in preclinical models.98 Ongoing clinical trials of gene-edited allogeneic T cell therapies are studying TRAC/B2M-knockout CD19 CAR T cells for B cell leukemia and lymphoma (ClinicalTrials.gov: NCT03166878 and NCT03229876), TRAC-knockout dual CD19/CD22 and CD19/CD20 CAR T cells for B cell leukemia (ClinicalTrials.gov: NCT03398967), and TCR-inserted CD19 CAR T cells for non-Hodgkin lymphoma and B cell ALL (ClinicalTrials.gov: NCT04649112 and NCT03666000).99 We anticipate continued advances in this arena with improved biological understandings regarding the optimal approaches to prevent cell rejection and improved technologies to enhance the efficiency of multiplex gene-edited immune cell populations.
Next-generation gene editing in the absence of double-strand DNA breaks
Technologies have evolved to exploit the gene targeting capacity of the CRISPR-Cas platform while avoiding the potential risks associated with DNA double-strand breaks. Simultaneously providing an inactivated Cas mutant that does not mediate DNA strand breaks (e.g., dCas) and an enzymatic moiety to induce a localized molecular change is providing a surfeit of opportunities for next-generation gene editing and modulation of gene transcription. CRISPR interference (CRISPRi) delivers dCas with a transcription repressor to selectively suppress gene transcription.100,101 CRISPRi can efficiently repress expression of single or multiple genes in mammalian cells, and its effects are reversible. In one report, CRISPRi mediated conditional suppression of PD-1 signaling in CAR T cells. Cells were engineered to express an anti-HER2 CAR, a tobacco etch virus (TEV) protease, and a PD-1 promoter region-targeting gRNA as well as a linker for activation of T cells (LAT) linked to dCas9-Krüppel-associated box (Krab) via a TEV-cleavable linker. Upon CAR T cell encounter with antigen, TEV cleaved the second complex, and thereby induced translocation of dCas9-Krab to the nucleus where it targeted the start site of the PD-1 gene and prevented PD-1 transcription. This synthetic circuit prevented PD-1 signaling and was associated with improved in vivo persistence and effectiveness against HER2-expressing oropharyngeal cancer xenografts.102
CRISPR activation (CRISPRa) selectively induces gene transcription via delivery of dCas with a transactivation moiety. Application of this technique has allowed mapping of functional enhancers that are candidates for therapeutic modulation.103 In the context of T cell editing for cancer immunotherapy, this approach could enable transient activation of transcriptional activators to enhance T cell effector function and/or to selectively induce expression of regulatory elements as a safety switch.
Short hairpin RNA (shRNA) technology has been applied to achieve gene silencing without knockout of the endogenous locus. Interleukin (IL)-6 shRNA engineered into CD19 CAR T cells (referred to as ssCART-19), intended to reduce severe CRS in CAR T cell therapy, showed gene silencing of IL-6 and decreased soluble IL-6 levels in vitro and in vivo.104 A clinical trial that treated 7 of 13 patients with ssCART-19 and the remaining with conventional CD19 CAR T cells observed that despite all patients in both treatment arms achieving complete response, ssCART-19 patients experienced lower CRS grade and lower serum IL-6 levels. In a phase 1/2a trial, shRNA technology has been integrated with other gene editing approaches in CD19 CAR T cells with TRAC-specific insertion of the CD19 CAR alongside a shRNA that suppresses expression of B2M for further mediation of alloreactivity (ClinicalTrials.gov: NCT03666000).105
Base editing, via a nucleotide deaminase linked to a DNA-binding protein, offers an approach to edit primary cells without double-strand breaks.36,106,107 This provides a theoretical advantage for clinical translation since risks of off-target genotoxicity are predicted to be diminished. Recent base editing in human T cells showed no evidence for detectable translocations compared to CRISPR-Cas9, which is associated with translocation frequencies that exceed 2%.108 In primary human T cells, adenine base editors have been evolved using a variant library to achieve 98%–99% target modification, maintained when multiplexed across three genomic loci.109 Multiplex base editing of TRAC, PDCD1, and B2M loci in CD19 CAR T cells was reported to deliver greater efficiency and result in decreased translocation frequency and fewer double-strand breaks compared to traditional CRISPR-Cas platforms.29 However, base editing can induce genome-wide off-target DNA mutations at mismatched DNA targets in the genome, bystander mutations within activity windows of base editors, and sgRNA-independent transcriptome-wide RNA deamination, and work is underway to overcome these challenges.104
Whereas base editors allow C>T or A>G conversions, the recently described method of prime editing can mediate all 12 nt substitutions as well as short insertions and short deletions. Thus far, prime editing using RNPs in primary T cells has demonstrated a proof of concept, but efficiency remains lower than in other platforms.110 In summary, gene editing in the absence of double-strand breaks offers a promising platform for safer editing compared to those that require double-strand breaks and induce associated genotoxicity, and these novel platforms are poised for improvements in efficiency and specificity that will enhance their value in clinical contexts.
