Abstract
Recent advances in genome editing technologies have magnified the prospect of single-dose cures for many genetic diseases. For most genetic disorders, precise DNA correction is anticipated to best treat patients. To install desired DNA changes with high precision, our laboratory developed base editors (BEs), which can correct the four most common single-base substitutions, and prime editors, which can install any substitution, insertion, and/or deletion over a stretch of dozens of base pairs. Compared to nuclease-dependent editing approaches that involve double-strand DNA breaks (DSBs) and often result in a large percentage of uncontrolled editing outcomes, such as mixtures of insertions and deletions (indels), larger deletions, and chromosomal rearrangements, base editors and prime editors often offer greater efficiency with fewer byproducts in slowly dividing or non-dividing cells, such as those that make up most of the cells in adult animals. Both viral and non-viral in vivo delivery methods have now been used to deliver base editors and prime editors in animal models, establishing that base editors and prime editors can serve as effective agents for in vivo therapeutic genome editing in animals. This review summarizes examples of in vivo somatic cell (post-natal) base editing and prime editing and prospects for future development.
Graphical abstract
Base editing and prime editing have enabled new paths to treat genetic disease by precise in vivo correction. Newby and Liu summarize published examples of in vivo base and prime editing. They review the mechanisms, capabilities, and limitations of these genome editing agents and discuss future prospects.
Introduction
More than 1 in 50 newborns are estimated to suffer from a heritable genetic disorder.1 Some gene therapies have started to address such diseases by delivering new copies of functional genes.2 However, gene therapy approaches typically cannot restore native gene regulation, can fail to maintain expression over time, and are unable to deliver very large genes. An ideal approach to treating genetic disease would permanently correct the causative disease mutation at the native site in the genome. No such therapies have yet reached patients, although some ongoing clinical trials disrupt targeted genomic loci with indels to yield therapeutic effects. For most genetic diseases, however, precise edits without mixtures of editing byproducts offer patients the best prospects to rescue the effects of a pathogenic mutation. Even dominant or gain-of-function mutations that can feasibly be treated by disruption of mutant sequences can also benefit from precise correction to restore endogenous expression of the healthy gene and minimize uncontrolled and undesired consequences of double-strand DNA breaks (DSBs).
Two classes of genome editing agents are particularly well-suited for directly correcting disease mutations because they can install programmable edits with relatively few uncontrolled indel outcomes (high product purity). These are (1) base editors (BEs),3, 4, 5 which use a programmable DNA binding protein such as a catalytically impaired CRISPR-Cas protein or a TALE repeat array to direct an adenine or cytidine deaminase to modify a targeted window of single-stranded DNA, resulting in C•G to T•A or A•T to G•C conversions; and (2) prime editors,6 which use a nuclease-impaired Cas protein to direct a reverse transcriptase that can replace or insert any desired sequence based on the information encoded in the co-delivered prime editor guide RNA (pegRNA). Base editors can correct transition mutations, the largest single class of human disease-causing mutations, accounting for ∼30% of known disease alleles. Prime editors are highly versatile and have been demonstrated to be capable of installing any base-to-base change as well as insertions of up to 44 base pairs and deletions of up to 80 base pairs. In theory, these features could permit prime editors to correct >89% of known human disease-causing mutations, excluding only those that involve aneuploidy, chromosomal rearrangement, or large duplications, insertions, or deletions.6
Direct genomic correction in the proper cell population at an appropriate intervention time has the potential to permanently cure genetic diseases. A major challenge to achieving this goal is the ability to safely deliver genome editing agents in sufficient quantities to a large enough fraction of relevant cells in vivo. The demonstration and optimization of genome editing in animal models is an important stepping stone on the path to the clinic. While many diseases do not yet have suitable delivery methods, some recent demonstrations have powerfully demonstrated potential approaches that can be adapted into clinical trials and human therapeutics.
In this review, we discuss three classes of mammalian cell genome editing agents that have been delivered in vivo and, within space limitations, present a summary of some relevant delivery methods for in vivo post-natal somatic cell genome editing. We also discuss preclinical demonstrations of therapeutic base editing and prime editing in mammalian models and prospects for future development.
Genome editing agent classes and therapeutic applicability
Integrases,7 recombinases,8 transposases,9 endonucleases such as meganucleases,10 and zinc finger nucleases (ZFNs)11 have all been demonstrated to introduce genomic changes in mammalian cells. However, because these technologies are not easy to reprogram to target any genomic site of interest, custom editor proteins must be generated through directed evolution, selection, or rational design in order to modify a desired genomic locus. Transcription activator-like effector nucleases (TALENs)12 are simple to design but not trivial to construct or clone. The generation of all these tools is typically time and resource intensive, and the resulting candidate editing agents can vary substantially in activity and specificity for the targeted site.
The discovery and subsequent development of CRISPR-Cas systems for genome editing13 greatly simplified the challenge of retargeting a nuclease to a genomic locus of interest. Cas9 was originally discovered as a component of an adaptive immune system in bacteria that cleaves bacteriophage DNA.14 The Cas9 protein can bind a guide RNA molecule that directs its nuclease activity to a target DNA sequence through simple guide RNA:DNA base pairing to the targeted site. Specificity of the ribonucleoprotein (RNP) complex can be reprogrammed by altering a short stretch of approximately 20 base pairs in the guide molecule. This method obviates the need for labor-intensive protein engineering or evolution that is typically required to retarget previous gene editing tools. Although simple rules for rational design of TALENs have been described,15,16 generating a TALEN to target a desired site requires synthesis of a multi-kilobase gene and production of a new protein for each target, rather than simply replacing 20 nucleotides of a small, co-delivered RNA as for Cas9.
The facile targeting enabled by Cas9 and the ease with which its nuclease activity can be inactivated while preserving its DNA-binding capability have enabled new tools that conduct other functions besides the introduction of DSBs. These include the first base editors and prime editors, as well as other Cas-guided effectors like transposases17,18 and domains that activate19 or repress20,21 transcription. CRISPR-free base editors that use TALE repeat arrays instead of Cas proteins to target DNA have also been developed, enabling precision gene editing in organelles that cannot be edited with CRISPR agents.22 Nucleases, base editors, and prime editors have already shown promising results in animal models, in some cases resulting in dramatic rescue of disease phenotypes following a one-time administration of the editing agent, as summarized below.
Nucleases
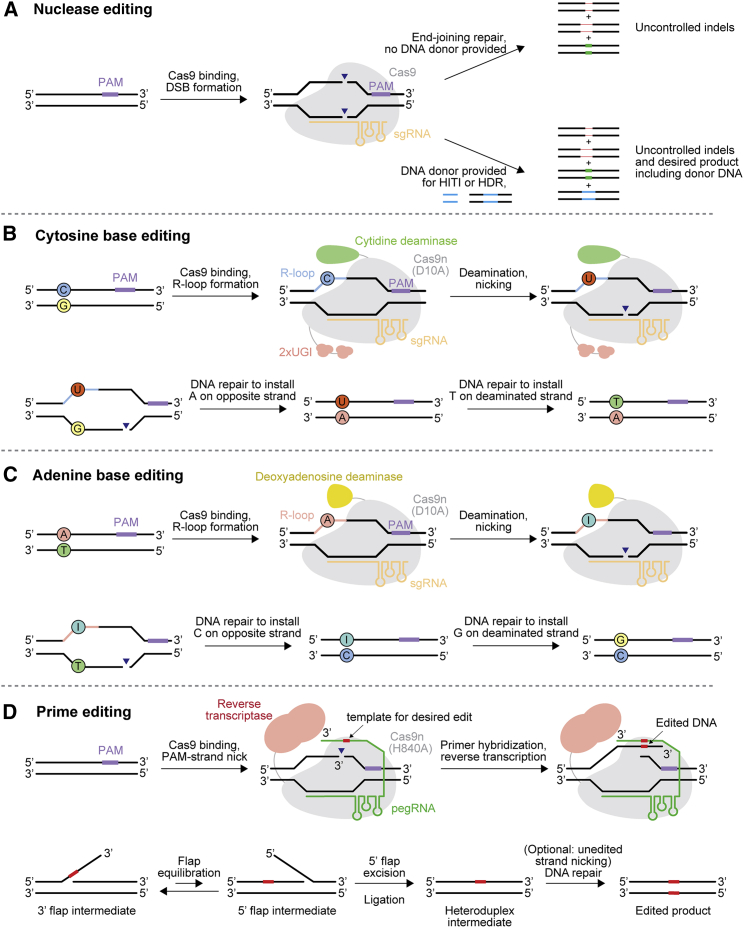
Nucleases edit the genome by the introduction of DSBs (Figure 1A). DSBs are typically repaired by non-homologous end joining (NHEJ)23,24 or microhomology-mediated end joining (MMEJ)25,26 processes, both of which result in small insertions and deletions (indels) (Figure 1A). End joining can yield perfect correction back to the pre-cut state, but such a perfectly corrected product can serve as a substrate once again for the nuclease. The set of indels resulting from NHEJ and MMEJ are often predictable and reproducible27, 28, 29 but are dictated by the surrounding sequence context and repair machinery. Thus, the specific outcomes of end-joining repair pathways have not been possible to control by researchers.
Figure 1.
Genome editing mechanisms of base editors, prime editors, and nucleases
(A) Nuclease-mediated editing. (B) Cytidine base editing. (C) Adenine base editing. (D) Prime editing. PAM, protospacer-adjacent motif; UGI, uracil glycosylase inhibitor domain; Cas9n, Cas9 nickase; sgRNA, single guide RNA; pegRNA, prime editor guide RNA; HITI, homology-independent targeted integration; HDR, homology-directed repair.
Large disruptions can also result from DSBs. Deletions or duplications spanning hundreds to thousands of base pairs,30,31 or loss of chromosome arms,32 have been reported as repair products following DSB introduction with frequencies of greater than 10% of resulting alleles. These outcomes are not detectable by standard amplicon sequencing that involves PCR amplification of a few hundred nucleotides around the target site but are detectable by long-read sequencing, whole-genome sequencing, or loss-of-heterozygosity analysis that involves comparing heterozygous single-nucleotide polymorphisms (SNPs) before and after editing. In addition, when multiple DSBs are produced simultaneously in different chromosomes, chromosomal translocation can result. The frequency of such translocation outcomes is about 0.5%–3% when two DSBs are simultaneously targeted by nucleases.33, 34, 35 However, a single nuclease could also cause translocations, particularly if off-target DSBs are also introduced. Extensive chromosomal rearrangements and copy number variations that occur between the cut site and the end of a chromosome, collectively called chromothripsis, as well as micronuclei composed of acentric chromosome arms separated by the DSB, have also been described as outcomes following DSBs.36 Although the capacity of nucleases to introduce unwanted genome changes that in principle could contribute to cancer should be considered in any potential therapy, they constitute relatively rare outcomes, and their potential clinical relevance is unknown.
Another potentially detrimental outcome following DSB formation by nucleases is the activation of the p53-mediated DNA damage response.37, 38, 39,206 Upon cellular detection of DSBs, p53 normally halts progression through the cell cycle until the damage is repaired or, if repair is not achieved, promotes the cell to undergo apoptosis.40 Dividing cells surviving nuclease-mediated genome editing may be enriched in cells that have inactivated their p53 DNA-damage response and continue to divide without proper checks for genome integrity,39 which could promote oncogenesis. Even in the absence of p53-inactivating mutations, a loss of edited cell fitness incurred by p53-induced cell cycle arrest could complicate nuclease-mediated therapeutics.
Nuclease-mediated editing with a donor DNA template
The most versatile application of nucleases for genome editing is the ability to insert, delete, or replace any desired sequence at a target site using homology-directed repair (HDR),41 templated by an exogenous donor DNA molecule (Figure 1A). The ability to conduct Cas9-mediated HDR in mammalian cells has advanced the study of human genetics and enabled the construction of a wide range of cell and animal models with modifications at genomic sites of interest. While highly impactful for biological research, HDR is limited in its therapeutic potential because it is inefficient in non-dividing or slowly dividing cells, such as those that make up most of the post-natal body. This inefficiency is at least partly due to the cell-cycle-dependent expression of cellular factors that mediate HDR, which are primarily expressed during S, G2, and M phases.42, 43, 44, 45 In addition, the requirement of a DNA donor template complicates the prospects for in vivo HDR, both due to the additional component that must be delivered and the potential for immune responses triggered by exogenous DNA. For these reasons, the application of HDR has been limited primarily to cultured cells, zygotes, and embryos. To our knowledge, the only reported examples of in vivo HDR in adult mammals have yielded relatively low editing efficiencies of 0.1%–6.5%.46, 47, 48, 49, 50, 51
An alternative nuclease-mediated method that can introduce targeted insertions in genomic DNA in non-dividing cells is homology-independent targeted integration (HITI).52 HITI relies on NHEJ to insert double-stranded DNA fragments provided in trans. Orientation of integration cannot be specified using HITI, and indels at each DSB site are also generated. The delivery of Cas9 nuclease and HITI donor DNA via a dual adeno-associated virus (AAV) system resulted in 4.5% restoration of Mertk expression levels in the mouse eye in a mouse model of retinitis pigementosa52 and permitted expression of an Abcd1 transgene in many tissues following systemic delivery in a mouse model of adrenoleukodystrophy.53
Ex vivo editing of cultured primary cells via HDR or HITI, followed by transplantation back into the patient, is a potential therapeutic strategy. HDR or HITI has been demonstrated in primary hematopoietic stem and progenitor cells (HSPCs),54, 55, 56, 57, 58, 59, 60,207,208 T cells,61,62 airway basal stem cells,63 induced pluripotent stem cells (iPSCs),64, 65, 66 and mesenchymal stromal cells,67 with efficiencies ranging from 10%–75%, which correspond to therapeutic levels of editing for some diseases. However, the majority of alleles that do not successfully incorporate the DNA donor are indel products resulting from NHEJ/MMEJ repair of DSBs without the involvement of the DNA donor, and heterozygous outcomes with one edited and one disrupted allele are common. Complete rescue of a genetic disease phenotype based on transplantation of cells edited by HDR or HITI is challenging due to incomplete editing, the difficulty of fully replacing endogenous cells with transplanted ones, and potentially lower fitness or engraftment durability of edited cells following DSBs.