Conclusions and perspectives
The success of CAR T cell immunotherapy for the treatment of B cell malignancies has opened the door to an ever-increasing array of therapeutic possibilities to use genetically engineered immune cell therapies to treat cancer, infection, autoimmunity, and beyond.19,111,112,113 Technological advances in gene editing are dovetailing with this success to exponentially increase possibilities to enhance immune cell potency via gene disruption and/or gene insertion.114,115 Future studies to better understand the biology and risks of gene editing and to increase efficiencies for multiplex genome engineering represent important and ongoing areas of study.116, 117, 118 While the promise of gene editing in the context of adoptive immune cell therapy remains high, very few clinical data are available regarding long-term safety and efficacy of gene editing in the context of immune cell therapies. We anticipate rapid evolution in technologies and efficiencies in the coming years, alongside increased biological understanding that will inform target selection, as well as increased numbers of clinical trials testing the safety and efficacy of gene editing platforms. Collectively, these advances are predicted to enhance outcomes for patients with cancer and other dread diseases.
Acknowledgments
This work was supported by a St. Baldrick’s/Stand Up 2 Cancer Pediatric Dream Team translational cancer research grant (SU2CAACR-DT1113 to C.L.M.). Stand Up 2 Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. C.L.M. is a member of the Parker Institute for Cancer Immunotherapy, which supports the Stanford University Cancer Immunotherapy Program. This work was also supported by the Virginia and D.K. Ludwig Fund for Cancer Research. T.M. received support from the Stanford Medical Scientist Training Program grant T32GM007365; the Stanford Interdisciplinary Graduate Fellowship; and the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program and the Stanford ChEM-H O’Leary-Thiry Graduate Fellowship.
Declaration of interests
C.L.M. has multiple patents pertinent to CAR T cells, is a cofounder of Lyell Immunopharma and Syncopation Life Sciences, which are developing CAR-based therapies, and consults for Lyell, NeoImmune Tech, Apricity, Nektar, Immatics, Mammoth Biosciences, and Ensoma. T.M. declares no competing interests.
References
- 1.June C.H., O’Connor R.S., Kawalekar O.U., Ghassemi S., Milone M.C. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 2.Kochenderfer J.N., Dudley M.E., Kassim S.H., Somerville R.P., Carpenter R.O., Stetler-Stevenson M., Yang J.C., Phan G.Q., Hughes M.S., Sherry R.M. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J. Clin. Oncol. 2015;33:540–549. doi: 10.1200/JCO.2014.56.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turtle C.J., Hanafi L.A., Berger C., Hudecek M., Pender B., Robinson E., Hawkins R., Chaney C., Cherian S., Chen X. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor–modified T cells. Sci. Transl. Med. 2016;8:355ra116. doi: 10.1126/scitranslmed.aaf8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majzner R.G., Theruvath J.L., Nellan A., Heitzeneder S., Cui Y., Mount C.W., Rietberg S.P., Linde M.H., Xu P., Rota C. CAR T cells targeting B7-H3, a pan-cancer antigen, demonstrate potent preclinical activity against pediatric solid tumors and brain tumors. Clin. Cancer Res. 2019;25:2560–2574. doi: 10.1158/1078-0432.CCR-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn N.F., Purdon T.J., van Leeuwen D.G., Lopez A.V., Curran K.J., Daniyan A.F., Brentjens R.J. CD40 ligand-modified chimeric antigen receptor T cells enhance antitumor function by eliciting an endogenous antitumor response. Cancer Cell. 2019;35:473–488.e6. doi: 10.1016/j.ccell.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra A.K., Kemler I., Dingli D. Preclinical development of CD126 CAR-T cells with broad antitumor activity. Blood Cancer J. 2021;11:3. doi: 10.1038/s41408-020-00405-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster S.J., Bishop M.R., Tam C.S., Waller E.K., Borchmann P., McGuirk J.P., Jäger U., Jaglowski S., Andreadis C., Westin J.R., JULIET Investigators Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 2019;380:45–56. doi: 10.1056/NEJMoa1804980. [DOI] [PubMed] [Google Scholar]
- 9.Park J.H., Rivière I., Gonen M., Wang X., Sénéchal B., Curran K.J., Sauter C., Wang Y., Santomasso B., Mead E. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiegel J.Y., Patel S., Muffly L., Hossain N.M., Oak J., Baird J.H., Frank M.J., Shiraz P., Sahaf B., Craig J. CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: A phase 1 trial. Nat. Med. 2021;27:1419–1431. doi: 10.1038/s41591-021-01436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee D.W., Kochenderfer J.N., Stetler-Stevenson M., Cui Y.K., Delbrook C., Feldman S.A., Fry T.J., Orentas R., Sabatino M., Shah N.N. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: A phase 1 dose-escalation trial. Lancet. 2015;385:517–528. doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishna S., Lowery F.J., Copeland A.R., Bahadiroglu E., Mukherjee R., Jia L., Anibal J.T., Sachs A., Adebola S.O., Gurusamy D. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science. 2020;370:1328–1334. doi: 10.1126/science.abb9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg S.A., Yannelli J.R., Yang J.C., Topalian S.L., Schwartzentruber D.J., Weber J.S., Parkinson D.R., Seipp C.A., Einhorn J.H., White D.E. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J. Natl. Cancer Inst. 1994;86:1159–1166. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg S.A., Packard B.S., Aebersold P.M., Solomon D., Topalian S.L., Toy S.T., Simon P., Lotze M.T., Yang J.C., Seipp C.A. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N. Engl. J. Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 15.Rohaan M.W., van den Berg J.H., Kvistborg P., Haanen J.B.A.G. Adoptive transfer of tumor-infiltrating lymphocytes in melanoma: A viable treatment option. J. Immunother. Cancer. 2018;6:102. doi: 10.1186/s40425-018-0391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins P.F., Morgan R.A., Feldman S.A., Yang J.C., Sherry R.M., Dudley M.E., Wunderlich J.R., Nahvi A.V., Helman L.J., Mackall C.L. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynn R.C., Weber E.W., Sotillo E., Gennert D., Xu P., Good Z., Anbunathan H., Lattin J., Jones R., Tieu V. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature. 2019;576:293–300. doi: 10.1038/s41586-019-1805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber E.W., Maus M.V., Mackall C.L. The emerging landscape of immune cell therapies. Cell. 2020;181:46–62. doi: 10.1016/j.cell.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouet P., Smih F., Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porteus M.H. A new class of medicines through DNA editing. N. Engl. J. Med. 2019;380:947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 22.Gaj T., Gersbach C.A., Barbas C.F., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rees H.A., Liu D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L., Wang J., Liu Y., Xie L., Su B., Mou D., Wang L., Liu T., Wang X., Zhang B. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 29.Murugan K., Babu K., Sundaresan R., Rajan R., Sashital D.G. The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol. Cell. 2017;68:15–25. doi: 10.1016/j.molcel.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan F., Wang W., Zhang J. CRISPR-Cas12 and Cas13: The lesser known siblings of CRISPR-Cas9. Cell Biol. Toxicol. 2019;35:489–492. doi: 10.1007/s10565-019-09489-1. [DOI] [PubMed] [Google Scholar]
- 31.Xu C., Zhou Y., Xiao Q., He B., Geng G., Wang Z., Cao B., Dong X., Bai W., Wang Y. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods. 2021;18:499–506. doi: 10.1038/s41592-021-01124-4. [DOI] [PubMed] [Google Scholar]