Nuclease-mediated disruption of target genomic loci
Most therapeutic demonstrations using nucleases are for cases in which disruption of target genomic sites can be therapeutic. Several of these demonstrations have involved in vivo genome editing. One such example is the treatment of a blindness disease mutation causing Leber congenital amaurosis type 10, which generates an aberrant splice donor in CEP290. In vivo sub-retinal delivery of a Cas9 nuclease via AAV was able to edit this genomic disease locus in mouse and non-human primate (NHP) models.68 Disruption of a mutated exon in Dmd corrected muscular dystrophy phenotypes in a mouse.69 Cas9-mediated in vivo knockout of Pcsk9 in the liver has been demonstrated as a potential treatment for hypercholesterolemia.70 Recently, an exciting phase I clinical report used Cas9 nuclease in vivo to disrupt transthyretin in the human liver for the treatment of hereditary transthyretin (ATTR) amyloidosis, showing good tolerance and efficient transthyretin knockdown in study participants,71 building on promising results achieved earlier in a mouse model.72
Ex vivo delivery of nucleases followed by transplantation of edited cells also has strong therapeutic potential, particularly in the blood, where engraftment is less of a barrier relative to solid organs. For example, sickle cell disease and β-thalassemia can both be rescued by upregulation of fetal hemoglobin, which is normally silenced around the time of birth. Nuclease-mediated introduction of indels in HSPCs at the fetal hemoglobin repressor BCL11A73,74 or at the fetal hemoglobin promoter75,76 can induce fetal hemoglobin expression and restore healthy red blood cells. Knockout of the HIV co-receptor CCR5 in T cells can protect against HIV infection.77,78 Furthermore, knockout of the checkpoint inhibitor PD-1 and other factors in T cells can permit enhanced anti-tumor activity,79, 80, 81 potentially improving CAR-T therapy.35
The high activity and facile programmability of Cas9 nuclease has enabled many nuclease-based therapeutic strategies. However, most genetic diseases cannot be addressed by indel introduction, and the poor efficiency of repair with a DNA donor in most therapeutically relevant cell types, as well as the potential drawbacks of making DSBs, emphasized the need for new gene editing technologies that overcome these limitations. To address these challenges, our laboratory developed base editors and prime editors.
Base editors
Base editors consist of DNA-modifying enzymes fused to a programmable DNA-targeting moiety. Our group, and Kondo and coworkers, reported the first cytosine base editors (CBEs) in 2016 using natural single-stranded DNA deaminase domains that convert cytosine nucleotides to uracil, which is read as a thymine during DNA replication, DNA repair, and transcription3, 5 (Figure 1B). In this manner, CBEs can convert C•G base pairs first to U•G base pairs and ultimately to T•A base pairs following DNA repair. To direct and limit the activity of deaminase to the desired genomic site, nuclease-impaired Cas9 was fused to single-strand cytidine deaminases. Once the guide RNA and catalytically inactivated Cas9 engage the target DNA, it creates a single-stranded stretch of genomic DNA (the R-loop), within which the deaminase converts cytosines within a small editing window into uracils. Cellular DNA repair machinery can resolve the resulting base mismatch toward the desired state or back to the starting state, but using a Cas9 nickase to nick the non-deaminated strand biases mismatch repair to replace that strand with a sequence templated from the edited one.3 In 2017, we reported the first adenine base editors (ABEs), which convert A•T base pairs to G•C base pairs4 (Figure 1C). Because natural deaminases that act on deoxyadenosine are not known, laboratory evolution of a deoxyadenosine deaminase was required to engineer ABEs. All ABEs described to date use this laboratory-evolved deoxyadenosine deaminase, or variants thereof.
Since the original description of base editors, more than 100 base editor variants have been developed to suit particular sequence contexts or to modulate the editing window, activity, or specificity of base editors. For a detailed discussion of these variants and how to choose the best one for a given application, we refer the reader to recent reviews.82,83 In 2020, our group, in collaboration with the Mougous and Mootha labs, reported base editors using TALE arrays to direct double-stranded DNA deaminases to enable CRISPR-free base editing in the nucleus and in mitochondria,22 where the delivery of guide RNAs has not yet been achieved. In addition, by promoting excision of the uracil intermediate generated during cytosine base editing and subsequent translesion DNA synthesis, C•G to G•C base editors have also been reported.84, 85, 86
Base editors are particularly attractive tools for genome editing because: (1) base editors do not rely on cellular HDR machinery, so they can install programmed edits with high efficiency even in non-dividing cells; (2) the editing outcome is particularly pure, typically with >10:1 ratios of desired editing products:indel byproducts; (3) base editing does not require DNA template delivery and can be performed entirely with mRNA or RNP agents; and (4) SNPs are the most common class of human disease-associated mutations, and base editors can theoretically correct >70% of disease-associated SNPs. ABEs are particularly applicable to the correction of human disease SNPs with the ability to correct approximately half of pathogenic point mutations.4 Furthermore, the translocations, large deletions, and p53 DNA damage responses that are observed when using nucleases are absent or greatly decreased when using base editors.31,33,87
The main limitation of base editors is the careful positioning required to place the target within the optimal editing window and exclude undesired bystander edits. The optimal editing window for original base editors is approximately nucleotides 4–7 of the protospacer, where nucleotide 1 is the most distant nucleotide from the protospacer-adjacent motif (PAM). Cas9 requires a PAM to bind its target sequence, and if the protospacer cannot be appropriately positioned, efficient base editing may not be possible. To address these limitations, many new Cas and deaminase variants have been described that have altered PAM specificities88, 89, 90, 91, 92 or editing windows.82,93, 94, 95, 96, 97, 98, 99 Some deaminases favor acting on nucleotides with particular sequence contexts,3,94,100 which can be useful to minimize editing of nearby bystander nucleotides. Adenine base editing has been reported to occasionally result in bystander cytosine deamination,101 but recent work has reported engineered deaminase variants that mitigate this phenomenon.102
An additional concern when using base editors, like any genome editing agent, for therapeutic applications is the potential for off-target editing. Off-target base editing can manifest as guide-independent, spurious editing of RNA103,104 or genomic DNA95,105,106 or as guide-dependent off-target editing at sites that are engaged by the RNP despite not perfectly matching the guide sequence.3,4 Although few detrimental effects of off-target editing have been described, the potential of spurious editing to contribute to oncogenesis is a major consideration for all therapeutic applications of genome editing. Many base editor variants have been described for both CBEs103,105,107,108 and ABEs95,97,104 that minimize or prevent off-target DNA or RNA editing while maintaining high on-target activity.
While in vivo base editing applications are discussed in detail below, ex vivo base editing of cells followed by transplantation is also an attractive potential therapeutic strategy. HSPCs can form the basis of autologous bone marrow transplantation and can be efficiently edited in 80% of alleles or more by electroporation of base editor mRNA87,109 or RNP.87,110 Because base editors do not introduce DSBs, they are a particularly useful tool for multiplex editing, whereas targeting multiple sites simultaneously with nucleases could cause toxicity due to the DNA damage response and frequent translocations arising from multiple DSBs in the same cell. In primary human T cells, over 90% editing at each of three sites was observed using CBE33 and ABE,97 and human HSPCs can be edited with greater than 80% efficiency at each of two sites.110 Base editing in human iPSCs, another cell type of interest for autologous transplantation, has also reached over 80% efficiency.111 Because autologous grafts can avoid immune complications, the ability to precisely modify a patient’s own cells for subsequent engraftment could enable more durable and safe treatments for genetic blood diseases.
Prime editors
To introduce precise DNA edits beyond those that can be installed by base editing without requiring DSBs, we reported prime editing (PE) in 2019.6 Prime editors are composed of an engineered reverse transcriptase fused to Cas9 nickase that introduces a nick in the R-loop at the target DNA site.6 The pegRNA contains a 3′ extension that anneals to the nicked target DNA strand. The annealed target DNA:pegRNA serves as a primer-template complex for reverse transcriptase, which polymerizes the desired sequence onto the nicked target DNA site as specified by a template encoded in the pegRNA. Cellular DNA repair then resolves the flap of edited DNA, resulting in permanent incorporation into the genome (Figure 1D). Prime editors were demonstrated to be able to install any single base-to-base change, deletions of at least 80 nucleotides, and insertions of at least 44 nucleotides.6
Prime editors are attractive tools for genome editing because they offer (1) high versatility relative to base editors while maintaining the advantages of avoiding DSBs, (2) pure editing outcomes with few indels relative to nuclease-mediated HDR, and (3) exquisitely precise editing with few to no off-target edits introduced,112, 113, 114, 115 likely due to the requirement of two additional DNA hybridization events (reverse-transcriptase priming and DNA flap resolution) beyond Cas9 binding that each provide opportunities to reject off-target sequences.
Using the simple two-component system of a prime editor and pegRNA (PE2), moderate editing activity of approximately 5%–20% is typically achieved with a particularly pure outcome yielding fewer than 1% indels. By delivering a second simple guide RNA that directs the same protein to nick the non-edited strand, but has no 3′ extension upon which the reverse transcriptase can act, DNA mismatch repair can be biased to favor resolving the heteroduplex intermediate toward the desired edited outcome (Figure 1D). While this “PE3” editing strategy that includes an additional nick can increase editing efficiencies 1.5- to 4-fold, it also has the potential to increase indel formation, often to approximately 10% of resulting alleles.6
Recently, our group, in collaboration with the Adamson and Weissman laboratories, conducted a pooled screen to identify DNA repair factors that counteract desired prime editing outcomes.116 We identified mismatch repair as a key antagonizing pathway, especially for small edits spanning no more than a few nucleotides. Inhibiting this pathway through the expression of an engineered MLH1 dominant negative gene resulted in both greater efficiency of the desired editing outcome and higher product purity (fewer undesired indel outcomes). We designated these new prime editing systems PE4 and PE5 when lacking or including a nicking guide RNA, respectively. In the same study, codon usage, nuclear localization signal (NLS) architectures, and Cas9 variants were also assessed, and an optimal “PEmax” architecture was established that further increased prime editing efficiencies. Separately, Kim and coworkers improved prime editing efficiency by fusing chromatin-modulating peptides to the editor, and Xue and coworkers also optimized NLS architecture and codon usage.117, 118
Besides the prime editor protein itself, the pegRNA architecture is also the subject of recent developments that improve PE outcomes. Our group, as well as Wang and colleagues, and others, observed that including a 3′ motif for pegRNAs can preserve their otherwise labile 3′ extension in the cell and confer greater editing activity.119, 120 Gao, Shendure, and their respective coworkers have reported dual pegRNA editing strategies that employ two pegRNAs to transcribe complementary sequences that improve editing outcomes.121, 122 The design of pegRNAs is a difficult process, so several computational tools have been developed to assist investigators,123, 124, 125, 126 including one that designs dual pegRNAs.121 In cultured cells, improvements to the editor protein (PE4max and PE5max) and the use of engineered pegRNAs (epegRNAs)120 synergize to yield a substantial boost in prime editing efficiencies, with typically 30%–60% of alleles edited to the desired outcome even in primary cells.116 Initially reported prime editors (PE2 and PE3) have already shown the ability to edit mouse,117,127,128 zebrafish,129 and Drosophila130 embryos with 10%–50% efficiency to generate animal models. In cultured hepatic progenitors, 2.3% prime editing was achieved, which was sufficient to extend the lifespan of an adult tyrosinemia mouse model after transplantation.131 We anticipate that the application of next-generation prime editors will further expand their utility both in vivo and ex vivo.