- 32.Paul B., Montoya G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020;43:8–17. doi: 10.1016/j.bj.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delhove J.M.K.M., Qasim W. Genome-edited T cell therapies. Curr. Stem Cell Rep. 2017;3:124–136. doi: 10.1007/s40778-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curinga G., Romano G., Sather B., Sommer K., Hale M., Khan I., Singh S., Song Y., Gwiazda K., Scharenberg A. 116. MegaTAL nucleases outperform TALENs in promoting homology-directed-gene modification in primary human T cells. Mol. Ther. 2015;23:S48. Suppl 1. [Google Scholar]
- 36.Antoniou P., Miccio A., Brusson M. Base and prime editing technologies for blood disorders. Front. Genome Ed. 2021;3:1. doi: 10.3389/fgeed.2021.618406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman T., Antovski P., Tebon P., Xu C., Ashammakhi N., Ahadian S., Morsut L., Khademhosseini A. Synthetic biology and tissue engineering: toward fabrication of complex and smart cellular constructs. Adv. Funct. Mater. 2020;30:1909882. [Google Scholar]
- 38.Kitada T., DiAndreth B., Teague B., Weiss R. Programming gene and engineered-cell therapies with synthetic biology. Science. 2018;359:eaad1067. doi: 10.1126/science.aad1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruder W.C., Lu T., Collins J.J. Synthetic biology moving into the clinic. Science. 2011;333:1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]
- 40.Tschumi B.O., Dumauthioz N., Marti B., Zhang L., Schneider P., Mach J.P., Romero P., Donda A. CART cells are prone to Fas-and DR5-mediated cell death. J. Immunother. Cancer. 2018;6:71. doi: 10.1186/s40425-018-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Künkele A., Johnson A.J., Rolczynski L.S., Chang C.A., Hoglund V., Kelly-Spratt K.S., Jensen M.C. Functional tuning of CARs reveals signaling threshold above which CD8+ CTL antitumor potency is attenuated due to cell Fas–FasL-dependent AICD. Cancer Immunol. Res. 2015;3:368–379. doi: 10.1158/2326-6066.CIR-14-0200. [DOI] [PubMed] [Google Scholar]
- 42.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung I.Y., Kim Y.Y., Yu H.S., Lee M., Kim S., Lee J. CRISPR/Cas9-mediated knockout of DGK improves antitumor activities of human T cells. Cancer Res. 2018;78:4692–4703. doi: 10.1158/0008-5472.CAN-18-0030. [DOI] [PubMed] [Google Scholar]
- 44.Giuffrida L., Sek K., Henderson M.A., Lai J., Chen A.X.Y., Meyran D., Todd K.L., Petley E.V., Mardiana S., Mølck C. CRISPR/Cas9 mediated deletion of the adenosine A2A receptor enhances CAR T cell efficacy. Nat. Commun. 2021;12:3236. doi: 10.1038/s41467-021-23331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y., Shao Q., Peng G. Exhaustion and senescence: Two crucial dysfunctional states of T cells in the tumor microenvironment. Cell. Mol. Immunol. 2020;17:27–35. doi: 10.1038/s41423-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Y., Li Y., Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792. doi: 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura H., Nose M., Hiai H., Minato N., Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 48.Odorizzi P.M., Pauken K.E., Paley M.A., Sharpe A., Wherry E.J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 2015;212:1125–1137. doi: 10.1084/jem.20142237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su S., Hu B., Shao J., Shen B., Du J., Du Y., Zhou J., Yu L., Zhang L., Chen F. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci. Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherkassky L., Morello A., Villena-Vargas J., Feng Y., Dimitrov D.S., Jones D.R., Sadelain M., Adusumilli P.S. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J. Clin. Invest. 2016;126:3130–3144. doi: 10.1172/JCI83092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017;7:737. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su S., Zou Z., Chen F., Ding N., Du J., Shao J., Li L., Fu Y., Hu B., Yang Y. CRISPR-Cas9-mediated disruption of PD-1 on human T cells for adoptive cellular therapies of EBV positive gastric cancer. OncoImmunology. 2016;6:e1249558. doi: 10.1080/2162402X.2016.1249558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beane J.D., Lee G., Zheng Z., Mendel M., Abate-Daga D., Bharathan M., Black M., Gandhi N., Yu Z., Chandran S. Clinical scale zinc finger nuclease-mediated gene editing of PD-1 in tumor infiltrating lymphocytes for the treatment of metastatic melanoma. Mol. Ther. 2015;23:1380–1390. doi: 10.1038/mt.