Genome editing agent delivery platforms
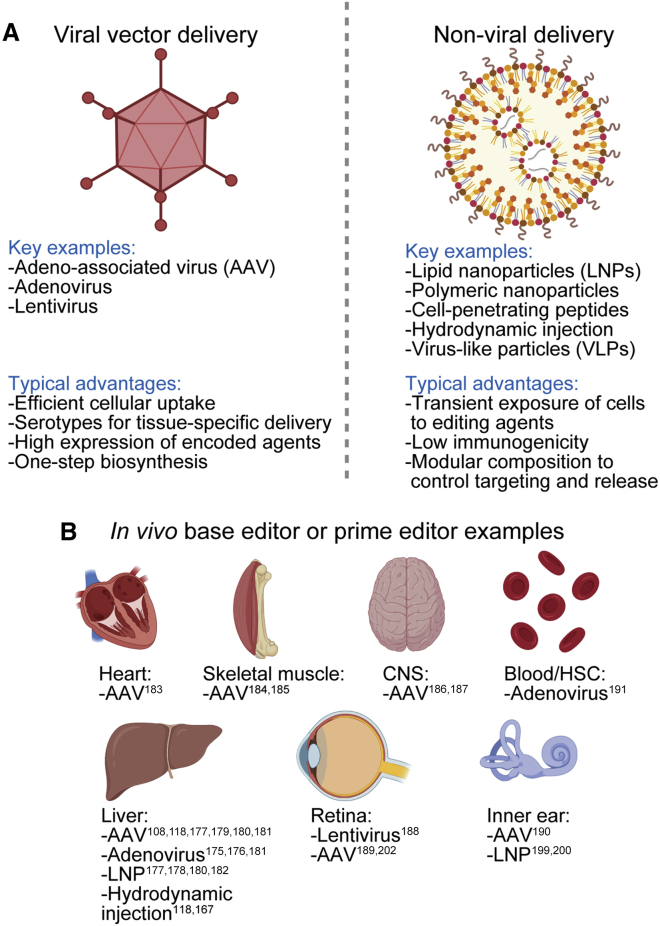
To edit cellular genomes effectively in vivo, appropriate vehicles must be used to deliver the editor to the relevant cell types (Figures 2A and 2B). A wide array of in vivo delivery methods have been described and reviewed elsewhere.132, 133, 134, 135 A selection of in vivo delivery modalities most relevant to gene editing are briefly summarized below.
Figure 2.
In vivo delivery options and demonstrations
(A) Key delivery options and their typical advantages. (B) Published reports of in vivo base editor and prime editor delivery to tissues. Images created using BioRender.com. CNS, central nervous system; HSC, hematopoietic stem cell.
Viral vectors
Viral vectors represent one such promising approach. Over millions of years of natural selection, viruses have evolved to readily infect cells and deliver their nucleic acid payload. Co-opting viruses to deliver genome editing agents has proven to be an effective method to achieve in vivo editing of cell types amenable to viral transduction. When using a viral vector for delivery, packaging limitations of the nucleic acid cargo are important constraints. The open reading frame encoding S. pyogenes Cas9, which is most commonly used for genome editing, is 4.1 kilobases (kb), base editors using the same targeting moiety are 4.8–5.6 kb, and prime editors are 6.4 kb. The addition of regulatory elements and a guide RNA cassette further increase the required cargo size.
Lentiviruses deliver RNA cargo that is reverse-transcribed and integrated into the host cell genome. Lentiviruses are most commonly used for ex vivo modification of cells, although in vivo delivery to the bone marrow, liver, and retina has also been demonstrated.136, 137, 138 Advantages of using a lentiviral vector for the delivery of genome editing agents are the moderate packaging capacity (∼8 kb) and the option to modulate tropism by exchanging the envelope glycoprotein, including the vesicular stomatitis virus G (VSVG) glycoprotein that confers broad tropism. Disadvantages include host immune responses against the virus, prolonged expression of a delivered editor when only transient expression is required, and the potential of the lentiviral genome to integrate in a detrimental location in the genome, dysregulating nearby genes.
Adenoviruses deliver DNA cargo that is transiently maintained episomally in the nucleus. Advantages of using an adenovirus to deliver a genome editing agent include the large packaging capacity (∼36 kb), the short duration of editor expression, which is desirable to minimize off-target editing, and the rarity of mutagenic genomic integration of the delivered DNA.139 The primary disadvantage of delivery via adenovirus is the potential of the vector to elicit a robust immune response in the patient, although the most recent generation of adenoviral vectors have greatly reduced this immunogenicity by removing endogenous viral genes from the delivered genome.140
AAVs deliver DNA cargo that is maintained episomally in the nucleus but persists for a longer duration compared to adenovirus; while expression from adenovirus is typically lost after less than 4 weeks, AAV expression can persist for years.141,142 AAV is attractive due to its particularly low immunogenicity, its clinical validation in both experimental and US Food and Drug Administration (FDA)-approved therapies,143 and the various natural and engineered serotypes that offer a variety of tissue-type specificities.144,145 Disadvantages of AAV for delivering genome editing agents include its small packaging capacity (∼4.7 kb, small enough that single-AAV production even of only S. pyogenes Cas9 together with one guide RNA and needed regulatory and accessory sequences is inefficient) and the long duration of expression, which is typically unnecessary and therefore undesired when delivering gene editing agents.
Non-viral platforms
A variety of synthetic delivery platforms have been engineered to permit transient delivery of DNA, RNA, or protein into cells without using viral components.146 While such methods typically do not reach the high transduction efficiency of evolved viral capsids, they may be less immunogenic, are typically not limited by the packaging capacity, and can minimize the duration of exposure of cells to editing activity when directly delivering genome editing agent mRNA or RNP.
Lipid nanoparticles (LNPs) are the most established platforms for the delivery of macromolecules including DNA, mRNA, and protein into cells, having been used for drug delivery since the 1990s147 and forming the vehicle for the first FDA-approved RNAi therapeutic in 2018.148 Ionizable cationic lipids complexed with the desired cargo enter cells first through endocytosis. Upon acidification of the endosome, these lipids become positively charged and interact with the negatively charged lipids that form the endosome membrane, leading to disruption of the membrane and delivery of the cargo into the cytoplasm.147 Modulating the components of lipid nanoparticles can confer additional functionality,149 including improving delivery to liver,150, 151, 152 neurons,153 lung and spleen,154, 155, 156 and tumors.157 Several polymeric nanoparticles have been developed to permit more controlled composition, size, and release of the cargo.149,158, 159, 160 Targeting moieties have been fused to the polymer constituents to direct cell specificity.161 Hybrid LNP-polymer nanoparticles have been developed with improved liver targeting and expression.162
Cell-penetrating peptides have been used to deliver macromolecules into cells,163 including Cas nucleases.164 While cell-penetrating peptides are not demonstrated to be compatible with systemic in vivo delivery of editors, they may be directly applied to epithelial cells in the lung165 or brain166 to achieve uptake and editing.
Hydrodynamic injection, involving the rapid addition of a large volume of solution to permeabilize cells, has been used to deliver genome editing agents in rodents.49,50,118,167 Although systemic delivery in humans is precluded due to the cardiac congestion resulting from quickly adding a large volume to the blood, local delivery of editor reagents to patients may be possible in some parts of the body.168, 169, 170
Virus-like particles can combine the benefits of viral and non-viral delivery platforms by using a viral capsid that evolved for efficient cell transduction to deliver a cargo of editor RNP.171, 172, 173, 174 Because RNP is quickly turned over, this method should minimize exposure time to the editing agent and reduce off-target editing.95,105 Thus, for cell types amenable to viral transduction, VLPs are particularly attractive delivery tools.
Therapeutic in vivo base editing
In vivo gene editing to treat patients suffering from diseases with a genetic component is a long-standing goal of modern medicine. While ex vivo editing can address some diseases where transplantation of edited cells is feasible, particularly in the blood, in vivo genome editing will likely be required to treat most genetic diseases. Such treatments could offer single-dose cures for genetic disease that restore endogenous regulation of the gene and require minimal burden on the patient. The programmable and pure outcomes generated by base editing and prime editing even in non-dividing cells means that they could be excellent platforms to correct disease-associated genotypes in vivo. While no such treatment has yet reached clinical trials, several therapeutic proofs of principle in mammalian animal models have been demonstrated that could provide a foundation for future clinical trials (Table 1; Figure 3).
Table 1.
Examples of therapeutic in vivo base editing and prime editing
| Disease model | Key organ (cell type) | Editing outcome | Editor variant | Delivery method | Publication date | Reference |
|---|---|---|---|---|---|---|
| Hypercholesterolemia in mice | liver (hepatocytes) | 28% editing, reduced PCSK9 protein expression | BE3 | adenovirus | September 2017 | 175 |
| Hypercholesterolemia in mice expressing human PCSK9 | liver (hepatocytes) | ∼20% editing, reduced PCSK9 protein expression | BE3 | adenovirus | January 2019 | 176 |
| Hypercholesterolemia in mice and macaque | liver (hepatocytes) | 67% editing in mouse and 28% editing in macaque, reduced PCSK9 protein expression | ABE7.10max | AAV8 and LNP encapsulating mRNA | May 2021 | 177 |
| Hypercholesterolemia in mice and macaque | liver (hepatocytes) | 70% editing in mouse and 67% editing in macaque, reduced PCSK9 protein expression | ABE8.8 m | LNP encapsulating mRNA | May 2021 | 178 |
| Hypercholesterolemia in mice | liver (hepatocytes) | 30% editing in mouse, reduced PCSK9 protein expression | tBE-V5-mA3 | AAV8 | May 2021 | 108 |
| Phenylketonuria in mice | liver (hepatocytes) | 10% editing after 4 weeks increasing to 25% after 26 weeks. Return of blood phenylalanine to normal levels. | SaKKH-BE3 | AAV8 | October 2018 | 179 |
| Phenylketonuria in mice | liver (hepatocytes) | 23% editing after 8 weeks using AAV. 19% editing 1 week after second LNP dose. Return of blood phenylalanine to normal levels. | SaKKH-BE3 | AAV8 and LNP encapsulating mRNA | February 2021 | 180 |
| Phenylketonuria in mice | liver (hepatocytes) | 10% editing using adenovirus to deliver PE3 to neonates. 2% editing when delivering PE2 to 5-week-old animals. <1% editing using AAV | PE2/PE3 lacking reverse transcriptase RNase H domain | AAV8 and adenovirus | August 2021 | 181 |
| Tyrosinemia in mice | liver (hepatocytes) | 9.5% editing and restoration of mouse body weight | optimized ABE6.3 | hydrodynamic injection of DNA | February 2019 | 167 |
| Tyrosinemia in mice | liver (hepatocytes) | 12.5% editing and restoration of mouse body weight | optimized ABE6.3 | LNP encapsulating mRNA | April 2020 | 182 |
| Alpha-1 antitrypsin deficiency in mice expressing human SERPINA1 | liver (hepatocytes) | 6.7% correction by hydrodynamic injection, 3% correction by dual AAV 10 weeks after treatment | improved PE3 | hydrodynamic injection of DNA and AAV8 | April 2021 | 118 |
| Hutchinson-Gilford progeria syndrome in mice expressing human progerin | heart (vascular smooth muscle cells) | 30% editing in heart, 20% in aorta. 2.4-fold increase in lifespan. | ABE7.10max-VRQR | AAV9 | January 2021 | 183 |
| Duchenne muscular dystrophy in mice | skeletal muscle (myofibers) | 3.3% local editing, 17% of local myofibers staining for restored dystrophin | ABE7.10 | AAV9 | April 2018 | 184 |
| Duchenne muscular dystrophy in mice | skeletal muscle (myofibers) | 35% local editing, 96% of local myofibers staining for restored dystrophin | ABE7.10max | AAV9 | April 2021 | 185 |
| Amyotrophic lateral sclerosis in mice harboring human SOD1-G93A | central nervous system (motor neurons) | 1.2% editing frequency, 11% increase in lifetime and 85% increase in duration between onset of late-stage disease and death | BE3 | AAV9 | January 2020 | 186 |
| Niemann-Pick disease in mice | central nervous system (Purkinje cells) | 48% editing in cortex, 42% in Purkinje cells, up to 59% in cortex at test site. 10% increase in lifetime. | BE3.9max | AAV9 | January 2020 | 187 |
| Leber congenital amaurosis | retina (retinal pigmented epithelium) | 15% base editing, restored visual function | ABE7.10max | lentivirus | October 2020 | 188 |
| Leber congenital amaurosis | retina (retinal pigmented epithelium) | 14% base editing, restored visual function. 89% editing in Rpe65 cDNA | ABE7.10max-SpCas9-NG | AAV9 | January 2021 | 189 |
| Recessive hearing loss in mice | inner ear (hair cells) | 2.3% bulk genomic correction, 33% cDNA correction. Greatly increased hearing at 4 weeks that slowly degenerated | AID-BE3.9max | AAV (Anc80 serotype) | June 2020 | 190 |
| Sickle cell disease/β-thalassemia in mice expressing human β-globin | blood (hematopoietic stem cells) | 30% editing following selection, 21% of blood β-like globins were fetal hemoglobin | ABE7.10max | adenovirus | February 2021 | 191 |
Figure 3.
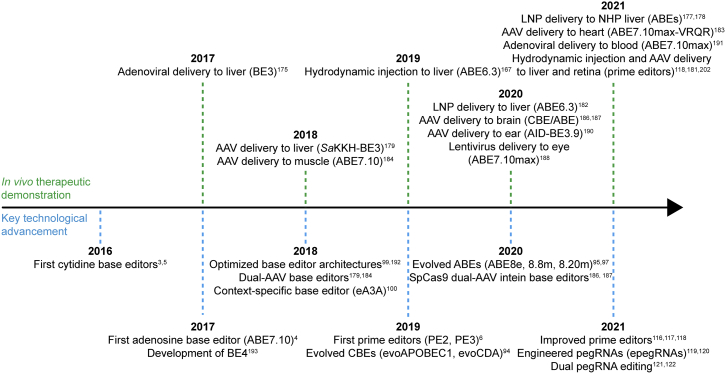
Timeline of base editor and prime editor development and applications
CBE, cytidine base editor; ABE, adenine base editor; AAV, adeno-associated virus; LNP, lipid nanoparticle; NHP, non-human primate.192,193
Liver
Due to the availability of multiple efficient liver delivery methods, the first and most efficient in vivo editing demonstrations have targeted diseases that can be treated by editing hepatocytes. Musunuru and coworkers first used an adenoviral vector to deliver the CBE “BE3” to the mouse liver and introduce a nonsense mutation in Pcsk9, disruption of which can treat hypercholesterolemia.175 This strategy, reported in 2017, yielded an average of 28% base editing in liver tissue, a 54% reduction in PCSK9 protein levels in blood, and 28% reduction in cholesterol levels 4 weeks after treatment. In 2019, Maresca and coworkers used a similar strategy that was demonstrated to knock out human PCSK9 in a humanized mouse model.176 This year, separate studies by Kathiresan, Schwank, and their respective coworkers demonstrated LNP delivery of ABE mRNA to disrupt PCSK9 in vivo in mice and cynomolgus monkeys via editing of a splice junction.178, 177 These studies achieved up to 66% base editing as measured from primate liver biopsies, resulting in a 90% reduction of PCSK9 protein in blood and 60% reduction in cholesterol.178 Schwank and coworkers177 also employed dual AAV delivery in mice, achieving 60% base editing in bulk liver177 (“bulk” meaning that cell populations were not separated or enriched from the tissue). However, the authors noted that LNP-mediated mRNA delivery is preferable to minimize the duration of editor exposure. Off-target editing analyses in both studies were remarkably clean, indicating that the LNP-mediated delivery of mRNA is a particularly efficient and precise editing strategy.
An additional study this year explored a creative strategy to reduce base editor off-target editing and applied this strategy in vivo to edit mouse Pcsk9. Chen and coworkers developed a base editor fused to an inhibitory domain that is removed at the target site by a co-localized split TEV protease.108 This new CBE variant, tBE-V5-mA3, did not yield observable off-target genomic or RNA editing in cell or animal models. It achieved 30% on-target editing to introduce a stop codon into liver Pcsk9 in 8-week-old mice when delivered in dual-AAV8 vectors. This treatment resulted in a decrease in PCSK9 protein serum levels by ∼80% and a decrease in serum cholesterol of ∼30%. Unlike most dual-AAV base editors, the architecture used in this study did not involve splitting Cas9 but rather kept full-length Cas9 on one vector with minimal regulatory elements and encoded the inhibited deaminase, split TEV protease, and guide RNAs on the other AAV. The guide RNAs included protein-binding sequences that recruited the deaminase or TEV protease.
In 2018, Schwank and coworkers developed a dual-AAV approach to deliver a variant S. aureus Cas9-BE3 to the mouse liver to correct a phenylketonuria mutation in vivo.179 To generate dual-AAV vectors, the base editor was split roughly in half with each end fused to a split intein, permitting reconstitution of the editor upon expression of both constructs. Dual-AAV BE3 treatment employing the AAV9 capsid restored phenylalanine levels in the blood to a normal range and conferred 10% editing in bulk liver at the target nucleotide 4 weeks after treatment, increasing to 25% after 26 weeks. A subsequent study by the same group this year established 10% editing in the same model conferred by in vivo LNP-mediated delivery of the S. aureus Cas9-BE3 mRNA (5-methoxyuridine modified).180 A second dose of LNP increased editing to 18%. Whole-genome sequencing of edited clones and RNA-seq in bulk tissue at the peak of editor expression following the delivery of base editors by either AAV or LNP detected no off-target editing in the genome or transcriptome in vivo of treated samples relative to unedited controls. Together, these demonstrations indicate that in vivo base editing in the liver can be efficient and reach therapeutic levels to correct disease phenotypes.
In 2019, Xue and coworkers demonstrated the treatment of a mouse model of tyrosinemia by hydrodynamic injection of ABE plasmid, resulting in approximately 10% editing in the bulk mouse liver and rescuing the weight loss phenotype associated with this model.167 The same study assessed LNP-mediated delivery of ABE mRNA but observed only 0.44% editing frequency, likely due to the unmodified mRNA employed. A subsequent study by Xue and coworkers using 5-methoxyuridine-modified nucleotides in place of uridine achieved 12.5% editing of the same tyrosinemia mutation.182 Notably, the study achieving the highest reported mRNA-mediated ABE editing in the liver (in PCSK9) used N1-methylpseudouridine in place of uridine.178
Heart
This year, our group, in collaboration with the Collins and Brown laboratories, reported the in vivo correction of a Hutchinson-Gilford progeria syndrome mutation by ABE in a mouse model.183 While the disease affects many tissues, the cause of death in humans and this mouse model is typically from cardiovascular disease.194 Delivery of S. pyogenes Cas9-ABEmax was conducted via dual-AAV vectors, each encoding half of the base editor with each end fused to a split intein (requiring a different architecture relative to the version described above, which used a Cas9 variant from a different bacterial species). In vivo editing using intravenous delivery of the AAV9 vector was optimal at postnatal day 14, yielding nearly 30% editing of the disease-causing mutation in LMNA in bulk heart and skeletal muscle, 20% editing of aorta and bone, and 55% editing of bulk liver 6 months after treatment. Editing resulted in restored vitality of model mice, health of the smooth muscle in the aorta, 49%–87% reduced expression of the mutant protein progerin in the heart and liver, and a 2.4-fold increase in median lifespan from 215 to 510 days. This constituted the largest benefit to health and lifespan that has been reported for this disease model and points to the potential power of in vivo base editor therapeutics.
Skeletal muscle
The first demonstration of in vivo base editing of the skeletal muscle was reported in 2018 by Kim and coworkers and involved the correction of a nonsense mutation in mouse Dmd, which models Duchenne muscular dystrophy.184 The authors developed a dual-AAV base editor architecture that relied on recombination of the two genomes following transduction and trans-splicing of the splicing donor tagged on the N-terminal vector to the splicing acceptor at the start of the C-terminal vector. Local injection of dual-AAVs employing the AAV9 capsid in the tibialis anterior muscle led to 3.3% genomic correction of the stop codon at 8 weeks after injection in bulk DNA extracted from the muscle. Immunofluorescence microscopy revealed that 17% of myofibers in the treated muscle showed evidence of restored expression of dystrophin, which is encoded by the Dmd gene.
This year, Olson and coworkers treated a separate mouse model of Duchenne muscular dystrophy involving the deletion of exon 51 using ABEmax.185 The authors employed the split-intein S. pyogenes Cas9 AAV architecture developed by our laboratory, which we observed to confer improved editing relative to trans-splicing AAVs.187 By disrupting the splice donor site of exon 50, the proper frame of dystrophin can be restored in the mouse disease model. Local injection of the dual-AAVs employing the AAV9 capsid in the tibialis anterior muscle led to 35% editing in genomic DNA extracted from the muscle 3 weeks after treatment. Immunofluorescence microscopy revealed that 96% of myofibers in the treated muscle showed restored expression of dystrophin. This improved editing efficiency relative to the 2018 study is likely due to the improved dual-AAV architecture as well as the improved base editor codon usage and NLS architecture employed. However, the authors note that the quantity of AAV employed in the recent study is above what could constitute a reasonable dose in human patients, and so additional optimization is required before moving into the clinic. Prior studies in mouse models indicate that even 4% restoration of dystrophin expression can improve muscular function and histopathology,195 and it is feasible that systemic delivery of dual-AAV base editors could surpass this level of restoration. Most myofibers contain multiple nuclei, so even a moderate editing efficiency could potentially still improve the function of a disproportionately large fraction of myofibers.
Central nervous system
Two demonstrations of in vivo base editing in the mouse central nervous system were reported in January 2020.187,186 Both made use of a split-intein dual-AAV-mediated approach, although the locations of the split site within S. pyogenes Cas9 differed between the two studies. Gaj and coworkers made use of a model mouse for amyotrophic lateral sclerosis (ALS) harboring approximately 25 copies of human SOD1 genes containing the disease-associated G93A mutation.186 The CBE BE3 was packaged into AAV9 capsids and injected into the lumbar cerebrospinal fluid of these mice to introduce stop codons in SOD1 to slow disease progression. The authors estimated that about 6.5% of spinal cord cells were transduced by both AAVs based on the expression of epitope tags, and sequencing revealed that 1.2% of reads contained the desired nonsense mutation. Immunofluorescence indicated that transduction was most efficient in astrocytes. Treated mice showed an 11% increase in mean survival relative to untreated controls and an 85% increase in the duration between the onset of late-stage disease and death.186
The second study demonstrating base editing in the central nervous system targeted correction of a disease mutation that models Niemann-Pick disease in mice,187 in addition to some neutral model sites that were used during optimization of our platform. By intracerebroventricular injection of neonatal mice with four different AAV capsid variants encoding a CBE, we observed 32%–50% editing in cortical cells and 0.5%–2.5% editing in cerebellar cells at the neutral model site. However, by enriching for transduced cells marked by GFP delivered via a third AAV, over 50% editing in the cerebellum was achieved. The transduced cerebellar cells were identified as Purkinje neurons using a mouse line with a reporter gene in that cell type. Similar results were also achieved when delivering an ABE.
Using the PHP.eB capsid, which has an enhanced ability to cross the blood-brain barrier in some mouse strains,145,196 we injected mice intravenously with dual-AAV CBEs targeting the DMNT1 test site at 9 weeks of age. Four weeks later, the target edit was measured in 59% of sequencing reads from bulk cortical tissue and 35% of reads from bulk cerebellar tissue.187 To our knowledge, this is the most efficient genome editing achieved in any bulk tissue from the central nervous system.
We assessed the ability of dual-AAV CBE injection to correct a model Niemann-Pick mutation in mice at the neonatal (P0) stage and in 4-week-old animals. We observed a 10% increase in lifespan for both cohorts. Bulk cortical editing was 48% and cerebellar editing was 0.3%, although enriching the transduced cerebellar (Purkinje) cells revealed 42% editing in that population, which is of particular importance for Niemann-Pick disease.197
Eye
As part of the same study in which we developed a split-intein dual-AAV architecture for base editor delivery, we also assessed editing a test locus in the mouse eye.187 Sub-retinal injection of dual-AAVs using the PHP.B capsid or Anc80 capsid to deliver CBE or ABE in 2-week-old mice resulted in 19%–26% editing of rod photoreceptor cells.
Separately, Palczewski and coworkers used a lentivirus to deliver ABEmax and correct a mutation in mouse Rpe65 modeling Leber congenital amaurosis.188 Sub-retinal injection of the lentivirus in 4-week-old mice resulted in an average of 15% base editing at the target genomic site and restored retinoid isomerase activity. Near-normal levels of visual function were achieved in treated mice. The same model was later treated by Kim, Bae, and coworkers using dual AAV delivery of an ABEmax variant in 3-week-old and 4-month-old mice.189 AAV-mediated editing resulted in 10%–14% editing of the genomic target and also restored visual function. Sequencing Rpe65 cDNA isolated from edited retina revealed an editing efficiency of ∼90% in Rpe65-expressing cells. The eye is a particularly attractive system for in vivo genome editing because of the potential for local sub-retinal delivery and the lack of debilitating immune responses against foreign material in the eye.198
Ear
We first assessed the delivery of base editors to the inner ear of mice in 2017.199 Mice at postnatal day 1 or 2 were injected with LNP-encapsulated CBE RNP targeting a test locus, achieving 1%–2% editing in the stria vascularis, approximately 1% editing in the modiolus, and 0.3%–0.6% editing in the organ of Corti. In a later study, we employed this approach to edit β-catenin in the inner ear and modulate Wnt signaling, inducing proliferation of the post-mitotic supporting cells and their differentiation into hair cells.200
In 2020, in collaboration with Chen, Holt, Kong, and their respective coworkers, we used the split-intein dual-AAV approach to correct a missense deafness mutation in Tmc1 in model mice, in which degeneration of inner ear hair cells is thought to prevent auditory stimulation.190 Model mice were injected at postnatal day 1. At day 14 post injection, bulk sequencing in the organ of Corti revealed 2.3% correction of the disease mutation. However, it is difficult to isolate only the hair cells of interest for sequencing. Because Tmc1 is expressed primarily in hair cells, we sequenced cDNA from extracted tissue and observed approximately 33% correction in all Tmc1 transcripts, indicating that we preferentially edited the target cell population. This editing resulted in greatly improved hearing at 4 weeks of age relative to untreated control mice. However, response thresholds increased by 6 weeks of age, indicating that this treatment slowed but did not stop degeneration of the hair cells and deafness.