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moon E.K., Ranganathan R., Eruslanov E., Kim S., Newick K., O’Brien S., Lo A., Liu X., Zhao Y., Albelda S.M. Blockade of programmed death 1 augments the ability of human T cells engineered to target NY-ESO-1 to control tumor growth after adoptive transfer. Clin. Cancer Res. 2016;22:436–447. doi: 10.1158/1078-0432.CCR-15-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pauken K.E., Godec J., Odorizzi P.M., Brown K.E., Yates K.B., Ngiow S.F., Burke K.P., Maleri S., Grande S.M., Francisco L.M. The PD-1 pathway regulates development and function of memory CD8+ T cells following respiratory viral infection. Cell Rep. 2020;31:107827. doi: 10.1016/j.celrep.2020.107827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weisberg S.P., Carpenter D.J., Chait M., Dogra P., Gartrell-Corrado R.D., Chen A.X., Campbell S., Liu W., Saraf P., Snyder M.E. Tissue-resident memory T cells mediate immune homeostasis in the human pancreas through the PD-1/PD-L1 pathway. Cell Rep. 2019;29:3916–3932.e5. doi: 10.1016/j.celrep.2019.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGowan E., Lin Q., Ma G., Yin H., Chen S., Lin Y. PD-1 disrupted CAR-T cells in the treatment of solid tumors: Promises and challenges. Biomed. Pharmacother. 2020;121:109625. doi: 10.1016/j.biopha.2019.109625. [DOI] [PubMed] [Google Scholar]
- 59.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 60.Shifrut E., Carnevale J., Tobin V., Roth T.L., Woo J.M., Bui C.T., Li P.J., Diolaiti M.E., Ashworth A., Marson A. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. 2018;175:1958–1971.e15. doi: 10.1016/j.cell.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cortez J.T., Montauti E., Shifrut E., Gatchalian J., Zhang Y., Shaked O., Xu Y., Roth T.L., Simeonov D.R., Zhang Y. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature. 2020;582:416–420. doi: 10.1038/s41586-020-2246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bandyopadhyay S., Valdor R., Macian F. Tle4 regulates epigenetic silencing of gamma interferon expression during effector T helper cell tolerance. Mol. Cell. Biol. 2014;34:233–245. doi: 10.1128/MCB.00902-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sowell R.T., Kaech S.M. Probing the diversity of T cell dysfunction in cancer. Cell. 2016;166:1362–1364. doi: 10.1016/j.cell.2016.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naluyima P., Lal K.G., Costanzo M.C., Kijak G.H., Gonzalez V.D., Blom K., Eller L.A., Creegan M., Hong T., Kim D. Terminal effector CD8 T cells defined by an IKZF2+IL-7R− transcriptional signature express FcγRIIIA, expand in HIV infection, and mediate potent HIV-specific antibody-dependent cellular cytotoxicity. J. Immunol. 2019;203:2210–2221. doi: 10.4049/jimmunol.1900422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D., Prager B.C., Gimple R.C., Aguilar B., Alizadeh D., Tang H., Lv D., Starr R., Brito A., Wu Q. CRISPR screening of CAR T cells and cancer stem cells reveals critical dependencies for cell-based therapies. Cancer Discov. 2021;11:1192–1211. doi: 10.1158/2159-8290.CD-20-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seki A., Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Georgiadis C., Rasaiyaah J., Gkazi S.A., Preece R., Etuk A., Christi A., Qasim W. Base-edited CAR T cells for combinational therapy against T cell malignancies. Leukemia. 2021 doi: 10.1038/s41375-021-01282-6. Published online May 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasaiyaah J., Georgiadis C., Preece R., Mock U., Qasim W. TCRαβ/CD3 disruption enables CD3-specific antileukemic T cell immunotherapy. JCI Insight. 2018;3:99442. doi: 10.1172/jci.insight.99442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooper M.L., Choi J., Staser K., Ritchey J.K., Devenport J.M., Eckardt K., Rettig M.P., Wang B., Eissenberg L.G., Ghobadi A. An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia. 2018;32:1970–1983. doi: 10.1038/s41375-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomes-Silva D., Srinivasan M., Sharma S., Lee C.M., Wagner D.L., Davis T.H., Rouce R.H., Bao G., Brenner M.K., Mamonkin M. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood. 2017;130:285–296. doi: 10.1182/blood-2017-01-761320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fleischer L.C., Raikar S.S., Moot R., Knight K.A., Doering C.B., Spencer H.T. Engineering CD5-targeted chimeric antigen receptors and edited T cells for the treatment of T-cell leukemia. Blood. 2017;130 doi: 10.1182/blood.V130.Suppl_1.1914.1914. [DOI] [Google Scholar]