Blood
Therapeutic gene delivery and genome editing of the blood has been achieved in humans via ex vivo modification followed by transplantation.74,201 However, an in vivo method to edit blood would be advantageous to eliminate the substantial risks associated with bone marrow transplantation. Earlier this year, Lieber and coworkers described in vivo delivery of base editors to hematopoietic stem cells (HSCs) followed by enrichment of edited cells in mice.191 They employed CBEs to install edits in the fetal hemoglobin promoter of mice containing the human β-globin locus with the goal of upregulating fetal hemoglobin, which can be therapeutic for sickle cell disease and β-thalassemia. Two adenoviral vectors were delivered to mice following chemically induced mobilization of HSCs into peripheral blood. One contained a CBE to disrupt a repressor binding site in the fetal hemoglobin promoter as well as a MGMTP140K selectable marker, the latter being flanked by inverted repeats to permit its integration. The second adenovirus contained the machinery to integrate the selectable marker. While the recombinase and base editor were only expressed transiently, the MGMTP140K marker was integrated into the genome of cells receiving both viruses and permitted selection for the transduced cells. At the endpoint 16 weeks after transduction, following four doses of O6-benzylguanine to selectively kill cells lacking MGMTP140K, 30% editing of the target site was achieved in blood, leading to 21% of human β-like globins being composed of fetal hemoglobin. This population was stable over 16 weeks following secondary transplantation.
The amount of fetal hemoglobin observed should prevent most symptoms of sickle cell disease and β-thalassemia, and this study provides a strong foundation for in vivo blood editing. However, it remains to be determined whether treatment with O6-benzylguanine or the standard bone marrow transplantation procedure leads to fewer side effects for patients. Methods permitting in vivo genome editing in the blood without requiring selection for edited cells or ablation of the patient’s hematopoietic system would be particularly attractive for future development.
Therapeutic in vivo prime editing
Although prime editors are capable of installing more versatile edits relative to base editors, they were developed much more recently—a few months before the start of the COVID-19 pandemic. So far, three studies have independently demonstrated in vivo prime editing, each using dual AAV vectors, and each selecting a different site to split the prime editor.118,202,181 Xue and coworkers first optimized the NLS architecture of prime editors, then conducted hydrodynamic injection of prime editor plasmids in a mouse model of alpha 1-antitrypsin deficiency leading to 6.7% correction of the SERPINA1 mutation.203 They developed a dual-AAV delivery platform for PE by splitting the editor into two pieces before Ser714 and inserting intein tags to drive protein splicing. Tail vein injection of dual-AAV8 vectors in 6-week-old mice conferred 3.1% correction of SERPINA1 in the liver 10 weeks after injection.118 In the same work, the authors also installed an oncogenic mutation by hydrodynamic injection of PE plasmids in adult mice harboring other genetic elements intended to induce cancer, resulting in an average of 10 tumors per mouse liver 25 days after injection.
Separately, Huang, Liang, and coworkers selected a prime editor split site before Ser1025 to place intein tags for a dual-AAV approach.202 They packaged each half into an AAV8 capsid and conducted subretinal injection to deliver the editor to 6-week-old wild-type mice targeting a test site, Dnmt1. Sequencing genomic DNA extracted from the retina 6 weeks after injection detected 1.7% prime editing when editor expression was placed under the CMV promoter and 0.6% editing when under an EF1α promoter.
Schwank and coworkers described the most recent adaptation of prime editors for in vivo delivery.181 They identified an optimal split site before Ser1153 for the insertion of inteins. They also optimized the NLS architecture and linker lengths between inteins and PE to maximize editing efficiency. Removing the RNase H domain of the reverse transcriptase in PE2 led to no loss of PE activity, and so the smaller construct was used in their in vivo delivery experiments. Intravenous injection of dual-AAV8 vectors in postnatal day 1 wild-type mice conferred 14% prime editing of the Dnmt1 test site in genomic DNA extracted from hepatocytes 4 weeks after injection. The same edit was achieved at 3.4% efficiency when dual-AAV8 vectors were injected into 5-week-old mice. To avoid the loss in editing activity associated with the split editor, the authors used adenovirus to deliver full-length PE2 (lacking only the RNase H domain) in mice. At the Dnmt1 test site, 58% prime editing was achieved in hepatocytes following injection in neonates and 36% editing when injecting in 5-week-old mice. The authors then tested these delivery methods for PE2 treatment in a mouse model of phenylketonuria. AAV delivery yielded less than 1% editing at the target site, but adenoviral delivery of PE2 led to correction of 5.6% of alleles when delivered to neonates and 2% of alleles when delivered to 5-week-old mice. The addition of a PE3 nicking guide yielded 10% editing in neonates. Prime editing from adenoviral treatment of neonates was sufficient to confer disease rescue in this mouse model.
We anticipate that the recent improvements to prime editing systems will substantially increase in vivo activity in future work.116, 117, 120 Prime editing was determined to be particularly efficient in HEK293T cells due to the lower activity of the mismatch repair pathway.116 In adult mammalian tissue, mismatch repair is likely to be a major blockade to prime editing, so the recent development of new PE4 and PE5 prime editing systems that evade this bottleneck should provide a substantial boost in editing activity, akin to what was observed in HeLa cells where endogenous mismatch repair is more active. The recently identified stabilizing pegRNA motifs119, 120 as well as further improvements to the codon usage, NLS architecture, and Cas9 domain116 are also likely to be impactful in increasing in vivo activity.
Several studies have used prime editor mRNA and RNP to introduce desired mutations in mouse117,127,128 and zebrafish129 embryos. In each case, edited embryos with editing frequencies of 3%–50% were readily obtained, in most cases with few byproducts. Success in these systems suggests that non-viral delivery of these materials to adult mammalian tissues may also be an effective strategy for prime editing. We anticipate that these developments, together with the continued advancement of prime editing systems,116, 117, 118, 119, 120 will result in many opportunities for therapeutic in vivo post-natal prime editing.
Future prospects
Among the three general, readily programmable classes of mammalian cell genome editing agents reported to date—nucleases, base editors, and prime editors—nucleases are ideally suited for gene disruption, base editors are particularly efficient at correcting the most common class of pathogenic mutations, and prime editors offer the greatest versatility for correcting the vast majority of disease-associated mutations. Within the past year, new generations of both base editors and prime editors have substantially increase their activity and targeting scope. When paired with suitable delivery methods for the cells of interest, base editors and prime editors have now been used to rescue animal models of genetic disease. Many of the demonstrated therapeutic strategies may also be applied to treat human patients suffering from genetic diseases. Additional genetic variants associated with disease are discovered each year, and facile targeting of editing agents could permit one-time personalized medicines that treat rare or even unique disease mutations, leveraging successful demonstrations at common disease mutations.
When delivering editors as DNA, as is common when using viral vectors, methods to limit the duration of exposure to the edit will also be desirable to minimize the possibility of off-target editing. Furthermore, the development of additional tools that do not require DSBs for in vivo use could provide new therapeutic modalities. Integrases or recombinases could permit the targeted installation of gene-sized inserts for gene augmentation approaches, and recently reported “CRISPRoff” methods could durably silence genes via self-perpetuating methylation.21
Appropriate delivery of editors to cell types that can impact disease phenotypes is vital for in vivo genome editing therapeutics. Recent advancements in both viral and non-viral delivery have demonstrated that in vivo genome editing in animals and in human patients can result in therapeutic outcomes. Nevertheless, more efficient delivery could further improve therapeutic prospects, and in vivo delivery remains challenging for many cell types relevant to disease treatment. The development or evolution of new delivery vehicles with improved potency and specificity for cell populations of interest, such as the brain, heart, bone marrow, retina, cochlea, lung, and intestine, would further advance the translation of genome editing technologies to treat or possibly cure genetic diseases. Simultaneously, investigators should take caution to avoid collateral germline editing, since human clinical germline editing raises serious ethical complications.204,205 Systemic delivery of base editors in AAV9 capsids did not detectably modify the mouse germline.187 New animal models that sensitively reveal the tissue type specificity of base editors and prime editor delivery will be valuable for speeding the development of enhanced delivery platforms.
Current data suggest that prime editors are particularly specific for their target sequences, with rarely observed guide-dependent editing at sites with homology to the protospacer.6,114 In addition, three independent laboratories have reported that stochastic guide-independent editing was not observed.112,113,115 A recent report that conducted whole-genome and whole-transcriptome sequencing in clonal cells after prime editing observed no off-target editing in either the genome or transcriptome and noted no effect in telomere integrity, RNA splicing, or gene expression.115 Off-target genome editing and RNA editing have been observed using certain base editors, although many base editor variants with minimized guide-dependent and guide-independent off-target activity have been developed to mitigate off-target editing.82,83,108
At present, there is no standard assay to assess the functional consequences or potential clinical relevance of off-target genome editing, and therefore it is difficult to meaningfully compare the off-target editing propensity among different combinations of gene editing agents, cell types, and delivery methods. The possibility that off-target editing could produce cancerous or pre-cancerous cells is the primary concern for long-term side effects of gene editing therapeutics. A pipeline that includes sequencing of all cancer-associated genes may be useful, particularly if a threshold for off-target editing was identified that distinguished a normal or acceptable rate of mutagenesis from one associated with an increased risk of cancer over a human lifetime.
Even without such a metric, base editors and prime editors have not yet been shown to induce cancer in animal models, and the relative risk of oncogenesis versus continuing to harbor mutations that cause serious genetic diseases in many cases may favor the careful application of the editing agent. Recent demonstrations of therapeutic in vivo base editing that delved deeply into the analysis of off-targets did not find evidence of undesired edits, while still observing strong evidence for the efficacy of such treatments.178,177,180 The lack of off-target in vivo editing may in part be due to the lower expression of editor that is achieved in vivo relative to in cultured cells where off-target editing has been observed.180
The rapid advancement in the capability, precision, programmability, and efficiency of genome editing agents over the past decade has yielded many new therapeutic possibilities to treat genetic diseases that were once considered incurable. Given the simple and modular targeting capabilities of current genome editing technologies, even personalized medicine for unique mutations is a feasible path. While a fanciful aspiration less than a decade ago, we anticipate that therapies to correct the root cause of genetic diseases will lead to long-lasting, single-dose cures and improved quality of life for many of the hundreds of millions of people impacted by pathogenic mutations.
Acknowledgments
This work was supported by US National Institutes of Health awards U01 AI142756, RM1 HG009490, R01 EB022376, and R35 GM118062 and the HHMI. G.A.N. was supported by a Helen Hay Whitney Postdoctoral Fellowship and the HHMI.
Declaration of interests
The authors have filed patent applications on genome editing technologies through the Broad Institute of MIT and Harvard. D.R.L. is a consultant and cofounder of Beam Therapeutics, Prime Medicine, Pairwise Plants, Editas Medicine, and Chroma Medicine, companies that use genome editing or genome engineering, including base editing, prime editing, and epigenetic modification.
Contributor Information
Gregory A. Newby, Email: gnewby@broadinstitute.org.
David R. Liu, Email: drliu@fas.harvard.edu.
References
- 1.Pyeritz, R.E., Korf, B.R., and Grody, W.W. (2019). Nature and Frequency of Genetic Disease. In Emery and Rimoin’s Principles and Practice of Medical Genetics and Genomics, Seventh Edition, B.R. Korf, R.E. Pyeritz, and W.W.Grody, eds. (Academic Press), pp. 47–51.