- 72.Hill L., Rouce R.H., Smith T.S., Yang L., Srinivasan M., Zhang H., Perconti S., Mehta B., Dakhova O., Randall J. CD5 CAR T-cells for treatment of patients with relapsed/refractory CD5 expressing T-cell lymphoma demonstrates safety and anti-tumor activity. Biol. Blood Marrow Transplant. 2020;26(Suppl):S237. [Google Scholar]
- 73.Strati P., Ahmed S., Furqan F., Fayad L.E., Lee H.J., Iyer S.P., Nair R., Nastoupil L.J., Parmar S., Rodriguez M.A. Prognostic impact of corticosteroids on efficacy of chimeric antigen receptor T-cell therapy in large B-cell lymphoma. Blood. 2021;137:3272–3276. doi: 10.1182/blood.2020008865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sachdeva M., Duchateau P., Depil S., Poirot L., Valton J. Granulocyte-macrophage colony-stimulating factor inactivation in CAR T-cells prevents monocyte-dependent release of key cytokine release syndrome mediators. J. Biol. Chem. 2019;294:5430–5437. doi: 10.1074/jbc.AC119.007558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sterner R.M., Sakemura R., Cox M.J., Yang N., Khadka R.H., Forsman C.L., Hansen M.J., Jin F., Ayasoufi K., Hefazi M. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood. 2019;133:697–709. doi: 10.1182/blood-2018-10-881722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodwin M., Lee E., Lakshmanan U., Shipp S., Froessl L., Barzaghi F., Passerini L., Narula M., Sheikali A., Lee C.M. CRISPR-based gene editing enables FOXP3 gene repair in IPEX patient cells. Sci. Adv. 2020;6:eaaz0571. doi: 10.1126/sciadv.aaz0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fraietta J.A., Nobles C.L., Sammons M.A., Lundh S., Carty S.A., Reich T.J., Cogdill A.P., Morrissette J.J.D., DeNizio J.E., Reddy S. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558:307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah N.N., Qin H., Yates B., Su L., Shalabi H., Raffeld M., Ahlman M.A., Stetler-Stevenson M., Yuan C., Guo S. Clonal expansion of CAR T cells harboring lentivector integration in the CBL gene following anti-CD22 CAR T-cell therapy. Blood Adv. 2019;3:2317–2322. doi: 10.1182/bloodadvances.2019000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Micklethwaite K.P., Gowrishankar K., Gloss B.S., Li Z., Street J.A., Moezzi L., Mach M.A., Sutrave G., Clancy L.E., Bishop D.C. Investigation of product derived lymphoma following infusion of piggyBac modified CD19 chimeric antigen receptor T-cells. Blood. 2021 doi: 10.1182/blood.2021010858. Published online May 11, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bishop D.C., Clancy L.E., Simms R., Burgess J., Mathew G., Moezzi L., Street J.A., Sutrave G., Atkins E., McGuire H.M. Development of CAR T-cell lymphoma in two of ten patients effectively treated with piggyBac modified CD19 CAR T-cells. Blood. 2021 doi: 10.1182/blood.2021010813. Published online May 19, 2021. [DOI] [PubMed] [Google Scholar]
- 82.Schambach A., Morgan M., Fehse B. Two cases of T cell lymphoma following Piggybac-mediated CAR T cell therapy. Mol. Ther. 2021;29:2631–2633. doi: 10.1016/j.ymthe.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen D.N., Roth T.L., Li P.J., Chen P.A., Apathy R., Mamedov M.R., Vo L.T., Tobin V.R., Goodman D., Shifrut E. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2020;38:44–49. doi: 10.1038/s41587-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pavani G., Amendola M. Targeted gene delivery: Where to land. Front. Genome Edit. 2021;2:36. doi: 10.3389/fgeed.2020.609650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dar H., Henderson D., Padalia Z., Porras A., Mu D., Kyungah M., Police S., Kalaitzidis D., Terrett J., Sagert J. Preclinical development of CTX120, an allogeneic CAR-T cell targeting Bcma. Blood. 2018;132(Suppl 1):1921. [Google Scholar]
- 86.Brown A.E., MacLeod D.T., Martin A.J., Wetzel K.J., Pham C.D., Antony J., McCreedy B., Bartsevich V.V., Nicholson M.G., Smith J. T cell receptor knockout efficiency utilizing an engineered meganuclease is influenced by stimulation conditions. J. Immunol. 2016;196(Suppl) 138.2. [Google Scholar]
- 87.Cai T., Galetto R., Gouble A., Smith J., Cavazos A., Konoplev S., Lane A.A., Guzman M.L., Kantarjian H.M., Pemmaraju N. Pre-clinical studies of anti-CD123 CAR-T cells for the treatment of blastic plasmacytoid dendritic cell neoplasm (BPDCN) Blood. 2016;128:4039. [Google Scholar]
- 88.Sommer C., Boldajipour B., Valton J., Galetto R., Bentley T., Sutton J., Ni Y., Leonard M., Van Blarcom T., Smith J. ALLO-715, an allogeneic BCMA CAR T therapy possessing an off-switch for the treatment of multiple myeloma. Blood. 2018;132(Suppl 1):591. [Google Scholar]
- 89.Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.Q., Reik A., Chu V., Paschon D.E., Zhang L., Kuball J. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mastaglio S., Genovese P., Magnani Z., Ruggiero E., Landoni E., Camisa B., Schiroli G., Provasi E., Lombardo A., Reik A. NY-ESO-1 TCR single edited stem and central memory T cells to treat multiple myeloma without graft-versus-host disease. Blood. 2017;130:606–618. doi: 10.1182/blood-2016-08-732636. [DOI] [PubMed] [Google Scholar]
- 91.Mullard A. Gene-editing pipeline takes off. Nat. Rev. Drug Discov. 2020;19:367–372. doi: 10.1038/d41573-020-00096-y. [DOI] [PubMed] [Google Scholar]
- 92.Poirot L., Philip B., Schiffer-Mannioui C., Le Clerre D., Chion-Sotinel I., Derniame S., Potrel P., Bas C., Lemaire L., Galetto R. Multiplex genome-edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75:3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 93.Qasim W., Zhan H., Samarasinghe S., Adams S., Amrolia P., Stafford S., Butler K., Rivat C., Wright G., Somana K. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017;9:eaaj2013. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 94.Benjamin R., Graham C., Yallop D., Jozwik A., Ciocarlie O., Jain N., Jabbour E.J., Maus M.V., Frigault M., Boissel N. Preliminary data on safety, cellular kinetics and anti-leukemic activity of UCART19, an allogeneic anti-CD19 CAR T-cell product, in a pool of adult and pediatric patients with high-risk CD19+ relapsed/refractory B-cell acute lymphoblastic leukemia. Blood. 2018;132(Suppl 1):896. [Google Scholar]
- 95.Benjamin R., Graham C., Yallop D., Jozwik A., Mirci-Danicar O.C., Lucchini G., Pinner D., Jain N., Kantarjian H., Boissel N., UCART19 Group Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: Results of two phase 1 studies. Lancet. 2020;396:1885–1894. doi: 10.1016/S0140-6736(20)32334-5. [DOI] [PubMed] [Google Scholar]
- 96.Hu Y., Zhou Y., Zhang M., Ge W., Li Y., Yang L., Wei G., Han L., Wang H., Zhang X. The safety and efficacy of a CRISPR/Cas9-engineered universal CAR-T cell product (CTA101) in patients with relapsed/refractory B-cell acute lymphoblastic leukemia. Blood. 2020;136(Suppl 1):52. [Google Scholar]
- 97.Lee J., Sheen J.H., Lim O., Lee Y., Ryu J., Shin D., Kim Y.Y., Kim M. Abrogation of HLA surface expression using CRISPR/Cas9 genome editing: A step toward universal T cell therapy. Sci. Rep. 2020;10:17753. doi: 10.1038/s41598-020-74772-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kagoya Y., Guo T., Yeung B., Saso K., Anczurowski M., Wang C.H., Murata K., Sugata K., Saijo H., Matsunaga Y. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to be used for adoptive T-cell therapy. Cancer Immunol. Res. 2020;8:926–936. doi: 10.1158/2326-6066.CIR-18-0508. [DOI] [PubMed] [Google Scholar]