- 2.Anguela X.M., High K.A. Entering the Modern Era of Gene Therapy. Annu. Rev. Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 3.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., Mochizuki M., Miyabe A., Araki M., Hara K.Y. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;353:aaf8729. doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 6.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calos M.P. The phiC31 integrase system for gene therapy. Curr. Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 8.Shultz J.L., Voziyanova E., Konieczka J.H., Voziyanov Y. A genome-wide analysis of FRT-like sequences in the human genome. PLoS ONE. 2011;6:e18077. doi: 10.1371/journal.pone.0018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Burnight E.R., Cooney A.L., Malani N., Brady T., Sander J.D., Staber J., Wheelan S.J., Joung J.K., McCray P.B., Jr. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. USA. 2013;110:E2279–E2287. doi: 10.1073/pnas.1305987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva G., Poirot L., Galetto R., Smith J., Montoya G., Duchateau P., Pâques F. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 2011;11:11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 12.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 13.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mojica F.J., Rodriguez-Valera F. The discovery of CRISPR in archaea and bacteria. FEBS J. 2016;283:3162–3169. doi: 10.1111/febs.13766. [DOI] [PubMed] [Google Scholar]
- 15.Miller J.C., Zhang L., Xia D.F., Campo J.J., Ankoudinova I.V., Guschin D.Y., Babiarz J.E., Meng X., Hinkley S.J., Lam S.C. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat. Methods. 2015;12:465–471. doi: 10.1038/nmeth.3330. [DOI] [PubMed] [Google Scholar]
- 16.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klompe S.E., Vo P.L.H., Halpin-Healy T.S., Sternberg S.H. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571:219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 18.Strecker J., Ladha A., Gardner Z., Schmid-Burgk J.L., Makarova K.S., Koonin E.V., Zhang F. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365:48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuñez J.K., Chen J., Pommier G.C., Cogan J.Z., Replogle J.M., Adriaens C., Ramadoss G.N., Shi Q., Hung K.L., Samelson A.J. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503–2519.e17. doi: 10.1016/j.cell.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mok B.Y., de Moraes M.H., Zeng J., Bosch D.E., Kotrys A.V., Raguram A., Hsu F., Radey M.C., Peterson S.B., Mootha V.K. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature. 2020;583:631–637. doi: 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Gent D.C., van der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 24.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sfeir A., Symington L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015;40:701–714. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seol J.H., Shim E.Y., Lee S.E. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. 2018;809:81–87. doi: 10.1016/j.mrfmmm.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Páleníková P., Khodak A., Kiselev V., Kosicki M. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018 doi: 10.1038/nbt.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V., Noble W.S., Shendure J. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019;47:7989–8003. doi: 10.1093/nar/gkz487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Y., Liu Z., Zhang Y., Chen M., Sui T., Lai L., Li Z. Large-Fragment Deletions Induced by Cas9 Cleavage while Not in the BEs System. Mol. Ther. Nucleic Acids. 2020;21:523–526. doi: 10.1016/j.omtn.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alanis-Lobato G., Zohren J., McCarthy A., Fogarty N.M.E., Kubikova N., Hardman E., Greco M., Wells D., Turner J.M.A., Niakan K.K. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2004832117. e2004832117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webber B.R., Lonetree C.L., Kluesner M.G., Johnson M.J., Pomeroy E.J., Diers M.D., Lahr W.S., Draper G.M., Slipek N.J., Smeester B.A. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019;10:5222. doi: 10.1038/s41467-019-13007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giannoukos G., Ciulla D.M., Marco E., Abdulkerim H.S., Barrera L.A., Bothmer A., Dhanapal V., Gloskowski S.W., Jayaram H., Maeder M.L. UDiTaS™, a genome editing detection method for indels and genome rearrangements. BMC Genomics. 2018;19:212. doi: 10.1186/s12864-018-4561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leibowitz M.L., Papathanasiou S., Doerfler P.A., Blaine L.J., Sun L., Yao Y., Zhang C.Z., Weiss M.J., Pellman D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet. 2021;53:895–905. doi: 10.1038/s41588-021-00838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 38.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24:939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 39.Enache O.M., Rendo V., Abdusamad M., Lam D., Davison D., Pal S., Currimjee N., Hess J., Pantel S., Nag A. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 2020;52:662–668. doi: 10.1038/s41588-020-0623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams A.B., Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016;6:a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 42.Yun M.H., Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleh-Gohari N., Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto Y., Kokuta T., Teshigahara A., Iijima K., Kitao H., Takata M., Tauchi H. Mitotic cells can repair DNA double-strand breaks via a homology-directed pathway. J. Radiat. Res. (Tokyo) 2021;62:25–33. doi: 10.1093/jrr/rraa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aylon Y., Liefshitz B., Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao Y., Wang L., Guo N., Wang S., Yang L., Li Y., Wang M., Yin S., Han H., Zeng L. Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J. Biol. Chem. 2018;293:6883–6892. doi: 10.1074/jbc.RA117.000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan Y., Ma Y., Li Q., Sun Z., Ma L., Wu L., Wang L., Zeng L., Shao Y., Chen Y. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol. Med. 2016;8:477–488. doi: 10.15252/emmm.201506039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huai C., Jia C., Sun R., Xu P., Min T., Wang Q., Zheng C., Chen H., Lu D. CRISPR/Cas9-mediated somatic and germline gene correction to restore hemostasis in hemophilia B mice. Hum. Genet. 2017;136:875–883. doi: 10.1007/s00439-017-1801-z. [DOI] [PubMed] [Google Scholar]
- 51.Stephens C.J., Lauron E.J., Kashentseva E., Lu Z.H., Yokoyama W.M., Curiel D.T. Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J. Control. Release. 2019;298:128–141. doi: 10.1016/j.jconrel.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong S.A., Seo J.H., Wi S., Jung E.S., Yu J., Hwang G.H., Yu J.H., Baek A., Park S., Bae S., Cho S.R. In vivo gene editing via homology-independent targeted integration for adrenoleukodystrophy treatment. Mol. Ther. 2021 doi: 10.1016/j.ymthe.2021.05.022. S1525-0016(21)00304-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson A.C., Dever D.P., Baik R., Camarena J., Hsu I., Charlesworth C.T., Morita C., Nakauchi H., Porteus M.H. Cas9-AAV6 gene correction of beta-globin in autologous HSCs improves sickle cell disease erythropoiesis in mice. Nat. Commun. 2021;12:686. doi: 10.1038/s41467-021-20909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cromer M.K., Camarena J., Martin R.M., Lesch B.J., Vakulskas C.A., Bode N.M., Kurgan G., Collingwood M.A., Rettig G.R., Behlke M.A. Gene replacement of α-globin with β-globin restores hemoglobin balance in β-thalassemia-derived hematopoietic stem and progenitor cells. Nat. Med. 2021;27:677–687. doi: 10.1038/s41591-021-01284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lattanzi A., Camarena J., Lahiri P., Segal H., Srifa W., Vakulskas C.A., Frock R.L., Kenrick J., Lee C., Talbott N. Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci. Transl. Med. 2021;13:eabf2444. doi: 10.1126/scitranslmed.abf2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Ravin S.S., Brault J., Meis R.J., Liu S., Li L., Pavel-Dinu M., Lazzarotto C.R., Liu T., Koontz S.M., Choi U. Enhanced homology-directed repair for highly efficient gene editing in hematopoietic stem/progenitor cells. Blood. 2021;137:2598–2608. doi: 10.1182/blood.2020008503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin J.J., Schröder M.S., Caiado F., Wyman S.K., Bray N.L., Bordi M., Dewitt M.A., Vu J.T., Kim W.T., Hockemeyer D. Controlled Cycling and Quiescence Enables Efficient HDR in Engraftment-Enriched Adult Hematopoietic Stem and Progenitor Cells. Cell Rep. 2020;32:108093. doi: 10.1016/j.celrep.2020.108093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lomova A., Clark D.N., Campo-Fernandez B., Flores-Bjurström C., Kaufman M.L., Fitz-Gibbon S., Wang X., Miyahira E.Y., Brown D., DeWitt M.A. Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells. 2019;37:284–294. doi: 10.1002/stem.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byambaa S., Uosaki H., Ohmori T., Hara H., Endo H., Nureki O., Hanazono Y. Non-viral ex vivo genome-editing in mouse bona fide hematopoietic stem cells with CRISPR/Cas9. Mol. Ther. Methods Clin. Dev. 2021;20:451–462. doi: 10.1016/j.omtm.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hale M., Lee B., Honaker Y., Leung W.H., Grier A.E., Jacobs H.M., Sommer K., Sahni J., Jackson S.W., Scharenberg A.M. Homology-Directed Recombination for Enhanced Engineering of Chimeric Antigen Receptor T Cells. Mol. Ther. Methods Clin. Dev. 2017;4:192–203. doi: 10.1016/j.omtm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiebking V., Lee C.M., Mostrel N., Lahiri P., Bak R., Bao G., Roncarolo M.G., Bertaina A., Porteus M.H. Genome editing of donor-derived T-cells to generate allogenic chimeric antigen receptor-modified T cells: Optimizing αβ T cell-depleted haploidentical hematopoietic stem cell transplantation. Haematologica. 2021;106:847–858. doi: 10.3324/haematol.2019.233882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaidyanathan S., Salahudeen A.A., Sellers Z.M., Bravo D.T., Choi S.S., Batish A., Le W., Baik R., de la O S., Kaushik M.P. High-Efficiency, Selection-free Gene Repair in Airway Stem Cells from Cystic Fibrosis Patients Rescues CFTR Function in Differentiated Epithelia. Cell Stem Cell. 2020;26:161–171.e4. doi: 10.1016/j.stem.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou J., Maeder M.L., Mali P., Pruett-Miller S.M., Thibodeau-Beganny S., Chou B.K., Chen G., Ye Z., Park I.H., Daley G.Q. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacków J., Guo Z., Hansen C., Abaci H.E., Doucet Y.S., Shin J.U., Hayashi R., DeLorenzo D., Kabata Y., Shinkuma S. CRISPR/Cas9-based targeted genome editing for correction of recessive dystrophic epidermolysis bullosa using iPS cells. Proc. Natl. Acad. Sci. USA. 2019:201907081. doi: 10.1073/pnas.1907081116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maxwell K.G., Augsornworawat P., Velazco-Cruz L., Kim M.H., Asada R., Hogrebe N.J., Morikawa S., Urano F., Millman J.R. Gene-edited human stem cell-derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med. 2020;12:eaax9106. doi: 10.1126/scitranslmed.aax9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srifa W., Kosaric N., Amorin A., Jadi O., Park Y., Mantri S., Camarena J., Gurtner G.C., Porteus M. Cas9-AAV6-engineered human mesenchymal stromal cells improved cutaneous wound healing in diabetic mice. Nat. Commun. 2020;11:2470. doi: 10.1038/s41467-020-16065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 69.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ding Q., Strong A., Patel K.M., Ng S.L., Gosis B.S., Regan S.N., Cowan C.A., Rader D.J., Musunuru K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gillmore J.D., Gane E., Taubel J., Kao J., Fontana M., Maitland M.L., Seitzer J., O’Connell D., Walsh K.R., Wood K. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 72.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 73.Wu Y., Zeng J., Roscoe B.P., Liu P., Yao Q., Lazzarotto C.R., Clement K., Cole M.A., Luk K., Baricordi C. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 2019;25:776–783. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 75.Traxler E.A., Yao Y., Wang Y.D., Woodard K.J., Kurita R., Nakamura Y., Hughes J.R., Hardison R.C., Blobel G.A., Li C., Weiss M.J. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat. Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Humbert O., Radtke S., Samuelson C., Carrillo R.R., Perez A.M., Reddy S.S., Lux C., Pattabhi S., Schefter L.E., Negre O. Therapeutically relevant engraftment of a CRISPR-Cas9-edited HSC-enriched population with HbF reactivation in nonhuman primates. Sci. Transl. Med. 2019;11:eaaw3768. doi: 10.1126/scitranslmed.aaw3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu L., Wang J., Liu Y., Xie L., Su B., Mou D., Wang L., Liu T., Wang X., Zhang B. CRISPR-Edited Stem Cells in a Patient with HIV and Acute Lymphocytic Leukemia. N Engl J Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 79.Liu X., Zhang Y., Cheng C., Cheng A.W., Zhang X., Li N., Xia C., Wei X., Liu X., Wang H. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 2017;27:154–157. doi: 10.1038/cr.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren J., Liu X., Fang C., Jiang S., June C.H., Zhao Y. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin. Cancer Res. 2017;23:2255–2266. doi: 10.1158/1078-0432.CCR-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rupp L.J., Schumann K., Roybal K.T., Gate R.E., Ye C.J., Lim W.A., Marson A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci. Rep. 2017;7:737. doi: 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang T.P., Newby G.A., Liu D.R. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. 2021;16:1089–1128. doi: 10.1038/s41596-020-00450-9. [DOI] [PubMed] [Google Scholar]