- 99.Pavlovic K., Tristán-Manzano M., Maldonado-Pérez N., Cortijo-Gutierrez M., Sánchez-Hernández S., Justicia-Lirio P., Carmona M.D., Herrera C., Martin F., Benabdellah K. Using gene editing approaches to fine-tune the immune system. Front. Immunol. 2020;11:570672. doi: 10.3389/fimmu.2020.570672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larson M.H., Gilbert L.A., Wang X., Lim W.A., Weissman J.S., Qi L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li L., Gao Y., Srivastava R., Wang W., Xiong Q., Fang Z., Pelayo A., Denson C., Goswami A., Harari-Steinfeld R. Lentiviral delivery of combinatorial CAR/CRISPRi circuit into human primary T cells is enhanced by TBK1/IKKϵ complex inhibitor BX795. J. Transl. Med. 2020;18:363. doi: 10.1186/s12967-020-02526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simeonov D.R., Gowen B.G., Boontanrart M., Roth T.L., Gagnon J.D., Mumbach M.R., Satpathy A.T., Lee Y., Bray N.L., Chan A.Y. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111–115. doi: 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kang L., Tang X., Xu N., Li M., Tan J., Qi W., Yu Z., Lou X., Xue S., Li X. shRNA-interleukin-6 modified CD19-specific chimeric antigen receptor T cell significantly improves the safety in acute lymphoblastic leukemia. Blood. 2019;134(Suppl 1):2621. [Google Scholar]
- 105.Shah B.D., Jacobson C.A., Solomon S., Jain N., Vainorius M., Heery C.R., He F.C., Reshef R., Herrera A.F., Akard L.P. Preliminary safety and efficacy of PBCAR0191, an allogeneic, off-the-shelf CD19-targeting CAR-T product, in relapsed/refractory (r/r) CD19+ NHL. J. Clin. Oncol. 2021;39:7516. 7516. [Google Scholar]
- 106.Jeong Y.K., Song B., Bae S. Current status and challenges of DNA base editing tools. Mol. Ther. 2020;28:1938–1952. doi: 10.1016/j.ymthe.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zeng J., Wu Y., Ren C., Bonanno J., Shen A.H., Shea D., Gehrke J.M., Clement K., Luk K., Yao Q. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 2020;26:535–541. doi: 10.1038/s41591-020-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Webber B.R., Lonetree C.L., Kluesner M.G., Johnson M.J., Pomeroy E.J., Diers M.D., Lahr W.S., Draper G.M., Slipek N.J., Smeester B.A. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019;10:5222. doi: 10.1038/s41467-019-13007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gaudelli N.M., Lam D.K., Rees H.A., Solá-Esteves N.M., Barrera L.A., Born D.A., Edwards A., Gehrke J.M., Lee S.J., Liquori A.J. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020;38:892–900. doi: 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- 110.Petri K., Zhang W., Ma J., Schmidts A., Lee H., Horng J.E., Kim D.Y., Kurt I.C., Clement K., Hsu J.Y. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00901-y. Published online April 29, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maldini C.R., Ellis G.I., Riley J.L. CAR T cells for infection, autoimmunity and allotransplantation. Nat. Rev. Immunol. 2018;18:605–616. doi: 10.1038/s41577-018-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zmievskaya E., Valiullina A., Ganeeva I., Petukhov A., Rizvanov A., Bulatov E. Application of CAR-T cell therapy beyond oncology: Autoimmune diseases and viral infections. Biomedicines. 2021;9:59. doi: 10.3390/biomedicines9010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aghajanian H., Kimura T., Rurik J.G., Hancock A.S., Leibowitz M.S., Li L., Scholler J., Monslow J., Lo A., Han W. Targeting cardiac fibrosis with engineered T cells. Nature. 2019;573:430–433. doi: 10.1038/s41586-019-1546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kleinstiver B.P., Sousa A.A., Walton R.T., Tak Y.E., Hsu J.Y., Clement K., Welch M.M., Horng J.E., Malagon-Lopez J., Scarfò I. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Azangou-Khyavy M., Ghasemi M., Khanali J., Boroomand-Saboor M., Jamalkhah M., Soleimani M., Kiani J. CRISPR/Cas: From tumor gene editing to T cell-based immunotherapy of cancer. Front. Immunol. 2020;11:2062. doi: 10.3389/fimmu.2020.02062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Perales M.A., Kebriaei P., Kean L.S., Sadelain M. Building a safer and faster CAR: Seatbelts, airbags, and CRISPR. Biol. Blood Marrow Transplant. 2018;24:27–31. doi: 10.1016/j.bbmt.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]