- 83.Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 84.Kurt I.C., Zhou R., Iyer S., Garcia S.P., Miller B.R., Langner L.M., Grünewald J., Joung J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2021;39:41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koblan L.W., Arbab M., Shen M.W., Hussmann J.A., Anzalone A.V., Doman J.L., Newby G.A., Yang D., Mok B., Replogle J.M. Efficient C•G-to-G•C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00938-z. Published online June 28, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen L., Park J.E., Paa P., Rajakumar P.D., Prekop H.T., Chew Y.T., Manivannan S.N., Chew W.L. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat. Commun. 2021;12:1384. doi: 10.1038/s41467-021-21559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newby G.A., Yen J.S., Woodard K.J., Mayuranathan T., Lazzarotto C.R., Li Y., Sheppard-Tillman H., Porter S.N., Yao Y., Mayberry K. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 2021;595:295–302. doi: 10.1038/s41586-021-03609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z., Liu D.R. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miller S.M., Wang T., Randolph P.B., Arbab M., Shen M.W., Huang T.P., Matuszek Z., Newby G.A., Rees H.A., Liu D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X., Ji P., Fan H., Dang L., Wan W., Liu S., Li Y., Yu W., Li X., Ma X. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun. Biol. 2020;3:62. doi: 10.1038/s42003-020-0796-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35:371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thuronyi B.W., Koblan L.W., Levy J.M., Yeh W.H., Zheng C., Newby G.A., Wilson C., Bhaumik M., Shubina-Oleinik O., Holt J.R., Liu D.R. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 2019;37:1070–1079. doi: 10.1038/s41587-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020;38:883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang T.P., Zhao K.T., Miller S.M., Gaudelli N.M., Oakes B.L., Fellmann C., Savage D.F., Liu D.R. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 2019;37:626–631. doi: 10.1038/s41587-019-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gaudelli N.M., Lam D.K., Rees H.A., Solá-Esteves N.M., Barrera L.A., Born D.A., Edwards A., Gehrke J.M., Lee S.J., Liquori A.J. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 2020;38:892–900. doi: 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- 98.Jiang W., Feng S., Huang S., Yu W., Li G., Yang G., Liu Y., Zhang Y., Zhang L., Hou Y. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res. 2018;28:855–861. doi: 10.1038/s41422-018-0052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zafra M.P., Schatoff E.M., Katti A., Foronda M., Breinig M., Schweitzer A.Y., Simon A., Han T., Goswami S., Montgomery E. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 2018;36:888–893. doi: 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gehrke J.M., Cervantes O., Clement M.K., Wu Y., Zeng J., Bauer D.E., Pinello L., Joung J.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018;36:977–982. doi: 10.1038/nbt.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim H.S., Jeong Y.K., Hur J.K., Kim J.S., Bae S. Adenine base editors catalyze cytosine conversions in human cells. Nat. Biotechnol. 2019;37:1145–1148. doi: 10.1038/s41587-019-0254-4. [DOI] [PubMed] [Google Scholar]
- 102.Jeong Y.K., Lee S., Hwang G.H., Hong S.A., Park S.E., Kim J.S., Woo J.S., Bae S. Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00943-2. Published online July 1, 2021. [DOI] [PubMed] [Google Scholar]
- 103.Grünewald J., Zhou R., Iyer S., Lareau C.A., Garcia S.P., Aryee M.J., Joung J.K. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 2019;37:1041–1048. doi: 10.1038/s41587-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rees H.A., Wilson C., Doman J.L., Liu D.R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 2019;5:eaax5717. doi: 10.1126/sciadv.aax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 107.Zhou C., Sun Y., Yan R., Liu Y., Zuo E., Gu C., Han L., Wei Y., Hu X., Zeng R. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571:275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 108.Wang L., Xue W., Zhang H., Gao R., Qiu H., Wei J., Zhou L., Lei Y.N., Wu X., Li X. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat. Cell Biol. 2021;23:552–563. doi: 10.1038/s41556-021-00671-4. [DOI] [PubMed] [Google Scholar]
- 109.Chu S.H., Packer M., Rees H., Lam D., Yu Y., Marshall J., Cheng L.I., Lam D., Olins J., Ran F.A. Rationally Designed Base Editors for Precise Editing of the Sickle Cell Disease Mutation. CRISPR J. 2021;4:169–177. doi: 10.1089/crispr.2020.0144. [DOI] [PubMed] [Google Scholar]
- 110.Zeng J., Wu Y., Ren C., Bonanno J., Shen A.H., Shea D., Gehrke J.M., Clement K., Luk K., Yao Q. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 2020;26:535–541. doi: 10.1038/s41591-020-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sürün D., Schneider A., Mircetic J., Neumann K., Lansing F., Paszkowski-Rogacz M., Hänchen V., Lee-Kirsch M.A., Buchholz F. Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors. Genes (Basel) 2020;11:E511. doi: 10.3390/genes11050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Habib O., Habib G., Hwang G.-H., Bae S. Comprehensive analysis of prime editing outcomes in human embryonic stem cells. bioRxiv. 2021 doi: 10.1101/2021.04.12.439533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin S., Lin Q., Luo Y., Zhu Z., Liu G., Li Y., Chen K., Qiu J.L., Gao C. Genome-wide specificity of prime editors in plants. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00891-x. Published online April 15, 2021. [DOI] [PubMed] [Google Scholar]
- 114.Kim D.Y., Moon S.B., Ko J.H., Kim Y.S., Kim D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res. 2020;48:10576–10589. doi: 10.1093/nar/gkaa764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gao R., Fu Z.-C., Li X., Wang Y., Wei J., Li G., Wang L., Wu J., Xue W., Huang X. No observable guide-RNA-independent off-target mutation induced by prime editor. bioRxiv. 2021 doi: 10.1101/2021.04.09.439109. [DOI] [Google Scholar]
- 116.Chen P.J., Hussmann J.A., Yan J., Knipping F., Ravisankar P., Chen P.-F. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021:184. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park S.J., Jeong T.Y., Shin S.K., Yoon D.E., Lim S.Y., Kim S.P., Choi J., Lee H., Hong J.I., Ahn J. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 2021;22:170. doi: 10.1186/s13059-021-02389-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu P., Liang S.Q., Zheng C., Mintzer E., Zhao Y.G., Ponnienselvan K., Mir A., Sontheimer E.J., Gao G., Flotte T.R. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat. Commun. 2021;12:2121. doi: 10.1038/s41467-021-22295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu Y., Yang G., Huang S., Li X., Wang X., Li G., Chi T., Chen Y., Huang X., Wang X. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res. 2021 doi: 10.1038/s41422-021-00520-x. Published online June 8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nelson J.W., Randolph P.B., Shen S.P., Everette K.A., Anzalone A.V., Newby G.A. Engineered pegRNAs that improve prime editing efficiency. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin Q., Jin S., Zong Y., Yu H., Zhu Z., Liu G., Kou L., Wang Y., Qiu J.L., Li J., Gao C. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021;39:923–927. doi: 10.1038/s41587-021-00868-w. [DOI] [PubMed] [Google Scholar]
- 122.Choi J., Chen W., Suiter C.C., Lee C., Chardon F.M., Yang W., Leith A., Daza R.M., Martin B., Shendure J. Precise genomic deletions using paired prime editing. bioRxiv. 2021 doi: 10.1101/2020.12.30.424891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hsu J.Y., Grünewald J., Szalay R., Shih J., Anzalone A.V., Lam K.C., Shen M.W., Petri K., Liu D.R., Joung J.K., Pinello L. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 2021;12:1034. doi: 10.1038/s41467-021-21337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hwang G.H., Jeong Y.K., Habib O., Hong S.A., Lim K., Kim J.S., Bae S. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res. 2021;49(W1):W499–W504. doi: 10.1093/nar/gkab319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Siegner S.M., Karasu M.E., Schröder M.S., Kontarakis Z., Corn J.E. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinformatics. 2021;22:101. doi: 10.1186/s12859-021-04034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Standage-Beier K., Tekel S.J., Brafman D.A., Wang X. Prime Editing Guide RNA Design Automation Using PINE-CONE. ACS Synth. Biol. 2021;10:422–427. doi: 10.1021/acssynbio.0c00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gao P., Lyu Q., Ghanam A.R., Lazzarotto C.R., Newby G.A., Zhang W., Choi M., Slivano O.J., Holden K., Walker J.A., 2nd Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol. 2021;22:83. doi: 10.1186/s13059-021-02304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu Y., Li X., He S., Huang S., Li C., Chen Y., Liu Z., Huang X., Wang X. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020;6:27. doi: 10.1038/s41421-020-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Petri K., Zhang W., Ma J., Schmidts A., Lee H., Horng J.E., Kim D.Y., Kurt I.C., Clement K., Hsu J.Y. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00901-y. Published online April 29, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bosch J.A., Birchak G., Perrimon N. Precise genome engineering in Drosophila using prime editing. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021996118. e2021996118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim Y., Hong S.A., Yu J., Eom J., Jang K., Yoon S., Hong D.H., Seo D., Lee S.N., Woo J.S. Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease. Cell Stem Cell. 2021;28:1614–1624.e5. doi: 10.1016/j.stem.2021.04.010. [DOI] [PubMed] [Google Scholar]
- 132.Yin H., Kauffman K.J., Anderson D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 133.Wei T., Cheng Q., Farbiak L., Anderson D.G., Langer R., Siegwart D.J. Delivery of Tissue-Targeted Scalpels: Opportunities and Challenges for In Vivo CRISPR/Cas-Based Genome Editing. ACS Nano. 2020;14:9243–9262. doi: 10.1021/acsnano.0c04707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xu X., Wan T., Xin H., Li D., Pan H., Wu J., Ping Y. Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 2019;21:e3107. doi: 10.1002/jgm.3107. [DOI] [PubMed] [Google Scholar]
- 135.Duan L., Ouyang K., Xu X., Xu L., Wen C., Zhou X., Qin Z., Xu Z., Sun W., Liang Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front. Genet. 2021;12:673286. doi: 10.3389/fgene.2021.673286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Richter M., Stone D., Miao C., Humbert O., Kiem H.P., Papayannopoulou T., Lieber A. In Vivo Hematopoietic Stem Cell Transduction. Hematol. Oncol. Clin. North Am. 2017;31:771–785. doi: 10.1016/j.hoc.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Milone M.C., O’Doherty U. Clinical use of lentiviral vectors. Leukemia. 2018;32:1529–1541. doi: 10.1038/s41375-018-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dalsgaard T., Cecchi C.R., Askou A.L., Bak R.O., Andersen P.O., Hougaard D., Jensen T.G., Dagnæs-Hansen F., Mikkelsen J.G., Corydon T.J., Aagaard L. Improved Lentiviral Gene Delivery to Mouse Liver by Hydrodynamic Vector Injection through Tail Vein. Mol. Ther. Nucleic Acids. 2018;12:672–683. doi: 10.1016/j.omtn.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tasca F., Wang Q., Gonçalves M.A.F.V. Adenoviral Vectors Meet Gene Editing: A Rising Partnership for the Genomic Engineering of Human Stem Cells and Their Progeny. Cells. 2020;9:E953. doi: 10.3390/cells9040953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X. Adenovirus-Mediated Gene Delivery: Potential Applications for Gene and Cell-Based Therapies in the New Era of Personalized Medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vassalli G., Büeler H., Dudler J., von Segesser L.K., Kappenberger L. Adeno-associated virus (AAV) vectors achieve prolonged transgene expression in mouse myocardium and arteries in vivo: a comparative study with adenovirus vectors. Int. J. Cardiol. 2003;90:229–238. doi: 10.1016/s0167-5273(02)00554-5. [DOI] [PubMed] [Google Scholar]
- 142.Chu D., Sullivan C.C., Weitzman M.D., Du L., Wolf P.L., Jamieson S.W., Thistlethwaite P.A. Direct comparison of efficiency and stability of gene transfer into the mammalian heart using adeno-associated virus versus adenovirus vectors. J. Thorac. Cardiovasc. Surg. 2003;126:671–679. doi: 10.1016/s0022-5223(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 143.Kuzmin D.A., Shutova M.V., Johnston N.R., Smith O.P., Fedorin V.V., Kukushkin Y.S., van der Loo J.C.M., Johnstone E.C. The clinical landscape for AAV gene therapies. Nat. Rev. Drug Discov. 2021;20:173–174. doi: 10.1038/d41573-021-00017-7. [DOI] [PubMed] [Google Scholar]
- 144.Pillay S., Zou W., Cheng F., Puschnik A.S., Meyer N.L., Ganaie S.S., Deng X., Wosen J.E., Davulcu O., Yan Z. Adeno-associated Virus (AAV) Serotypes Have Distinctive Interactions with Domains of the Cellular AAV Receptor. J. Virol. 2017;91 doi: 10.1128/JVI.00391-17. e00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Piotrowski-Daspit A.S., Glaze P.M., Saltzman W.M. Debugging the genetic code: non-viral in vivo delivery of therapeutic genome editing technologies. Curr. Opin. Biomed. Eng. 2018;7:24–32. doi: 10.1016/j.cobme.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cullis P.R., Hope M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ma C.C., Wang Z.L., Xu T., He Z.Y., Wei Y.Q. The approved gene therapy drugs worldwide: from 1998 to 2019. Biotechnol. Adv. 2020;40:107502. doi: 10.1016/j.biotechadv.2019.107502. [DOI] [PubMed] [Google Scholar]
- 149.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jiang L., Park J.S., Yin L., Laureano R., Jacquinet E., Yang J., Liang S., Frassetto A., Zhuo J., Yan X. Dual mRNA therapy restores metabolic function in long-term studies in mice with propionic acidemia. Nat. Commun. 2020;11:5339. doi: 10.1038/s41467-020-19156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hajj K.A., Melamed J.R., Chaudhary N., Lamson N.G., Ball R.L., Yerneni S.S., Whitehead K.A. A Potent Branched-Tail Lipid Nanoparticle Enables Multiplexed mRNA Delivery and Gene Editing In Vivo. Nano Lett. 2020;20:5167–5175. doi: 10.1021/acs.nanolett.0c00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qiu M., Glass Z., Chen J., Haas M., Jin X., Zhao X., Rui X., Ye Z., Li Y., Zhang F., Xu Q. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2020401118. e2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rungta R.L., Choi H.B., Lin P.J., Ko R.W., Ashby D., Nair J., Manoharan M., Cullis P.R., Macvicar B.A. Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain. Mol. Ther. Nucleic Acids. 2013;2:e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wei T., Cheng Q., Min Y.L., Olson E.N., Siegwart D.J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020;11:3232. doi: 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu S., Cheng Q., Wei T., Yu X., Johnson L.T., Farbiak L., Siegwart D.J. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR-Cas gene editing. Nat. Mater. 2021;20:701–710. doi: 10.1038/s41563-020-00886-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liang C., Li F., Wang L., Zhang Z.K., Wang C., He B., Li J., Chen Z., Shaikh A.B., Liu J. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials. 2017;147:68–85. doi: 10.1016/j.biomaterials.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 158.Zelmer C., Zweifel L.P., Kapinos L.E., Craciun I., Güven Z.P., Palivan C.G., Lim R.Y.H. Organelle-specific targeting of polymersomes into the cell nucleus. Proc. Natl. Acad. Sci. USA. 2020;117:2770–2778. doi: 10.1073/pnas.1916395117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sun H., Erdman W., 3rd, Yuan Y., Mohamed M.A., Xie R., Wang Y., Gong S., Cheng C. Crosslinked polymer nanocapsules for therapeutic, diagnostic, and theranostic applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12:e1653. doi: 10.1002/wnan.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lee K., Conboy M., Park H.M., Jiang F., Kim H.J., Dewitt M.A., Mackley V.A., Chang K., Rao A., Skinner C. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang Y., Shahi P.K., Xie R., Zhang H., Abdeen A.A., Yodsanit N., Ma Z., Saha K., Pattnaik B.R., Gong S. A pH-responsive silica-metal-organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome-editing machineries. J. Control. Release. 2020;324:194–203. doi: 10.1016/j.jconrel.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Prieve M.G., Harvie P., Monahan S.D., Roy D., Li A.G., Blevins T.L., Paschal A.E., Waldheim M., Bell E.C., Galperin A. Targeted mRNA Therapy for Ornithine Transcarbamylase Deficiency. Mol. Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie J., Bi Y., Zhang H., Dong S., Teng L., Lee R.J., Yang Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020;11:697. doi: 10.3389/fphar.2020.00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Del’Guidice T., Lepetit-Stoffaes J.P., Bordeleau L.J., Roberge J., Théberge V., Lauvaux C., Barbeau X., Trottier J., Dave V., Roy D.C. Membrane permeabilizing amphiphilic peptide delivers recombinant transcription factor and CRISPR-Cas9/Cpf1 ribonucleoproteins in hard-to-modify cells. PLoS ONE. 2018;13:e0195558. doi: 10.1371/journal.pone.0195558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Krishnamurthy S., Wohlford-Lenane C., Kandimalla S., Sartre G., Meyerholz D.K., Théberge V., Hallée S., Duperré A.M., Del’Guidice T., Lepetit-Stoffaes J.P. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 2019;10:4906. doi: 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Park H., Oh J., Shim G., Cho B., Chang Y., Kim S., Baek S., Kim H., Shin J., Choi H. In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci. 2019;22:524–528. doi: 10.1038/s41593-019-0352-0. [DOI] [PubMed] [Google Scholar]
- 167.Song C.Q., Jiang T., Richter M., Rhym L.H., Koblan L.W., Zafra M.P., Schatoff E.M., Doman J.L., Cao Y., Dow L.E. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 2020;4:125–130. doi: 10.1038/s41551-019-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Suda T., Liu D. Hydrodynamic gene delivery: its principles and applications. Mol. Ther. 2007;15:2063–2069. doi: 10.1038/sj.mt.6300314. [DOI] [PubMed] [Google Scholar]
- 169.Sendra L., Herrero M.J., Aliño S.F. Translational Advances of Hydrofection by Hydrodynamic Injection. Genes (Basel) 2018;9:E136. doi: 10.3390/genes9030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Le Guen Y.T., Le Gall T., Midoux P., Guégan P., Braun S., Montier T. Gene transfer to skeletal muscle using hydrodynamic limb vein injection: current applications, hurdles and possible optimizations. J. Gene Med. 2020;22:e3150. doi: 10.1002/jgm.3150. [DOI] [PubMed] [Google Scholar]
- 171.Lyu P., Wang L., Lu B. Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing. Life (Basel) 2020;10:366. doi: 10.3390/life10120366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Montagna C., Petris G., Casini A., Maule G., Franceschini G.M., Zanella I., Conti L., Arnoldi F., Burrone O.R., Zentilin L. VSV-G-Enveloped Vesicles for Traceless Delivery of CRISPR-Cas9. Mol. Ther. Nucleic Acids. 2018;12:453–462. doi: 10.1016/j.omtn.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mangeot P.E., Risson V., Fusil F., Marnef A., Laurent E., Blin J., Mournetas V., Massouridès E., Sohier T.J.M., Corbin A. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019;10:45. doi: 10.1038/s41467-018-07845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kaczmarczyk S.J., Sitaraman K., Young H.A., Hughes S.H., Chatterjee D.K. Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. USA. 2011;108:16998–17003. doi: 10.1073/pnas.1101874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Chadwick A.C., Wang X., Musunuru K. In Vivo Base Editing of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) as a Therapeutic Alternative to Genome Editing. Arterioscler. Thromb. Vasc. Biol. 2017;37:1741–1747. doi: 10.1161/ATVBAHA.117.309881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Carreras A., Pane L.S., Nitsch R., Madeyski-Bengtson K., Porritt M., Akcakaya P., Taheri-Ghahfarokhi A., Ericson E., Bjursell M., Perez-Alcazar M. In vivo genome and base editing of a human PCSK9 knock-in hypercholesterolemic mouse model. BMC Biol. 2019;17:4. doi: 10.1186/s12915-018-0624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rothgangl T., Dennis M.K., Lin P.J.C., Oka R., Witzigmann D., Villiger L., Qi W., Hruzova M., Kissling L., Lenggenhager D. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat. Biotechnol. 2021;39:949–957. doi: 10.1038/s41587-021-00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Musunuru K., Chadwick A.C., Mizoguchi T., Garcia S.P., DeNizio J.E., Reiss C.W., Wang K., Iyer S., Dutta C., Clendaniel V. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593:429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 179.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Häberle J., Matos J., Robinson M.D. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 180.Villiger L., Rothgangl T., Witzigmann D., Oka R., Lin P.J.C., Qi W., Janjuha S., Berk C., Ringnalda F., Beattie M.B. In vivo cytidine base editing of hepatocytes without detectable off-target mutations in RNA and DNA. Nat. Biomed. Eng. 2021;5:179–189. doi: 10.1038/s41551-020-00671-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Böck D., Rothgangl T., Villiger L., Schmidheini L., Mathis N., Ioannidi E., Kreutzer S., Kontarakis Z., Rimann N., Grisch-Chan H.M. Treatment of a metabolic liver disease by in vivo prime editing in mice. bioRxiv. 2021 doi: 10.1101/2021.08.17.456632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jiang T., Henderson J.M., Coote K., Cheng Y., Valley H.C., Zhang X.O., Wang Q., Rhym L.H., Cao Y., Newby G.A. Chemical modifications of adenine base editor mRNA and guide RNA expand its application scope. Nat. Commun. 2020;11:1979. doi: 10.1038/s41467-020-15892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Koblan L.W., Erdos M.R., Wilson C., Cabral W.A., Levy J.M., Xiong Z.M., Tavarez U.L., Davison L.M., Gete Y.G., Mao X. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ryu S.M., Koo T., Kim K., Lim K., Baek G., Kim S.T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 185.Chemello F., Chai A.C., Li H., Rodriguez-Caycedo C., Sanchez-Ortiz E., Atmanli A., Mireault A.A., Liu N., Bassel-Duby R., Olson E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021;7:eabg4910. doi: 10.1126/sciadv.abg4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lim C.K.W., Gapinske M., Brooks A.K., Woods W.S., Powell J.E., Zeballos C M.A., Winter J., Perez-Pinera P., Gaj T. Treatment of a Mouse Model of ALS by In Vivo Base Editing. Mol. Ther. 2020;28:1177–1189. doi: 10.1016/j.ymthe.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Levy J.M., Yeh W.H., Pendse N., Davis J.R., Hennessey E., Butcher R., Koblan L.W., Comander J., Liu Q., Liu D.R. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 2020;4:97–110. doi: 10.1038/s41551-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Suh S., Choi E.H., Leinonen H., Foik A.T., Newby G.A., Yeh W.H., Dong Z., Kiser P.D., Lyon D.C., Liu D.R., Palczewski K. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng. 2021;5:169–178. doi: 10.1038/s41551-020-00632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jo D.H., Jang H.-K., Cho C.S., Han J.H., Ryu G., Jung Y., Bae S., Kim J.H. Therapeutic adenine base editing corrects nonsense mutation and improves visual function in a mouse model of Leber congenital amaurosis. bioRxiv. 2021 doi: 10.1101/2021.01.07.425822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yeh W.H., Shubina-Oleinik O., Levy J.M., Pan B., Newby G.A., Wornow M., Burt R., Chen J.C., Holt J.R., Liu D.R. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020;12:eaay9101. doi: 10.1126/scitranslmed.aay9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li C., Georgakopoulou A., Mishra A., Gil S., Hawkins R.D., Yannaki E., Lieber A. In vivo HSPC gene therapy with base editors allows for efficient reactivation of fetal γ-globin in β-YAC mice. Blood Adv. 2021;5:1122–1135. doi: 10.1182/bloodadvances.2020003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Koblan L.W., Doman J.L., Wilson C., Levy J.M., Tay T., Newby G.A., Maianti J.P., Raguram A., Liu D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018;36:843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Varga R., Eriksson M., Erdos M.R., Olive M., Harten I., Kolodgie F., Capell B.C., Cheng J., Faddah D., Perkins S. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA. 2006;103:3250–3255. doi: 10.1073/pnas.0600012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.van Putten M., Hulsker M., Young C., Nadarajah V.D., Heemskerk H., van der Weerd L., ’t Hoen P.A., van Ommen G.J., Aartsma-Rus A.M. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB J. 2013;27:2484–2495. doi: 10.1096/fj.12-224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-Linked Protein LY6A Drives AAV-PHP.B Transport across the Blood-Brain Barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ko D.C., Milenkovic L., Beier S.M., Manuel H., Buchanan J., Scott M.P. Cell-autonomous death of cerebellar purkinje neurons with autophagy in Niemann-Pick type C disease. PLoS Genet. 2005;1:81–95. doi: 10.1371/journal.pgen.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Keino H., Horie S., Sugita S. Immune Privilege and Eye-Derived T-Regulatory Cells. J. Immunol. Res. 2018;2018:1679197. doi: 10.1155/2018/1679197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rees H.A., Komor A.C., Yeh W.H., Caetano-Lopes J., Warman M., Edge A.S.B., Liu D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017;8:15790. doi: 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yeh W.H., Chiang H., Rees H.A., Edge A.S.B., Liu D.R. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 2018;9:2184. doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Esrick E.B., Lehmann L.E., Biffi A., Achebe M., Brendel C., Ciuculescu M.F., Daley H., MacKinnon B., Morris E., Federico A. Post-Transcriptional Genetic Silencing of BCL11A to Treat Sickle Cell Disease. N. Engl. J. Med. 2021;384:205–215. doi: 10.1056/NEJMoa2029392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhi S., Chen Y., Wu G., Wen J., Wu J., Liu Q., Li Y., Kang R., Hu S., Wang J. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol. Ther. 2021 doi: 10.1016/j.ymthe.2021.07.011. S1525-0016(21)00365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carlson J.A., Rogers B.B., Sifers R.N., Finegold M.J., Clift S.M., DeMayo F.J., Bullock D.W., Woo S.L. Accumulation of PiZ alpha 1-antitrypsin causes liver damage in transgenic mice. J. Clin. Invest. 1989;83:1183–1190. doi: 10.1172/JCI113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lander E.S., Baylis F., Zhang F., Charpentier E., Berg P., Bourgain C., Friedrich B., Joung J.K., Li J., Liu D. Adopt a moratorium on heritable genome editing. Nature. 2019;567:165–168. doi: 10.1038/d41586-019-00726-5. [DOI] [PubMed] [Google Scholar]
- 205.The National Academies of Sciences, Engineering, and Medicine . The National Academies Press; 2017. Human Genome Editing: Science, Ethics, and Governance. [PubMed] [Google Scholar]
- 206.Schiroli G., Conti A., Ferrari S. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell. 2019;24:551–565.e8. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C., Gregory P.D., van der Burg M., Gentner B., Montini E., Lombardo A., Naldini L. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ferrari S., Jacob A., Beretta S. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat Biotechnol. 2020;38:1298–1308. doi: 10.1038/s41587-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]